Abstract
Nectar may contain many secondary metabolites that are commonly toxic and bitter-tasting. It has been hypothesized that such bitter-tasting secondary metabolites might keep the nectar exclusive to only a few pollinators. To test this hypothesis, we examined functional changes of bitter taste receptor genes (Tas2rs) in a species of nectar-feeding bird (Anna's hummingbird) by comparing these genes with those from two closely related insect-feeding species (chimney swift and chuck-will's widow). We previously identified a larger number of Tas2rs in the hummingbird than in its close insectivorous relatives. In the present study, we demonstrate higher sensitivity and new functions in the hummingbird Tas2r gene copies generated by a lineage-specific duplication, which has been shaped by positive selection. These results suggest that the bitter taste may lead to increased sensitivities and specialized abilities of the hummingbird to detect bitter-tasting nectar. Moreover, this study potentially supports the hypothesis that bitter-tasting nectar may have been specialized for some pollinators, thus enforcing plant–pollinator mutualism.
Keywords: Tas2r, bitter taste, pollinator, nectar, gene duplication
1. Introduction
Bitterness is one of the five basic tastes in vertebrates, which typically functions as a natural defence to prevent the consumption of bitter-tasting toxins such as plant secondary metabolites and insect defensive secretions [1]. Bitter taste in vertebrates is mediated by a group of G protein-coupled receptors encoded by the type 2 taste receptor genes (Tas2rs) [2], the number of which ranges from 0 in the dolphin to 51 in the frog [3]. In general, the number of putatively functional Tas2rs in a vertebrate species is positively correlated with the abundance of potential toxins in its diet, supporting the hypothesis that dietary toxins have shaped the diversity of the Tas2r gene repertoire in vertebrates [3,4]. Although differences in feeding ecology could partially explain changes in Tas2r gene number in vertebrates, additional factors must be involved, because mismatches between feeding ecology and taste receptor evolution have also been identified [5–7].
Plant–pollinator mutualisms describe interactions whereby pollinators acquire food, and flowers achieve higher reproductive success through the spread of pollen by pollinators; such interactions are prevalent in nature [8]. Nectar is a sugar-rich liquid excreted from nectaries, and may contain many secondary metabolites that are commonly toxic and bitter-tasting, as a form of defence to some floral visitors [9,10]. Current techniques are available for the detection of very small traces of bitter-tasting compounds in nectar [11], and a number of studies have demonstrated that bitter-tasting metabolites are common in nectar, such as alkaloids, phenolics and non-protein amino acids [10]. One hypothesis to explain the purpose of such bad-tasting compounds is that these bitter-tasting secondary metabolites might keep the nectar exclusive to only a few pollinators [11,12]. To test this hypothesis, we examined functional changes of bitter taste receptor genes (Tas2rs) in a species of nectar-feeding bird (Anna's hummingbird, Calypte anna) and compared them to those of two closely related insect-feeding species (chimney swift, Chaetura pelagica, and chuck-will's widow, Caprimulgus carolinensis) [13].
Following the release of avian genomes [13], we previously identified all complete and intact Tas2r sequences of 48 bird species [4]. A species-specific Tas2r gene duplication (n = 5) was found to have occurred in the Anna's hummingbird, which carries a larger Tas2r gene repertoire (n = 6) compared with its two close relatives, the chimney swift (n = 4) and the chuck-will's widow (n = 3). Both of these species are non-pollinators that mostly feed on insects [4]. In the present study, we conducted selective pressure analysis and a cell-based functional assay to test (i) whether Tas2r copies generated by the hummingbird-specific duplication have undergone positive selection and (ii) if these copies produce functional divergence.
2. Material and methods
(a). Sequence data
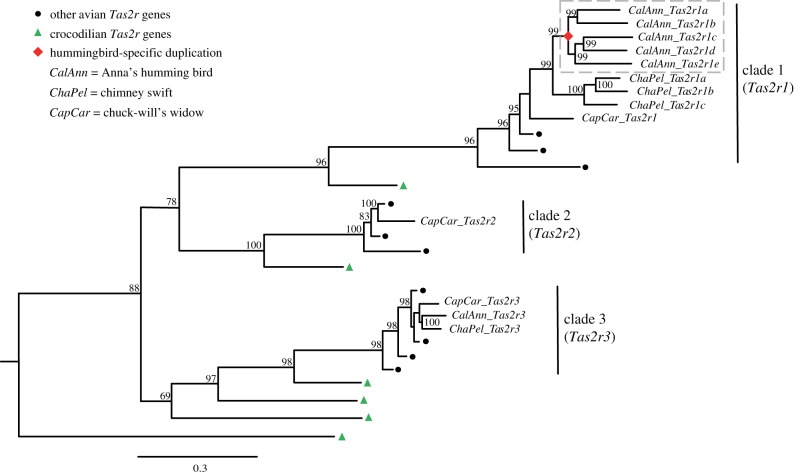
All complete and intact Tas2rs from birds were taken from our previous work [4]. The simplified tree of avian Tas2rs (figure 1), which highlighted the evolutionary history of Tas2rs from the hummingbird and its two close relatives (chimney swift and chuck-will's widow), was modified from figure 2 of our earlier publication [4]. For simplicity, we renamed all 13 Tas2rs from the three bird species studied here in order of appearance (figure 1).
