Abstract
Purpose
The interactions between lifestyle and genetic factors play an important role in obesity development. Mutations in melanocortin-4-receptor (MC4R) gene are one of the most common cause of monogenic obesity, however, the functional effects of polymorphic variants near MC4R gene in general populations remain uncertain. The aim of our study was to analyze whether the common single nucleotide polymorphisms (SNPs) of MC4R gene influence the food preferences, physical activity, body fat content and distribution, as well as fasting and postprandial energy expenditure and substrates utilization.
Methods
We genotyped previously identified MC4R SNPs: rs17782313, rs633265, rs1350341, rs12970134 in 927 subjects, who underwent anthropometric, total body fat content, visceral (VAT) and subcutaneous adipose tissue (SAT) measurements, and daily physical activity and dietary intake analysis. In randomly selected 47 subjects the energy expenditure, carbohydrate and lipid utilizations were evaluated in fasting state and after high-carbohydrate and control meals intake.
Results
We found the significant associations between studied SNPs of MC4R gene and VAT and VAT/SAT ratio. Moreover, the GG genotype carriers of rs1350341, who had the lowest VAT accumulation (p = 0.012), presented higher relative increase in postprandial carbohydrate utilization (p = 0.013, p = 0.024).
Conclusions
We have observed that common SNPs of the MC4R gene influence the body fat content and distribution, as well as relative increase in postprandial carbohydrate utilization. We believe that our study may help to understand better the impact of MC4R gene on obesity development, and to help to provide personalized prevention/treatment strategies to fight against obesity and its metabolic consequences.
Keywords: Melanocortin-4-receptor MC4R gene, Nutrigenetic, Postprandial carbohydrate utilization, Body fat accumulation, Visceral fat content, Obesity
Introduction
The prevalence of obesity is rapidly increasing worldwide, and it has become an important clinical problem [1]. This multifactorial disorder is a major risk factor of type 2 diabetes mellitus (T2DM), hypertension, hyperlipidemia, cardiovascular diseases and the increase in the obesity development can lead to further morbidity and mortality [2, 3]. The large-scale genome-wide association studies (GWAS) and meta-analyzes have revealed over 52 new loci, including single nucleotide polymorphisms (SNPs) in or near the fat mass and obesity-associated (FTO) gene, melanocortin-4 receptor (MC4R) gene, Peroxisome proliferator-activated receptor gamma (PPAR), brain-derived neurotrophic factor (BDNF), neuronal growth regulator 1 (NEGR1), BTB/POZ domain-containing protein (KCTD15), and many other genes associated with BMI, waist circumference and/or waist–hip ratio (WHR) [4]. Some of the SNPs may be associated with the weight gain due to larger amounts of consumed food, due to deprivation of postprandial satiety, as well as due to deprivation of energy expenditure and/or substrate utilization [5–10]. In our previous studies, we have noticed that some of the metabolic changes may appear only in postprandial state, and that postprandial metabolism can be dependent on the meal content [11, 12], as well as on the carried genetic variants [13, 14]. It is beyond any doubts that genetic factors predispose to obesity, however, obesity should not be considered only as a genetic disorder. The increasing prevalence of obesity and T2DM in the modern environment generally can be contributed by the excessive energy intake, and/or by changes in dietary habits dependent on the age, as well as by diminished physical activity, but also it is already known, that these diseases may be influenced by interaction between lifestyle and genetic factors [6, 15, 16], which can modulate the impact of the environment on each individual’s risk.
The MC4R gene mutations are one of the most common form of monogenic obesity [4, 17, 18], however the functional effects of polymorphic variants in general population remain uncertain. The MC4Rs are expressed in several sites in the brain, including hypothalamus, forebrain and hindbrain, which had been implicated in central energy balance regulation. The activation of MC4Rs, as a part of melanocortin system, increases energy expenditure, insulin sensitivity and it has been implicated in the food intake regulation. Rich in MC4Rs are also amygdala and latheral hypothalamus, the important centers of taste perception, which connect hypothalamus with mesolimbic reward system. Moreover, melanocortin system signaling in the latheral hypothalamus has been shown to affect the response to a high-fat diet [19]. Some of the rare MC4R gene mutations are associated with binge eating, excessive hunger, meal choices, food-seeking behaviors and hyperphagia, as well as with influencing the energy expenditure [19–22]. The GWAS have identified also some common genetic variants near the MC4R gene (rs17782313, rs17700633), which are related to the increased fat mass content, body weight, obesity [17] and T2DM [23], but it is still unclear whether dietary factors may influence the relation between some genetic variants and obesity. Only few studies have been attempted so far and results are conflicting. Some authors suggest that SNPs 12970134 influences energy and dietary fat intake [24, 25], as well as eating behaviors [26], while other authors suggest that rs12970134 nor rs17782313 near the MC4R gene do not influence food intake, nor preferences for specific food items [21]. It is already known that interactions between MC4R gene with diet play a significant role in obesity and T2DM development [27], but the mechanisms by which different variants of SNPs near MC4R gene may influence the metabolic changes that lead to obesity, have not been known particularly yet. Therefore an early assessment and detection of any possible dependencies between the MC4R risk alleles and diet, and next the modifications of their dietary patterns based on these results, can be an efficient strategy to prevent obesity and its metabolic consequences. To our knowledge, associations between SNPs near MC4R gene and possible metabolic expressions, such as energy expenditure, have been investigated only by Kring et al. [28], and only for rs17782313, but substrates utilization after meal intake, dependently on various SNPs near MC4R gene, have not been studied yet.
The aim of our study was to analyze whether some of the common genetic variants near MC4R gene, which have associated with obesity in GWAS, may influence the body fat content, body fat distribution, visceral fat accumulation, as well as dietary intake, physical activity, fasting and postprandial energy expenditure and substrates utilization, after meals with different macronutrient content. This trial was registered at www.clinicaltrials.gov as NCT03792685.
Materials and methods
Participants
We genotyped previously identified MC4R SNPs: rs17782313, rs633265, rs1350341, rs12970134 in 927 subjects (the mean age 40.22 ± 0.47 years, mean BMI 28.17 ± 0.22 kg/m2, 473 men and 454 women). The 597 of individuals were overweight/obese, with BMI ≥ 25 kg/m2 (the mean age 44.20 ± 0.58 years; mean BMI 31.37 0.24 kg/m2), and 330 were healthy volunteers with normal weight, BMI < 25 kg/m2 (mean age 33.08 ± 0.64 years, mean BMI 22.39 ± 0.11 kg/m2). Participants were sent by primary care physicians as an apparently healthy people, and were recruited for a cohort study, described previously [14, 29]. The clinical characteristics of the studied population, determined by investigated genotypes, are reported in Tables 1, 2, 3, and 4.
Table 1.
