Abstract
In the context of plant–pathogen and plant–mutualist interactions, the underlying molecular bases associated with host colonization have been extensively studied. However, it is not the case for non-mutualistic beneficial interactions or associative symbiosis with plants. Particularly, little is known about the transcriptional regulations associated with the immune tolerance of plants towards beneficial microbes. In this context, the study of the Burkholderia rice model is very promising to describe the molecular mechanisms involved in associative symbiosis. Indeed, several species of the Burkholderia sensu lato (s.l.) genus can colonize rice tissues and have beneficial effects; particularly, two species have been thoroughly studied: Burkholderia vietnamiensis and Paraburkholderia kururiensis. This study aims to compare the interaction of these species with rice and especially to identify common or specific plant responses. Therefore, we analyzed root colonization of the rice cultivar Nipponbare using DsRed-tagged bacterial strains and produced the transcriptomes of both roots and leaves 7 days after root inoculation. This led us to the identification of a co-expression jasmonic acid (JA)-related network exhibiting opposite regulation in response to the two strains in the leaves of inoculated plants. We then monitored by quantitative polymerase chain reaction (qPCR) the expression of JA-related genes during time course colonization by each strain. Our results reveal a temporal shift in this JA systemic response, which can be related to different colonization strategies of both strains.
Keywords: RNAseq, endophyte, symbiosis, Burkholderia, rice, jasmonic acid
Introduction
Plant microbiome is nowadays extensively studied, as it represents a huge potential for agriculture. Numerous studies describe the importance of microbes for plant’s nutrient supply and resistance to diseases and pests (Finkel et al., 2017). Therefore, microbes are potential solutions to do the transition to a sustainable agriculture while maintaining yield (Busby et al., 2017). Especially, some rhizobacteria have been shown to have tremendous beneficial effects on plant growth (Hayat et al., 2010) and resistance to pathogens (Beneduzi et al., 2012). These beneficial effects are induced through hormonal modulations following the colonization of plant roots (Vacheron et al., 2013) and inner tissues for endophytes (Hardoim et al., 2008) as well as systemic regulations of immunity (Pieterse et al., 2014). However, the perception of microbe-associated molecular patterns (MAMPs) generally leads to an immune response called MAMPs-triggered immunity (MTI) characterized by the synthesis of antimicrobial compounds (Jones and Dangl, 2006). In the same way pathogens suppress plant immunity, and beneficial microbes are able to escape (Trdá et al., 2015) or modulate (Zamioudis and Pieterse, 2012) the immune response of plants cells. Interestingly, the suppression of MTI by both pathogenic bacteria (Millet et al., 2010) and beneficial fungi (Jacobs et al., 2011) is commonly mediated via the jasmonic acid (JA) signaling pathway. However, the global physiological response and especially the transcriptional regulations induced by plants during the interaction with beneficial bacteria are not well described.
The interaction between plants and rhizospheric or endophytic bacteria-forming associative symbioses has been studied in several species belonging to the genera Azoarcus, Azospirillum, Herbaspirillum, Acetobacter, Gluconacetobacter, Bacillus, Phyllobacterium, Pseudomonas, and (Para-)Burkholderia (Ahemad and Kibret, 2014). Most studies on these models focused on the bacterial response to the interaction with its host plant (Shidore et al., 2012; Coutinho et al., 2015; Sheibani-Tezerji et al., 2015), while relatively few studies analyzed the transcriptional response of plants. Nonetheless, several studies described plants’ transcriptional regulations including lowering of defense (Bordiec et al., 2011; Rekha et al., 2018), hormonal signaling (Drogue et al., 2014; Rekha et al., 2018), developmental reprogramming (Paungfoo-Lonhienne et al., 2016), and iron homeostasis (Brusamarello-Santos et al., 2019; Stringlis et al., 2018).
Within the diversity of bacteria interacting with plants, the Burkholderia sensu lato (s.l.) genus of Betaproteobacteria stands out for several reasons. It contains plant pathogenic species (B glumae, Burkholderia gladioli, and Burkholderia plantarii) (Maeda et al., 2006), N2-fixing nodule-forming species in association with tropical legumes (Gyaneshwar et al., 2011) as well as species-forming associative symbiosis particularly with cereals such as rice (Coutinho et al., 2013; Govindarajan et al., 2008). Also, a phylogenetic separation discriminates the Burkholderia sensu stricto (s.s.) genus, which contains animal or plants pathogens as well as human opportunists, and the Paraburkholderia genus, which contains mainly plant-associated and environmental species (Sawana et al., 2014; Estrada-de los Santos et al., 2016). Also, most recent phylogenetic refinements of Burkholderia s.l. taxa defined other genera—Caballeronia, Robbsia, Trinickia, and Mycetohabitans—which are supported by both differences in genomic and ecological features (Estrada-de los Santos et al., 2018).
Interestingly, two strains of the Burkholderia s.l. genus able to fix N2 have been described as growth promoters in rice. Paraburkholderia kururiensis M130 (hereafter Pk) is a beneficial rice endophyte (Mattos et al., 2008) related to environmental and plant-beneficial strains (Kaur et al., 2017). Burkholderia vietnamiensis TVV75T (hereafter Bv) is a rice-associated species having positive effect on yield (Trân Van et al., 2000; Govindarajan et al., 2008), which belongs to the Burkholderia cepacia complex, a complex of species that can cause serious risks to cystic fibrosis patients (Vial et al., 2011). In order to decipher how rice perceives these two beneficial strains belonging to genera with contrasted ecologic backgrounds, we studied the transcriptional responses of rice during the establishment of the interaction. Our aim was to identify plant physiological processes and potentially key genes involved in beneficial rice-rhizobacteria interactions and also differentially regulated by each strain. We first analyzed the colonization patterns of Pk and Bv on the Oryza sativa Nipponbare genotype. We then analyzed the root and leaf transcriptional responses to the bacterial colonization by RNAseq. This led us to the identification of a co-expression JA-related network. Therefore, we monitored, throughout the establishment of the interaction, the expression of JA-related genes by reverse transcriptase–quantitative polymerase chain reaction (RT-qPCR).
Materials and Methods
Plants and Bacterial Cultivation
O. sativa L. ssp. japonica cv Nipponbare was used in this study. For all experiments, seeds were dehusked and sterilized as follows: 70% ethanol for 10 min and 9.6% NaClO supplemented with 1% Tween 20 for 30 min. Treated seeds were rinsed twice with sterile distilled water, twice with 2% thiosulfate solution, and finally four times with sterile distilled water. To confirm surface sterilization, 100 µl of the last rinsing solution was plated in tryptic soy agar (TSA) medium (Sigma-Aldrich). Seeds were then put in sterile distilled water at 28°C for 24 h and transferred on 8% H2O agar plate for 30 h. Homogeneously germinated seeds were transferred to sterile magenta boxes (SPL Lifesciences Co. Ltd) containing 150 ml of autoclaved perlite and 200 ml of sterile hydroponic medium (recipe in Supplementary Table 1). Plants were grown in a growth chamber (16 h light; 8 h dark; 28°C; 70% humidity).
All bacterial strains (listed in Supplementary Table 2) were cultured as follows: Glycerol stocks (20%) of bacterial cells conserved at −80°C were plated in low-salt lysogeny broth (LB) (Sigma-Aldrich) agar plates and incubated for 72 h at 28°C. Liquid low salt LB medium of 20 ml was then inoculated in 50-ml Falcon tubes and incubated for 16 h under agitation (180 rpm) at 28°C. Overnight culture of 500 µl was inoculated in fresh liquid medium for 2 h. Bacterial cells were then centrifuged for 5 min at 4,000 rpm and resuspended in sterile distilled water. Each plantlet was inoculated with 107 bacterial cells 4 days after sowing in hydroponic system.
Bacterial Transformation
P. kururiensis M130 and B. vietnamiensis TVV75 cells were transformed by electroporation with the pIN29 plasmid (Vergunst et al., 2010). The plasmid chosen to transform the strains, pIN29, comprises a chloramphenicol resistance gene as well as the DsRed gene under the control of a constitutive TAC promoter. After 24 h of incubation of selective medium low salt LB Cm (200 μg·ml−1) at 28°C, the most fluorescent colonies were selected.
Rice Root Colonization Assays
The roots of plants were harvested at 1, 7, and 14 days postinoculation (dpi), weighted, and grinded in sterile water with a sterile ceramic bead using a FastPrep-24™ 5G at 6 m·s−1 for 40 s. The solution was then diluted and inoculated in low-salt LB selective medium containing 200 μg·ml−1 of chloramphenicol and incubated at 28°C for 24 h. Colony-forming units were then enumerated. The size of the root-associated bacterial population was measured during two independent experiments, each comprising nine plants. In order to measure the size of the endophytic population, the inoculated rice roots were surface disinfected for 1 min using a solution of 1% chloramine T (Sigma-Aldrich) supplemented with 0.1% Tween 20. Roots were then rinsed six times with sterile water. Controls of disinfection were performed by plating rinsing water in TSA medium (Sigma-Aldrich) overnight. Surface-disinfected roots were then treated as previously described. The size of the endophytic population was measured on five plants.
Microscopy
All microscopic observations of the bacterial colonization were restricted to the primary root in order to compare the colonization patterns on roots that have been in contact with the bacterial population for the same amount of time. Primary roots were harvested at 7 and 14 dpi and mounted between slide and slips cover and directly examined with the microscopes. Epifluorescence observations were performed using a Nikon Eclipse Ni-E microscope. Confocal Laser Scanning observations were performed using a Zeiss LSM880 confocal microscope.
RNA Extraction
For the analysis of root and leaf transcriptional profiles, both plants’ tissues were harvested at 6 h postinoculation (hpi), 1 dpi, 7 dpi, or 14 dpi with live bacterial cells. Each biological replicate consisted of five pooled root system or five pooled last mature leaves harvested from a single hydroponic system. For each time point and each inoculated strain, three biological replicates were harvested. Roots and leaves of untreated plants were collected at the same time points. After harvest, samples were snap-frozen in liquid nitrogen and stored at −80°C.
Rice roots were homogenized in liquid nitrogen using cooled mortar and pestle. Rice leaves were grinded using a TissueLyser II (Retsch) set to 30 Hz for 30 s. Total RNA extraction using TRI-reagent (Sigma) was performed according to manufacturer’s instructions. All samples were treated with DNase I (Ambion) and purified using the RNA Clean & Concentrator kit (Zymo) according to manufacturer’s instructions. The integrity and quality of the total RNA were confirmed using a NanoDrop™ 1000 spectrophotometer (Thermo Fisher) and a 2100 BioAnalyzer (Agilent).
RNA Sequencing and Mapping of Reads
Quality of RNA was checked by determining the RNA Integrity Number (RIN) with a Fragment Analyzer (Agilent). For the library preparation, samples with a RIN value > 6 were used. Eighteen RNA libraries were prepared using an Illumina TruSeq stranded mRNA sample preparation kit by MGX-Montpellier GenomiX core facility (MGX) France (https://www.mgx.cnrs.fr/). Library construction and sequencing were performed as described in Karmakar et al. (2019) on an Illumina HiSeq 2500. The quantitative and qualitative validation of the library was performed by qPCR, Roche LightCycler 480, and a Fragment Analyzer (Agilent) using a Standard Sensitivity NGS kit. Quality control and assessment of raw Illumina reads in FASTQ format were done by FastQC software (version 0.11.5) to obtain per base quality, Guanine-Cytosine (GC) content, and sequence length distribution. Clean reads were obtained by removing the low-quality reads, adapters, and poly-N-containing reads by using Trimmomatic v0.36 software (Bolger et al., 2014). RNAseq reads were aligned to the IRGSP 1.0 version of the rice genome using HISAT2 v2.0.5.1 (Kim et al., 2015). The number of reads mapped to each gene locus was counted using HTSEq-count v0.6.0 (Anders et al., 2015).
Differential Gene Expression and Gene Ontology (GO) Term Enrichment Analysis
DESeq2 v3.7 (Love et al., 2014) was used to calculate differential gene expression between non-inoculated and inoculated conditions. All genes having an adjusted p-value inferior to 0.01 were considered as significantly differentially expressed. All functional enrichment analyses were performed using g:Profiler (version e95_eg42_p13_f6e58b9) with g:SCS multiple testing correction method applying significance threshold of 0.05 (Reimand et al., 2007; Raudvere et al., 2019).
