Abstract
There is a wide variety of extracellular vesicles (EVs) that differ in size and cargo composition. EVs isolated from human plasma or serum carry lipid, protein, and RNA cargo that provides insights to the regulation of normal physiological processes, and to pathological states. Specific populations of EVs have been proposed to contain protein and RNA cargo that are biomarkers for neurologic and systemic diseases. Although there is a considerable amount of evidence that circulating lipids are biomarkers for multiple disease states, it not clear if these lipid biomarkers are enriched in EVs, or if specific populations of EVs are enriched for particular classes of lipid. A highly reproducible workflow for the analysis of lipid content in EVs isolated from human plasma or serum would facilitate this area of research. Here we optimized an MS/MSALL workflow for the untargeted analysis of the lipid content in EVs isolated from human serum. A simple sequential ultracentrifugation protocol isolated three distinct types of serum EVs that were identified based on size, targeted protein, and untargeted lipidomic analyses. EVs in the upper and middle fractions were approximately 140 nm in diameter, while EVs in the pellet were approximately 110 nm in diameter. EVs in the upper most buoyant fractions contained the highest concentration of lipids, were enriched with phospholipids, and immunopositive for the cytoskeletal markers actin, α-actinin, and the mitochondrial protein mitofillin, but negative for the typical EV markers CD63, TSG101, and flotillin. A central fraction of EVs was devoid of cytoskeletal and mitochondrial markers, and positive for CD63, and TSG101, but negative for flotillin. The EV pellet contained no cytoskeletal or mitochondrial markers, but was positive for CD63, TSG101, and flotillin. The EV pellet contained the lowest concentration of most lipids, but was enriched with ceramide. These results provided new insights into the lipid composition of EVs isolated from serum using a simple ultracentrifugation isolation method suitable for lipidomic analysis by mass spectrometry.
Keywords: Extracellular vesicle, exosome, lipidomics, lipid, serum, blood
Introduction
Communication between cells and organs involving the secretion of membrane vesicles (collectively called extracellular vesicles or EVs) has attracted a great deal of interest,1–3 and our understanding of roles for this form of cellular communication in normal physiology and pathology is rapidly increasing. There are multiple types of EVs that are released by distinct mechanisms,4 and there is evidence that the stimulus used to evoke EV shedding modifies the cargo.5,6 EVs are composed of a lipid bilayer, transmembrane proteins, cytosolic proteins, and RNA.1 Surface proteins of EVs appear to regulate targeting and capture by recipient cells, which can then incorporate EV cargo, resulting in modifications of the recipient cells’ physiological state.7,8 The possibility of using EVs, or subpopulations of EVs as biomarkers for disease, or as molecular targets to be therapeutically regulated are active areas of research.9,10
Various “omic” and targeted techniques have been applied to EVs to determine the composition of protein and miRNA cargo.11,12 This information has been broadly applied to understand regulatory and pathological roles for EVs, and particular cargo have been proposed as surrogate measures for a wide variety of disease activity13–17 (also see Roy et al.18 for a recent review). Compared with protein and miRNA, the lipid components of EVs have been relatively less studied, especially in biological fluids. The vast majority of EV lipidomic studies have been conducted on EVs isolated from tissue culture.19–21 The concentration and protein/miRNA composition of circulating EVs has been associated with various physiological and pathological conditions, and there is increasing evidence that the cargo EVs originating from abnormal tissues may serve as biomarkers for disease status.9,22,23 Human blood contains a complex mixture of EVs originating from multiple tissues, making the isolation of well-defined EV fractions a challenging task.24 Circulating EVs are gaining popularity as a source for biomarkers that could be used to diagnose and/or track the trajectory of diseases including Alzheimer’s,25 HIV-associated neurocognitive disorders,26 Parkinson’s disease,27 cancers,9,28 and kidney disease.29 Very little is currently known about the lipidomic composition of circulating EVs. As many of these lipid components are biologically active, a better understanding of the lipid components of EVs isolated from serum samples will be beneficial for the discovery of novel biomarkers.
The study of pathways and networks for lipid metabolism and signaling is a rapidly expanding research field.30,31 The essential roles of lipidomics in identifying the biochemical mechanisms of lipid metabolism, investigating the functions of genes of interest, discovering novel biomarkers, and evaluating drug efficacy are becoming increasingly recognized.32 Mass spectrometry (MS)-based lipidomics has become one of the predominant approaches because of its high throughput, qualitative and quantitative abilities in lipid detection.33 MS-based lipidomics strategies can be divided into untargeted and targeted approaches, each with their own advantages and limitations. Targeted lipidomics focuses on the analysis of a priori defined lipid species. Although these methods can be fully quantitative, these approaches are limited in scope. Direct infusion34 or liquid chromatography (LC)-based information-dependent acquisition (IDA) protocols have been developed for untargeted approaches. These methods preselect a list of candidate precursor ions in real time based on survey MS data and user-defined information-dependent criteria to identify all lipid species that have these defined precursors. Although more comprehensive than targeted approaches, IDA approaches can be compromised by reproducibility and MS/MS data quality.35 Recently, alternative data-independent MS/MS acquisition (DIA) methods for untargeted lipidomics analyses have received considerable attention.36,37 In contrast to IDA, DIA methods conduct MS/MS for all precursors simultaneously, thereby increasing the coverage of observable molecules and reducing the identification of false negatives.38 Among available DIA methods, the sequential precursor ion fragmentation (MS/MSALL) technique of stepping through a predefined mass range in small increments is rather promising owing to its speed, simplicity, and high-quality MS/MS data. This MS/MSALL approach is bias-free and delivers high-quality product ion spectra, even in the absence of an MS precursor ion signal. The generated MS/MS data can further be directly applied toward batch library searching or spectral MS2-level algorithms for lipid molecular profiling workflow. We believe the MS/MSALL method is well suited to deconvolute the complex lipid compositions of EVs. In this study, we optimized this untargeted lipidomic strategy for a highly reproducible characterization of lipids in EVs fractionated from human serum.
Methods
Human serum
Human serum samples were collected from 10 healthy volunteers. Subject demographics are shown in Online Supplemental Table S1. Five men and five women were enrolled with average age of 37.3 years. Seven of the subjects were Caucasian and three were Asian. The collection and use of human samples was approved by The Johns Hopkins School of Medicine Institutional Review Board, in accordance with the WMA declaration of Helsinki. Serum samples were individually analyzed.
