Abstract
Background and purpose
Nanogels (NGs) are promising drug delivery tools but are typically limited to hydrophilic drugs. Many potential new drugs are hydrophobic. Our study systematically investigates amphiphilic NGs with varying hydrophobicity, but similar colloidal features to ensure comparability. The amphiphilic NGs used in this experiment consist of a hydrophilic polymer network with randomly distributed hydrophobic groups. For the synthesis we used a new synthetic platform approach. Their amphiphilic character allows the encapsulation of hydrophobic drugs. Importantly, the hydrophilic/hydrophobic balance determines drug loading and biological interactions. In particular, protein adsorption to NG surfaces is dependent on hydrophobicity and critically determines circulation time. Our study investigates how network hydrophobicity influences protein binding, biocompatibility and cellular uptake.
Methods
Biocompatibility of the NGs was examined by WST-1 assay in monocytic-like THP-1 cells. Serum protein corona formation was investigated using dynamic light scattering and two-dimensional gel electrophoresis. Proteins were identified by liquid chromatography-tandem mass spectrometry. In addition, cellular uptake was analyzed via flow cytometry.
Results
All NGs were highly biocompatible. The protein binding patterns for the two most hydrophobic NGs were very similar to each other but clearly different from the hydrophilic ones. Overall, protein binding was increased with increasing hydrophobicity, resulting in increased cellular uptake.
Conclusion
Our study supports the establishment of structure–property relationships and contributes to the accurate balance between maximum loading capacity with low protein binding, optimal biological half-life and good biocompatibility. This is an important step to derive design principles of amphiphilic NGs to be applied as drug delivery vehicles.
Keywords: adjustable amphiphilic nanogels, tuneable hydrophilic/hydrophobic balance, biocompatibility, cellular uptake, protein corona, THP-1 cells
Introduction
Nanogels (NGs) as cross-linked polymeric nanoparticles (NPs) are emerging as flexible, versatile drug carriers,1 being interesting for many applications ranging from cancer therapy to nanoantibiotics.2–8 However, conventional NGs exhibit one major drawback: their overall hydrophilicity dramatically limits their application for hydrophobic drugs.
Amphiphilic NGs, based on a hydrophilic polymer matrix containing hydrophobic groups, recently emerged as new carriers for hydrophobic drugs.9–11 Accurately tuning delivery properties can be achieved by changing the network’s hydrophilic/hydrophobic balance. In general, increasing hydrophobicity enables higher loading capacities and more sustained release profiles,12 but may lead to several undesired effects such as decreased systemic circulation time and reduced biocompatibility.13,14 In blood, plasma proteins selectively adsorb to NG surfaces, forming a specific protein corona. The identity and amount of bound proteins are mainly determined by surface characteristics with hydrophobicity and surface charge being the most important ones.13–19 The corona represents the actual bio-nano-interface, mediates cellular interactions and depicts the biological identity of a NP.20,21 Hydrophobic surfaces favor binding of opsonins, specific plasma proteins, that label NPs for uptake and removal by the reticuloendothelial system (RES).22,23,24 Consequently, blood circulation time is dramatically reduced, limiting the therapeutic potential of the NPs.25 In contrast, hydrophilic surface moieties protect NPs from opsonization and prevent cellular uptake by the RES, described as “stealth effect”.26–29
Amphiphilic NGs combine aspects from hydrophobic and hydrophilic surfaces in one single colloidal system. Thus, their protein interactions and biocompatibility are far less understood. It becomes obvious that structural optimization for drug delivery applications requires a careful balancing, as increasing hydrophobicity leads to increased loading capacities of hydrophobic cargoes but at the other side also results in a reduced half-life and biocompatibility. To correlate specific hydrophilic/hydrophobic network compositions to loading/release profiles and to NG behavior in biological systems, systematic investigations are needed.
However, up to now such systematic studies are highly challenging since conventional synthetic approaches to generate amphiphilic NGs also change the colloidal features, such as size, size distribution and morphology. The protein corona is also dependent on NP size, curvature and shape.30–32 Consequently, it would be impossible to discriminate whether a different NG behavior results from altered size or modified amphiphilicity.
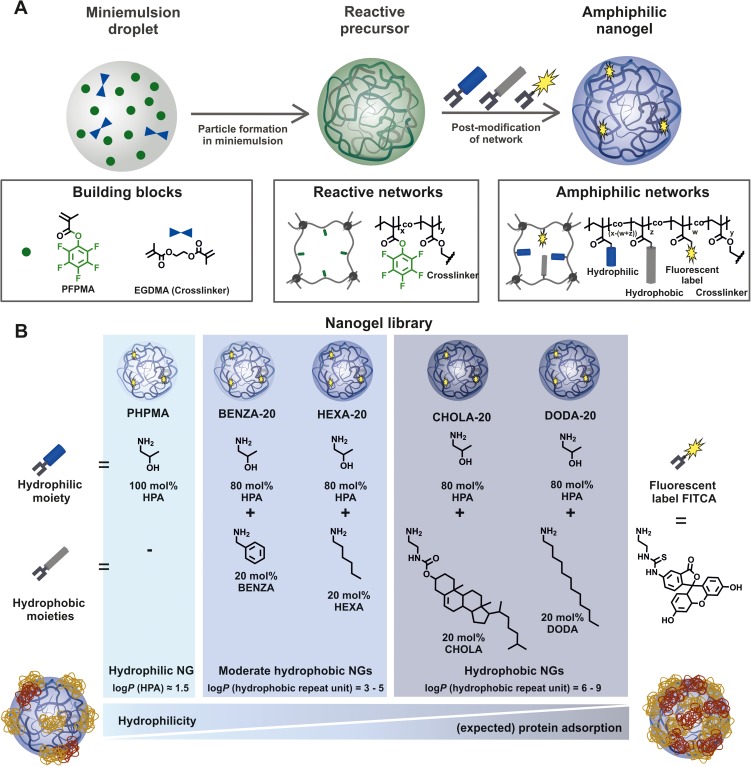
Thus, a new synthetic approach is required. We have recently developed a synthetic platform approach based on the functionalization of reactive precursor particles with hydrophilic and hydrophobic moieties,12,33 allowing to tuning of amphiphilicity while providing a majority of similar colloidal features. Therefore, we now have access to a library of comparable NGs with varying network composition but similar colloidal properties. All NGs are based on an amphiphilic network of poly(methacrylamide) copolymers containing 80 mol-% of hydrophilic 2-hydroxypropylamin (HPA) groups but 20 mol-% of different hydrophobic amides, ie, benzyl, hexyl, cholesteryl and dodecyl moieties referred to as BENZA-20, HEXA-20, CHOLA-20 and DODA-20.
This allows us, for the first time, to systematically investigate the influence of network composition on the interaction with biological systems. Human monocytic like THP-1 cells are used as a model system for human monocytes, which are a main component of the RES. We investigated biocompatibility and cellular uptake in THP-1 cells. Protein binding was analyzed by two-dimensional gel electrophoresis (2DE) and liquid chromatography-tandem mass spectrometry (LC-MS/MS) combined with the Top 3 approach for quantification.34,35 To the best of our knowledge, this is the first systematic investigation on highly comparable amphiphilic NGs with different hydrophobicities, their specific protein coronas and the resulting effects in terms of cellular uptake and biocompatibility. In combination with our previously determined loading capacities and release profiles,12 these results are important to establish structure–property and structure–activity relationships.
Materials and methods
Synthesis of amphiphilic NGs
All reagents were purchased from commercial sources and used without further purification, unless otherwise stated. Anhydrous solvents were obtained from a MB-SPS-800 (MBraun, Germany) solvent purification system and ultrapure water (MQ) from a LaboStar Pro UV 2 (Evoqua, Germany) water system. Pentafluorophenyl methacrylate (PFPMA),36,37 poly(pentafluorophenyl methacrylate) (PPFPMA),38 CHOLA39,40 and [2-(aminoethyl)-carbamothioyl]-5-aminofluorescein (FITCA)41 and post-modification of PPFPMA12,42 were prepared according to literature procedures. Moisture or air-sensitive reactions were carried out in dry glassware under nitrogen atmosphere. Dialysis was performed in benzoylated cellulose dialysis tubes (width: 32 mm, molecular weight cutoff 2000 g/mol, Sigma-Aldrich, Germany).
Reactive precursor particles
Amphiphilic NGs were synthesized as recently described.12 First, reactive PPFPMA precursor particles were synthesized via miniemulsion polymerization. Briefly, an oil phase consisting of 10 g PFPMA, 0.4 g ethylene glycol dimethacrylate (EGDMA, 5 mol-% w.r.t PFPMA), hexadecane (0.44 g) and 195 mg 2,2′-azobis(2-methylpropionitrile) (AIBN) was dispersed in an aqueous phase of sodium dodecyl sulfate (SDS) dissolved in deionized (DI) water (1.25 mg/mL, in 200 mL DI water, 2.5 wt-% w.r.t PFPMA) by pre-sonication in a sonication bath (Elmasonic P30 H, Elma, Germany) for 10 mins. Full dispersion was achieved by ultrasonication with a Digital Sonifier SFX 550 (Branson, Germany). The emulsion was purged with nitrogen for 10 mins before the reaction was allowed to proceed for 24 hrs at 70°C. Particle dispersions were purified via repeated centrifugation-washing-redispersion steps and freeze-dried for storage.
Amphiphilic NGs
Post-modification of reactive precursor particles was carried out on dispersed PPFPMA particles (400 mg, 1.59 mmol w.r.t monomer units of PPFPMA particles, 1.0 eq) in dimethylformamide (DMF) by adding different molar ratios of amine functionalized moieties (3.0 eq w.r.t. monomer units) and triethylamine (TEA) (660 µL, 4.77 mmol, 3.0 eq w.r.t. monomer units), as summarized in Table S1.