The anthropometric and body composition characteristics of rs17782313 genotypes
| rs17782313 | C/C | C/T | T/T | p value |
|---|---|---|---|---|
| N (women/men) | 27 (10/17) | 316 (145/171) | 584 (299/285) | |
| Genotype frequency | 0.029 | 0.341 | 0.630 | > 0.05 |
| BMI (kg/m2) | 30.05 ± 1.27 | 28.47 ± 0.37 | 27.90 ± 0.27 | 0.14 |
| WHR | 0.94 ± 0.02 | 0.93 ± 0.01 | 0.92 ± 0.00 | 0.15 |
| Fat mass (kg) | 29.94 ± 2.51 | 25.98 ± 0.82 | 25.76 ± 0.57 | 0.32 |
| Fat mass (%) | 32.52 ± 1.53 | 29.72 ± 0.62 | 30.31 ± 0.43 | 0.36 |
| Visceral fat (cm3) | 166.61 ± 30.02 | 105.10 ± 5.90 | 94.04 ± 3.24 | < 0.001 |
| Visceral fat (%) | 42.28 ± 3.56 | 37.09 ± 0.89 | 35.63 ± 0.49 | 0.024 |
| Subcutaneous fat (cm3) | 200.89 ± 22.34 | 156.43 ± 5.28 | 160.16 ± 4.07 | 0.09 |
| Subcutaneous fat (%) | 57.72 ± 3.56 | 62.91 ± 0.90 | 64.28 ± 0.51 | 0.033 |
| Visceral/subcutaneous fat ratio | 0.86 ± 0.12 | 0.69 ± 0.04 | 0.60 ± 0.02 | 0.001 |
Table 2.
The anthropometric and body composition characteristics of rs633265 genotypes
| rs633265 | G/G | G/T | T/T | p value |
|---|---|---|---|---|
| N (women/men) | 317 (161/156) | 472 (233/239) | 138 (60/78) | |
| Genotype frequency | 0.342 | 0.509 | 0.149 | > 0.05 |
| BMI (kg/m2) | 27.63 ± 0.35 | 28.36 ± 0.31 | 28.43 ± 0.52 | 0.25 |
| WHR | 0.92 ± 0.01 | 0.92 ± 0.00 | 0.94 ± 0.01 | 0.22 |
| Fat mass (kg) | 25.13 ± 0.72 | 26.27 ± 0.69 | 26.30 ± 1.11 | 0.49 |
| Fat mass (%) | 29.92 ± 0.57 | 30.34 ± 0.51 | 29.93 ± 0.80 | 0.83 |
| Visceral fat (cm3) | 93.14 ± 4.27 | 96.95 ± 4.22 | 120.98 ± 9.58 | 0.007 |
| Visceral fat (%) | 35.40 ± 0.67 | 36.20 ± 064 | 38.48 ± 1.33 | 0.07 |
| Subcutaneous fat (cm3) | 160.70 ± 5.38 | 155.26 ± 4.43 | 172.35 ± 8.78 | 0.18 |
| Subcutaneous fat (%) | 64.60 ± 0.67 | 63.66 ± 0.67 | 61.61 ± 1.32 | 0.097 |
| Visceral/subcutaneous fat ratio | 0.60 ± 0.02 | 0.64 ± 0.02 | 0.72 ± 0.05 | 0.05 |
Table 3.
The anthropometric and body composition characteristics of rs12970134 genotypes
| rs12970134 | A/A | A/G | G/G | p value |
|---|---|---|---|---|
| N (women/men) | 41 (18/23) | 363 (172/191) | 523 (265/258) | |
| Genotype frequency | 0.044 | 0.392 | 0.564 | > 0.05 |
| BMI (kg/m2) | 30.63 ± 1.10 | 28.32 ± 0.34 | 27.88 ± 029 | 0.03 |
| WHR | 0.95 ± 0.02 | 0.93 ± 0.00 | 0.92 ± 0.00 | 0.12 |
| Fat mass (kg) | 31.58 ± 2.21 | 25.78 ± 0.73 | 25.72 ± 0.62 | 0.03 |
| Fat mass (%) | 33.90 ± 1.31 | 29.86 ± 0.56 | 30.20 ± 0.47 | 0.07 |
| Visceral fat (cm3) | 152.13 ± 25.21 | 101.52 ± 4.93 | 95.29 ± 3.63 | 0.002 |
| Visceral fat (%) | 38.29 ± 3.36 | 36.90 ± 0.76 | 35.73 ± 0.53 | 0.31 |
| Subcutaneous fat (cm3) | 213.25 ± 16.35 | 154.61 ± 4.66 | 161.10 ± 4.48 | 0.003 |
| Subcutaneous fat (%) | 61.72 ± 3.36 | 63.10 ± 0.76 | 64.17 ± 0.56 | 0.37 |
| Visceral/subcutaneous fat ratio | 0.76 ± 0.12 | 0.68 ± 0.03 | 0.60 ± 0.02 | 0.02 |
Table 4.
The anthropometric, body composition, daily physical activity, energy and macronutrients intake characteristics of rs1350341 genotypes
| rs1350341 | A/A | A/G | G/G | p value |
|---|---|---|---|---|
| N (women/men) | 138 (62/76) | 457 (232/225) | 304 (160/144) | |
| Genotype frequency | 0.154 | 0.508 | 0.338 | > 0.05 |
| BMI (kg/m2) | 28.54 ± 0.53 | 28.49 ± 0.32 | 27.76 ± 0.36 | 0.28 |
| WHR | 0.93 ± 0.01 | 0.93 ± 0.00 | 0.92 ± 0.01 | 0.35 |
| Fat mass (kg) | 26.64 ± 1.11 | 26.66 ± 0.71 | 25.62 ± 0.74 | 0.58 |
| Fat mass (%) | 30.35 ± 0.79 | 30.69 ± 0.52 | 30.45 ± 0.58 | 0.99 |
| Visceral fat (cm3) | 122.28 ± 9.70 | 99.50 ± 4.39 | 95.26 ± 4.47 | 0.012 |
| Visceral fat (%) | 38.55 ± 1.35 | 36.41 ± 0.66 | 35.36 ± 0.69 | 0.07 |
| Subcutaneous fat (cm3) | 174.06 ± 8.82 | 158.01 ± 4.57 | 164.45 ± 5.57 | 0.22 |
| Subcutaneous fat (%) | 61.54 ± 1.34 | 63.45 ± 0.69 | 64.64 ± 0.69 | 0.09 |
| Visceral/subcutaneous fat ratio | 0.72 ± 0.05 | 0.65 ± 0.03 | 0.60 ± 0.02 | 0.046 |
| Energy intake (kcal/day) | 1758.72 ± 73.86 | 1813.25 ± 43.21 | 1901.84 ± 58.94 | 0.27 |
| Protein intake (g/day) | 80.19 ± 3.13 | 83.22 ± 1.87 | 87.95 ± 2.58 | 0.13 |
| Protein intake (% of energy) | 18.8 ± 0.01 | 18.9 ± 0.00 | 19.2 ± 0.00 | 0.75 |
| Fat intake (g/day) | 61.65 ± 3.32 | 62.86 ± 2.52 | 68.26 ± 2.72 | 0.26 |
| Fat intake (% of energy) | 30.6 ± 0.01 | 30.2 ± 0.00 | 31.7 ± 0.01 | 0.13 |
| Carbohydrate intake (g/day) | 227.58 ± 9.59 | 231.04 ± 5.19 | 242.31 ± 8.00 | 0.36 |
| Carbohydrate intake (% of energy) | 48.1 ± 0.01 | 47.8 ± 0.01 | 47.4 ± 0.01 | 0.75 |
| Physical activity (MET) | 9971 ± 635 | 9653 ± 377 | 8939 ± 486 | 0.35 |
Anthropometric measurements
The weight and height were measured by the trained researchers in a standardized way [30]. All individuals underwent body weight and body composition analysis: body fat content (bioelectrical impedance analysis, InBody 220, Biospace, Korea), visceral abdominal adipose tissue (VAT) and subcutaneous abdominal adipose tissue (SAT) by a bioelectrical impedance analysis (Maltron 920-2 BioScan, Maltron International Ltd, UK). Body Mass Index (BMI) was calculated as body weight in kilograms divided by the square of height in meters. A waist circumference was measured midway between the iliac crest and the lower costal margin and hip circumference was measured at the maximum protrusion of the gluteal region. Waist-Hip Ratio was calculated as the waist circumference in centimeters divided by the hip circumference in centimeters.