Isolation of EVs
EVs were isolated from normal human serum by differential ultracentrifugation as described previously.39,40 Serum (200 µL) was diluted to 600 µL with phosphate buffered saline (PBS) and centrifuged at 2700 × g for 15 min at 20°C to remove large debris, then ultracentrifuged at 10,000 × g for 30 min at 4°C to remove larger microvesicles and apoptotic bodies. The resulting supernatant was ultracentrifuged at 100,000 × g for 3 h at 4°C. A total of seven fractions were collected in a top-down fashion (F1–F6) ending with the bottom pellet (F7) (Figure 1(a)). The size and number of EVs in all seven fractions were determined by nanoparticle tracking analysis (described in more detail below). The top (F1), middle (F5), and bottom (pellet) were extracted for lipidomic analyses.
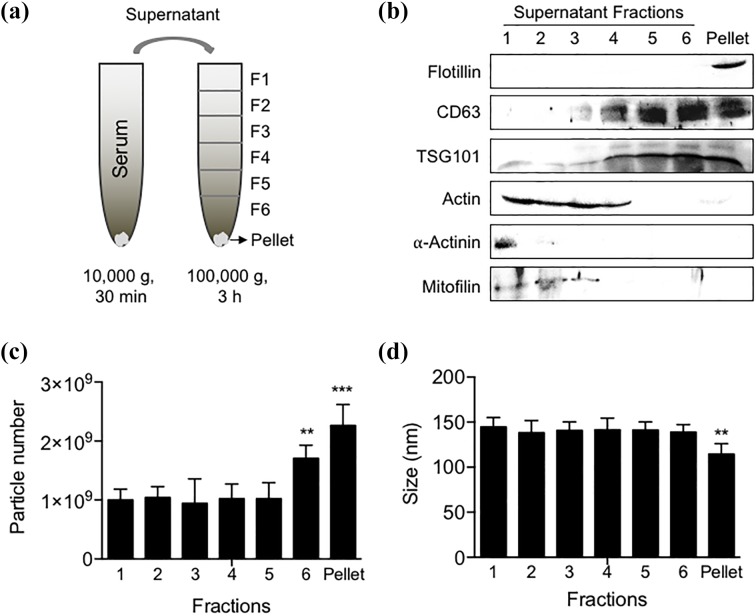
Figure 1.
Isolation and characterization of EV fractions isolated from human serum. (a) A differential ultracentrifugation procedure was used to isolate six fractions and a high-density pellet of EVs from human serum. (b) Representative Western blot showing the immunoreactivity of isolated serum fractions for the indicated proteins that include the lipid raft-associated protein flotillin, the tetraspan protein CD63, the ESCRT-1 protein TSG101, the cytoskeletal proteins actin, α-actinin, and the mitochondrial protein mitofillin. (c) EV size and (d) number measured by nanoparticle tracking analysis for each fraction. Data are mean ± SD. EVs: extracellular vesicles; SD: standard deviation.
Nanoparticle tracking analysis
Size and number of EVs were quantified using a ZetaView Nanoparticle Tracker (Particle Metrix GmBH, Meerbusch, Germany), and corresponding ZetaVeiw software (8.03.04.01). A nanosphere size standard (100 nm diameter; Thermo Scientific; Waltham, MA) was used to calibrate the instrument prior to readings. Instrument pre-acquisition parameters were set to a temperature of 23°C, sensitivity of 65, frame rate of 30 frames per second (fps), shutter speed of 100, and a laser pulse duration equal to that of shutter duration. Post-acquisition parameters were set to a minimum brightness of 25, maximum size of 200 pixels, and a minimum size of 10 pixels. For each sample 1 mL of diluted EVs were injected into the sample-carrier cell and the particle count was measured at five positions, with two cycles of reading per position. The cell was washed with PBS after every sample. The mean size and concentration of EVs/mL (±SEM) was calculated from four replicate experiments. The coefficient of variation (CV) as determined from a pooled sample was 4.8% for size and 4.4% for concentration of EVs.
EV and serum extraction
EVs and serum samples were extracted to obtain a crude lipid fraction. In brief, 200 µL of the pellet enriched with EVs, or 45 µL of an EVs-depleted serum fraction (the remained serum after removal of exosomes by ultracentrifugation) or 15 µL of original serum were gently mixed in a glass vial with appropriate volume of ultrapure water to 1 mL. Then 2.9 mL methanol:dichloromethane (2:0.9, v/v) that included internal standards to identify 12 classes of lipids was added to form a monophasic solution. To obtain a biphasic mixture, 1 mL of ultrapure water and 0.9 mL dichloromethane were added, and the resultant mixture incubated at 4°C for 30 min then centrifuged (10 min, 3000 × g, 4°C) to separate the organic and aqueous phases. Then, 1 mL of the organic phase containing a crude lipid extract was transferred to a 2 mL glass vial and stored at −20°C. Prior to analysis, 0.5 mL of the organic layer extract was dried using a nitrogen evaporator (Organomation Associates, Inc., Berlin, Massachusetts, USA) then resuspended in 120 µL of running solvent (dichloromethane:methanol (1:1) with 5 mM ammonium acetate) containing 5 mg/mL of the ceramide (C17:0) as an internal standard.
Materials
Internal standards included (Online Supplemental Table S2) 1,2-dilauroyl-sn-glycero-3-phosphocholine (PC 12:0/12:0), 1,2-dilauroyl-sn-glycero-3-phospho-l-serine (sodium salt) (PS 12:0/12:0, PS C12), 1,2-dilauroyl-sn-glycero-3-phosphoethanolamine (PE 12:0/12:0, PE C12), 1,2-dilauroyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (PG 12:0/12:0, PG C12), 1,2-dilauroyl-sn-glycero-3-phosphate (sodium salt) (PA 12:0/12:0, PA C12), cholesteryl-d7 palmitate (cholesterol-d7 ester 16:0, cholesterol ester d7), N-lauroyl-d-erythro-sphingosine (C12 ceramide d18:1/12:0, Cer C12), N-heptadecanoyl-d-erythro-sphingosine (C17 ceramide d18:1/17:0, Cer C17), N-lauroyl-d-erythro-sphingosylphosphorylcholine (12:0 SM d18:1/12:0 SM C12), 1,3-dihexadecanoyl glycerol (d5) [1,3-16:0 DG (d5), DG d5], 1,3(d5)-dihexadecanoyl-2-octadecanoyl-glycerol [TG d5-(16:0/18:0/16:0), TG d5], d-galactosyl-β-1,1′ N-lauroyl-d-erythro-sphingosine [C12 galactosyl(β) ceramide (d18:1/12:0), GlcCer C12], d-lactosyl-β-1,1′ N-dodecanoyl-d-erythro-sphingosine [lactosyl (β) C12 ceramide, LacCer C12] were supplied by Avanti Polar Lipids (Alabaster, Alabama, USA). Atmospheric pressure chemical ionization (APCI) positive calibration solution (P/N: 4460131) was purchased from AB SCIEX (Concord, Ontario, Canada). Standard stock solutions of Cer C12:0 (5 mg/mL), Cer C17:0 (5 mg/mL), PE C12:0 (12.5 mg/mL), PA C12:0 (7.25 mg/mL), PG C12:0 (12.5 mg/mL), DG d5 (0.5 mg/mL), TG d5 (0.5 mg/mL), LacCer C12:0 (1.25 mg/mL), and GlcCer C12:0 (2.5 mg/mL) were dissolved in dichloromethane:methanol 1:1 (v/v), SM C12:0 (10 mg/mL), PC C12:0 (25 mg/mL), and PS C12:0 (10 mg/mL) were dissolved in methanol, cholesterol ester d7 was dissolved in dichloromethane and stored at −20°C. Ultrapure water was used in all experiments (resistivity >18 MΩ cm).