Quantification of mRNA Levels Using RT-qCPR
cDNA was produced from 350 ng of DNase-treated total RNA using the SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). The cDNA reaction was diluted five times before qPCR using MESA BLUE qPCR Master Mix for SYBR® assay (Eurogentec) on an Mx3005P qPCR system (Agilent Technologies). The relative expression level was calculated according to Pfaffl (2001). Three independent samples were analyzed for each condition, and each sample was assayed in triplicate. Primers used are listed in Supplementary Table 3.
Results
Analysis of Root Colonization
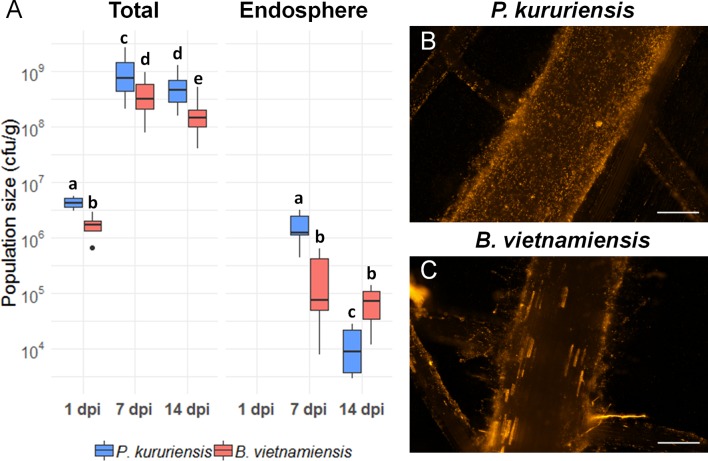
We used hydroponic culture of rice plants, grown in axenic condition and inoculated with DsRed-tagged strains, to monitor the bacterial colonization at 1, 7, and 14 dpi. First, the root’s colonization was measured by counting the bacterial populations on the rhizoplan and endosphere (see Materials and Methods). The roots of rice plants were rapidly colonized by both bacterial strains (Figure 1A). The populations of Pk and Bv reach a median value of 4.16 × 106 and 1.73 × 106 cfu·g−1, respectively, at 1 dpi. At 7 dpi, the maximum measured size of the bacterial population is reached for both strains: 7.49 × 108 and 3.18 × 108 cfu·g−1 for Pk and Bv, respectively. Then, between 7 and 14 dpi, the size of the total root-associated population decreases for each strain, reaching a median value of 4.6 × 108 and 1.46 × 108 cfu·g−1 for Pk and Bv, respectively. Between the same time points, the variation of the endophytic population size differs between the two strains. Indeed, the endophytic population size of Pk decreases from 1.25 × 106 to 1.17 × 104 cfu·g−1 between 7 and 14 dpi, while the median number of endophytic Bv cells remain stable with 7.34 × 104 and 7.5 × 104 cfu·g−1 at 7 and 14 dpi, respectively.
Figure 1.
Colonization of the roots of hydroponically grown rice plants by Bv and Pk. (A) Population dynamics of DsRed-tagged Bv and Pk associated with rice roots and inside the plant roots. The data reported are the median of bacterial population size from 18 plants and two independent experiments for rhizosphere compartment and five plants for the endophytic compartment. The letters indicate the significance groups in each compartment according to post-hoc tests on a generalized linear model. Epifluorescence microscopy pictures of the colonization of rice primary roots at 7 days postinoculation by Pk (B) and Bv (C) DsRed cells. White bars represent 200 µm. Bv, Burkholderia vietnamiensis TVV75T; Pk, Paraburkholderia kururiensis M130.
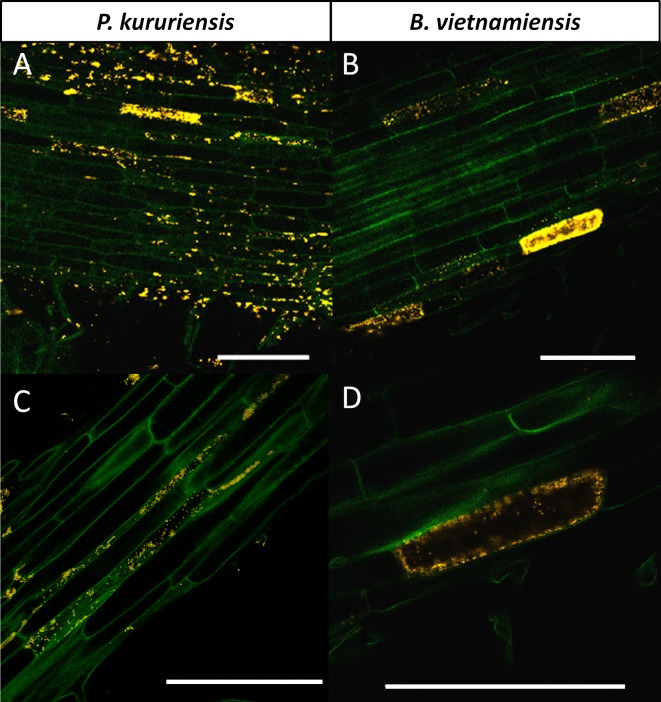
Microscopic observations of the primary roots of inoculated rice plants demonstrated that both bacterial strains tagged with the DsRed gene colonized the root surface after inoculation (Figures 1B, C). Moreover specific zones were more densely colonized by bacteria such as the surface of root hairs and the emergence of lateral roots (Supplementary Figure 1). By comparing the colonization of the two strains, differences can be observed in the way they colonized the surface of the primary root. Several epidermal plant cells seemed colonized intracellularly by the tagged Bv cells, while the phenomenon was observed less frequently in Pk-colonized roots (Figure 1C; Supplementary Figure 1). Observations by confocal microscopy revealed that for Pk, the highly colonized epidermal cells appear to be only colonized on their surface as the whole outline of epidermal cells as well as in intercellular spaces (Figures 2A, C), while for Bv, most of the observations show that bacterial cells were able to cross the cell wall and were observed in the cytoplasm of the cell (Figures 2B, D). Thus, both strains colonized the roots of the Nipponbare cultivar both externally and endophytically but through apparently different intensities and entry roads. We then wondered if the host plant induces contrasted transcriptional regulations associated with the differential colonization pattern of each strain.
Figure 2.
Endophytic colonization of the roots of hydroponically grown rice plants by Bv and Pk. Confocal microscopy observations of Pk and Bv DsRed-tagged cells colonizing the inner tissues of rice roots at 14 days postinoculation. (A) Pk cells colonizing the surface and the intercellular spaces of epidermic cells. (B) Multiple epidermic cells colonized by Bv cells. (C) Apoplastic colonization by Pk cells. (D) Rice root epidermic intracellular colonization by Bv cells. White bars represent 100 µm. Bv, Burkholderia vietnamiensis TVV75T; Pk, Paraburkholderia kururiensis M130.
Transcriptional Response of Rice to Bacterial Inoculation
In order to identify changes in O. sativa transcriptome in response to both strains, we performed RNAseq on leaves and roots of non-inoculated controls, Pk-inoculated and Bv-inoculated plants at 7 dpi. We chose this time point to avoid the initial plant defense burst due to a bacterial inoculation in hydroponic system (hours to 1 dpi) and allow an advanced colonization stage of the roots but without any visible developmental effect such as increased growth. To confirm that the inoculated and non-inoculated control plants were at the same developmental stage, we measured the dry weight of the plants and could not detect any significant impact of the inoculation on biomass production (Supplementary Figure 2) nor on plant height. Therefore, we assume that the differences between the transcriptomes of inoculated plants and non-inoculated controls should be related to bacterial colonization of the roots rather than a developmental impact of inoculation.
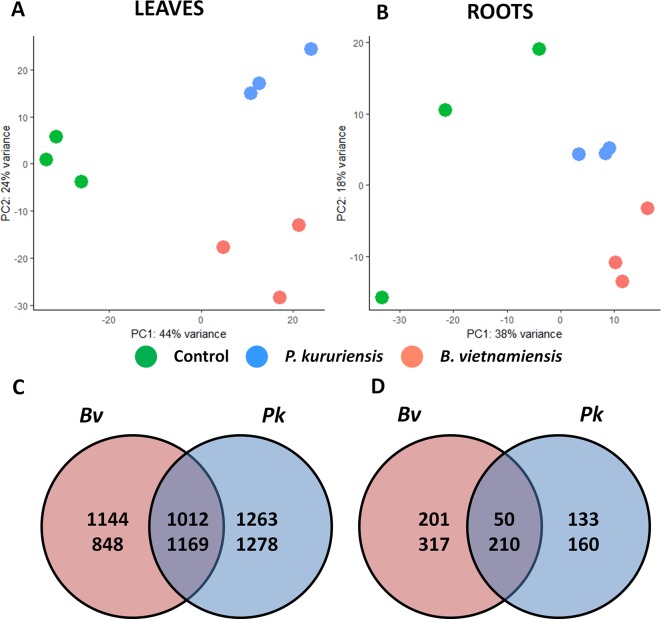
A total of 843 million reads were sequenced with an average of 47 million reads per sample (Table 1). An average of 68% of reads was uniquely mapped to the O. sativa genome per sample. A principal component analysis also discriminates the transcriptome of non-inoculated plants compared with the inoculated ones as well as the plant responses to each bacterial strain (Figures 3A, B). Differential expression analysis yields a total of 4,951 and 5,275 significantly (p < 0.01) differentially expressed genes (DEGs) in response to Bv and Pk, respectively. Comparing leaf and root transcriptomes reveals that there are five and eight times more DEGs detected in leaves in response to Bv and Pk, respectively, than in roots (Figures 3C, D). When comparing the response to Pk and Bv, a large proportion of DEGs are commonly regulated in leaves (around 50% in response to both strains) contrarily to roots in which the transcriptional regulation appears to be more specific to each inoculated strain. Indeed, only 33% of the DEGs in response to Bv are also differentially expressed in response to Pk. The proportions of commonly up-regulated DEGs in roots represent 20% and 27% of DEGs in response to Bv and Pk, respectively.
Table 1.
Summary of RNAseq data generated for rice transcriptomes.
| Sample | Organ | Condition | Total Number of Reads | Number of Uniquely Mapped Reads | Proportion of Mapped Reads (%) |
|---|---|---|---|---|---|
| LC1 | Leaves | Control | 38,533,867 | 26,161,511 | 68 |
| LC2 | Leaves | Control | 44,792,837 | 29,952,254 | 67 |
| LC3 | Leaves | Control | 33,331,123 | 21,989,917 | 66 |
| LK1 | Leaves | Paraburkholderia kururiensis | 46,895,520 | 32,677,603 | 70 |
| LK2 | Leaves | P. kururiensis | 65,261,206 | 42,077,779 | 64 |
| LK3 | Leaves | P. kururiensis | 46,707,369 | 33,553,685 | 72 |
| LV1 | Leaves | Burkholderia vietnamiensis | 43,862,789 | 31,855,527 | 73 |
| LV2 | Leaves | B. vietnamiensis | 41,034,781 | 27,022,594 | 66 |
| LV3 | Leaves | B. vietnamiensis | 49,608,083 | 35,455,520 | 71 |
| RC1 | Roots | Control | 38,522,573 | 27,000,000 | 70 |
| RC2 | Roots | Control | 50,820,144 | 35,400,000 | 70 |
| RC3 | Roots | Control | 44,458,436 | 30,800,000 | 69 |
| RK1 | Roots | P. kururiensis | 49,275,052 | 33,600,000 | 68 |
| RK2 | Roots | P. kururiensis | 52,634,407 | 34,700,000 | 66 |
| RK3 | Roots | P. kururiensis | 49,754,162 | 30,900,000 | 62 |
| RV1 | Roots | B. vietnamiensis | 51,314,965 | 34,700,000 | 68 |
| RV2 | Roots | B. vietnamiensis | 39,023,620 | 25,800,000 | 66 |
| RV3 | Roots | B. vietnamiensis | 57,406,096 | 39,300,000 | 68 |
Figure 3.
Comparative analysis of leaves and roots transcriptomes in response to Bv and Pk colonization. Principal component analysis of the normalized number of reads mapped per gene in leaves (A) and in roots (B). Number of genes regulated in leaves (C) and in roots (D) following bacterial inoculation, FDR < 0.01. The numbers on the upper part correspond to the number of genes up-regulated, inversely for the number of down-regulated genes. Bv, Burkholderia vietnamiensis TVV75T; Pk, Paraburkholderia kururiensis M130; FDR, false discovery rate.
