Abstract
Bacterial conjugation is one of the most important mechanisms for spread of antibiotic resistance among bacteria. We have previously demonstrated that cefotaxime (CTX) exposure up-regulates expression of Type-IV conjugation transfer genes, and that this leads to increased transfer of a blaCTX–M–1 encoding IncI1 resistance plasmid pTF2 in Escherichia coli. To elucidate the underlying mechanisms, a search for genes that are essential for the up-regulated expression of the transfer (tra) genes in the presence of CTX was undertaken. We constructed a reporter gene-fusion strain MG1655/pTF2 ΔtraF:lacZ where the promoter region of the traF-gene of the plasmid pTF2 was fused with a lacZ on the native plasmid. Random mutagenesis mediated by Tn5 transposon was carried out in the strain, and seven genes (rfaH, yhiN, waaP, waaQ, gnd, pgl, and ISEcp1) were identified where insertion prevented CTX-induced up regulation of traF. Site-specific mutagenesis was carried out, and for all seven mutants, gene deletions abolished the CTX induced up-regulation of traF, and the increased conjugation transfer of the plasmid in the presence of CTX was no longer observed. In addition, the deletion of the genes also abolished CTX induced expression of the blaCTX–M–1 gene. Our results suggested that through CTX induced induction of the identified genes, blaCTX–M–1 expression increased, which led to up-regulation of traF and plasmid transfer. These data reveal that a number of chromosomally encoded genes contribute to the antibiotic induced up-regulation of the conjugation machinery of plasmids, and such genes may be future targets to prevent antibiotic induced spread of resistance plasmids.
Keywords: Escherichia coli, antibiotic induced conjugation, cefotaxime, blaCTX–M–1 resistance plasmid, transfer genes
Introduction
Conjugation allows bacteria to transfer genetic material from one cell to another via cell to cell contact (Llosa et al., 2002). It has been recognized as one of most important contributors for dissemination of antimicrobial resistance genes (Bennett, 2008). Conjugative plasmids-transfer in Gram-negative bacteria requires the expression of transfer (tra) genes involved in DNA transfer and replication, and in mating pair formation (De La Cruz et al., 2010). tra genes encode relaxases which are required for processing the DNA and accessory proteins, which are able to recognize the origin of transfer (oriT) and to cut the DNA molecule at the nic site (Koraimann and Wagner, 2014). Plasmid-conjugation is primarily mediated by Type IV secretion systems (T4SSs), which are multi-protein complexes located in the membrane of the cell and which are able to support the donor and recipient mating-pair (Frost and Koraimann, 2010).
A substantial amount of data suggests that sub-inhibitory concentrations of antibiotics may significantly increase the conjugation transfer frequency both in vitro and in the animal gut (Barr et al., 1986; Stevens et al., 1993; Whittle et al., 2002; Bahl et al., 2004; Feld et al., 2008; Aminov, 2011; Lu et al., 2017). In our previous study, we designed an experimental setup for measurement of conjugation frequency in which we could separate conjugation rate from the power of selection by the antibiotics. We showed that the transfer frequency of the plasmid pTF2 in Escherichia coli MG1655 in an antibiotic free environment was increased significantly when the donor was pre-grown in broth containing cefotaxime (CTX) (Moller et al., 2017). However, the underlying mechanism remains to be determined.
TraF, an essential component of the E. coli T4SS, is responsible for the processing of pilus assembly, which is essential in the formation of mating apparatus and for conjugative plasmid transfer (Waters et al., 1992; Huang et al., 2019). Previous studies showed that modification of any region of traF abolished pilus synthesis, resulting in a loss of conjugative function (Lento et al., 2016). In the current study we used this gene fused to a lacZ reporter gene to identify genes not directly involved in the plasmid transfer mechanism and involved in the CTX-induced increased conjugative transfer of the ESBL encoding plasmid. We used a genetic screen, where random insertional mutagenesis was performed in E. coli MG1655/pTF2 with a lacZ reporter gene fused to the traF promoter on the plasmid. Our results identified six chromosomally encoded genes and one plasmid encoded gene involved in the CTX induced plasmid transfer mechanism. Such genes may be future targets to prevent antibiotic induced spread of resistance plasmids.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) media was used in experiments for growth of bacteria. Bacterial strains were grown at 37°C except for strains containing the temperature sensitive plasmids, pKD46 and pCP20, which were grown at 30°C. When appropriate, media were supplemented with antibiotics (Sigma, Copenhagen, Denmark) including 20 mg/L gentamicin (Gem); 50 mg/L kanamycin (Kam); 25 mg/L chloramphenicol (Cap); 10 mg/L Trimethoprim (Tmp) and 2–512 mg/L cefotaxime (CTX). The β-galactosidase chromogenic indicator 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal) was used at a concentration of 80 mg/L.
TABLE 1.
Bacterial strains and plasmids used in this study.
