Abstract
Human intervention, pre-human climate change (or a combination of both), as well as genetic effects, contribute to species extinctions. While many species from oceanic islands have gone extinct due to direct human impacts, the effects of pre-human climate change and human settlement on the genomic diversity of insular species and the role that loss of genomic diversity played in their extinctions remains largely unexplored. To address this question, we sequenced whole genomes of two extinct New Zealand passerines, the huia (Heteralocha acutirostris) and South Island kōkako (Callaeas cinereus). Both species showed similar demographic trajectories throughout the Pleistocene. However, the South Island kōkako continued to decline after the last glaciation, while the huia experienced some recovery. Moreover, there was no indication of inbreeding resulting from recent mating among closely related individuals in either species. This latter result indicates that population fragmentation associated with forest clearing by Maōri may not have been strong enough to lead to an increase in inbreeding and exposure to genomic erosion. While genomic erosion may not have directly contributed to their extinctions, further habitat fragmentation and the introduction of mammalian predators by Europeans may have been an important driver of extinction in huia and South Island kōkako.
Keywords: genomic erosion, glaciations, decline, extinction
1. Introduction
Species declines and extinctions are complex and multifactorial [1,2]. Two paradigms have been proposed in conservation biology [3]. The first paradigm focuses on how extrinsic factors, such as climate fluctuations or human activities, contribute to population decline and extinction. While the role of humans in the extinction of species over the past 500 years is well recognized [4], past climate changes were also major drivers of population declines and extinctions [5,6]. However, the relative impact of human activities and climate on biodiversity are still intensely debated and these impacts may well vary among species [1,2].
As a consequence of declines in population size and geographical distribution from these extrinsic factors, small and isolated populations can then be exposed to intrinsic threats. This is why a second paradigm, which instead focuses on intrinsic demographic and genetic effects, is also central to conservation biology [3]. The role that detrimental genetic effects play in the long-term persistence of populations is well known [7,8]. Such detrimental effects can be referred to as genomic erosion, which reduces species viability through drift, inbreeding and increase in genetic load [7,9]. Recent empirical data on extinct woolly mammoths (Mammuthus primigenius; [10,11]), endangered gorilla (Gorilla beringei sp.; [12]) and crested ibis (Nipponia nippon; [13]) have shown that severe population declines expose populations to genomic erosion. Moreover, species that have experienced long-term, pre-human decline in effective population size (Ne) may be more vulnerable to human-induced declines and to genomic erosion as was suggested for the critically endangered Sumatran rhinoceros (Dicerorhinus sumatrensis; [14]). Similarly, several avian species on the IUCN Red List of Threatened Species have experienced long-term, pre-human population reductions Ne [15], further highlighting the link between long-term population decline and higher exposure to genomic erosion.
Species from oceanic islands recently colonized by humans are particularly vulnerable to human disturbance due to their small census size and Ne and their limited ability to alter their range in response to anthropogenic pressures [9]. Moreover, island populations have experienced higher extinction rates compared with mainland species [16]. In fact, even though islands represent only 5.3% of the surface of the earth, they have hosted 75% of the known vertebrate extinctions over the past 500 years [17] due to both habitat modification, over-hunting and the introduction of non-native mammalian predators [18].
As a case in point, New Zealand endemic fauna has experienced two major extinction events in association with Maōri (ca 1360 CE; [19]) and European settlement (ca 1800 CE; [20]). These extinctions have been attributed to direct human interference such as habitat destruction [18] or hunting (e.g. moa, [21]; Megadyptes waitaha [22]). Moreover, because New Zealand endemics evolved in the absence of mammalian predators and because a large proportion of its avifauna is flightless, the accidental or deliberate introduction of mammals has been an important driver of species decline [20]. The colonization of New Zealand by the ancestors of its endemic species from a small number of founders, their evolution in a confined geographical area as well as pre-human climate fluctuations may have reduced the genetic diversity of species well before human settlement (e.g. kea, Nestor notabilis; [15,23]). It is thus possible that New Zealand avian species characterized by historically low genetic diversity may have been more easily exposed to genomic erosion following the human-induced declines of the last 800 years [24]. Yet, to date, the effect of pre-human climate fluctuations and of Maōri settlement on the genome-wide diversity of insular avian species in New Zealand remains largely unexplored. Understanding these effects would allow us to determine whether genomic erosion contributed to their extinction.