The reaction was allowed to proceed at 50°C for 24 hrs. Afterward, the particles were purified by extensive dialysis first against DMF (1 week) and subsequently against DI water (1 week) and MQ (1 week). The particles were then freeze-dried for storage upon use. In this study, the following five NGs were used: poly(N-(2-hydroxypropyl)methacrylamide) (PHPMA) as hydrophilic control, bearing no hydrophobic groups and variants that each contained 80 mol-% hydrophilic HPA and 20 mol-% of a different hydrophobic group, ie, benzylamine (BENZA-20), hexylamine (HEXA-20), an amine-functionalized cholesteryl group (CHOLA-20) and dodecylamine (DODA-20). Estimated differences in NG hydrophobicities were calculated using http://www.molinspiration.com resulting in the theoretical logarithmic partition coefficients (logP).
FITC-labeled amphiphilic NGs
The synthesis of the fluorescence labeled NGs was carried out analog to the post-modification of the unlabeled NGs but in a two-step approach. Briefly, freeze-dried PPFPMA particles (2.00 g, 7.93 mmol w.r.t monomer units of PPFPMA particles, 1.0 eq) were dispersed in 400 mL DMF by short treatment in a sonication bath (Elmasonic P30 H, Elma). The particles were swollen overnight in DMF before amine functionalized fluorescein (FITCA) (71 mg, 0.16 mmol, 0.02 eq w.r.t monomer units) and TEA (3.3 mL, 23.8 mmol, 3 eq w.r.t. monomer units) were added and heated to 50°C. After 24 hrs, 30 mL of the dispersion (150 mg PPFPMA-FITC particles) were reacted for another 24 hrs with different ratios of amine functionalized moieties (3.0 eq w.r.t. monomer units), as summarized in Table S2.
Afterward, the NGs were purified by extensive dialysis against DMF and subsequently against DI water and MQ. The NGs were then freeze-dried and obtained as pale-yellow powder. They can be stored and redispersed in DI water or PBS by vortex and short sonication treatment. Characterization of the NGs via dynamic light scattering(DLS) measurement after redispersion in DI water is shown in Table 1.
Table 1.
Characterization of the physico-chemical properties of the NGs
| Nanogels | Particle diameter Dh [nm] (from dls) | Size difference Δdh [nm] | Zeta potential Ζ [mv] | |||
|---|---|---|---|---|---|---|
| Type | Sample | MQ | CCM | MQ | PBS | |
| Hydrophilic | PHPMA | 210±45 | 225±5 | 15 | −15.7 | −12.4 |
| Moderate hydrophobic | BENZA-20 | 185±35 | 235±25 | 50 | −9.4 | −5.8 |
| HEXA-20 | 170±65 | 255±10 | 85 | −8.8 | −5.1 | |
| Hydrophobic | CHOLA-20 | 170±40 | 220±5 | 50 | −13.4 | −7.0 |
| DODA-20 | 180±70 | 280±5 | 100 | −18.7 | −19.7 | |
Notes: NGs were dispersed in MQ, PBS or CCM by vortexing, followed by 30-min sonication at RT. DLS was performed after 2 hrs. Particle sizes are denoted as the hydrodynamic diameter and the respective standard deviation.
Abbreviations: NGs, nanogels; DLS, dynamic light scattering; MQ, ultrapure water; CCM, complete cell culture medium (RPMI 1640 containing 10% FBS); RT, room temperature.
NG dispersion
NGs were dispersed in MQ or PBS by vortexing, followed by 30 min sonication at room temperature (RT).
Characterization of the NGs
The success of the post-modification reaction was monitored by attenuated total reflection- Fourier-transform infrared spectroscopy (ATR-FTIR spectroscopy) on freeze-dried particles as described previously.12 The size of the NGs after post-modification was determined by DLS using a nicomp nano Z3000 (Particle Sizing Systems, USA) at a fixed scattering angle of 173°. The measurements were carried out at RT. In order to measure zeta potential, NGs were measured at 200 µg/mL in MQ or PBS at 25°C using Zetasizer Nano ZS (Malvern Instruments GmbH, Germany). A pre-equilibration time of 2 mins was used, attenuator and voltage were selected automatically and three runs of 30 measurements were used.
The size of the NGs in the dry state was investigated via transmission electron microscopy (TEM). Samples were prepared by applying a 10 μL droplet of the NG dispersion (1 mg/mL in MQ) on a carbon-coated copper grid (400 meshes, Quantifoil Micro Tools GmbH, Großlöbichau, Germany) for 45 s. The supernatant was removed with filter paper. This process was repeated 10 times and the grids were allowed to dry in air overnight. The TEM samples were measured afterward using the TEM mode of a Hitachi Scanning Electron Microscope (SU8030, Hitachi High-Technologies Corporation, Tokyo, Japan) with a working voltage of 30.0 kV at different magnifications. In order to evaluate the size and the size distribution of the NGs in the dried state, the diameter of 500 NGs each sample was determined with the software ImageJ (version 1.52e).
Measurement of the water contact angle
Polymer films were produced via spin-coating of polymer solutions on glass slides (Ø 25 mm, Neolab, Germany) with a spin processor WS-659MZ-23NPPB (Laurell Technologies Corporation, USA). Polymers solutions (1.5 wt-% in CHCl3 with a few drops of MeOH, if needed) were dropped on the objects and coated at 11,000 rounds per minute (rpm) for 40 s. Polymer films were dried under vacuum (RT, 90 mins). Static contact angle measurements were performed using sessile drop method on a contact angle system OCA 20 (DataPhysics Instruments, Germany) and processed with SCA20 (Version 3.12.11, DataPhysics Instruments). Two microliters MQ were dropped on the substrate and allowed to equilibrate (20 s, RT). The contact angles of 18 measurements from three coating samples were determined and averaged.
Cell culture
The human monocytic like cell line THP-1 was obtained from DSMZ (German Collection of Microorganisms and Cell Cultures, Germany) and cultivated in RPMI 1640 complete cell culture medium (CCM) containing 10% FBS, 2 mM L-Glutamine, 100 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), 100 units/mL penicillin, 0.1 mg/mL streptomycin in an incubator (37°C, 5% CO2). Cell culture medium, FBS and additives were obtained from PAN Biotech, Germany. Cells were counted using a Casy TTC (Roche Diagnostics GmbH, Germany).
Cell viability
Cells were seeded in 96-well plates (TPP, Germany) at 15,000 cells/100 μL CCM, incubated for 24 hrs (37°C, 5% CO2), treated with NGs (final concentrations 20, 75 and 100 µg/mL) and incubated for additional 24 cells were washed twice with PBS. Next, WST-1 solution (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate, Roche, Switzerland) was added according to manufacturer instruction and incubated for another 2 hrs (37°C, 5% CO2). Absorbance was measured using a GENios plate reader at 450 nm (TECAN, Switzerland).
Protein corona analysis
The NGs were dispersed at 300 µg/mL in 5 mL CCM (with 10% FBS gold) in sterile glass vials containing magnetic stir bar by stirring (700 rpm, 30 mins, RT). NGs were loaded on top of 4.5 mL 1.5 M sucrose (except PHPMA, which was loaded on 1.25 M sucrose) and pelleted via ultracentrifugation (17,900 g, 2.5 hrs at RT). CCM without NGs served as control and was treated similarly. Pellets were washed twice with PBS (17,900 g, 30 mins at RT). Samples were divided into two equal parts, one for two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) analysis and one for LC-MS/MS analysis.
2D-PAGE analysis of the protein corona
Unless otherwise stated, chemicals were purchased from Carl Roth GmbH (Germany). Proteins were eluted in 500 µL 2D lysis buffer (7 M urea, 2 M thiourea, 4% chaps, 2% pharmalyte pH 4–7, 1% dithiothreitol (DTT) and protease inhibitors). Total protein content was measured using the 2D Quant kit (GE Healthcare, Germany). For the isoelectric focusingsamples (500 µL) were loaded on nonlinear IPG strips (24 cm ImmobilineTM DrySrip pH 4–7 (NL), GE Healthcare) and equilibrated for 1 hr in equilibration buffer EB1 (360 mg/mL urea, 24 mg/mL SDS and 50.4 mM/mL Tris HCl, pH 8.6). Active rehydration and focusing were performed (15 hrs at 30 V, 1.5 hrs at 200 V, 1 hr at 500 V, 13.5 hrs gradient 500–1000 V, 3 hrs gradient 1000–8000 V and 6 hrs at 8000 V) with the GE Ettan IPGphor 3 (GE Healthcare). Afterward proteins were reduced (1% DTT in EB1, 15 mins) and then alkylated (4% iodoacetamide in EB1, 15 mins). Stripes were transferred onto 12.5% SDS-polyacrylamide gels. Electrophoresis was carried out with the GE Ettan DALTtwelve System Separation Unit (GE Healthcare). Gels were fixed (30% ethanol, 10% acetic acid in water) and stained with ruthenium II tris (bathophenanthroline disulfate) chelate (0.4 µM RU-II and 20% ET in MQ).43 Gels were scanned with a VersaDoc 4000MP imaging system (Bio-Rad, Germany) using excitation λ=473 nm/detection λ=610 nm and analyzed with Delta2D version 4.6 (Decodon, Germany). Each sample was analyzed in three independent replicates.