| Strains | Genotype | References |
| MG1655/pTF2 | E. coli MG1655 + blaCTX–M–1 containing IncI1 plasmid pTF2 (AmpR) | Kjeldsen et al., 2015 |
| J53-2 | E. coli, RifR | Appelbaum et al., 1972 |
| ATCC® 25922 | E. coli Reference strain | Ceri et al., 1999 |
| GL100 | MG1655/pTF2 ΔtraF:lacZ, CTXR | This work |
| GL101 | MG1655 rfaH:Tn5/pTF2 ΔtraF:lacZ, CTXR, TmpR | This work |
| GL102 | MG1655 yhiN:Tn5/pTF2 ΔtraF:lacZ, CTXR, TmpR | This work |
| GL103 | MG1655 waaP:Tn5/pTF2 ΔtraF:lacZ, CTXR, TmpR | This work |
| GL104 | MG1655 waaQ:Tn5/pTF2 ΔtraF:lacZ, CTXR, TmpR | This work |
| GL105 | MG1655 gnd:Tn5/pTF2 ΔtraF:lacZ, CTXR, TmpR | This work |
| GL106 | MG1655 pgl:Tn5/pTF2 ΔtraF:lacZ, CTXR, TmpR | This work |
| GL107 | MG1655/pTF2 ΔtraF:lacZ ISEcp1:Tn5, CTXR, TmpR | This work |
| GL111 | MG1655 ΔrfaH/pTF2, CTXR | This work |
| GL112 | MG1655 ΔyhiN/pTF2, CTXR | This work |
| GL113 | MG1655 ΔwaaP/pTF2, CTXR | This work |
| GL114 | MG1655 ΔwaaQ/pTF2, CTXR | This work |
| GL115 | MG1655 Δgnd/pTF2, CTXR | This work |
| GL116 | MG1655 Δpgl/pTF2, CTXR | This work |
| GL117 | MG1655/pTF2 ΔISEcp1, CTXR | This work |
| Plasmids | ||
| pKD46 | reppSC101ts GemR ParaBADγβ exo | Doublet et al., 2008 |
| pKD4 | repR6K γAmpR FRT KamR FRT | Datsenko and Wanner, 2000 |
| pCP20 | reppSC101ts AmpR CapR cI857 λPR | Cherepanov and Wackernagel, 1995 |
| pCE36 | repR6K γ KamR FRT lacZY this | Ellermeier et al., 2002 |
Antimicrobial Susceptibility Testing
The minimal inhibitory concentrations (MIC) of CTX was determined using the broth microdilution methods using 0–512 mg/L by 2-fold dilution increases, and using the control strain E. coli ATCC® 25922 following the CLSI guidelines M100-S25 as previous described (Wayne and Clinical and Laboratory Standard Institute [CLSI], 2015).
Construction of lacZ Reporter Fusions
A LacZ reporter fusion MG1655/pTF2 ΔtraF:lacZ was created using the λ Red recombination method as previously described (Datsenko and Wanner, 2000; Ellermeier et al., 2002). Using the primers traF-F and traF-R, a PCR fragment of the kanamycin cassette (KamR) from plasmid pKD4 was amplified using PhusionTM Hot Start II DNA polymerase (ThermoFisher Scientific) and purified using the GeneJET PCR Purification Kit (ThermoFisher Scientific) and the fragment was introduced into MG1655/pTF2 harboring pKD46 by electroporation, to exchange the traF gene with the kanamycin cassette. The kanamycin resistant cassette was removed using plasmid pCP20, and the lacZ transcriptional fusion plasmid pCE36 was integrated into the tyrosine DNA recombinase (FLP) recombination target sequence at the deleted traF locus (Datsenko and Wanner, 2000; Ellermeier et al., 2002). Competent cells for electroporation was prepared by washing three times with ice-cold water and electroporation buffer (10% glycerol) at an optical density of 0.6 (OD600). 200 ng DNA was mixed with 50 μL of competent cells for electroporation at 25 μF, 200Ω, and 2.5 kV. Primer sequences can be seen in Supplementary Table S1.
Transposon Mutant Library Generation
A transposon library was generated by electroporation of the EZ-Tn5TM transposome into MG1655/pTF2 ΔtraF:lacZ. EZ-Tn5TM transposome complexes were formed between an EZ-Tn5TM transposon (Epicenter) and EZ-Tn5TM transposase (Epicenter), carrying a trimethoprim resistance marker to serve as a selection marker for transposon mutants. One μL of the EZ-Tn5TM transposome was used for electroporation and bacteria were plated on LB agar plates containing 10 mg/L Tmp. Totally, 2 × 104 transposon mutants were separated on the plates. The transposon library was stored as 30% glycerol stocks at −80°C.
Screening of Transposon-Library
The transposon-library was plated on LB agar plates containing CTX (32 mg/L) and X-gal (80 mg/L) and screened for white/light blue colonies, corresponding to absence of CTX induced up-regulation of traF.
β-Galactosidase Assay
β-galactosidase assays were carried out according to the method of Miller (1972). Overnight cultures were diluted 100 fold in LB broth and allowed to grow at 37°C. Two experiments were performed: (i) β -galactosidase activity at different optical density: MG1655/pTF2 ΔtraF:lacZ cultures with and without CTX were grown at 37°C and 2 mL samples were collected at OD600 = 0.1, 0.3, 0.5, 0.7, 0.9, 1.1, 1.3, and 1.5; (ii) β -galactosidase assay of wild-type (MG1655/pTF2 ΔtraF:lacZ) and transposon-mutants at OD600 = 0.5: MG1655/pTF2 ΔtraF:lacZ and the seven transposon-mutants were grown with and without CTX to OD600 = 0.5 and 2 mL samples were collected. All samples were immediately cooled down and centrifuged for 3 min at 8000 rpm. Pellets were resuspended in 1 mL 100 mM Z-buffer (PH 7.0), permeabilized by adding 25 μL 0.1% (W/V) SDS and 50 μL chloroform and mixed by vortexing and incubated 5 min at room temperature. β-galactosidase assay was performed using 200 μL ONPG (4 mg/mL) (o-nitrophenyl-β-D-galactopyranoside) in Z-buffer. Samples were incubated at room temperature and when color change was observed, the reactions were terminated by the addition of 500 μL of 1 M Na2CO3. OD420 and OD550 was measured, and activity calculated in Miller-units = 1000 × (OD420-(1.75 × OD550))/(T × V × OD600). Data correspond to three independent assays conducted in duplicate, and all values are the mean ± S.D.
Transposon Site Identification
Genomic DNAs were isolated from mutants using the MasterPureTM complete DNA purification Kit (Epicenter) according to the instructions of the supplier. The identification of the transposon insertion site was done by whole genomic sequencing in an Illumina MiSeq (Illumina, Inc., San Diego, CA, United States) at a 300-bp paired-end-read format. Sequencing reads were de novo assembled using the SPAdes v.3.5.0 (Bankevich et al., 2012). Transposon insertions sites were identified using BLAST in CLC Main Workbench 8.0.0 (CLC bio, Denmark), and the locations of the transposon inserts were determined by a blastn comparison with the sequence of E. coli K-12 MG1655 (accession number U00096.3) (Altschul et al., 1990).