Figure 1.
The simplified Bayesian gene tree of bird Tas2rs. Node support values represent Bayesian posterior probabilities shown as percentages, among which values lower than 50% were not shown. (Online version in colour.)
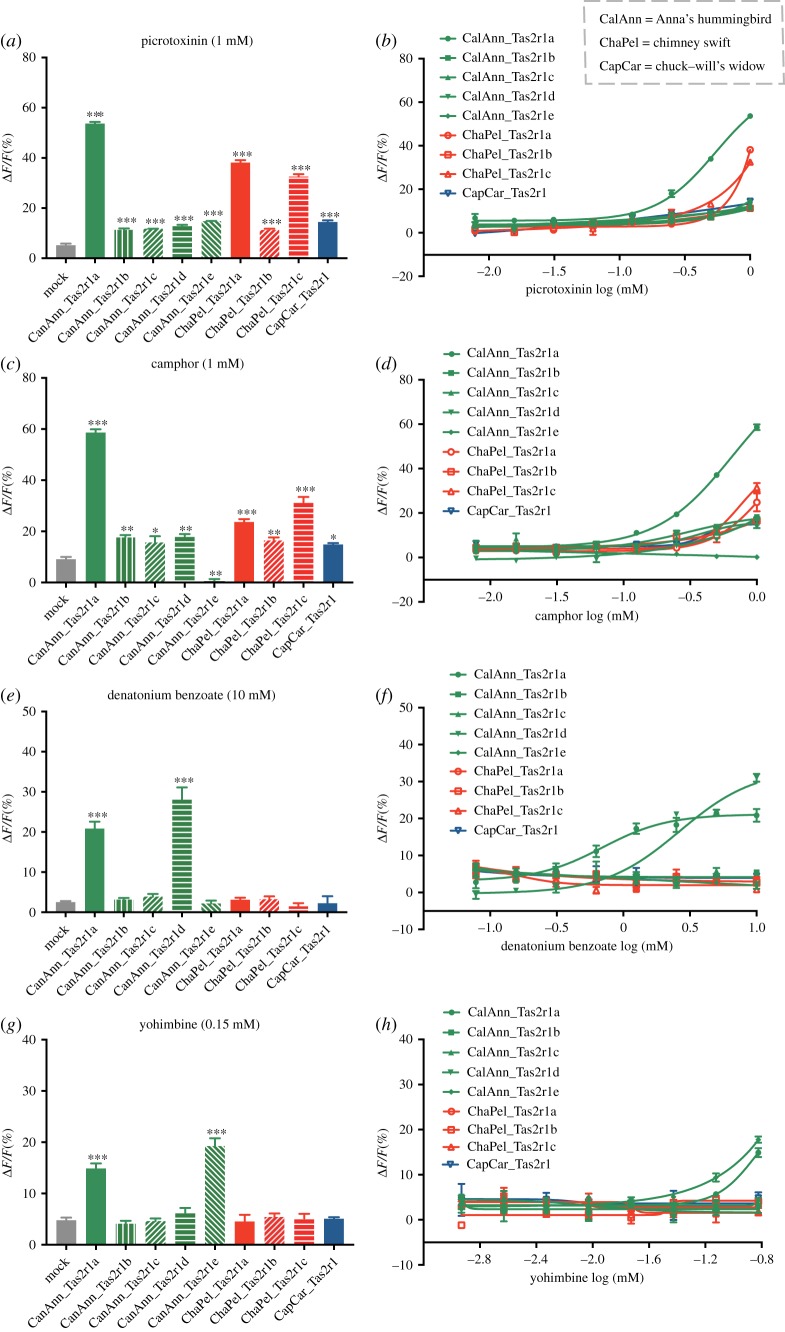
Figure 2.
