Abstract
Background:
In recent years, sepsis-induced acute respiratory distress syndrome (ARDS) has remained a major clinical challenge for patients in intensive care units. While some progress has been reported over the years, the pathogenesis of ARDS still needs to be further expounded.
Methods:
In the present study, gene set enrichment analysis, differentially expressed genes analysis, short time-series expression miner, protein–protein interaction (PPI) networks, module analysis, hypergeometric test, and functional enrichment analysis were performed in whole blood gene expression profiles of sepsis and induced-sepsis ARDS to explore the molecular mechanism of sepsis-induced ARDS.
Results:
Further dysregulated genes in the process evolving from healthy control through sepsis to sepsis-induced ARDS were identified and organized into 10 functional modules based on their PPI networks. These functional modules were significantly involved in cell cycle, ubiquitin mediated proteolysis, spliceosome, and other pathways. MYC, STAT3, LEF1, and BRCA1 were potential transcription factors (TFs) regulating these modules. A TF-module-pathway global regulation network was constructed. In particular, our findings suggest that MYC and STAT3 may be the key regulatory genes in the underlying dysfunction of sepsis-induced ARDS. Receiver operating characteristic curve analysis showed the core genes in the global regulation network may be biomarkers for sepsis or sepsis-induced ARDS.
Conclusions:
We found that MYC and STAT3 may be the key regulatory genes in the underlying dysfunction of sepsis-induced ARDS.
The reviews of this paper are available via the supplementary material section.
Keywords: acute respiratory distress syndrome, sepsis, sepsis-induced ARDS
Introduction
Sepsis is a type of fatal syndrome caused by the entrance of bacteria or fungi into the blood to infect the human immune and coagulation systems,1,2 with an estimated 30 million new cases and 6 million deaths in the world each year.3 One of the reasons for the high fatality rate of sepsis patients is that they tend to evolve into acute respiratory distress syndrome (ARDS). The incidence of sepsis-induced ARDS is still increasing,4 and has higher mortality rates than ARDS induced by other causes.5
To date, the mechanism of sepsis-induced ARDS has been studied and some progress has been made. Some neutrophil-related pathways may be involved in early sepsis-induced ARDS6; the α-AR signal transduction pathway also plays a role in ARDS7; and IQGAP1 may affect the pulmonary microvascular barrier, cell connection, and the cytoskeleton and related pathways involving in sepsis-induced ARDS.8 The above studies have deepened our understanding of the mechanism of sepsis-induced ARDS; however, only a few genes have been elucidated in a single study, and the mechanism of sepsis-induced ARDS has not been expounded comprehensively.
The present study identified further deregulated genes (FDGs) in the evolving process from healthy control through sepsis to sepsis-induced ARDS. According to these FDGs, a protein–protein interaction network was constructed and modular analysis performed. The prediction of potential transcription factors (TFs) regulating functional modules helps us observe the transcriptional regulation of sepsis-induced ARDS from the global level. We found that MYC and STAT3 may be the key regulatory genes in the underlying dysfunction of sepsis-induced ARDS.
Materials and methods
Data processing
We downloaded GSE32707 of the whole blood gene expression profiles from the website of the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/),9 which included 58 sepsis samples, 31 sepsis-induced ARDS samples, 21 systemic inflammatory response syndrome (SIRS) samples, and 34 normal controls. The normalizeBetweenArrays function in the limma package was used to normalize the gene expression profile. This gene expression profile was based on GPL10558.10 If a gene corresponds to multiple probes, the average expression value of these probes is the expression value of the gene; 21 SIRS samples were removed in the present study.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed using the normalized gene expression profile to explore the biological process (BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to sepsis and sepsis-induced ARDS.11 GSEA was carried out using JAVA software, which can be found on the official website (http://software.broadinstitute.org/gsea/index.jsp). Gene sets c5.bp.v6.2.symbols.gmt and c2.cp.kegg.v6.2.symbols.gmt were used as the reference gene sets.12 A nominal value of p < 0.05 was considered as statistically significant.
Differentially expressed gene analysis and short time-series expression miner
Compared with control samples, differentially expressed genes (DEGs) in sepsis samples and sepsis-induced samples were screened using the limma package in R. The fold changes (FCs) in expression of individual genes were calculated, and genes with |log2FC| > 1 and p adjusted by the false discovery rate (P.adj) < 0.05 were considered as significant. If a DEG was further upregulated or downregulated (|logFCsepsis-induced ARDS versus control| > |logFCsepsis versus control|) in sepsis-induced ARDS compared with sepsis, that gene was considered a FDG in the process evolving from healthy control through sepsis to sepsis-induced ARDS. FDGs could be organized into different clusters based on expression patterns using short time-series expression miner (STEM).13
Construction of protein–protein interaction networks and modular analysis
Based on the STRING database (https://string-db.org/),14 protein–protein interaction (PPI) networks of FDGs were constructed, and only interactions with a combined score >500 were retained. Cytoscape software was reserved for network visualization.15 The ClusterONE plug-in of Cytoscape software was used to perform modular analysis of the PPI network (Minimum = 30).16 The FDGs were organized into different functional modules using ClusterOne. The ClusterProfiler package17 in R was used to performed functional enrichment analysis for these functional modules. Functional enrichment analysis contained BP and KEGG pathway. p adjusted by the false discovery rate (P.adj) < 0.05 was considered as significant.