Targeted Deletion Mutagenesis
Site specific gene deletion in MG1655/pTF2 was done by insertion of kanamycin cassettes by the Lambda red recombinase system (Doublet et al., 2008). Insertions were confirmed by PCR. The kanamycin cassette was then removed from the seven KamR mutants using pCP20 as previously described (Doublet et al., 2008). Primers used for generating and confirming mutations are listed in Supplementary Table S1.
RNA Extraction and RT-qPCR
Single colonies of MG1655/pTF2 and the seven deletion mutants were grown overnight in LB media at 37°C. The cultures were diluted 1000 fold and grown with and without 1/2 MIC of CTX to OD600 = 0.5. A FastPrep cell disrupter system (Qbiogene, Illkirch, France) and RNeasy Mini Kit (Qiagen, Sollentuna, Sweden) was used to extract total RNA. RNA quantity was determined by NanoDrop 1000 spectrophotometer (Thermo Scientific, Hvidovre, Denmark). Genomic DNA was removed by TURBOTM DNase kit (2 U/μL) (Ambion, Life Technologies, Nearum, Denmark). Purified RNA was reverse-transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Naerum, Denmark). RT-qPCR was performed using FastStart Essential DNA Green Master (Roche, Hvidovre, Denmark) and a LightCycler 96 (Roche, Hvidovre, Denmark) as described by Pfaffl (2001). gapA and nusG, which have previously been validated, were used as reference genes (Kjeldsen et al., 2015). RT-qPCR was performed twice on separate biological samples and the results were calculated by the 2–ΔΔCt method. Primer sequences can be seen in Supplementary Table S1.
Growth Experiment
Growth of MG1655/pTF2 and deletion mutants was evaluated without and with 1/2 MIC CTX. Growth curves were obtained in biological triplicate using the automated microbiology growth curve analysis system Bioscreen CTM (Oy Growth Curves Ab Ltd, Finland). A final volume of 200 μL LB broth was inoculated with cells from overnight cultures to a final cell density of 5 × 105 cfu/mL, using a SensititreTM Nephelometer (Thermo ScientificTM, Roskilde, Denmark) with a 0.5 McFarland turbidity standard. The OD600 was measured every 15 min with continuous shaking for 24 h at 37°C. OD values of blank samples were subtracted from sample OD values at the respective time points before analyzing the data.
Conjugation Experiments
MG1655/pTF2 and deletion mutants were used as donors and E. coli J53-2 as recipient strain in the conjugation experiments aiming to determine the effect of targeted genes deletion on conjugational transfer rate. MG1655/pTF2 and its mutants were grown in LB media without and with 1/2 MIC of CTX to exponential phase (OD600 = 0.5). Antibiotics were removed by a washing steps and conjugation was performed by mixing donor and recipient strain in a 1:1 ratio on filters (0.22 μM, Millipore, Copenhagen, Denmark) on LB agar plates at 37°C for 30 and 60 min as previously described (Moller et al., 2017). The bacterial material was washed from the filters using isotonic NaCl and plated on LB agar plates containing 2 mg/L CTX (for quantifying donor + transconjugants) or 50 mg/L rifampicin and 2 mg/L CTX (quantifying transconjugants only) and incubated overnight at 37°C. The conjugation frequency was calculated as transconjugants divided by number of donors. The conjugation experiments were performed in three biological duplicates with three technical replicates each.
Statistical Analyses
Statistical analysis was performed using the GraphPad Prism (GraphPad Software) version 7.03. Comparisons of gene expression and conjugation frequencies with and without antibiotics were performed by student’s t-test with Welch’s correction. A P-value of ≤0.05 was considered statistically significant.
Results
Identifying Genes Involved in CTX-Induced traF Expression
Our previous work have reported that the transfer genes and proteins involved in conjugation of the blaCTX–M–1 plasmid pTF2 were significantly up-regulated when E. coli MG1655/pTF2 was treated with 1/2 MIC (126 mg/L) concentrations of CTX during growth (Moller et al., 2017). In order to identify genes involved in the mechanism by which CTX influence the Type IV secretion system (T4SS) and hence conjugation, MG1655/pTF2 ΔtraF:lacZ, containing a LacZ reporter fused to the traF promoter, was constructed. Growing this strain with CTX on X-gal plates led to dark blue colonies, revealing high expression of traF during CTX exposure. A transposon library of MG1655/pTF2 ΔtraF:lacZ with random Tmp-resistant Tn5 transposon insertions was screened on CTX and X-gal for lighter blue colonies, to identify genes involved in the CTX induced traF induction. A total of 14 light blue colonies were isolated.
In order to identify the transposon insert site, whole genome sequencing was performed on these 14 light blue mutants. Totally eight different insertion sites were identified, six of them located on the chromosome [rfaH (one isolate), yhiN (one isolate), waaP (six isolates in two different positions), waaQ (two identical isolates), gnd (one isolate), and pgl (two identical isolates)] and one on the plasmid [ISEcp1 (one isolate)] (Table 2 and Supplementary Figure S1).
TABLE 2.
CTX responsive genes and their products.
| Gene | Product | Transposon insertion site∗ | Gene access no. |
| rfaH | Transcription antiterminator | 82/83 | M94889.1 |
| yhiN | Putative FAD/NAD(P)-binding oxidoreductase | 42/43 | NC_000913.3 |
| waaP | LPS core heptose (I) kinase | 434/435 683/684 |
NC_000913.3 |
| waaQ | LPS core heptosyltransferase III | 607/608 | NC_000913.3 |
| gnd | 6-phosphogluconate dehydrogenase | 757/758 | NC_000913.3 |
| pgl | 6-phosphogluconolactonase | 850/851 | NC_000913.3 |
| ISEcp1 | ISEcp1 transposase | 973/974 | KJ563250 |
∗Numbers refer to the base-position of the gene, between which the transposon has inserted.