Functional divergence of Tas2r1 receptors in Anna's hummingbird and its two close relatives. HEK293 cells transiently transfected with an avian Tas2r receptor and a Gα16-gust44 were assayed for their responses to four compounds. (a,c,e,g) Quantitative analysis of responses of avian Tas2r1 receptors to picrotoxinin (1 mM), camphor (1 mM), denatonium benzoate (10 mM) and yohimbine (0.15 mM) (mean ± s.e., *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA). (b,d,f,h) Dose-dependent responses of avian Tas2r1 receptors to picrotoxinin, camphor, denatonium benzoate and yohimbine. All data were fitted using GraphPad Prism 7.
(b). Selective pressure analysis
To test whether the hummingbird-specific gene duplication has undergone positive selection, we estimated the ratio (ω) of non-synonymous to synonymous substitutions (dN/dS) using the Codeml program in PAML [14]. The improved branch-site model [15] was used to detect whether positive selection has affected additional sites along with the hummingbird-specific gene duplication. Thus, the foreground branches were set as all branches connecting the hummingbird (alternative model); the corresponding null model was the same as the alternative model, except ω of the foreground branches was fixed at 1 [15,16]. The gene tree used for the selection test was taken from our previous study [4], but all genes from clades 2 and 3 (figure 1) were excluded, due to high sequence divergence between clades. For comparison, we additionally used two pairs of site-specific models in PAML to examine the five copies of hummingbird Tas1rs in clade 1 (figure 1). The first pair includes M1a and M2a; M1a assumes two classes of sites, one is constrained by 0 < ω < 1 and the other is fixed at ω = 1; M2a is an extension of M1a, with an extra class of sites allowing ω > 1. The second pair includes M8a and M8; M8a assumes a beta distribution for ω among sites with 0 < ω < 1, but allows an extra class of sites to have ω = 1; M8 is same to M8a, but allows the extra class of sites to have ω > 1 [17]. Likelihood-ratio tests were used to identify significant evidence of positive selection by comparing two competing models [18].
(c). Functional assays
Information on the chemicals used in this study is presented in electronic supplementary material, table S1. Our cell-based functional assays were carried out as previously described [19,20], see also supplementary methods in the electronic supplementary material.
3. Results
(a). Tas2r phylogeny and nomenclature
All avian Tas2rs formed three major clades in the gene tree (figure 1) [4]. For the three bird species studied here, clade 1 contained the most genes: five in the hummingbird, three in the chimney swift and one in the chuck-will's widow. Clade 2 included only one gene (chuck-will's widow), and clade 3 consisted of three genes, one from each of the three species (figure 1). Gene duplication events were observed in clade 1 only: five copies in the hummingbird have been generated at the origin of this species, while three copies have occurred at the origin of the chimney swift (figure 1). For convenience, we renamed genes in clade 1 as Tas2r1, those in clade 2 as Tas2r2 and those in clade 3 as Tas2r3; multiple copies from one species in clade 1 were named alphabetically in order of appearance in the tree (figure 1). For clarity, the renamed gene sequences and their previous names are listed in electronic supplementary material, dataset S1.
(b). Positive selection on duplicated Tas2r genes
To test whether Tas2r1 duplicates specific to the hummingbird have undergone positive selection, we used the improved branch-site model [14]. We identified a small proportion of sites (5.7%) with a signature of positive selection (ω = 6.226, p = 1.55 × 10−6; electronic supplementary material, table S2) in the dataset consisting of all 48 bird species from clade 1 (figure 1). Furthermore, seven positively selected sites were detected by this test (electronic supplementary material, table S2). In addition, the two site models M2a and M8 allowing positive selection are significantly different from M1a and M8a, respectively, suggesting that positive selection must have acted on the hummingbird Tas2rs (electronic supplementary material, table S3). M2a identified 10 positively selected sites, which are a subset of sites detected by M8 (electronic supplementary material, table S3). Notably, six sites that are putatively under positive selection were identified by both site models (M2a and M8) and the branch-site model (electronic supplementary material, tables S2 and S3), and four of them (91K, 92I, 181L and 271K) are located in extracellular loops that may mediate ligand recognition and receptor activation (electronic supplementary material, figure S1) [21].