Prediction of potential TFs for modules by hypergeometric test
Based on the interaction of human TFs with their target genes in the TRRUST v2 database,18 the hypergeometric test was used to predict potential TFs regulating functional modules. The hypergeometric test was performed using the igraph package (https://igraph.org/r/) in R. A p value of <0.05 was considered as statistically significant. In addition, Pearson correlation between TF and its target gene expression was explored. A TF-module-pathway global transcriptional regulation network was constructed.
Identification of core genes and receiver operating characteristic curve analysis
In the TF-module-pathway global transcriptional regulation network, according to the semantic similarities of gene ontology (GO) terms used for gene annotation, we rank the gene inside the interactome by the average functional similarities between the gene and its interaction partners. Genes with a higher average functional similarity are considered as the more crucial genes.19 In the present study, 10 genes with largest average functional similarity, and the TFs, were considered as core genes in the TF-module-pathway global transcriptional regulatory network. The average functional similarities for genes were calculated using the GOsemsim package in R.20 In addition, receiver operating characteristic (ROC) curve analysis was carried out to access the diagnostic value of these genes for sepsis or sepsis-induced ARDS. ROC curve analysis was performed using the pROC package in R.21
Results
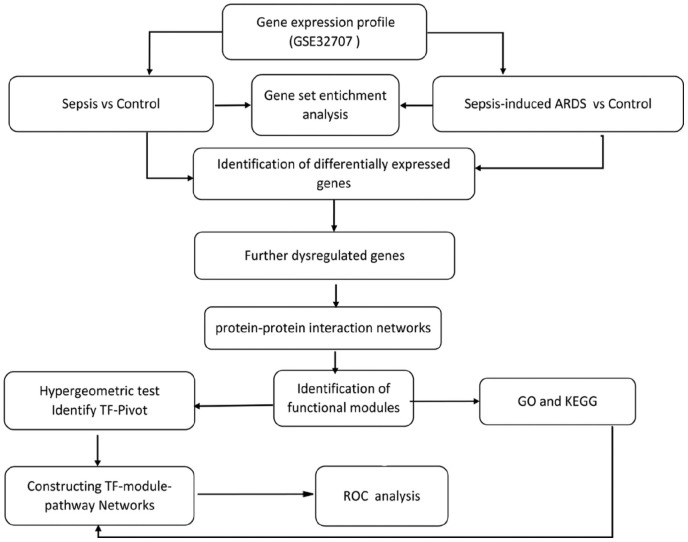
In the present study, GSEA was performed. FDGs were identified in the evolving process leading from healthy control through sepsis to sepsis-induced ARDS. A PPI network of FDGs was constructed and used in modular analysis. Functional enrichment analysis was performed for the functional modules. A hypergeometry test was performed to identify potential TFs of modules. A TF-module-pathway global transcriptional regulatory network was constructed. ROC curve analysis was used to access potential diagnostic markers (Figure 1).
Figure 1.
Flow chart of this study.
Sepsis and sepsis-induced ARDS are significantly enriched in pathogen-related biological processes and pathways
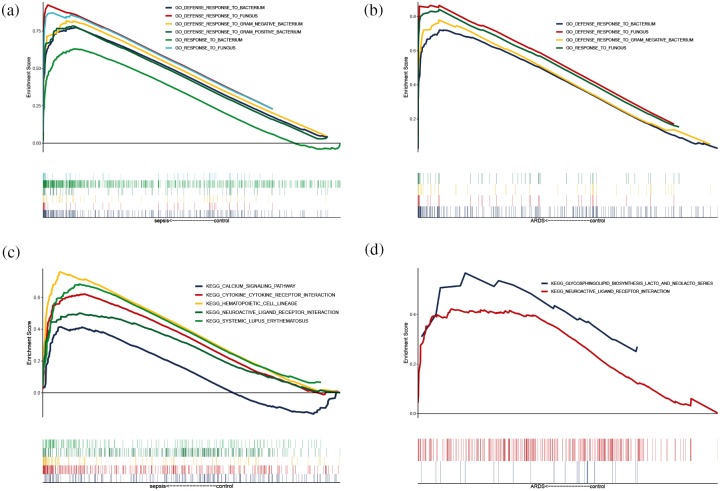
GSEA results for BP enrichment analysis suggested ‘Response to fungus’, ‘Defense response to fungus’, ‘Defense response to gram negative bacterium’, and ‘Defense response to bacterium’ were significantly enriched in the sepsis samples (Figure 2a), while ‘Response to fungus’, ‘Defense response to gram negative bacterium’, and ‘Defense response to bacterium’ were significantly enriched in the sepsis-induced ARDS samples (Figure 2b).