The physical activity and dietary intake analyses
In all of the volunteers the daily physical activity was estimated by an International Physical Activity Questionnaire-Long Form (IPAQ-LF), which is a self-administered questionnaire and it has acceptable validity to assess the levels of physical activity [31]. The results were expressed as MET (metabolic equivalent)-min per week (MET level × minutes of activity × events per week). The 3-day food diaries analyses were performed in a group of 623 subjects, as previously described [32, 33]. Portions of food were estimated by comparing with color photographs for each portion size (albums), as well as subjects were asked to weight food if possible. The daily energy, carbohydrates, fats and proteins intake were analyzed using Dieta 4 software.
The pre- and post-meal tests
In the randomly selected 47 healthy subjects the standardized meal tests were performed, accordingly to previously described procedures [11–13, 34]. Taking into consideration the fact that investigated factors may be characterized by sexual dimorphism [35], only male participants were included in the study group. All subjects were free from the prediabetes states, T2DM, endocrine, renal, hepatic and gastrointestinal disorders, as well as without any treatments that might affect the results. A group of 47 men was chosen in order to fulfill inclusion criteria. Since the overall sample size was bigger that 47, a 47-combination without repetition from 350 was used to select the desired sample (350 men fulfilled the conditions). The subjects were instructed to avoid coffee, alcohol and excessive physical exercise on the day before each test, and to maintain their regular lifestyle throughout the study. Briefly, after an overnight fast, the subjects arrived at the laboratory and were positioned in bed in the quiet room with thermoneutral conditions (22–25 °C) for at least 30 min of rest. Resting energy expenditure and substrates utilization were determined by a computed open-circuit indirect calorimetry- the noninvasive method recommended to measure REE [36], based on resting oxygen uptake and resting carbon dioxide production, by a ventilated canopy Vmax Encore 29n System (Viasys HealthCare, Yorba Linda, CA, USA), which is one of the most valid gas analysis system [37]. Next, subjects received a standardized high-carbohydrate (HC) meal (300 ml, Nutridrink Fat Free, Nutricia Poland, Warsaw, Poland), which provided 450 kcal (89.3% of energy from carbohydrate, 10.7% of energy from protein, and 0% of energy from fat). Moreover, after 1–2 weeks, the 24 individuals who agreed to participate the second meal challenge test, received an isocaloric (450 kcal) control (C) meal (360 ml, Cubitan, Nutricia Poland, Warsaw, Poland), which provided 45.1% of energy from carbohydrate, 29.7% of energy from protein, and 25.2% of energy from fat. Subjects were asked to consume the entire meal within 10 min. During meal test only 150 ml of water was allowed to consume. Fasting (0 min) and postprandial (60, 120, 180, 240 min) energy expenditure, carbohydrate and fat utilization were evaluated for a further 4h after meal intake. The measurements were performed for 30 min and were expressed as kcal or mg/min/ffm (fat free mass).
Genetic analyses
DNA was extracted from the peripheral blood leukocytes using a classical salting out method. All SNPs were genotyped by TaqMan SNP technology from ready to use human assays library (Applied Biosystems, USA) using a high throughput genotyping system—OpenArray from Life Technologies (USA). SNPs analysis was performed in duplicate following the manufacturer’s instructions. As a negative control, we used a sample without template. The negative control was helpful for measuring any false positive signal caused by contamination. No significant deviation from Hardy Weinberg equilibrium was observed for either of the SNPs in this study (all p > 0.05).
Statistical analysis
In order to check whether the genotypes’ frequencies did not differ significantly (in the statistical manner), i.e. reflected the populational distribution, a series of proportions’ tests were conducted. Statistically significant differences between groups determined by genotypes were estimated either by the Kruskal–Wallis test or with the use of one-way ANOVA depending on fulfilling the following assumptions: homogeneity of variances, approximately normal distribution within groups. One-way ANOVA was carried out when both conditions were accomplished. Selected quantitative features, that constituted the subject of the study, were considered as dependent variables. To apprise which genotypes caused the particular test to be significant at a significance level 0.05, post-hoc analysis was performed applying either the Wilcoxon rank-sum test or the t-test (for all pairwise comparisons). Assumption of homogeneity of variances was checked with the Levene test, while the normality was verified with the Shapiro–Wilk test. Due to the problem of multiple testing false discovery rate (FDR) p value adjustment was used [38]. For all calculations the R software environment [39] was employed.
Results
The anthropometric and body composition characteristics of studied group by genotypes are presented in Tables 1, 2, 3, and 4. The genotype frequencies were compared with those expected under Hardy–Weinberg equilibrium (p < 0.05). The characteristic of the meal-test substudy population is presented in Table 5. The genotypes frequencies in the meal-test substudy population are presented in Table 6. The selected group reflects the population (in terms of genotypes’ distribution), since none of the test gave a statistically significant result (Table 7).
Table 5.
The characteristic of the meal-test substudy population
| rs12970134 | A/A | A/G | G/G | p value |
|---|---|---|---|---|
| Age (years) | 36.02 ± 0 | 36.32 ± 1.95 | 38.12 ± 2.26 | 0.55 |
| BMI (kg/m2) | 28.25 ± 0 | 27.32 ± 1.17 | 30.30 ± 1.62 | 0.1 |
| Fat mass (%) | 30.00 ± 0 | 21.45 ± 1.56 | 28.18 ± 1.77 | 0.01 |
| rs17782313 | C/C | C/T | T/T | p value |
|---|---|---|---|---|
| Age (years) | NA | 35.38 ± 2.07 | 28.45 ± 2.00 | 0.29 |
| BMI (kg/m2) | NA | 27.75 ± 1.33 | 29.64 ± 1.43 | 0.35 |
| Fat mass (%) | NA | 22.44 ± 1.81 | 26.88 ± 1.65 | 0.08 |
| rs633265 | G/G | G/T | T/T | p value |
|---|---|---|---|---|
| Age (years) | 40.66 ± 3.09 | 37.15 ± 1.53 | 26.85 ± 1.0 | 0.01 |
| BMI (kg/m2) | 27.96 ± 1.25 | 29.43 ± 1.48 | 29.21 ± 4.48 | 0.67 |
| Fat mass (%) | 25.33 ± 1.60 | 25.71 ± 1.73 | 21.74 ± 6.33 | 0.38 |
| rs1350341 | A/A | A/G | G/G | p value |
|---|---|---|---|---|
| Age (years) | 26.85 ± 1.0 | 37.15 ± 1.53 | 40.66 ± 3.09 | 0.01 |
| BMI (kg/m2) | 29.21 ± 4.48 | 29.43 ± 1.48 | 27.96 ± 1.25 | 0.67 |
| Fat mass (%) | 21.74 ± 6.33 | 25.71 ± 1.73 | 25.33 ± 1.60 | 0.38 |
Table 6.