In order to identify the biological processes transcriptionally regulated by the colonization of both bacterial strains, a GO enrichment analysis was carried out on the commonly regulated DEGs (intersections in Figures 3C, D). Supplementary Figure 3 provides a visualization of the enriched GO terms from the commonly up-regulated or down-regulated DEGs in leaves and roots (complete list of enriched GO terms available in Supplementary Table 4). Three main biological processes are commonly regulated during the interaction with both strains in leaves and roots: response to stimuli, and metabolic and also developmental processes. First, stress-related genes are enriched in the commonly DEGs. Indeed, in leaves, the “response to abiotic stimulus” as well as the “response to oxidative stress” terms are enriched in the up-regulated DEGs, whereas defense-related GO terms are enriched in the down-regulated DEGs in both leaves and roots. Also, several hormone-related GO terms are enriched in the DEGs in both leaves and roots. In leaves, GO terms related to auxins and abscisic acid (ABA) response are enriched in the up-regulated genes, while in roots, the up-regulated DEGs are enriched in cytokinines (CK), brassinosteroids (BR), and ethylene response terms. Finally, in roots, down-regulated DEGs are enriched in gibberellic acid (GA) and salicylic acid (SA) response-related terms. The analysis also revealed that metabolic processes are transcriptionally regulated in response to the interaction with both strains: In leaves, the up-regulated DEGs are enriched in the “photosynthesis” and “translation” terms, while the down-regulated DEGs are enriched in “starch metabolic process” and “membrane lipid catabolic process” terms. Furthermore, in leaves, the “iron ion homeostasis” term is enriched in up-regulated DEGs, while the “metal ion transport” term is enriched in the down-regulated DEGs of roots. Although the inoculation of both strains did not significantly impact the biomass of rice plants (Supplementary Figure 2), several development-related GO terms are enriched in the DEGs in both leaves and roots. First, in leaves, among others, the “anatomical structure development” term is enriched in the up-regulated DEGs, and the “glucan biosynthetic process” term is enriched in the down-regulated DEGs, which correspond to genes implicated in cellulose and callose synthesis. Finally, root transcriptional response is also enriched in development-related GO terms such as the “xylem development” term enriched in the up-regulated DEGs and the “lignin biosynthetic process” term enriched in the down-regulated DEGs.
Additionally, in order to identify the biological processes specifically induced during the interaction with each strain, we performed a GO term enrichment analysis on the DEGs specifically regulated by each strain (Supplementary Figures 4 and 5; Supplementary Tables 5 and 6). This analysis revealed that leaves of plants inoculated with Pk up-regulate genes related to biosynthetic process and translation, while the interaction with Bv induces the up-regulation of genes coding for components of the photosystem II and also the down-regulation of protein folding genes (Supplementary Figure 4, genes listed in Supplementary Table 7). Interestingly, the up-regulated DEGs in response to each strain are enriched in one hormonal signaling pathway. Indeed, the interaction with Pk induced the up-regulation of JA-related genes while cytokinin-related genes are enriched in the up-regulated DEGs in the leaves of Bv-inoculated plants. The analysis of the specific root transcriptome also revealed processes specifically induced by each strain. Particularly, the response to Pk in roots encompasses the down-regulation of oxidative stress response-related genes and also chitin catabolic process, while the interaction with Bv induced the down-regulation of genes involved in defense, JA signaling, and response to stimuli (Supplementary Figure 5, corresponding genes in Supplementary Table 8).
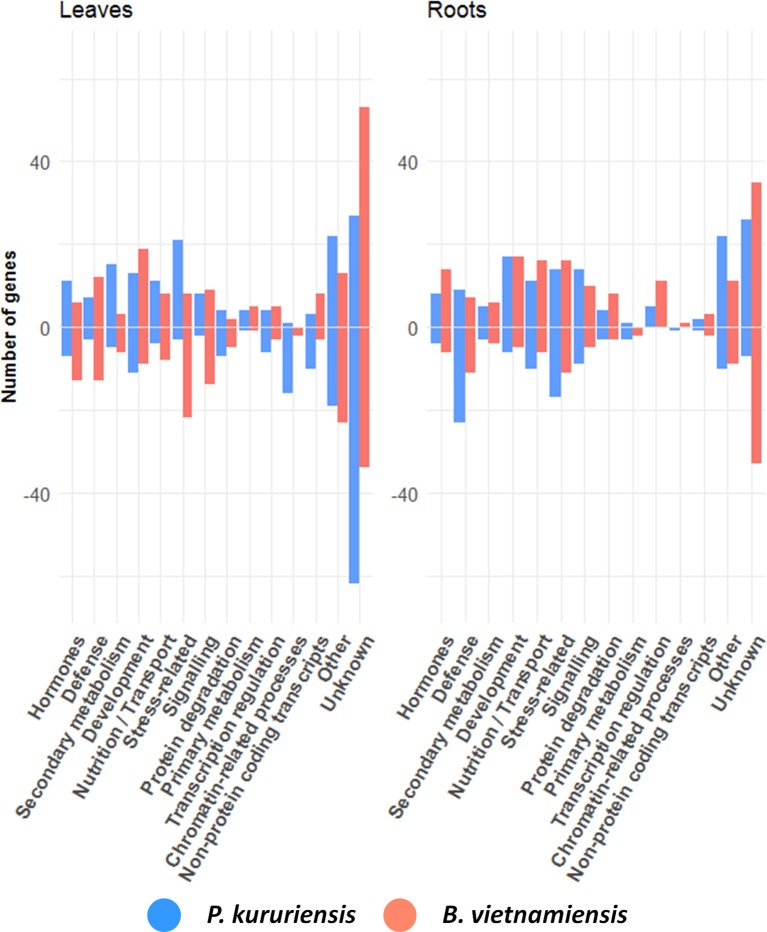
In order to deepen the transcriptome analysis, a functional categorization of the top 200 up-regulated and down-regulated DEGs identified in response to each strain was carried out using all available databases for rice annotation (UniProt, Kyoto Encyclopedia of Genes and Genomes (KEGG), RAP-DB, and Oryzabase). The proportion of genes related to each category is presented in Figure 4 and exemplifies rice-specific responses to the two bacterial strains. In leaves, more stress-related genes as well as genes involved in secondary metabolism are up-regulated in response to Pk compared with the response to Bv. Also, in response to Pk, 16 genes related to chromatin remodeling are down-regulated compared with two in response to Bv. Inversely, the inoculation of Bv induced the down-regulation in leaves of 22 stress-related genes, whereas only three are down-regulated in response to Pk. In roots, twice as much transcription regulation and protein degradation-related genes are up-regulated in response to Bv than in response to Pk. Conversely, approximately twice as much “nutrition/transport” and signaling-related genes are down-regulated in response to Pk than in response to Bv.
Figure 4.
Functional categories of rice DEGs upon Bv and Pk colonization. Number of DEGs related to functional categories among the top 200 up-regulated and down-regulated DEGs in response to each strain. For each graph, positive or negative numbers correspond to the number of up-regulated or down-regulated genes, respectively. DEGs, differentially expressed genes; Bv, Burkholderia vietnamiensis TVV75T; Pk, Paraburkholderia kururiensis M130.
In order to identify plant key genes that could be related to the observed patterns of root colonization, we focused our analysis on defense (leaves in Table 2, roots in Table 3) and hormone-related genes (leaves in Table 4, roots in Table 5). Indeed, these processes are enriched among the strain-specific DEGs (Supplementary Figures 4 and 5) and could pinpoint differences in the physiological response of rice following the perception of each strain.
Table 2.
Defense-related differentially expressed genes (DEGs) in leaves in response to Paraburkholderia kururiensis and Burkholderia vietnamiensis. Presented genes are part of the top 200 up-regulated and down-regulated significantly DEGs [false discovery rate (FDR) < 0.01]; all results for leaf transcriptome can be found in Supplementary Table 6.
| Gene ID | Log2 fold change | Gene Symbol/Trait Ontology | RAP Annotation | |
|---|---|---|---|---|
| P. kururiensis | B. vietnamiensis | |||
| Os05g0537100 | 1.70 | 2.44 | WRKY7 | WRKY7 |
| Os07g0539900 | −2.17 | −1.59 | PR2 | Similar to beta-1,3-glucanase-like protein |
| Os02g0118875 | −1.89 | −1.80 | NB-ARC | NB-ARC domain-containing protein |
| Os01g0940800 | −1.74 | −1.52 | Gns6 | Similar to beta-1,3-glucanase precursor |
| Os10g0124400 | −1.54 | −1.67 | NB-ARC | Hypothetical conserved gene |
| Os05g0492600 | −1.84 | −1.59 | YR48 | Similar to NBS-LRR type resistance protein |
| Os01g0731100 | 2.69 | PR | Similar to pathogen-related protein | |
| Os02g0759400 | 2.57 | RING1/Disease resistance | Zinc finger, RING/FYVE/PHD-type domain-containing protein | |
| Os02g0787300 | 1.88 | MAPKK4 | Mitogen-activated protein kinase kinase 4, defense response | |
| Os02g0626100 | 1.70 | PAL1 | Similar to phenylalanine ammonia-lyase | |
| Os02g0181300 | 1.66 | WRKY71 | WRKY transcription factor, defense response | |
| Os12g0520200 | 1.56 | PAL3 | Similar to phenylalanine ammonia-lyase | |
| Os01g0855600 | 1.55 | HS1 | Similar to Hs1pro-1 protein | |
| Os11g0227200 | −1.57 | LRR/Disease resistance | Similar to NBS-LRR disease resistance protein homologue | |
| Os02g0570400 | −1.61 | KSL7/Disease resistance | Similar to Ent-kaurene synthase 1A | |
| Os09g0474000 | −1.67 | TFX1/Disease resistance | bZIP transcription factor, bZIP-1 domain-containing protein | |
| Os01g0859500 | −1.73 | LG2 | Similar to basic leucine zipper protein (Ligueless2) | |
| Os05g0375400 | 2.21 | PR2, OsEGL1 | Beta-glucanase precursor | |
| Os03g0320600 | 1.98 | VQ11 | VQ domain-containing protein | |
| Os08g0170200 | 1.88 | Disease resistance protein domain-containing protein | ||
| Os04g0462500 | 1.75 | GF14b, GF14b, 14-3-3b | Similar to 14-3-3-like protein GF14-6 | |
| Os04g0680800 | 1.64 | MLO4, MLO4 | Mlo-related protein family protein | |
| Os11g0505300 | 1.63 | STV11, SOT1 | Sulfotransferase, resistance to rice stripe virus | |
| Os12g0468300 | 1.63 | NB-ARC | Similar to NB-ARC domain-containing protein, expressed | |
| Os02g0251900 | 1.61 | VQ7 | Similar to tobacco rattle virus-induced protein variant 2 | |
| Os04g0118800 | 1.54 | NB-ARC | NB-ARC domain-containing protein | |
| Os01g0837000 | 1.54 | NPR4, NPR4 | Ankyrin repeat containing protein | |
| Os11g0604900 | 1.51 | NB-ARC | Similar to NB-ARC domain-containing protein | |
| Os01g0289600 | −1.46 | WRKY9 | Similar to WRKY transcription factor 9 | |
| Os01g0382000 | −1.47 | PR1b | Similar to pathogenesis-related protein PRB1-2 precursor | |
| Os08g0235800 | −1.49 | WRKY25 | Similar to WRKY transcription factor 25 | |
| Os10g0569400 | −1.50 | Rir1a | RIR1a protein precursor | |
| Os08g0386200 | −1.55 | WRKY69 | WRKY transcription factor 69 | |
| Os05g0427400 | −1.64 | PAL4 | Similar to phenylalanine ammonia-lyase | |
| Os08g0173600 | −1.86 | PROPEP4/Anti herbivore response | Conserved hypothetical protein | |
| Os07g0117900 | −1.90 | NB-ARC | NB-ARC domain-containing protein | |
| Os06g0244000 | −2.09 | AAMT | Similar to anthranilic acid methyltransferase 3 | |
| Os04g0205200 | −2.09 | NB-ARC | NB-ARC domain-containing protein | |
| Os03g0195100 | −2.16 | ALD1 | Putative aminotransferase, response against blast fungus | |
| Os12g0154800 | −2.31 | GLP12-2 | Germin-like protein 12-2, disease resistance | |
| Os12g0154700 | −2.91 | GLP12-1 | Germin-like protein 12-1, disease resistance | |
Table 3.