Here, we examine the long-term response to climate change and the recent effects of human settlement on the genome-wide diversity of two extinct forest passerines from the Callaeidae family or New Zealand wattlebirds [25,26], the huia (Heteralocha acutirostris) and South Island kōkako (Callaeas cinereus). Huia were common throughout the North Island but went extinct in 1907, whereas South Island kōkako were only found in the South Island and were declared extinct in the 1960s [27]. Using demographic reconstructions, we show that these species had similar responses to habitat change during the last glaciation. Moreover, inbreeding coefficients were not consistent with genomic erosion close to the time of extinction. Our data thus suggest that further habitat fragmentation and the introduction of mammalian predators by Europeans may have been the main drivers of the extinction of these two species.
2. Material and methods
(a). DNA extraction, library preparation and sequencing
We extracted DNA from toepads for one huia (Heteralocha acutirostris) and one South Island kōkako (Callaeas cinereus) collected in 1886 and 1849, respectively (electronic supplementary material, table S1). We built one deep-sequencing library per bird following Meyer & Kircher [28] and sequenced them on HiseqX lanes (see electronic supplementary material). All laboratory procedures were conducted in a dedicated historical DNA laboratory. We took appropriate precautions to minimize the risk of contamination of historical samples [29].
(b). Data processing
After trimming adapters, we mapped the raw genomic data to a de novo assembly for the North Island kōkako (Callaeas wilsoni; https://b10k.genomics.cn/) using BWA 0.7.13 aln [30]. We removed duplicates, realigned bam files around indels and filtered them for mapping quality. We then called variants, filtered them for base quality, depth and proximity to indels. Finally, we masked repeats and CpG sites from bam and vcf files to limit possible biases from DNA damage (see electronic supplementary material).
(c). Data analysis
We first used the Pairwise Sequentially Markovian Coalescent (PSMC 0.6.5) [31] to infer temporal changes in the effective population sizes (Ne) of huia and South Island kōkako. Secondly, we used mlRho v.2.7 [32] to estimate population mutation rates (θ), which approximates expected heterozygosity under the infinite sites model. Finally, we identified runs of homozygosity (ROH) and estimated individual inbreeding coefficients (FROH) using a sliding-window approach [33] (see electronic supplementary material).
3. Results
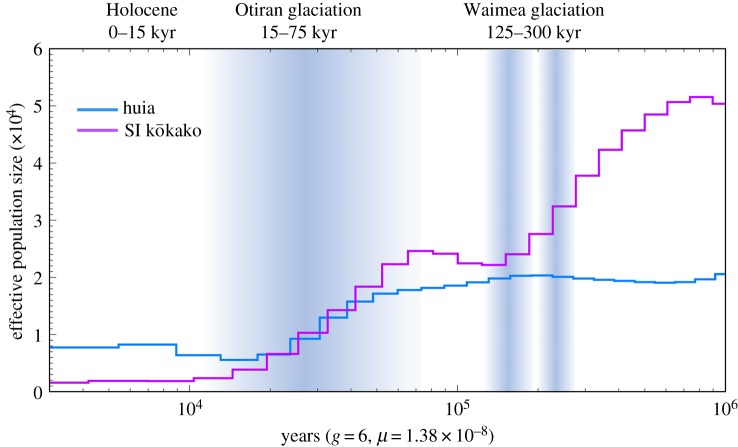
Demographic reconstruction based on 10× and 14× coverage genomes in huia and South Island kōkako, respectively (electronic supplementary material, figure S1 and table S1) and corrected for low coverage showed broadly similar Ne trajectories (figure 1; electronic supplementary material, figures S2 and S3). However, while huia showed a nearly stable Ne between 1 Myr and 100 kyr BP, South Island kōkako experienced a severe decline dating back to approximately 700 kyr BP. Both species experienced a 2- to 10-fold decline in Ne coinciding with the last glaciation approximately 60–70 kyr BP (figure 1). While the Ne of both species was estimated at approximately 6000 at the Last Glacial Maximum (LGM) approximately 20 kyr BP, huia Ne seems to have increased slightly to approximately 8000 after the LGM, whereas South Island kōkako Ne declined to less than 2000 (figure 1).