Untargeted lipid analysis by MS/MSALL
Crude lipid extracts were analyzed by MS/MSALL using a TripleTOF™ 5600 (AB SCIEX, Redwood City, California, USA) mass spectrometer. Samples (50 µL) were introduced into the mass spectrometer system by direct infusion using a DuoSpray electrospray ionization source and autosampler (Shimadzu; Kyoto, Japan) at the flow rate of 7 µL/min and run in duplicate in the positive ion mode. The running solvent is the mixture of the dichloromethane and methanol (1:1, v/v) with 5 mM ammonium acetate. The instrument was operated at a mass resolution of 30,000 for time of flight (TOF) MS scan and 15,000 for product ion scan in the high sensitivity mode, and automatically calibrated every 10-sample injections using APCI positive calibration solution delivered via a calibration delivery system (AB SCIEX). The source parameters include ion source gases 15 lbf/in2 (GS1), 20 lbf/in2 (GS2), curtain gas 30 lbf/in2, temperature 150°C, positive ion spray voltage +5200 V, declustering potential at 80 V, and collision energy at 10 V. An initial TOF MS scan provided an overview of the total lipid content at an accumulation time of 5 s. Precursor ions were selected by sequential 1 Da mass steps from 200.050 to 1200.050 m/z. The analytes in each 1 Da step were then introduced into the collision chamber and fragments were produced by collision-induced dissociation and identified by TOF with a scan range of 100–1500 m/z (accumulation time of 300 ms). The collision energy for each MS/MS step was 40 eV. A pooled sample containing extracts from all groups was repeatedly run eight times for lipid identification and subsequent selection of targeted lipids. All data were acquired using Analyst 1.7 TF (AB SCIEX, Concord, Ontario, Canada).
Data processing and analysis
The TOF MS and MS/MSALL data obtained from each sample run and pooled samples were post-aligned to the internal standards using Analyst 1.7 TF with mass error less than 5 ppm. Broad lipid assignments and identifications were conducted on the aligned data of pooled samples with LipidView™ software (v1.3 Beta, AB SCIEX, Concord, Ontario, Canada). The identification of the lipid species is based on the matched pairs of precursor and fragment to the in silico tandem MS database of LipidView™. To ensure that the identified lipids were reliable and reproducible we only included lipids whose peaks of MS/MS fragments appeared in seven of the eight pooled runs, and had a CV value less than 20%. Lipids meeting these criteria were included in the targeted list to identify these lipids in experimental samples. The calculation of CV values and the identification of lipids meeting criteria were conducted using a customized program in MATLAB (version R2017a; MathWorks). A targeted processing method was generated based on the targeted lipid list, and used to probe the experimental sample data set using MultiQuant software (version 3.0, AB SCIEX, Concord, Ontario, Canada). For the relative quantification, the resultant peak intensities of each characteristic fragment were corrected by the internal standard for the corresponding class of lipid, and each sample duplicate was averaged for further analysis. The intensities of isomeric triacylglyceride (TAG) species were calculated individually based on the each neutral loss of fatty acid (FA) moieties, instead of the sum of all the FA fragment ion intensities. Peak intensities of 0 were replaced with the minimum detectable intensity that was calculated by dividing 0.01 by the average intensity value of the internal standard for the corresponding class of lipid.
Average molar ratios were calculated as follows: The relative intensity of each lipid species was normalized to the intensity of the internal standard for the respective class of lipid in the F1, F5, and pellet fractions. The relative intensities for each lipid in F1 and F5 were compared to the corresponding lipid in the pellet fraction and expressed as a ratio. The resulting values obtained for each lipid species were then expressed as a fraction of the total lipid content in the respective class of lipid to obtain the average molar ratio for each lipid species by class. This is an estimate of the molar ratio, as the relative intensity for each lipid species is proportional to its molar concentration.
Effects of the CV threshold on precision
In exploratory studies, we examined the relationship of the CV to instrument precision by investigating the intraday and interday precision of the selected features during repeated runs with three different CV thresholds. Intraday precision was determined by measuring the relative standard deviation (RSD) of the relative intensity of each targeted lipid species in six injections during an 8-h MS run. The interday precision was determined by measuring the RSD of the relative intensity for each targeted lipid species in six injections conducted over three consecutive days (two injections/day). We used these data to identify a CV threshold that provided the most robust lipid identifications with optimal precision.
Nonparametric analysis
The relative peak intensity for each lipid species across all fractions was subjected to z-score normalization by subtracting the mean intensity of the lipid from all values, and dividing this value by the standard deviation. This produces scaled intensity values with a mean of 0 and a standard deviation of 1. z-Score normalization was necessary for visualization and comparison due to large differences in lipid intensity depending on the class and species of lipid. The z-scored values for each fraction were then averaged and converted into a heat map using Graphpad Prism 7 without hierarchical clustering.
Western blotting
The protein amount in each fraction and pellet were measured by bicinchoninic acid protein assay. Equal amount of protein (20 μg) from each fraction and pellet were resolved by 10% SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Bio-Rad; Hercules, CA). Nonspecific binding sites were blocked with 5% (w/v) milk in TBS containing 0.1% Tween 20 (TBS-T). After blocking, blots were incubated overnight with the primary polyclonal antibody flotillin 1 (1:1000; Abcam), CD63 (1:200; Santa Cruz Biotechnology), TSG101 (1:1000; BD Biosciences), actinin-4 (1:1000; Gentex), and mitofilin (1:5000; Thermo Fisher Scientific). After washes with TBS-T, blots were incubated for 2 h with the appropriate IgG horseradish peroxidase–linked secondary antibody (1:1000; Cell Signaling Technology) and developed by enhanced chemiluminescence. Image analysis was performed using a G: BOX imaging system (Syngene).
