Abstract
The goal of this work was to investigate the molecular profiles and metastasis markers in Chinese patients with gastric carcinoma (GC). In total, we performed whole exome sequencing (WES) on 74 GC patients with tumor and adjacent normal formalin-fixed, paraffin-embedded (FFPE) tissue samples. The mutation spectrum of these samples showed a high concordance with TCGA and other studies on GC. PTPRT is significantly associated with metastasis of GC, suggesting its predictive role in metastasis of GC. Patients carrying BRCA2 mutations tend not to metastasize, which may be related to their sensitivity to chemotherapy. Mutations in MACF1, CDC27, HMCN1, CDH1 and PDZD2 were moderately enriched in peritoneal metastasis (PM) samples. Furthermore, we found two genomic regions (1p36.21 and Xq26.3) were associated with PM of GC, and patients with amplification of 1p36.21 and Xq26.3 have a worse prognosis (P = 0.002, 0.01, respectively). Our analysis provides GC patients with potential markers for single and combination therapies.
Subject terms: Cancer genomics, Metastasis
Introduction
GC is one of the most common cancers and a leading cause of cancer death worldwide1, with a 5-year survival rate of about 30%2. The highest incidence is in East Asia, Central and Eastern Europe, and South Africa3. Surgery and chemotherapy are the mainstay treatments of GC, but nearly 20% GC patients develop peritoneal metastasis (PM), which is the most common form of metastasis of GC4. PM can lead to bowel obstruction and formation of massive amounts of malignant ascites, resulting in a poor prognosis5–7.
Several studies have used next generation sequencing strategies to determine the mutation spectrum of GC, and many significantly mutated driver genes have been identified, such as TP53, ARID1A, PIK3CA, and others8–11. However, most of these studies focus on the mutation profiles of GC and are mainly based on fresh frozen (FF) samples, but FF tissue has limited availability; therefore, little is known about the metastasis mechanism of GC, including peritoneal metastasis12. Formalin-fixing paraffin-embedding (FFPE) has been a standard sample preparation method for decades, and they are useful resources for cancer studies. There are many efforts to develop strategies to use FFPE specimens in cancer research, and several studies confirmed the technical feasibility13–15. However, these studies mainly use next-generation sequencing (NGS) target region panels, and WES has rarely been reported in studies with a large sample size.
Some factors are considered to be associated with the risk of PM, such as younger age, female gender, advanced T- and N-stage et al.7. Chemokines genes, such as CXCL12 and VEGF, have been reported to be elevated in the development of PM6. Takeno et al. identified a 22-gene expression profile which is associated with PM16. Zhang et al. reported a case of GC with matched primary cancer and peritoneal metastatic tissue, identified several genes especially mutated in PM cancer17. These studies find some genes or clinical features which may plays a role in the prognosis of PM, but the molecular mechanisms by which GC undergoes PM are not completely elucidated yet.
In this study, we first performed WES on 74 FFPE samples of GC based on the BGIseq-500 platform, compared the molecular profiles of Chinese southern GC patients with TCGA and other cohorts, and then investigated the markers of metastasis of GC. We found that mutations in several genes and copy number variations (CNVs) of two genomic regions are associated with metastasis of GC, which can be further validated in large-scale studies.
Results
Patient characteristics
A total of 74 GC patients (hereinafter referred to as Fujian cohort) from Fujian Province, China, with complete clinical follow-up information were sequenced, including 32(43%) intestinal-type, 28(38%) diffuse-type,10(14%) mixed-type and 4(5%) indeterminate adenocarcinomas; 28 (38%) of the patients were less than 60 years of age, 46 (62%) were more than 60 years of age. The majority of the subjects were male (52,70%), and the remaining 22 (30%) were female. In all 7 (9%) were stage I, 8 (11%) stage II, 51 (68%) stage III, and 9 (12%) stage IV; and 57 (77%) patients had metastasis in a follow-up exam, of which 26 (35%) patients had peritoneal metastasis. The clinical characteristics and statistics were list in Table 1 and Supplementary Table S1.
Table 1.
The clinical characteristics and statistics of GC cases included in this study (N = 74).
