Summary
Precise temporal control of gene expression in neuronal progenitors is necessary for correct regulation of neurogenesis and cell fate specification. However, the cellular heterogeneity of the developing CNS has posed a major obstacle to identifying the gene regulatory networks that control these processes. To address this, we used single cell RNA-sequencing to profile ten developmental stages encompassing the full course of retinal neurogenesis. This allowed us to comprehensively characterize changes in gene expression that occur during initiation of neurogenesis, changes in developmental competence, and specification and differentiation of each major retinal cell type. We identify NFI transcription factors (Nfia, Nfib, and Nfix) as selectively expressed in late RPCs, and show they control bipolar interneuron and Müller glia cell fate specification and promote proliferative quiescence.
eToc blurb:
We use single-cell RNA-Seq analysis to comprehensively profile gene expression during mouse retinal development. We find major differences between early and late-stage, as well as primary and neurogenic, progenitors. We also find that NFI factors control cell cycle exit and generation of late-born cell types.
Introduction
Neural progenitor cells (NPCs) of the central nervous system (CNS) undergo stereotypical, stage-dependent transitions during neurogenesis. These include transition from a slowly proliferating neuroepithelial cells to actively proliferating neurogenic progenitors (Martynoga et al., 2012; Schmidt et al., 2013); transition from symmetric proliferative to asymmetric neurogenic, and ultimately to terminal neurogenic division modes n (Homem et al., 2015; Taverna et al., 2014); and changes in developmental competence, or the ability of NPCs to give rise to different subtypes of neurons and glia (Cayouette et al., 2013; Kohwi and Doe, 2013; Okano and Temple, 2009). While the molecular mechanisms that control these processes in the vertebrate nervous system mostly remain unclear, the retina is a tractable system for studying them, owing to its greater accessibility and lower cellular heterogeneity than other CNS regions. The major retinal cell types show a stereotyped and birth order, with retinal ganglion cells born first and Mϋller glia born last (Figure 1A). Classic studies using retroviral-mediated sparse labeling of retinal progenitors in rodents (Turner and Cepko, 1987; Turner et al., 1990), and more recent studies using live imaging (Gomes et al., 2010; He et al., 2012) showed that retinal progenitor cells (RPCs) are multipotent, with changes in clone composition and size controlled largely by intrinsic mechanisms. This has raised several unanswered questions (Cepko, 2014). First, how heterogeneous are RPCs, and do they exhibit any cell fate biases? Second, are competence transitions gradual or discrete, and how are they controlled? Third, what genes direct an individual progenitor to exit the cell cycle and differentiate as a specific subtype of neuron or glia?
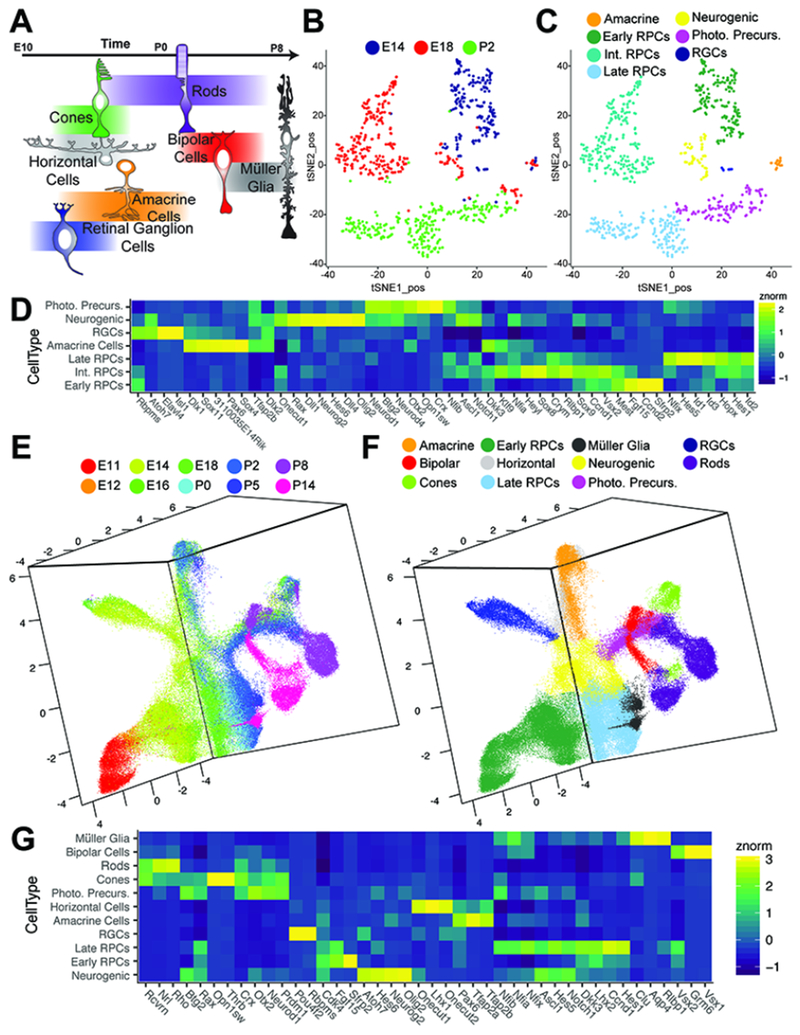
Figure 1. Single cell RNA-sequencing of the developing retina.
(A) Schematic of retinal cell birth order. (B-C) tSNE-dimension reduction of gene expression profiles from Chx10:GFP-positive RPCs via a modified Smart-Seq2 protocol, with cells labeled by (B) age or (C) cell type as determined by marker gene expression. (D) Heatmap of marker genes identified through differential expression analysis with respect to cell type in Smart-Seq2 samples. (E-F) UMAP-dimension reduction of droplet-based single cell RNA-sequencing of developing mouse retina, with doublets and extra-retinal cells removed. Samples are colored by (E) age or (F) annotated cell type as determined by marker gene expression. (G) Heatmap of differentially expressed genes in annotated cell types in 10× samples. Abbreviations: Int. RPCs – Intermediate RPCs; Photo. Precurs. – Photoreceptor Precursors; RGCs – Retinal Ganglion Cells.
Past work provides some insights into each of these questions. Differential expression analysis has identified genes with dynamic expression amongst RPCs, both during neurogenesis and at specific developmental stages (Blackshaw et al., 2004; Boije et al., 2008; Dixit et al., 2014; Farhy et al., 2013; James et al., 2004; Laboissonniere et al., 2017; Mochizuki et al., 2014; Trimarchi et al., 2008). With few exceptions, the functional significance of this heterogeneity is unknown. Cell lineage analysis of RPCs expressing individual transcription factors shows limited, and often temporally-regulated, cellular fate restriction (Brzezinski et al., 2011; Brzezinski et al., 2012; Emerson et al., 2013; Hafler et al., 2012; Jusuf et al., 2012), but the relationship between RPC heterogeneity and cell fate determination remains unclear. Furthermore, although several transcription factors are known to regulate RPC competence (Brzezinski et al., 2012; Elliott et al., 2008; Mattar et al., 2015), their contribution is either modest, or affects only retinal ganglion cell specification. Little is known about the transcriptional regulatory networks initiating retinal neurogenesis and controlling changes in mode of RPC division.
To comprehensively define changes in gene expression associated with competence transitions, regulation of neurogenic divisions, and cell fate specification, we conducted single cell RNA-Seq analysis of the developing mouse retina. We isolated single retinal cells at time points ranging from prior to initiation of neurogenesis through to its completion, including cells committed to each major cell type. Previous large-scale expression profiling studies of retinal development have been limited in their scope owing to their use of bulk dissected material (Aldiri et al., 2017; Blackshaw et al., 2001; Blackshaw et al., 2004; Hoshino et al., 2017). Past single cell expression profiling studies in retina either analyzed only small numbers of cells (Laboissonniere et al., 2017; Mullally et al., 2016; Trimarchi et al., 2008), or profiled mature cells (Macosko et al., 2015; Shekhar et al., 2016).
In this study, we examine the dynamics of gene expression within retinal cells to investigate the specifics of RPC competence, neurogenesis, and temporal cell fate specification. Using single cell RNA-sequencing (scRNA-seq), we can reconstruct developmental trajectories of distinct retinal cell fates and identify the genes and gene networks that influence RPC competence, retinal neurogenesis, and cell fate specification during development. We use these data to identify candidate genes that regulate cell fate commitment and retinal neurogenesis, and we show that NFI transcription factors regulate generation of late-born retinal cells and cell cycle exit. This work advances our understanding of retinal development, and provides a template for investigation of temporal patterning in all areas of the developing CNS.
Results
Examination of Retinal Progenitor Cell Heterogeneity via Smart-Seq Analysis
We first performed single cell RNA-sequencing on FACS-isolated Chx10-GFP (+) mouse RPCs (Rowan and Cepko, 2004), using an adapted Smart-Seq2 protocol (Chevee et al., 2018) at embryonic (E) days 14 and 18, and postnatal (P) day 2, which correspond to early, intermediate and late stages of retinal neurogenesis, respectively (Figure 1B). Analysis of 747 individual cells (Figure S1A–D) revealed three major clusters expressing canonical RPC markers (e.g. Ccnd1, Cdk4, Pax6; Figure S1F). Clusters primarily correspond to each time point sampled (Figure 1B; Figure S1E) when plotted using 2-D t-stochastic neighbor embedding (tSNE) analysis (van der Maaten and Hinton, 2008) using genes displaying high expression variance across all cells (Table S1). We resolved these clusters into discrete groups distinguished by expression of genes marking cell cycle phases, with subclusters specific to G1/S (e.g. Pcna,Ccne2) and G2/M phase (e.g. Ccnbl, Ube2c), respectively (Figure S1G). As reported, (Kowalczyk et al., 2015; Liu et al., 2017), co-expression of transcripts marking multiple phases is observed, identifying cells transitioning between cell cycle phases (Figure S1G). A much smaller cluster, which included cells from each age, expressed both genes associated with active proliferation (Cdk4) and multiple neurogenic bHLH factors (e.g. Atoh7, Olig2, Neurog2, Neurod1; Figure S1H). Finally, many P2 cells do not express cell cycle regulators, and expressed markers of immature photoreceptors (e.g. Crx), while smaller clusters of postmitotic cells expressed genes specific to immature amacrine (Tfap2b) and retinal ganglion cells (Pou4f2, Isl1; Figure S1I). We annotated teach individual cell based on transcriptional profiles of transcripts enriched within individual clusters (Figure 1C).
Differential gene expression testing across primary RPCs - all RPCs excluding neurogenic cells - identifies 1195 genes (q-val < 1.0 e-10, mean expression > 1.0 RNA copies per cell; Table S2) with significant differential expression amongst progenitor populations across development. However, within individual ages, we do not observe subclusters amongst these RPCs, except for those defined by markers of cell cycle phase. Conversely, differential expression testing across annotated cell types identified 4754 genes (q-val < 1.0 e-10; Table S3), including transcription factors that promote retinal neurogenesis (Atoh7, Neurog2, Ascl1) and photoreceptor specification (Neurod1, Crx) (Figure 1D). Genetic lineage analysis has suggested RPCs expressing neurogenic bHLH factors such as Atoh7, Olig2, and Neurog2 are substantially more likely to undergo terminal neurogenic divisions (Brzezinski et al., 2011; Brzezinski et al., 2012; Hafler et al., 2012). Together, these results indicate RPCs undergo significant transcriptional changes across developmental time, consistent with a change in developmental competence, and that both cell cycle phase and neurogenic potential influence the transcriptional heterogeneity of RPCs. This dataset also provides an unbiased, high-depth analysis of gene expression in RPCs and a subset of postmitotic neural precursors, at multiple timepoints during retinal neurogenesis.
Droplet-based scRNA-Seq reveals the full transcriptional landscape of mouse retinal development.
We next sought to profile retinal development more comprehensively using droplet-based single cell RNA sequencing, which can analyze more cells and time points. We profiled 120,804 single cells from whole retinas at 10 select developmental time points, ranging from prior to the onset of neurogenesis (E11) through terminal fate specification (P14), using the 10× Genomics Chromium 3’ v2 platform (PN-120223) (Figure S2A). Libraries were sequenced to a mean depth of ~110,220,000 reads per library, corresponding to a mean UMI count of 2099.75 and 1153.43 genes per cell (Figure S2B–E). Preliminary clustering and cell type annotation was performed on single cell profiles from individual timepoints using a modified Monocle dpFeature workflow (Qiu et al., 2017) (Figure S3–S4). All time points were then aggregated into a single dataset for further analyses. Using 3290 high-variance genes across all cells (Table S4), we established a reduced three-dimensional representation of the developing retina using UMAP (McInnes and Healy, 2018) (Figure S2F–G; Movie 1). A second round of clustering (Figure S2H) and cell type annotation was performed in which doublets and extra-retinal cells were identified and removed (Figure 1E–F; Figure S2I; Movie 2).