Annotation of lipid species
Lipid species were annotated as previously defined.41 For example, glycerophospholipid and glycerolipid species were annotated by “sum composition,” and denoted as <lipid class><total number of C in FA moieties>:<total number of double bonds in FA moieties> (e.g. PC 34:1). Sphingolipid species were annotated by “sum composition,” and denoted as <lipid class><total number of C in the long-chain base and FA moiety>:<total number of double bonds in the long-chain base and FA moiety>; <total number of OH groups in the long-chain base and FA moiety> (e.g. Cer 34:1;2, SM 35:1;2). Plasmalogen species were annotated by “sum composition,” and denoted as <lipid class><O-><total number of C in FA moieties>: <total number of double bonds in FA moieties> (e.g. PC O-36:1). Molecular species glycerophospholipid were annotated by “molecular species composition,” and denoted as <lipid class><number of C in the first FA moiety>:<number of double bonds in the second FA moiety>/<number of C in the second FA moiety>:<number of double bonds in the second FA moiety> (e.g. PG 16:0/16:0). Molecular species sphingolipid annotated by “molecular species composition” were denoted as <lipid class><d/t><total number of C in the long-chain base><total number of double bonds in the long-chain base>/<total number of C in FA moiety><total number of double bonds in FA moiety> (e.g. Cer d18:1/16:0, the “d” and “t” designations used in shorthand notation of sphingolipids refer to 1,3 dihydroxy and 1,3,4-trihydroxy long-chain bases, respectively).
Results
Isolation of EVs
A stepwise ultracentrifugation technique was used to isolate EV fractions. Western blotting was first applied to analyze the characteristic proteins in each fraction. Fraction 1 was immunopositive for the cytoskeletal protein actin, and the mitochondrial protein mitofillin. Fractions 2 and 3 were positive for actin and mitofillin. Fraction 3 was immunopositive for typical EV markers that included the tetraspan protein CD63, the ESCT-1 complex protein TSG101, and were negative for the cytoskeletal protein α-actinin, and the mitochondrial protein mitofillin, suggesting an absence of contaminating cellular debris (with the potential exception of fraction 4 that was immunopositive for actin). Fractions 5, 6 were immunopositive for CD63, TSG101 and negative for actin, α-actinin and mitofillin. The lipid raft-associated protein flotillin was only detected in the pellet, and this fraction was also positive for CD63, TSG101, but negative for actin, α-actinin, and mitofillin (Figure 1(b)). The number of particles in fractions 1–5 were similar at approximately 1 × 109 particles, the particle number increased in fraction 6, and was highest at 2.3 × 109 ± 3.2 × 108 particles in the pellet (Figure 1(c)). Particle size was strikingly similar in fractions 1–5 at approximately 140 nm, and smallest in the pellet with a particle size of 116.3 ± 12.8 nm (Figure 1(d)). Based on these findings, we conducted a lipidomic analysis on the top fraction (F1) that did not contain any of the typical EV markers, the middle fraction (F5) that contained typical EV markers, but did not contain markers of cellular contamination, and the bottom fraction (pellet) that contained the typical EV markers and the lipid raft marker flotillin.
Validation of reproducible lipid features
We first extracted a pooled serum sample consisting of small aliquots of each experimental sample (F1, F5, and the pellet) to identify features that are highly reproducible for the subsequent analysis of individual samples. A pooled extract from each of F1, F5, and the pellet were subjected to MS analysis with eight sequential injections using the identical direct infusion MS/MSALL DIA method applied to experimental samples. A broad lipid assignment and identification was conducted on the aligned data of pooled samples with LipidView™ software. This initial method identifies all features that appear at least once in the eight runs. To ensure that we are only conducting further analysis on consistently identified features, only those features present in at least seven of the eight pooled runs, with a CV value of less than 20% are included for further analysis. Using these criteria, 4024 of the initially identified features were reduced to 597 highly reproducible features. These highly reproducible features were used to create a targeted lipids list in LipidView™, and this targeted list was used to probe the experimental data set. These targeted lipid lists were used to analyze the lipid data of each fraction of serum.
To further confirm the CV threshold, we explored the relationship of CV to instrument precision by investigating the intraday and interday precision of the selected features during repeated runs. A CV threshold of 10% showed the best precision with 88% of the interday, and 91% of the intraday features showing an RSD less than 20%. However, only 219 features met these criteria. Increasing the CV threshold to 20% only slightly reduced precision, with 78% of the interday, and 80% of the intraday features showing an RSD <20%, but the number of feature meeting criteria increased to 597. Increasing the CV threshold to 30% resulted in a large reduction of precision, with 48% of the interday and 55% of intraday features showing an RSD <20%, with 1400 features meeting criteria. Without any threshold for the selection of features 55% of interday and 48% of intraday features had an RSD <20% (Online Supplemental Table S3), with 4024 identified features. We used a 20% CV threshold for this study, as it provided optimal precision and feature coverage.
From an initial 597 features that met criteria, a total of 422 of lipid features were confirmed using the LipidView™ database after removal of unlikely lipid species (i.e. FA side chain with a carbon number less than 12, and additional fragments that each identified the same lipid). The identified lipid classes included glycerophospholipid/lyso-glycerophospholipid species (PA/PA O/LPA/LPA O, PC/PC O/LPC/LPC O, PE/PE O/LPE/LPE O, PG/PG O, PS/PS O/LPS/LPS O), sphingolipid species (Cer, HexCer/Hex2Cer, SM), cholesteryl ester (CE), and glycerolipid species (DAG, TAG). The detailed information of each lipid feature including the sum composition, molecular composition, precursor and product ions are listed in Online Supplemental Table S4.