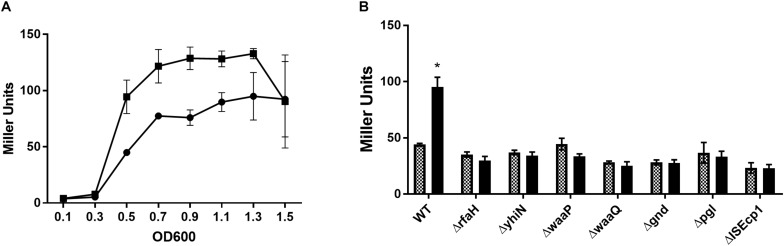
To confirm the reduced expression from the traF promoter, a β-galactosidase assay was performed, with and without CTX, using MG1655/pTF2 ΔtraF:lacZ and the 7 transposon mutants. Measuring the changes in expression during growth, showed that the β-galactosidase level in MG1655/pTF2 ΔtraF:lacZ was increased significantly from OD600 = 0.5 by CTX treatment (Figure 1A). Measuring the LacZ expression from MG1655/pTF2 ΔtraF:lacZ and the seven transposon-mutants at OD600 = 0.5 revealed that CTX did not induce the traF promoter in the mutants, confirming the results from the transposon screen (Figure 1B).
FIGURE 1.
β-galactosidase activity increases in MG1655/pTF2 ΔtraF:lacZ during CTX exposure, but not in the transposon-mutants. (A) LacZ expression of MG1655/pTF2 ΔtraF:lacZ with (square) and without (Circle) CTX during growth. (B) LacZ expression of MG1655/pTF2 ΔtraF:lacZ (WT) and the seven transposon-mutants without (squared bars) and with 1/2 MIC CTX (black bars) exposure at OD600 = 0.5. All experiments were carried out in triplicate, and each value is presented as the average plus standard deviation. The star indicates statistical significance between antibiotic treated and untreated at level: ∗P ≤ 0.05.
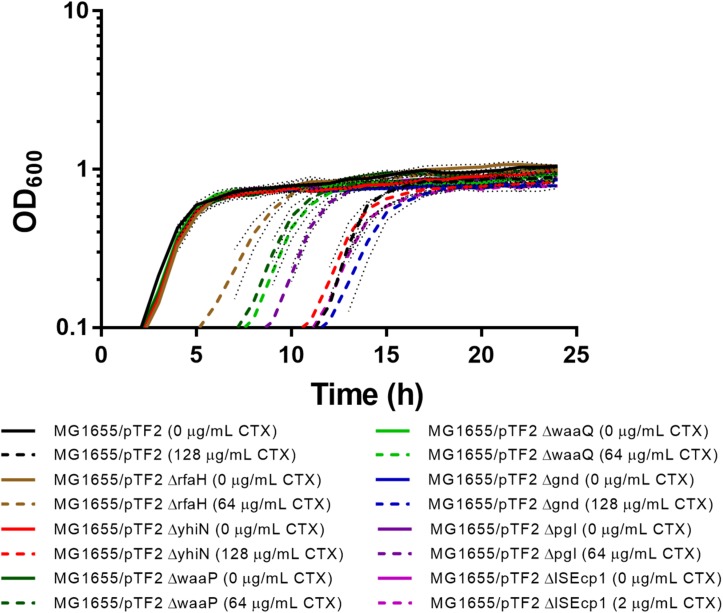
Individual deletions of the seven identified genes were constructed in MG1655/pTF2. In order to investigate whether the deletions had an impact on CTX resistance and on bacterial growth, the MIC of CTX for the mutants was determined and the growth pattern of the strains with and without 1/2 MIC CTX was investigated. The MIC of CTX for ΔyhiN and Δgnd corresponded to the MIC of the wild-type (MG1655/pTF2), while MIC for ΔrfaH, ΔwaaP, ΔwaaQ, and Δpgl decreased two fold from 256 mg/L to 128 mg/L and ΔISEcp1 decreased seven fold from 256 mg/L to 4 mg/L. Without CTX exposure, the strains showed a similar growth pattern, however, when exposed to CTX (1/2 MIC of the corresponding strain), the ΔrfaH, ΔwaaP, ΔwaaQ, and Δpgl showed decreased length of lag phase compared to MG1655/pTF2 (Figure 2). However, since all expression and conjugation experiments were performed with bacteria grown to the same OD, the variations in lag-phase were not expected to influence the obtained results.
FIGURE 2.
Growth phenotypes of MG1655/pTF2 and its mutants with and without CTX. All strains were grown without and with CTX (1/2 MIC of the corresponding strain). Three independent replicates were performed and the data shown represent the mean and dots represent standard deviations.
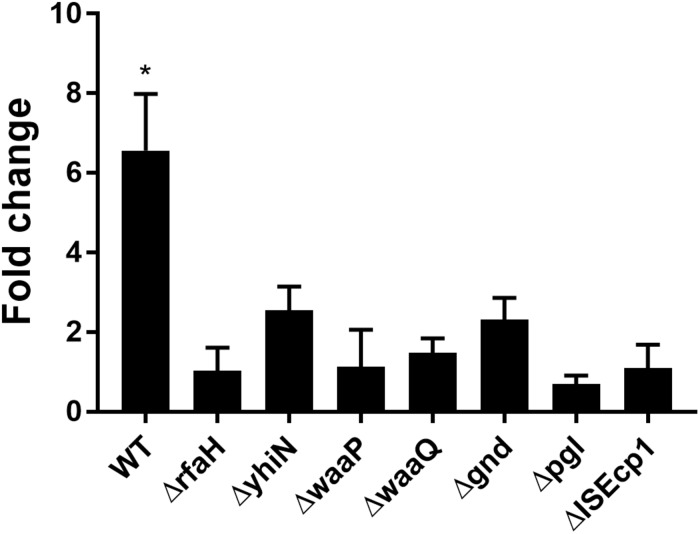
To confirm the importance of the genes in CTX induced traF regulation, we used RT-qPCR analysis, and found a significant up-regulation of traF (6.1-fold, t-test, P = 0.01) when the wild-type strain (MG1655/pTF2) was treated with CTX. In contrast, and in support of the β-galactosidase assay results, none of the deletion mutants showed significant CTX induced traF up-regulation (Figure 3).
FIGURE 3.
CTX induced expression of the traF gene in WT (MG1655/pTF2) and deletion mutants. Strains were grown with 1/2 MIC CTX or without. Data are presented as fold change in CTX induced expression relative to expression levels without CTX. Three independent replicates including two technical replicates each were performed. The data shown represents the mean and the error bars represent standard deviations. The expression data was normalized to gapA and nusG. The stars indicate statistical significance between the two conditions at level: ∗P ≤ 0.05.