The resulting representation contains a core manifold consisting of primary RPC at all ages between E11 and P8 that express canonical RPC markers (Pax6, Vsx2, Lhx2, etc; Figure 1G). We also observe a population of proliferating (Ccnd1-positive) cells expressing multiple neurogenic bHLH genes (Olig2, Neurog2), and show reduced expression of Vsx2 and Lhx2 compared to other RPCs (Figure 1G). This population corresponds to the neurogenic RPC population identified in the Smart-Seq analysis (Figure 1C–D), and is seen between E12 and P8 (Figure 1E). The neurogenic population is adjacent to, and extends from, primary RPCs (Figure 1F). Trajectories of differentiating cells corresponding to all major retinal neuronal subtypes, with the exception of horizontal cells, can be seen emerging as separate branches from this population of neurogenic RPCs. A branch corresponding to differentiating Müller glial precursors, in contrast, emerges from the primary RPC cluster. The proximity of Müller glia and primary RPCs is consistent with the cell populations exhibiting overlapping gene expression profiles (Blackshaw et al., 2004; Nelson et al., 2011; Roesch et al., 2008). Closer examination identified cells with high levels of horizontal cell-specific (Lhx1; Figure S5A) (Liu et al., 2000; Poche et al., 2007) or horizontal cell-enriched genes (Prox1; Figure S5B) (Dyer et al., 2003) present within the presumptive amacrine trajectory. Recursive analysis of the amacrine trajectory identified both starburst amacrine and horizontal cells as separate populations distinct from other developing amacrine cells (Figure S5C–D).
Within the reduced dimensional embedding, most cells are distributed along a single contiguous manifold, indicating we have profiled most key transitions that occur during mouse retinal development, with no significant discontinuity in coverage across this process. Cell-type classification, number and proportions of annotated cell types are listed in Table S5 and Figure S2J–L. The timing and sequential progression of major cell type trajectories is consistent with previous studies (Young, 1985a, b), highlighting the initiation of neurogenesis at E12; the generation of the majority of retinal ganglion cells, presumptive cones and horizontal cells between E14 and E16; the specification of amacrine cells in the late embryonic and early postnatal time-points; a burst of rod photoreceptor differentiation during the early postnatal periods; and specification and differentiation of bipolar cells and Müller glia from P5 onwards. Examination of the proportions of primary and neurogenic RPCs and gliogenic cells revealed a relatively stable proportion of neurogenic cells captured during retinal neurogenesis from E14-P8 (Figure S2M). However, in the absence of lineage tracing data, we are unable to measure the developmental changes in cellular division mode (i.e. symmetric proliferative, asymmetric neurogenic, symmetric terminal divisions).
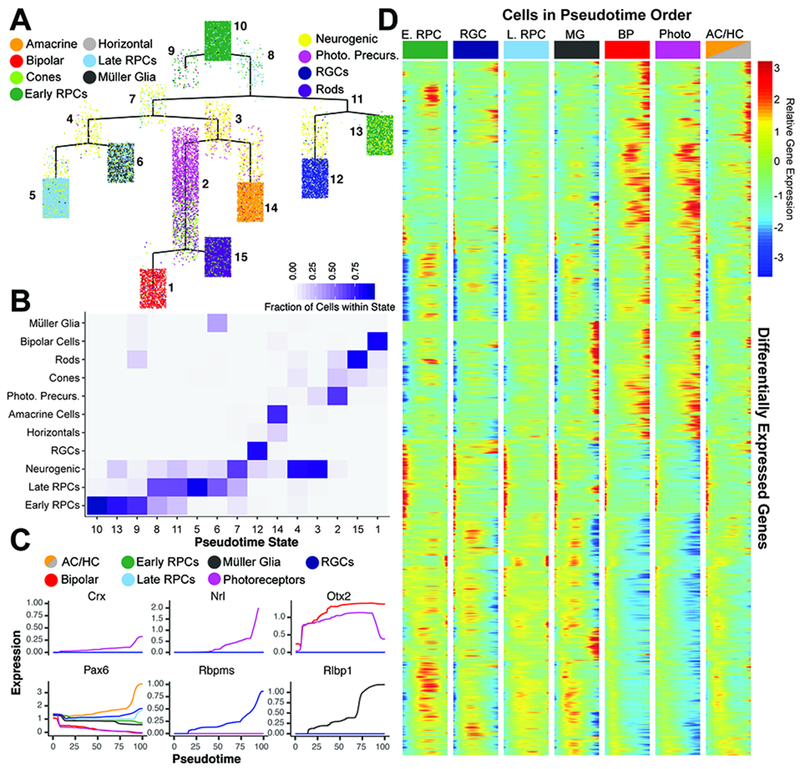
To identify genes that show dynamic regulation during retinal development, we performed a pseudotemporal analysis using 3259 high variance genes in neuroretinal cells only, excluding both annotated extraretinal cells and doublets (Trapnell et al., 2014) (Table S6). Due to matrix size limitations of dependent algorithms, we performed the pseudotime analysis on ~32,000 cells randomly sampled across the entire dataset. The subset of cells used for pseudotemporal analysis accurately reflected cell proportions within the full dataset with respect to developmental age and celltype (Figure S6A–C). Using this approach, we were able to reconstruct a tree that reflects known temporal ordering of cell fate specification within the retina, and displays terminal branches that are both comprised of single cell types and reflect the known developmental trajectories of these cell (Figure 2A–B; S6D–F). For example, rods and bipolar cells (pseudotime states 15 and 1, respectively) derive from a common pseudotime branch (Figure 2A;C), consistent with these cell types arising from Otx2-positive progenitors (Koike et al., 2007; Wang et al., 2014).
Figure 2. Pseudotime analysis reveals temporal regulation of transcript expression during differentiation of retinal cell types.
(A) Complex pseudotime tree of retinal cells colored by annotated celltype. Pseudotime state identity is numbered. (B) Heatmap of pseudotemporal states reveals the individual cell type proportions within each state. (C) Known marker gene expression across pseudotime colored by cell type designation of each trajectory. (D) Branched heatmap faceted by terminal pseudotime state corresponding to annotated cell type. Expression of all 7,487 differentially expressed transcripts across pseudotime starting from pseudotime value of 0 (left side of each column; top of the complex tree in panel A), and following a continuous path down the tree towards each terminal branch (right side of each column. Abbreviations: AC/HC – Amacrine Cells/Horizontal Cells; BP - Bipolar Cells; E. RPC – Early Primary RPC; L. RPC – Late Primary RPC; MG - Müller Glia; Photo - Photoreceptors; Photo. Precurs. - Photoreceptor Precursors; RGCs – Retinal Ganglion Cells.
Differential gene expression analysis across pseudotime identified a total of 7487 genes with significant changes in gene expression during retinal development (Figure 2D; Table S7 q-val < 1e-5). This list includes known markers of maturing cell types (Figure 2C). Recursive analysis of major branch points in the complex pseudotime tree identified genes that exhibit significant differential expression during specification and maturation of individual cell types. Using this strategy, we were able to identify genes expressed during early, intermediate and late stages of differentiation of postmitotic precursors of retinal ganglion cells, amacrine/horizontal cells, photoreceptor cells, bipolar cells and Müller glia (Figure S7–S12). Temporal- and trajectory-appropriate expression of all known transcription factors that regulate differentiation of individual retinal cell types was observed, with Isl1 and Pou4f2 detected as early markers of RGCs (Li et al., 2014), and Otx2 for immature photoreceptors and bipolar cells (Baas et al., 2000), for instance. Recursive analysis of the amacrine/horizontal cell trajectory further identified genes that mark developing amacrine, starburst amacrine, and horizontal cells (Figure S5E–F; Figure S13–14).
Temporally dynamic changes in gene expression within RPCs
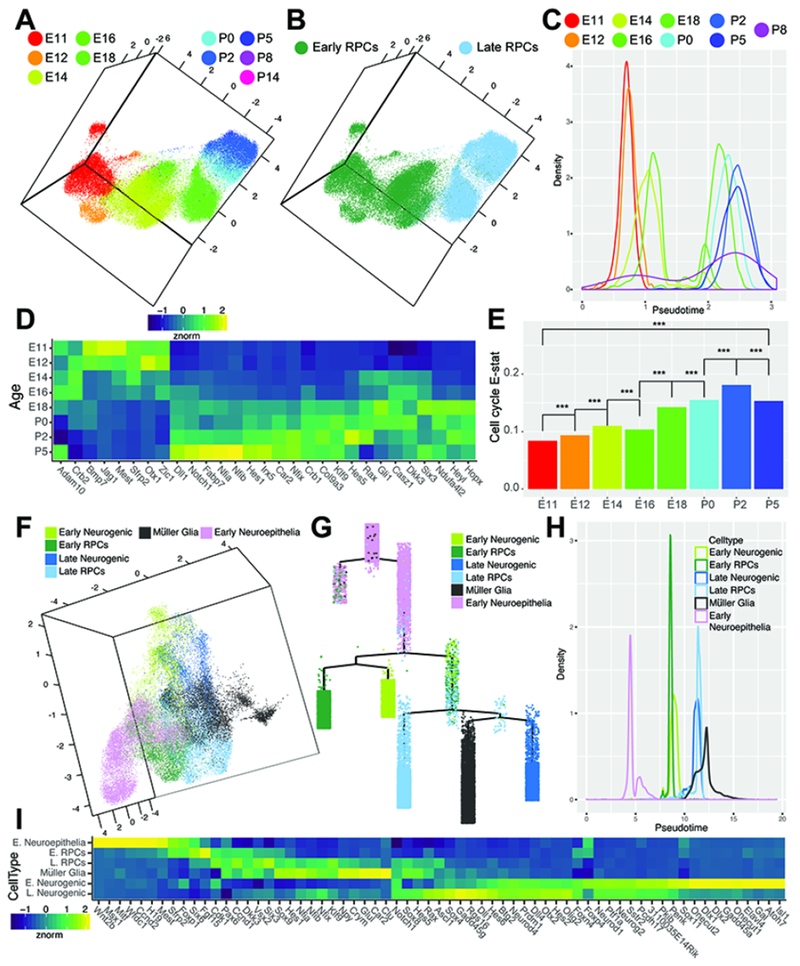
To characterize the transcriptional changes within RPCs across developmental time, as well as the changes in gene expression within progenitors likely to commit to the neurogenic or gliogenic fractions of cells, we performed a pseudotemporal analysis on subsets of specific cell types. We first addressed the changes in gene expression across development in the RPC subset. UMAP representations and pseudotime analyses using a set of 1763 high variance genes across the subsetted RPCs revealed a temporal progression from early (E11) through late (P8) time-points. We observed a clear segregation of RPCs occurring between E16 and E18. A clustering solution for the cells in UMAP space agrees with this classification, and allowed us to delineate stable classes of early and late RPCs (Figure 3A–C; Figure 1F). Cells from both E16 and E18 timepoints were stratified across this divide, indicating that this does not reflect a batch effect (Figure 3C). The stratification of early vs late RPCs matches the end of the interval in which early-born retinal cell types (RGCs, horizontal cells, cone photoreceptors and GABAergic amacrine cells) are generated (Voinescu et al., 2009; Young, 1985a, b)(Figure 1A). We identified 3291 genes that show significant differential expression across the RPC pseudotime (qval <1e-20, Figure 3D). We identify previously established markers of both early (Fgf15 and Sfrp2) and late (Crym and Car2) RPCs (Blackshaw et al., 2004) (Figure 1G) and a host of previously unidentified markers of early versus late RPCs, including the late RPC-enriched NFI transcription factors (Nfia, Nfib, and Nfix; Figure 3D).
Figure 3. Identification of candidate transcripts in the regulation of RPC competence, neurogenesis and gliogenesis.
(A-B) UMAP dimension reduction of subsetted primary RPCs colored by (A) age or (B) cell type. (C) Density plot of cells across RPC pseudotime analysis colored by age. (D) Heatmap of differentially expressed genes across RPC pseudotime, displaying normalized transcript enrichment in cells grouped by age. (E) EVAsc analysis of relative dissimilarity of canonical cell cycle genes within RPCs assessed at individual ages. (F) UMAP-dimension reduction displaying the classification of early neuroepithelial cells, early and late primary RPCs, early and late neurogenic, and gliogenic cells identified in pseudotemporal analyses. (G) Complex pseudotime tree of RPCs, neurogenic and gliogenic cells, highlighting designation of early neuroepithelial cells, early and late RPCs, early and late neurogenic cells, and Müller glia. (H) Density plot of RPC, neurogenic, gliogenic, and early neuroepithelial cells across pseudotime. (I) Heatmap of differentially expressed genes across RPC/neurogenic/gliogenic pseudotime, displaying normalized enrichment across all cells within the designated cell-type. *** indicates p<0.001 from EVAsc analysis. Abbreviations: E. Neuroepithelia - Early Neuroepithelial Cells; E. RPCs - Early Primary Retinal Progenitor Cells; L. RPCs - Late Primary Retinal Progenitor Cells; E. Neurogenic - Early Neurogenic Cells; L. Neurogenic - Late Neurogenic Cells.
To further examine the heterogeneity of RPCs across development, we applied Expression Variation Analysis in single cells (EVAsc) to quantify the relative dissimilarity in transcriptional profiles among RPCs from distinct developmental time points (Afsari et al., 2014; Davis-Marcisak et al., 2018). Variation in expression of cell cycle genes (Figure 3E) and the FGF-signaling pathway (Figure S15C) increases over developmental time, while variation in the Wnt and Notch pathways decreases over time (Figure S15C). The increased variance in cell cycle gene expression in late-stage RPCs likely serves as a proxy signal for the increase in cell cycle length in RPCs seen over the course of development, as previously seen in rat retina (Alexiades and Cepko, 1996).