| Characteristics | No. (%) | PM | Non-PM | P valuea |
|---|---|---|---|---|
| Age | 0.112 | |||
| <60 | 28 (37.8) | 13 | 15 | |
| >=60 | 46 (62.2) | 13 | 33 | |
| Sex | 0.885 | |||
| Male | 52 (70.3) | 18 | 34 | |
| Female | 22 (29.7) | 8 | 14 | |
| Lauren type | 0.232b | |||
| Intestinal | 32 (43.2) | 9 | 23 | |
| Diffuse | 28 (37.8) | 12 | 16 | |
| Mixed | 10 (13.5) | 3 | 7 | |
| Indeterminatec | 4 (5.4) | 2 | 2 | |
| Tumor stage | 0.046d,e | |||
| Stage I | 7 (9.5) | 0 | 7 | |
| Stage II | 7 (9.5) | 2 | 5 | |
| Stage III | 51 (68.9) | 20 | 31 | |
| Stage IV | 9 (12.1) | 4 | 5 | |
| Differentiation | 0.018 | |||
| Poor | 51 (68.9) | 23 | 28 | |
| Well | 20 (27) | 3 | 17 | |
| Indeterminatec | 3 (4.1) | 0 | 3 | |
aDerived from a χ2 test unless otherwise specified. bIntestinal-subtype versus diffuse-subtype. cIndeterminate samples not include for difference tests dstage II & III versus stage I. eDerived from a Fisher’s exact test.
Genomic mutations of Chinese GC patients
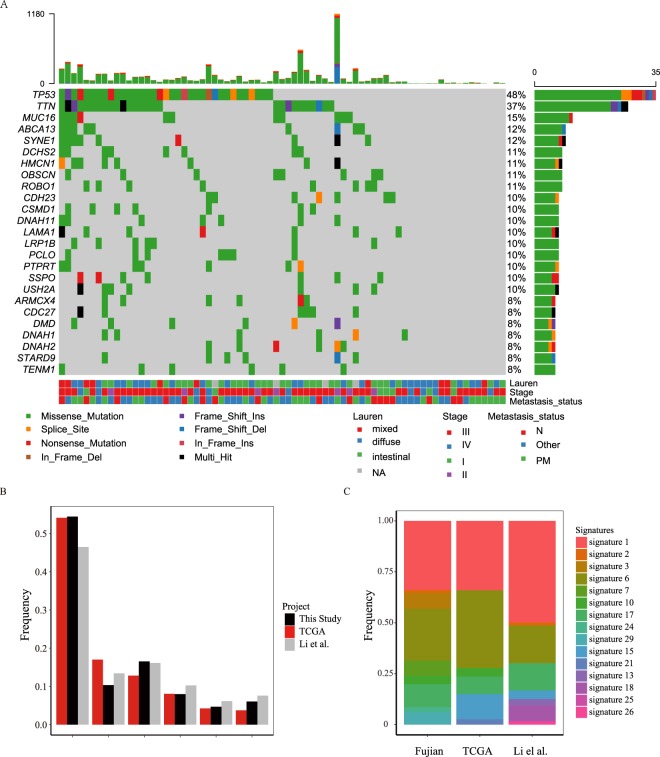
A total of 11,118 mutations were detected in this study, the mean number of somatic mutations per patient was 150 (range from 0 to 1517) (Supplementary Table S2). Somatic SNVs (sSNVs) and indels (sIndels) accounted for 95.4% and 4.6% of the mutations, respectively. Of the mutations, 3,066 (27.6%) were synonymous, 6,857 (61.7%) missense, 463 (4.2%) nonsense (stopgain), 9 (0.1%) stoploss, 212 (1.9%) splice site, 452 (4.1%) were frameshift indels, and 59 (0.5%) were in-frame indels. Several cancer-related genes were frequently mutated in Fujian cohort, such as TP53(37/74), LRP1B(8/74), PTPRT(7/74), and ARID1A(5/74), consistent with previous studies on GC10,11,18 (Fig. 1A, Supplementary Table S3). Notably, all of the ARID1A-mutated samples carried wildtype TP53 (P = 0.027), the mutation pattern had been reported in previous studies but was more pronounced in this study8,9. We randomly selected 36 mutation sites for mass spectrometry validation, and 34 (94.4%) of them were verified as somatic mutations (Supplementary Table S4).
Figure 1.
The mutation spectrum of GC in this study and the comparation with other studies. (A) Heat map showing somatic mutation profiles of cancer genes in this study. Left, the number of samples with mutations in a given gene. Top, the clinical type of samples and mutations burden of each sample. (B) The six classes of base substitution in three studies. (C) The mutation signatures in three studies.
The mutations in the exon and splice site regions of other two datasets, TCGA (download from https://cancergenome.nih.gov/) and Li et al.18 were used for further comparative analysis(Supplementary Table S1). For point mutations, C > T, G > A transitions accounted for 54.4% of the sSNVs, and the ratio of the 6 types of base substitution is similar to the studies of TCGA and Li et al. (Fig. 1B). We further found that the spectrum of flanking nucleotides surrounding the mutated base was highly concordant between our results and the other two datasets (Supplementary Fig. S1A). The context-dependent mutational patterns of these three datasets were then identified using mSignatureDB (http://tardis.cgu.edu.tw/msignaturedb/) to explore the heterogeneity of mutagenic processes in GC and its diagnostic potential19. The results showed that prevalence of signatures 1, 6, and 17 were similar in the three studies, accounting for the majority of mutational processes (Fig. 1C). While signature 1 and 6 are related to spontaneous deamination of 5-methylcytosine and DNA mismatch repair, respectively, which results in C > T transitions and predominantly occurs at NpCpG trinucleotides20,21, other signatures specific to a study may be due to other endogenous mutational processes, treatment, or environment22.