Lipidomic analysis of EV fractions
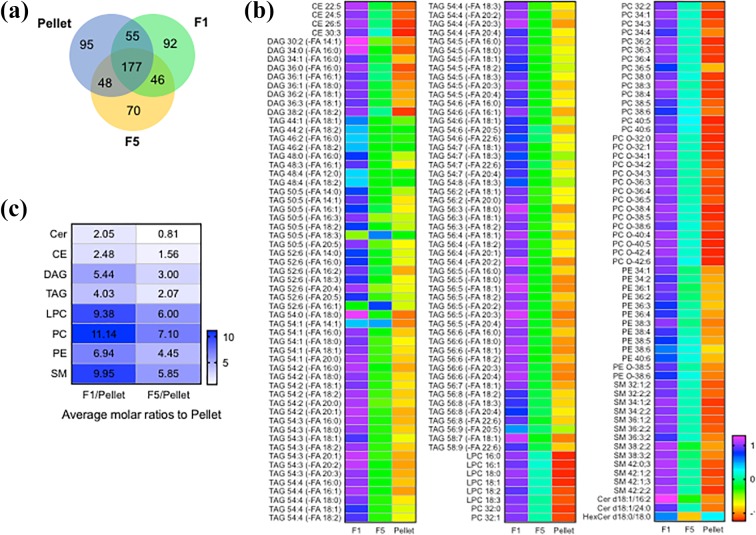
We next conducted a lipidomic analysis of fractions 1 (F1), 5 (F5), and the pellet using equal numbers of particles from each fraction to normalize the amount of input material. The total number of lipid features meeting criteria for selection in F1 (n = 370), F2 (n = 341), and the pellet (n = 375) were similar. Fractions F1, F5, and the pellet contained 177 shared lipid species (Figure 2(a)), and there were 106 lipid features unique to F1, 85 unique to F5, and 110 unique to the pellet (Figure 2(a)). The pellet contained the largest number of unique ceramides (n = 12), compared with F1 (n = 4), or F5 (n = 6). In contrast, F1 contained the largest number of unique phospholipids (n = 41), compared with F5 (n = 19), and the pellet (n = 21). The FA composition of TAGs in F1 was largely unsaturated, F5 contained a mixture of saturated and unsaturated species, and the pellet contained largely saturated FAs (Table 1). Comparing lipids that were consistently detected in F1, F5, and the pellet, there was a clear visual separation where F1 contained the highest concentration of nearly all lipid species, F5 was intermediate, and the pellet contained the lowest concentration of most lipid species (Figure 2(b)). A total of 53 (out of 422) independent lipid species were significantly different between the F1, F5, and the pellet (Table 2). The lipid content of F1 was generally higher than F5 or the pellet, consistent with the buoyancy of this fraction. Comparing the average molar ratios for all lipid species by class we found that F1/pellet contained the highest concentration of all lipid classes followed by F5/pellet (with the exception of ceramides) (Figure 2(c)). These data demonstrate that F1 and F5 are enriched for all lipid classes with the exception of ceramides that are enriched in the pellet.
Figure 2.
Lipid composition of F1, F5, and the pellet isolated from serum fractions. (a) Venn diagram showing the distribution of common and unique lipids identified in the indicated fraction. (b) z-Score normalized heat map showing the relative concentrations of shared lipids in the three fractions. The relative intensities of each lipid in specific class of lipid were normalized to the corresponding internal standard for each lipid class. (c) Average molar ratios for the indicated classes of lipid in F1 and F5 compared with the pellet.
Table 1.
The unique lipids features identified from the pooled F1, F5, and pellet fractions of the serum exosomes isolation by ultracentrifugation.
| F1 | F5 | Pellet |
|---|---|---|
| CE 20:1 | CE 22:2 | CE 18:4 |
| CE 22:6 | Cer d18:1/22:0 | CE 24:2 |
| Cer d18:0/14:1 | DAG 30:0 (-FA 14:0) | CE 32:6 |
| Cer d18:1/18:0 | DAG 32:0 (-FA 16:0) | CE 34:4 |
| Cer d18:1/24:1 | DAG 32:2 (-FA 16:1) | CE 34:5 |
| DAG 32:1 (-FA 16:0) | DAG 36:4 (-FA 18:2) | Cer d18:1/14:0 |
| DAG 34:2 (-FA 14:1) | DAG 40:5 (-FA 20:1) | Cer d18:0/16:0 |
| DAG 34:2 (-FA 16:1) | HexCer d18:1/16:0 | Cer d18:0/16:3 |
| DAG 40:4 (-FA 20:1) | HexCer d18:1/24:0 | DAG 32:0 (-FA 14:0) |
| HexCer d18:2/24:1 | HexCer d18:1/26:0 | DAG 34:0 (-FA 14:0) |
| LPC 20:4 | LPC 20:4 | DAG 38:2 (-FA 18:1) |
| LPC 20:5 | LPC O-26:0 | DAG 38:3 (-FA 18:1) |
| LPC 30:0 | LPE O-24:0 | DAG 38:3 (-FA 18:3) |
| LPC O-16:0 | PA 44:3 | DAG 38:4 (-FA 18:2) |
| LPC O-16:1 | PA O-16:0/16:0 | DAG 40:1 (-FA 20:0) |
| LPE 18:1 | PC 32:3 | DAG 40:4 (-FA 20:0) |
| LPS 28:1 | PC O-20:3 | DAG 40:5 (-FA 20:0) |
| LPS O-30:3 | PC O-20:4 | HexCer d18:1/16:3 |
| PA 40:4 | PC O-22:6 | HexCer d18:1/18:0 |
| PA O-40:4 | PC O-24:0 | LPC O-20:0 |
| PA O-48:2 | PC O-44:6 | LPE 26:0 |
| PA O-48:3 | PE 34:0 | LPE O-30:4 |
| PC 30:0 | PE O-36:1 | LPG 24:0 |
| PC 30:3 | PE O-36:3 | LPG 24:1 |
| PC 34:5 | PG 36:1 | LPS 16:3 |
| PC 36:1 | PG 36:5 | LPS 20:3 |
| PC 40:1 | PG O-36:6 | LPS O-18:0 |