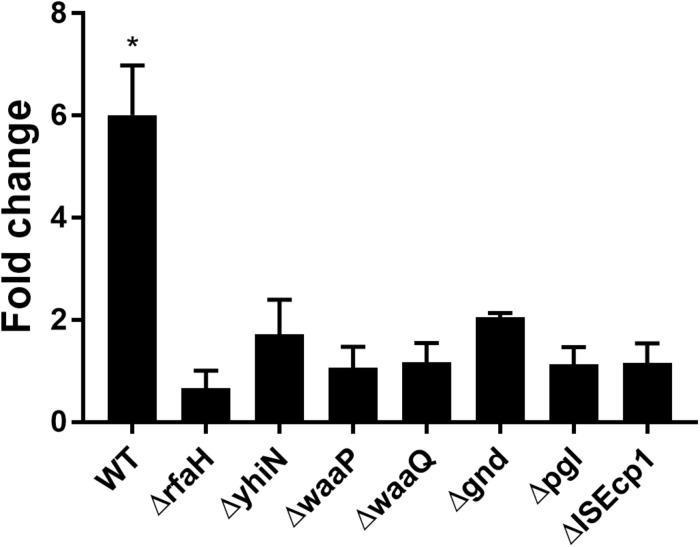
CTX-Induced blaCTX–M–1 Expression
We have previously reported that the induction of transfer gene-expression and increased conjugation frequency by treatment with CTX is dependent on the presence of the antibiotic resistance gene blaCTX–M–1 (Moller et al., 2017). To investigate whether the identified seven genes also work in a CTX-M-1 dependent manner to regulate the CTX-induced increased conjugation, we performed RT-qPCR. We found that the blaCTX–M–1 gene expression was significantly up-regulated (6.56- fold, t-test, P = 0.03) in the wild-type strain during CTX treatment, whereas CTX exposure had limited, non-significant, effect on blaCTX–M–1 expression in the seven knock-out mutants (Figure 4) and the seven isolated transposon-mutants (data not shown). Expression of blaCTX–M–1 was not significantly different between the mutants and the wild-type, when the strains were not exposed to CTX (Supplementary Figure S2).
FIGURE 4.
CTX induced expression of blaCTX–M–1 in WT (MG1655/pTF2) and deletion mutants. Strains were grown with 1/2MIC CTX or without. Data are presented as fold change in CTX induced expression relative to expression levels without CTX. Two independent replicates including two technical replicates each were performed. The data shown represents the mean and the error bars represent standard deviations. The expression data was normalized to two validated reference genes, gapA and nusG. The stars indicate statistical significance at level: ∗P ≤ 0.05.
CTX-Induced Expression of Selected Gene in Wild-Type and ΔrfaH Mutant
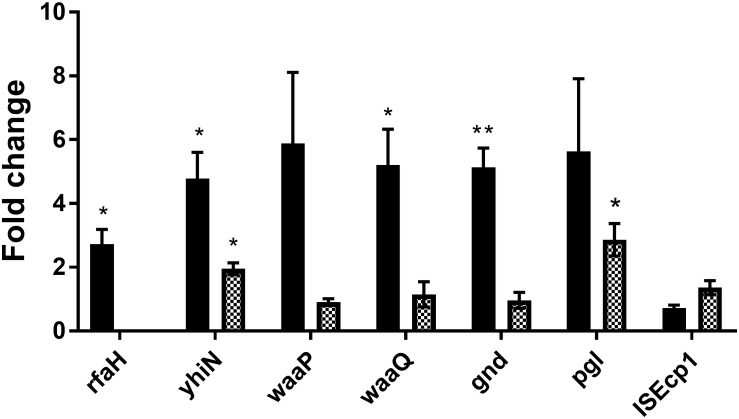
In order to investigate whether the genes identified by transposon mutagenesis were regulating tra gene expression independent of CTX or not, RT-qPCR was performed to investigate the expression of the seven genes in the wild-type strain, comparing expression between CTX treated and untreated wild-type. Significant up-regulation of rfaH (2.7- fold, t-test, P = 0.03), yhiN (4.8- fold, t-test, P = 0.02), waaQ (5.2- fold, t-test, P = 0.03) and gnd (5.1- fold, t-test, P = 0.006) was observed, when the wild-type was treated with 1/2 MIC of CTX during growth. For the waaP and pgl genes, a non-significant, 5.9- and 5.6- fold (t-test, P > 0.05) increase in gene expression was observed. The data showed that CTX do up-regulate the expression of these genes, except the ISEcp1 (Figure 5, black bars). In addition, we investigated the expression level of the individual genes in the rfaH mutant background, to evaluate a possible regulatory link between RfaH and the remaining genes. Only yhiN (2.0- fold, t-test, P = 0.01) and pgl (2.9- fold, t-test, P = 0.03) showed significant up-regulation when ΔrfaH was treated with CTX during growth, however, not to the level observed in the wild-type (Figure 5, squared bars).
FIGURE 5.
CTX induces expression of six genes involved in antibiotic induced traF expression in MG1655/pTF2. MG1655/pTF2 (Black bars) and the ΔrfaH (squared bars) were grown with 1/2 MIC CTX or without. Data are presented as fold change in CTX induced expression relative to expression levels without CTX. Three independent replicates including two technical replicates each were performed. The data shown represents the mean and the error bars represent standard deviations. The expression data was normalized to gapA and nusG. The stars indicate statistical significance at different levels: ∗P ≤ 0.05, ∗∗P ≤ 0.01.
CTX-Induced Conjugative Plasmid Transfer
In order to investigate whether the deletion of the seven genes would affect the number of CTX-induced conjugation events, conjugation experiments was performed with the wild-type (MG1655/pTF2) and the mutants, pre-grown with or without CTX. The data showed that the CTX treatment significantly increased plasmid transfer frequency in MG1655/pTF2 with 25.0- and 41.0- fold (t-test, P ≤ 0.05) after 30 and 60 min of cell contact, relative to conjugation without pre-growth in the presence of CTX (Table 3). In contrast, all of the mutants revealed a decreased CTX-induced plasmid transfer compared to the wild-type. We did not obtain any transconjugants for ΔISEcp1 probably due to transfer frequencies below detection limit.