We also found that the recurrently mutated genes in our study were similar to TCGA and Li et al., and the overlap between these three studies is about 50% (Supplementary Fig. S1B,C). Some cancer-related genes that have been reported in other populations (Hong Kong and Russian) were also found frequently mutated in our cohorts, including TP53, LRP1B, PTPRT, ARID1A, FAT4, FAT1, and APC10,11,18,23. We found several genes especially mutated in each subtype of GC. NUP214(2/32, means 2 samples mutated among 32 samples, similarly hereinafter), APC (2/32), PRDM16(3/32), and SMAD4 (2/32) altered more than once in intestinal-subtype tumors, but not in diffuse-type tumors. DBX3(2/28), MYH9(2/28), and AFF3(2/28) altered only in diffuse-type tumors (Fig. 1A). Notably, RHOA and CDH1 are genes tend to frequently mutated in diffuse-type GC according to previous studies10, in this study, however, these two genes were mutated only once in diffuse-type tumors, while not in intestinal tumors.
Patients that were older (age >= 60) have significantly higher TMB (P = 0.0021) and TNB (P = 0.034) than younger patients (age < 60) (Supplementary Fig. S2A,B). and male patients tend to carry more mutations than female patients (P = 0.034), but the difference in TNB was not significant (P = 0.82) (Supplementary Fig. S2C,D). We didn’t find any significant difference of mutation burden between intestinal-type and diffuse-type tumors.
Somatic copy number variations of Chinese GC patients
An analysis of copy number alterations of these 74 samples showed that most chromosome arms had undergone copy number gain or loss, with frequent amplified regions including 1q, 6p, 7, 8q, 13q, 20 (frequencies from 12% to 64%), and frequent losses observed on chromosomes 4, 14q, 18q, 19, 21q, 22q (frequencies from 16% to 43%) (Supplementary Fig. S3A). These overall somatic copy number variant (sCNV) patterns are consistent with previously published studies on GC10,11,23,24. We identified 156 focal amplifications and 69 focal deletions, in well-known oncogenes, such as ERBB2, CCNE1, KRAS, MYC, EGFR, and CDK6, and cancer-related genes such as GATA4, GATA6, CD44 and ZNF217 (Supplementary Table S5). Some tumor suppressor genes were identified in focal deleted regions, such as CDKN2A, FAT1 and SMAD4 (Supplementary Fig. S3B). These results are consistent with other studies such as TCGA and Wang et al.10,11. Through analyzing the sCNV at different subtypes of GC, we found that sCNVs occurred more frequently in intestinal-type and diffuse-type than mixed-type tumors (Supplementary Fig. S3C). Especially, there are 134 cancer-related genes amplified or deleted in intestinal-type tumors, much more than in diffuse-type (40) and mixed-type (4) tumors. Overall, we found 155 cancer genes amplified or deleted in our samples, in which half of them (78 genes) have been reported by TCGA or Wang et al. (Supplementary Fig. S3D, Table S6), the other half (77 cancer genes) with sCNVs identified in this study could be further confirmed for their involvement in the development of GC.