Strikingly, Notch pathway gene expression increased steadily over the course of retinal development in RPCs, forming a smooth temporal gradient. This is particularly clear when canonical Notch target genes such as Hes1 and Hes5 are examined (Figure 3D; Figure S15A), and is consistent with the known role of Notch signaling in driving specification of late-born Müller glia (de Melo et al., 2016a; de Melo et al., 2016b; Jadhav et al., 2006). The increase in expression levels of Notch pathway components across late RPCs observed within the dataset is consistent with the EVAsc analyses, which suggests a decreased heterogeneity of pathway components amongst RPCs as development progresses (Figure S15C).
Transcriptional co-activator proteins (Basp1) and known cell cycle regulators (Kpna2, Kif2a) display enriched expression in embryonic RPCs at times of increased neurogenesis (E14-E18) and a burst of expression at P5, during a period of substantial terminal divisions. A limited number of Müller glia-expressed genes are upregulated in late postnatal primary RPCs (P2-P5; Cd9, Sat1; Figure S15B), coincident with the onset of gliogenesis. IA number of genes implicated in inhibiting cell cycle progression also were upregulated in early postnatal (late-)RPCs (Oaz1, Pebp1; Figure S15B; P0-P2) (Al-Mulla et al., 2011; Wang and Jiang, 2014). The functional significance of altered Oaz1 and Pebp1 expression may reflect the increased cell cycle length seen in late-stage RPCs (Alexiades and Cepko, 1996).
We next investigated if these changes in gene expression seen across pseudotime in RPCs might translate into functional differences in regulation of neurogenesis and gliogenesis. We subset the data to all RPCs (both neurogenic and primary RPCs) or gliogenic cells and again performed pseudotemporal analysis on ~32,000 sampled cells. Analysis of the resulting complex pseudotemporal hierarchy confirmed separate early and late progenitor populations, as well as the gliogenic population. However, three additional populations of cells were identified based on differential expression of genes across pseudotime states. An early neuroepithelial progenitor cell population emerged, comprised of cells primarily from the earliest (E11-E14) time points and prior to the onset of RPC neurogenesis (Figure 3F–H; Figure S15D–F). Genes with enriched expression within this subset of cells include those enriched in ciliary margin and/or retinal pigmented epithelium (Mitf, Ccnd2,Msx1, H19), and reflect genes expressed after early eye-field specification (Blackshaw et al., 2004 ; Cho and Cepko, 2006; Liu et al., 2007) (Figure 3H).
The subdivision of the primary RPC population into early and late RPCs around E16-E18 was paralleled by an early and late population of neurogenic RPCs. Several genes are selectively expressed in either early (Gadd45a, Sox11, Elavl3, Gal) or late-stage (Gadd45g, Rgs16) neurogenic cells (Figure 3I). Transcription factors that control specification of early-born cell types such as RGCs (Sox11, Atoh7), horizontal cells (Onecut1/2) and GABAergic amacrine cells (Dlx1/2) were selectively expressed in early neurogenic RPCs (de Melo et al., 2005; Emerson et al., 2013; Jiang et al., 2013)(Figure 3I). Likewise, genes that specify late-born cell types (Prdm1, Otx2, Ascl1) were enriched in the late-neurogenic fraction (Brzezinski et al., 2010; Katoh et al., 2010; Nelson et al., 2009; Nishida et al., 2003) (Figure 3I). These transcriptional differences suggest a sharp functional distinction between early and late neurogenic cells that derives in part from transcriptional differences established in early vs late primary RPCs, and refects changes in progenitor competence over the course of retinal development.
Gene co-expression and reuse modeled by scCoGAPS identifies distinct cellular states and developmental transitions
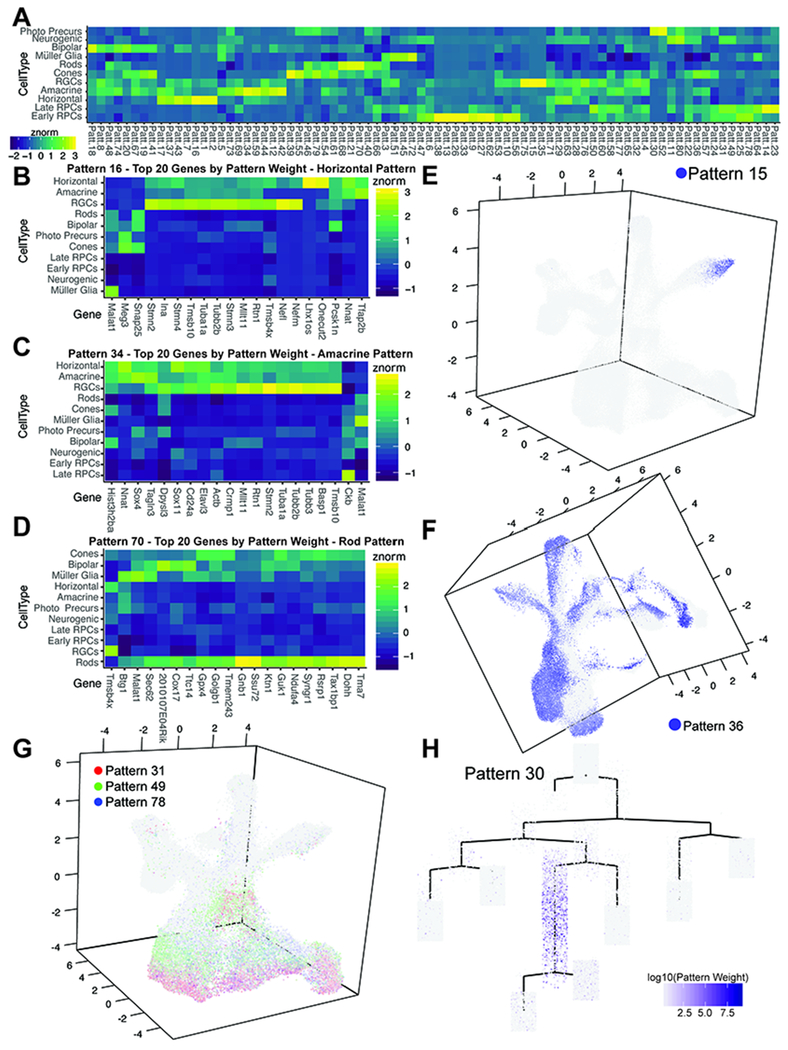
While our pseudotemporal analyses identified developmental trajectories associated with most major retinal cell types, it was unable to resolve more closely-related cell types or trajectories. These included differentiation of horizontal cells from amacrine cells and the differences between immature cone and rod photoreceptors. To identify gene signatures of discrete cell populations, including those not resolved by our embedding and pseudotemporal analyses, we applied scCoGAPS (Stein-O’Brien et al., 2018) to learn patterns of co-regulated gene expression. Unlike other methods which learn gene signatures on a subset of cells, and then project the entire dataset (Macosko et al., 2015), scCoGAPS uses an ensemble-based approach across multiple sets of cells to learn these signatures in parallel. Using scCoGAPS, we identified 80 independent patterns of gene usage across the full time course of retinal development, including patterns that distinguished differentiating horizontal cells from amacrine cells, and cones from rods (Figure 4A). Individual patterns reflect cell type specification and capture key developmental transitions, including populations such as neurogenic RPCs and photoreceptor precursors (Stein-O’Brien et al., 2018)(Figure 4A). Corresponding gene weights for each of the 80 patterns are indicated in Table S8. While patterns can distinguish cell types and correlated well, for example with annotations of horizontal cells (Patterns 1,2, and 16; Figure 4A), many genes with high gene weights within individual patterns showed expression across multiple cell types (Figure 4B–D). Additional patterns are also associated with mature RGCs (Pattern 15; Figure 4E), or recover other phenotypic features of these data, such as sex (Pattern 36; Figure 4F). These data show that gene reuse across cell types can be modeled using scCoGAPS. In addition, combinations of gene expression can better distinguish cell types and states with closely related gene expression profiles, thereby providing an improvement over the use of a limited number of marker genes to classify, especially in contexts with a high-degree of gene reuse (Cleary et al., 2017).
Figure 4. scCoGAPS analysis on single cell RNA-sequencing samples reveals patterns of gene expression within developmental processes.
(A) Correlation heatmap of pattern weight with cell types. (B-D) Heatmaps of the top 20 genes by gene weight of patterns (B) 16, (C) 34, and (D) 70 that correspond to Horizontal Cells, Amacrine Cells, Rod Photoreceptors, respectively. (E-F) Examples of graphical representations of pattern weights of individual cells within the UMAP-dimension reduction. (E) Pattern 15 marks the terminal trajectory of RGCs. (F) Pattern 36, with pattern marker Xist, highlights sex of origin. Combinations of patterns can be used to assess developmental processes such as (G) the influence of cell cycle phase on RPC clustering. (H) scCoGAPs pattern weights of retinal cells plotted within the complex pseudotime (Figure 2A) highlights photoreceptor/bipolar cell precursors in Pattern 30, using Otx2 as a pattern marker. Abbreviations: Photo Precurs - Photoreceptor Precursors.
Visualization of multiple, temporally-regulated patterns can characterize continuous biological processes, such as the progression of progenitors through the cell cycle (Figure 4G). Plotting pattern weights on pseudotime representations (Figure S16) highlights the association of patterns with developmental transitions (Figure 4H; photoreceptor/bipolar cell precursors with high Otx2 expression). The application of scCoGAPS to these single cell RNA-Seq data also captured technical features as well. Combinations of biologically incompatible patterns (e.g. two patterns for distinct mature cell types within the same cell) were used as one criteria for identifying and excluding doublet cells from further analysis (Stein-O’Brien et al., 2018).
NFI transcription factors regulate specification of late-born retinal cell types and drive cell cycle exit
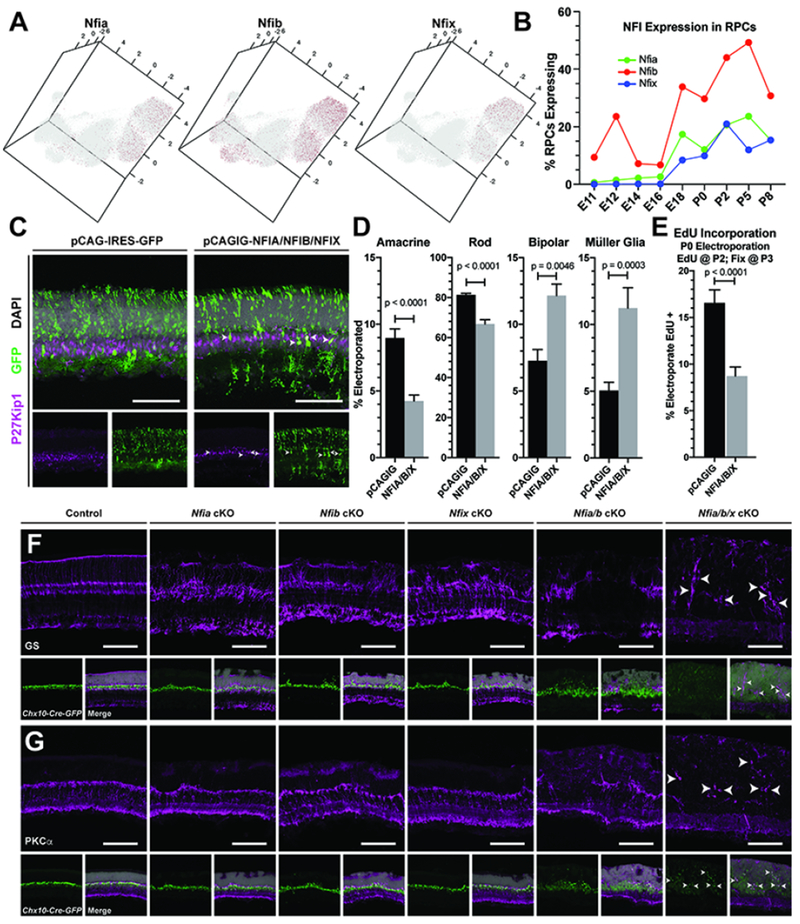
A consistent theme in our differential expression, pseudotime, and scCoGAPS analyses was the identification of the NFI factors Nfia/b/x, as having high expression and high gene weights in cells/patterns associated with late-stage RPCs, bipolar cells and Müller glia (Figure 5A–B; Figure S17). We also observed expression of Nfia/b/x within the amacrine cell trajectory, and high levels of Nfia transcript expression in presumptive amacrine cells via in situ hybridization; consistent with previous reports (Keeley and Reese, 2018; Figure S17A–D). Our previous studies indicate that overexpression of NFIA alone results in an increase in the fraction of cells that show radial morphology, but that these radial cells fail to express markers of mature Müller glia (de Melo et al., 2016a). As Nfia/b/x display enriched and overlapping expression within late RPCs in our single-cell RNA-seq dataset, and NFI factors are required for gliogenesis in cortex and spinal cord (Deneen et al., 2006; Kang et al., 2012; Matuzelski et al., 2017; Nagao et al., 2016), we hypothesized that NFI transcription factors may play a similar role in mediating the transition from neurogenesis to gliogenesis in the developing retina. Based on these observations, we chose to further investigate the function of the NFI transcription factors within late-stage RPCs.