Figure 1.
PSMC for huia and South Island kōkako scaled using a generation time of 6 years [34,35], a substitution rate of 1.38 × 10−8 substitution/site/generation inferred from [36] and a uniform false negative rate (uFNR) of 40%. (Online version in colour.)
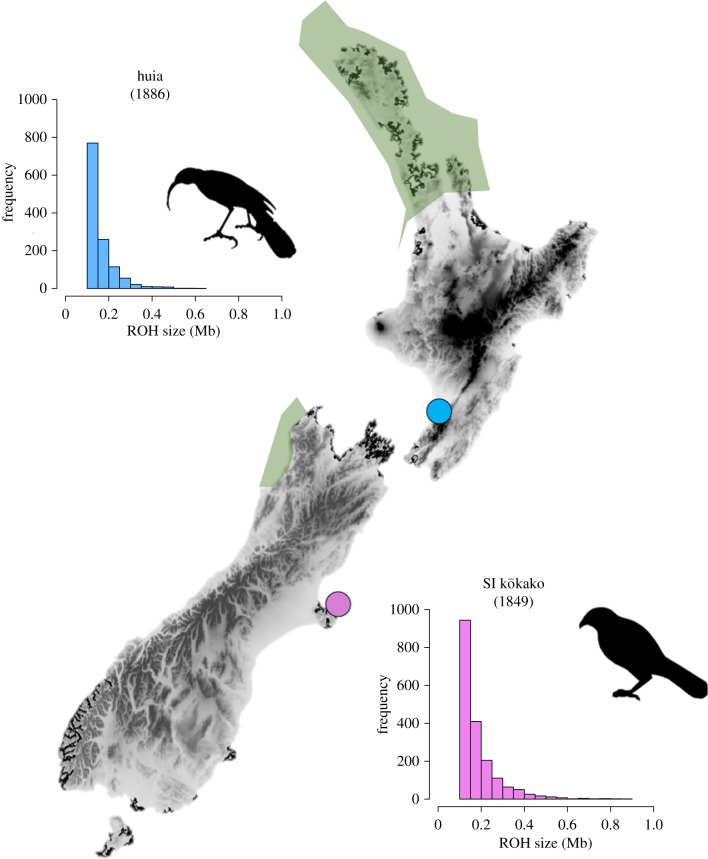
Both species showed similar levels of genome-wide heterozygosity, estimated at 0.94–1 SNPs per thousand base pairs (table 1). While the inbreeding coefficient was higher in South Island kōkako (FROH = 0.32) compared to huia (FROH = 0.19), the majority of the ROH identified were less than 1 Mb in both species (figure 2).
Table 1.
Heterozygosity per 1000 bp (θ) and inbreeding estimates (FROH). θ = population mutation rate which approximates heterozygosity under the infinite sites model.
| species | θ | θ (95% CI) | FROH > 100 kb | FROH > 1 Mb |
|---|---|---|---|---|
| huia | 0.944 | 0.942–0.947 | 0.187 | 0 |
| South Island kōkako | 1 | 0.998–1 | 0.319 | 0 |
| rifleman | 1.67a | n.a. | n.a. | n.a. |
aEstimated as SNP rate per 103 bases [37].
Figure 2.
Sampling locations of huia and South Island kōkako museum skins. Green-shaded areas depict forest refugia during the LGM ca 22 000 years BP, after Alloway et al. [38]. Bar plots depict the distribution of ROHs > 100 kb. (Online version in colour.)
4. Discussion
Using complete genomes, we examined the long-term response to climate change and tested the hypothesis that habitat modification associated with Maōri settlement impacted the genome-wide diversity of huia and South Island kōkako prior to their extinction.