Metastasis-associated driving genes in GC
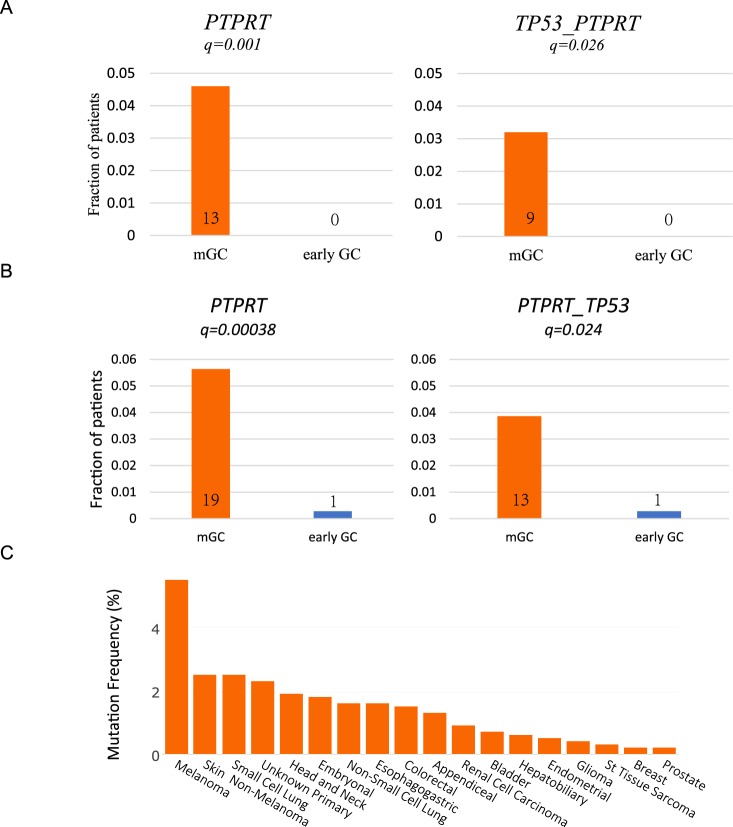
PTPRT has recently been reported to be closely related to early metastasis of colorectal cancer. In this study, we found PTPRT mutated in 7 patients (7/74), and metastasis occurred in six out of seven patients in clinical follow-up, suggesting that PTPRT may be associated with metastasis of GC. Because of the small number of samples in this study, we validated it with two published independent queues. We analyzed 620 GC patients from the MSK-Impact25 and GENIE26 prospective sequencing studies, including 280 patients with metastatic GC (Primary Stage IV or metastasis) and 340 patients with early GC (Primary, stage I-III). We then evaluated the relationship between the driving gene module and metastasis tendency in GC. It is noteworthy that, after correcting multiple hypothesis tests, we found that in this independent data set, the combination of TP53 and PTPRT (q = 0.026), or PTPRT (q = 0.001) mutations alone were enriched in metastasis compared with early GC (Fig. 2A, Supplementary Table S7), similar to those reported by Zheng Hu et al. in colorectal cancer27. When the three cohorts were merged, the sample size increased to 694 (PTPRT mutation carriers increased to 20), and the correlation between mutation carriers and metastasis was more significant (Fig. 2B, q = 0.024 and q = 0.00038, respectively). We further analyzed the mutations of PTPRT in 10,000 metastatic cancers published by MSK-impact in 201728.It was found that PTPRT has a considerable mutation frequency in many metastatic cancers, such as melanoma, small cell lung cancer, head and neck carcinoma, suggesting that PTPRT may be involved in multiple cancers (Fig. 2C).
Figure 2.
Association of mutations in TP53 and PTPRT with GC metastasis. (A) Association of TP53 and PTPRT gene mutations with metastasis in an independent cohort of 620 GC patients. (B) The correlation between TP53 and PTPRT mutations and metastasis after integration of Fujian, MSK-impact and GENIE cohorts. (C) The frequency of PTPRT mutations in metastasis cancers in the MSK-impact pan-cancer study.
In addition, we found that BRCA2 mutation tended to enrich in non-metastatic samples (q = 0.02, Supplementary Table S7). Li et al. have reported that BRCA2 mutation is associated with better prognosis of GC18. Evidence suggests that BRCA2 mutation is related with improved platinum-based chemotherapy response and prolonged survival in patients with ovarian cancer29. It has been suggested that BRCA2 mutation is associated with increased survival because of the reduced ability of BRCA2 mutated cancer cells to repair damaged DNA caused by chemotherapy. Based on the above results, we speculate that GC patients with BRCA2 mutation may benefit from chemotherapy to reduce the metastasis of tumors.
Genomic alterations associated with PM
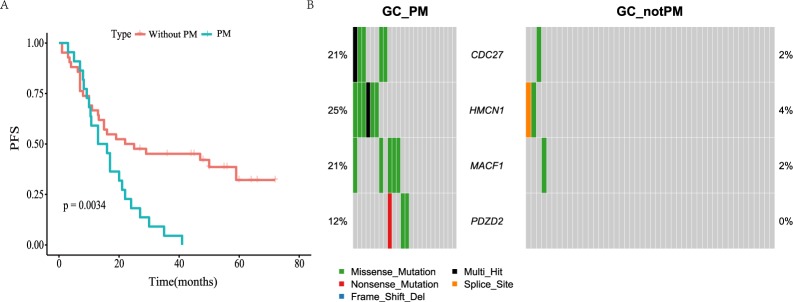
PM is the main form of metastasis of GC, and is an important cause of morbidity and mortality of GC patients. The patients with PM had a worse prognosis than those without PM in this study (P = 0.0034, Fig. 3A). In Fujian cohort, we found that the patients with advanced stage tend to occur PM (Stage II&III-VS-Stage I, P = 0.046), and poorly differentiated GC were more likely to develop PM (Table 1, P = 0.015). In addition, we found that the younger patients (age < 60) tend to occur PM than older patients (age >= 60), although not statistically significant (P = 0.112).
Figure 3.