Figure 5. NFI factors regulate specification of late retinal cell fates.
(A) Expression of NFI factors within the UMAP dimension reduction of all RPCs. (B) Graph of the percentage of RPCs expressing NFI factors within the scRNA-seq dataset. (C) P14 retinas stained for the Müller glia marker P27Kip1 after in vivo electroporations at P0 of either control (pCAGIG) or pooled pCAGIG-NFIA, NFIB, and NFIX constructs. (D) Quantification of the proportion of electroporated cells that co-label with markers for amacrine, photoreceptor, bipolar and Müller glial cells after in vivo electroporation of pCAGIG control or pCAGIG-NFIA/B/X. (E) Quantification of the proportion of electroporated cells that incorporated EdU after a 24-hour pulse from P2-P3. (F-G) Chx10-Cre mediated loss of function of Nfia, Nfib, Nfix, Nfia and Nfib, or Nfia, Nfib, Nfix. (F) Disruption of retinal organization and loss of Müller glia is marked by glutamine synthetase staining (GS). (G) Loss of rod bipolar cells cells is seen with PKCα staining, a. Arrowheads indicate remaining Müller glia (F) and bipolar cells (G). p-values are the result of unpaired t-tests on cell counts with data presented as mean ± SEM. Cell counts were performed on 2 or more sections from 5-8 animals per condition. Scale bars - 100μm
We used in vivo electroporation to overexpress NFIA/B/X genes in P0 retina (de Melo and Blackshaw, 2018), and at P14 saw an increase in both bipolar cells and Müller glia -- the two last-born major retinal cell types -- along with a complementary reduction in the fraction of rod photoreceptors and amacrine cells (Figure 5C–D). This increase in specification of the latest-born retinal cell fates was accompanied by a decrease in incorporation of EdU in NFIA/B/X-overexpressing cells compared to control electroporated cells, when measured using a 24hr EdU pulse/chase from P2 to P3 (Figure 5E). Although we cannot rule out that the decrease in EdU incorporation results from cell cycle lengthening or arrest, we suggest that these combined results favor an interpretation by which NFI factors promote both cell cycle exit and bipolar/glial cell specification in the postnatal retina.
To further address the function of the NFI factors in the developing retina, we generated conditional loss-of-function alleles of each Nfi gene (Campbell et al., 2008; Hsu et al., 2011) in RPCs by using the Chx10-Cre line (Rowan and Cepko, 2004). In addition to single mutants for each gene, we also generated Nfia/b double and Nfia/b/x triple mutants, and assessed the retinal phenotype of each mutant line. Loss of function of either Nfia, Nfib or Nfix in RPCs led to a disruption of Müller glial marker staining and breaks in the retinal outer limiting membrane (OLM) (Figure 5F; Figure S18B). These phenotypes were amplified in Nfia/b double mutant mice, exhibiting a more pronounced loss of Müller glial markers (glutamine synthetase; GS; Figure 5D; p27, Lhx2; Figure S18) and a severe disruption of the OLM, which led to major defects in retinal lamination (Figure S19B). These animals also showed a reduction in bipolar cell markers including Pkca, Vsx2, and Isl1 (Figure 5G; Figure S18). Nfia/b/x triple mutants showed a nearly complete loss of both Müller glial and bipolar cell markers (Figure 5F–G), although other retinal cell-specific markers were present but with altered expression patterns due to disruption of retinal lamination, which likely results from the absence of Müller glia (Byrne et al., 2013; de Melo et al., 2016b) (Figure S18).
To determine if NFI factors are required for bipolar and Müller glial cell specification, or if they selectively regulate the survival of these cell types, we measured apoptotic cell death in conjunction with immunostaining for mature cell types in P7 retina. TUNEL-positive cells were increased in Nfia/b/x triple mutant compared to control retinas (Figure S19D). Co-labeling of TUNEL-positive cells with markers of amacrine cells (Pax6), photoreceptors (recoverin), or rod bipolar cells (PKCa) indicated a small (1-3 cells/section) but significant increase in the number of apoptotic bipolar cells and photoreceptors in Nfia/b/x mutant retinas compared to control (0-1 cell/section) (Figure S19D). However, we could not directly measure glial cell death, due to a lack of availability of strictly Müller glia-specific markers at P7. While we cannot rule out a potential role of NFI factors in bipolar and Müller glial cell survival, we believe the small increases in bipolar cell death cannot account for the near total loss of bipolar cells seen a week later at P14. Therefore, we support a role of the NFI transcription factors in control of bipolar Müller glial cell specification. Taken together, these results demonstrate that Nfia/b/x are necessary and sufficient for specification and differentiation of late-born retinal cell types -- Müller glia and bipolar cells -- within the postnatal retina.
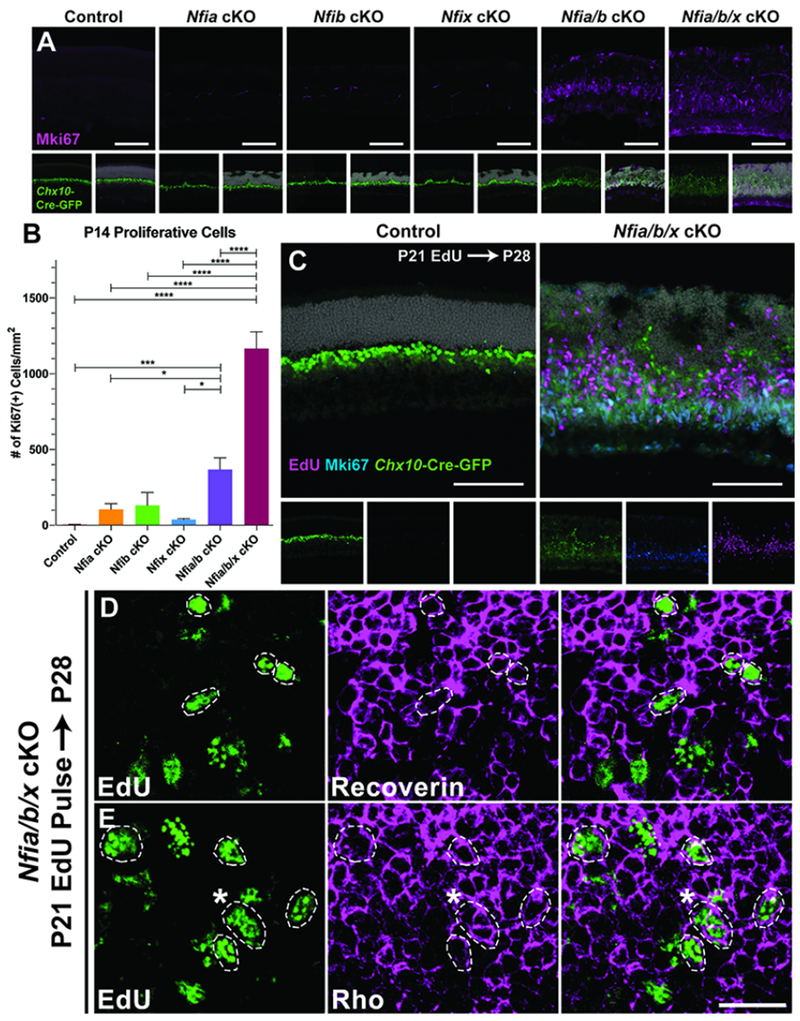
In addition to the significant loss of bipolar and Müller glial cells, persistent, weak Chx10-GFP-Cre reporter expression was seen in both Nfia/b double and Nfia/b/x triple mutant retinas, similar to transgene expression levels in RPCs. Based on this, and previous reports implicating Nfix in proliferative quescience (Martynoga et al., 2013), we hypothesized that the NFI mutants may retain proliferating RPCs. To address this, we analyzed the RPC marker Mki67 in the mutant retinas. Proliferation persisted in P14 Nfia/b double mutants Nfia/b/x triple mutant retinas, well beyond when RPCs are normally present (Figure 6A–B). Proliferating cells were abundant even as late as P28 (Figure 6C, Figure S19H), and were accompanied by a thickening of the retina (Figure S19F). Although a small, yet significant, number of proliferating microglia are seen in mutant retinas (Figure S19C+G), most proliferative cells co-label with Chx10-GFP-Cre transgene, indicating that they are of retinal origin (Figure 6C). However, they are practically devoid of other canonical RPC markers such as Pax6 and Vsx2, and also fail to express RPC-specific genes that are also required for Müller glial development such as Lhx2 (de Melo et al., 2016b; Figure S18). Consistent with the activation of retinal microglia, remaining Müller glial cells within the retinas of Nfia/b double or Nfia/b/x triple mutants have become reactive, exhibiting increased expression of glial fibrillary activating protein (GFAP; Figure S19A). Co-labeling of P7 Chx10-GFP-positive cells with TUNEL indicated that many, but not all, of these ectopic RPCs may be undergoing cell death (Figure S19E). However, as Chx10-GFP also is expressed within bipolar cells, we cannot exclude a scenario where some of the TUNEL/Chx10-GFP-positive cells are in fact dying bipolar cells.
Figure 6. Loss of NFI factors results in sustained RPC proliferation and neurogenesis.
(A) Proliferative cells were detected using anti-Ki67. (B) Quantification of Ki67+ cells normalized to area of the assessed retina; p < 0.0001 (C) P21 EdU injections chased to P28 and co-stained with Ki67 in control and Nfia/b/x triple mutants. (D-E) Confocal images of P28 retinas injected with EdU at P21 and co-labeling with (D) recoverin or (E) rhodopsin, indicates differentiation of ectopic RPCs into photoreceptors (circled nuclei). Asterisk in panel E indicates marker colocalization. Cell counts were performed on 3 or more sections from 3-5 animals per condition. Statistics are results of a one-way ANOVA followed by Tukey’s multiple comparison test with data presented as mean ± SEM. * - p < 0.05; *** - p < 0.001; **** - p < 0.0001. Scale bars: A and C - 100μm; D-E - 20μm.
To examine the extent of differentiation of this ectopic population of proliferative cells within the retina, we pulsed control and Nfia/b/x triple mutants with EdU at P21 and chased labeled cells at P28. EdU labeling indicates that ~15% of proliferating cells in P21 retina give rise to recoverin-positive photoreceptor-like cells by P28 (Figure 6D; Figure S19I). Co-labeling of EdU with rhodopsin (Figure 6E) is consistent with ongoing generation of rod photoreceptors. However, a proportion (~7%) of EdU labeled cells maintain expression of the proliferative marker Mki67 (Figure 6C; Figure S19I). Therefore, retinal thickening seen in Nfia/b/x triple mutants can, at least in part, be attributed to the continuous proliferation and differentiation of retinal cells. Although we cannot rule out a developmental delay resulting from NFI transcription factor loss of function, we favor an interpretation that suggests expression of Nfia/b/x in late-stage RPCs not only directs specification of Müller glial and bipolar cells on late-stage RPCs, but also promotes cell cycle exit.
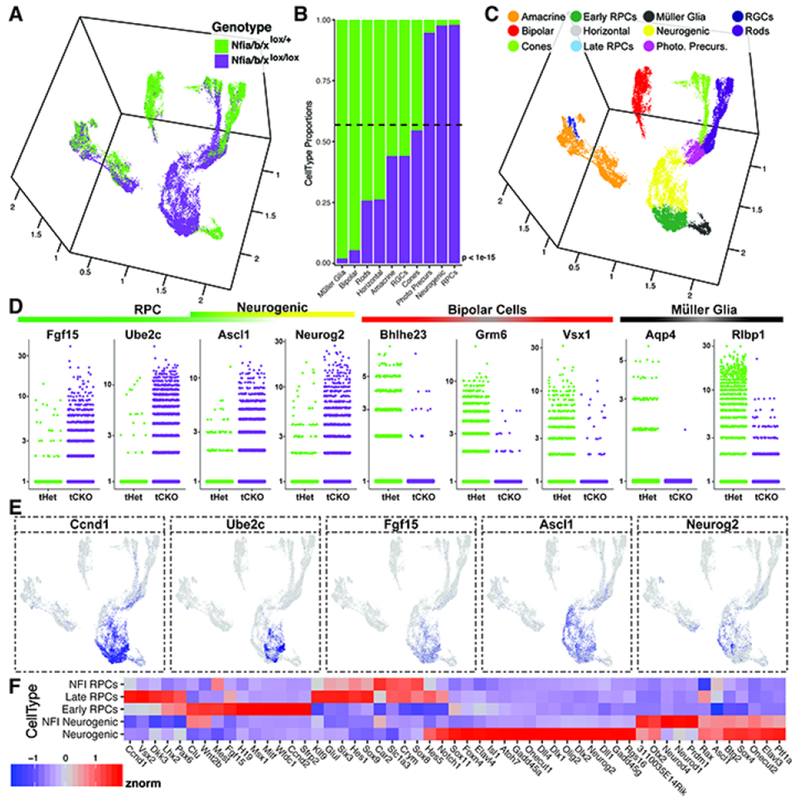
To further analyze Nfia/b/x loss-of-function mutants, we performed single-cell RNA-sequencing on P14 Chx10-GFP-Cre;Nfialox/+;Nfiblox/+;Nfixlox/+ control and Chx10-GFP-Cre;Nfialox/lox;Nfiblox/lox;Nfixlox/lox mutant retinas, performing biological replicates for both genotypes (Figure S20). Using the same bioinformatic workflow as the developmental single-cell RNA-sequencing dataset, we obtained a reduced-dimensional space using UMAP and annotated cell types based on marker gene expression within clusters of cells, removing annotated doublets and extra-retinal cells within final visualizations (Figure 7, Figure S20), which demonstrated that few Müller glial and bipolar cells in remained in Nfia/b/x mutant retinas (Figure 7B–D). Many RPCs, neurogenic cells and photoreceptor precursors were also seen in mutant retinas (Figure 7B–E), consistent with previous observations of ongoing proliferation and photoreceptor generation (Figure 6).