Defense-related differentially expressed genes (DEGs) in roots in response to Paraburkholderia kururiensis and Burkholderia vietnamiensis. Presented genes are part of the top 200 up-regulated and down-regulated significantly DEGs [false discovery rate (FDR) < 0.01]; all results for root transcriptome can be found in Supplementary Table 7.
| Gene ID | Log2 fold change | Gene Symbol/Trait Ontology | RAP Annotation | |
|---|---|---|---|---|
| P. kururiensis | B. vietnamiensis | |||
| Roots | ||||
| Os08g0332600 | 1.07 | 0.74 | NB-ARC | Disease resistance protein domain-containing protein |
| Os08g0189500 | −1.15 | −1.57 | OsGLP8-6 | Germin-like protein 8-6, disease resistance |
| Os08g0231400 | −1.15 | −1.62 | GLP8-12 | Germin-like protein 8-12, Disease resistance |
| Os10g0537800 | −1.76 | −1.80 | AP77 | Peptidase aspartic, catalytic domain-containing protein |
| Os12g0500500 | −1.72 | −1.84 | NB-ARC | NB-ARC domain-containing protein |
| Os01g0508500 | −2.07 | −1.89 | RH2/NRR repressor homolog 2 | Conserved hypothetical protein |
| Os02g0807900 | −1.12 | −2.17 | WAK21 | Conserved hypothetical protein |
| Os09g0417600 | −1.92 | −2.64 | WRKY76 | WRKY transcription factor, transcriptional repressor |
| Os03g0195100 | −1.91 | −2.65 | ALD1 | Putative aminotransferase, response against blast fungus |
| Os09g0417800 | −2.49 | −2.66 | WRKY62 | WRKY transcription factor, transcriptional repressor |
| Os01g0508100 | −2.07 | −2.83 | RH3/NRR repressor homolog 3 | Ferritin/ribonucleotide reductase-like family protein |
| Os06g0105100 | −2.75 | −3.16 | WRKY86 | Similar to legumain |
| Os05g0247800 | −3.47 | −3.74 | XIP | Glycoside hydrolase, family 18 protein |
| Os08g0332600 | 1.07 | NB-ARC | Disease resistance protein domain-containing protein | |
| Os08g0446200 | 0.75 | OsPEPR1 | Similar to receptor-like protein kinase precursor | |
| Os12g0448900 | 0.74 | PIOX, RalphaO | Fatty acid alpha-dioxygenase family | |
| Os09g0127300 | 0.71 | CCR17 | NAD(P)-binding domain-containing protein | |
| Os12g0199100 | 0.70 | NB-ARC | NB-ARC domain-containing protein | |
| Os07g0273700 | 0.69 | WRKY123 | Disease resistance protein domain-containing protein | |
| Os01g0269800 | 0.68 | NB-ARC | NB-ARC domain-containing protein | |
| Os03g0411100 | 0.64 | OsHAP2E | Heme activator protein, biotic and abiotic resistances | |
| Os11g0152700 | 0.43 | OsbZIP79 | Similar to transcription factor HBP-1b(C38) (fragment) | |
| Os02g0626400 | 0.41 | PAL8 | Phenylalanine ammonia-lyase (EC 4.3.1.5) | |
| Os01g0713200 | −1.14 | Gns10 | Similar to beta-glucanase | |
| Os02g0629800 | −1.15 | OsPR12/DEFL7 | Similar to defensin precursor | |
| Os05g0554000 | −1.19 | MATE2 | Similar to cDNA clone:001-123-D07, full insert sequence | |
| Os12g0555200 | −1.20 | PBZ1 | Similar to Probenazole-inducible protein PBZ1 | |
| Os05g0247100 | −1.24 | HI-XIP/Insect resistance | Similar to glycosyl hydrolases family 18 | |
| Os07g0129200 | −1.26 | PR1a | Pathogenesis-related 1a protein | |
| Os07g0127500 | −1.31 | PR1-72 | Similar to PR-1a pathogenesis related protein (Hv-1a) precursor | |
| Os02g0570700 | −1.37 | Cyp71Z7 | Cytochrome P450 family protein | |
| Os01g0947000 | −1.42 | Disease resistance | Similar to beta-1,3-glucanase precursor | |
| Os02g0605900 | −1.45 | CHIT6 | Similar to chitinase (EC 3.2.1.14) A | |
| Os12g0437800 | −1.62 | Sci1, PR6 | Similar to MPI | |
| Os11g0700900 | −1.67 | C10923/Chitinase | Glycoside hydrolase | |
| Os01g0940700 | −1.85 | PR2, Glu1 | Similar to glucan endo-1,3-beta-glucosidase | |
| Os07g0127700 | −1.98 | Disease resistance | Similar to pathogenesis-related protein class 1 | |
| Os12g0628600 | −1.99 | PR5 | Similar to thaumatin-like pathogenesis-related protein 3 precursor | |
| Os07g0663700 | −2.19 | SDR110C-MI3/Disease resistance | Similar to oxidoreductase | |
| Os08g0518900 | −2.27 | C10122/Disease resistance | Chitinase (EC 3.2.1.14) | |
| Os03g0130300 | −2.54 | DEF8 | Similar to Cp-thionin | |
| Os11g0701600 | −2.84 | Chitinase | Glycoside hydrolase, catalytic domain-containing protein | |
| Os04g0316200 | −2.92 | Gnk2-domain protein | Protein of unknown function DUF26 domain-containing protein | |
| Os11g0701800 | −3.16 | OsRIXI/Disease resistance | Chitinase (EC 3.2.1.14) III | |
| Os07g0129300 | −3.67 | PR1a | Similar to PR1a protein | |
| Os11g0701000 | −4.67 | Chib3H-c | Class III chitinase homologue (OsChib3H-c) | |
| Os04g0289500 | 3.58 | PR1 | Allergen V5/Tpx-1-related domain-containing protein | |
| Os12g0491800 | 2.15 | KSL10 | Similar to Ent-kaurene synthase 1A | |
| Os08g0374000 | 0.99 | OsBetvI | Bet v I allergen family protein | |
| Os10g0163040 | 0.96 | NB-ARC | Similar to Blast resistance protein | |
| Os08g0261000 | 0.91 | NB-ARC | NB-ARC domain-containing protein | |
| Os09g0356000 | 0.91 | SIRK1 | Similar to OsD305 | |
| Os01g0859500 | 0.79 | LG2 | Similar to Basic leucine zipper protein (Liguleless2) | |
| Os09g0517200 | −1.65 | NB-ARC | Hypothetical conserved gene | |
| Os05g0478700 | −1.67 | WRKY84 | Hypothetical conserved gene | |
| Os03g0335200 | −1.68 | WRKY79 | Similar to WRKY1 (WRKY transcription factor 17) | |
| Os11g0117500 | −1.75 | WRKY40 | DNA-binding WRKY domain-containing protein | |
| Os06g0649000 | −1.81 | WRKY28 | PAMP-responsive transrepressor | |
| Os04g0635500 | −1.93 | Blast/SA induced | Similar to Wound induced protein (fragment) | |
| Os11g0154500 | −2.09 | Nac17/Disease Resistance | Blast disease-responsive transcription factor, disease resistance | |
| Os11g0462500 | −2.66 | NB-ARC | Similar to NB-ARC domain-containing protein | |
| Os05g0248200 | −4.65 | Chitinase | Glycoside hydrolase, family 18 protein | |
| Os04g0375300 | −6.34 | rNBS56 | Similar to NBS-LRR protein (fragment) | |
| Os06g0279900 | −7.11 | NB-ARC | Similar to NB-ARC domain-containing protein, expressed | |
Table 4.
Hormone-related differentially expressed genes (DEGs) in leaves in response to Paraburkholderia kururiensis and Burkholderia vietnamiensis. Presented genes are part of the top 200 up-regulated and down-regulated significantly DEGs (false discovery rate (FDR) < 0.01); all results for leaf transcriptome can be found in Supplementary Table 6.
| Gene ID | Log2 fold change | Gene Symbol/Function | RAP Annotation | |
|---|---|---|---|---|
| P. kururiensis | B. vietnamiensis | |||
| Gibberellic acid | ||||
| Jasmonate | ||||
| Auxin | ||||
| Ethylene | ||||
| Abscisic acid | ||||
| Cytokinine | ||||
| Brassinosteroid | ||||
| Os06g0729400 | 1.85 | 2.74 | GASR8 | Similar to gibberellin-regulated protein 2 precursor |
| Os09g0470500 | −1.49 | Hox4 | Homeodomain leucine zipper protein | |
| Os03g0352450 | −1.60 | INO80, CHR732/GA signaling | Hypothetical conserved gene | |
| Os08g0560000 | −1.66 | GA20ox7 | Similar to gibberellin 20 oxidase 2 | |
| Os01g0757200 | 2.11 | GA2ox3 | GA 2-oxidase3, GA metabolism | |
| Os06g0110000 | 1.59 | KAO | Similar to DWARF3 (fragment) | |
| Os02g0570400 | −1.61 | KS7 | Similar to Ent-kaurene synthase 1A | |
| Os04g0301500 | 1.88 | −1.65 | RERJ1 | Helix-loop-helix DNA-binding domain-containing protein |
| Os03g0767000 | 2.11 | AOS1 | Allene oxide synthase (CYP74A1), Biosynthesis of jasmonic acid (JA) | |
| Os01g0705700 | 1.84 | MYL1 | Similar to transcription factor ICE1 (Inducer of CBF expression 1) | |
| Os10g0392400 | 1.49 | JAZ12 | Tify domain-containing protein | |
| Os05g0439100 | 1.46 | MYC7E | Similar to transcription factor MYC7E (fragment) | |
| Os03g0181100 | −1.97 | JAZ10 | Tify domain-containing protein | |
| Os01g0643300 | 2.44 | 2.29 | PIN3A | Auxin efflux carrier protein, auxin transport |
| Os12g0601300 | 2.81 | IAA30 | Similar to auxin-responsive protein (Aux/IAA) (fragment) | |
| Os07g0182400 | 1.93 | IAA24 | AUX/IAA protein family protein | |
| Os02g0523800 | 1.72 | IPK2 | Inositol polyphosphate kinase, auxin signaling | |
| Os06g0323100 | −1.53 | IAA Methylase | Similar to H1005F08.18 protein | |
| Os06g0671600 | −1.69 | Small auxin-up RNA 26 | Beta tubulin, autoregulation binding site domain-containing protein | |
| Os05g0528600 | −2.02 | YUCCA2 | Flavin monooxygenase-like enzyme, auxin biosynthesis | |
| Os09g0522000 | 2.94 | −1.91 | DREB1B | Similar to dehydration-responsive element-binding protein 1B |
| Os09g0451000 | 2.19 | 1.64 | ACO2 | ACC oxidase, ethylene biosynthesis |
| Os04g0572400 | 4.19 | DREB1E | Similar to CRT/DRE binding factor 1 | |
| Os02g0677300 | 3.41 | DREB1G | Similar to CRT/DRE binding factor 1 | |
| Os10g0562900 | 2.87 | DERF12 | Non-protein coding transcript | |
| Os08g0474000 | 2.64 | DERF3 | Similar to AP2 domain-containing protein RAP2.6 (fragment) | |
| Os02g0654700 | 2.27 | BIERF3 | AP2/ERF family protein, abiotic stress response | |
| Os09g0522200 | 1.83 | DREB1A | DRE-binding protein 1A | |
| Os07g0155600 | −1.53 | EIN2 | Nramp ion-transporter family protein, ethylene signaling pathway | |
| Os07g0410300 | 4.78 | ERF136 | Conserved hypothetical protein | |
| Os07g0410700 | 1.93 | DERF2 | Similar to Ethylene-responsive element binding protein 1 | |
| Os01g0797600 | 1.66 | BIERF2 | AP2/ERF family protein, stress signaling | |
| Os02g0676800 | −1.94 | ERF20 | Similar to dehydration responsive element binding protein 1E (DREB1E protein) | |
| Os06g0493100 | −2.31 | bphi008a | Conserved hypothetical protein | |
| Os02g0636600 | −2.27 | −1.81 | GEM/ABA-related | GRAM domain-containing protein |
| Os02g0703600 | 3.07 | ABA8OX1 | Similar to abscisic acid 8′-hydroxylase 1 | |
| Os07g0569100 | 1.86 | REM4.1 | Remorin protein, coordination of interlink between ABA and BR signaling | |
| Os07g0281800 | −1.57 | ABA synthesis | Similar to aldehyde oxidase-2 | |
| Os02g0182100 | −1.80 | RR24 | B-type response regulator, cytokinin signaling | |
| Os11g0143300 | −1.71 | RR9 | A-type response regulator, cytokinin signaling | |
| Os01g0952500 | −1.77 | RR4 | A-type response regulator, cytokinin signaling | |
| Os08g0460600 | −1.89 | CKX11 | Similar to cytokinin dehydrogenase 11 | |
| Os01g0197100 | 1.72 | dwf2, SMG11 | Cytochrome P450, brassinosteroids biosynthesis, Regulation of plant architecture | |
| Os09g0409950 | −1.48 | BRI1-interacting protein 120 | Hypothetical conserved gene | |
| Strigolactone | ||||
| Os01g0935400 | 1.62 | AtLBO ortholog | 2OG-Fe(II) oxygenase domain-containing protein | |
| Salicylic acid | ||||
| Os04g0581100 | −2.01 | S3H/SA conjugation | 2OG-Fe(II) oxygenase domain-containing protein | |
Table 5.