Genomic alterations and their prognostic significance associated with PM. (A) Kaplan-Meier plots for DFS in PM and not-PM patients. (B) Genes which enriched in PM patients. Fisher exact test, P < 0.05 and q < 0.1 (two GC samples without mutations were not considered in this analysis).
To determine if there are mutation of genes specifically associated with PM, we compared the mutation pattern of genes between the PM group and Non-PM group, identified 3 genes (Fisher exact test, p < 0.05, q-value < 0.1), including CDC27, MACF1 and HMCN1, which showed moderate enrichment in the PM group (Fig. 3B, Supplementary Table S8). Among them, MACF1 is a pan-cancer driver gene, which is related to cell adhesion function4. Interesting, we also found the gene PDZD2, which was previously supposed to involved in the early stages of prostate tumorigenesis, was especially altered both in this study (Fig. 3B, mutated in three PM sample) and the PM tissue of the GC patient in Zhang et al.’s study17. Similarly, we studied the relationship between PM and gene mutation in MSK-Impact cohort. There were 88 samples of PM in MSK-Impact cohort, and CDH1 gene was enriched in PM samples (Supplementary Table S8). However, the MSK-Impact panel does not contain the four genes (CDC27, MACF1, PDZD2 and HMCN1) mentioned above, so the relationship between the mutations of these genes and PM needs to be studied in a larger sample size.
Furthermore, we found that the amplification of several regions is enriched in PM patients, and two of them (1p36.21 and Xq26.3) are associated with a worse prognosis (Supplementary Fig. S4A,B). Interestingly, the 1p36.21 region contains a gene family named PRAME (preferentially expressed antigen of melanoma), which is expressed in many cancers and was functions in reproductive tissues during development30. PRAME had been reported as an independent marker for metastasis in uveal melanoma, indicating the value as a marker in the process of PM31. The Xq26.3 region contains a gene family named CT45 (the Cancer/Testis Antigen Family 45), which is especially overexpressed in various cancer types. It had been reported that the family member CT45A1 in breast cancer and lung cancer can act as a proto-oncogene to trigger tumorigenesis and cancer metastasis32,33. However, in the TCGA cohort, we did not find significant correlation between the amplification of these two regions and clinical prognosis. The reason may be the population difference between TCGA and Chinese, or the difference of other characteristics. Through the analysis of copy number alteration of PM samples, we found that there were significant deletions in many genomic regions of PM samples, involving many cancer genes and immune related genes (such as B2M, RHOA, IFNE, JAK1/2, etc.). KEGG pathway enrichment analysis showed that many cancer pathways, immune related pathways, such as JAK-STAT signaling pathway, Cell cycle, WNT signaling and Antigen processing and presentation pathway, were damaged (Table 2). However, this pattern is not obvious in non-PM samples.
Table 2.
Enriched cancer-related pathway of peritoneal metastasis GC samples by GSEA analysis.
| Gene Set Name (Pathway) | # Genes in Overlap (k) | # Genes in Gene Set (K) | q-value |
|---|---|---|---|
| Cytokine-cytokine receptor interaction | 51 | 265 | 3.05E-17 |
| JAK-STAT signaling pathway | 37 | 155 | 8.49E-16 |
| Regulation of autophagy | 17 | 35 | 6.91E-13 |
| Chemokine signaling pathway | 34 | 189 | 3.97E-11 |
| Natural killer cell mediated cytotoxicity | 27 | 137 | 7.66E-10 |
| Colorectal cancer | 17 | 62 | 1.40E-08 |
| Antigen processing and presentation | 19 | 88 | 8.22E-08 |
| Pathways in cancer | 35 | 325 | 1.01E-05 |
| Cell cycle | 18 | 124 | 7.34E-05 |
| MAPK signaling pathway | 28 | 267 | 1.32E-04 |
| Focal adhesion | 23 | 199 | 1.53E-04 |
| T cell receptor signaling pathway | 14 | 108 | 1.33E-03 |
| B cell receptor signaling pathway | 11 | 75 | 1.91E-03 |
| Cardiac muscle contraction | 11 | 78 | 2.62E-03 |
| P53 signaling pathway | 10 | 68 | 3.16E-03 |
| VEGF signaling pathway | 10 | 76 | 6.52E-03 |
| Cell adhesion molecules (CAMs) | 14 | 133 | 7.56E-03 |
| Non-small cell lung cancer | 8 | 54 | 8.41E-03 |
| WNT signaling pathway | 15 | 150 | 8.41E-03 |
Discussion
Overall, the GC mutation profiles of Fujian cohort are similar to those of TCGA and other cohorts. The feasibility of WES sequencing strategy based on FFPE and BGI-seq platform for cancer research was confirmed by comparative analysis of different queues.