LC-MS/MS
In solution digestion
Unless otherwise stated, chemicals were purchased from Carl Roth GmbH. NG pellets were reconstituted in 50 µL of denaturation buffer (6 M urea/2 M thiourea in 10 mM HEPES, pH 8.0), reduced for 30 mins at RT with 1 µL of 0.5 M DTT in 50 mM of ammonium bicarbonate (ABC) and alkylated for 30 mins at RT in the dark by adding 1 µL of 0.5 M iodoacetamide in ABC. Subsequently, 2 µL of Trypsin/Lys-C Mix (Promega, USA) at 0.5 µg/µL in ABC was added and incubated for 4 hrs at RT. The urea concentration was decreased to <2 M by addition of 150 µL of ABC for trypsin digestion overnight at RT. The digestion was stopped by acidification with 200 µL of 5% acetonitrile (ACN), 3% trifluoroacetic acid (TFA). Tryptic peptides were desalted with stage tips.44
Liquid chromatography-electrospray ionization-tandem mass spectroscopy (LC-ESI-MS/MS)
Unless otherwise stated, all devices were purchased from Thermo Scientific (USA) and all chemicals were purchased from Carl Roth GmbH. Desalted peptides were reconstituted in 20 μL of 0.1% (v/v) TFA, 5% (v/v) ACN and 4 µL were analyzed by a reversed-phase capillary nano-liquid chromatography system (Ultimate 3000), connected to an Orbitrap Velos mass spectrometer via a nanospray. The LC system was flexion source equipped with a stainless-steel emitter. Samples were injected and concentrated on a trap column (PepMap100 C18, 3 μm, 100 Å, 75 μm i.d. ×2 cm), equilibrated with 0.05% TFA, 2% ACN in water. After switching the trap column inline, LC separations were performed on a capillary column (Acclaim PepMap100 C18, 2 μm, 100 Å, 75 μm i.d. ×25 cm) at an eluent flow rate of 300 nL/min using a linear gradient of 3–50% B in 50 mins. Mobile phase A contained 0.1% formic acid (FA) in water, and mobile phase B contained 0.1% FA in ACN. Mass spectra were acquired in a data-dependent mode utilizing a single MS survey scan with a resolution of 60,000 in the Orbitrap, and MS/MS scans of the 20 most intense precursor ions in the linear trap quadrupole. The MS survey range was m/z 350–1500. The dynamic exclusion time (for precursor ions) was set to 60 s and automatic gain control was set to 1×106 and 5.000 for Orbitrap-MS and LTQ-MS/MS scans, respectively.
Protein identification and label-free quantification of protein abundance
MS and MS/MS data from each LC/MS run were analyzed using the MaxQuant software (Version 1.6.0.16). Identification of proteins was performed using the MaxQuant-implemented Andromeda peptide search engine against a reference proteome database of Bos taurus (Bovine/Uniprot proteome ID: UP000009136, Version October 1, 2017). Initial maximum precursor and fragment mass deviations were set to 7 ppm and 0.5 Da, respectively. Variable modification (methionine oxidation and N-terminal acetylation) and fixed modification (cysteine carbamidomethylation) were set for the search and trypsin with a maximum of two missed cleavages were chosen for searching. The minimum peptide length was set to 7 amino acids and the false discovery rate for peptide and protein identification was set to 0.01. Relative protein abundance was determined using the Top 3 approach. Top 3 intensities of the identified proteins were calculated by summing up the intensities of the three most intense unique peptides for each protein.34,35 Table S3 summarizes the quantification of NG protein coronas.
Cellular uptake of FITC-labeled NGs in THP-1 cells
Data for physico-chemical characterization of the FITC-labeled NGs are included in Table S4. NGs were sonicated in a Sonorex RK510 water bath (30 mins, RT). Subsequently, 40 µg/mL of the pre-incubated NGs (2 hrs, RT, CCM containing 10% FBS) were incubated with THP-1 cells for 2 hrs (37°C, 5% CO2). After washing with PBS three times (500 g, 5 mins), uptake was analyzed using a FACS Aria III (BD Biosciences, Germany), equipped with a blue laser (488 nm), a fluorescence detector (582/42 nm) and a 70 µm nozzle. An example for the differentiation panels we used during the flow cytometry is given in Figure S1.
Results
Physico-chemical characterization of the NGs
In order to investigate the influence of NG hydrophobicity on protein interaction, biocompatibility and cellular uptake, a library of amphiphilic NGs (Figure 1) with varying hydrophobicities but similar colloidal properties was synthesized. For this, we used our recently developed synthetic platform approach which is based on the functionalization of reactive precursor particles with hydrophilic and hydrophobic moieties (Figure 1A).12 Well-defined reactive precursor particles with narrow size distribution were obtained from miniemulsion polymerization (respective DLS curve can be found in Figure S2). The colloidal features of these precursor particles, ie, crosslinking density, size and size distribution, are then translated into the different amphiphilic NGs equally. Table 1 summarizes particles sizes and the respective standard deviation, as a measure for particle size dispersity (DLS curves are included in Figure S2). To further demonstrate the similarity in colloidal features between precursor particles and the amphiphilic NGs, TEM images of PPFPMA and NGs were compared (TEM images and statistical evaluations of PPFPMA precursors and CHOLA-20 NGs as representative samples for the amphiphilic NGs are included in Figure S3). Following this approach (Figure 1A), we now have access to a library of comparable NGs with varying network composition, including a hydrophilic NG (PHPMA) and four different amphiphilic NGs (Figure 1B) with each containing 80 mol-% hydrophilic HPA in combination with 20 mol-% of a specific hydrophobic group, ie, benzylamine (BENZA-20), hexylamine (HEXA-20), an amine-functionalized cholesteryl (CHOLA-20) or dodecylamine (DODA-20). Having demonstrated equal molar incorporation of the specific hydrophobic groups (20 mol-% each), we assessed hydrophobicity of the different NGs. First, the logarithmic partition coefficients (logP) of the resulting hydrophilic/hydrophobic methacrylamide repeating units on the network copolymer were calculated allowing for a first classification of the five NGs into three groups: 1) hydrophilic, 2) moderate hydrophobic and 3) hydrophobic (Figure 1B). The hydrophilic group only contains PHPMA (logP =1.52) while BENZA-20 (logP =3.18) and HEXA-20 (logP =4.23) were assigned to the moderately hydrophobic NGs. DODA-20 (logP =7.26) and CHOLA-20 (logP =8.86) represent the hydrophobic NGs.
Figure 1.
A reactive precursor particle approach gives access to a NG library with varying hydrophobicity and similar colloidal features.
Notes: Schematic synthesis of the NGs is depicted (A) based on precursor particles being subsequently amine functionalized (B). We used HPA that translates to PHPMA and four modified variants that each contained 80 mol-% HPA and 20 mol-% of a different hydrophobic group, ie hexylamine (HEXA-20), benzylamine (BENZA-20), linear dodecylamine (DODA-20) as well as an amine functionalized cholesteryl group (CHOLA-20).
Abbreviations: NG, nanogel; NGs, nanogels; HPA, 2-hydroxypropylamine; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide); FITCA, amine functionalized fluorescein; EGDMA, ethylene glycol dimethacrylate.
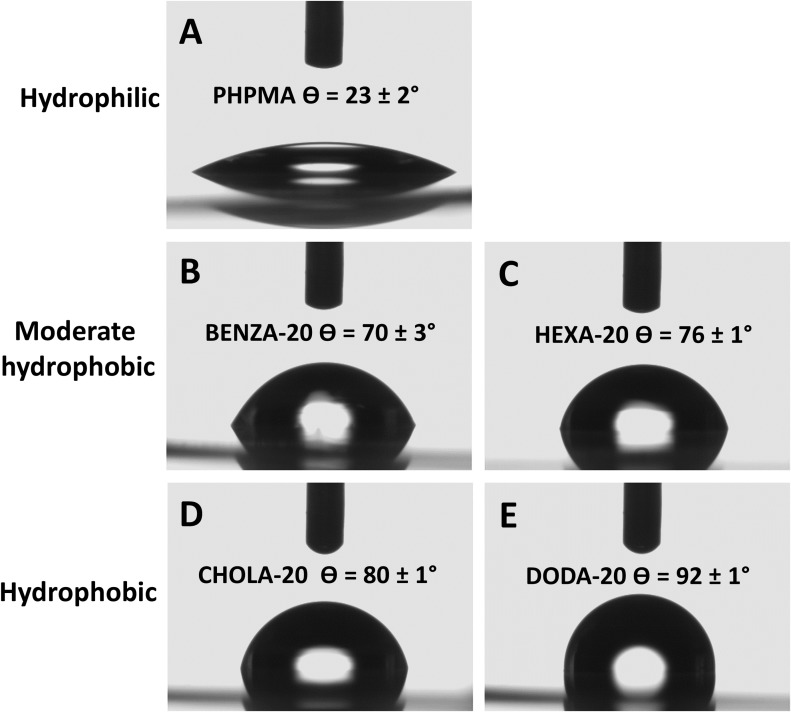
To reinforce these theoretical values, the hydrophobicity of each NG was examined by static contact angle measurements using the sessile drop method. However, as contact angle measurements on dried NP films could be misleading due to capillary forces at interparticulate cavities,45 the measurements were carried out on linear polymer analogs, confirming the above-described classification (Figure 2). The hydrophilic PHPMA (Figure 2A) has a contact angle of θ 23±2°. BENZA-20 (Figure 2B) and HEXA-20 (Figure 2C) of the moderate hydrophobic group showed θ 70±3° and θ 76±1°, respectively. The largest contact angles were found for the members of the hydrophobic group, ie, CHOLA-20 (Figure 2D) and DODA-20 (Figure 2E) with θ 80±1° and θ 92±1°.
Figure 2.
Contact angle measurement allows for the categorization of the NGs.
Notes: We could differentiate three groups: with PHPMA (A) as hydrophilic, BENZA-20 (B) and HEXA-20 (C) as moderate hydrophobic and CHOLA-20 (D) and DODA-20 (E) as hydrophobic.
Abbreviations: NGs, nanogels; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
In addition, loading and release experiments with Nile red as hydrophobic model cargo have been carried out previously to determine the influence of the polymer composition on the internal network hydrophobicity.12 The incorporation of 20 mol-% of hydrophobic groups increased the loading capacity in the order: BENZA-20<HEXA-20<DODA-20<CHOLA-20. The loading capacity of CHOLA-20 was increased threefold compared to PHPMA while the corresponding release rate was significantly reduced.12 These results not only fortify the classification based on hydrophobicity but also demonstrate the high potential of the amphiphilic NGs as nanocarriers for hydrophobic compounds. However, for evaluating the therapeutic potential the interactions of the NGs with biological systems should be determined next.