Figure 2.
GSEA results. (a) Biological processes significantly enriched in sepsis samples. (b) BPs significantly enriched in sepsis induced ARDS. (c) KEGG pathways significantly enriched in sepsis. (d) KEGG pathways significantly enriched in sepsis-induced ARDS.
ARDS, acute respiratory distress syndrome; BP, biological processes; GSEA, gene set enrichment analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
In the GSEA for KEGG pathway, ‘Calcium signaling pathway’, ‘Cytokine-cytokine receptor interaction’, ‘Hematopoietic cell lineage’, ‘Neuroactive ligand receptor interaction’, and ‘Systemic lupus erythematosus’ were significantly enriched in the sepsis samples (Figure 2c), while ‘Glycosphingolipid biosynthesis lacto and neolacto series’, and ‘Neuroactive ligand receptor interaction’ were significantly enriched in the sepsis induced-ARDS group (Figure 2d). These results indicated that, after the progress of sepsis leads to ARDS, the body’s reactions change, and these reactions can be reflected in whole blood gene expression patterns.
FDGs in sepsis-induced ARDS compared with sepsis
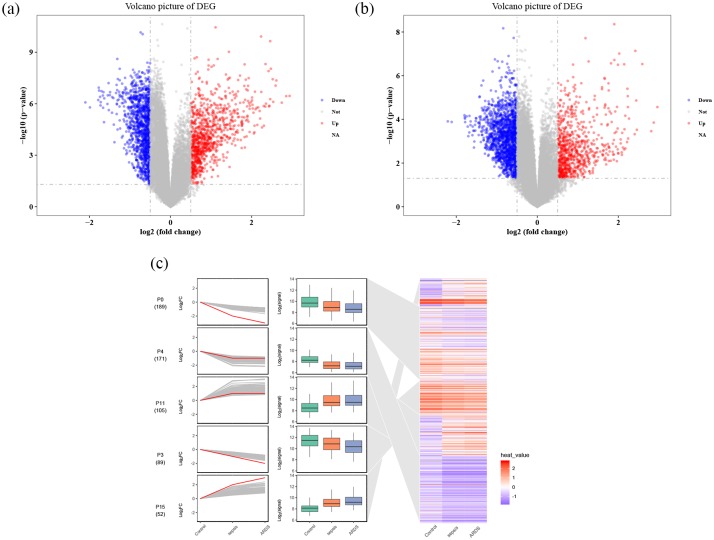
Compared with control samples, a total of 8354 DEGs in sepsis were identified, of which 5025 are downregulated and 3329 upregulated (Figure 3a). A total of 7859 DEGs, 4596 of which are downregulated and 3263 upregulated, were found in sepsis-induced ARDS compared with control samples (Figure 3b). Among the 5633 genes that were common DEGs in sepsis and sepsis-induced ARDS compared with controls, 3521 DEGs were identified as FDGs. STEM identified 15 gene profiles (clusters), of which 2 gene profiles (clusters) were significantly upregulated and three significantly downregulated in the process of evolving from healthy control through sepsis to sepsis-induced ARDS.
Figure 3.
DEG analysis and STEM. (a) DEGs in sepsis samples, red represents upregulated genes, blue represents downregulated genes, and gray represents no significantly DEGs. (b) DEGs in sepsis-induced ARDS samples. (c) STEM figure of DEGs, red is the fitted curve, which means gene cluster in sepsis-induced ARDS cut trend, and the blue area means gene cluster in the positions of the heat.
ARDS, acute respiratory distress syndrome; DEG, differentially expressed genes; STEM, short time-series expression miner.