Metastasis is a major cause of cancer-related death in cancer patients, but the molecular determinants of this process are largely unknown. We found several genes related to metastasis of GC, and PTPRT mutation can be used as a specific marker of metastasis. In addition, we found that patients with BRCA2 mutation had fewer metastases, which may be related to the benefit of chemotherapy in patients with BRCA2 mutation. In the Fujian cohort, two samples carried mutations in BRCA2. One of the patients with missense mutations (NO592779, well differentiated, who received chemotherapy after surgery) had no metastasis during clinical follow-up and had a progression-free survival of 26 months. However, another patient (NO573713, poorly differentiated) carried a missense mutation and a frameshift deletion of BRCA2, which progressed and metastasis 15 months after surgery. It is noteworthy that this patient’s mutation load is the highest in the Fujian cohort and carries a series of loss of function mutations of tumor suppressor genes (Supplementary Table S9 and 10).
PM can lead to bowel obstruction or malignant ascites, resulting in a poor prognosis and decline in the quality of life, so it is important to identify risk factors for PM. Mutations in CDH1, MACF1, CDC27 and HMCN1 were associated with PM, but this need to be confirmed in a larger cohort. We also found 2 regions, 1p36.21 and Xq26.3, that are amplified in PM patients, and associated with a poorer outcome. Importantly, two gene families of these amplified regions, PRAME and CT45, had been reported to overexpressed in various cancer types and associated with cancer metastasis6,9,17,31, indicating their potential as biomarkers for PM in GC patients. 9p21.3 contains several cancer genes and immune genes, which are significantly deleted in peritoneal metastasis samples. Previous studies have suggested that the absence of this region is associated with worse prognosis and limited benefit from immunotherapy34,35. On the other hand, at the CNV level of all cancer-related genes, we found that ERBB2 (also known as HER2) amplified samples were more likely to have no peritoneal metastasis (q = 0.043, Supplementary Table S11), which might be a better treatment for HER2-amplified patients, such as HER2 inhibitors25. As the sample size of our study is limited, further studies should be conducted to confirm these findings.
Materials and Methods
Patient cohort
This study was approved by the Ethical Committee of the Union Medical College Hospital Affiliated of Fujian Medical University and carried out according to the approved guidelines. All patients signed informed consent prior to their enrollment. In total, 300 cases with sufficient clinical pathological information were provided;155 of which with pathological paraffin blocks were selected for WES. Samples of cancer and adjacent normal tissues were taken from each case at the same time, a total of 6 FFPE sections with size of 10 μm in 1 cm × 1 cm and tumor content of more than 50% were selected. Of the selected samples, 74 were successful for subsequent library construction and sequencing (Supplementary Table S1). GC data of MSK-impact (n = 204) and GENIE (n = 416) were downloaded from http://synapse.org/genie and cBioPortal (id = egc_msk_2017), respectively.
WES library construction and next-generation sequencing
The genomic DNA of FFPE samples was randomly fragmented and the size of the library fragments was mainly distributed between 150 bp and 250 bp. The end repair of DNA fragments was performed, and an “A” base was added at the 3’-end of each strand. Adapters were then ligated to both ends of the end repaired dA tailed DNA fragments for amplification and sequencing. Size-selected DNA fragments were amplified by ligation-mediated PCR, purified, and whole-exome capture was performed using the BGI Human All Exon V4 kit. Captured products were then circularized. Rolling circle amplification (RCA) was performed to produce DNA Nanoballs (DNBs). Each resulting qualified captured library was then loaded on BGISEQ-500 platform and pair-end 50 bp or pair-end 100 bp sequencing was conducted for each captured library. We sequenced an average of 1,533,107,107 reads for each sample, after reads quality filtering and duplication removing, the sequencing depths for FFPE tumors and corresponding normal tissues were 117X and 92X on averages, respectively.
Identification of somatic mutations
The sequencing data processing and variants detection pipeline is shown in Supplementary Fig. S5. Reads containing sequencing adapters and low-quality reads were removed using SOAPnuke software36. Then the high-quality data of each sample was mapped to the human HG19 reference genome and the duplicate reads were removed with Edico software (http://edicogenome.com/dragen-bioit-platform). To ensure accurate variant calling, local realignment around Indels and base quality score recalibration was performed using GATK37,38. Then the sequencing depth and coverage for each sample were calculated based on the alignments, and samples with low coverage or depth were re-sequenced on the same library to achieve enough sequencing depth.