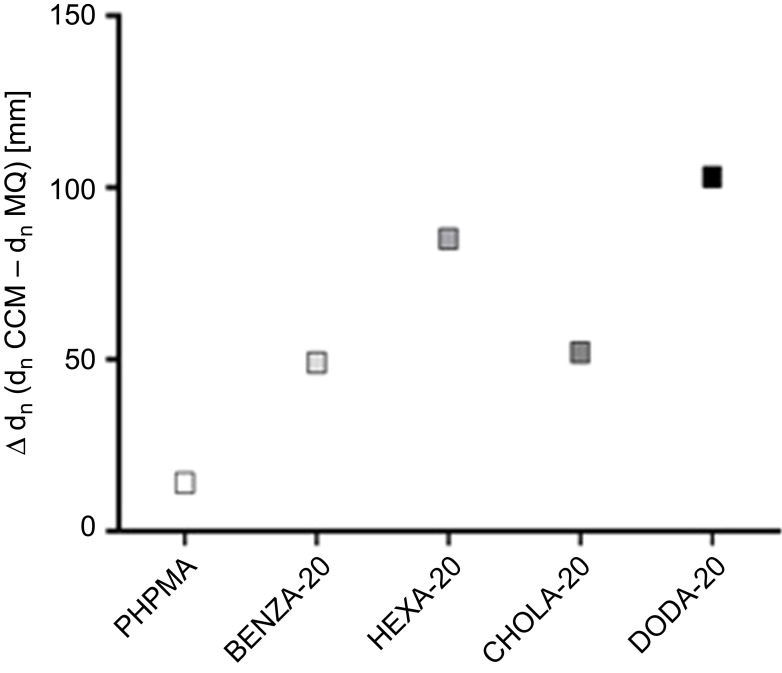
Increasing hydrodynamic diameters of the NGs upon incubation in physiological medium as assessed via comparative DLS measurements already indicate formation of protein coronas. Table 1 summarizes the size increases as differences of the hydrodynamic diameters in CCM and MQ (Δdh), which is also graphically depicted in Figure 3. Size increases are largely consistent with reported changes due to protein corona formation.46 In addition, the zeta potential in MQ and PBS was determined (Table 1). Neutral to slightly negative values in MQ and PBS suggest that colloidal stability of the NGs can mainly be attributed to neutral PHPMA segments on the particle surface.
Figure 3.
Illustration of size differences (Δdh) between incubation of NGs in MQ and CCM.
Notes: NGs were dispersed in MQ and CCM by vortexing, followed by 30 min sonication at RT. DLS was performed after 2 hrs.
Abbreviations: NGs, nanogels; MQ, ultrapure water; CCM, complete cell culture medium (RPMI 1640 containing 10% FBS); RT, room temperature; DLS, dynamic light scattering; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
Biocompatibility of NGs
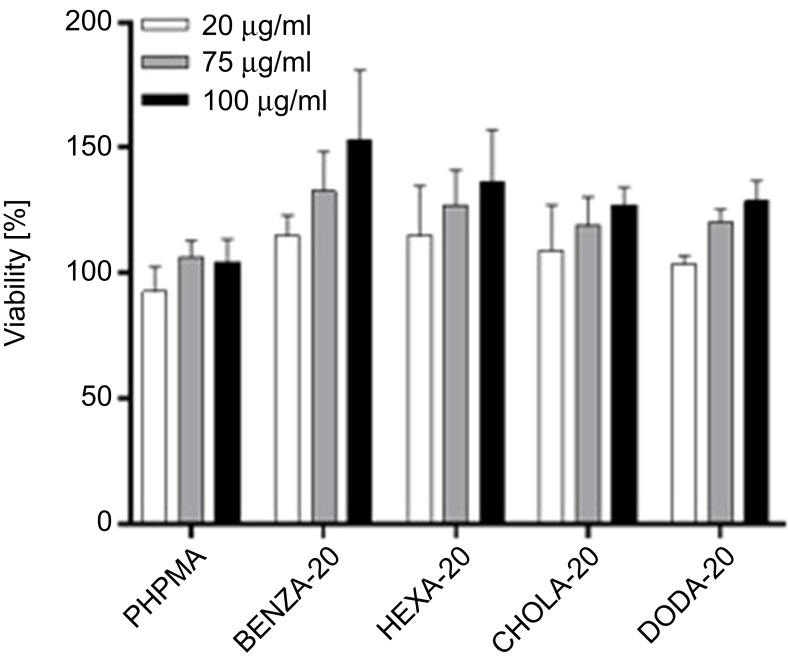
PHPMA, which is the main component (80 mol-%) of the amphiphilic NGs, is known to be biocompatible.42,47 However, the amphiphilic networks also contain additional 20 mol-% of hydrophobic groups, which may influence the interaction of the NGs with lipid membranes and cause some toxicity. Previously, we have already demonstrated that amphiphilic NGs with varying amphiphilicity showed no cytotoxic effect on normal human keratinocytes, suggesting good biocompatibility.12 Reasoned by the fact that monocytes are a main component of the RES, we assessed the biocompatibility in the monocytic like human cell line THP-1 (Figure 4). All NGs were found to be biocompatible, which is in line with our previous study.
Figure 4.
Viability of monocytic like THP-1 cells shows good biocompatibility of the NGs with different network hydrophobicities.
Notes: Depicted are mean values of three biological repeats and standard error of mean (SEM).
Abbreviation: NGs, nanogels.
Investigation of protein corona of NGs by 2DE
To further investigate the protein corona that is suggested by the observed size increase by DLS measurements in CCM, the protein binding of all NGs was examined using 2DE. The resulting 2D gel images obtained after 30 min incubation in CCM revealed significant qualitative and quantitative differences between the coronas of all NGs. Each NG displayed a unique and individual spot pattern (Figure 5, Figures S4A–E), where also the amount and intensity of spots varied significantly. While the hydrophilic NG PHPMA showed the lowest number of protein spots, considerable increased spot numbers were observed with increasing hydrophobicity. Hence, the protein binding was far less prominent for PHPMA (Figure 5A) compared to all other NGs. In particular, when compared to the moderate hydrophobic NG BENZA-20 (Figure 5B) and the hydrophobic DODA-20 (Figure 5C), the observed increase in spot number and intensity supports the assumption of an increased protein binding with increased hydrophobicity. The spot patterns of all NGs were analyzed and compared to PHPMA, serving as a benchmark. All protein spots with significantly increased intensity compared to PHPMA (ie, intensity increased >1.5, p<0.05, n=3) were taken into account. Largest differences were found for one of the most hydrophobic NGs, ie DODA-20, where in total 49 significantly increased protein spots compared to PHPMA were detected (Figure 5D). Concerning the number of spots being different from PHPMA the NGs can be ranked as such: DODA-20>CHOLA-20=BENZA-20>HEXA-20.
Figure 5.
Differences in the protein coronas are illustrated using the spot patterns of the obtained 2DE analysis from representative NGs.
Notes: NGs (300 µg/mL) were incubated in CCM for 30 mins. Bound proteins were eluted and analyzed using 2DE analysis. Depicted are the resulting representative 2D gels for (A) PHPMA, (B) BENZA-20 and (C) DODA-20. All other 2D images are depicted in Figure S4. In comparison to blank control (no NGs), labels indicate significantly modified proteins within the respective NGs. Similarities and differences regarding the number of significant spots (ie, intensity increased >1.5, p<0.05, n=3) in each corona relative to PHPMA as a benchmark are visualized in a Venn diagram (D).
Abbreviations: 2DE, two-dimensional gel electrophoresis; NGs, nanogels; CCM, complete cell culture medium (RPMI 1640 containing 10% FBS); PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
Protein binding patterns between the two hydrophobic NGs showed a high degree of similarity, ie, DODA-20 and CHOLA-20 had 16 protein spots in common. Whereas DODA-20 had only 11 and 6 spots in common with the moderate hydrophobic NGs BENZA-20 and HEXA-20, respectively. Similarities and differences in the protein coronas are visualized in a Venn diagram based on the spot numbers (Figure 5D). It becomes obvious that the number of significant spots follows roughly the same trend as the NG hydrophobicity. When comparing the two NGs within the respective group (ie, moderate hydrophobic and hydrophobic), it is remarkable that the slightly less hydrophobic NG in the respective cluster each has a higher number of pronounced spots. This suggests that the formation and composition of the protein corona is not only dependent on the overall hydrophobicity but may also depend on the molecular structure of the hydrophobic groups.
Identification of the corona proteins via LC-MS/MS
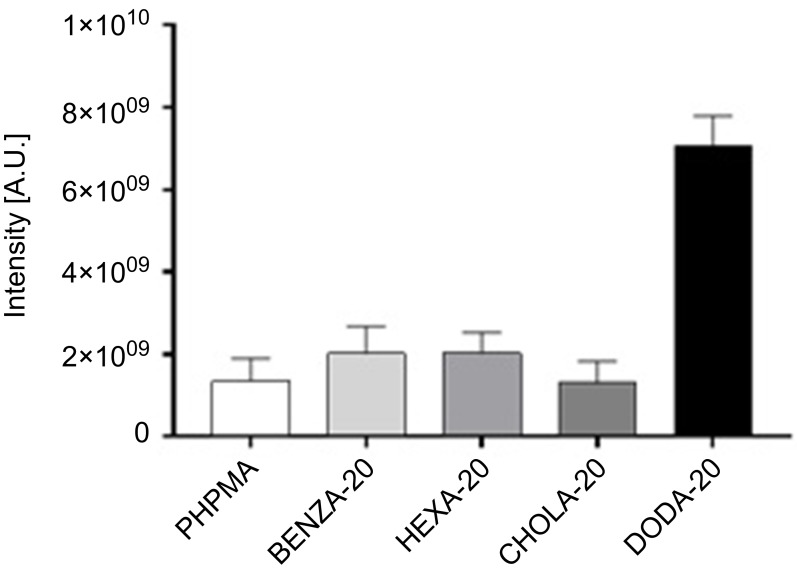
Having demonstrated qualitative differences in the protein corona, quantitative investigations of protein binding were conducted using LC-MS/MS using the Top 3 approach (Table S3). Again, DODA-20 showed the highest protein binding, while all the other NGs showed similar protein binding (Figure 6). Interestingly, total bound protein does not necessarily match with the total number of different proteins identified in each corona. This can occur when a single protein binds to a NG with an increased amount, while several proteins bind to another NG in smaller amounts. For example, the highest number of different individual proteins was found in HEXA-20 (Table 2) where 111 different proteins could be identified, while slightly fewer proteins (105) could be identified on DODA-20, despite the far higher amount of total bound proteins.
Figure 6.
Comparison of the amount of bound proteins in the corona of the respective NGs using the Top 3 approach.