Demographic reconstructions indicated very similar responses to glaciations between species with a reduction in Ne shortly after the onset of the last glaciation and little to no recovery at the end of the LGM approximately 14–22 kyr BP [38]. This pattern is very similar to another forest passerine, the rifleman (Acanthisitta chloris; [15]). However, the rifleman had a much higher Ne > 20 000 at the end of the LGM [15]. While the signal of long-term population decline could indicate limited migration between subpopulations [39], this decline in Ne is consistent with a severe reduction in forest cover in the southern North Island and the South Island [40,41]. With the exception of extensive forest tracts mostly confined to the northern parts of the North Island and some isolated forest patches in the South Island (figure 2; [38,42]), most of New Zealand's vegetation was characterized by extensive grassland and shrublands at the LGM [38,40,41]. Being both forest species, huia and South Island kōkako were thus most likely restricted to such forest refugia, as was the case for other forest species [43–45]. Yet, it is unclear why both species had a similar Ne at the LGM while the forest refugium was smaller in the South Island compared with the North Island [40] and why South Island kōkako had a lower Ne compared to huia after the LGM when both species should have experienced a similar population expansion. However, this latter difference in Ne may be due to the limitation of the PSMC to estimate recent Ne (less than 10 000 BP) [31,46].
While fossil preservation can be affected by temperature, humidity or acidity, an abundance of fossils from the Early to Late Holocene deposits of forest species (e.g. kaka, N. meridionalis; pigeon, Hemiphaga novaeseelandiae; parakeets, Cyanoramphus sp.; [47,48]) suggests that demographic expansion occurred as species tracked their habitat after the LGM [40,41]. Conversely, open-habitat species like the alpine kea (N. notabilis) seem to have experienced a decline in Ne or at least lack of post-glacial demographic recovery, as their range became restricted to alpine areas [15,23]. Because both huia and South Island kōkako were forest dwellers, they should have also experienced population expansion after the LGM. In fact, Ne estimates of approximately 30–90 000 huia prior to human arrival in New Zealand based on rapidly evolving mitochondrial sequences suggest that post-glacial recovery could have occurred in these species [49]. However, because the number of recombination events is limited over the recent past and because of the lag time between demographic expansion and increase in Ne, PSMC lacks the power to detect recent population fluctuations (less than 10 000 BP) [31,46]. Moreover, the reliability of Ne estimates can be affected by coverage, the proportion of missing data and the uncertainty about substitution rates (electronic supplementary material, figures S2 and S3; [31,46]). These estimates should be thus interpreted with caution. Nevertheless, in spite of these limitations, the overall long-term decrease in Ne in both species is consistent with that of extant species classified as endangered on the IUCN Red List of Threatened Species [15,50]. Conversely, the rifleman had a higher heterozygosity [37] and Ne, which is consistent with their least concern conservation status [50].
While the relatively important declines in Ne through time in both species could have made them more vulnerable to genomic erosion, inbreeding (FROH) was low in both species and mostly comprised fragments less than 1 Mb. This indicates that the observed inbreeding was the result of background relatedness (i.e. short ROH caused by random mating in a large population that results in pairing of distant relatives) and not of recent mating among related individuals [51]. While 40% of forest had been cleared by Maōri between the thirteenth and nineteenth centuries [52–54], this result suggests that habitat fragmentation prior to the 1850s may not have been severe enough to reduce gene flow among populations and to increase inbreeding in huia and South Island kōkako populations. Because European settlement had just started at the time of sampling of these museum skins ca 1860–1880 [53], forest habitat may still have allowed large populations to thrive. In fact, previous results based on historical microsatellite data from the same period did not show evidence for population subdivision in huia [49].
Although our data do not show evidence for genomic erosion, future temporal comparison of historical genomes spanning the time of European settlement to the extinction of these species (i.e. huia: 1907; South Island kōkako: 1960s) could indicate whether genomic erosion associated with human-induced bottlenecks contributed to their extinction [55]. This may be especially relevant to South Island kōkako, which went extinct in the 1960s. Assuming a generation time of 6 years [34,35], a period of 100 years corresponds to 17 kōkako generations, which may have been enough for small and fragmented populations to accumulate genetic load. For instance, a population decline dating back to 20 and 100 years ago for the gorilla [12] and crested ibis [13], respectively, led to increases in inbreeding and genetic load. Moreover, numerous extant avian species in New Zealand have lost a large proportion of their historical genetic diversity and may also have accumulated genetic load, with severe consequences for their viability (e.g. kākāpō, Strigops habroptilus; [56,57]; saddleback, Philesturnus sp.; South Island robin, Petroica australis [58,59]). Conversely, huia went extinct in 1907 [27], 21 years after the study skin was sampled, which corresponds to around three generations [35,49]. It is thus likely that huia experienced a rapid decline and extinction resulting mostly from further forest clearance and the introduction of mammalian predators by Europeans, without genetic erosion contributing markedly to their extinction [60].