SSNVs and sIndels were detected using the MuTect39 and Varscan2 software40, respectively. Then these mutations (sSNVs and sIndels) were annotated with ANNOVAR41 and followed by several filtering steps to remove potential false positives and obtain reliable results. For MuTect, in addition to the build-in filters, the following filtering criteria were applied: (1) total read count in tumor and normal DNA >= 10; (2) mutation allele fraction >=10% and >=5 reads that support this mutation; (3) mutation site is at least five bases away from the end of the read; (4) the SNV was not encompassed in short repeat regions; (5) presence of variant on both strands and the distribution of reads supporting this variant on the two strand is not biased; (6) the frequency of variant is less than 0.5% at 1,000 Genomes (1000G) database (http://www.1000genomes.org), Exome Sequencing Project (ESP) 6500 database (http://evs.gs.washington.edu/EVS) or Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org). For Varscan2, in addition to the built-in filters, the following filtering criteria were applied: (1) coverage >= 10 in normal DNA and coverage >=10 in tumor DNA; (2) variant frequency >=15%; (3) the Indel was not encompassed in short repeat regions; (4) the frequency of Indel is less than 0.5% at 1,000 Genomes (1000G) database, Exome Sequencing Project (ESP) 6500 database and Exome Aggregation Consortium (ExAC) database. The final mutation results were list in Supplementary Table S3.
SCNVs were detected by the CNV workflow tools within GATK4 (https://github.com/broadinstitute/gatk). The FFPE normal samples were used as control to identify tumor-specific genomic alterations. Then the copy-number segment data was used as input to the GISTIC242 to detect recurrently amplified or deleted genomic regions. GSITIC2 analysis was performed using the default parameters.
Confirmation of mutations
36 mutation sites, containing 21 cancer gene mutations and 15 mutations in PM samples specific genes were randomly selected for mass spectrometry validation. In total, 34 mutations were validated by the MassARRAY platform (including mutations that not been detected before, such as mutations in NUP107), with a 94% validation rate. We considered validation a success when both the tumor and normal genotype generated by MassARRAY platform were the same as the sequencing result, and failure if the genotype called by mass spectrometry was not the same as sequencing.
Neoantigen prediction
SSNV mutations were used to predict neoantigens by NetMHC, NetMHCpan, PickPocket, PSSMHCpan and SMM43. The poor-quality peptides were removed according to two criteria: (1) IC50 < 500 in at least in three tools; (2) MT score < WT score for each peptide.
Statistical methods
A Wilcoxon test was used to analyze the significance of the association of PM associated genes, patient age and patient gender. The Fisher exact test was used to analyze the significance of associations of the number of gene mutations with PM and not-PM. All tests were two-sided, and statistical significance was set at p < 0.05 or q < 0.05 if applicable. All statistical analyses were performed with RStudio software (Version 3.5.1)
Supplementary information
Acknowledgements
We thank the department of pathology at Union Medical College Hospital Affiliated to Fujian Medical University for their assistance in sample and data collection. We also thank Lei Chen and Michael Dean for their constructive advices on the manuscript. We would like to thank Yun Zhao, Xuehui Tang and Lei Ge for their administrative support. This work was supported by the Critical Patented Project of The Science & Technology Bureau of Fujian Province, China (grant number 2013YZ0002-2), the Joint Project of the Natural Science and Health Foundation of Fujian Province, China (grant number 2015J01397), the Science, Technology and Innovation Commission of Shenzhen Municipality under grant No.GJHZ20170314152701465 and No.JCYJ20170303151334808.
Author Contributions
Q.C., X.-Q.Z. and C.C. conceived the experiments. X.-Q.Z., C.-M.S., Q.-L.L. and Y.Z. conducted the experiments. C.C., X.-C.H., Z.-Y.Z., Z.-K.Z., J.-W.Z., B.L. S.-Y.L. and S.Q. analyzed the results. F.W., R.-P.W. and Z.-Q.H. provided patient specimens and conducted histopathological examinations. C.C. and X.-C.H. wrote the manuscript. All authors reviewed the manuscript.
Data Availability
The data reported in this study are available in the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number CNP0000159).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chao Chen, Chunmei Shi and Xiaochun Huang contributed equally.
Contributor Information
Xiuqing Zhang, Email: zhangxq@genomics.cn.
Qiang Chen, Email: cqiang8@189.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50171-7.