Hormone-related differentially expressed genes (DEGs) in roots in response to Paraburkholderia kururiensis and Burkholderia vietnamiensis. Presented genes are part of the top 200 up-regulated and down-regulated significantly DEGs [false discovery rate (FDR) < 0.01]; all results for root transcriptome can be found in Supplementary Table 7.
| Gene ID | Log2 fold change | Gene Symbol/Function | RAP Annotation | |
|---|---|---|---|---|
| P. kururiensis | B. vietnamiensis | |||
| Jasmonate | ||||
| Auxin | ||||
| Cytokinine | ||||
| Ethylene | ||||
| Brassinosteroids | ||||
| Gibberellic acid | ||||
| Abscisic acid | ||||
| Salicylic acid | ||||
| Os01g0370000 | 0.87 | 0.96 | OPR9 | NADH:flavin oxidoreductase/NADH oxidase |
| Os05g0362100 | 0.57 | JA synthesis | Similar to protein MFP-b | |
| Os08g0509100 | −2.07 | LOX8 | Similar to lipoxygenase, chloroplast precursor (EC 1.13.11.12) | |
| Os03g0181100 | −1.62 | JAZ10 | Tify domain-containing protein | |
| Os03g0741100 | −1.93 | bHLH148/JA responsive | Basic helix-loop-helix transcription factor | |
| Os03g0180800 | −2.01 | JAZ9 | TIFY domain-containing transcriptional regulator | |
| Os02g0228900 | 0.58 | IAA7 | Similar to auxin-responsive protein IAA18 | |
| Os11g0523800 | 0.49 | ARF1 | Similar to isoform 3 of auxin response factor 23 | |
| Os02g0305950 | −2.92 | SAUR PROTEIN 7 | Similar to calmodulin binding protein | |
| Os08g0529000 | −7.15 | PIN5b | Auxin efflux carrier, auxin homeostasis | |
| Os04g0673300 | 0.66 | 0.94 | RR6 | A-type response regulator, cytokinin signaling |
| Os04g0556500 | −1.35 | −1.64 | cZOGT1 | cis-zeatin-O-glucosyltransferase |
| Os07g0449700 | 1.55 | RR7 | A-type response regulator, cytokinin signaling | |
| Os10g0483500 | 0.66 | CKX3 | FAD linked oxidase, N-terminal domain-containing protein | |
| Os08g0460600 | 1.29 | CKX11 | Similar to cytokinin dehydrogenase 11 | |
| Os11g0143300 | 1.02 | RR9 | A-type response regulator, cytokinin signaling | |
| Os12g0139400 | 1.00 | RR10 | A-type response regulator, cytokinin signaling | |
| Os02g0830200 | 0.81 | RR3 | A-type response regulator, cytokinin signaling | |
| Os05g0591600 | −1.76 | LOGL8/CK synthesis | Similar to carboxy-lyase | |
| Os06g0573900 | 1.31 | 2.12 | ACO | Similar to 1-aminocyclopropane-1-carboxylic acid oxidase |
| Os02g0771600 | 1.03 | 1.06 | ACO3 | Similar to 1-aminocyclopropane-1-carboxylate oxidase |
| Os04g0610900 | 0.77 | 0.93 | CTR3 | Similar to EDR1 |
| Os03g0341000 | −1.79 | −1.99 | ERF66 | Similar to AP2 domain-containing protein RAP2.2 |
| Os07g0674800 | −2.11 | −3.02 | ERF67 | Similar to AP2 domain transcription factor EREBP |
| Os01g0313300 | −1.36 | −2.79 | ERF68 | Similar to EREBP-3 protein (fragment) |
| Os07g0575000 | 1.69 | ERF6 | ERF, DNA-binding domain-containing protein | |
| Os05g0155200 | 0.49 | ERS2 | Similar to ethylene receptor | |
| Os05g0361700 | −1.07 | ERF61 | Similar to EREBP-2 protein (fragment) | |
| Os06g0586000 | −1.31 | ETH responsive | Conserved hypothetical protein | |
| Os03g0183200 | 1.25 | ERF69 | Similar to AP2 domain-containing protein, expressed | |
| Os05g0316800 | 1.03 | ERF56 | Similar to ethylene-responsive transcription factor 9 | |
| Os10g0390800 | 0.75 | EBL1 | Similar to ethylene-responsive transcription factor 3 (EREBP-3) | |
| Os09g0522000 | −2.71 | DREB1B | Similar to dehydration-responsive element-binding protein 1B | |
| Os02g0677300 | −3.39 | DREB1G | Similar to CRT/DRE binding factor 1 | |
| Os11g0289700 | 1.28 | BR synthesis | Cytochrome P450 family protein | |
| Os04g0641700 | −1.32 | ILI1 | Similar to H0423H10.4 protein | |
| Os09g0441400 | −1.02 | BR synthesis | Similar to elicitor-inducible cytochrome P450 | |
| Os05g0560900 | −1.15 | −1.73 | GA2ox8 | Similar to gibberellin 2-beta-dioxygenase |
| Os06g0729400 | 0.55 | GASR8 | Similar to Gibberellin-regulated protein 2 precursor | |
| Os05g0158600 | −1.44 | GA2ox1 | Similar to OsGA2ox1 | |
| Os03g0437200 | −1.99 | −1.67 | Bsr-d1 | Abscisic acid-induced antioxidant defense |
| Os02g0255500 | 0.42 | 0.72 | PYL3 | Similar to extensin (fragment) |
| Os04g0581100 | −2.06 | S3H/SA conjugation | 2OG-Fe(II) oxygenase domain-containing protein | |
| Os11g0259700 | 2.02 | SA Conjugation | Similar to SAM-dependent carboxyl methyltransferase | |
| Strigolactones | ||||
| Os03g0203200 | 0.69 | dwf14 | Strigolactone receptor, strigolactone perception, regulation of shoot branching | |
In roots, 13 defense-related genes are commonly regulated, while 33 and 18 are specifically Pk and Bv regulated (Table 3). Additionally, 12 hormone-related genes (implicated in JA, CK, ethylene, GA, and ABA syntheses or signaling) are commonly regulated in response to both strains, while 14 and 19 hormone-related genes are Pk and Bv regulated, respectively (Table 5). The latest encompasses genes implicated in auxins, BR, SA, and strigolactone synthesis, or signaling. In leaves, 41 defense-related genes are regulated, six being commonly regulated, while 11 and 24 are specifically induced by Pk and Bv, respectively (Table 2). Also hormone-related DEGs are detected in leaves; six genes implicated in GA, JA, auxins, ethylene and ABA synthesis, transport, or signaling are commonly regulated. Finally, in response to each strain, 20 hormone-related specific DEGs are detected. Interestingly, only the interaction with Pk induced the regulation of BR- and SL-related genes, while only Pv induced the regulation of a SA-related gene. Other hormonal pathways, such as CK, ABA, ethylene, JA, auxins, and GA, are modulated by each strain but induced the regulation of different genes (Table 4).
Validation of RNAseq Data by qCPR
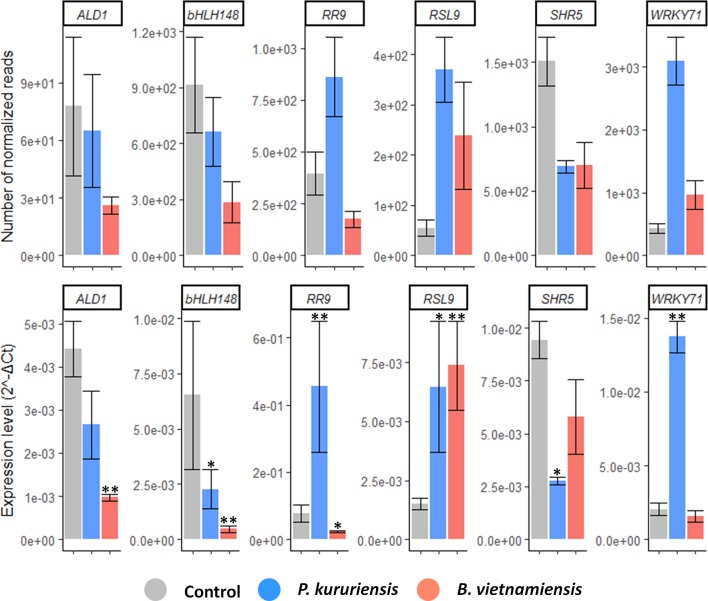
To validate the RNAseq data, we selected genes implicated in defense, hormone signaling, or development regulated at 7 dpi by one or both strains (Supplementary Table 9). We measured the level of expression of the selected genes by qRT-PCR in an independently conducted experiment. According to RNAseq analysis, in leaves, two defense-related genes are specifically regulated by each strain: ALD1 is specifically down-regulated in response to Bv, and WRKY71 is specifically up-regulated in response to Pk. Also, two hormone-related genes were detected as specifically regulated by each strain; namely, bHLH148, a JA signaling component, is down-regulated in roots inoculated with Bv; while RR9, a CK signaling component, is up-regulated in the leaves of Pk-inoculated plants. Finally, in roots, RSL9 is up-regulated and SHR5 is down-regulated in response to both strains. Gene expression changes obtained through qPCR analysis demonstrated a pattern similar to RNAseq (Figure 5) except for the over-expression of RR9 in response to Pk, which was not detected as significant by DESeq2.
Figure 5.
Confirmation of genes regulated upon Bv and Pk colonization. Gene expression quantified by RNAseq (top) and quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR; bottom). For RNAseq, the data presented are the mean count of normalized reads. For qRT-PCR, transcript levels were normalized to that of the reference gene EF 1α (Os03g0177400). Stars represent significant differences of mean expression compared with the control condition according to post-hoc test on a generalized linear model. * corresponds to p < 0.05 and ** to p < 0.0001. Error bars represent the standard deviation (n = 3). Bv, Burkholderia vietnamiensis TVV75T; Pk, Paraburkholderia kururiensis M130; DEGs, differentially expressed genes.
Temporal Analysis of Strain-Specific Marker Genes
Following the confirmation of strain-specific transcriptional regulations, we wanted to identify genes that are differentially expressed in both conditions but with opposite regulations. Among all DEGs detected in roots and leaves, only three DEGs are potential differential markers: RERJ1 (Os04g0301500), a JA-responsive gene; ATL15 (Os01g0597600), a putative amino acid transporter; and DREB1B (Os09g0522000), a drought-responsive gene. Noteworthy, these three genes are only detected as DEGs in leaves and follow the same pattern of expression, as they are up-regulated in response to Pk and down-regulated in response to Bv.
Among these three genes, two of them, ATL15 and RERJ1, are part of a co-expression network recovered from RiceFREND database (Sato et al., 2013), which contains four JA-related genes (JAZ6, 10, 12, and AOS1) (Figure 6A). As JA is one of the main phytohormones implicated in defense (Pozo et al., 2004), we further wanted to know if the whole co-expression network could act as a differential marker of the response to each bacterial strain. In order to describe the transcriptional regulation of this co-expression network throughout the establishment of the interaction, we produced a transcriptional kinetic of rice leaves tissues at 6 hpi, 1 dpi, 7 dpi, and 14 dpi and analyzed all six genes by qRT-PCR.
Figure 6.