TABLE 3.
Fold changes of CTX-induced (1/2 MIC) increased conjugation transfer frequency, relative to untreated strains, using MG1655/pTF2 (WT) and the mutants as donors and J53-2 as recipient.
| Donor | Conjugation time (min) | Conjugation transfer frequency (Control)a | Conjugation transfer frequency (CTX)a | Plasmid transfer fold differenceb |
| WT | 30 | 3.90 × 10–5 ± 1.05 × 10–5 | 9.75 × 10–4 ± 1.46 × 10–4 | 25.00∗∗ |
| 60 | 3.00 × 10–4 ± 0.49 × 10–4 | 1.23 × 10–2 ± 0.34 × 10–2 | 41.00∗ | |
| ΔrfaH | 30 | 1.13 × 10–5 ± 0.32 × 10–5 | 0.87 × 10–5 ± 0.26 × 10–5 | 0.77 |
| 60 | 1.14 × 10–4 ± 0.12 × 10–4 | 1.06 × 10–4 ± 0.39 × 10–4 | 0.93 | |
| ΔyhiN | 30 | 2.48 × 10–5 ± 0.96 × 10–5 | 3.64 × 10–4 ± 0.39 × 10–4 | 14.68∗∗ |
| 60 | 5.25 × 10–4 ± 1.24 × 10–4 | 2.86 × 10–3 ± 0.60 × 10–3 | 4.50∗ | |
| ΔwaaP | 30 | 2.33 × 10–5 ± 0.65 × 10–5 | 3.46 × 10–4 ± 0.72 × 10–4 | 14.85∗ |
| 60 | 3.29 × 10–4 ± 0.79 × 10–4 | 3.09 × 10–3 ± 0.97 × 10–3 | 9.57 | |
| ΔwaaQ | 30 | 2.97 × 10–5 ± 1.04 × 10–5 | 3.23 × 10–5 ± 0.67 × 10–5 | 1.09 |
| 60 | 2.73 × 10–4 ± 0.60 × 10–4 | 4.47 × 10–3 ± 1.44 × 10–3 | 16.37 | |
| Δgnd | 30 | 2.44 × 10–5 ± 0.64 × 10–5 | 3.16 × 10–4 ± 0.65 × 10–4 | 12.95∗ |
| 60 | 3.59 × 10–4 ± 0.88 × 10–4 | 5.47 × 10–3 ± 0.4707 × 10–3 | 15.24∗∗ | |
| Δpgl | 30 | 1.16 × 10–5 ± 0.24 × 10–5 | 1.26 × 10–4 ± 0.24 × 10–4 | 10.86∗ |
| 60 | 2.27 × 10–4 ± 0.48 × 10–4 | 2.31 × 10–3 ± 0.41 × 10–3 | 10.18∗ |
aThe results shown are from two biological replicates with three technical replicates each, the data shown represent the mean ± standard deviation. bThe stars indicate statistical significance at level: ∗P ≤ 0.05, ∗∗P ≤ 0.01.
Discussion
The contribution of antibiotics as a stimulating factor to the promotion of conjugation transfer has previously been investigated (Barr et al., 1986; Bahl et al., 2004; Feld et al., 2008; Lu et al., 2017; Moller et al., 2017). However, despite our current knowledge that antibiotics can increase conjugation frequency, it still remains unclear which mechanisms are involved in this phenomenon. We have previously obtained evidence that 1/2 MIC concentrations of CTX affects expression of the conjugation apparatus of plasmid pTF2, as tra-genes were significantly up-regulated at both the transcriptional and translational level (Moller et al., 2017). traF is one of several T4SS proteins involved in pilus assembly and essential for plasmid transfer (Frost et al., 1994). In the current study, a traF:lacZ reporter gene-fusion was constructed, to enable a screening for genes which affect the antibiotic induced up-regulation of traF expression. Random insertional mutagenesis mediated by Tn5 transposon was carried out in the reporter strain and in total seven genes were identified (rfaH, yhiN, waaP (rfaP), waaQ (rfaQ), gnd, and pgl and the pTF2 plasmid-encoded insertion sequence ISEcp1) where knock-out by transposon insertion abolished the CTX induced up-regulation of traF.
RfaH is a transcriptional antiterminator, which activates operons encoding lipopolysaccharide (LPS) core components, pili, toxins, capsules and antibiotic biosynthesis in Enterobacteriaceae (Sanderson and Stocker, 1981; Bailey et al., 2000; Nandymazumdar and Artsimovitch, 2015). Furthermore, reduced plasmid transfer has been shown for a Salmonella Typhimurium rfaH mutant (Sanderson and Stocker, 1981). A possible explanation for CTX induced up-regulation of traF is therefore that CTX treatment increases the expression of rfaH, and this activates the expression of pilus encoding genes, such as traF. A previous study has shown RfaH binding to an ops element in the promoter region of the tra operon on the F plasmid (Bailey et al., 1997). Sequence analysis revealed an ops element in the tra promoter region on pTF2 (IncI1 plasmid), supporting direct RfaH regulation of traF expression. Furthermore, we showed that CTX induce rfaH and traF expression, however, the latter not in a rfaH knock out background, and thus that the CTX induction of the traF expression is dependent on RfaH. The waaP and waaQ genes, which are regulated by RfaH, play important roles in LPS biosynthesis and the formation of a stable outer membrane (Yethon et al., 1998; Belogurov et al., 2009). Mutations in the waa locus can significantly alter outer membrane permeability and hypersensitivity to detergents and hydrophobic antibiotics (Schnaitman and Klena, 1993). gnd and pgl are two enzymes of the oxidative branch of the hexose monophosphate shunt in the pathway of glucose metabolism (Kupor and Fraenkel, 1969). It is unknown how gnd and pgl regulate tra expression, but gnd is adjacent to a RfaH-regulated transcription unit, suggesting RfaH affects expression of gnd (Belogurov et al., 2009). The yhiN gene is a putative FAD/NAD(P) binding oxidoreductase with unknown function. It is part of the RpoS regulon, indicating an importance in the stress response (Vijayakumar et al., 2004). Previously published results have shown that high levels of RpoS affect conjugative transfer in Pseudomonas knackmussii (Miyazaki et al., 2012). The blaCTX–M genes are often associated with ISEcp1-like elements (Karim et al., 2001; Baraniak et al., 2002; Chanawong et al., 2002; Poirel et al., 2003). These elements contain putative -35 and -10 promoter regions within the 3′ end of ISEcp1 affecting the expression level of the blaCTX–M gene (Karim et al., 2001; Poirel et al., 2003).