The genotypes distribution in the meal-test substudy population participated in the high-carbohydrate (HC) and control (C) meal tests
| rs12970134 | A/A | A/G | G/G |
|---|---|---|---|
| N (HC/C) | 1/1 | 23/11 | 23/12 |
| Genotype frequency (HC/C) | 0.021/0.041 | 0.489/0.459 | 0.490/0.500 |
| rs17782313 | C/C | C/T | T/T |
|---|---|---|---|
| N (HC/C) | 0/0 | 20/11 | 27/13 |
| Genotype frequency (HC/C) | NA | 0.426/0.458 | 0.574/0.542 |
| rs633265 | G/G | G/T | T/T |
|---|---|---|---|
| N (HC/C) | 14/7 | 26/14 | 7/3 |
| Genotype frequency (HC/C) | 0.298/0.292 | 0.553/0.583 | 0.149/0.125 |
| rs1350341 | A/A | A/G | G/G |
|---|---|---|---|
| N (HC/C) | 7/3 | 26/14 | 14/7 |
| Genotype frequency (HC/C) | 0.149/0.125 | 0.553/0.583 | 0.298/0.292 |
Table 7.
The comparison of genotypes frequencies in general study cohort and meal test substudy population
| rs12970134 | rs1350341 | rs17782313 | rs633265 |
|---|---|---|---|
| AA: 6.81e−01 | AA: 6.77e−01 | CC: 4.27e−01 | GG: 1.00e + 00 |
| AG: 5.11e−01 | AG: 4.85e−01 | CT: 6.33e−01 | GT: 6.85e−01 |
| GG: 7.66e−01 | GG: 8.79e−01 | TT: 1.00e + 00 | TT: 5.89e−01 |
The carriers of genotype CC in rs17782313 presented the highest visceral fat accumulation (Table 1), and post-hoc analyses showed significant differences between CC and TT variants (p = 0.028). Between CC and CT carriers, as well as between CT and TT we noticed some trends (p = 0.059 and p = 0.101, respectively). The carriers of CC genotype showed also higher VAT/SAT ratio (Table 1). In post-hoc analyses we have noticed significant differences between CC and TT genotypes (p = 0.048), as well as significant differences between carriers of CT and TT variants (p = 0.013). Between CC and CT genotypes we did not find any significant differences (p = 0.211). We have not noticed any associations between rs17782313 and energy intake, dietary intake, physical activity, energy expenditure, nor substrate utilizations during meal challenge tests (data not shown).
The highest VAT content was observed also in carriers of TT genotype in rs633265 (Table 2). In the post-hoc analysis the significant differences were noted between GG and TT genotypes (p = 0.009), as well as between GT and TT variants (p = 0.023). Between GG and GT genotypes we did not find any differences (p = 0.526). The TT carriers presented also strong tendency to the highest VAT/SAT ratio (p = 0.05). We have not noticed any associations between rs633265 and energy intake, dietary intake, physical activity, energy expenditure, nor substrate utilizations during meal tests (data not shown).
Analysis dependently on rs12970134 genotypes showed that the homozygous carriers of allele A presented the highest BMI, total body fat mass, visceral and subcutaneous fat content, as well as VAT/SAT ratio (Table 3). The post-hoc analysis showed significant differences in BMI between AA and GG variants (p = 0.01), as well as between AA and AG genotypes carriers (p = 0.032). Between AG and GG variants differences were not significant (p = 0.32). The AA genotype carriers presented the highest total body fat content (Table 3), and significant differences were observed between AA and GG variants (p = 0.010), as well as between AA and AG (p = 0.012), but not between AG and GG genotypes carriers (p = 0.95). We have noted also the differences in body fat distribution and VAT content between AA and GG genotypes (p = 0.035). Between AA and AG genotypes the differences in post-hoc analysis were not so pronounced, but we have noticed some trends (p = 0.06). Between AG and GG variants we did not find any significant differences (p = 0.309). The carriers of AA genotype presented also the highest SAT content (Table 3). In the post-hoc analysis we noticed the differences between AA and GG genotypes (p = 0.002), as well as between AA and AG variants carriers (p = 0.0008). Between AG and GG variants we did not find any significant differences (p = 0.32). The AA genotype carriers presented also the highest VAT/SAT ratio (Table 3), however the post-hoc analysis showed significant differences only between AG and GG variants (p = 0.032). Between AA and GG genotypes carriers, as well as between AA and AG, we did not notice any significant differences (p = 0.18 and p = 0.47, respectively). We have not observed any associations between rs12970134 and energy intake, dietary intake, physical activity, energy expenditure, nor substrate utilizations during meal tests (data not shown).
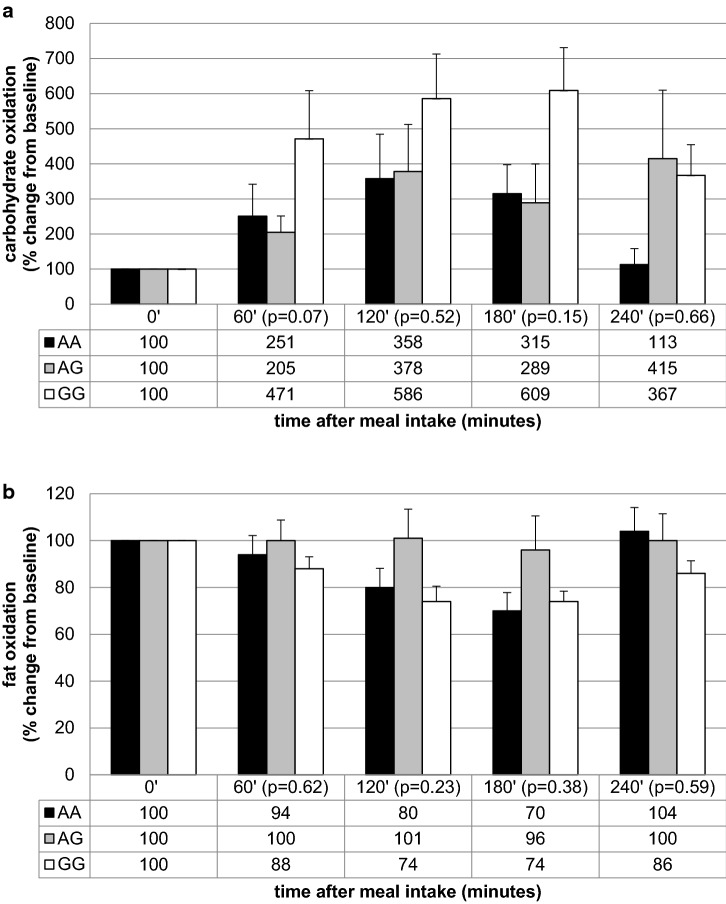
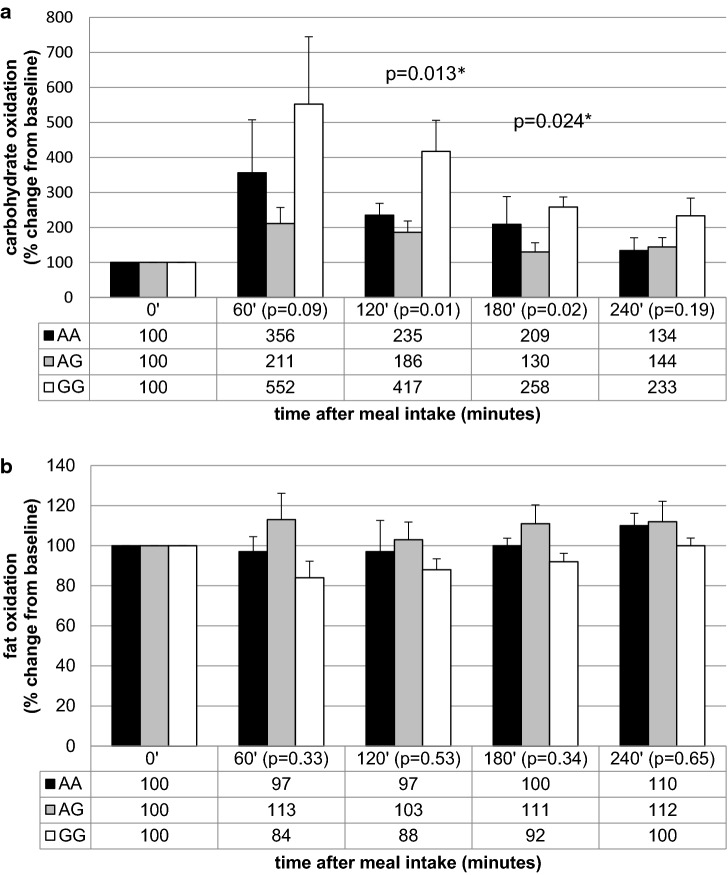
Further analysis showed that the homozygous carriers of allele A in rs1350341 presented the highest VAT accumulation (Table 4) and the significant differences we have observed between carriers of AA and GG variants (p = 0.01), as well as between AA and AG genotypes (p = 0.03). Between AG and GG variants the differences were not significant (p = 0.5). The carriers of AA genotype presented also the highest VAT/SAT ratio (p = 0.046, Table 4), and the significant differences in VAT/SAT ratio were noted between AA and GG variants carriers (p = 0.02). Between AA and AG carriers, as well as between AG and GG we noticed just some trends (p = 0.17 and p = 0.13, respectively). We observed also a tendency for higher SAT accumulation in GG genotype carriers (p = 0.09, Table 4). The results from the meal challenge tests showed that he GG genotype carriers presented significantly lower fasting carbohydrate utilization and higher fasting fat oxidation (Table 8). When we analyzed the percentage change from the baseline, we found that homozygous men of G allele presented the higher relative increase in carbohydrate utilization after both meals intake (Figs. 1a, 2a). We did not observe any differences in the percentage change of postprandial fat oxidation (Figs. 1b, 2b). We have not noted any differences in energy expenditure at fasting state, nor in the postprandial change of energy expenditure after both meals intake (data not shown).