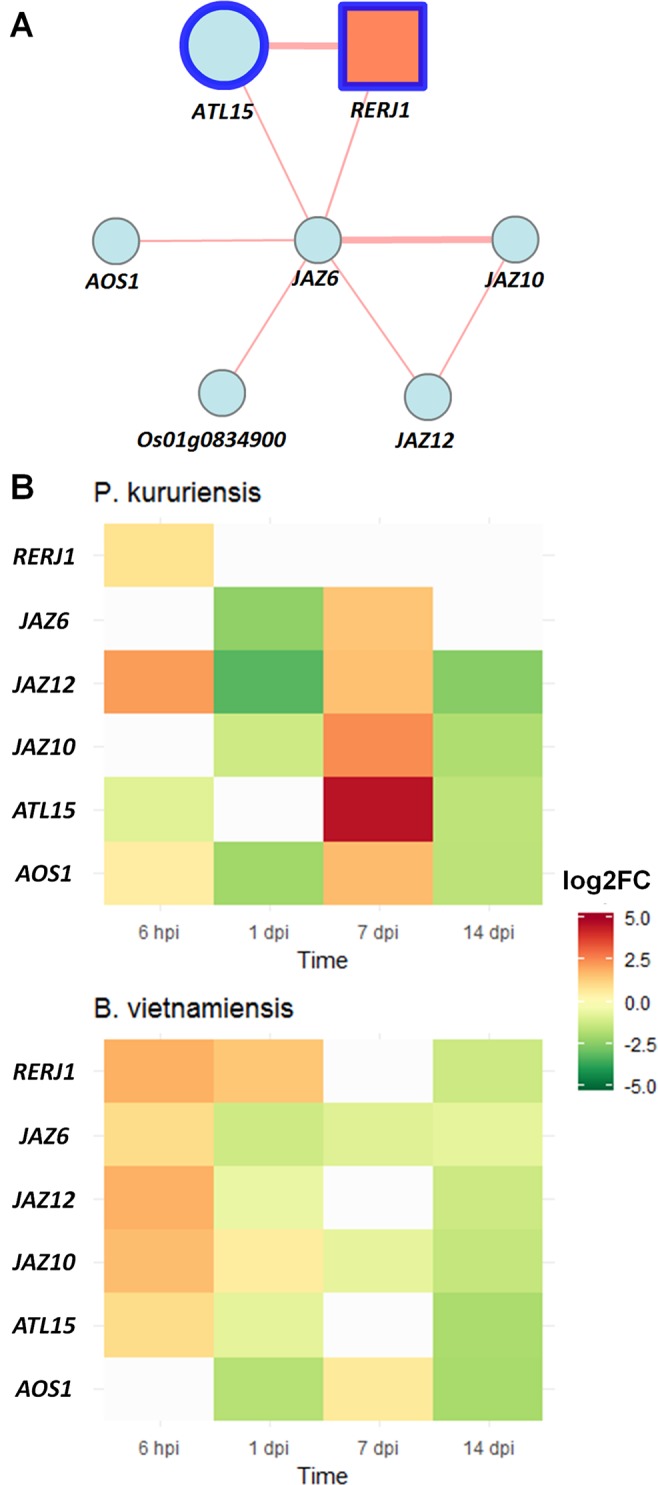
JA-related co-expression network transcriptional regulations induced by Pk and Bv root colonization. (A) Co-expression network of differential markers (circled in blue); edges’ width is proportional to the co-expression ratio between each gene according to RiceFREND database. (B) Gene expression dynamics of the co-expression network in response to Pk and Bv quantified by qRT-PCR. Transcript levels were normalized to that of reference gene EF 1α (Os03g0177400). Values between 0.5 and −0.5 are colored in gray. Data presented are the mean of log2 fold change (n = 3). JA, jasmonic acid; Bv, Burkholderia vietnamiensis TVV75T; Pk, Paraburkholderia kururiensis M130.
First, at 7 dpi, we can confirm that depending on the inoculated strains, the regulation of the co-expression network is opposite (Figure 6; Supplementary Table 10). On the one hand, JAZ6, JAZ10, JAZ12, ATL15, and AOS1 are up-regulated following the inoculation with Pk; and on the other hand, JAZ6 and JAZ10 are down-regulated in response to Bv. Interestingly, and in sharp contrast to the 7 dpi response, for the short-term response (6 hpi), we observed the up-regulation of five out of the six genes of the co-expression network in rice leaves after inoculation with Bv, whereas only JAZ12 is up-regulated in response to Pk. Then, the whole network appears to decline for the rest of the kinetics in response to Bv. Eventually, at 14 dpi, the expression levels of the six genes are quite comparable between the two conditions being all down-regulated compared with non-inoculated controls.
Discussion
The aim of this study was to describe the transcriptional regulations induced by rice following the perception of naturally associated beneficial bacteria. Additionally, we took advantage of the particular phylogenetic organization of the Burkholderia s.l. genus to compare the responses of plants with two closely related beneficial species with different phylogenetic backgrounds in terms of ecology.
P. kururiensis M130 and B. vietnamiensis TVV75 Differentially Colonize Roots of O. sativa Cultivar Nipponbare
We first observed that both bacteria were able to efficiently colonize the rice Nipponbare roots. The amount of culturable bacterial cells associated with rice roots appears to be coherent with the literature, as Compant et al. (2010) described that generally a range between 107 and 109 cfu·g−1 of root fresh weight is found colonizing roots both externally and internally. The same goes for the endophytic population, which generally ranges between 105 and 107 cfu·g−1. The decrease of the bacterial population size between 7 and 14 dpi may be due to the increase of root biomass by the formation of newly emerged roots, which are not importantly colonized at least in the time of our experiments. However, by comparing the colonization of the two strains, two main differences can be observed in the dynamic of root colonization. First, Pk forms a significantly bigger population while colonizing rice roots surface than does Bv at every time postinoculation. Second, the dynamic of the endophytic population of both strains is very different, as the number of endophytic Pk cells declines between 7 and 14 dpi which is not the case for Bv. This could be due to the fact that the endophytic colonization by Pk is restricted by the plant throughout time in contrast to Bv, which maintains its population size. From this observation, two hypotheses can be proposed: Either the plant is not able to control the colonization by Bv, or this strain is more efficiently colonizing the newly emerged roots.
Both the maturation zone and the area of lateral root emergence were identified as hotspots for Pk and Bv colonization. A similar area of colonization was identified for Paraburkholderia phytofirmans strain PsJN in grape plants (Vitis vinifera L.) (Compant et al., 2005) as well as for B. vietnamiensis MGK3, which also intensively colonizes the same areas of the roots of rice plants (Govindarajan et al., 2008). Root exudates, which contain essential nutrients for microbes, are released in the lateral root emergence zones (Badri and Vivanco, 2009). This may aid colonization and allow the possible entry of bacteria via mechanisms such as “crack entry” into the internal tissues (Hardoim et al., 2008).
Both strains were observed massively colonizing the maturation zone, on both the outside and even the inside of some root hair cells (Supplementary Figure 1). The accumulation of bacterial cells in the root hair zone has already been described for Pk (Mattos et al., 2008), and it was proposed to be a common hotspot for rhizobacterial colonization (Compant et al., 2010), as it is correlated to a higher local exudate concentration (Gamalero et al., 2004). Apparent intracellular colonization of epidermal cells was observed for both species but more frequently for Bv. In the case of Bv, the fact that the vacuole is still intact when observing through the colonized epidermic cell offers evidence that the cell is still living (Figure 2D). Also, as the bacterial cells seem to have passed through the cell wall, the fluorescence signal appears to be cytoplasmic. Intracellular colonization of rice root epidermal cells by bacteria was also observed during the interaction with Azoarcus BH72 (Reinhold-Hurek et al., 2006) as well as during the colonization of ryegrass roots by Paraburkholderiabryophila Ha185 (Hsu et al., 2018). Furthermore, intracellular colonization of root hair cells was also observed by Prieto et al. (2011). They showed by confocal microscopy that Pseudomonas putida PICP2 and Pseudomonas fluorescens PICF7 are able to colonize root hair cells of olive trees (Olea euroapea L.) and subsequently move into epidermal cells. This observation led them to propose a new route of entry in intern plant tissues for endophytic bacteria, which starts with the colonization of single root hairs.
Root Colonization Induces More Transcriptional Regulations in Aerial Parts Than in Roots
The analysis of root and leaf transcriptomes revealed that colonization of rice roots by both strains induced more transcriptional regulations in leaves than in roots (Figure 3B). This could be due to the fact that the inoculated plants were harvested at 7 dpi and therefore at an established state of the interaction between bacteria and rice. In contrast, previous studies revealed that the inoculation of beneficial rhizobacteria induced the regulation of at least 1,000 genes in rice roots at earlier time points after inoculation (Drogue et al., 2014; Brusamarello-Santos et al., 2019; Rekha et al., 2018). Moreover, compared with the only study that analyzed the leaf transcriptional response of rice to the inoculation by beneficial rhizobacteria that retrieved only 2,414 DEGs at early stages of the interaction (Wu et al., 2018), when our study identified data of at least 4,000 DEGs in leaves in response to both strains.
Roots Trigger Contrasted Transcriptional Response Depending on the Inoculated Strain
In response to the colonization by both strains, transcriptional reprogramming of defense-related genes occurs in rice roots (Table 3). Only 13 of them appeared commonly regulated in the same way. First, three WRKY (62, 76, and 86),genes are commonly down-regulated; interestingly, those three genes are negative regulator of defense (Peng et al., 2008; Yokotani et al., 2013)—putatively for WRKY86 (Choi et al., 2017). Also, two negative regulators of NH1, the rice NPR1 homolog of the major SA response regulator (Yuan et al., 2007; Chern et al., 2012), namely, RH2 and RH3, are also down-regulated (Chern et al., 2014). Taken together, as these genes are described as negative regulators of defense, their down-regulation may reflect a common defense response towards root colonizing bacteria.
The remaining 51 genes were differentially regulated in response to each strain (33 responding to Pk and 18 to Bv, respectively). Indeed, roots colonized with Pk induce the down-regulation of 11 pathogenesis-related (PR) genes as well as five chitinases and two xylanase inhibitors. One of the down-regulated PR genes is PBZ1 (Os12g0555200), and it is described as a defense marker in leaves, but it is also up-regulated in response to root invasion by Magnaporthe oryzae (Marcel et al., 2010). Moreover, bZIP79, which is described as a phytoalexin synthesis suppressor (Miyamoto et al., 2015), is up-regulated. On the other hand, roots colonization by Bv induce the down-regulation of only one chitinase, whereas two PR genes are up-regulated, of which one gene, BetvI, is targeted by parasitic nematode to suppress root defense (Chen et al., 2018). Furthermore, four WRKY genes of which two are thought to encode negative regulators (WRKY28 (Chujo et al., 2013) and WRKY79 (Choi et al., 2017)) and WRKY40, which is up-regulated in striga-resistant rice roots (Swarbrick et al., 2008), are here down-regulated. Taken together, these results show that the colonization of rice roots by Pk is associated with a down-regulation of PR genes, while the colonization by Bv is associated with the down-regulation of defense-suppressing WRKY genes. This transcriptional regulation of defense-related genes could be the consequence of the more invasive colonization pattern of Bv cells described above.
As previously described (Vacheron et al., 2013; Drogue et al., 2014; Rekha et al., 2018), the inoculation of beneficial rhizobacteria induces important hormone-related transcriptional regulation in rice roots (Table 5). Similarly, the inoculation of Pk and Bv induced the regulation of genes encoding for signaling components of CK and ethylene. Particularly, both bacterial strains induced the up-regulation of two genes encoding for ACO, which are enzymes implicated in the synthesis of ethylene (Ravanbakhsh et al., 2018). Interestingly, in response to the inoculation of other beneficial bacteria, genes coding for ACOs were down-regulated in rice roots (Drogue et al., 2014; Brusamarello-Santos et al., 2019). Also, important transcriptional regulations of ethylene responsive factors (ERFs) occur in response to both strains. In the same way, cytokinine signaling is impacted as both conditions induce the up-regulation of at least two RR genes and one cytokinine oxidase encoding gene (Tsai et al., 2012); however, it is not the same genes that are regulated by each bacterium.
Systemic Responses Associated With Root Inoculation Suggest a Time Shift in Defense Response Between the Two Species
Interestingly, the RNAseq analysis revealed that the inoculation of rhizobacteria induced major transcriptional regulations in leaves (Tables 2 and 4; Supplementary Table 5). As previously described (Persello-Cartieaux et al., 2003; Verhagen et al., 2004; Campos-Soriano et al., 2012), the colonization of beneficial microbes induces transcriptional regulations of defense-related genes in the leaves of the inoculated plants. Both strains induced the up-regulation of WRKY7 as well as the down-regulation of three putative R genes and two PR genes. Pk induced the up-regulation of WRKY71, which is known to confer enhanced disease resistance to Xoo (Liu et al., 2007) and the down-regulation of TFX1, which is known as a susceptibility gene for Xoo (Sugio et al., 2007). On the other hand, Bv induces the up-regulation of five R genes: three (NB-ARC, MLO4, and STV11) (Wang et al., 2014), NPR4, as well as the down-regulation of three WRKY genes (9, 25, and 69), of which two are SA responsive (Liu et al., 2007; Choi et al., 2017), and also the down-regulation of ALD1, a basal immunity regulator needed for the accumulation of SA (Jung et al., 2016).