Figure 7. ScRNA-seq analysis of Nfia/b/x mutants.
(A) UMAP dimension reduction of scRNA-seq on P14 Nfia/b/x triple conditional knockouts and heterozygous controls, colored by genotype. (B) Genotype comparisons of the proportions of cell types obtained from scRNA-seq experiments. (C) UMAP dimension reduction of NFI mutant and control scRNA-seq experiments colored by annotated cell type. (D) Jitter plots of the cellular expression of marker genes of primary RPCs, neurogenic cells, bipolar cells, and Müller glia, faceted by genotype. (E) UMAP dimension reduction plots of cellular expression of primary RPC markers (Ccnd1, Ube2c, Fgf15, Ascl1) and markers of neurogenic cells (Ascl1 and Neurog2). Cellular expression is colored on a scale of low (grey) to high (blue) expression. (F) Heatmap displaying the relative expression levels of transcripts within Nfia/b/x mutant primary and neurogenic RPCs compared to early and late primary and neurogenic wild-type RPCs from the developmental dataset. Abbreviations: Photo. Precurs. - Photoreceptor Precursors; tHet - triple Nfia/b/x Heterozygous Chx10-Cre (+); tCKO - triple Nfia/b/x conditional knock-out. P-values (B) are the result of a chi square test for proportions of cells by genotype.
Nfia/b/x mutant primary and neurogenic RPCs show key changes in gene expression relative to wild-type late-stage RPCs. First, neurogenic mutant RPCs show high levels of Otx2, Neurod1, and Prdml expression, consistent with ongoing selective generation of rod photoreceptors (Figure 7F). In mutant primary RPCs, we observe reduced expression of transcription factors necessary for maintenance of RPC identity and late cell type specification, such as Rax, Pax6, and Vsx2, and Notch pathway genes Notchl and Hes5 (Figure 7E), while mutant neurogenic RPCs show lower expression of the Notch ligands Dll1 and Dll4. These data suggest that NFI factors may act in RPCs to activate genes that promote Müller glial and bipolar cell specification, while repressing genes that promote the generation of rod photoreceptors.
Discussion:
This study represents the first large-scale scRNA-Seq-based analysis across the full course of neurogenesis in any mammalian CNS region. The large number of individual cells profiled alretinal neurogenesis and cell type specification. This represents a major advance beyond previous work and has broad implications for studies of neural development. Previous expression profiling studies of developing retina, even when supplemented with large-scale in situ hybridization, could not provide interpretable expression data at the cellular level (Blackshaw et al., 2001; Blackshaw et al., 2004; Hoshino et al., 2017). Likewise, previous single cell gene expression profiling of retinal progenitors has been performed, but profiled only on a few hundred cells using semi-quantitative microarray-based analysis (Trimarchi et al., 2008). ScRNA-Seq studies of neurogenesis in other brain regions profiled only a few thousand cells, at many fewer developmental ages (La Manno et al., 2016; Mayer et al., 2018; Nowakowski et al., 2017; Zhong et al., 2018), and thus do not fully capture the transcriptional changes associated with the initiation of neurogenesis, changes in development competence, and loss of proliferative potential.
Previous studies have suggested that RPCs which express neurogenic bHLH factors such as Neurog2, Atoh7 and Olig2 may be biased to generate specific neuronal subtypes (Brzezinski et al., 2011; Brzezinski et al., 2012; Hafler et al., 2012). This study identifies a clear transcriptional distinction between the uncommitted population of primary RPCs, and an RPC population expressing genes that include neurogenic bHLH factors. Significant transcriptional differences are also observed between early and late stages in both primary and neurogenic RPCs, coincident with changes in RPC competence. Since most of the RPC transcriptome is temporally dynamic, we suggest that the developmental age of an individual RPC can be inferred using these data. Nonetheless, beyond the categories described here, we see no clear evidence for molecularly distinct RPC subtypes at individual ages, supporting a stochastic model of cell fate specification during retina cell fate specification (Gomes et al., 2010; He et al., 2012).
Since these data represent a snapshot in time for each individual cell, neither division mode or cell cycle duration can be assessed. However, the increase in variation of expression of cell cycle genes within RPCs at individual ages as development progresses implies an increase in cell cycle length. We also observe a progressive increase in Notch signaling. These transitions may reflect changes that occur gradually and continuously over the course of neurogenesis, such as the shift between symmetric and asymmetric modes of cell division (Livesey and Cepko, 2001), and the increase in retinal cell cycle length (Alexiades and Cepko, 1996). In contrast, competence transitions appear to be more discrete, with sharper distinctions between early versus late populations of primary and neurogenic RPCs cells. Likewise, transitions between early neuroepithelial cells and proliferating RPCs are sharp, as are transitions between the primary and neurogenic RPC population.
Both supervised pseudotime and unsupervised scCoGAPS identified hundreds of candidates for genes that regulate developmental competence transitions and/or postmitotic differentiation of individual retinal cell types. Many of these genes show dynamic and complex expression patterns, and they are expressed in discrete temporal windows in multiple cell types. Such complex expression profiles are more the rule than the exception, and they are identifiable from scCoGAPS patterns. As a consequence, functional analysis of such genes needs to be conducted and interpreted carefully. Whereas scCoGAPS finds patterns shared across cells, additional techniques are neededl to quantify the relative heterogeneity of cells at specific developmental ages. Here, we apply the EVAsc statistic (Afsari et al., 2014; Davis-Marcisak et al., 2018) to quantify differences in transcriptional heterogeneity of discrete gene sets as a measure of relative pathway dysregulation amongst cells aggregated by a common feature -- in our case, by developmental age.
This analysis identified NFI transcription factors as candidate regulators of temporal patterning in the developing retina. NFI factors are essential for astrocyte specification in both mice and humans (Deneen et al., 2006; Glasgow et al., 2014; Kang et al., 2012; Subramanian et al., 2011; Tchieu et al., 2019), as well as subpopulations of radial glia (Shu et al., 2003). In the spinal cord, Nfia and Nfib drive astrocyte specification by cooperatively activating transcription of Nfix (Matuzelski et al., 2017). Loss of function of Nfia, Nfib or Nfix individually each leads to defects in Müller glial differentiation and disruption of retinal morphology. However, Nfia/b double or Nfia/b/x triple mutants show that NFI factors regulate the generation of not only Müller glia, but also late-born bipolar neurons. In addition, NFI factors are also necessary for cell cycle exit in late-stage RPCs.
Previous studies of the role of NFI factors in control of cell proliferation have demonstrated that their role is complex and cell type-dependent. While Nfix inhibits proliferation of both ES-derived neuroepithelial cells in culture (Martynoga et al., 2013) and postmitotic neural stem cells of the SVZ (Heng et al., 2015), Nfia promotes astrocyte proliferation (Glasgow et al., 2013). Chromatin immunoprecipitation followed by DNA-sequencing (ChIP-Seq) of NFIB in both prostate and lung cancer cells indicates that NFIB target sites are associated with genes that regulate cell cycle progression (Denny et al., 2016; Grabowska et al., 2016). The combined observations of intermediate cell expansion and an increased severity of prostate cancer following NFIB loss in humans (Grabowska et al., 2016) and ChIP-Seq data suggests that the NFI transcription factors may directly regulate expression of genes controlling proliferative potential. However, as NFIA overexpression promotes proliferation within human glioblastoma cells (Lee et al., 2014), further insight into the mechanisms by which the NFI transcription factors regulate RPC proliferation will require a detailed examination of the direct targets of NFI factors in the developing retina. This dual role in regulating both proliferative quiescence and cell fate specification has not been reported in the developing CNS, but may represent a widely used mechanism for generating appropriate numbers of late-born cells.
Analysis of Nfia/b/x mutants using scRNA-Seq has suggested mechanisms by which NFI transcription factors control both cell fate specification and proliferative quiescence. Expression of multiple cell-cycle genes confirmed the presence of ongoing proliferation within Nfia/b/x mutant retinas. The observation that the expression of RPC-specifying transcription factors (Pax6,Lhx2, Vsx2, Rax) is reduced in the absence of Nfia/b/x expression further suggests the importance of these transcription factors in specifying retinal cell fate. Consistent with our observations of reduced Lhx2 and Rax expression in NFI mutant RPCs, selective loss of function of Vsx2, Lhx2 or Rax in RPCs resulted in loss of late-born cell types, including bipolar cells and Müller glia (de Melo et al., 2016b; Livne-Bar et al., 2006; Rodgers et al., 2018). However, unlike previous studies in which selective deletion of Lhx2 or Rax in RPCs results in cell cycle exit and precocious differentiation (de Melo et al., 2016b; Rodgers et al., 2018), we observe maintenance of RPC proliferation.
The cell diversity of other CNS regions are considerably higher than retina, and their development far less well characterized. ScRNA-Seq, in conjunction with both unsupervised pattern identification and supervised pseudotime analysis like that described here, will provide the potential to understand the gene regulatory networks that give rise to the immense diversity of cell types within the CNS.
STAR METHODS:
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by Lead Contact Seth Blackshaw (sblack@jhmi.edu).
Experimental Model and Subject Details
Timed pregnant CD-1 mice used for droplet-based single cell RNA sequencing, in situ hybridization, and electroporation were purchased from Charles River Laboratories. For Smart-Seq2 analysis, Chx10-Cre-GFP mice were used (Rowan and Cepko, 2004). Nfialox/lox(see below); Nfiblox/lox (Hsu et al., 2011), and NfiXox/lox (Campbell et al., 2008) mice were crossed to Chx10-Cre-GFP to generate RPC-specific loss of function mutants of these genes. Nfialox/oxmice were generated in the Roswell Park Gene Targeting and Transgenic Shared Resource using heterozygous targeted ES cells from EUCOMM project 38437 (KOMP). Mice containing the targeted allele were crossed with Flp-deleter mice (Jackson Labs Stock No. 009086) to excise the lacZ reporter and generate a floxed allele of Nfia. Mice were housed in a climate-controlled pathogen free facility, on a 14 hour-10 hour light/dark cycle (07:00 lights on-19:00 lights off). All experimental procedures were preapproved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine. For all single cell experiments, dissociated cells were obtained from pools of retinas from both male and female pups as determined visually (where applicable) or through Xist expression within the datasets. All single-cell experiments displayed relatively equal representation of each sex within the dataset. Developmental ages of animals for single-cell experiments include E11, E12, E14, E16, E18, P0, P2, P5, P8, and P14. All immunohistochemistry was performed at P14 or P28, as noted within the text. Cell death assays were performed on P7 retinas.
Method Details
Tissue Dissociation
Eyes were enucleated from animals and retinas dissected in fresh and cold 1× PBS, using eyes from approximately one litter of animals for each sample in order to ensure appropriate numbers of cells were captured for downstream analyses. Dissected retinas were then transferred to 200μl of cold HBSS per retina (P14) or an approximate equivalent amount of tissue for younger ages. An equivalent amount of Papain solution (for 1ml - 700μl reagent grade water, 100μl of freshly prepared 50mM L-Cysteine (Sigma), 100μM 10mM EDTA, 10μM 60mM 2-mercaptoethanol (Sigma), and Papain added to 1mg/ml (Worthington)) was added and incubated at 37°C for 10 minutes, with slight trituration performed every 2 minutes. 600μl of Neurobasal Media supplemented with 10% FBS was added for every 400μl of dissociation solution, and samples were further dissociated with gentle pipetting. Samples were subjected to DNAse treatment (5μl DNAseI (RNAse free Recombinant DNAseI; Roche) for every 1ml of dissociation solution; 5 minutes at 37°C). Cells were then pelleted through centrifugation for 5 minutes at 300 RCF. Liquid was carefully aspirated off the cell pellet, followed by resuspension of the pellet in 1-5ml Neurobasal media with 1% FBS, depending on required concentration of cells in suspension. Cellular aggregates were removed by straining cells through a 50μm filter.
Single cell library preparation
Smart-Seq2 analysis was performed on individual sorted Chx10-Cre-GFP (+) RPCs isolated by FACS into 96-well plates, and processed as previously described (Chevee et al., 2018). Single cell suspensions for 10× libraries were loaded onto the 10× Genomics Chromium Single Cell system using the v2 chemistry per manufacturer’s instructions (Zheng et al., 2017). Approximately 17,000 live cells were loaded per sample in order to capture transcripts from roughly 10,000 cells. Estimations of cellular concentration and live cells in suspension was made through Trypan Blue staining and use of the Countess II cell counter (ThermoFisher). Single cell RNA capture and library preparations were performed according to manufacturer’s instructions. Sample libraries were sequenced on the NextSeq 500 (Illumina).