Notes: Proteins in the NG coronas were identified using LC-MS/MS and abundancies were determined using the Top 3 approach. The intensity for each protein is obtained by summing up the intensities of the three most intense unique peptides for that protein. Based on that, total protein intensities for each NG were determined.
Abbreviations: NGs, nanogels; LC-MS/MS, liquid chromatography-tandem mass spectrometry; A.U., arbitrary unit; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
Table 2.
Overview on number of corona proteins identified via LC-MS/MS compared to the hydrophilic NG PHPMA
| Nanogels | Number of identified proteins in the corona | |||
|---|---|---|---|---|
| Type | Sample | Total (specific for this corona) |
Comparison to PHPMA | |
| Decreased (intensity factor <0.5) |
Increased (intensity factor >1.5) |
|||
| Moderate hydrophobic | BENZA-20 | 88 (1) | 18 | 32 |
| HEXA-20 | 111 (4) | 19 | 62 | |
| Hydrophobic | CHOLA-20 | 104 (4) | 9 | 62 |
| DODA-20 | 105 (4) | 11 | 58 | |
Notes: Given is the total number of all identified proteins for the respective NG (total). The number of proteins found only on this NG is given in brackets. The number of proteins with a lower (<0.5) or higher (>1.5) intensity compared to PHPMA are listed in the following columns.
Abbreviations: LC-MS/MS, liquid chromatography-tandem mass spectrometry; NG, nanogel.
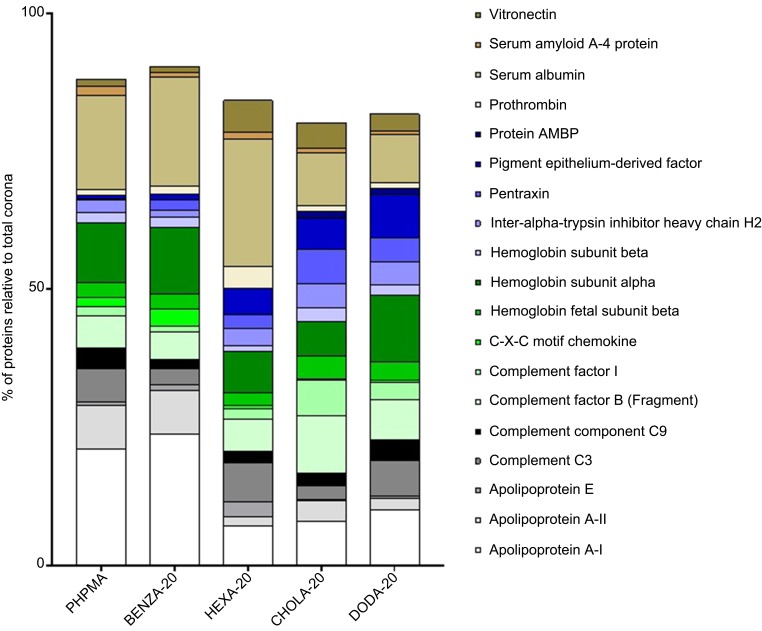
In order to address this context more precisely, the next step was to compare the specific composition of the protein coronas of the NGs with each other. Figure 7 shows the summary of the 15 most abundant proteins across all NGs, which make up at least 78% of the total coronas. As our NGs only differ in hydrophobicity while colloidal properties are largely similar, we can directly observe the influence of hydrophobicity on the protein corona composition. By applying a uniform color scheme, differences in the corona composition can be easily visualized. It is quite self-evident that the NGs have different influences on the corona compositions. We also wanted to investigate whether our groups (moderate hydrophobic and hydrophobic) are similar with regard to their protein corona. Therefore, the obtained protein intensities of the Top 3 approach were used. We compared the composition of the different protein coronas of each NG relative to PHPMA. As a result, a classification of the respective protein into the group, increased or decreased compared to PHPMA, could be made. The results of the NGs with BENZA-20 and HEXA-20 as representatives of the moderate hydrophobic and CHOLA-20 and DODA-20 as members of the hydrophobic group were then compared. The visualization is depicted as cluster analysis (Figure 8) and clearly shows the increased (red) and decreased (green) proteins of the respective corona compared to PHPMA. The protein coronas of DODA-20 and CHOLA-20 are very similar to each other and distinctly distinguishable to the coronas of BENZA-20 and HEXA-20. This can be seen by the very similar color pattern. The majority of proteins (one box each) display the same behavior with respect to increased or decreased intensity compared to PHPMA within the hydrophobic group. Thus, the hydrophobicity of the individual NGs seems to act as an important factor for the composition of the protein corona.
Figure 7.
Impact of the various NGs upon the relative distribution of the most abundant proteins among all protein coronas.
Notes: Depicted are the 15 most abundant proteins of each single protein corona (summarized overall), which were identified via LC-MS/MS and relative percentages were calculated using the Top 3 approach. Results for each protein are given in percentage of the total bound proteins in that corona. Remaining proteins (residual % up to 100) are not depicted.
Abbreviations: NGs, nanogels; LC-MS/MS, liquid chromatography-tandem mass spectrometry; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
Figure 8.
Cluster analysis for comparison of the NG corona proteins shows the grouping of the NGs regarding similarity within the protein corona.
Notes: Similarities and differences in the protein coronas of the NGs are visualized using a HCA. Each protein (every rectangle stands for a different identified protein) is compared to PHPMA, which serves as a benchmark particle (values are normalized) and values are transformed into a heat map. Red shows a higher intensity and green a lower intensity compared to PHPMA.
Abbreviations: NGs, nanogels; HCA, hierarchical cluster analysis; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
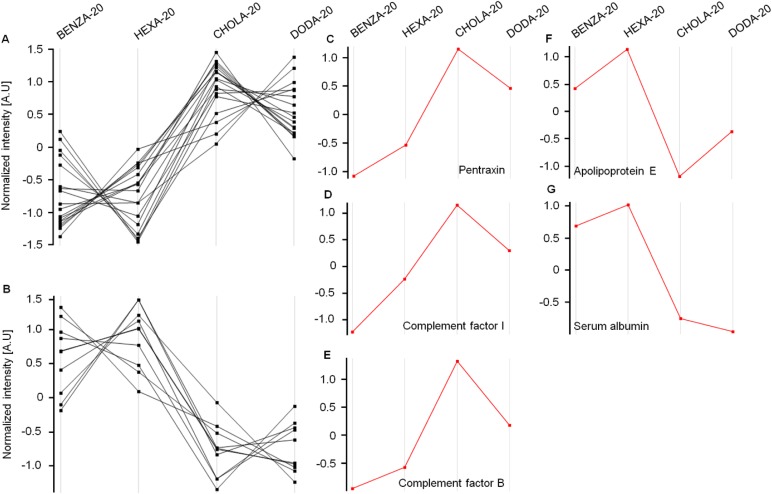
Subsequently, we were able to detect a number of different proteins that were significantly reduced (factor <0.5) or increased (factor >1.5) regarding their intensities compared to PHPMA (Table 2). Protein intensities were normalized using PHPMA as a reference, log(2) transformed and the Z-score was calculated. Profiles of altered proteins were then compared between the two groups of amphiphilic NGs (moderate hydrophobic group versus hydrophobic group). We found proteins that behaved alike, eg, being rather low in intensity on the moderate hydrophobic NGs (BENZA-20 and HEXA-20) and increased on the hydrophobic ones (CHOLA-20 and DODA-20) or vice versa (Figure 9A and B, Table 3). For example, proteins such as pentraxin, complement factor I and complement factor B (Figure 9C–E) bound stronger to the more hydrophobic NGs (DODA-20 and CHOLA-20). Apolipoprotein E and serum albumin, on the other hand, were found in much lower amounts (Figure 9F and G). Thus, we can conclude that the hydrophobicity of the NGs significantly influenced the amount and the identity of proteins in the corona.
Figure 9.
The comparison, with regard to their intensities, of normalized expression profiles of the identified corona proteins supports the grouping of the investigated NGs.
Notes: Protein intensities were normalized (PHPMA as reference), log(2) transformed and the Z-score was calculated using Perseus Version 1.6.1.3. The expression profiles were then examined for similarities. With increasing hydrophobicity, corona proteins of the two groups of NGs, namely moderate hydrophobic (BENZA-20+HEXA-20) and hydrophobic (DODA-20+CHOLA-20), display either an increase (A) or a decrease (B) in their expression profiles. Pentraxin (C), complement factor I (D) and complement factor B (E) are depicted as examples for proteins with an increased intensity among the hydrophobic NGs compared to the moderately hydrophobic NGs. Apolipoprotein E (F) and serum albumin (G) on the other hand show a decreased intensity among the hydrophobic NGs compared to the moderate NGs.
Abbreviations: NGs, nanogels; A.U., arbitrary unit; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
Table 3.
Comparative assessment of protein coronas
| Expression pattern | Protein | Accession number |
|---|---|---|
| A Increased protein adsorption with increasing hydrophobicity of the NGs |
Complement factor B (Fragment) | Q3KUS7 |
| Pentraxin | C4T8B4 | |
| Haemoglobin fetal subunit beta | P02081 | |
| Complement factor I | F1N4M7 | |
| Haemoglobin subunit beta | P02070 | |
| Plasma serine protease inhibitor | Q9N2I2 | |
| Haptoglobin (Zonulin) | Q2TBU0 | |
| Thrombospondin-1 | Q28178 | |
| Alpha-2-macroglobulin | Q7SIH1 | |
| Apolipoprotein B | E1BNR0 | |
| Histidine-rich glycoprotein | F1MKS5 | |
| Alpha-1-antiproteinase | P34955 | |
| Alpha-1B-glycoprotein | Q2KJF1 | |
| Actin, cytoplasmic 2 | P63258 | |
| Uncharacterized protein (F1MI18) | F1MI18 | |
| Hemopexin | Q3SZV7 | |
| Coagulation factor XII | P98140 | |
| Fetuin-B | Q58D62 | |
| B Decreased protein adsorption with increasing hydrophobicity of the NGs |
Serum albumin | A0A140T897 |
| Prothrombin | P00735 | |
| Serum amyloid A-4 protein | Q32L76 | |
| Apolipoprotein E | Q03247 | |
| Alpha-fetoprotein | Q3SZ57 | |
| ApoN protein | Q2KIH2 | |
| Carboxypeptidase B2 | Q2KIG3 | |
| Apolipoprotein C-IV | Q3SYR5 | |
| ECM1 protein | A5PJT7 | |
| Alpha-2-antiplasmin (Alpha-2-AP) | P28800 |
Notes: The intensities of the adsorbed corona proteins identified by LC-MS/MS were normalized and the Z-score was determined with Perseus (Version 1.6.1.3). Proteins with increasing (A) or decreasing (B) profiles depending on the hydrophobicity of NGs have been grouped and are shown here.