(c). Functional divergence of duplicated Tas2r genes
To determine whether duplication events of avian Tas2rs were accompanied by functional divergence, we conducted cell-based functional assays on all Tas2r1 receptors in clade 1 (figure 1) from the hummingbird and its two close relatives. We expressed all nine Tas2r1 receptors individually in HEK293 cells (PeakRapid) by transiently transfecting a Tas2r1 construct along with a coupling chimeric G protein, Gα16-gust44. We tested these Tas2r1 receptors for their responses toward 24 commercially available bitter tasting compounds that have been shown to activate human Tas2r receptors (electronic supplementary material, table S1) [22]. Activation of each avian Tas2r1 receptor was monitored by their relative fluorescence changes (ΔF/F). Mock-transfected cells were used as negative controls.
After screening all of the 24 bitter compounds (electronic supplementary material, table S1), we found that various Tas2r1 receptors differed in the number of bitter compounds that elicited a response (electronic supplementary material, table S4). Specifically, some receptors seemed to be broadly tuned, such as CalAnn_Tas2r1a and CapCar_Tas2r1, which showed relatively wide-range responses toward six and five compounds, respectively. On the other hand, other receptors appeared to be activated by a narrow range of tuning (electronic supplementary material, figures S2 and S3). Based on the chemical screening results, we found that all nine of the Tas2r1 receptors tested were able to recognize picrotoxinin, with different degrees of sensitivity; CalAnn_Tas2r1a showed the highest level of response intensity (figure 2a,b). CalAnn_Tas2r1a also appeared to be the most sensitive, followed by ChaPel_Tas2r1a and ChaPel_Tas2r1c, while the others showed relatively low sensitivities (figure 2b). All of the receptors were also triggered by camphor, with the exception of CalAnn_Tas2r1e, which showed no response at any concentrations tested. CalAnn_Tas2r1a seemed to have the highest sensitivity to camphor, whereas the sensitivities of ChaPel_Tas2r1c and ChaPel_Tas2r1a are the second and third highest, respectively (figure 2c,d). Overall, functional differences were apparent among the five copies of Anna's hummingbird Tas2r1, and also between hummingbird Tas2r1 copies and their outgroups, reflected by their different degrees of sensitivity in response to the examined bitter compounds.
Intriguingly, both CalAnn_Tas2r1a and CalAnn_Tas2r1d were responsive to denatonium benzoate, while other receptors showed no response at any concentrations tested (figure 2e,f). Similarly, yohimbine only elicited a response in the two receptors (CalAnn_Tas2r1a and CalAnn_Tas2r1e), but did not activate others (figure 2g,h). These results suggest that the gene duplication events in the hummingbird may have led to new gene functions, for example, the detection of denatonium benzoate and yohimbine. To test whether this potentially new function is also displayed in non-Tas2r1 and non-hummingbird receptors, we additionally performed calcium mobilization assays for CalAnn_Tas2r3, CapCar_Tas2r2, CapCar_Tas2r3 and ChaPel_Tas2r3 (figure 1). Our results confirmed that none of these receptors were responsive to these two bitter compounds at any of the concentrations tested (electronic supplementary material, figure S4). Therefore, we conclude that the ability to detect denatonium benzoate and yohimbine is likely a novel function specific to the hummingbird Tas2r1 receptors.
4. Discussion
Several hypotheses about the possible functions of toxic and bitter-tasting nectar have been proposed. One of which is the pollinator fidelity hypothesis, which assumes that specialized pollinators would be less repelled by toxic nectar than generalists [11,12]. In other words, this hypothesis argues that toxic nectar may maintain exclusivity to specific pollinators. In this study, we examined functional changes of bitter taste receptor genes (Tas2rs) in a nectar-feeding bird by comparing them with two closely related insect-feeding birds. Our results showed different levels of sensitivity and new functions in the hummingbird Tas2r gene copies, which may have been shaped by positive selection. These results suggest that the bitter taste may confer increased sensitivities and specialized abilities of the hummingbird to detect bitter-tasting nectar. In addition, this study potentially supports the hypothesis that toxic and bitter-tasting nectar may have played an important role in maintaining exclusivity to specific pollinators.