Sepsis-induced ARDS may result from multiple functional modules
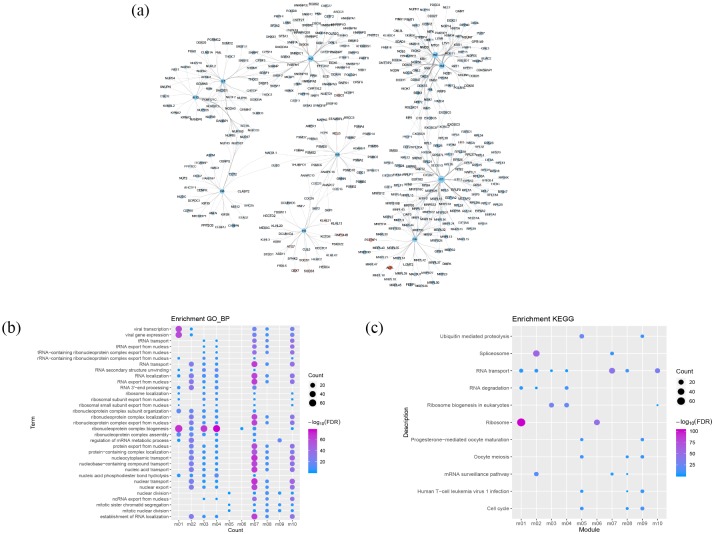
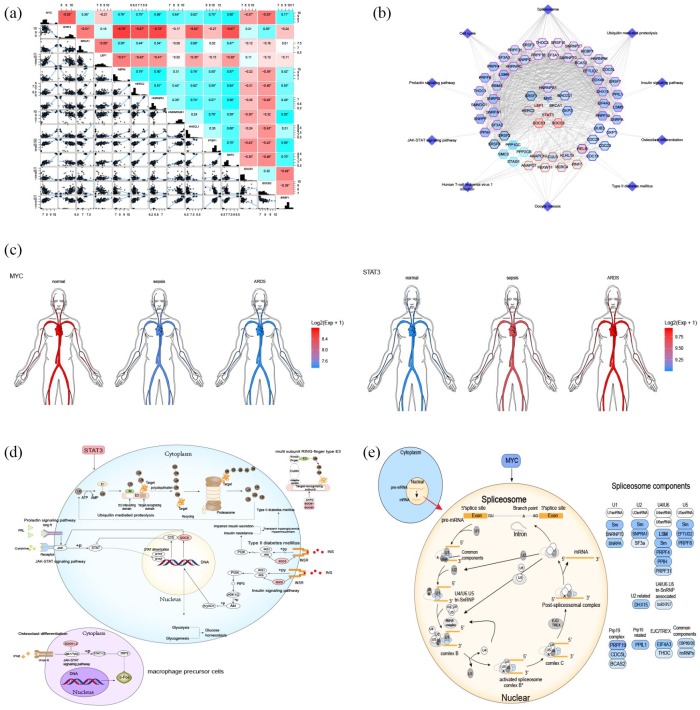
After constructing a PPI network with 3521 FDGs, 2060 nodes and 23,395 edges were obtained. A total of 435 module-genes formed 10 functional modules (Figure 4a) identified by ClusterONE. Enrichment analysis was carried out on these functional modules. More than four functional modules, such as viral transcription, viral gene expression, and ribonucleoprotein complex biogenesis, were significantly enriched in some BPs (Figure 4b). Two or more functional modules were involved in a few pathways, such as splicesome, RNA transport, cell cycle, and ubiquitin mediated proteolysis pathways. These may be key BPs and pathways in the process of evolving from healthy control through sepsis to sepsis-induced ARDS.
Figure 4.
Modular analysis of PPI networks and functional enrichment analysis. (a) Modules and module-related genes, the colors map for log2fold change of DEG. (b) BP with more than four functional modules significantly enriched. (c) KEGG pathways with more than one functional modules enriched.
BP, biological process; DEG, differentially expressed genes; PPI, protein–protein interaction; KEGG, Kyoto Encyclopedia of Genes and Genomes.
The global regulatory landscape of sepsis-induced ARDS
The results of hypergeometric tests suggested that MYC, STAT3, LEF1, and BRCA1 were potential TFs for modules. Correlation analysis (Figure 5a) confirmed that there was a significant correlation between these four TFs and their target genes. A TF-module-pathway network was constructed (Figure 5b), providing a preliminarily demonstration of the global transcriptional regulatory landscape of sepsis-induced ARDS. MYC was further downregulated and STAT3 was further upregulated in the process of evolving from healthy control through sepsis to sepsis-induced ARDS (Figure 5c). Six STAT3 pathways may be involved in sepsis-induced ARDS (Figure 5d), of which the Prolactin signaling pathway and the JAK-STAT signaling pathway are inhibited by upregulated suppressor of cytokine signaling-1 (SOCS). Osteoclast differentiation means that interferon can inhibit c-Fos proto-oncogene protein by binding with receptors and then entering cells via the JAK-STAT signaling pathway. However, since the JAK-STAT signaling pathway is inhibited, the inhibitory effect on c-Fos of osteoclast differentiation is also weakened. The insulin signaling pathway and SOCS in the rise of type II diabetes mellitus (T2DM) may also cause an imbalance of blood glucose levels and the aggravation of T2DM. This may be related to stress hyperglycemia.
Figure 5.
Potential key TFs in sepsis-induced ARDS. (a) TFs correlated with target genes; (b) Network diagram of TF-module-pathway related to sepsis-induced ARDS. The center of the network is TFs, the color of gene nodes represents log2fold change, and the color at the edge of gene nodes represents modules. (c) Expression of STAT3 and MYC in human circulation. (d) STAT3-related KEGG pathways. (e) MYC-related KEGG pathways.