In conclusion, our results indicate a severe reduction in huia and South Island kōkako Ne as a result of long-term climate change. However, while our data do not allow us to detect very recent bottlenecks associated with human activities, the low inbreeding coefficients close to extinction of these species suggest that Maōri settlement did not lead to an increase in inbreeding in huia and South Island kōkako. Consequently, neither species seems to have been exposed to genomic erosion at the time of European arrival. While temporal comparison of historical genomes in South Island kōkako are required to properly examine the role of genomic erosion in the extinction of the species, it seems likely that huia went extinct rapidly through the combined effects of forest clearance and mammalian predation.
Supplementary Material
Acknowledgements
We thank Anita Gamauf (Vienna Museum, Austria), Mark Adams (Natural History Museum), Alan Tennyson (Te Papa, New Zealand), Emma Burns (Otago Museum, New Zealand) and Matt Rayner (Auckland Museum, Auckland, New Zealand) for lending museum skins. We also thank Guojie Zhang and the Bird 10 000 Genome Project (B10 K) (https://b10k.genomics.cn/) for access to the genome assembly. We acknowledge support from the Uppsala Multidisciplinary Centre for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. Sequencing was performed by the Swedish National Genomics Infrastructure (NGI) at the Science for Life Laboratory, which is supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation.
Data accessibility
Raw fastq reads are deposited at the European Nucleotide Archive (ENA), accession number (PRJEB33922) [60].
Authors' contributions
N.D. and L.D. conceived the study. M.K. and O.K. performed DNA extractions of museum skins. N.D. and J.v.S. performed library preparation for deep-sequencing. B.C.R. provided blood samples for reference genome sequencing. N.D. analysed the data and wrote the manuscript. All authors contributed to the final version of the manuscript, approved it for publication and agree to be accountable for its contents.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by FORMAS (2015-676) to L.D.; the Swiss National Science Foundation to N.D. (P2SKP3_165031 and P300PA_177845); University of Otago PBRF grants to N.D., M.K. and B.C.R.
References
- 1.Barnosky AD, Koch PL, Feranec RS, Wing SL, Shabel AB. 2004. Assessing the causes of late Pleistocene extinctions on the continents. Science 306, 70–75. ( 10.1126/science.1101476) [DOI] [PubMed] [Google Scholar]
- 2.Koch PL, Barnosky D. 2014. Late Quaternary extinctions: state of the debate. Annu. Rev. Ecol. Evol. Syst. 37, 215–250. ( 10.1146/annurev.ecolsys.34.011802.132415) [DOI] [Google Scholar]
- 3.Caughley G. 1994. Directions in conservation biology. J. Anim. Ecol. 63, 215–244. ( 10.2307/5542) [DOI] [Google Scholar]
- 4.Dirzo R, Young HS, Galetti M, Ceballos G, Isaac NJB, Collen B. 2014. Defaunation in the Anthropocene. Science 345, 401–406. ( 10.1126/science.1251817) [DOI] [PubMed] [Google Scholar]
- 5.Lorenzen ED, et al. 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479, 359–364. ( 10.1038/nature10574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper A, Turney C, Hughen KA, Brook BW, McDonald HG, Bradshaw CJA. 2015. Abrupt warming events drove Late Pleistocene Holarctic megafaunal turnover. Science 349, 602–606. ( 10.1126/science.aac4315) [DOI] [PubMed] [Google Scholar]
- 7.Frankham R. 2005. Genetics and extinction. Biol. Conserv. 126, 131–140. ( 10.1016/j.biocon.2005.05.002) [DOI] [Google Scholar]
- 8.Frankham R. 1996. Relationship of genetic variation to population size in wildlife. Conserv. Biol. 10, 1500–1508. ( 10.1046/j.1523-1739.1996.10061500.x) [DOI] [Google Scholar]
- 9.Frankham R. 1997. Do island populations have less genetic variation than mainland populations? Heredity 78, 311–327. ( 10.1038/sj.hdy.6880980) [DOI] [PubMed] [Google Scholar]
- 10.Palkopoulou E, et al. 2015. Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr. Biol. 25, 1395–1400. ( 10.1016/j.cub.2015.04.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers RL, Slatkin M. 2017. Excess of genomic defects in a woolly mammoth on Wrangel island. PLoS Genet. 13, e1006601 ( 10.1371/journal.pgen.1006601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Valk T, Díez-del-Molino D, Marques-Bonet T, Guschanski K, Dalén L.. 2019. Historical genomes reveal the genomic consequences of recent population decline in eastern gorillas. Curr. Biol. 29, 165–170.e6. ( 10.1016/j.cub.2018.11.055) [DOI] [PubMed] [Google Scholar]