Deletions of the seven genes were performed in MG1655/pTF2 to evaluate their involvement in CTX induced increased plasmid transfer. For the wild-type MG1655/pTF2 we observed a significant CTX induction of expression of traF and blaCTX–M–1, and a significant induction of plasmid transfer. In contrast, we saw that the mutants were hampered in CTX-induced traF and blaCTX–M–1 expression and plasmid transfer. Results confirmed that increased blaCTX–M–1 gene expression is necessary for increased conjugation transfer frequency. We have previously shown that exposing a blaCTX–M–1 mutant of MG1655/pTF2 to CTX did not lead to induced tra gene expression and plasmid transfer (Moller et al., 2017). Furthermore, when exposed to CTX, the ΔISEcp1 mutant expressed very low levels of blaCTX–M–1, as seen from the MIC, and was unable to induce traF expression. ISEcp1-like elements have been shown to contain promoter sequences for high level expression of blaCTX–M β-lactamase genes (Poirel et al., 2003); probably explaining why we observed limited CTX induced expression of blaCTX–M–1. The ISEcp1 most likely is not part of the CTX induced conjugation pathway, instead the deletion simply affects blaCTX–M–1 expression, supported by the severely reduced MIC, and the lack of CTX induced traF and blaCTX–M–1 expression.
Our results showed that the deletion of the genes resulted in reduced CTX induction of plasmid transfer compared to the wild-type (MG1655/pTF2), confirming that the rfaH, yhiN, waaP, waaQ, gnd and pgl, genes are involved in the CTX induced increased pTF2 plasmid conjugative transfer.
In order to investigate how the seven identified genes contribute to CTX induced traF expression, we measured whether the genes themselves were regulated by CTX. We found that the expression of the rfaH, yhiN, waaQ, and gnd genes in the wild-type indeed were up-regulated significantly in the presence of CTX, and the expression of waaP and pgl was also up-regulated, although not significant. Only expression of ISEcp1 was not affected by CTX. This CTX-induced up-regulation of waaP, waaQ, and gnd disappeared in an rfaH mutant background, and the CTX induced expression of yhiN and pgl was decreased in the ΔrfaH strain compared to the expression levels in the wild-type background. These results support that RfaH is central not only in the regulation of these five genes, but also in the CTX induction of conjugation. Thus our current model is that CTX induce blaCTX–M–1 expression as well as rfaH expression. This affects tra expression directly as well as through changed expression of the waaP, waaQ, gnd, yhiN, and pgl genes, which affect blaCTX–M–1 expression and hence traF expression.
Conclusion
In conclusion, six genes involved in CTX induced increased conjugation have been identified. Further experiments are needed to uncover the role of these genes in the pathways by which bacteria sense CTX and signals induced tra expression.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
GL has performed all the experiments. LT, JO, and GL have participated in the design of the study and have participated in the article preparation. All authors have approved the final article.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. GL was supported by a Ph.D. fellowship from the China Scholarship Council.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02203/full#supplementary-material
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Aminov R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. 10.3389/fmicb.2011.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum P. C., Trichardt P. F., Krizsanovich K., Coetzee J. N. Hugon. (1972). Abortive transuction of motility in proteus and providence strains. J. Gen. Microbiol. 70 361–364. 10.1099/00221287-70-2-361 [DOI] [PubMed] [Google Scholar]
- Bahl M. I., Sorensen S. J., Hansen L. H., Licht T. R. (2004). Effect of tetracycline on transfer and establishment of the tetracycline-inducible conjugative transposon Tn916 in the guts of gnotobiotic rats. Appl. Environ. Microbiol. 70 758–764. 10.1128/aem.70.2.758-764.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M. J., Hughes C., Koronakis V. (1997). RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26 845–851. 10.1046/j.1365-2958.1997.6432014.x [DOI] [PubMed] [Google Scholar]
- Bailey M. J., Hughes C., Koronakis V. (2000). In vitro recruitment of the RfaH regulatory protein into a specialised transcription complex, directed by the nucleic acid ops element. Mol. Gen. Genet. 262 1052–1059. 10.1007/pl00008648 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak A., Fiett J., Hryniewicz W., Nordmann P., Gniadkowski M. (2002). Ceftazidime-hydrolysing CTX-M-15 extended-spectrum beta-lactamase (ESBL) in Poland. J. Antimicrob. Chemother. 50 393–396. 10.1093/jac/dkf151 [DOI] [PubMed] [Google Scholar]
- Barr V., Barr K., Millar M. R., Lacey R. W. (1986). Beta-lactam antibiotics increase the frequency of plasmid transfer in Staphylococcus aureus. J. Antimicrob. Chemother. 17 409–413. 10.1093/jac/17.4.409 [DOI] [PubMed] [Google Scholar]
- Belogurov G. A., Mooney R. A., Svetlov V., Landick R., Artsimovitch I. (2009). Functional specialization of transcription elongation factors. EMBO J. 28 112–122. 10.1038/emboj.2008.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 153(Suppl. 1), S347–S357. 10.1038/sj.bjp.0707607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceri H., Olson M. E., Stremick C., Read R. R., Morck D., Buret A. (1999). The calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37 1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanawong A., M’zali F. H., Heritage J., Xiong J. H., Hawkey P. M. (2002). Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People’s Republic of China. Antimicrob. Agents Chemother. 46 630–637. 10.1128/aac.46.3.630-637.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. P., Wackernagel W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158 9–14. 10.1016/0378-1119(95)00193-a [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standard Institute [CLSI] (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Wayne, PA: Clinical and Laboratory Standard Institute. [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz F., Frost L. S., Meyer R. J., Zechner E. L. (2010). Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34 18–40. 10.1111/j.1574-6976.2009.00195.x [DOI] [PubMed] [Google Scholar]