ARDS, acute respiratory distress syndrome; KEGG, Kyoto Encyclopedia of Genes and Genomes; TFs, transcription factors.
It is worth mentioning that ubiquitin-mediated proteolysis is involved in the ubiquitination process, in which many types of E3 enzymes recognize target proteins. The genes involved in the TF-module-network play a role in multi-subunit ring-finger type E3 enzymes, which change the way substrates are transferred, thus playing a specific role in the deterioration of ARDS.
The spliceosome pathway, which is regulated by MYC, is used to adjust molecular transcription levels (Figure 5e). The spliceosome is not a simple, stable complex, but a dynamic family of particles that congregates on the mRNA precursor, helping to fold it into a conformation that enables transesterification. Downregulated MYC regulates the downregulation of five genes encoding small nuclear ribonucleoproteins U1, U2, U4, U5, and U6 in the splicing body, as well as several splicing-body-related proteins and common components. It causes abnormal levels of molecular transcription and inhibits mRNA synthesis. Our findings indicate that its effect on sepsis-induced ARDS progression may warrant further study.
The core genes of global landscape regulation can be used as potential biomarkers for sepsis or sepsis-induced ARDS
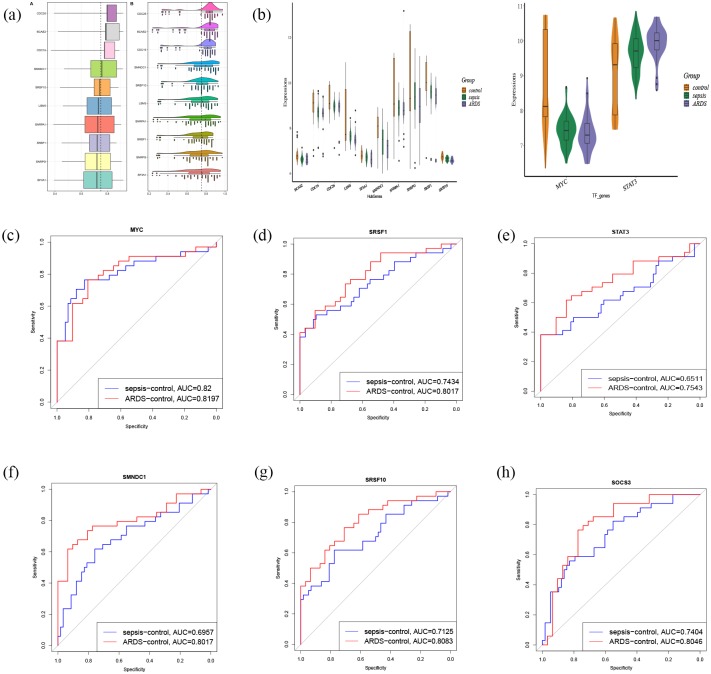
The 10 genes with the largest average functional similarity in the TF-module-pathway network are: CDC26, BCAS2, CDC16, SMNDC1, SRSF10, LSM5, SNRPA1, SRSF1, SNRPG, and SF3A1 (Figure 6a). All of these were further downregulated, and none of them were further upregulated, in the process of going from healthy control through sepsis to sepsis-induced ARDS (Figure 6b). ROC curve analysis suggests that MYC, SRSF1, STAT3, SMNPC1, SRSF10, and SOCS3 may be potential biomarkers (Figure 6c–h) for sepsis or sepsis-induced ARDS.
Figure 6.
Core genes and TFs and ROC curve analysis. (a) The 10 genes with the largest average functional similarity. (b) Expression of core genes and two TFs. (c–h) ROC curve analysis for 10 core genes and two TFs.
ROC, receiver operating characteristic; TFs, transcription factors.
Discussion
With limited improvement in the management of sepsis over the past decade,22 it is imperative to further elucidate the mechanisms of sepsis and sepsis-induced ARDS and to seek new treatments to combat these conditions. In the present study, based on the whole-blood gene expression profiles of sepsis and sepsis-induced ARDS, we identified FDGs in the process of evolving from healthy control through sepsis to sepsis-induced ARDS. The FDGs were organized into 10 functional modules based on their PPI networks. Enrichment analysis indicated that the modules were significantly involved in ubiquitin-mediated proteolysis, cell cycle, the spliceosome, and the insulin signaling pathway. Unsurprisingly, ubiquitin-mediated proteolysis has been reported to be associated with ARDS,23 while ribosome biogenesis in eukaryotes,24 and the spliceosome might be involved with sepsis.1 We also found that some pathways may also be associated with sepsis-induced ARDS, such as progesterone-mediated oocyte maturation, and the ribosome pathway. Based on PPI and modular analysis constructing FDGs, plus additional verification using STEM, some gene profiles (clusters) were confirmed to be significantly correlated with the sepsis-induced ARDS process. These results suggest that sepsis-induced ARDS causes dysregulation of multiple functional modules of the body, as reflected in the whole blood gene expression profile. Potential TFs of regulatory function modules were predicted, and a TF-module-pathway global transcriptional regulatory network was constructed to help reveal the molecular mechanism of sepsis-induced ARDS for further study. In addition, based on our constructed TF-module-pathway global transcriptional regulatory network, we found that the core genes in the network may be a potential marker for sepsis or sepsis-induced ARDS.