It is also in this organ only that differential markers have been detected, some of which designated the JA signaling pathway as a putative marker of the interaction with the two bacterial species. Also, interestingly, in contrast with the regulation of JA-related genes, several GA-related genes seem to be down-regulated in response to Pk and up-regulated in response to Bv. This antagonism between JA and GA has been described (Yang et al., 2012; Yimer et al., 2018) and proposed as one of the ways for plants to fine-tune the balance between growth and defense (Huot et al., 2014). Nonetheless, this apparent contrast in terms of JA-related genes transcriptional regulation at 7 dpi may only be a consequence of a delayed JA systemic signaling in response to Pk compared with the response to Bv, which induced the up-regulation of the co-expression network as soon as 6 hpi. As JA-induced plant defenses have been proposed to contribute to the restriction of endophytic colonization in grasses (Miché et al., 2006), this temporal shift in the induction of JA-related genes between the responses to the two bacterial strains could be associated with a delayed JA-induced defense signal. We propose the following interpretation: the perception of Bv induces very early (6 hpi) the up-regulation of JA-related genes in leaves to restrict the colonization, whereas in response to Pk, this JA signal happens at 7 dpi and results in the decrease of the bacterial population (Figure 1A).
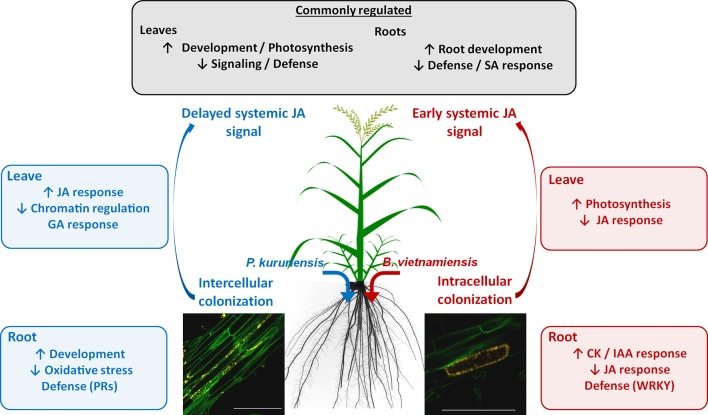
The comparison of the interactions between rice and Pk and between rice and Bv revealed important differences in the process of root colonization and rice transcriptional regulations induced by each strain, which we summarize in Figure 7. First, the numerous intracellular colonization of root epidermic cells by Bv resembles a pathogen infection compared with the apoplastic colonization observed for Pk and other beneficial endophytes (McCully, 2001; Reinhold-Hurek et al., 2007). However, such intracellular colonization by beneficial endophytes has already been observed (Reinhold-Hurek and Hurek, 1998). Also, the specific root response to Pk is characterized by the down-regulation of gene coding for chitinase and PR proteins, while Bv colonization specifically induced the down-regulation of several WRKY genes (Table 3). Moreover, Bv specifically induced the down-regulation of JA signaling genes (Supplementary Figure 5; Supplementary Table 6) in addition to the common SA-related down-regulation in roots. Therefore, the strategies to circumvent the immune system of roots appear to be different between the two strains. Finally, the fact that the root inoculation of Bv induced the up-regulation of JA-related genes in leaves in only 6 hpi compared with the delayed similar signal in response to Pk colonization at 7 dpi also supports the fact that Bv induces a response similar to that of pathogens. Indeed, the rice root colonization by M. oryzae induced the up-regulation of JA-related genes in leaves at 3 and 4 dpi (Marcel et al., 2010). All these elements support the fact that Bv appears to have a much more invasive colonization strategy in terms of both patterns and modulation of the plant immune system. This statement is in accordance with the opportunistic and pathogenic background of the Burkholderia s.s. genus (Eberl and Vandamme, 2016). Consequently, it would be of interest to analyze the response of the cultivar Nipponbare to a larger diversity of plant-associated Burkholderia and Paraburkholderia species to investigate if the differences observed between the two strains can be extrapolated to other species of the respective clades.
Figure 7.
Local and systemic transcriptional regulations of rice in response to Pk and Bv root colonization. Common responses include the up-regulation of development-related genes and the down-regulation of defense-related genes in both leaves and roots. Strain-specific regulations in roots encompass the down-regulation of different defense-related genes: While Pk induced the down-regulation of pathogenesis-related (PR) genes, Bv induced the down-regulation of WRKY transcription factors. Root colonization also induced a systemic up-regulation of jasmonic acid-related (JA) genes in leaves, however, not at the same time for each strain: While Bv induced this signal at early stages of the interaction, Pk induced a transient delayed systemic up-regulation of JA-related genes. Bv, Burkholderia vietnamiensis TVV75T; Pk, Paraburkholderia kururiensis M130.
Another interesting conclusion from this study is the importance of the JA signaling in the interaction with beneficial rhizobacteria. As previously discussed, this major component of plant defense classically associated with the resistance to herbivores and necrotrophic pathogens appears to be involved in a larger diversity of biotic interactions (Thaler, 2004). We have demonstrated that there is a temporal delay in the induction of JA-related genes in leaves following the colonization by Pk comparatively to the response to Bv. It would be of interest to investigate the role of JA in the establishment of these interactions in terms of colonization level and plant defense status using JA-deficient mutants.
Data Availability
These sequence data for this study can be found in the EMBL database under accession number PRJEB31936.
Author Contributions
EK, PC, and LM contributed to the conception and design of the study. EK, AW, IR, CB, and AK performed data collection and analysis. EK wrote the first draft. PC and LM contributed to manuscript revision. All authors read and approved the submitted version.
Funding
The authors acknowledge the CGIAR Research Program on Rice Agri-food Systems (RICE) and the MIC-CERES Project (FC Project ID 2013-1888; AF Project ID 1301-003, jointly supported by Agropolis Fondation through the “Investissements d’avenir” programme with reference number ANR-10-LABX-0001-01, and Fondazione Cariplo) for funding. EK and AW were supported by a fellowship from the French Ministry of Higher Education, Research and Innovation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Annette Vergunst for providing the pIN29 plasmid. We would like to thank Antony Champion and Trang Hieu Nguyen for providing the JAZ qPCR primers.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01141/full#supplementary-material
References
- Ahemad M., Kibret M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J. King Saud Univ. - Sci. 26, 1–20. 10.1016/j.jksus.2013.05.001 [DOI] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri D. V., Vivanco J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32, 666–681. 10.1111/j.1365-3040.2009.01926.x [DOI] [PubMed] [Google Scholar]
- Beneduzi A., Ambrosini A., Passaglia L. M. P. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. 10.1590/S1415-47572012000600020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordiec S., Paquis S., Lacroix H., Dhondt S., Ait Barka E., Kauffmann S., et al. (2011). Comparative analysis of defence responses induced by the endophytic plant growth-promoting rhizobacterium Burkholderia phytofirmans strain PsJN and the non-host bacterium Pseudomonas syringae pv. pisi in grapevine cell suspensions. J. Exp. Bot. 62, 595–603. 10.1093/jxb/erq291 [DOI] [PubMed] [Google Scholar]
- Brusamarello-Santos L. C. C., Alberton D., Valdameri G., Camilios-Neto D., Covre R., Lopes K., de P., et al. (2019). Modulation of defence and iron homeostasis genes in rice roots by the diazotrophic endophyte Herbaspirillum seropedicae. Sci. Rep. 9, 10573. 10.1038/s41598-019-45866-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby P. E., Soman C., Wagner M. R., Friesen M. L., Kremer J., Bennett A., et al. (2017). Research priorities for harnessing plant microbiomes in sustainable agriculture. PLOS Biol. 15, e2001793. 10.1371/journal.pbio.2001793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Soriano L., García-Martínez J., Segundo B. S. (2012). The arbuscular mycorrhizal symbiosis promotes the systemic induction of regulatory defence-related genes in rice leaves and confers resistance to pathogen infection. Mol. Plant Pathol. 13, 579–592. 10.1111/j.1364-3703.2011.00773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Hu L., Sun L., Lin B., Huang K., Zhuo K., et al. (2018). A novel Meloidogyne graminicola effector, MgMO237, interacts with multiple host defence-related proteins to manipulate plant basal immunity and promote parasitism. Mol. Plant Pathol. 19, 1942–1955. 10.1111/mpp.12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M., Bai W., Ruan D., Oh T., Chen X., Ronald P. C. (2014). Interaction specificity and coexpression of rice NPR1 homologs 1 and 3 (NH1 and NH3), TGA transcription factors and negative regulator of resistance (NRR) proteins. BMC Genomics 15, 461. 10.1186/1471-2164-15-461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M., Bai W., Sze-To W. H., Canlas P. E., Bartley L. E., Ronald P. C. (2012). A rice transient assay system identifies a novel domain in NRR required for interaction with NH1/OsNPR1 and inhibition of NH1-mediated transcriptional activation. Plant Methods 8, 6. 10.1186/1746-4811-8-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N. Y., Lee E., Lee S. G., Choi C. H., Park S. R., Ahn I., et al. (2017). Genome-wide expression profiling of oswrky superfamily genes during infection with Xanthomonas oryzae pv. oryzae using real-time PCR. Front. Plant Sci. 8, 1628. 10.3389/fpls.2017.01628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T., Miyamoto K., Shimogawa T., Shimizu T., Otake Y., Yokotani N., et al. (2013). OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol. Biol. 82, 23–37. 10.1007/s11103-013-0032-5 [DOI] [PubMed] [Google Scholar]
- Compant S., Clément C., Sessitsch A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. 10.1016/j.soilbio.2009.11.024 [DOI] [Google Scholar]
- Compant S., Reiter B., Sessitsch A., Nowak J., Clement C., Ait Barka E. (2005). Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderiasp. strain PsJN. Appl. Environ. Microbiol. 71, 1685–1693. 10.1128/AEM.71.4.1685-1693.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho B. G., Licastro D., Mendonça-Previato L., Cámara M., Venturi V. (2015). Plant-influenced gene expression in the rice endophyte Burkholderia kururiensis M130. Mol. Plant-Microbe Interact. 28, 10–21. 10.1094/MPMI-07-14-0225-R [DOI] [PubMed] [Google Scholar]
- Coutinho B. G., Passos D., Silva D., Previato J. O., Mendonça-Previato L., Venturi V. (2013). Draft genome sequence of the rice endophyte Burkholderia kururiensis M130. Genome Announc. 1, e00225–e00212. 10.1128/genomeA.00225-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogue B., Sanguin H., Chamam A., Mozar M., Llauro C., Panaud O., et al. (2014). Plant root transcriptome profiling reveals a strain-dependent response during Azospirillum–rice cooperation. Front. Plant Sci. 5, 607. 10.3389/fpls.2014.00607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl L., Vandamme P. (2016). Members of the genus Burkholderia: good and bad guys. F1000Res. 5(F1000 Faculty Rev):1007. 10.12688/f1000research.8221.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-de los Santos P., Rojas-Rojas F. U., Tapia-Garcia E. Y., Vasquez-Murrieta M. S., Hirsch A. M. (2016). To split or not to split: an opinion on dividing the genus Burkholderia. Ann. Microbiol. 66, 1303–1314. 10.1007/s13213-015-1183-1 [DOI] [Google Scholar]
- Estrada-de los Santos P., Palmer M., Chávez-Ramírez B., Beukes C., Steenkamp E., Briscoe L., et al. (2018). Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 9, 389. 10.3390/genes9080389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel O. M., Castrillo G., Herrera Paredes S., Salas González I., Dangl J. L. (2017). Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163. 10.1016/j.pbi.2017.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalero E., Lingua G., Giusy Caprì F., Fusconi A., Berta G., Lemanceau P. (2004). Colonization pattern of primary tomato roots by Pseudomonas fluorescens A6RI characterized by dilution plating, flow cytometry, fluorescence, confocal and scanning electron microscopy. FEMS Microbiol. Ecol. 48, 79–87. 10.1016/j.femsec.2003.12.012 [DOI] [PubMed] [Google Scholar]
- Govindarajan M., Balandreau J., Kwon S.-W., Weon H.-Y., Lakshminarasimhan C. (2008). Effects of the inoculation of Burkholderia vietnamensis and related endophytic diazotrophic bacteria on grain yield of rice. Microb. Ecol. 55, 21–37. 10.1007/s00248-007-9247-9 [DOI] [PubMed] [Google Scholar]
- Gyaneshwar P., Hirsch A. M., Moulin L., Chen W.-M., Elliott G. N., Bontemps C., et al. (2011). Legume-nodulating Betaproteobacteria: diversity, host range, and future prospects. Mol. Plant-Microbe Interact. 24, 1276–1288. 10.1094/MPMI-06-11-0172 [DOI] [PubMed] [Google Scholar]