| PC 40:4 | PG O-38:1 | LPS O-18:2 |
| PC 42:5 | PG O-38:6 | LPS O-30:0 |
| PC O-32:2 | PG O-40:2 | PA 38:5 |
| PC O-34:4 | PS 36:4 | PA O-48:1 |
| PC O-36:1 | PS 48:3 | PA O-48:2 |
| PC O-38:0 | SM 32:3;2 | PA O-48:3 |
| PE 36:5 | SM 40:1;2 | PC O-42:5 |
| PE 38:0 | SM 42:3;2 | PC O-44:3 |
| PE 40:4 | TAG 42:2 (-FA 16:0) | PE 38:1 |
| PE 40:5 | TAG 44:1 (-FA 14:1) | PE O-38:0 |
| PE O-34:1 | TAG 44:4 (-FA 14:0) | PE O-46:6 |
| PE O-40:4 | TAG 46:3 (-FA 14:1) | PG 20:2/20:2 |
| PE O-40:5 | TAG 50:5 (-FA 16:0) | PG O-20:0/20:0 |
| PE O-42:6 | TAG 50:6 (-FA 12:0) | PS 32:2 |
| PE O-46:5 | TAG 50:8 (-FA 18:2) | PS 36:4 |
| PG 38:6 | TAG 52:6 (-FA 18:1) | PS 36:6 |
| PG 48:5 | TAG 52:7 (-FA 16:2) | PS 42:6 |
| PG 48:6 | TAG 54:0 (-FA 16:0) | PS O-28:1 |
| PG O-18:1/18:1 | TAG 54:0 (-FA 20:0) | PS O-36:1 |
| PG O-38:0 | TAG 54:2 (-FA 20:2) | PS O-38:0 |
| PS 28:0 | TAG 54:4 (-FA 20:1) | PS O-38:1 |
| PS 36:5 | TAG 54:6 (-FA 16:2) | PS O-46:0 |
| PS O-38:6 | TAG 54:8 (-FA 20:5) | PS O-48:6 |
| PS O-48:6 | TAG 54:9 (-FA 20:5) | SM 34:1;2 |
| SM 44:1;2 | TAG 56:1 (-FA 20:0) | SM 46:1;2 |
| SM 44:2;2 | TAG 56:2 (-FA 16:0) | SM 48:1;2 |
| SM 46:2;2 | TAG 56:2 (-FA 22:1) | TAG 36:1 (-FA 12:1) |
| SM 48:1;2 | TAG 56:4 (-FA 18:3) | TAG 38:0 (-FA 18:0) |
| TAG 42:3 (-FA 18:3) | TAG 56:6 (-FA 22:6) | TAG 38:2 (-FA 16:0) |
| TAG 44:1 (-FA 12:1) | TAG 56:7 (-FA 18:3) | TAG 40:0 (-FA 12:0) |
| TAG 44:2 (-FA 14:1) | TAG 56:7 (-FA 20:3) | TAG 40:1 (-FA 16:1) |
| TAG 44:2 (-FA 16:1) | TAG 58:10 (-FA 20:5) | TAG 40:2 (-FA 14:0) |
| TAG 44:3 (-FA 12:0) | TAG 58:4 (-FA 18:1) | TAG 40:2 (-FA 18:2) |
| TAG 44:3 (-FA 16:1) | TAG 58:4 (-FA 18:2) | TAG 42:0 (-FA 12:0) |
| TAG 44:4 (-FA 18:2) | TAG 58:5 (-FA 18:1) | TAG 42:0 (-FA 18:0) |
| TAG 46:2 (-FA 12:1) | TAG 58:6 (-FA 18:1) | TAG 42:1 (-FA 14:0) |
| TAG 46:2 (-FA 14:1) | TAG 58:7 (-FA 18:3) | TAG 42:2 (-FA 12:0) |
| TAG 46:3 (-FA 16:1) | TAG 58:8 (-FA 22:6) | TAG 42:2 (-FA 18:1) |
| TAG 48:5 (-FA 12:1) | TAG 60:10 (-FA 20:4) | TAG 42:6 (-FA 16:0) |
| TAG 48:5 (-FA 20:4) | TAG 60:10 (-FA 22:6) | TAG 44:1 (-FA 12:0) |
| TAG 48:5 (-FA 16:3) | TAG 60:3 (-FA 24:0) | TAG 44:2 (-FA 14:0) |
| TAG 50:5 (-FA 20:3) | TAG 60:9 (-FA 20:4) | TAG 44:3 (-FA 14:1) |
| TAG 50:6 (-FA 18:2) | TAG 60:9 (-FA 22:6) | TAG 44:3 (-FA 16:0) |
| TAG 50:6 (-FA 22:6) | TAG 44:5 (-FA 14:0) | |
| TAG 50:7 (-FA 18:1) | TAG 46:3 (-FA 12:1) | |
| TAG 52:6 (-FA 22:6) | TAG 46:3 (-FA 14:0) | |
| TAG 52:8 (-FA 22:6) | TAG 48:0 (-FA 24:0) | |
| TAG 54:3 (-FA 18:3) | TAG 48:4 (-FA 14:1) | |
| TAG 54:4 (-FA 22:1) | TAG 48:4 (-FA 16:1) | |
| TAG 54:7 (-FA 20:3) | TAG 48:4 (-FA 18:1) | |
| TAG 54:8 (-FA 22:6) | TAG 48:5 (-FA 12:0) | |
| TAG 56:0 (-FA 20:0) | TAG 48:5 (-FA 16:0) | |
| TAG 56:10 (-FA 22:6) | TAG 48:5 (-FA 18:2) | |
| TAG 56:3 (-FA 16:0) | TAG 50:5 (-FA 20:1) | |
| TAG 56:3 (-FA 22:1) | TAG 54:6 (-FA 20:4) | |
| TAG 56:4 (-FA 20:0) | TAG 56:1 (-FA 16:0) | |
| TAG 56:6 (-FA 20:5) | TAG 56:3 (-FA 20:0) | |
| TAG 58:1 (-FA 16:0) | TAG 56:3 (-FA 20:2) | |
| TAG 58:1 (-FA 20:0) | TAG 56:4 (-FA 16:0) | |
| TAG 58:5 (-FA 18:2) | TAG 56:4 (-FA 20:4) | |
| TAG 58:7 (-FA 20:4) | TAG 56:7 (-FA 20:2) | |
| TAG 58:8 (-FA 20:2) | TAG 58:11 (-FA 20:5) | |
| TAG 58:8 (-FA 20:4) | TAG 58:3 (-FA 18:2) | |
| TAG 60:3 (-FA 18:2) | TAG 58:5 (-FA 16:0) | |
| TAG 58:6 (-FA 16:0) | ||
| TAG 58:7 (-FA 20:2) | ||
| TAG 60:4 (-FA 20:0) | ||
| TAG 60:5 (-FA 20:5) |
CE: cholesterol ester; Cer: ceramide; DAG: diacylglyceride; HexCer: hexosylceramide; Hex2Cer: dihexosylceramide; PA: phosphatidic acid; LPA: lysophosphatidic acid; PC: phosphatidylcholine; LPC: lysophosphatidylcholine; PE phosphatidyletanolamine; LPE: lysophosphatidyletanolamine; PG phosphatidylglycerol; LPG: lysophosphatidylglycerol; PS: phosphatidylserine; LPS: lysophosphatidylserine; SM: sphingomyelin; TAG: triacylglyceride; FA: fatty acid; O-: ether-linked lipids.