MYC, STAT3, LEF1, and BRCA1 were differentially expressed TFs in sepsis and sepsis-induced ARDS, while MYC and STAT3 were FDGs from healthy controls through sepsis to sepsis-induced ARDS. Of the STAT3 pathways involved, the prolactin signaling pathway regulates main biological functions such as reproduction, growth and development, and the immune response, while the JAK-STAT signaling pathway regulates the natural immune response, which is the main mechanism regulating various cytokines and growth factors,25 allowing the occurrence of disease if they are suppressed. Interestingly, multiple genes involved in ubiquitination are regulated by TFs, and we can conclude that protein ubiquitination plays an important role in sepsis-induced ARDS. Ubiquitination is crucial for physiological processes such as cell survival, differentiation, natural and adaptive immunity, etc.26 Ubiquitin-like proteins are involved in almost all cellular processes, including in the pathogenesis of disease in two different ways: increasing the production of disease promoters, and decreasing the production of disease inhibitors.27 In the cascade process of E1, E2, and E3, E3 plays a key role in the selection of substrates and the coordination of E2-ubiquitin and the binding of specific ubiquitin bonds to substrates. Different types of E3 enzymes can also mediate PPIs and cell signal transmission. RING and HECT are included in the different types of E3 enzymes involved in this study. E3 is a huge family of enzymes, that function mainly to catalyze ubiquitin transfer. Talking about differences, catalyzed RING E3 ubiquitin transfers to the substrate directly from E2-ubiquitin, while HECT, which contains a catalytic cysteine, first receives ubiquitin from the E2-ubiquitin to form E3-ubiquitin thioester intermediates, and ubiquitin is then transferred to the substrate.28 In the upregulated SOCS-controlled pathway regulated by STAT3, the resistance to sugar degradation and insulin resistance play a role, so as to upset blood glucose level and worsen T2DM. Sepsis can induce a series of metabolic disorders, one of which is hyperglycemia.29 In the osteoclast differentiation pathway regulated by STAT3, the inhibitory effect of C-Fos is weakened. C-Fos regulatory protein is considered to be the regulation factor of cell proliferation, differentiation, and transformation, which is also associated with the death of apoptotic cells. Sepsis can increase the transcription and translation of C-Fos,30 but other studies confirm that C-Fos might be not important for cell apoptosis,31 so the role of C-Fos in sepsis-induced ARDS requires further study. The involvement of MYC in the spliceosome pathway regulates molecular transcription levels. Regulating the selection of splicing sites is an important mechanism to control human genetic information, which proves the principle that diseases caused by missplicing events can eventually be cured.32 Downregulated MYC leads to the downregulation of gene expression and suppression of transcription throughout the whole body, and this abnormality is the basis of many human diseases, or is related to the severity of such diseases.33 Therefore, studying the role of MYC in sepsis-induced ARDS may provide new therapeutic approaches.
Although our study provides some new insights into sepsis-induced ARDS, there are still several notable limitations. Firstly, the sample size of this study was relatively small. In particular, whether these core genes can be used as biomarkers for sepsis or sepsis-induced ARDS need to be validated in a larger data set. Secondly, our study was based on bioinformatics methods that proposed potential molecular mechanisms of sepsis-induced ARDS, which need to be verified in molecular experiments. In addition, because there is no data of ARDS induced by other causes in GSE32707, whether our findings are applicable to ARDS induced by other causes remains unclear and needs further study.
In conclusion, we constructed a preliminarily TF-module-pathway global transcriptional regulatory network for sepsis-induced ARDS based on FDGs in the process of evolving from healthy control through sepsis to sepsis-induced ARDS. MYC and STAT3 may be key regulatory genes in the underlying dysfunction of sepsis-induced ARDS.