- Hardoim P. R., van Overbeek L. S., van Elsas J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471. 10.1016/j.tim.2008.07.008 [DOI] [PubMed] [Google Scholar]
- Hayat R., Ali S., Amara U., Khalid R., Ahmed I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. 10.1007/s13213-010-0117-1 [DOI] [Google Scholar]
- Hsu P.-C. L., O’Callaghan M., Condron L., Hurst M. R. H. (2018). Use of a gnotobiotic plant assay for assessing root colonization and mineral phosphate solubilization by Paraburkholderia bryophila Ha185 in association with perennial ryegrass (Lolium perenne L.). Plant Soil 425, 43–55. 10.1007/s11104-018-3633-6 [DOI] [Google Scholar]
- Huot B., Yao J., Montgomery B. L., He S. Y. (2014). Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287. 10.1093/mp/ssu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S., Zechmann B., Molitor A., Trujillo M., Petutschnig E., Lipka V., et al. (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 156, 726–740. 10.1104/pp.111.176446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature. 444, 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Jung G. Y., Park J. Y., Choi H. J., Yoo S.-J., Park J.-K., Jung H. W. (2016). A rice gene homologous to Arabidopsis AGD2-LIKE DEFENSE1 participates in disease resistance response against infection with Magnaporthe oryzae. Plant Pathol. J. 32, 357–362. 10.5423/PPJ.NT.10.2015.0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar K., Kundu A., Rizvi A. Z., Dubois E., Severac D., Czernic P., et al. (2019). Transcriptomic analysis with the progress of symbiosis in ‘crack-entry’ legume Arachis hypogaea highlights its contrast with ‘infection thread’ adapted legumes. Mol. Plant-Microbe Interact. 32, 271–285. 10.1094/MPMI-06-18-0174-R [DOI] [PubMed] [Google Scholar]
- Kaur C., Selvakumar G., Ganeshamurthy A. N. (2017). “Burkholderia to Paraburkholderia: the journey of a plant-beneficial-environmental bacterium,” in Recent advances in applied microbiology (Singapore: Springer Singapore; ), 213–228. 10.1007/978-981-10-5275-0_10 [DOI] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12, 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Bai X., Wang X., Chu C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979. 10.1016/j.jplph.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Shinohara H., Kiba A., Ohnishi K., Furuya N., Kawamura Y., et al. (2006). Phylogenetic study and multiplex PCR-based detection of Burkholderia plantarii, Burkholderia glumae and Burkholderia gladioli using gyrB and rpoD sequences. Int. J. Syst. Evol. Microbiol. 56, 1031–1038. 10.1099/ijs.0.64184-0 [DOI] [PubMed] [Google Scholar]
- Marcel S., Sawers R., Oakeley E., Angliker H., Paszkowski U. (2010). Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell. 22, 3177–3187. 10.1105/tpc.110.078048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattos K. A., Pádua V. L. M., Romeiro A., Hallack L. F., Neves B. C., Ulisses T. M. U., et al. (2008). Endophytic colonization of rice Oryza sativa L. by the diazotrophic bacterium Burkholderia kururiensis and its ability to enhance plant growth. An. Acad. Bras. Cienc. 80, 477–493. 10.1590/S0001-37652008000300009 [DOI] [PubMed] [Google Scholar]
- McCully M. E. (2001). Niches for bacterial endophytes in crop plants: a plant biologist’s view. Funct. Plant Biol. 28, 983. 10.1071/PP01101 [DOI] [Google Scholar]
- Miché L., Battistoni F., Gemmer S., Belghazi M., Reinhold-Hurek B. (2006). Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Mol. Plant-Microbe Interact. 19, 502–511. 10.1094/MPMI-19-0502 [DOI] [PubMed] [Google Scholar]
- Millet Y. A., Danna C. H., Clay N. K., Songnuan W., Simon M. D., Werck-Reichhart D., et al. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell. 22, 973–990. 10.1105/tpc.109.069658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K., Nishizawa Y., Minami E., Nojiri H., Yamane H., Okada K. (2015). Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J. Plant Physiol. 173, 19–27. 10.1016/j.jplph.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C., Lonhienne T. G. A., Yeoh Y. K., Donose B. C., Webb R. I., Parsons J., et al. (2016). Crosstalk between sugarcane and a plant-growth promoting Burkholderia species. Sci. Rep. 6, 37389. 10.1038/srep37389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Bartley L. E., Chen X., Dardick C., Chern M., Ruan R., et al. (2008). OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 1, 446–458. 10.1093/mp/ssn024 [DOI] [PubMed] [Google Scholar]
- Persello-Cartieaux F., Nussaume L., Robaglia C. (2003). Tales from the underground: molecular plant–rhizobacteria interactions. Plant Cell Environ. 26, 189–199. 10.1046/j.1365-3040.2003.00956.x [DOI] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C. M. J., Zamioudis C., Berendsen R. L., Weller D. M., Van Wees S. C. M., Bakker P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52. 10.1146/annurev-phyto-082712-102340 [DOI] [PubMed]
- Pozo M. J., Van Loon L. C., Pieterse C. M. J. (2004). Jasmonates—signals in plant–microbe interactions. J. Plant Growth Regul. 23, 211–222. 10.1007/s00344-004-0031-5 [DOI] [Google Scholar]
- Prieto P., Schilirò E., Maldonado-González M. M., Valderrama R., Barroso-Albarracín J. B., Mercado-Blanco J. (2011). Root hairs play a key role in the endophytic colonization of olive roots by Pseudomonas spp. with biocontrol activity. Microb. Ecol. 62, 435–445. 10.1007/s00248-011-9827-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., et al. (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198. 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanbakhsh M., Sasidharan R., Voesenek L. A. C. J., Kowalchuk G. A., Jousset A. (2018). Microbial modulation of plant ethylene signaling: ecological and evolutionary consequences. Microbiome 6, 52. 10.1186/s40168-018-0436-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J., Kull M., Peterson H., Hansen J., Vilo J. (2007). g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–W200. 10.1093/nar/gkm226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B., Hurek T. (1998). Life in grasses: diazotrophic endophytes. Trends Microbiol. 6, 139–144. 10.1016/S0966-842X(98)01229-3 [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B., Krause A., Leyser B., Miché L., Hurek T. (2007). “The rice apoplast as a habitat for endophytic N2-fixing bacteria,” in The apoplast of higher plants: compartment of storage, transport and reactions (Dordrecht: Springer Netherlands; ), 427–443. 10.1007/978-1-4020-5843-1_30 [DOI] [Google Scholar]
- Reinhold-Hurek B., Maes T., Gemmer S., Van Montagu M., Hurek T. (2006). An endoglucanase is involved in infection of rice roots by the not-cellulose-metabolizing endophyte Azoarcus sp. strain BH72. Mol. Plant-Microbe Interact. 19, 181–188. 10.1094/MPMI-19-0181 [DOI] [PubMed] [Google Scholar]
- Rekha K., Kumar R. M., Ilango K., Rex A., Usha B. (2018). Transcriptome profiling of rice roots in early response to Bacillus subtilis (RR4) colonization. Botany 96, 749–765. 10.1139/cjb-2018-0052 [DOI] [Google Scholar]
- Sato Y., Namiki N., Takehisa H., Kamatsuki K., Minami H., Ikawa H., et al. (2013). RiceFREND: a platform for retrieving coexpressed gene networks in rice. Nucleic Acids Res. 41, D1214–D1221. 10.1093/nar/gks1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawana A., Adeolu M., Gupta R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderiagen. nov. harboring environmental species. Front. Genet. 5, 429. 10.3389/fgene.2014.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheibani-Tezerji R., Rattei T., Sessitsch A., Trognitz F., Mitter B. (2015). Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. MBio 6, e00621–e00615. 10.1128/mBio.00621-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shidore T., Dinse T., Öhrlein J., Becker A., Reinhold-Hurek B. (2012). Transcriptomic analysis of responses to exudates reveal genes required for rhizosphere competence of the endophyte Azoarcus sp. strain BH72. Environ. Microbiol. 14, 2775–2787. 10.1111/j.1462-2920.2012.02777.x [DOI] [PubMed] [Google Scholar]
- Stringlis I. A., Proietti S., Hickman R., Van Verk M. C., Zamioudis C., Pieterse C. M. J. (2018). Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 93, 166–180. 10.1111/tpj.13741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A., Yang B., Zhu T., White F. F. (2007). Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAgamma1 and OsTFX1 during bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 104, 10720–10725. 10.1073/pnas.0701742104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbrick P. J., Huang K., Liu G., Slate J., Press M. C., Scholes J. D., et al. (2008). Global patterns of gene expression in rice cultivars undergoing a susceptible or resistant interaction with the parasitic plant Striga hermonthica. New Phytol. 179, 515–529. 10.1111/j.1469-8137.2008.02484.x [DOI] [PubMed] [Google Scholar]
- Thaler J. S. (2004). The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 135, 530–538. 10.1104/pp.104.041566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trân Van V., Berge O., Ngô Kê S., Balandreau J., Heulin T. (2000). Repeated beneficial effects of rice inoculation with a strain of Burkholderia vietnamiensis on early and late yield components in low fertility sulphate acid soils of Vietnam. Plant Soil 218, 273–284. 10.1023/A:1014986916913 [DOI] [Google Scholar]
- Trdá L., Boutrot F., Claverie J., Brulé D., Dorey S., Poinssot B. (2015). Perception of pathogenic or beneficial bacteria and their evasion of host immunity: pattern recognition receptors in the frontline. Front. Plant Sci. 6, 219. 10.3389/fpls.2015.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-C., Weir N. R., Hill K., Zhang W., Kim H. J., Shiu S.-H., et al. (2012). Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 158, 1666–1684. 10.1104/pp.111.192765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron J., Desbrosses G., Bouffaud M.-L., Touraine B., Moënne-Loccoz Y., Muller D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4, 356. 10.3389/fpls.2013.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van V. T., Berge O., Balandreau J., Kê S. N., Heulin T. (1996). Isolation and nitrogenase activity of Burkholderia vietnamiensis, a nitrogen-fixing bacterium associated with rice Oryza sativa L on a sulphate acid soil of Vietnam. Agronomie 8, 479–491. 10.1051/agro:19960802 [DOI] [Google Scholar]
- Vergunst A. C., Meijer A. H., Renshaw S. A., O’Callaghan D. (2010). Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infect. Immun. 78, 1495–1508. 10.1128/IAI.00743-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen B. W. M., Glazebrook J., Zhu T., Chang H.-S., van Loon L. C., Pieterse C. M. J. (2004). The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol. Plant-Microbe Interact. 17, 895–908. 10.1094/MPMI.2004.17.8.895 [DOI] [PubMed] [Google Scholar]
- Vial L., Chapalain A., Groleau M.-C., Déziel E. (2011). The various lifestyles of the Burkholderia cepacia complex species: a tribute to adaptation. Environ. Microbiol. 13, 1–12. 10.1111/j.1462-2920.2010.02343.x [DOI] [PubMed] [Google Scholar]
- Wang Q., Liu Y., He J., Zheng X., Hu J., Liu Y., et al. (2014). STV11 encodes a sulphotransferase and confers durable resistance to rice stripe virus. Nat. Commun. 5, 4768. 10.1038/ncomms5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Peng X., Yang M., Zhang W., Dazzo F. B., Uphoff N., et al. (2018). Rhizobia promote the growth of rice shoots by targeting cell signaling, division and expansion. Plant Mol. Biol. 97, 507–523. 10.1007/s11103-018-0756-3 [DOI] [PubMed] [Google Scholar]
- Yang D.-L., Yao J., Mei C.-S., Tong X.-H., Zeng L.-J., Li Q., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109, E1192–E1200. 10.1073/pnas.1201616109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimer H. Z., Nahar K., Kyndt T., Haeck A., Van Meulebroek L., Vanhaecke L., et al. (2018). Gibberellin antagonizes jasmonate-induced defense against Meloidogyne graminicola in rice. New Phytol. 218, 646–660. 10.1111/nph.15046 [DOI] [PubMed] [Google Scholar]
- Yokotani N., Sato Y., Tanabe S., Chujo T., Shimizu T., Okada K., et al. (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64, 5085–5097. 10.1093/jxb/ert298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Zhong S., Li Q., Zhu Z., Lou Y., Wang L., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5, 313–324. 10.1111/j.1467-7652.2007.00243.x [DOI] [PubMed] [Google Scholar]
- Zamioudis C., Pieterse C. M. J. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant-Microbe Interact. 25, 139–150. 10.1094/MPMI-06-11-0179 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
These sequence data for this study can be found in the EMBL database under accession number PRJEB31936.