Library preprocessing
Sequencing output was processed through the Cell Ranger 1.2.1 or Cell Ranger 2.1.0 mkfastq and count pipelines using default parameters. Reads were quantified using the mouse reference index provided by 10× Genomics (refdata-cellranger-mm10 v1.2.0). Raw count matrices for individual runs were manually aggregated and cells were given unique, sample-specific cell identifiers to prevent duplication of non-unique barcodes across samples. Raw data were processed through the Cell Ranger pipeline and raw counts were aggregated together for input into the Monocle2 R/Bioconductor platform (Trapnell et al., 2014).
Coarse assignment of cell type at individual time points
tSNE-dimension reduction was performed on the top principal components learned from high variance genes in cells captured at individual timepoints. Mclust version 5.4 (Scrucca et al., 2016) was used to cluster cells in tSNE-space at which point cell type identity of clusters was assigned based on expression of known marker genes for either retinal or non-retinal tissue.
Cell normalization, identification of high variance genes, and differential testing
After coarse annotation of cells from individual time points, the 10× data were manually aggregated to create a comprehensive dataset. Initial cell type designation was used to aid in supervising downstream analyses. To identify genes with higher variation than expected, we first normalized for sequencing depth using the Waddington-OT transformation to transcript copies per 10,000 (CPT) (Schiebinger et al., 2017). To identify high-variance genes, a generalized additive model (MGCV R package (Wood, 2011)) was fit to the log2 mean CPT versus a cubic spline fit to the log2 coefficient of variation (BCV) across all genes with detectable expression in at least 5 cells (Figure S3). Genes with residuals to this fit greater than or equal to 1.5 were chosen as ‘high-variance’ genes and the log2 CPT or CPC values for these selected genes were used as input for downstream analyses as appropriate. All differential expression tests were performed across all expressed genes using the Monocle2 VGAM likelihood ratio test (Trapnell et al., 2014). In all cases, the number of genes detected in each cell was included in both the full and reduced models as a nuisance parameter.
Visualization of global cellular state(s)
Dimensionality reduction and visualization for the aggregate 10× data was performed using Uniform Manifold Approximation and Projection (UMAP) (McInnes and Healy, 2018) for all cells passing QC. Briefly the first 20 principle components of the log10(CPT+1) of the high-variance genes was used as input for the python implementation of the UMAP algorithm with the following additional parameters: min_dist = 0.3, n_neighbors=30, random_state=1, n_components=3, metric=“canberra”. The resulting 3 dimensional embedding was imported into R and visualized using the rgl package.
Clustering analysis and final cell type assignment
Clustering on the UMAP embedding was performed using k-means clustering on UMAP coordinates. Cell type assignment was informed by both marker gene expression and previous coarse cell type annotation from clustering performed on the individual ages. Clustering analysis of the amacrine trajectory to delineate amacrine and horizontal cells was performed using k-means clustering on the largeVis coordinates.
Pseudotime
Monocle pseudotemporal analysis was performed on the high variance gene set derived from the subsets of cells being analyzed, altering the dimension parameter to refine resulting trees to reflect both known biology and terminal states comprised largely of single cell types. For pseudotemporal analysis performed on the subset of RPCs, the following additional parameters were used during dimension reduction: tol = 1.0e-8, iambda=400*ncol (CellDataSet). The root state was identified as the state that contained the majority of cells with the earliest developmental age for each individual analysis. Genes with significant expression changes as a function of pseudotime were identified using the Monocle differential gene test, using a multiple-testing corrected q-value cutoff of 1.0e-5. Cell type identity of individual pseudotime states were assigned based on the cell type identity of the majority of cells within a given branch. BEAM tests were performed on most major branch points of the cellular hierarchy using all default parameters with the exception of the dimensionality of the embedding.
Pattern discovery - scCoGAPS
scCoGAPS and PatternMarker analysis was performed using the R/Bioconductor package CoGAPS version 3.0.0 as described (Stein-O’Brien et al., 2018). Briefly, the expression matrix of high variance genes was subset into 200 sets of cells for parallelization. A sampling scheme, using expertly curated cell type annotations, was used in order to ensure representation of rare cell types in each set. A static ratio of cell types was established to reflect biological prevalence and diversity of each cell type while allowing for adequate representation. Cells were then sampled with replacement to ensure the necessary numbers to maintain this ratio in all sets. The resulting 200 sets of 1500 cells each were then run in parallel over a range of dimensionalizations. Consensus amplitude signatures were derived by a matching algorithm designed to ensure robustness of signatures (Star Methods). Pattern weights for all the cells were then learned in parallel from these signatures to ensure reciprocity across all of the sets. The PatternMarker statistic was calculated as previously described (Stein-O’Brien et al., 2017). The entire pipeline has been compiled into the scCoGAPS function found within the CoGAPS package starting at version 3.0.0.
Pathway dysregulation analysis
We quantify pathway dysregulation using EVA from the R/Bioconductor package GSReg version 1.17.0 (Afsari et al., 2014). Briefly, EVA computes the Kendal-τ dissimilarity between transcriptional profiles of genes in a pathway for all cells in one group and compares their expected dissimilarity to that computed for all the cells in another group using U-theory statistics. This statistic quantifies relative pathway dysregulation between cells in these two conditions. Because the Kendal-τ dissimilarity is rank-based, it is robust to normalization and read depth, but ill-defined for missing values. To address this, we imputed scRNA-Seq data from RPCs with MAGIC version 0.1.0 (Python) prior to analysis (Davis-Marcisak et al., 2018; Tang et al., 2016). EVA is applied to gene sets for the cell cycle from GeneGlobe pathways, FGF pathway from the reactome pathway database, NOTCH pathway from the hallmark gene sets, and WNT pathway from the hallmark gene sets.
in situ hybridization
Whole heads (E12-P5) or enucleated eyes of animals were placed directly into Tissue-Tek OCT media (VWR) and frozen and stored at −80°C prior to sectioning. Section RNA in situ hybridization was performed as previously described (Blackshaw et al., 2004). Briefly, sections were air dried on slides and then fixed in 4% paraformaldehyde in PBS for 10 minutes. Slides are then washed in PBS and incubated in acetylation solution (.1M triethanolamine hydrochloride + .27% v/v acetic anhydride) for 10 minutes. Following additional washed in PBS, slides are placed in hybridization solution (50% formamide v/v, 5× saline-sodium citrate buffer (SSC), 5× Denhardt’s Solution, 250μg/ml Yeast tRNA, 500μg/ml Sperm DNA) for 2 hours. Slides are then incubated in hybridization solution supplemented with DIG-labeled antisense probes overnight at 70°C in a humidified chamber using siliconized coverslips. Slides are then washed in 5× SSC at 65°C until coverslips fall off, followed by two washes in .2× SSC at 65°C for 30 minutes each and one 5 mi nute wash in .2× SSC at room temperature. Slides are washed in .1M Tris pH 7.5; .15M NaCl and placed into antibody blocking buffer (.1M Tris pH7.5; .15M NaCl, 5% heat inactivated sheep sherum) for 1 hour at room temperature. Slides are then incubated overnight in blocking buffer supplemented with anti-DIG-AP antibody (1:5000; Roche). Antibody solution is washed off using three 5 minute washes in 1M Tris pH 7.5; .15M NaCl. Slides are then washed in .1M Tris pH 9.5; .1M NaCl; .05M MgCl2 once for 5 minutes. Colorimetric reactions for antibody detection are performed in the dark using 1M Tris pH 9.5; .1M NaCl; .05M MgCl2 solution supplemented with .337 mg/ml NBT and .175mg/ml BCIP. Reactions are stopped by placing slides in 10 mM Tris pH7.5 and 50 mM EDTA.
Immunohistochemistry and EdU staining
Eyes were enucleated from animals and placed in cold 4% paraformaldehyde for 1 hour. Retinas were then dissected and placed into 30% sucrose in PBS overnight at 4°C, after which they were mounted in Tissue-Tek OCT media (VWR) and frozen prior to sectioning. Immunohistochemistry was performed as previously described (de Melo et al., 2016a). Slides are air dried and then placed directly into blocking solution (1× PBS, 5% Horse serum, .2% Triton X-100, .02% sodium azide, .1% BSA w/v) for two hours. Slides are placed in primary antibody diluted in blocking solution overnight at 4°C. Slides are then washed in 1× PBS plus .05% tri ton thrice for 5 minutes each. Primary antibodies are detected through incubation using fluorescently-tagged secondary antibodies diluted 1:500 in blocking buffer for 2 hours in the dark. Slides are then washed in 1× PBS plus .05% triton and then nuclei are counterstained using DAPI (1:5000 in 1× PBS plus .05% triton). Slides are then coverslipped using Vectashield (Vector Labs).
For adult EdU analyses, animals were injected with 50 mg/kg EdU (10 mM in saline) at P21 and euthanized at P28. Developmental EdU injections were performed through injection of 20ul of 10mM EdU in saline into P2 mouse pups and euthanized 24 hours later. EdU staining was performed using the Click-IT EdU AlexaFluor 647 imaging kit (Invitrogen) per manufacturer’s instructions, with slides placed into blocking steps for the immunohistochemistry protocol directly after EdU detection. Nuclei were counterstained with DAPI (1:5000) and coverslipped using Vectashield (Vector Labs). Immunohistochemical data shown was imaged and photographed on either the BZ-X700 microscope (Keyence) or using a LSM 700 confocal (Zeiss).
Cell Death Assay
Retinal sections were stained using the in situ cell death Detection kit, TMR red (12156792910, Roche Applied Sciences) according to the manufacturer’s instructions. Briefly, retinal sections were washed three times for 5 minutes in PBS and permeabilized for 2 minutes in freshly prepared cold 0.1% Triton X-100 and 0.1% sodium citrate solution. The slides were then washed twice with PBS before being resuspended in the TUNEL reaction mixture for 1 hour in a humidified chamber at 37°C. Slides were washed with PBS before continuing with the immunohistochemistry staining protocol. Nuclei were counterstained with DAPI (1:5000) and cover slipped using Vectashield hardset antifade mounting media (Vector Labs). Slides were imaged using the BZ-X700 microscope (Keyence).
Cell Counts
All cell counts were performed by investigators blinded to the experimental condition/genotype. Subsets of images were counted multiple times to ensure reproducibility across experimental conditions and replicates. For P3 electroporation/EdU and P14 electroporation experiments, images displaying less than 50 or 100 electroporated cells per field of view, respectively, were excluded from downstream analyses to avoid over-representation of changes in cell type proportions from small numbers of cells.
Quantification and Statistical Analysis
Differential gene tests on the single cell datasets were performed using all expressed genes within the Monocle2 VGAM likelihood ratio test (Trapnell et al., 2014).
All bar graph data is shown as mean ± SEM, with statistical analyses were done with Prism 7 (GraphPad). Comparisons conducted were Student’s t tests or one-way ANOVAs with a Tukey’s multiple comparisons correction. p < 0.05 was considered as statistically significant. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001
Data and Software Availability:
Single cell RNA-Seq count data are available for direct download at https://github.com/gofflab/developing_mouse_retina_scRNASeq. Interactive queries of individual gene expression patterns can be performed at http://mouse.retina.gofflab.org. Raw sequencing data is deposited with the NCBI Short Read Archive and Gene Expression Omnibus under accession number GSE118614.
Supplementary Material
Movie 1 – UMAP-dimension reduction of droplet-based single cell RNA-sequencing of single developing mouse retinal cells with samples colored by developmental age. Related to Figure 1.
Table S5 - 10× cell type features. Related to Figure 1E–G and Figure S2J–M.
Table S6 - High variance genes across 10× retinal cells. Related to Figure 2.
Table S7 - Genes displaying differential expression across retina pseudotime. Related to Figure 2D.
Table S8 - Gene weights within scCoGAPS patterns. Related to Figure 4.
Movie 2 – UMAP-dimension reduction of droplet-based single cell RNA-sequencing of single developing mouse retinal cells with samples colored by annotated cell type as determined by marker gene expression in clustered cells. Extra-retinal and doublet cells have been removed. Related to Figure 1F.