Birds generally have fewer Tas2rs compared to most other vertebrates [4]. In fact, some birds do not even possess any functional Tas2rs [23]. It has been hypothesized that herbivorous and insectivorous birds may require a higher number of Tas2rs than carnivorous birds that feed on non-insect animals, because plant products contain more toxins than animal tissues, and insects release toxic defensive secretions [4]. However, no significant differences have been found in the number of Tas2rs between nectarivorous and insectivorous birds [4]. Thus, it is surprising that the Anna's hummingbird—which mainly feeds on nectar—has a relatively higher number of Tas2rs than its two close insectivorous relatives. This increased number was the result of a lineage-specific duplication, leading to the production of five Tas2r copies (figure 1) [4]. The five Tas2r copies showed varying sensitivities to picrotoxinin and camphor, as well as unique detection of denatonium benzoate and yohimbine, as demonstrated by our cell-based assays (figure 2). Intriguingly, Tas2r copies of the insectivorous chimney swift also showed relatively higher sensitivities to bitterness (figure 2b,d), which could be linked to the bitter-tasting defensive secretions in insects [24]. As a result, we cannot rule out the possibility that high sensitivities to bitterness are also needed in insectivorous birds. With the exception of denatonium benzoate, the three bitter compounds (picrotoxinin, camphor and yohimbine) are all naturally occurring chemicals extracted from plants. However, none of them are likely to exist in nectar other than camphor, which was isolated from the floral component [25]. Thus, future in-depth studies of bitter compounds in nectar distributed around hummingbird habitats would provide a better understanding of how hummingbirds use bitter taste to identify nectar in nature. Moreover, all the 24 bitter compounds tested in our assays have been verified to activate human Tas2rs [22], but only six, five and two are able to activate the hummingbird, chuck-will's widow, and chimney swift Tas2rs, respectively (figure 2 and electronic supplementary material, table S4). Thus, there is an apparent bias in bitter compounds that activate bird and human Tas2rs. Despite such limitations, the enhanced sensitivities to some bitter compounds and novel functions to detect new bitter compounds suggest that the hummingbird may use the bitter taste to detect nectar that have bitter-tasting secondary metabolites. Although it is long believed that the bitter taste is a natural defence against the ingestion of bitter-tasting and potentially toxic foods in the wild [1], the functional enhancement and expansion of the hummingbird bitter receptors indicates that this nectar-feeding bird may prefer some bitter compounds compared to their insect-feeding relatives. This hypothesis is supported by a previous study, which showed that hummingbirds prefer nectar with a low-nicotine concentration [26]. Similarly, low-toxin nectar preference was also found in other pollinators such as honeybees [27]. As such, the bitter taste may be useful to help the hummingbird recognize specific and toxic nectar which non-pollinators may not be able to detect, thus enforcing plant–pollinator mutualism. It should be noted that our cell-based assays are in vitro tests, which cannot replace in vivo tests, although both types of tests remain a consistent trend on a qualitative level [28]. Moreover, the selection for bitterness in birds could be more complicated than the only relation with nectar. Regardless, our results should stimulate behavioural studies in the future to test whether nectar-feeding animals can prefer the bitter-tasting secondary metabolites in low concentrations from their nectar foods.
Supplementary Material
Acknowledgements
We thank Bing-Jun Wang for his excellent assistance with the plasmid construction.
Data accessibility
Tas2r gene sequences used in our cell-based functional assays are provided in electronic supplementary material, dataset S1.
Authors' contributions
H.Z. conceived and designed the research and experiments; Y.W. performed the experiments; Y.W., H.J. and H.Z. analysed the data; P.J. supervised the cell-based functional assays; and Y.W. and H.Z. wrote the manuscript. All authors edited and approved the final version of the manuscript and agree to be held accountable for the content therein.
Competing interests
We declare we have no competing interests.
Funding
This study was supported by National Natural Science Foundation of China (nos. 31672272 and 31722051).