Table 2.
Shared lipid features that were significantly different in abundance between EV fractions.a
| Lipid name | F1 Avg (SD) |
F5 Avg (SD) |
EV Avg (SD) |
p Values | ||
|---|---|---|---|---|---|---|
| F1 − EV | F5 − EV | F1 − F5 | ||||
| CE 30:3 | 12.54 (2.65) | 8.27 (1.74) | 1.44 (0.71) | <0.0001 | <0.0001 | <0.0001 |
| Cer d18:1/16:2 | 0.80 (0.07) | 0.47 (0.04) | 0.33 (0.04) | <0.0001 | <0.0001 | <0.0001 |
| Cer d18:1/24:0 | 0.13 (0.04) | 0.08 (0.04) | 0.01 (0.01) | <0.0001 | <0.001 | <0.05 |
| DAG 16:0/20:0/0:0 | 8.69 (2.09) | 4.70 (1.52) | 0.95 (0.49 | <0.001 | 0.09264 | 0.2094 |
| DAG 18:2/20:0/0:0 | 106.64 (22.2) | 62.12 (19.93) | 10.65 (6.45) | <0.0001 | <0.0001 | <0.0001 |
| HexCer d18:0/18:0 | 0.03 (0.01) | 0.01 (0.01) | 0.03 (0.01) | 0.547458 | <0.001 | <0.001 |
| LPC 16:0 | 1.06 (0.23) | 0.67 (0.11) | 0.11 (0.06) | <0.0001 | <0.0001 | <0.0001 |
| LPC 18:0 | 0.59 (0.12) | 0.37 (0.05) | 0.06 (0.04) | <0.0001 | <0.0001 | <0.0001 |
| LPC 18:1 | 0.36 (0.09) | 0.23 (0.05) | 0.04 (0.03) | <0.0001 | <0.0001 | <0.05 |
| LPC 18:2 | 0.69 (0.24) | 0.45 (0.15) | 0.07 (0.05) | <0.0001 | <0.0001 | <0.0001 |
| PC 34:1 | 17.05 (2.95) | 10.92 (2.35) | 1.79 (1.32) | <0.0001 | <0.0001 | <0.0001 |
| PC 36:2 | 20.24 (3.59) | 13.00 (2.39) | 2.61 (1.85) | <0.0001 | <0.0001 | <0.0001 |
| PC 36:3 | 13.52 (3.27) | 8.34 (2.21) | 1.10 (0.80) | <0.0001 | <0.0001 | <0.0001 |
| PC 36:4 | 15.09 (3.51) | 9.70 (2.57) | 1.28 (0.90) | <0.0001 | <0.0001 | <0.0001 |
| PC 38:3 | 3.37 (0.69) | 2.01 (0.49) | 0.29 (0.16) | <0.0001 | <0.0001 | <0.05 |
| PC 38:4 | 6.93 (1.75) | 4.39 (1.12) | 0.54 (0.31) | <0.0001 | <0.0001 | <0.01 |
| PC 38:5 | 2.45 (0.98) | 1.66 (0.54) | 0.20 (0.12) | <0.001 | <0.001 | 0.24255 |
| PC 38:6 | 2.25 (0.98) | 1.59 (0.69) | 0.18 (0.14) | <0.001 | <0.01 | 0.32440 |
| PC O-32:0 | 0.17 (0.03) | 0.11 (0.02) | 0.02 (0.01) | <0.001 | <0.01 | 0.26278 |
| PC O-32:1 | 0.18 (0.04) | 0.11 (0.03) | 0.02 (0.01) | <0.001 | <0.01 | 0.24071 |
| PC O-34:1 | 0.54 (0.10) | 0.33 (0.07) | 0.05 (0.03) | <0.0001 | <0.0001 | <0.001 |
| PC O-34:2 | 0.77 (0.20) | 0.46 (0.12) | 0.07 (0.03) | <0.0001 | <0.0001 | <0.0001 |
| PC O-34:3 | 0.63 (0.21) | 0.38 (0.13) | 0.06 (0.03) | <0.0001 | <0.0001 | <0.0001 |
| PC O-36:3 | 0.61 (0.15) | 0.36 (0.09) | 0.05 (0.03) | <0.0001 | <0.0001 | <0.0001 |
| PC O-36:4 | 1.24 (0.29) | 0.74 (0.20) | 0.10 (0.05) | <0.0001 | <0.0001 | <0.0001 |
| PC O-36:5 | 0.82 (0.25) | 0.50 (0.17) | 0.07 (0.03) | <0.0001 | <0.0001 | <0.0001 |
| PC O-38:4 | 0.72 (0.12) | 0.46 (0.09) | 0.06 (0.04) | <0.0001 | <0.0001 | <0.0001 |
| PC O-38:5 | 1.04 (0.21) | 0.66 (0.15) | 0.09 (0.06) | <0.0001 | <0.0001 | <0.0001 |
| PC O-38:6 | 0.46 (0.14) | 0.30 (0.10) | 0.04 (0.02) | <0.0001 | <0.0001 | <0.01 |
| PC O-40:5 | 0.18 (0.03) | 0.11 (0.02) | 0.02 (0.01) | <0.01 | <0.01 | 0.287674 |
| PE 34:2 | 0.03 (0.01) | 0.02 (0.01) | 0.001 (0.01) | <0.0001 | <0.001 | 0.07888 |
| PE 36:1 | 0.02 (0.01) | 0.01 (0.01) | 0.001 (0.01) | <0.01 | <0.05 | 0.09374 |
| PE 36:2 | 0.10 (0.05) | 0.06 (0.02) | 0.02 (0.01) | <0.0001 | <0.0001 | <0.0001 |
| PE 36:3 | 0.04 (0.02) | 0.02 (0.01) | 0.01 (0.01) | <0.0001 | <0.0001 | <0.05 |
| PE 36:4 | 0.03 (0.01) | 0.02 (0.01) | 0.01 (0.01) | <0.001 | <0.001 | 0.178920 |
| PE 38:4 | 0.05 (0.02) | 0.03 (0.01) | 0.001 (0.01) | <0.0001 | <0.0001 | <0.01 |
| PE O-38:5 | 0.01 (0.01) | 0.01 (0.01) | 0.001 (0.01) | <0.0001 | <0.0001 | <0.01 |
| PE O-38:6 | 0.01 (0.01) | 0.00 (0.01) | 0.001 (0.01) | <0.001 | <0.001 | 0.2377739 |
| SM 34:1;2 | 10.76 (2.26) | 6.55 (1.70) | 0.92 (0.63) | <0.0001 | <0.0001 | <0.0001 |
| SM 36:1;2 | 1.82 (0.46) | 1.10 (0.36) | 0.18 (0.12) | <0.0001 | <0.0001 | <0.05 |
| SM 38:2;2 | 6.11 (1.83) | 2.67 (1.56) | 0.56 (0.42) | <0.0001 | <0.0001 | <0.0001 |
| SM 42:1;2 | 1.99 (0.42) | 1.18 (0.33) | 0.19 (0.12) | <0.0001 | <0.0001 | <0.05 |
| SM 42:2;2 | 5.37 (1.00) | 3.16 (0.72) | 0.47 (0.28) | <0.0001 | <0.0001 | <0.0001 |
| TAG 54:2 (-FA 18:1) | 2.10 (0.68) | 1.03 (0.33) | 0.64 (0.59) | <0.0001 | 0.05789 | <0.0001 |
| TAG 54:2 (-FA 18:2) | 0.17 (0.04) | 0.08 (0.03) | 0.05 (0.04) | 0.693309 | 0.87924 | 0.737423 |
| TAG 54:3 (-FA 18:0) | 1.52 (0.38) | 0.74 (0.25) | 0.43 (0.42) | <0.001 | 0.13283 | <0.01 |
| TAG 54:3 (-FA 18:1) | 10.05 (4.53) | 4.57 (1.85) | 2.77 (3.39) | <0.0001 | <0.0001 | <0.0001 |
| TAG 54:3 (-FA 18:2) | 1.58 (0.40) | 0.75 (0.23) | 0.44 (0.43) | <0.0001 | 0.1276712 | <0.001 |
| TAG 54:4 (-FA 18:1) | 10.32 (4.87) | 4.68 (2.04) | 3.01 (4.07) | <0.0001 | <0.0001 | <0.0001 |
| TAG 54:4 (-FA 18:2) | 5.84 (2.40) | 2.68 (1.05) | 1.70 (2.10) | <0.0001 | <0.0001 | <0.0001 |