Abbreviations: LC-MS/MS, liquid chromatography-tandem mass spectrometry; NGs, nanogels.
Cellular uptake of NGs
Finally, we investigated how changes in protein binding are reflected by an altered cellular uptake in THP-1 cells. For this, we used analogous amphiphilic NGs but containing covalently attached FITC as fluorescent label, which facilitated easy quantification using flow cytometry based on a methodology recently established.48 The respective NGs are denoted as PHPMA-FITC, BENZA-20-FITC, HEXA-20-FITC, DODA-20-FITC and CHOLA-20-FITC. The physico-chemical characterization of the FITC-labeled NGs is summarized in Table 4. NGs were pre-incubated for 2 hrs in CCM such that a protein corona can form and were then incubated with THP-1 cells for 2 hrs. Figure 10 shows clear variations in the cellular uptake with PHPMA-FITC being taken up least, while the more hydrophobic NGs, ie, CHOLA-20-FITC, showed an enhanced uptake. All four NGs were significantly (p<0.0005) increased in uptake compared to the control NG PHPMA-FITC. Furthermore, it was observed that the uptake of the hydrophobic NG CHOLA-20-FITC in THP-1 cells compared to both moderate hydrophobic NGs (BENZA-20-FITC and HEXA-20-FITC) is also significantly (p<0.0005) increased. For the second representative of the hydrophobic group, DODA-20-FITC, this was only the case in comparison to BENZA-20-FITC (p<0.0005). These results correlate well with the increased amount of protein adsorption and demonstrate that increasing hydrophobicity led to an increase in cellular uptake in THP-1 cells, emphasizing the correlation between protein corona and cellular uptake.
Figure 10.
Flow cytometry data reveal a correlation between the uptake of NGs in human monocytic like THP-1 cells and the hydrophobicity of NGs.
Notes: NGs were pre-incubated for 2 hrs with CCM containing 10% FBS. After 2 hrs of incubation with THP-1 cells in an incubator (37°C, 5% CO2) uptake was measured using flow cytometry. Mean fluorescence values including SEM (n=3 biological repeats) as bars with significance (*P<0.0005) are depicted. All four NGs were significantly (p<0.0005) increased in uptake versus the control PHPMA (not depicted).
Abbreviations: NGs, nanogels; CCM, complete cell culture medium (RPMI 1640 with 10% FBS); SEM, standard error of mean; A.U., arbitrary unit; PHPMA, poly(N-(2-hydroxypropyl)methacrylamide).
Discussion
Here, we investigated protein binding, biocompatibility and cellular uptake of amphiphilic NGs. This new family of NGs is based on a cross-linked amphiphilic copolymer network allowing for solubilization of hydrophobic drugs in the interior. As previously shown, network hydrophobicity can be modified to accurately tailor loading and release profiles,12 demonstrating the high potential of these new carriers for medical applications. However, accurately determining their therapeutic potential requires a more detailed investigation of the interactions of the NGs with biological systems. In particular, the adsorbed protein corona is of importance as this links to rapid blood clearance and limits the therapeutic potential of the NGs.
We prepared a library of well-defined amphiphilic NGs with varying hydrophobicity (Figure 1) but similar colloidal features, ie, crosslinking density, size, size distribution, thus enabling true comparability between different NGs. Even though polydispersity can play a role in interaction of particles with biological systems, our concept allows the reduction of this effect to a systematic influence that is assumed to contribute to all samples equally. As can be seen in Table 1, only slight deviations in particle sizes and size distributions of NGs can be observed, which are within the margin of error for DLS. Only the hydrophilic PHPMA NG is larger than the hydrophobically modified ones. This is suggested to stem from a higher network hydration, resulting in a greater extent of swelling. The monocytic like human cell line THP-1 was chosen as a cell model, as monocytes are a main component of the RES being responsible for fast uptake and clearance. At first, biocompatibility was assessed as amphiphilicity may represent a risk factor due to unfavorably interactions with lipid membranes.49,50,51 All NGs were highly biocompatible (Figure 4), which is in line with our previous experiments on normal human keratinocytes.12 The increased viability of the cells to more than 100% can be attributed to several factors. It may also indicate a general possible influence of NPs on colorimetric assays52 or also on the mitochondrial reductase system. However, these experiments strengthen our assumption that the highly hydrophilic nature of PHPMA as the main network component translates to the amphiphilic NGs and ensures good biocompatibility.
Next, we focused on the determination of protein binding in physiological fluids as the resulting protein corona determines the biological identity of the NGs in vivo.53,54 Protein adsorption is largely determined by surface charge and hydrophobicity.17,55 Our study confirmed that similarities in protein binding profiles were well correlated with the estimated hydrophobicity of the NGs. The increase of hydrodynamic diameters (Table 1 and Figure 3) indicates differences in corona formation with varying NG hydrophobicity. While the truly hydrophilic PHPMA NG shows only a marginal size increase, the more hydrophobic NGs show significantly increased sizes, suggesting increased protein adsorption. These findings correlate very well with the results from 2DE (Figure 5, Figures S4A–E), confirming a significantly increased number and intensity of spots for the moderate hydrophobic NG BENZA-20 and the hydrophobic NG DODA-20 compared to PHPMA. Similar results were obtained by LC-MS/MS investigations (Figure 6), showing an increase of protein adsorption for the hydrophobic NG DODA-20. While similar trends have been observed for other NP systems,17,18 our study revealed that for NGs with comparable hydrophobicity but different functional hydrophobic moieties (ie, BENZA-20 vs HEXA-20 and DODA-20 vs CHOLA-20) the increase in protein adsorption varied significantly. This becomes especially obvious by comparing DODA-20 and CHOLA-20. Even though both hydrophobic repeating units in the network show similar logP-values (7.3 and 8.9), the size increase for DODA-20 was double (Δs=100 nm) compared to CHOLA-20 (Δs=50 nm) and intensity of total bound proteins was almost four times higher for DODA-20 compared to CHOLA-20 (Figure 6). Thus, protein binding does not appear to be governed by overall hydrophobicity alone, but is influenced by the molecular structure, as well.
We also examined the quantitative composition of the protein coronas, which typically is unique for different particles.56 However, it is assumed that specific protein binding profiles also share commonalities that may result in similar biological consequences. It is of high interest to identify specific particle characteristics that define the occurrence of particular protein corona commonalities, which in turn will help to determine a well-defined biological response. Here we focused on the influence of NG network composition (and corresponding surface hydrophobicity) on the process of opsonization, which is responsible for removal of the particles by uptake into cells of the RES. This is mediated by binding of specific proteins, called opsonins. While there are also opsonins triggering hemolysis and thrombogenicity, adsorption of opsonins belonging to the complement system leads to rapid clearance of NPs from the systemic blood cycle via complement receptor-mediated phagocytosis.57
Thus, we focused on correlating network hydrophobicity with adsorption of these types of opsonins. Up to now, this task was highly challenging since varying the network hydrophobicity also resulted in changes to the colloidal features which can also cause changes in protein adsorption profiles. Our synthetic platform approach, which generates well-defined NGs with similar colloidal features, represents a unique opportunity to systematically study the influence of network composition of protein binding.
Interestingly, we found a higher overlap among the hydrophobic NGs. Among the protein spots that were significantly increased in intensity (2DE, Figure 5D) compared to PHPMA (factor >1.5), the hydrophobic NGs CHOLA-20 and DODA-20 have a total of 16 spots in common. If we consider the group of the moderately hydrophobic NGs (BENZA-20 and HEXA-20), we only find six protein spots in common. Similarities between comparable structures (Figure 1B) are smaller. We find only six common protein spots in HEXA-20 and DODA-20 (linear moieties) and nine in BENZA-20 and CHOLA-20 (non-linear moieties). This suggests that the NG hydrophobicity has a greater influence on the corona composition than, for example, the structure. This relationship is also evident in the cluster analysis of the protein corona (Figure 8), which confirms an increased similarity between the hydrophobicities of the NGs than between similar structures, strengthening the impression from the 2D gels.
For a deeper understanding, we took a closer look at the 15 most abundant proteins for each corona (Figure 7). We identified proteins with different adsorption characteristics in the group of the moderate hydrophobic NGs (BENZA-20 and HEXA-20) compared to the hydrophobic group (CHOLA-20 and DODA-20). To improve visualization, we normalized the expression profiles of the intensities of the proteins to the reference particle PHPMA (Figure 9). The increase (Figure 9A) or decrease (Figure 9B) of protein adsorption with increasing hydrophobicity is now clearly visible. Interestingly, we found an enrichment of complement activation components such as pentraxin (Figure 9C), complement factor I (Figure 9D) and B (Figure 9E) for the most hydrophobic NGs, ie, CHOLA-20 and DODA-20. In comparison, the moderate hydrophobic NGs BENZA-20 and HEXA-20 show lower binding of these complement activation factors. Pentraxins are plasma proteins with a pentameric structure that serve as recognition molecules for foreign substances (eg, antigens) and label them for activation of the innate immune system. They also interact with the classical cascade of the complement system through binding of complement component 1q (C1q) as well as Fc receptors to activate immune responses.58 With complement factors I and B two other interesting candidates were found in the corona of the hydrophobic NGs. Complement factor I plays a role in the regulation of the complement cascade, while complement factor B is involved in the activation of the alternative pathway of complement activation.59,60 These opsonin corona components could drastically reduce blood retention time and thus the biological half-life, hampering medical application.