Table S4 - High variance genes used for UMAP dimension reduction on 10× samples. Related to Figure 1E–F and Figure S2F–I.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies: | ||
| goat anti-Brn3 (1:200) | Santa Cruz Biotechnology | Cat. # SC-6026; RRID:AB_673441 |
| mouse anti calbindin-D-28K; Calb1 (1:200) | Sigma-Aldrich | Cat. # C9848 (clone CB-955); RRID:AB_476894 |
| sheep anti-Chx10 (1:500) | Exalpha Biologicals | Cat. # X1180P; RRID:AB_2314191 |
| rabbit anti-GFP (1:1000) | Thermo Fisher Scientific | Cat. # A6455; RRID:AB_221570 |
| mouse anti-Glutamine synthetase; Glul (1:200) | BD Biosciences | Cat. # 610518 (Clone6); RRID:AB_397880 |
| mouse anti-Islet1 (1:200) | Developmental Studies Hybridoma Bank |
Cat. # 40.2D6; RRID:AB_528315 |
| mouse anti-Ki67 (1:200) | BD Biosciences | Cat. # 550609 (Clone B56); RRID:AB_393778 |
| rabbit anti-Lhx2 (1:1000) | generated in house with Covance; (de Melo et al., 2012) | RRID:AB_2783882 |
| mouse anti-P27Kip1 (1:500) | BD Transduction Labs | Cat. # 610241 (clone57/Kip1/p27); RRID:AB_397636 |
| mouse anti-Pax6 (1:200) | Developmental Studies Hybridoma Bank |
Cat. # Pax6a.a 1-223; RRID:AB_528427 |
| mouse anti-Nfia/b (1:200) | CDI Labs | Cat. # R1356.1.2C6; RRID:AB_2618885 |
| mouse anti-Tfap2a (1:200) | Abnova | Cat. # Clone 2G5; RRID:AB_490092 |
| rabbit anti-Recoverin (1:200) | Millipore | Cat. # AB5585; Lot LV1480447; RRID:AB_2253622 |
| mouse anti-Pkca (1:200) | Millipore | Cat. # Clone M4; RRID:AB_2284233 |
| rabbit anti-GFAP (1:500) | DakoCytomation | Cat. # Z0334; RRID:AB_10013382 |
| mouse anti-Rho4D2 (Rhodopsin; 1:200) | Dr. Robert Molday; (Laird and Molday, 1988) | RRID:AB_2315273 |
| rabbit anti-b-catenin (Ctnnb1; 1:200) | Sigma | Cat. # C2206; RRID:AB_476831 |
| rabbit anti-Iba1 (1:400) | Wako | Cat. # 019-19741; RRID:AB_839504 |
| donkey anti rabbit 488 (1:500) | Jackson ImmunoResearch | Cat. # 711-485-152; RRID:AB_2492289 |
| goat anti-rabbit 555 (1:500) | Thermo Fisher Scientific | Cat. # A-21428; RRID:AB_141784 |
| donkey anti-rabbit 594 (1:500) | Thermo Fisher Scientific | Cat. # A-21207; RRID:AB_141637 |
| goat anti-rabbit 633 (1:500) | Thermo Fisher Scientific | Cat. # A-21070; RRID:AB_2535731 |
| donkey anti-rabbit 647 (1:500) | Jackson ImmunoResearch | Cat. # 711-605-152; RRID:AB_2492288 |
| donkey anti-mouse 555 (1:500) | Thermo Fisher Scientific | Cat. # A-31570; RRID:AB_2536180 |
| donkey anti-mouse 594 (1:500) | Jackson ImmunoResearch | Cat. # 715-585-150; RRID:AB_2340854 |
| donkey anti-mouse 647 (1:500) | Jackson ImmunoResearch | Cat. # 715-605-150; RRID:AB_2340862 |
| donkey anti-goat 594 (1:500) | Thermo Fisher Scientific | Cat. # A-11058; RRID:AB_2534105 |
| donkey anti-goat 633 (1:500) | Thermo Fisher Scientific | Cat. # A-21082; RRID:AB_2535739 |
| donkey anti-sheep 568 (1:500) | Thermo Fisher Scientific | Cat. # A-21099; RRID:AB_2535753 |
| donkey anti-sheep 647 (1:500) | Jackson ImmunoResearch | Cat. # 713-605-003; RRID:AB_2340750 |
| Critical Commercial Assays: | ||
| Chromium Single Cell 3’ Library & Gel Bead Kit v2 | 10× Genomics | Cat. # 120237 |
| Chromium i7 Multiplex Kit | 10× Genomics | Cat. # 120262 |
| Chromium Single Cell A Chip Kit | 10× Genomics | Cat. # 1000009 |
| Chromium Controller & Accessory Kit | 10× Genomics | Cat. # 120223 |
| Click-iT EdU Alexa Fluor 647 Imaging Kit | Thermo FIsher Scientific | Cat. # C10340 |
| In Situ Cell Death Detection Kit, TMR red | Sigma | Cat. # 12156792910 |
| Experimental Models: Organisms/Strains | ||
| Mouse:CD1.Tg(Chx10-EGFP/cre/-ALPP)2Clc | Dr. Connie Cepko; (Rowan and Cepko, 2004) | RRID:MGI:3838985 |
| Mouse:Nfiafl/fl | Dr. Richard Gronostajski; This study |
N/A |
| Mouse:Nfibfl/fl | Dr. Richard Gronostajski; (Hsu et al., 2011) | N/A |
| Mouse:Nfixfl/fl | Dr. Richard Gronostajski; (Campbell et al., 2008) |
N/A |
| Deposited Data: | ||
| Retinal Development Smart-Seq2 Data | This Paper | GEO: GSE118614 |
| Retinal Development 10× Data | This Paper | GEO: GSE118614 |
| P14 Nfia/b/x Mutant and control retina 10× Data | This Paper | GEO: GSE118614 |
| Software: | ||
| Cell Ranger - 2.1.0 | 10× Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| GraphPad Prism 7 | GraphPad | www.graphpad.com |
| Monocle2 | Cole Trapnell | http://cole-trapnell-lab.github.io/monocle-release/docs/ |
| R version 3.4.1 | The R project | https://www.r-project.org/ |
| scCoGAPS | Bioconductor; This Paper | https://bioconductor.org/packages/release/bioc/hyml/CoGAPS.html |
| EVAsc | (Davis-Marcisak et al., 2018) | https://bioconductor.org/packages/release/bioc/html/GSReg.html |
| MAGIC | (van Dijk et al., 2018) |
https://github.com/DpeerLab/magic, https://github.com/KrishnaswamyLab/magic |
| umap | (McInnes and Healy, 2018) | https://github.com/lmcinnes/umap |
| largeVis | (Tang et al., 2016) | https://github.com/lferry007/LargeVis |
| Mclust | (Scrucca et al., 2016) | https://cran.r-project.org/web/packages/mclust/index.html |
Highlights:
Mouse retinal neurogenesis was profiled using single-cell RNA-Seq.
Early and late-stage retinal progenitors are distinct.
Primary and neurogenic retinal progenitors are distinct.
NFI factors control cell cycle exit and generation of late-born retinal cell types.
Acknowledgements:
This work was supported by grants from the NIH (R01EY020560 and U01EY027267 to SB, F32EY024201 and K99EY027844 to BSC, K08EY027093 to FR, R01CA177669 and U01CA212007 to EJF), NYSTEM awards C026429, C030133 and C30290GG (RMG), the Chan-Zuckerberg Initiative DAF (2018-183445 to LAG, and 2018-183444 to EJF), the Johns Hopkins University Catalyst (EF & LAG) and Discovery awards (EJF), and the Johns Hopkins University School of Medicine Synergy Award (SB, LAG, & EJF). The authors would like to thank C.A. Berlinicke and D.J. Zack for assistance with FACS analysis, the Johns Hopkins P30 Center for Neuroscience Research (NS050274), use of the 10× Genomics Single Cell system through the Genetic Resources Core Facility; the Johns Hopkins Institute of Genetic Medicine, Baltimore, MD, and the Hopkins Microarray and Deep Sequencing Core for use of the 10× Genomics Single Cell system and assistance with library sequencing; and J. Nathans, A. Kolodkin, R. Bremner, W. Yap, A. McCallion, and D.W. Kim for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
References:
- Afsari B, Geman D, and Fertig EJ (2014). Learning dysregulated pathways in cancers from differential variability analysis. Cancer Inform 13, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mulla F, Bitar MS, Al-Maghrebi M, Behbehani AI, Al-Ali W, Rath O, Doyle B, Tan KY, Pitt A, and Kolch W (2011). Raf kinase inhibitor protein RKIP enhances signaling by glycogen synthase kinase-3beta. Cancer Res 71, 1334–1343. [DOI] [PubMed] [Google Scholar]
- Aldiri I, Xu B, Wang L, Chen X, Hiler D, Griffiths L, Valentine M, Shirinifard A, Thiagarajan S, Sablauer A, et al. (2017). The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis. Neuron 94, 550–568 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiades MR, and Cepko C (1996). Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn 205, 293–307. [DOI] [PubMed] [Google Scholar]
- Baas D, Bumsted KM, Martinez JA, Vaccarino FM, Wikler KC, and Barnstable CJ (2000). The subcellular localization of Otx2 is cell-type specific and developmentally regulated in the mouse retina. Brain Res Mol Brain Res 78, 26–37. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Fraioli RE, Furukawa T, and Cepko CL (2001). Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell 107, 579–589. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, et al. (2004). Genomic analysis of mouse retinal development. PLoS Biol 2, E247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boije H, Edqvist PH, and Hallbook F (2008). Temporal and spatial expression of transcription factors FoxN4, Ptf1a, Prox1, Isl1 and Lim1 mRNA in the developing chick retina. Gene Expr Patterns 8, 117–123. [DOI] [PubMed] [Google Scholar]
- Brzezinski J.A.t., Kim EJ, Johnson JE, and Reh TA (2011). Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development 138, 3519–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski J.A.t., Lamba DA, and Reh TA (2010). Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 137, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski J.A.t., Prasov L, and Glaser T (2012). Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev Biol 365, 395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne LC, Khalid F, Lee T, Zin EA, Greenberg KP, Visel M, Schaffer DV, and Flannery JG (2013). AAV-mediated, optogenetic ablation of Muller Glia leads to structural and functional changes in the mouse retina. PLoS One 8, e76075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CE, Piper M, Plachez C, Yeh YT, Baizer JS, Osinski JM, Litwack ED, Richards LJ, and Gronostajski RM (2008). The transcription factor Nfix is essential for normal brain development. BMC Dev Biol 8, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette M, Mattar P, and Harris WA (2013). Progenitor competence: genes switching places. Cell 152, 13–14. [DOI] [PubMed] [Google Scholar]
- Cepko C (2014). Intrinsically different retinal progenitor cells produce specific types of progeny. Nat Rev Neurosci 15, 615–627. [DOI] [PubMed] [Google Scholar]
- Chevee M, Robertson JJ, Cannon GH, Brown SP, and Goff LA (2018). Variation in Activity State, Axonal Projection, and Position Define the Transcriptional Identity of Individual Neocortical Projection Neurons. Cell Rep 22, 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, and Cepko CL (2006). Wnt2b/beta-catenin-mediated canonical Wnt signaling determines the peripheral fates of the chick eye. Development 133, 3167–3177. [DOI] [PubMed] [Google Scholar]
- Cleary B, Cong L, Cheung A, Lander ES, and Regev A (2017). Efficient Generation of Transcriptomic Profiles by Random Composite Measurements. Cell 171, 1424–1436 e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Marcisak EF, Orugunta P, Stein-O’Brien G, Puram SV, Torres ER, Hopkins A, Jaffee EM, Favorov AV, Afrasi B, Goff LA, et al. (2018). Expression variation analysis for tumor heterogeneity in single-cell RNA-sequencing data. Bioarxiv 479287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J, and Blackshaw S (2018). In Vivo Electroporation of Developing Mouse Retina. Methods Mol Biol 1715, 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J, Clark BS, and Blackshaw S (2016a). Multiple intrinsic factors act in concert with Lhx2 to direct retinal gliogenesis. Scientific reports 6, 32757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J, Du G, Fonseca M, Gillespie LA, Turk WJ, Rubenstein JL, and Eisenstat DD (2005). Dlx1 and Dlx2 function is necessary for terminal differentiation and survival of late-born retinal ganglion cells in the developing mouse retina. Development 132, 311–322. [DOI] [PubMed] [Google Scholar]
- de Melo J, Miki K, Rattner A, Smallwood P, Zibetti C, Hirokawa K, Monuki ES, Campochiaro PA, and Blackshaw S (2012). Injury-independent induction of reactive gliosis in retina by loss of function of the LIM homeodomain transcription factor Lhx2. Proc Natl Acad Sci U S A 109, 4657–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J, Zibetti C, Clark BS, Hwang W, Miranda-Angulo AL, Qian J, and Blackshaw S (2016b). Lhx2 Is an Essential Factor for Retinal Gliogenesis and Notch Signaling. J Neurosci 36, 2391–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, and Anderson DJ (2006). The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52, 953–968. [DOI] [PubMed] [Google Scholar]
- Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, Gruner BM, Chiou SH, Schep AN, Baral J, Hamard C, et al. (2016). Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell 166, 328–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Tachibana N, Touahri Y, Zinyk D, Logan C, and Schuurmans C (2014). Gene expression is dynamically regulated in retinal progenitor cells prior to and during overt cellular differentiation. Gene Expr Patterns 14, 42–54. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, and Oliver G (2003). Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet 34, 53–58. [DOI] [PubMed] [Google Scholar]
- Elliott J, Jolicoeur C, Ramamurthy V, and Cayouette M (2008). Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron 60, 26–39. [DOI] [PubMed] [Google Scholar]