References
- 1.Hohenberger P, Gretschel S. Gastic cancer. The Lancet. 2003;362:305–315. doi: 10.1016/s0140-6736(03)13975-x. [DOI] [PubMed] [Google Scholar]
- 2.Guggenheim DE, Shah MA. Gastric cancer epidemiology and risk factors. J Surg Oncol. 2013;107:230–236. doi: 10.1002/jso.23262. [DOI] [PubMed] [Google Scholar]
- 3.Kong X, Wang JL, Chen HM, Fang JY. Comparison of the clinicopathological characteristics of young and elderly patients with gastric carcinoma: a meta analysis. J Surg Oncol. 2012;106:346–352. doi: 10.1002/jso.23004. [DOI] [PubMed] [Google Scholar]
- 4.Bailey MH, Tokheim C, Porta-Pardo E. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;173:371–385. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montori G, et al. The treatment of peritoneal carcinomatosis in advanced gastric cancer: state of the art. International journal of surgical oncology. 2014;2014:912418. doi: 10.1155/2014/912418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mura G, Verdelli B. The features of peritoneal metastases from gastric cancer. Journal of Cancer Metastasis and Treatment. 2016;2:365. doi: 10.20517/2394-4722.2016.19. [DOI] [Google Scholar]
- 7.Thomassen I, et al. Peritoneal carcinomatosis of gastric origin: a population-based study on incidence, survival and risk factors. International journal of cancer. 2014;134:622–628. doi: 10.1002/ijc.28373. [DOI] [PubMed] [Google Scholar]
- 8.Wang K, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nature genetics. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 9.Zang ZJ, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 10.Adam JB, Vesteinn T, Ilya S, Sheila MR. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadd AG, et al. Targeted, high-depth, next-generation sequencing of cancer genes in formalin-fixed, paraffin-embedded and fine-needle aspiration tumor specimens. J Mol Diagn. 2013;15:234–247. doi: 10.1016/j.jmoldx.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, et al. Profiling cancer gene mutations in clinical formalin-fixed, paraffin-embedded colorectal tumor specimens using targeted next-generation sequencing. Oncologist. 2014;19:336–343. doi: 10.1634/theoncologist.2013-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frampton GM, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeno A, et al. Gene expression profile prospectively predicts peritoneal relapse after curative surgery of gastric cancer. Annals of surgical oncology. 2010;17:1033–1042. doi: 10.1245/s10434-009-0854-1. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, et al. Whole genome and transcriptome sequencing of matched primary and peritoneal metastatic gastric carcinoma. Sci Rep. 2015;5:13750. doi: 10.1038/srep13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen K, et al. Mutational landscape of gastric adenocarcinoma in Chinese: implications for prognosis and therapy. Proc Natl Acad Sci USA. 2015;112:1107–1112. doi: 10.1073/pnas.1422640112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang PJ, et al. mSignatureDB: a database for deciphering mutational signatures in human cancers. Nucleic Acids Res. 2018;46:D964–D970. doi: 10.1093/nar/gkx1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeifer G. P. mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- 21.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087 e2073. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng N, et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673–684. doi: 10.1136/gutjnl-2011-301839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura Y, et al. Genetic alterations in 102 primary gastric cancers by comparative genomic hybridization: gain of 20q and loss of 18q are associated with tumor progression. Mod Pathol. 2004;17:1328–1337. doi: 10.1038/modpathol.3800180. [DOI] [PubMed] [Google Scholar]
- 25.Janjigian YY, et al. Genetic Predictors of Response to Systemic Therapy in Esophagogastric Cancer. Cancer discovery. 2018;8:49–58. doi: 10.1158/2159-8290.CD-17-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium TAPG. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer discovery. 2017;7:818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, B. et al. Factors associated with peritoneal metastasis in non-serosa-invasive gastric cancer: a retrospective study of a prospectively-collected database. BMC Cancer13 (2013). [DOI] [PMC free article] [PubMed]
- 28.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature medicine. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang D, Khan S, Sun Y, Zhang W. Association of BRCA1 and BRCA2 Mutations With Survival, Chemotherapy Sensitivity, and Gene Mutator Phenotype in Patients With Ovarian Cancer. JAMA. 2011;306:1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Z, et al. PRAME is critical for breast cancer growth and metastasis. Gene. 2016;594:160–164. doi: 10.1016/j.gene.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Field MG, et al. PRAME as an Independent Biomarker for Metastasis in Uveal Melanoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:1234–1242. doi: 10.1158/1078-0432.CCR-15-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang B, et al. CT45A1 acts as a new proto-oncogene to trigger tumorigenesis and cancer metastasis. Cell death & disease. 2014;5:e1285. doi: 10.1038/cddis.2014.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang F, Tang S, Guo X, Yang C, Jia K. CT45A1 siRNA silencing suppresses the proliferation, metastasis and invasion of lung cancer cells by downregulating the ERK/CREB signaling pathway. Molecular medicine reports. 2017;16:6708–6714. doi: 10.3892/mmr.2017.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong S, et al. The genomic landscape of Epstein-Barr virus-associated pulmonary lymphoepithelioma-like carcinoma. Nature communications. 2019;10:3108. doi: 10.1038/s41467-019-10902-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang W, et al. Comprehensive Genomic Profiling Identifies Novel Genetic Predictors of Response to Anti-PD-(L)1 Therapies in Non-Small Cell Lung Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2019;25:5015–5026. doi: 10.1158/1078-0432.CCR-19-0585. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7:1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu G, et al. PSSMHCpan: a novel PSSM-based software for predicting class I peptide-HLA binding affinity. GigaScience. 2017;6:1–11. doi: 10.1093/gigascience/gix017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this study are available in the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number CNP0000159).