Supplemental Material
Supplemental material, Author_response_to_reviewers_comments_v.2_ for Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS by Jianfeng Zhang, Yifeng Luo, Xiaoling Wang, Jieyun Zhu, Qian Li, Jihua Feng, Dan He, Zhimei Zhong, Xiaowen Zheng, Junyu Lu, Donghua Zou and Jiefeng Luo in Therapeutic Advances in Respiratory Disease
Supplemental Material
Supplemental material, Author_response_to_reviewer_comments_v.1 for Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS by Jianfeng Zhang, Yifeng Luo, Xiaoling Wang, Jieyun Zhu, Qian Li, Jihua Feng, Dan He, Zhimei Zhong, Xiaowen Zheng, Junyu Lu, Donghua Zou and Jiefeng Luo in Therapeutic Advances in Respiratory Disease
Supplemental Material
Supplemental material, Reviewer_1_v.1 for Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS by Jianfeng Zhang, Yifeng Luo, Xiaoling Wang, Jieyun Zhu, Qian Li, Jihua Feng, Dan He, Zhimei Zhong, Xiaowen Zheng, Junyu Lu, Donghua Zou and Jiefeng Luo in Therapeutic Advances in Respiratory Disease
Supplemental Material
Supplemental material, Reviewer_1_v.2 for Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS by Jianfeng Zhang, Yifeng Luo, Xiaoling Wang, Jieyun Zhu, Qian Li, Jihua Feng, Dan He, Zhimei Zhong, Xiaowen Zheng, Junyu Lu, Donghua Zou and Jiefeng Luo in Therapeutic Advances in Respiratory Disease
Supplemental Material
Supplemental material, Reviewer_2_v.1 for Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS by Jianfeng Zhang, Yifeng Luo, Xiaoling Wang, Jieyun Zhu, Qian Li, Jihua Feng, Dan He, Zhimei Zhong, Xiaowen Zheng, Junyu Lu, Donghua Zou and Jiefeng Luo in Therapeutic Advances in Respiratory Disease
Footnotes
Author contributions: JZ and DZ performed the majority of experiments and analyzed the data. YL, XW, JZ, QL, JF, DH, ZZ, XZ, and JL performed the molecular investigations. JZ and JL designed and coordinated the research. JZ and DZ wrote the paper. All authors read and approved the final manuscript.
Funding: This study was supported by the National Natural Science Foundation of China (Grant No: 81960343), the Key Research and Development project of Guangxi (Grant No: Guike AB17195002), Guangxi Natural Science Foundation (Grant No: 2017GXNSFAA198249), the Scientific Research Funding from Population and Family Planning Commission of Guangxi Zhuang Autonomous Region (Grant No: S2017009), the funding from Department of Education of Guangxi Zhuang Autonomous Region (Grant No: 2017JGA159) and the High-Level Medical Expert Training Program of Guangxi “139” Plan Funding (Grant No: G201903027).
Conflict of interest statement: The authors declare that there is no conflict of interest.
ORCID iD: Donghua Zou  https://orcid.org/0000-0002-3891-9472
https://orcid.org/0000-0002-3891-9472
Supplemental material: The reviews of this paper are available via the supplementary material section.
Availability of data and material: The datasets generated during the current study are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32707)
Contributor Information
Jianfeng Zhang, Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic China.
Yifeng Luo, Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic China.
Xiaoling Wang, Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic China.
Jieyun Zhu, Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic China.
Qian Li, Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic China.
Jihua Feng, Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic China.
Dan He, Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic China.
Zhimei Zhong, Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic China.
Xiaowen Zheng, Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic China.
Junyu Lu, Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic China.
Donghua Zou, Department of Neurology, The Fifth Affiliated Hospital of Guangxi Medical University, No 89 Qixing Road, Nanning, Guangxi 530022, People’s Republic China.
Jiefeng Luo, Department of Neurology, The Second Affiliated Hospital of Guangxi Medical University, No 166 Daxuedong Road, Nanning, Guangxi 530007, People’s Republic China; Department of Emergency, The Second Affiliated Hospital of Guangxi Medical University, Nanning 530007, People’s Republic China.