- Emerson MM, Surzenko N, Goetz JJ, Trimarchi J, and Cepko CL (2013). Otx2 and Onecut1 promote the fates of cone photoreceptors and horizontal cells and repress rod photoreceptors. Dev Cell 26, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhy C, Elgart M, Shapira Z, Oron-Karni V, Yaron O, Menuchin Y, Rechavi G, and Ashery-Padan R (2013). Pax6 is required for normal cell-cycle exit and the differentiation kinetics of retinal progenitor cells. PLoS One 8, e76489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Laug D, Brawley VS, Zhang Z, Corder A, Yin Z, Wong ST, Li XN, Foster AE, Ahmed N, et al. (2013). The miR-223/nuclear factor I-A axis regulates glial precursor proliferation and tumorigenesis in the CNS. J Neurosci 33, 13560–13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Zhu W, Stolt CC, Huang TW, Chen F, LoTurco JJ, Neul JL, Wegner M, Mohila C, and Deneen B (2014). Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci 17, 1322–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes FL, Zhang G, Carbonell F, Correa JA, Harris WA, Simons BD, and Cayouette M (2010). Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development 138, 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska MM, Kelly SM, Reese AL, Cates JM, Case TC, Zhang J, DeGraff DJ, Strand DW, Miller NL, Clark PE, et al. (2016). Nfib Regulates Transcriptional Networks That Control the Development of Prostatic Hyperplasia. Endocrinology 157, 1094–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafler BP, Surzenko N, Beier KT, Punzo C, Trimarchi JM, Kong JH, and Cepko CL (2012). Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc Natl Acad Sci U S A 109, 7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Zhang G, Almeida AD, Cayouette M, Simons BD, and Harris WA (2012). How variable clones build an invariant retina. Neuron 75, 786–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng YH, Zhou B, Harris L, Harvey T, Smith A, Horne E, Martynoga B, Andersen J, Achimastou A, Cato K, et al. (2015). NFIX Regulates Proliferation and Migration Within the Murine SVZ Neurogenic Niche. Cereb Cortex 25, 3758–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem CC, Repic M, and Knoblich JA (2015). Proliferation control in neural stem and progenitor cells. Nat Rev Neurosci 16, 647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Ratnapriya R, Brooks MJ, Chaitankar V, Wilken MS, Zhang C, Starostik MR, Gieser L, La Torre A, Nishio M, et al. (2017). Molecular Anatomy of the Developing Human Retina. Dev Cell 43, 763–779 e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Osinski J, Campbell CE, Litwack ED, Wang D, Liu S, Bachurski CJ, and Gronostajski RM (2011). Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev Biol 354, 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav AP, Cho SH, and Cepko CL (2006). Notch activity permits retinal cells to progress through multiple progenitor states and acquire a stem cell property. Proc Natl Acad Sci U S A 103, 18998–19003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James J, Das AV, Rahnenfuhrer J, and Ahmad I (2004). Cellular and molecular characterization of early and late retinal stem cells/progenitors: differential regulation of proliferation and context dependent role of Notch signaling. J Neurobiol 61, 359–376. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Ding Q, Xie X, Libby RT, Lefebvre V, and Gan L (2013). Transcription factors SOX4 and SOX11 function redundantly to regulate the development of mouse retinal ganglion cells. J Biol Chem 288, 18429–18438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusuf PR, Albadri S, Paolini A, Currie PD, Argenton F, Higashijima S, Harris WA, and Poggi L (2012). Biasing amacrine subtypes in the Atoh7 lineage through expression of Barhl2. J Neurosci 32, 13929–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, et al. (2012). Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 74, 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Omori Y, Onishi A, Sato S, Kondo M, and Furukawa T (2010). Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J Neurosci 30, 6515–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, and Doe CQ (2013). Temporal fate specification and neural progenitor competence during development. Nat Rev Neurosci 14, 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike C, Nishida A, Ueno S, Saito H, Sanuki R, Sato S, Furukawa A, Aizawa S, Matsuo I, Suzuki N, et al. (2007). Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol 27, 8318–8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, and Regev A (2015). Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res 25, 1860–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. (2016). Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580 e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laboissonniere LA, Martin GM, Goetz JJ, Bi R, Pope B, Weinand K, Ellson L, Fru D, Lee M, Wester AK, et al. (2017). Single cell transcriptome profiling of developing chick retinal cells. J Comp Neurol 525, 2735–2781. [DOI] [PubMed] [Google Scholar]
- Laird DW, and Molday RS (1988). Evidence against the role of rhodopsin in rod outer segment binding to RPE cells. Invest Ophthalmol Vis Sci 29, 419–428. [PubMed] [Google Scholar]
- Lee JS, Xiao J, Patel P, Schade J, Wang J, Deneen B, Erdreich-Epstein A, and Song HR (2014). A novel tumor-promoting role for nuclear factor IA in glioblastomas is mediated through negative regulation of p53, p21, and PAI1. Neuro Oncol 16, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Wu F, Ruonala R, Sapkota D, Hu Z, and Mu X (2014). Isl1 and Pou4f2 form a complex to regulate target genes in developing retinal ganglion cells. PLoS One 9, e92105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Xu S, Wang Y, Mazerolle C, Thurig S, Coles BL, Ren JC, Taketo MM, van der Kooy D, and Wallace VA (2007). Ciliary margin transdifferentiation from neural retina is controlled by canonical Wnt signaling. Dev Biol 308, 54–67. [DOI] [PubMed] [Google Scholar]
- Liu W, Wang JH, and Xiang M (2000). Specific expression of the LIM/homeodomain protein Lim-1 in horizontal cells during retinogenesis. Dev Dyn 217, 320–325. [DOI] [PubMed] [Google Scholar]
- Liu Z, Lou H, Xie K, Wang H, Chen N, Aparicio OM, Zhang MQ, Jiang R, and Chen T (2017). Reconstructing cell cycle pseudo time-series via single-cell transcriptome data. Nature communications 8, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ., and Cepko CL. (2001). Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci 2, 109–118. [DOI] [PubMed] [Google Scholar]
- Livne-Bar I, Pacal M, Cheung MC, Hankin M, Trogadis J, Chen D, Dorval KM, and Bremner R (2006). Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc Natl Acad Sci U S A 103, 4988–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Drechsel D, and Guillemot F (2012). Molecular control of neurogenesis: a view from the mammalian cerebral cortex. Cold Spring Harb Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Mateo JL, Zhou B, Andersen J, Achimastou A, Urban N, van den Berg D, Georgopoulou D, Hadjur S, Wittbrodt J, et al. (2013). Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev 27, 1769–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P, Ericson J, Blackshaw S, and Cayouette M (2015). A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron 85, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuzelski E, Bunt J, Harkins D, Lim JWC, Gronostajski RM, Richards LJ, Harris L, and Piper M (2017). Transcriptional regulation of Nfix by NFIB drives astrocytic maturation within the developing spinal cord. Dev Biol 432, 286–297. [DOI] [PubMed] [Google Scholar]
- Mayer C, Hafemeister C, Bandler RC, Machold R, Batista Brito R, Jaglin X, Allaway K, Butler A, Fishell G, and Satija R (2018). Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, and Healy J (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv (2018) 1802.03426. [Google Scholar]
- Mochizuki Y, Iida A, Lyons E, Kageyama R, Nakauchi H, Murakami A, and Watanabe S (2014). Use of cell type-specific transcriptome to identify genes specifically involved in Muller glia differentiation during retinal development. Dev Neurobiol 74, 426–437. [DOI] [PubMed] [Google Scholar]
- Mullally M, Albrecht C, Horton M, Laboissonniere LA, Goetz JJ, Chowdhury R, Manning A, Wester AK, Bose Q, and Trimarchi JM (2016). Expression Profiling of Developing Zebrafish Retinal Cells. Zebrafish 13, 272–280. [DOI] [PubMed] [Google Scholar]
- Nagao M, Ogata T, Sawada Y, and Gotoh Y (2016). Zbtb20 promotes astrocytogenesis during neocortical development. Nature communications 7, 11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Ray CA, Hayashi T, Bermingham-McDonogh O, and Reh TA (2009). Acheate-scute like 1 (Ascl1) is required for normal delta-like (Dll) gene expression and notch signaling during retinal development. Dev Dyn 238, 2163–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Ueki Y, Reardon S, Karl MO, Georgi S, Hartman BH, Lamba DA, and Reh TA (2011). Genome-wide analysis of Muller glial differentiation reveals a requirement for Notch signaling in postmitotic cells to maintain the glial fate. PLoS One 6, e22817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, and Furukawa T (2003). Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci 6, 1255–1263. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, and Temple S (2009). Cell types to order: temporal specification of CNS stem cells. Curr Opin Neurobiol 19, 112–119. [DOI] [PubMed] [Google Scholar]
- Poche RA, Kwan KM, Raven MA, Furuta Y, Reese BE, and Behringer RR (2007). Lim1 is essential for the correct laminar positioning of retinal horizontal cells. J Neurosci 27, 14099–14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers HM, Huffman VJ, Voronina VA, Lewandoski M, and Mathers PH (2018). The role of the Rx homeobox gene in retinal progenitor proliferation and cell fate specification. Mech Dev 151, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, and Cepko CL (2008). The transcriptome of retinal Muller glial cells. J Comp Neurol 509, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, and Cepko CL (2004). Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol 271, 388–402. [DOI] [PubMed] [Google Scholar]
- Schiebinger G, Jian S, Tabaka M, Cleary B, Subramanian V, Solomon A, Liu S, Lin S, Berube P, Lee L, et al. (2017). Reconstruction of developmental landscapes by optimal-transport analysis of single-cell gene expression sheds light on cellular reprogramming. [Google Scholar]
- Schmidt R, Strahle U, and Scholpp S (2013). Neurogenesis in zebrafish - from embryo to adult. Neural Dev 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca L, Fop M, Murphy TB, and Raftery AE (2016). mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. The R Journal 8, 205–233. [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single Cell Transcriptomics. Cell 166, 1308–1323 e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Butz KG, Plachez C, Gronostajski RM, and Richards LJ (2003). Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci 23, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein-O’Brien GL, Carey JL, Lee WS, Considine M, Favorov AV, Flam E, Guo T, Li S, Marchionni L, Sherman T, et al. (2017). PatternMarkers & GWCoGAPS for novel data-driven biomarkers via whole transcriptome NMF. Bioinformatics 33, 1892–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein-O’Brien GL, Clark BS, Sherman T, Hu Q, Zibetti C, Sealfon R, Wolf A, Liu S, Qian J, Colantuoni C, et al. (2018). Decomposing cell identity for transfer learning across platforms, tissues and species. bioRxiv, 395004. [Google Scholar]
- Subramanian L, Sarkar A, Shetty AS, Muralidharan B, Padmanabhan H, Piper M, Monuki ES, Bach I, Gronostajski RM, Richards LJ, et al. (2011). Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc Natl Acad Sci U S A 108, E265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Liu J, Zhang M, and Mei Q (2016). Visualizing large-scale and high-dimensional data Paper presented at: Proceedings of the 25th International Conference on World Wide Web (International World Wide Web Conferences Steering Committee). [Google Scholar]
- Taverna E, Gotz M, and Huttner WB (2014). The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol 30, 465–502. [DOI] [PubMed] [Google Scholar]
- Tchieu J, Calder EL, Guttikonda SR, Gutzwiller EM, Aromolaran KA, Steinbeck JA, Goldstein PA, and Studer L (2019). NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells. Nat Biotechnol 37, 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, and Cepko CL (2008). Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS ONE 3, e1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, and Cepko CL (1987). A common progenitor for neurons and glia persists in rat retina late in development. Nature 328, 131–136. [DOI] [PubMed] [Google Scholar]
- Turner DL, Snyder EY, and Cepko CL (1990). Lineage-independent determination of cell type in the embryonic mouse retina. Neuron 4, 833–845. [DOI] [PubMed] [Google Scholar]
- van der Maaten LJP, and Hinton GE (2008). Visualizing Data using t-SNE. Journal of Machine Learning Reserach 9, 2579–2605. [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. (2018). Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729 e727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinescu PE, Kay JN, and Sanes JR (2009). Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J Comp Neurol 517, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sengel C, Emerson MM, and Cepko CL (2014). A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina. Dev Cell 30, 513–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, and Jiang L (2014). Effects of ornithine decarboxylase antizyme 1 on the proliferation and differentiation of human oral cancer cells. Int J Mol Med 34, 1606–1612. [DOI] [PubMed] [Google Scholar]
- Wood SN (2011). Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. Journal of the Royal Statistical Society (B) 73, 3–36. [Google Scholar]
- Young RW (1985a). Cell differentiation in the retina of the mouse. Anat Rec 212, 199–205. [DOI] [PubMed] [Google Scholar]
- Young RW (1985b). Cell proliferation during postnatal development of the retina in the mouse. Brain Res 353, 229–239. [DOI] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nature communications 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1 – UMAP-dimension reduction of droplet-based single cell RNA-sequencing of single developing mouse retinal cells with samples colored by developmental age. Related to Figure 1.
Table S5 - 10× cell type features. Related to Figure 1E–G and Figure S2J–M.
Table S6 - High variance genes across 10× retinal cells. Related to Figure 2.
Table S7 - Genes displaying differential expression across retina pseudotime. Related to Figure 2D.
Table S8 - Gene weights within scCoGAPS patterns. Related to Figure 4.
Movie 2 – UMAP-dimension reduction of droplet-based single cell RNA-sequencing of single developing mouse retinal cells with samples colored by annotated cell type as determined by marker gene expression in clustered cells. Extra-retinal and doublet cells have been removed. Related to Figure 1F.
Table S4 - High variance genes used for UMAP dimension reduction on 10× samples. Related to Figure 1E–F and Figure S2F–I.