- Doublet B., Douard G., Targant H., Meunier D., Madec J. Y., Cloeckaert A. (2008). Antibiotic marker modifications of lambda Red and FLP helper plasmids, pKD46 and pCP20, for inactivation of chromosomal genes using PCR products in multidrug-resistant strains. J. Microbiol. Methods 75 359–361. 10.1016/j.mimet.2008.06.010 [DOI] [PubMed] [Google Scholar]
- Ellermeier C. D., Janakiraman A., Slauch J. M. (2002). Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290 153–161. 10.1016/s0378-1119(02)00551-6 [DOI] [PubMed] [Google Scholar]
- Feld L., Schjorring S., Hammer K., Licht T. R., Danielsen M., Krogfelt K., et al. (2008). Selective pressure affects transfer and establishment of a Lactobacillus plantarum resistance plasmid in the gastrointestinal environment. J. Antimicrob. Chemother. 61 845–852. 10.1093/jac/dkn033 [DOI] [PubMed] [Google Scholar]
- Frost L. S., Ippen-Ihler K., Skurray R. A. (1994). Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58 162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Koraimann G. (2010). Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. 5 1057–1071. 10.2217/fmb.10.70 [DOI] [PubMed] [Google Scholar]
- Huang H., Liao J., Zheng X., Chen Y., Ren H. (2019). Low-level free nitrous acid efficiently inhibits the conjugative transfer of antibiotic resistance by altering intracellular ions and disabling transfer apparatus. Water Res. 158 383–391. 10.1016/j.watres.2019.04.046 [DOI] [PubMed] [Google Scholar]
- Karim A., Poirel L., Nagarajan S., Nordmann P. (2001). Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201 237–241. 10.1016/s0378-1097(01)00276-2 [DOI] [PubMed] [Google Scholar]
- Kjeldsen T. S., Overgaard M., Nielsen S. S., Bortolaia V., Jelsbak L., Sommer M., et al. (2015). CTX-M-1 beta-lactamase expression in Escherichia coli is dependent on cefotaxime concentration, growth phase and gene location. J. Antimicrob. Chemother. 70 62–70. 10.1093/jac/dku332 [DOI] [PubMed] [Google Scholar]
- Koraimann G., Wagner M. A. (2014). Social behavior and decision making in bacterial conjugation. Front. Cell Infect. Microbiol. 4:54. 10.3389/fcimb.2014.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupor S. R., Fraenkel D. G. (1969). 6-phosphogluconolactonase mutants of Escherichia coli and a maltose blue gene. J. Bacteriol. 100 1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lento C., Ferraro M., Wilson D., Audette G. F. (2016). HDX-MS and deletion analysis of the type 4 secretion system protein TraF from the Escherichia coli F plasmid. FEBS Lett. 590 376–386. 10.1002/1873-3468.12066 [DOI] [PubMed] [Google Scholar]
- Llosa M., Gomis-Ruth F. X., Coll M., De La Cruz Fd F. (2002). Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45 1–8. 10.1046/j.1365-2958.2002.03014.x [DOI] [PubMed] [Google Scholar]
- Lu Y., Zeng J., Wang L., Lan K., E S., Wang L., et al. (2017). Antibiotics Promote Escherichia coli-Pseudomonas aeruginosa Conjugation through inhibiting Quorum Sensing. Antimicrob. Agents Chemother. 61:e01284-17. 10.1128/AAC.01284-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. (1972). Assay of B-galactosidase in: Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Miyazaki R., Minoia M., Pradervand N., Sulser S., Reinhard F., Van Der Meer J. R. (2012). Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet. 8:e1002818. 10.1371/journal.pgen.1002818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller T. S. B., Liu G., Boysen A., Thomsen L. E., Luthje F. L., Mortensen S., et al. (2017). Treatment with cefotaxime affects expression of conjugation associated proteins and conjugation transfer frequency of an IncI1 plasmid in Escherichia coli. Front. Microbiol. 8:2365. 10.3389/fmicb.2017.02365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandymazumdar M., Artsimovitch I. (2015). Ubiquitous transcription factors display structural plasticity and diverse functions: NusG proteins - Shifting shapes and paradigms. Bioessays 37 324–334. 10.1002/bies.201400177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Decousser J. W., Nordmann P. (2003). Insertion sequence ISEcp1B is involved in expression and mobilization of a bla(CTX-M) beta-lactamase gene. Antimicrob. Agents Chemother. 47 2938–2945. 10.1128/aac.47.9.2938-2945.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Stocker B. A. (1981). Gene rfaH, which affects lipopolysaccharide core structure in Salmonella typhimurium, is required also for expression of F-factor functions. J. Bacteriol. 146 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C. A., Klena J. D. (1993). Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57 655–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A. M., Shoemaker N. B., Li L. Y., Salyers A. A. (1993). Tetracycline regulation of genes on Bacteroides conjugative transposons. J. Bacteriol. 175 6134–6141. 10.1128/jb.175.19.6134-6141.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar S. R., Kirchhof M. G., Patten C. L., Schellhorn H. E. (2004). RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J. Bacteriol. 186 8499–8507. 10.1128/jb.186.24.8499-8507.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters V. L., Strack B., Pansegrau W., Lanka E., Guiney D. G. (1992). Mutational analysis of essential IncP alpha plasmid transfer genes traF and traG and involvement of traF in phage sensitivity. J. Bacteriol. 174 6666–6673. 10.1128/jb.174.20.6666-6673.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle G., Shoemaker N. B., Salyers A. A. (2002). Characterization of genes involved in modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 184 3839–3847. 10.1128/jb.184.14.3839-3847.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yethon J. A., Heinrichs D. E., Monteiro M. A., Perry M. B., Whitfield C. (1998). Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 273 26310–26316. 10.1074/jbc.273.41.26310 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.