- 13.Feng S, et al. 2019. The genomic footprints of the fall and recovery of the crested ibis. Curr. Biol. 29, 340–349.e7. ( 10.1016/j.cub.2018.12.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mays HL, et al. 2018. Genomic analysis of demographic history and ecological niche modeling in the endangered Sumatran rhinoceros Dicerorhinus sumatrensis. Curr. Biol. 28, 70–76. ( 10.1016/j.cub.2017.11.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadachowska-Brzyska K, Li C, Smeds L, Zhang G, Ellegren H. 2015. Temporal dynamics of avian populations during Pleistocene revealed by whole-genome sequences. Curr. Biol. 25, 1375–1380. ( 10.1016/j.cub.2015.03.047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankham R. 1998. Inbreeding and extinction: island populations. Conserv. Biol. 12, 665–675. ( 10.1046/J.1523-1739.1998.96456.X) [DOI] [Google Scholar]
- 17.Tershy BR, Shen KW, Newton KM, Holmes ND, Croll DA. 2015. The importance of islands for the protection of biological and linguistic diversity. Bioscience 65, 592–597. ( 10.1093/biosci/biv031) [DOI] [Google Scholar]
- 18.Blackburn TM, Cassey P, Duncan RP, Evans KL, Gaston KJ. 2004. Avian extinction and mammalian introductions on oceanic islands. Science 305, 1955–1958. ( 10.1126/science.1101617) [DOI] [PubMed] [Google Scholar]
- 19.Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH. 2008. Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc. Natl Acad. Sci. USA 105, 7676–7680. ( 10.1073/pnas.0801507105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan RP, Blackburn TM. 2004. Extinction and endemism in the New Zealand avifauna. Glob. Ecol. Biogeogr. 13, 509–517. ( 10.1111/j.1466-822X.2004.00132.x) [DOI] [Google Scholar]
- 21.Holdaway RN, Allentoft ME, Jacomb C, Oskam CL, Beavan NR, Bunce M. 2014. An extremely low-density human population exterminated New Zealand moa. Nat. Commun. 5, 5436 ( 10.1038/ncomms6436) [DOI] [PubMed] [Google Scholar]
- 22.Boessenkool S, Austin JJ, Worthy TH, Scofield P, Cooper A, Seddon PJ, Waters JM. 2009. Relict or colonizer? Extinction and range expansion of penguins in southern New Zealand. Proc. R. Soc. B 276, 815–821. ( 10.1098/rspb.2008.1246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dussex N, Rawlence NJ, Robertson BC. 2015. Ancient and contemporary DNA reveal a pre-human decline but no population bottleneck associated with recent human persecution in the kea (Nestor notabilis). PLoS ONE 10, e0118522 ( 10.1371/journal.pone.0118522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamieson IG, Wallis GP, Briskie JV. 2006. Inbreeding and endangered species management: is New Zealand out of step with the rest of the world? Conserv. Biol. 20, 38–47. ( 10.1111/J.1523-1739.2006.00282.X) [DOI] [PubMed] [Google Scholar]
- 25.Barker FK, Cibois A, Schikler P, Feinstein J, Cracraft J. 2004. Phylogeny and diversification of the largest avian radiation. Proc. Natl Acad. Sci. USA 101, 11 040–11 045. ( 10.1073/pnas.0401892101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepherd LD, Lambert DM. 2007. The relationships and origins of the New Zealand wattlebirds (Passeriformes, Callaeatidae) from DNA sequence analyses. Mol. Phylogenet. Evol. 43, 480–492. ( 10.1016/j.ympev.2006.12.008) [DOI] [PubMed] [Google Scholar]
- 27.Heather B, Robertson H, Onley D. 2005. The field guide to the birds of New Zealand. Auckland, New Zealand: Viking. [Google Scholar]
- 28.Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, pdb.prot5448 ( 10.1101/pdb.prot5448) [DOI] [PubMed] [Google Scholar]
- 29.Knapp M, Clarke AC, Horsburgh KA, Matisoo-Smith EA. 2012. Setting the stage—building and working in an ancient DNA laboratory. Ann. Anat. 194, 3–6. ( 10.1016/j.aanat.2011.03.008) [DOI] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595. ( 10.1093/bioinformatics/btp698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496. ( 10.1038/nature10231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haubold B, Pfaffelhuber P, Lynch M. 2010. MlRho—a program for estimating the population mutation and recombination rates from shotgun-sequenced diploid genomes. Mol. Ecol. 19, 277–284. ( 10.1111/j.1365-294X.2009.04482.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. ( 10.1086/519795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Double M, Murphy S. 2000. Genetic variation within and among populations of North Island kokako. Science and Research Internal Report 176. Wellington, New Zealand: Department of Conservation.