Table 8.
The association between MC4R rs1350341 genotypes and fasting energy expenditure, carbohydrate and fat utilization, before high-carbohydrate (HC) and control (C) meals intake
| rs1350341 | A/A | A/G | G/G | p value |
|---|---|---|---|---|
| HC meal | ||||
| N | 7 | 26 | 14 | |
| Fasting energy expenditure (kcal/min/ffm) | 0.019 ± 0.001 | 0.019 ± 0.001 | 0.018 ± 0.000 | 0.76 |
| Fasting carbohydrate utilization (mg/min/ffm) | 0.61 ± 0.13 | 0.97 ± 0.15 | 0.34 ± 0.06 | 0.01 |
| Fasting fat utilization (mg/min/ffm) | 1.34 ± 0.11 | 1.17 ± 0.08 | 1.35 ± 0.06 | 0.26 |
| C meal | ||||
| N | 3 | 14 | 7 | |
| Fasting energy expenditure (kcal/min/ffm) | 0.019 ± 0.001 | 0.018 ± 0.001 | 0.019 ± 0.000 | 0.59 |
| Fasting carbohydrate utilization (mg/min/ffm) | 0.57 ± 0.18 | 0.92 ± 0.15 | 0.40 ± 0.08 | 0.07 |
| Fasting fat utilization (mg/min/ffm) | 1.34 ± 0.06 | 1.10 ± 0.06 | 1.39 ± 0.07 | 0.01 |
Fig. 1.
Association of MC4R rs1350341 genotypes with relative increase in carbohydrate (a) and fat (b) oxidation after HC meal
Fig. 2.
Association of MC4R rs1350341 genotypes with relative increase in carbohydrate (a) and fat (b) oxidation after C meal intake. *Post-hoc analysis for differences between GG and AG genotypes
For all of the investigated SNPs, in the meal-test substudy population, we did not notice any significant differences between carriers of different genotypes in fasting glucose concentrations, insulin levels, HOMA-IR, VAT, SAT contents, nor VAT/SAT ratios (data not shown), and we did not observe any differences in the calculated area under the curves (AUCs) of postprandial energy expenditure, carbohydrate and fat utilization levels (data not shown).
Discussion
The MC4R is a key regulator of energy balance through functionally divergent central melanocortin neuronal pathways, influencing food intake and energy expenditure [19]. Our study showed that all of the investigated SNPs near the MC4R gene are associated with body fat distribution, and some of them with body fat content.
The higher visceral fat accumulation we have noticed in men carrying allele C in rs17782313, and the highest VAT was observed in the homozygous subjects. The allele C carriers presented also significantly higher VAT/SAT ratio, what means that more of the body fat is deposited viscerally. What is interesting, we have noticed associations between rs17782313 and body fat distribution despite the lack of any significant associations between this common SNP and BMI, WHR, as well as total body fat content. Qi et al. [24] and Zobel et al.[40] showed that rs17782313 near MC4R gene is associated with higher BMI, what was not observed in our study, maybe due to too small study sample. However, the higher waist circumference and abdominal obesity in carriers of C alleles, which were observed in the mentioned studies, might be associated with higher visceral fat accumulation, what has been confirmed by our results. The increase in visceral fat content is a major risk factor for type 2 diabetes mellitus, and Qi et al. [24] noted that the allele-C was associated with the 14% (2–32%) increased risk of T2DM. Moreover, the visceral fat accumulation, as well as diet, may lead to the insulin resistance (IR) development, and moreover, the authors found that CC genotype women presented higher energy, total fat (and percentage of energy from total fat) and protein intake [24]. The other authors [41] observed an association of the C-allele with a higher prevalence of snacking. Valladares et al. [42] found an association between rs17782313 genetic variants and the scores of enjoyment of food subscale, satiety responsiveness subscale, but without any differences in emotional over- nor under-eating, food fussiness, food responsiveness in children. Moreover, in a detailed examination authors noted that a very low satiety responsiveness scores, and very high scores for the enjoyment of food subscale accompanied the CC genotype, what may lead to over-eating. In our population we did not find any associations between rs17782313 genotypes and energy intake, dietary intake nor physical activity, and also study by Hasselbalch et al. [21] did not confirm those findings. It is worth to notice that in that study only women were included and, as it was observed, the sex differences may influence the associations between genetic variations and dietary intake. We have not noted any associations between rs17782313 and energy expenditure, nor substrate utilizations during meal tests, what is consistent with Kring et al. [28] study results, in which authors did not find any significant associations between energy expenditure and rs17782313.
The highest visceral fat content and strong tendency to the highest VAT/SAT ratio we have observed also in carriers of TT genotype in rs633265. What is also worth no notice, we have observed the differences in fat distribution without any significant differences in the percentage of total body fat content, neither in BMI nor WHR. Grant et al. [43] found that rs633265 was associated with childhood obesity in European American, what we did not observed in our adult European population, however we found that it influences the body fat distribution.