Since CHOLA-20 and DODA-20 NGs contain several components of this complement system in their protein coronas, we expect that these NGs will be cleared much faster compared to the moderate hydrophobic NGs BENZA-20, HEXA-20 and the hydrophilic PHPMA NG. Indeed, this expectation is supported by our cell uptake experiments. We could clearly show that increasing hydrophobicity and the resulting specific adsorption of opsonins leads to an increased cellular uptake in human monocytic like THP-1 cells (Figure 10). This is in line with other reports, showing that in general an increased hydrophobicity often leads to a much faster uptake of NPs.13,17,24 In our study, we were able to demonstrate that the protein adsorption also crucially depends on the molecular structure of the respective functional hydrophobic groups. As demonstrated for CHOLA-20 and DODA-20 NGs of comparable hydrophobicity (logP around 7–8), the amount of protein adsorption varied significantly. However, composition of the protein coronas showed commonalities, translating to a similar cellular uptake. Our platform approach resulting in NGs with varying hydrophilicity but comparable colloidal features allowed the accurate identification of this difference in protein binding patterns on the molecular composition of amphiphilic NGs for the first time. These results show the importance of not only tuning drug delivery vehicles with respect to one aspect (eg, high loading capacities) but rather investigating them in their entirety to account for opposing effects. For the amphiphilic NGs used here, this means the hydrophilic/hydrophobic balance needs to be accurately adjusted to enable high drug loading and desirable release kinetics while providing an optimum biological half-life.
Conclusion
In this study, amphiphilic NGs with tuneable hydrophobicity but similar colloidal features were used to systematically examine the influence of network composition and hydrophobicity on the interactions with biological systems. We investigated not only biocompatibility and protein binding but also cellular uptake of the NGs in human monocytic like THP-1 cells. It could be shown that increasing network hydrophobicity increases interactions with proteins. For example, DODA-20, as one of the most hydrophobic representative NGs used in this study, showed a much higher protein intensity in LC-MS/MS compared to PHPMA, being the most hydrophilic NGs. Strikingly, it was also observed that for DODA-20 and CHOLA-20 NGs of similar hydrophobicity, significant differences in the amount of adsorbed proteins could be detected. This suggests a strong additional influence of the molecular structure of the hydrophobic groups on protein binding. This was confirmed as well using 2DE gels.
In addition to these qualitative investigations, quantitative studies showed that the composition of the protein corona also varied in dependence on the hydrophobicity. Specifically, a higher amount of opsonins was determined for the protein coronas of the hydrophobic NGs CHOLA-20 and DODA-20 compared to the moderate NGs BENZA-20 and HEXA-20 as well as to the hydrophilic NG PHPMA. These findings correlated well with the increased uptake of these NGs into THP-1 cells. Thus, hydrophobicity and molecular structure of the respective hydrophobic groups not only influenced protein binding (both qualitative and quantitative), but also the interactions of the NGs with cells of the RES. Therefore, these results, together with our recent investigation on the loading and release of a hydrophobic model compound, allow the design of optimal drug carriers systems for hydrophobic drugs. For this, the nanocarrier’s network amphiphilicity needs to be balanced with respect to maximum loading capacity on one side and low protein binding on the other side, resulting in an increased biological half-life. As an example, the moderate hydrophobic BENZA-20 NGs represent a compromise between the enhanced loading capacity compared to PHPMA NGs and moderate cellular uptake in monocytic THP-1 cells. To further exploit the benefits of CHOLA-20 NGs, namely high loading capacity and slow release kinetics, the protein binding should be reduced to gain an increased biological half-life. In this regard, amphiphilic NGs with a covalent bound PEG-shell will be synthesized for further studies.
Acknowledgments
The authors thank their institutions for supporting this project. Furthermore, we would like to acknowledge the assistance of the Core Facility BioSupraMol supported by the Deutsche Forschungsgemeinschaft (DFG). This work was supported by Bundesinstitut für Risikobewertung (BfR; SFP 1322-502) and by the Collaborative Research Center (CRC) 1112 with funding for instrumentation. We acknowledge additional financial support for consumables from Verband der Chemischen Industrie eV (VCI) and Focus Area Nanoscale of the Freie Universität Berlin. A Gruber especially thanks the CRC 1112 and Focus area Nanoscale for scholarships to finance her PhD.
Disclosure
The authors have no affiliation or financial involvement with any organization or entity with a financial interest. Andrea Haase reports grants from German Federal Institute for Risk Assessment, during the conduct of the study. The authors report no other conflicts of interest in this work.
References
- 1.Klinger D, Landfester K. Stimuli-responsive microgels for the loading and release of functional compounds: fundamental concepts and applications. Polymer. 2012;53(23):5209–5231. doi: 10.1016/j.polymer.2012.08.053 [DOI] [Google Scholar]
- 2.Huh AJ, Kwon YJ. “Nanoantibiotics”: a new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J Control Release. 2011;156(2):128–145. doi: 10.1016/j.jconrel.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 3.Kar M, Fechner L, Nagel G, Glitscher E, Noe Rimondino G, Calderón M. Chapter 12 responsive nanogels for anti-cancer therapy In: Vashist A, Kaushik AK, Ahmad S, Nair M, editors. Nanogels for Biomedical Applications. London: The Royal Society of Chemistry; 2018:210–260. doi: 10.1039/9781788010481 [DOI] [Google Scholar]
- 4.Molina M, Asadian-Birjand M, Balach J, Bergueiro J, Miceli E, Calderón M. Stimuli-responsive nanogel composites and their application in nanomedicine. Chem Soc Rev. 2015;44(17):6161–6186. [DOI] [PubMed] [Google Scholar]
- 5.Molina M, Wedepohl S, Miceli E, Calderon M. Overcoming drug resistance with on-demand charged thermoresponsive dendritic nanogels. Nanomedicine (Lond). 2017;12(2):117–129. doi: 10.2217/nnm-2016-0308 [DOI] [PubMed] [Google Scholar]
- 6.Shatsberg Z, Zhang X, Ofek P, et al. Functionalized nanogels carrying an anticancer microRNA for glioblastoma therapy. J Control Release. 2016;239:159–168. doi: 10.1016/j.jconrel.2016.08.029 [DOI] [PubMed] [Google Scholar]
- 7.Guo B, Zhao J, Wu C, et al. One-pot synthesis of polypyrrole nanoparticles with tunable photothermal conversion and drug loading capacity. Colloids Surf B Biointerfaces. 2019;177:346–355. doi: 10.1016/j.colsurfb.2019.02.016 [DOI] [PubMed] [Google Scholar]
- 8.Wu C, Zhao J, Hu F, et al. Design of injectable agar-based composite hydrogel for multi-mode tumor therapy. Carbohydr Polym. 2018;180:112–121. doi: 10.1016/j.carbpol.2017.10.024 [DOI] [PubMed] [Google Scholar]
- 9.Schulte B, Rahimi K, Keul H, Demco DE, Walther A, Möller M. Blending of reactive prepolymers to control the morphology and polarity of polyglycidol based microgels. Soft Matter. 2015;11(5):943–953. doi: 10.1039/c4sm02116a [DOI] [PubMed] [Google Scholar]
- 10.Tiwari R, Heuser T, Weyandt E, Wang B, Walther A. Polyacid microgels with adaptive hydrophobic pockets and ampholytic character: synthesis, solution properties and insights into internal nanostructure by cryogenic-TEM. Soft Matter. 2015;11(42):8342–8353. doi: 10.1039/c5sm01327e [DOI] [PubMed] [Google Scholar]
- 11.Wischke C, Kruger A, Roch T, et al. Endothelial cell response to (co)polymer nanoparticles depending on the inflammatory environment and comonomer ratio. Eur J Pharm Biopharm. 2013;84(2):288–296. doi: 10.1016/j.ejpb.2013.01.025 [DOI] [PubMed] [Google Scholar]
- 12.Gruber A, Işık D, Fontanezi BB, Böttcher C, Schäfer-Korting M, Klinger D. A versatile synthetic platform for amphiphilic nanogels with tunable hydrophobicity. Polym Chem. 2018;9(47):5572–5584. doi: 10.1039/C8PY01123K [DOI] [Google Scholar]
- 13.Simon J, Wolf T, Klein K, Landfester K, Wurm FR, Mailander V. Hydrophilicity regulates the stealth properties of polyphosphoester-coated nanocarriers. Angew Chem Int Ed Engl. 2018;57(19):5548–5553. doi: 10.1002/anie.201800272 [DOI] [PubMed] [Google Scholar]
- 14.Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv Drug Deliv Rev. 2009;61(6):428–437. doi: 10.1016/j.addr.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cedervall T, Lynch I, Foy M, et al. Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew Chem Int Ed Engl. 2007;46(30):5754–5756. doi: 10.1002/anie.200700465 [DOI] [PubMed] [Google Scholar]
- 16.Cedervall T, Lynch I, Lindman S, et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci U S A. 2007;104(7):2050–2055. doi: 10.1073/pnas.0608582104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gessner A, Waicz R, Lieske A, Paulke BR, Mäder K, Müller RH. Nanoparticles with decreasing surface hydrophobicities: influence on plasma protein adsorption. Int J Pharm. 2000;196(2):245–249. [DOI] [PubMed] [Google Scholar]
- 18.Lindman S, Lynch I, Thulin E, Nilsson H, Dawson KA, Linse S. Systematic investigation of the thermodynamics of HSA adsorption to N-iso-propylacrylamide/N-tert-butylacrylamide copolymer nanoparticles. Effects of particle size and hydrophobicity. Nano Lett. 2007;7(4):914–920. doi: 10.1021/nl062743+ [DOI] [PubMed] [Google Scholar]