- 35.Armstrong DP, Davidson RS, Perrott JK, Roygard J, Buchanan L. 2005. Density-dependent population growth in a reintroduced population of North Island saddlebacks. J. Anim. Ecol. 74, 160–170. ( 10.1111/j.1365-2656.2004.00908.x) [DOI] [Google Scholar]
- 36.Smeds L, Qvarnström A, Ellegren H. 2016. Direct estimate of the rate of germline mutation in a bird. Genome Res. 26, 1211–1218. ( 10.1101/gr.204669.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang G, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1321. ( 10.1126/science.1251385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alloway BV, et al. 2007. Towards a climate event stratigraphy for New Zealand over the past 30 000 years (NZ-INTIMATE project). J. Quat. Sci. 22, 9–35. ( 10.1002/jqs.1079) [DOI] [Google Scholar]
- 39.Mazet O, Rodríguez W, Grusea S, Boitard S, Chikhi L. 2016. On the importance of being structured: instantaneous coalescence rates and human evolution-lessons for ancestral population size inference? Heredity 116, 362–371. ( 10.1038/hdy.2015.104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGlone MS, Newnham RM, Moar NT. 2010. The vegetation cover of New Zealand during the Last Glacial Maximum: do pollen records under-represent woody vegetation? Terra Aust. 32, 49–68. ( 10.22459/ta32.11.2010.04) [DOI] [Google Scholar]
- 41.McGlone MS. 1995. Lateglacial landscape and vegetation change and the younger dryas climatic oscillation in New Zealand. Quat. Sci. Rev. 14, 867–881. ( 10.1016/0277-3791(95)00068-2) [DOI] [Google Scholar]
- 42.Newnham RM, Lowe DJ, Williams PW. 1999. Quaternary environmental change in New Zealand: a review. Prog. Phys. Geogr. 23, 567–610. ( 10.1177/030913339902300406) [DOI] [Google Scholar]
- 43.Trewick SA, Wallis GP. 2001. Bridging the ‘beech-gap’: New Zealand invertebrate phylogeography implicates Pleistocene glaciation and Pliocene isolation. Evolution 55, 2170–2180. ( 10.1111/j.0014-3820.2001.tb00733.x) [DOI] [PubMed] [Google Scholar]
- 44.Wallis GP, Trewick SA. 2009. New Zealand phylogeography: evolution on a small continent. Mol. Ecol. 18, 3548–3580. ( 10.1111/j.1365-294X.2009.04294.x) [DOI] [PubMed] [Google Scholar]
- 45.Worthy TH, Holdaway RN. 1994. Quaternary fossil faunas from caves in Takaka Valley and on Takaka Hill, northwest Nelson, South Island, New Zealand. J. R. Soc. New Zeal. 24, 297–391. ( 10.1080/03014223.1994.9517474) [DOI] [Google Scholar]
- 46.Nadachowska-Brzyska K, Burri R, Smeds L, Ellegren H. 2016. PSMC analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula flycatchers. Mol. Ecol. 25, 1058–1072. ( 10.1111/mec.13540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Worthy TH, Holdaway RN. 1996. Quaternary fossil faunas, overlapping taphonomies, and palaeofaunal reconstruction in North Canterbury, South Island, New Zealand. J. R. Soc. New Zeal. 26, 275–361. ( 10.1080/03014223.1996.9517514) [DOI] [Google Scholar]
- 48.Worthy TH, Holdaway RN. 1995. Quaternary fossil faunas from caves on Mt Cookson, North Canterbury, South Island, New Zealand. J. R. Soc. New Zeal. 25, 333–370. ( 10.1080/03014223.1995.9517494) [DOI] [Google Scholar]
- 49.Lambert DM, Shepherd LD, Huynen L, Beans-Picón G, Walter GH, Millar CD. 2009. The molecular ecology of the extinct New Zealand huia. PLoS ONE 4, 1–10. ( 10.1371/journal.pone.0008019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.IUCN. 2015. IUCN Red List of Threatened Species. http://www.iucnredlist.org (accessed 27 July 2015).