When we analyzed data dependently on genetic variants in rs12970134, we have noted that the homozygous carriers of allele A presented the highest BMI, total body fat mass, visceral and subcutaneous fat content, as well as VAT/SAT ratio. Our results are consistent with Zobel et al. [40] findings, which showed that the minor A-allele of rs12970134 was associated with overweight, obesity, morbid obesity, as well as abdominal obesity. We did not find any associations between genotypes of rs12970134 with dietary intake nor with physical activity (data not shown). Also Hasselbalch et al. [21] did not notice any associations between this genetic variants and total food intake, even if they found a positive association with intake of energy from the whole grain.
When we analyzed our results dependently on rs1350341 genotypes, we noted that the homozygous carriers of allele A presented the highest visceral fat accumulation, even if we did not find any differences in total body fat content, BMI nor WHR. The carriers of AA genotype presented also the highest VAT/SAT ratio. We have observed also tendency for higher SAT accumulation in GG genotype carriers, what may be a possible way of protection against visceral fat deposition [44]. The GG genotype carriers presented the lowest visceral fat content, and conversely, the homozygotes for the risk allele A, which presented the highest VAT accumulation and VAT/SAT ratio. The results obtained from the food diaries, and physical questionnaires were not statistically significant, and we observed just some tendencies, maybe because these data was collected from the questionnaires, and most probably to evaluate any potential associations and to minimize the possible estimating errors, the larger study sample would be needed, what is a major limitation of our study.
The analysis of results from the meal-test substudy showed that in fasting state the GG genotype carriers presented significantly lower carbohydrate utilization and higher fat oxidation. We analyzed the diet-induced thermogenesis and postprandial substrate utilization, but because we had noticed the different values at fasting state, therefore we decided to analyze the results as a percentage change from the baseline (the relative increase from the fasting state). We found that homozygous men of G allele presented higher relative increase in carbohydrate utilization after both meals. The homozygous A allele carriers were younger, but we have noted significantly differences or trends between AG and GG variants carriers, most probably due to too small numbers in AA genotype variant group, what is also a limitation of our study. A low allele frequencies in general population, and therefore in our study population, could be the limiting factors, and we must be cautious when extrapolating our results. We did not observe any differences in the percentage change of postprandial fat oxidation, nor in the fasting and postprandial energy expenditure. Observed in our experiment the highest relative increase in carbohydrate utilization after meal intake in GG genotype carriers (rs1350341), may protect against conversion glucose into the fat, as a result of de novo lipogenesis [45, 46], what was not noted in the AA genotype men. It can be an explanation why the carriers of AA genotype presented diminished ability to glucose utilization after meal intake, what could appear due to the higher VAT content and possible impaired insulin sensitivity, or on the other hand, it can also lead to exceed fat accumulation. Moreover, because we did not find any differences in the total body fat content, and we have noted some trends to the lower SAT content in AA genotype men, we can hypothesize that maybe the carriers of this SNP may have a lesser subcutaneous lipid storage capacity, what can be a reason of visceral fat accumulation and it’s clinical consequences, such as metabolic syndrome [47], or the higher risk of T2DM, what has been shown for carriers of this SNP in rs1350341 near the MC4R gene [24]. Some of the investigated SNPs are located close to the non-coding regions of DNA, and the potential associations between adiposity-related traits and non-coding variations are discussed [48–50]. The non-coding regions are very interesting and intriguing, but molecular mechanisms for these associations have yet to be clarified, with remembering that they can be in strong LD with causal variants of coding genes regions [48, 50]. The further genomic work is needed to experimentally determine the role of the non-coding DNA regions, taking into consideration also the gene–gene and gene-environment interactions.
Conclusions
In conclusion, in the present study, we have examined the associations between the single-nucleotide polymorphisms near the MC4R gene and body fat content, body fat distribution, food intake, physical activity and metabolic differences in postprandial state. To our knowledge, the association between these common MC4R genetic variants and postprandial energy expenditure and substrate utilization has not been examined previously. We found the significant associations of investigated SNPs with body fat distribution, and we observed that investigated SNPs near MC4R gene may modulate postprandial metabolism. The GG (in rs1350341) genotype subjects, who presented the lowest visceral fat accumulation, presented also significantly higher relative increase in postprandial carbohydrate utilization, than AA genotype carriers. These observations deserve further investigations. We hope that our study may help to understand the pathways that control body fat deposition in humans, and to provide personalized treatment and prevention strategies to fight against obesity and its consequences, such as T2DM.
Acknowledgements
The study was founded by the Polish Ministry of Science and Higher Education (4774/B/P01/2009/37).
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Ethical standards
The study was conducted according to the guidelines laid down in the Declaration of Helsinki. The study protocol was approved by the local Ethics Committee of the Medical University of Bialystok (Poland), and written informed consents were obtained from all participants.
References
- 1.Kelly T, Yang W, Chen CS, Reynolds K, He J. Global burden of obesity in 2005 and projections to 2030. Int J Obes (Lond) 2008;32(9):1431–1437. doi: 10.1038/ijo.2008.102. [DOI] [PubMed] [Google Scholar]
- 2.Mottillo S, Filion KB, Genest J, Joseph L, Pilote L, Poirier P, Rinfret S, Schiffrin EL, Eisenberg MJ. The metabolic syndrome and cardiovascular risk a systematic review and meta-analysis. J Am Coll Cardiol. 2010;56(14):1113–1132. doi: 10.1016/j.jacc.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 3.Guh DP, Zhang W, Bansback N, Amarsi Z, Birmingham CL, Anis AH. The incidence of co-morbidities related to obesity and overweight: a systematic review and meta-analysis. BMC Public Health. 2009;9:88. doi: 10.1186/1471-2458-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loos RJ. Genetic determinants of common obesity and their value in prediction. Best Pract Res Clin Endocrinol Metab. 2012;26(2):211–226. doi: 10.1016/j.beem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Wardle J, Carnell S, Haworth CM, Farooqi IS, O'Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab. 2008;93(9):3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- 6.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359(24):2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- 7.Cecil J, Dalton M, Finlayson G, Blundell J, Hetherington M, Palmer C. Obesity and eating behaviour in children and adolescents: contribution of common gene polymorphisms. Int Rev Psychiatry. 2012;24(3):200–210. doi: 10.3109/09540261.2012.685056. [DOI] [PubMed] [Google Scholar]