- 19.Luck M, Paulke BR, Schroder W, Blunk T, Muller RH. Analysis of plasma protein adsorption on polymeric nanoparticles with different surface characteristics. J Biomed Mater Res. 1998;39(3):478–485. [DOI] [PubMed] [Google Scholar]
- 20.Pearson RM, Juettner VV, Hong S. Biomolecular corona on nanoparticles: a survey of recent literature and its implications in targeted drug delivery. Front Chem. 2014;2:108. doi: 10.3389/fchem.2014.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strojan K, Leonardi A, Bregar VB, Krizaj I, Svete J, Pavlin M. Dispersion of nanoparticles in different media importantly determines the composition of their protein corona. PLoS One. 2017;12(1):e0169552. doi: 10.1371/journal.pone.0169552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones SW, Roberts RA, Robbins GR, et al. Nanoparticle clearance is governed by Th1/Th2 immunity and strain background. J Clin Invest. 2013;123(7):3061–3073. doi: 10.1172/JCI66895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh N, Park JH. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int J Nanomedicine. 2014;9(Suppl 1):51–63. doi: 10.2147/IJN.S26592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owens DE, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307(1):93–102. doi: 10.1016/j.ijpharm.2005.10.010 [DOI] [PubMed] [Google Scholar]
- 25.Schottler S, Landfester K, Mailander V. Controlling the stealth effect of nanocarriers through understanding the protein corona. Angew Chem Int Ed Engl. 2016;55(31):8806–8815. doi: 10.1002/anie.201602233 [DOI] [PubMed] [Google Scholar]
- 26.Allen TM, Hansen CB, de Menezes DEL. Pharmacokinetics of long-circulating liposomes. Adv Drug Deliv Rev. 1995;16(2):267–284. doi: 10.1016/0169-409X(95)00029-7 [DOI] [Google Scholar]
- 27.Amoozgar Z, Yeo Y. Recent advances in stealth coating of nanoparticle drug delivery systems. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4(2):219–233. doi: 10.1002/wnan.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Essa S, Rabanel JM, Hildgen P. Characterization of rhodamine loaded PEG-g-PLA nanoparticles (NPs): effect of poly(ethylene glycol) grafting density. Int J Pharm. 2011;411(1–2):178–187. doi: 10.1016/j.ijpharm.2011.02.039 [DOI] [PubMed] [Google Scholar]
- 29.Reddy KR, Wright TL, Pockros PJ, et al. Efficacy and safety of pegylated (40-kd) interferon alpha-2a compared with interferon alpha-2a in noncirrhotic patients with chronic hepatitis C. Hepatology (Baltimore, Md). 2001;33(2):433–438. doi: 10.1053/jhep.2001.21747 [DOI] [PubMed] [Google Scholar]
- 30.García-Álvarez R, Hadjidemetriou M, Sánchez-Iglesias A, Liz-Marzán LM, Kostarelos K. In vivo formation of protein corona on gold nanoparticles. The effect of their size and shape. Nanoscale. 2018;10(3):1256–1264. doi: 10.1039/c7nr08322j [DOI] [PubMed] [Google Scholar]
- 31.Lundqvist M, Augustsson C, Lilja M, et al. The nanoparticle protein corona formed in human blood or human blood fractions. PLoS One. 2017;12(4):e0175871. doi: 10.1371/journal.pone.0175871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman M, Laurent S, Tawil N, Yahia LH, Mahmoudi M. Nanoparticle and protein corona In: Protein-Nanoparticle Interactions: The Bio-Nano Interface. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013:21–44. [Google Scholar]
- 33.Fleischmann C, Gopez J, Lundberg P, et al. A robust platform for functional microgels via thiol-ene achemistry with reactive polyether-based nanoparticles. Polym Chem. 2015;6(11):2029–2037. doi: 10.1039/C4PY01766H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva JC, Denny R, Dorschel C, et al. Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol Cell Proteomics. 2006;5(4):589–607. doi: 10.1074/mcp.M500321-MCP200 [DOI] [PubMed] [Google Scholar]
- 35.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5(1):144–156. doi: 10.1074/mcp.M500230-MCP200 [DOI] [PubMed] [Google Scholar]
- 36.Blazejewski J-C, Hofstraat JW, Lequesne C, Wakselman C, Wiersum UE. Formation of monomeric halogenoaryl acrylates in the presence of hindered pyridine bases. J Fluor Chem. 1998;91(2):175–177. doi: 10.1016/S0022-1139(98)00222-X [DOI] [Google Scholar]
- 37.Eberhardt M, Mruk R, Zentel R, Theato P. Synthesis of Pentafluorophenyl(meth)acrylate Polymers: New Precursor Polymers for the Synthesis of Multifunctional Materials. European Polymer Journal 2005;41(7):1569–1575. doi: 10.1016/j.eurpolymj.2005.01.025 [DOI] [Google Scholar]
- 38.Eberhardt M, Théato P. RAFT polymerization of pentafluorophenyl methacrylate: preparation of reactive linear diblock copolymers. Macromol Rapid Commun. 2005;26(18):1488–1493. doi: 10.1002/(ISSN)1521-3927 [DOI] [Google Scholar]
- 39.Kearns MD, Donkor AM, Savva M. Structure-transfection activity studies of novel cationic cholesterol-based amphiphiles. Mol Pharm. 2008;5(1):128–139. doi: 10.1021/mp700131c [DOI] [PubMed] [Google Scholar]
- 40.Nie Y, Gunther M, Gu Z, Wagner E. Pyridylhydrazone-based PEGylation for pH-reversible lipopolyplex shielding. Biomaterials. 2011;32(3):858–869. doi: 10.1016/j.biomaterials.2010.09.032 [DOI] [PubMed] [Google Scholar]
- 41.Gentsch R, Pippig F, Nilles K, et al. Modular approach toward bioactive fiber meshes carrying oligosaccharides. Macromolecules. 2010;43(22):9239–9247. doi: 10.1021/ma101607a [DOI] [Google Scholar]
- 42.Gibson MI, Fröhlich E, Klok H-A. Postpolymerization modification of poly(pentafluorophenyl methacrylate): synthesis of a diverse water-soluble polymer library. J Polym Sci A Polym Chem. 2009;47(17):4332–4345. doi: 10.1002/pola.v47:17 [DOI] [Google Scholar]
- 43.Rabilloud T, Strub J-M, Luche S, Girardet JL, van Dorsselaer A, Lunardi J. Ruthenium II tris (bathophenanthroline disulfonate), a powerful fluorescent stain for detection of proteins in gel with minimal interference in subsequent mass spectrometry analysis. Proteome. 2000;1(1):1–14. doi: 10.1007/s102160000002 [DOI] [Google Scholar]
- 44.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- 45.Staufenbiel S, Merino M, Li W, et al. Surface characterization and protein interaction of a series of model poly[acrylonitrile-co-(N-vinyl pyrrolidone)] nanocarriers for drug targeting. Int J Pharm. 2015;485(1–2):87–96. doi: 10.1016/j.ijpharm.2015.02.072 [DOI] [PubMed] [Google Scholar]
- 46.Walkey CD, Chan WC. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem Soc Rev. 2012;41(7):2780–2799. doi: 10.1039/c1cs15233e [DOI] [PubMed] [Google Scholar]
- 47.Lammers T, Ulbrich K. HPMA copolymers: 30 years of advances. Adv Drug Deliv Rev. 2010;62(2):119–121. doi: 10.1016/j.addr.2009.12.004 [DOI] [PubMed] [Google Scholar]
- 48.Salvati A, Nelissen I, Haase A, et al. Quantitative measurement of nanoparticle uptake by flow cytometry illustrated by an interlaboratory comparison of the uptake of labelled polystyrene nanoparticles. NanoImpact. 2018;9:42–50. doi: 10.1016/j.impact.2017.10.004 [DOI] [Google Scholar]
- 49.Lee K, Yu Y. Lipid bilayer disruption induced by amphiphilic Janus nanoparticles: the non-monotonic effect of charged lipids. Soft Matter. 2019. doi: 10.1039/C8SM02525H [DOI] [PubMed] [Google Scholar]
- 50.Jones MC, Jones SA, Riffo-Vasquez Y, et al. Quantitative assessment of nanoparticle surface hydrophobicity and its influence on pulmonary biocompatibility. J Control Release. 2014;183:94–104. doi: 10.1016/j.jconrel.2014.03.022 [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Chen X, Gu N. Computational investigation of interaction between nanoparticles and membranes: hydrophobic/hydrophilic effect. J Phys Chem B. 2008;112(51):16647–16653. doi: 10.1021/jp8051906 [DOI] [PubMed] [Google Scholar]
- 52.Ong KJ, MacCormack TJ, Clark RJ, et al. Widespread nanoparticle-assay interference: implications for nanotoxicity testing. PLoS One. 2014;9(3):e90650. doi: 10.1371/journal.pone.0090650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. What the cell “sees” in bionanoscience. J Am Chem Soc. 2010;132(16):5761–5768. doi: 10.1021/ja910675v [DOI] [PubMed] [Google Scholar]
- 54.Gunawan C, Lim M, Marquis CP, Amal R. Nanoparticle–protein corona complexes govern the biological fates and functions of nanoparticles. J Mater Chem B. 2014;2(15):2060–2083. doi: 10.1039/c3tb21526a [DOI] [PubMed] [Google Scholar]
- 55.Jo DH, Kim JH, Lee TG, Kim JH. Size, surface charge, and shape determine therapeutic effects of nanoparticles on brain and retinal diseases. Nanomedicine. 2015;11(7):1603–1611. doi: 10.1016/j.nano.2015.04.015 [DOI] [PubMed] [Google Scholar]
- 56.Corbo C, Molinaro R, Parodi A, Toledano Furman NE, Salvatore F, Tasciotti E. The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery. Nanomedicine (Lond). 2016;11(1):81–100. doi: 10.2217/nnm.15.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol Pharm. 2008;5(4):487–495. doi: 10.1021/mp800032f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du Clos TW. Pentraxins: structure, function, and role in inflammation. ISRN Inflamm. 2013;2013:379040. doi: 10.1155/2013/379040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roversi P, Johnson S, Caesar JJ, et al. Structural basis for complement factor I control and its disease-associated sequence polymorphisms. Proc Natl Acad Sci U S A. 2011;108(31):12839–12844. doi: 10.1073/pnas.1102167108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwaeble WJ, Ali YM, Sim RB. Chapter 15 - the roles and contributions of the complement system in the pathophysiology of autoimmune diseases A2 - rose, noel R In: Mackay IR, editor. The Autoimmune Diseases. 5th ed. Boston: Academic Press; 2014:217–227. [Google Scholar]