- 51.Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. 2012. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 91, 275–292. ( 10.1016/j.ajhg.2012.06.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McWethy DB, Whitlock C, Wilmshurst JM, McGlone MS, Li X. 2009. Rapid deforestation of South Island, New Zealand, by early Polynesian fires. The Holocene 19, 883–897. ( 10.1177/0959683609336563) [DOI] [Google Scholar]
- 53.McWethy DB, et al. 2010. Rapid landscape transformation in South Island, New Zealand, following initial Polynesian settlement. Proc. Natl Acad. Sci. USA 107, 21 343–21 348. ( 10.1073/pnas.1011801107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perry GLW, Wilmshurst JM, McGlone MS, McWethy DB, Whitlock C. 2012. Explaining fire-driven landscape transformation during the Initial Burning Period of New Zealand's prehistory. Glob. Chang. Biol. 18, 1609–1621. ( 10.1111/j.1365-2486.2011.02631.x) [DOI] [Google Scholar]
- 55.Díez-del-Molino D, Sánchez-Barreiro F, Barnes I, Gilbert MTP, Dalén L. 2017. Quantifying temporal genomic erosion in endangered species. Trends Ecol. Evol. 33, 1–10. ( 10.1016/j.tree.2017.12.002) [DOI] [PubMed] [Google Scholar]
- 56.Dussex N, von Seth J, Robertson BC, Dalén L.. 2018. Full mitogenomes in the critically endangered kākāpō reveal major post-glacial and anthropogenic effects on neutral genetic diversity. Genes 9, 220 ( 10.3390/genes9040220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bergner LM, Dussex N, Jamieson IG, Robertson BC. 2016. European colonization, not Polynesian arrival, impacted population size and genetic diversity in the critically endangered New Zealand kākāpō. J. Hered. 107, 593–602. ( 10.1093/jhered/esw065) [DOI] [PubMed] [Google Scholar]
- 58.Sutton JT, Robertson BC, Jamieson IG. 2015. MHC variation reflects the bottleneck histories of New Zealand passerines. Mol. Ecol. 24, 362–373. ( 10.1111/mec.13039) [DOI] [PubMed] [Google Scholar]
- 59.Taylor SS, Jamieson IG, Wallis GP. 2007. Historic and contemporary levels of genetic variation in two New Zealand passerines with different histories of decline. J. Evol. Biol. 20, 2035–2047. ( 10.1111/j.1420-9101.2007.01362.x) [DOI] [PubMed] [Google Scholar]
- 60.Dussex N, von Seth J, Knapp M, Kardailsky O, Robertson BC, Dalén L.. 2019. Data from: Complete genomes of two extinct New Zealand passerines show responses to climate fluctuations but no evidence for genomic erosion prior to extinction ENA Digital Repository (PRJEB33922). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Dussex N, von Seth J, Knapp M, Kardailsky O, Robertson BC, Dalén L.. 2019. Data from: Complete genomes of two extinct New Zealand passerines show responses to climate fluctuations but no evidence for genomic erosion prior to extinction ENA Digital Repository (PRJEB33922). [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Raw fastq reads are deposited at the European Nucleotide Archive (ENA), accession number (PRJEB33922) [60].