- 8.Haupt A, Thamer C, Staiger H, Tschritter O, Kirchhoff K, Machicao F, Häring HU, Stefan N, Fritsche A. Variation in the FTO gene influences food intake but not energy expenditure. Exp Clin Endocrinol Diabetes. 2009;117(4):194–197. doi: 10.1055/s-0028-1087176. [DOI] [PubMed] [Google Scholar]
- 9.Goran MI. Genetic influences on human energy expenditure and substrate utilization. Behav Genet. 1997;27(4):389–399. doi: 10.1023/a:1025644215744. [DOI] [PubMed] [Google Scholar]
- 10.McCaffery JM, Papandonatos GD, Peter I, Huggins GS, Raynor HA, Delahanty LM, Cheskin LJ, Balasubramanyam A, Wagenknecht LE, Wing RR, AHEAD GSoL. Group LAR Obesity susceptibility loci and dietary intake in the Look AHEAD Trial. Am J Clin Nutr. 2012;95(6):1477–1486. doi: 10.3945/ajcn.111.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adamska-Patruno E, Ostrowska L, Golonko A, Pietraszewska B, Goscik J, Kretowski A, Gorska M. Evaluation of energy expenditure and oxidation of energy substrates in adult males after intake of meals with varying fat and carbohydrate content. Nutrients. 2018;10(5):627. doi: 10.3390/nu10050627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamska E, Ostrowska L, Gościk J, Waszczeniuk M, Krętowski A, Górska M. Intake of meals containing high levels of carbohydrates or high levels of unsaturated fatty acids induces postprandial dysmetabolism in young overweight/obese men. Biomed Res Int. 2015;2015:147196. doi: 10.1155/2015/147196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adamska E, Kretowski A, Goscik J, Citko A, Bauer W, Waszczeniuk M, Maliszewska K, Paczkowska-Abdulsalam M, Niemira M, Szczerbinski L, Ciborowski M, Gorska M. The type 2 diabetes susceptibility TCF7L2 gene variants affect postprandial glucose and fat utilization in non-diabetic subjects. Diabetes Metab. 2017 doi: 10.1016/j.diabet.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Kretowski A, Adamska E, Maliszewska K, Wawrusiewicz-Kurylonek N, Citko A, Goscik J, Bauer W, Wilk J, Golonko A, Waszczeniuk M, Lipinska D, Hryniewicka J, Niemira M, Paczkowska M, Ciborowski M, Gorska M. The rs340874 PROX1 type 2 diabetes mellitus risk variant is associated with visceral fat accumulation and alterations in postprandial glucose and lipid metabolism. Genes Nutr. 2015;10(2):454. doi: 10.1007/s12263-015-0454-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barness LA, Opitz JM, Gilbert-Barness E. Obesity: genetic, molecular, and environmental aspects. American Journal of Medical Genetics Part A. 2007;143A(24):3016–3034. doi: 10.1002/ajmg.a.32035. [DOI] [PubMed] [Google Scholar]
- 16.Bienertová-Vasků J, Bienert P, Forejt M, Tomandl J, Brázdová Z, Vasků A. Genotype x nutrient association of common polymorphisms in obesity-related genes with food preferences and time structure of energy intake. Br J Nutr. 2010;103(3):352–359. doi: 10.1017/S0007114509991887. [DOI] [PubMed] [Google Scholar]
- 17.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, Berndt SI, Jacobs KB, Chanock SJ, Hayes RB, Bergmann S, Bennett AJ, Bingham SA, Bochud M, Brown M, Cauchi S, Connell JM, Cooper C, Smith GD, Day I, Dina C, De S, Dermitzakis ET, Doney AS, Elliott KS, Elliott P, Evans DM, Sadaf Farooqi I, Froguel P, Ghori J, Groves CJ, Gwilliam R, Hadley D, Hall AS, Hattersley AT, Hebebrand J, Heid IM, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Herrera B, Hinney A, Hunt SE, Jarvelin MR, Johnson T, Jolley JD, Karpe F, Keniry A, Khaw KT, Luben RN, Mangino M, Marchini J, McArdle WL, McGinnis R, Meyre D, Munroe PB, Morris AD, Ness AR, Neville MJ, Nica AC, Ong KK, O'Rahilly S, Owen KR, Palmer CN, Papadakis K, Potter S, Pouta A, Qi L, Randall JC, Rayner NW, Ring SM, Sandhu MS, Scherag A, Sims MA, Song K, Soranzo N, Speliotes EK, Syddall HE, Teichmann SA, Timpson NJ, Tobias JH, Uda M, Vogel CI, Wallace C, Waterworth DM, Weedon MN, Willer CJ, Wraight Yuan X, Zeggini E, Hirschhorn JN, Strachan DP, Ouwehand WH, Caulfield MJ, Samani NJ, Frayling TM, Vollenweider P, Waeber G, Mooser V, Deloukas P, McCarthy MI, Wareham NJ, Barroso I, Kraft P, Hankinson SE, Hunter DJ, Hu FB, Lyon HN, Voight BF, Ridderstrale M, Groop L, Scheet P, Sanna S, Abecasis GR, Albai G, Nagaraja R, Schlessinger D, Jackson AU, Tuomilehto J, Collins FS, Boehnke M, Mohlke KL, Prostate L, C.lorectal, and Ovarian (PLCO) Cancer Screening Trial. KORA. Study NH. Initiative DG. Study S. Consortium WTCC. FUSION Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranadive SA, Vaisse C. Lessons from extreme human obesity: monogenic disorders. Endocrinol Metab Clin North Am. 2008;37(3):733–751. doi: 10.1016/j.ecl.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adan RA, Tiesjema B, Hillebrand JJ, la Fleur SE, Kas MJ, de Krom M. The MC4 receptor and control of appetite. Br J Pharmacol. 2006;149(7):815–827. doi: 10.1038/sj.bjp.0706929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branson R, Potoczna N, Kral JG, Lentes KU, Hoehe MR, Horber FF. Binge eating as a major phenotype of melanocortin 4 receptor gene mutations. N Engl J Med. 2003;348(12):1096–1103. doi: 10.1056/NEJMoa021971. [DOI] [PubMed] [Google Scholar]
- 21.Hasselbalch AL, Angquist L, Christiansen L, Heitmann BL, Kyvik KO, Sørensen TI. A variant in the fat mass and obesity-associated gene (FTO) and variants near the melanocortin-4 receptor gene (MC4R) do not influence dietary intake. J Nutr. 2010;140(4):831–834. doi: 10.3945/jn.109.114439. [DOI] [PubMed] [Google Scholar]
- 22.Farooqi IS, O'Rahilly S. Monogenic obesity in humans. Annu Rev Med. 2005;56:443–458. doi: 10.1146/annurev.med.56.062904.144924. [DOI] [PubMed] [Google Scholar]
- 23.Xi B, Takeuchi F, Chandak GR, Kato N, Pan HW, Zhou DH, Pan HY, Mi J, Consortium A-TD Common polymorphism near the MC4R gene is associated with type 2 diabetes: data from a meta-analysis of 123,373 individuals. Diabetologia. 2012;55(10):2660–2666. doi: 10.1007/s00125-012-2655-5. [DOI] [PubMed] [Google Scholar]
- 24.Qi L, Kraft P, Hunter DJ, Hu FB. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum Mol Genet. 2008;17(22):3502–3508. doi: 10.1093/hmg/ddn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalilitehrani A, Qorbani M, Hosseini S, Pishva H. The association of MC4R rs17782313 polymorphism with dietary intake in Iranian adults. Gene. 2015;563(2):125–129. doi: 10.1016/j.gene.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Song J, Yang Y, Chawla NV, Ma J, Wang H. Rs12970134 near MC4R is associated with appetite and beverage intake in overweight and obese children: a family-based association study in Chinese population. PLoS ONE. 2017;12(5):e0177983. doi: 10.1371/journal.pone.0177983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koochakpoor G, Hosseini-Esfahani F, Daneshpour MS, Hosseini SA, Mirmiran P. Effect of interactions of polymorphisms in the Melanocortin-4 receptor gene with dietary factors on the risk of obesity and Type 2 diabetes: a systematic review. Diabet Med. 2016;33(8):1026–1034. doi: 10.1111/dme.13052. [DOI] [PubMed] [Google Scholar]