References
- 1. Ma J, Chen C, Barth AS, et al. Lysosome and cytoskeleton pathways are robustly enriched in the blood of septic patients: a meta-analysis of transcriptomic data. Mediators Inflamm 2015; 2015: 984825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dean E. Sepsis. Nurs Stand 2016; 30: 15. [DOI] [PubMed] [Google Scholar]
- 3. Gotur DB. Sepsis in a panorama: what the cardiovascular physician should know. Methodist Debakey Cardiovasc J 2018; 14: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eworuke E, Major JM, Gilbert McClain LI. National incidence rates for acute respiratory distress syndrome (ARDS) and ARDS cause-specific factors in the United States (2006–2014). J Crit Care 2018; 47: 192–197. [DOI] [PubMed] [Google Scholar]
- 5. Bellani G, Laffey JG, Pham T, et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA 2016; 315: 788–800. [DOI] [PubMed] [Google Scholar]
- 6. Kangelaris KN, Prakash A, Liu KD, et al. Increased expression of neutrophil-related genes in patients with early sepsis-induced ARDS. Am J Physiol Lung Cell Mol Physiol 2015; 308: L1102–L1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lyu X, Cong Z, Li D, et al. Effect mechanism of alpha-adrenoceptor on sepsis-induced acute respiratory distress syndrome. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 2018; 30: 83–87. [DOI] [PubMed] [Google Scholar]
- 8. Zhang W, Fang Q. Research progress of IQGAP1 in the pathogenesis of ARDS induced by sepsis. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 2017; 29: 647–650. [DOI] [PubMed] [Google Scholar]
- 9. Dolinay T, Kim YS, Howrylak J, et al. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am J Respir Crit Care Med 2012; 185: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liberzon A, Birger C, Thorvaldsdottir H, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015; 1: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 2006; 7: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45: D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods 2012; 9: 471–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han H, Cho JW, Lee S, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res 2018; 46: D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han Y, Yu G, Sarioglu H, et al. Proteomic investigation of the interactome of FMNL1 in hematopoietic cells unveils a role in calcium-dependent membrane plasticity. J Proteomics 2013; 78: 72–82. [DOI] [PubMed] [Google Scholar]
- 20. Yu G, Li F, Qin Y, et al. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 2010; 26: 976–978. [DOI] [PubMed] [Google Scholar]
- 21. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011; 12: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sagy M, Al-Qaqaa Y, Kim P. Definitions and pathophysiology of sepsis. Curr Probl Pediatr Adolesc Health Care 2013; 43: 260–263. [DOI] [PubMed] [Google Scholar]
- 23. Blackwell TS, Christman JW. The role of nuclear factor-kappa B in cytokine gene regulation. Am J Respir Cell Mol Biol 1997; 17: 3–9. [DOI] [PubMed] [Google Scholar]
- 24. Nawa Y, Kawahara K, Tancharoen S, et al. Nucleophosmin may act as an alarmin: implications for severe sepsis. J Leukoc Biol 2009; 86: 645–653. [DOI] [PubMed] [Google Scholar]
- 25. Yan Z, Gibson SA, Buckley JA, et al. Role of the JAK/STAT signaling pathway in regulation of innate immunity in neuroinflammatory diseases. Clin Immunol 2018; 189: 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med 2014; 20: 1242–1253. [DOI] [PubMed] [Google Scholar]
- 27. Etzioni A, Ciechanover A, Pikarsky E. Immune defects caused by mutations in the ubiquitin system. J Allergy Clin Immunol 2017; 139: 743–753. [DOI] [PubMed] [Google Scholar]
- 28. Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol 2016; 17: 626–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirasawa H, Oda S, Nakamura M. Blood glucose control in patients with severe sepsis and septic shock. World J Gastroenterol 2009; 15: 4132–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barke RA, Brady PS, Roy S, et al. The possible inhibitory role of the leucine-zipper DNA binding protein c-fos in the regulation of hepatic gene expression after sepsis. Surgery 1992; 112: 412–418. [PubMed] [Google Scholar]
- 31. Gajate C, Alonso MT, Schimmang T, et al. C-Fos is not essential for apoptosis. Biochem Biophys Res Commun 1996; 218: 267–272. [DOI] [PubMed] [Google Scholar]
- 32. Novoyatleva T, Tang Y, Rafalska I, et al. Pre-mRNA missplicing as a cause of human disease. Prog Mol Subcell Biol 2006; 44: 27–46. [DOI] [PubMed] [Google Scholar]
- 33. Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011; 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Author_response_to_reviewers_comments_v.2_ for Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS by Jianfeng Zhang, Yifeng Luo, Xiaoling Wang, Jieyun Zhu, Qian Li, Jihua Feng, Dan He, Zhimei Zhong, Xiaowen Zheng, Junyu Lu, Donghua Zou and Jiefeng Luo in Therapeutic Advances in Respiratory Disease
Supplemental material, Author_response_to_reviewer_comments_v.1 for Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS by Jianfeng Zhang, Yifeng Luo, Xiaoling Wang, Jieyun Zhu, Qian Li, Jihua Feng, Dan He, Zhimei Zhong, Xiaowen Zheng, Junyu Lu, Donghua Zou and Jiefeng Luo in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_1_v.1 for Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS by Jianfeng Zhang, Yifeng Luo, Xiaoling Wang, Jieyun Zhu, Qian Li, Jihua Feng, Dan He, Zhimei Zhong, Xiaowen Zheng, Junyu Lu, Donghua Zou and Jiefeng Luo in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_1_v.2 for Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS by Jianfeng Zhang, Yifeng Luo, Xiaoling Wang, Jieyun Zhu, Qian Li, Jihua Feng, Dan He, Zhimei Zhong, Xiaowen Zheng, Junyu Lu, Donghua Zou and Jiefeng Luo in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_2_v.1 for Global transcriptional regulation of STAT3- and MYC-mediated sepsis-induced ARDS by Jianfeng Zhang, Yifeng Luo, Xiaoling Wang, Jieyun Zhu, Qian Li, Jihua Feng, Dan He, Zhimei Zhong, Xiaowen Zheng, Junyu Lu, Donghua Zou and Jiefeng Luo in Therapeutic Advances in Respiratory Disease