References
- 1.Yarmolinsky DA, Zuker CS, Ryba NJP. 2009. Common sense about taste: from mammals to insects. Cell 139, 234–244. ( 10.1016/j.cell.2009.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. 2000. T2Rs function as bitter taste receptors. Cell 100, 703–711. ( 10.1016/s0092-8674(00)80706-0) [DOI] [PubMed] [Google Scholar]
- 3.Li D, Zhang J. 2014. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol. Biol. Evol. 31, 303–309. ( 10.1093/molbev/mst219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K, Zhao H. 2015. Birds generally carry a small repertoire of bitter taste receptor genes. Genome Biol. Evol. 7, 2705–2715. ( 10.1093/gbe/evv180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H, Zhang J. 2012. Mismatches between feeding ecology and taste receptor evolution: an inconvenient truth. Proc. Natl Acad. Sci. USA 109, E1464 ( 10.1073/pnas.1205205109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao H, Xu D, Zhang S, Zhang J. 2012. Genomic and genetic evidence for the loss of umami taste in bats. Genome Biol. Evol. 4, 73–79. ( 10.1093/gbe/evr126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng P, Zheng JS, Rossiter SJ, Wang D, Zhao H. 2014. Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol. Evol. 6, 1254–1265. ( 10.1093/gbe/evu095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kearns CA, Inouye DW, Waser NM. 1998. Endangered mutualisms—the conservation of plant–pollinator interactions. Annu. Rev. Ecol. Syst. 29, 83–112. ( 10.1146/annurev.ecolsys.29.1.83) [DOI] [Google Scholar]
- 9.Lüttge U. 1977. Nectar composition and membrane transport of sugars and amino acids: a review on the present state of nectar research. Apidologie 8, 305–319. ( 10.1051/apido:19770402) [DOI] [Google Scholar]
- 10.Baker HG. 1977. Non-sugar chemical constituents of nectar. Apidologie 8, 349–356. ( 10.1051/apido:19770405) [DOI] [Google Scholar]
- 11.Alder LS. 2001. The ecological significance of toxic nectar. Oikos 91, 409–420. ( 10.1034/j.1600-0706.2000.910301.x) [DOI] [Google Scholar]
- 12.Rhoades DF, Bergdahl JC. 1981. Adaptive significance of toxic nectar. Am. Nat. 117, 798–803. ( 10.1086/283765) [DOI] [Google Scholar]
- 13.Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1320–1331. ( 10.1126/science.1253451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479. ( 10.1093/molbev/msi237) [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917. ( 10.1093/oxfordjournals.molbev.a004148) [DOI] [PubMed] [Google Scholar]
- 17.Wong WS, Yang Z, Goldman N, Nielsen R. 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168, 1041–1051. ( 10.1534/genetics.104.031153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anisimova M, Bielawski JP, Yang Z. 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 18, 1585–1592. ( 10.1093/oxfordjournals.molbev.a003945) [DOI] [PubMed] [Google Scholar]
- 19.Lei W, Ravoninjohary A, Li X, Margolskee RF, Reed DR, Beauchamp GK, Jiang P. 2015. Functional analyses of bitter taste receptors in domestic cats (Felis catus). PLoS ONE 10, e0139670 ( 10.1371/journal.pone.0139670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiao H, Wang Y, Zhang L, Jiang P, Zhao H. 2018. Lineage-specific duplication and adaptive evolution of bitter taste receptor genes in bats. Mol. Ecol. 27, 4475–4488. ( 10.1111/mec.14873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peeters MC, van Westen GJ, Li Q, IJzerman AP. 2011. Importance of the extracellular loops in G protein-coupled receptors for ligand recognition and receptor activation. Trends Pharmacol. Sci. 32, 35–42. ( 10.1016/j.tips.2010.10.001) [DOI] [PubMed] [Google Scholar]
- 22.Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. 2010. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses 35, 157–170. ( 10.1093/chemse/bjp092) [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Li J, Zhang J. 2015. Molecular evidence for the loss of three basic tastes in penguins. Curr. Biol. 25, R141–R142. ( 10.1016/j.cub.2015.01.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Despres L, David JP, Gallet C. 2007. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 22, 298–307. ( 10.1016/j.tree.2007.02.010) [DOI] [PubMed] [Google Scholar]
- 25.Jerković I, Kuś PM. 2014. Terpenes in honey: occurrence, origin and their role as chemical biomarkers. RSC Adv. 4, 31 710–31 728. ( 10.1039/c4ra04791e) [DOI] [Google Scholar]
- 26.Kessler D, Bhattacharya S, Diezel C, Rothe E, Gase K, Schottner M, Baldwin IT. 2012. Unpredictability of nectar nicotine promotes outcrossing by hummingbirds in Nicotiana attenuata. Plant J. 71, 529–538. ( 10.1111/j.1365-313X.2012.05008.x) [DOI] [PubMed] [Google Scholar]
- 27.Thomson JD, Draguleasa MA, Tan MG. 2015. Flowers with caffeinated nectar receive more pollination. Arthropod Plant Interact. 9, 1–7. ( 10.1007/s11829-014-9350-z) [DOI] [Google Scholar]
- 28.Cheled-Shoval S, Behrens M, Korb A, Di Pizio A, Meyerhof W, Uni Z, Niv MY. 2017. From cell to beak: in-vitro and in-vivo characterization of chicken bitter taste thresholds. Molecules 22, 821 ( 10.3390/molecules22050821) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tas2r gene sequences used in our cell-based functional assays are provided in electronic supplementary material, dataset S1.