- 28.Kring SI, Holst C, Toubro S, Astrup A, Hansen T, Pedersen O, Sørensen TI. Common variants near MC4R in relation to body fat, body fat distribution, metabolic traits and energy expenditure. Int J Obes (Lond) 2010;34(1):182–189. doi: 10.1038/ijo.2009.215. [DOI] [PubMed] [Google Scholar]
- 29.Adamska E, Waszczeniuk M, Gościk J, Golonko A, Wilk J, Pliszka J, Maliszewska K, Lipińska D, Milewski R, Wasilewska A, Citko A, Nikołajuk A, Ostrowska L, Krętowski A, Górska M. The usefulness of glycated hemoglobin A1c (HbA1c) for identifying dysglycemic states in individuals without previously diagnosed diabetes. Adv Med Sci. 2012;57(2):296–301. doi: 10.2478/v10039-012-0030-x. [DOI] [PubMed] [Google Scholar]
- 30.Nagy E, Vicente-Rodriguez G, Manios Y, Béghin L, Iliescu C, Censi L, Dietrich S, Ortega FB, De Vriendt T, Plada M, Moreno LA, Molnar D, Group HS Harmonization process and reliability assessment of anthropometric measurements in a multicenter study in adolescents. Int J Obes (Lond) 2008;32(Suppl 5):S58–65. doi: 10.1038/ijo.2008.184. [DOI] [PubMed] [Google Scholar]
- 31.Hagströmer M, Oja P, Sjöström M. The International Physical Activity Questionnaire (IPAQ): a study of concurrent and construct validity. Public Health Nutr. 2006;9(6):755–762. doi: 10.1079/phn2005898. [DOI] [PubMed] [Google Scholar]
- 32.Adamska E, Ostrowska L, Maliszewska K, Citko A, Waszczeniuk M, Przystupa W, Majewski R, Wasilewska A, Milewski R, Krytowski A, Górska M. Differences in dietary habits and food preferences of adults depending on the age. Rocz Panstw Zakl Hig. 2012;63(1):73–81. [PubMed] [Google Scholar]
- 33.Ostrowska L, Witczak K, Adamska E. Effect of nutrition and atherogenic index on the occurrence and intensity of insulin resistance. Pol Arch Med Wewn. 2013;123(6):289–296. doi: 10.20452/pamw.1774. [DOI] [PubMed] [Google Scholar]
- 34.Adamska-Patruno E, Ostrowska L, Goscik J, Pietraszewska B, Kretowski A, Gorska M. The relationship between the leptin/ghrelin ratio and meals with various macronutrient contents in men with different nutritional status: a randomized crossover study. Nutr J. 2018;17(1):118. doi: 10.1186/s12937-018-0427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J, Varghese RT, Zhou L, Vella A, Jensen MD. Glucose tolerance and free fatty acid metabolism in adults with variations in TCF7L2 rs7903146. Metabolism. 2017;68:55–63. doi: 10.1016/j.metabol.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Achamrah N, Jésus P, Grigioni S, Rimbert A, Petit A, Déchelotte P, Folope V, Coëffier M. Validity of predictive equations for resting energy expenditure developed for obese patients: impact of body composition method. Nutrients. 2018;10(1):63. doi: 10.3390/nu10010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper JA, Watras AC, O'Brien MJ, Luke A, Dobratz JR, Earthman CP, Schoeller DA. Assessing validity and reliability of resting metabolic rate in six gas analysis systems. J Am Diet Assoc. 2009;109(1):128–132. doi: 10.1016/j.jada.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamin Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57(1):289–300. [Google Scholar]
- 39.Team RC (2012) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org/
- 40.Zobel DP, Andreasen CH, Grarup N, Eiberg H, Sørensen TI, Sandbaek A, Lauritzen T, Borch-Johnsen K, Jørgensen T, Pedersen O, Hansen T. Variants near MC4R are associated with obesity and influence obesity-related quantitative traits in a population of middle-aged people: studies of 14,940 Danes. Diabetes. 2009;58(3):757–764. doi: 10.2337/db08-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stutzmann F, Cauchi S, Durand E, Calvacanti-Proença C, Pigeyre M, Hartikainen AL, Sovio U, Tichet J, Marre M, Weill J, Balkau B, Potoczna N, Laitinen J, Elliott P, Järvelin MR, Horber F, Meyre D, Froguel P. Common genetic variation near MC4R is associated with eating behaviour patterns in European populations. Int J Obes (Lond) 2009;33(3):373–378. doi: 10.1038/ijo.2008.279. [DOI] [PubMed] [Google Scholar]
- 42.Valladares M, Domínguez-Vásquez P, Obregón AM, Weisstaub G, Burrows R, Maiz A, Santos JL. Melanocortin-4 receptor gene variants in Chilean families: association with childhood obesity and eating behavior. Nutr Neurosci. 2010;13(2):71–78. doi: 10.1179/147683010X12611460763643. [DOI] [PubMed] [Google Scholar]
- 43.Grant SF, Bradfield JP, Zhang H, Wang K, Kim CE, Annaiah K, Santa E, Glessner JT, Thomas K, Garris M, Frackelton EC, Otieno FG, Shaner JL, Smith RM, Imielinski M, Chiavacci RM, Li M, Berkowitz RI, Hakonarson H. Investigation of the locus near MC4R with childhood obesity in Americans of European and African ancestry. Obesity (Silver Spring) 2009;17(7):1461–1465. doi: 10.1038/oby.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porter SA, Massaro JM, Hoffmann U, Vasan RS, O'Donnel CJ, Fox CS. Abdominal subcutaneous adipose tissue: a protective fat depot? Diabetes Care. 2009;32(6):1068–1075. doi: 10.2337/dc08-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minehira K, Bettschart V, Vidal H, Vega N, Di Vetta V, Rey V, Schneiter P, Tappy L. Effect of carbohydrate overfeeding on whole body and adipose tissue metabolism in humans. Obes Res. 2003;11(9):1096–1103. doi: 10.1038/oby.2003.150. [DOI] [PubMed] [Google Scholar]
- 46.Flatt JP. Conversion of carbohydrate to fat in adipose tissue: an energy-yielding and therefore, self-limiting process. J Lipid Res. 1970;11(2):131–143. [PubMed] [Google Scholar]
- 47.Serra MC, Ryan AS, Goldberg AP. Reduced LPL and subcutaneous lipid storage capacity are associated with metabolic syndrome in postmenopausal women with obesity. Obes Sci Pract. 2017;3(1):106–114. doi: 10.1002/osp4.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans DS, Calton MA, Kim MJ, Kwok PY, Miljkovic I, Harris T, Koster A, Liu Y, Tranah GJ, Ahituv N, Hsueh WC, Vaisse C. Genetic association study of adiposity and melanocortin-4 receptor (MC4R) common variants: replication and functional characterization of non-coding regions. PLoS ONE. 2014;9(5):e96805. doi: 10.1371/journal.pone.0096805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muller YL, Thearle MS, Piaggi P, Hanson RL, Hoffman D, Gene B, Mahkee D, Huang K, Kobes S, Votruba S, Knowler WC, Bogardus C, Baier LJ. Common genetic variation in and near the melanocortin 4 receptor gene (MC4R) is associated with body mass index in American Indian adults and children. Hum Genet. 2014;133(11):1431–1441. doi: 10.1007/s00439-014-1477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Mägi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segrè AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpeläinen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proença C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grässler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jørgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, König IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaløy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimäki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O'Donnell CJ, O'Rahilly S, Ong KK, Oostra B, Paré G, Parker AN, Perola M, Pichler I, Pietiläinen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstråle M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tönjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kähönen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Grönberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O'Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ, MAGIC. Consortium P Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]