| TAG 54:5 (-FA 18:1) | 4.07 (1.91) | 1.91 (0.81) | 1.23 (1.51) | <0.0001 | <0.01 | <0.0001 |
| TAG 54:5 (-FA 18:2) | 5.73 (3.02) | 2.68 (1.21) | 1.77 (2.22) | <0.0001 | <0.001 | <0.0001 |
| TAG 54:6 (-FA 20:5) | 2.58 (1.70) | 1.31 (0.72) | 0.03 (0.02) | <0.0001 | <0.0001 | <0.0001 |
CE: cholesterol ester; Cer: ceramide; DAG: diacylglyceride; HexCer: hexosylceramide; Hex2Cer: dihexosylceramide; PA: phosphatidic acid; LPA: lysophosphatidic acid; PC: phosphatidylcholine; LPC: lysophosphatidylcholine; PE phosphatidyletanolamine; LPE: lysophosphatidyletanolamine; PG phosphatidylglycerol; LPG: lysophosphatidylglycerol; PS: phosphatidylserine; LPS: lysophosphatidylserine; SM: sphingomyelin; TAG: triacylglyceride; FA: fatty acid; O-: ether-linked lipids; SD: standard deviation; ANOVA: analysis of variance; EV: extracellular vesicle.
aData are mean ± SD. ANOVA with Bonferroni correction for multiple comparisons.
Discussion
Disease-associated modifications in plasma lipid content have been reported in Alzheimer’s disease, HIV infection, Parkinson’s disease, cerebrovascular damage, and a number of different metabolic disorders.42–59 Although the direct infusion MS/MSALL approach has been applied in the lipidomics analysis of clinical studies, there are potential issues with reproducibility that need to be optimized. One potential issue is the reliable identification of lipid species. After MS/MS analysis, lipid identification and quantification software such as LipidView™ identify thousands of potential lipid species from samples. However, a fairly large proportion of the lipid species identified and/or the peak intensity assigned to any particular identification are not reproducible when samples are repeatedly analyzed several times. Presumably, these variations result from inconsistencies in the ionization process of sample with complex matrices. The identification of a reliable lipid species list for further data analysis is critical for obtaining accurate and reproducible experimental results in studies using clinical samples. One of the commonly used solutions in LC-based lipidomics is the “80% rule,”60,61 in which a lipid feature will be maintained if it has a nonzero measurement value in at least 80% of the sample data set. Using this rule, missing values caused by slight shifts in analyte peaks are reduced. However, this strategy does not consider inconsistencies in the ionization efficiency of lipid species contained in a complex matrix. These inconsistencies can produce variable peak intensities that are used for quantitation. To increase the consistency of peak intensities for quantitative analyses we defined a CV for each of the identified lipid species and removed ones that were not reliably quantified. Briefly, a pooled sample is subjected to direct infusion MS/MSALL analysis for eight consecutive runs, and the data applied to the LipidView™ for lipid identification. Only the lipid species that appeared with nonzero values seven of eight runs, with a calculated CV of <20% were retained for further analysis.
Here we developed and optimized an untargeted MS-based lipidomic method for the analysis of EV lipid content in human serum. A simple commonly used differential ultracentrifugation technique was used to separate serum that produced several distinct EV fractions. The upper three fractions contained larger EVs that were negative for the typical EV markers, CD63 and TSG101, but were positive for the cytoskeletal protein actin, and contained the highest lipid content. These data are consistent with a more buoyant EV that likely originated from a membrane bubbling or an apoptotic event. Fraction 4 appears to be a transitional fraction, and contains CD63, TSG101, and actin. The lower fractions 5 and 6 contained CD63, TSG101, but were negative for actin, and mitofillin. They contained an intermediate lipid content compared with fraction 1 and the pellet. These EVs are likely a mixture of cellular blebs, apoptotic bodies, and some exosomes. The pellet contained CD63, TSG101 and was the only fraction immunopositive for flotillin. The pellet contained the lowest concentration of lipids, and the highest concentration of ceramide, consistent with the high protein/lipid ratio found in exosomes.62
There are several caveats to this method that should be noted. Although ultracentrifugation seems to greatly enrich the EVs content, the composition of plasma or serum is complex and contains multiple types of lipoproteins with different sizes and densities. High-density (d) lipoproteins (HDL, d = 1.063–1.21 g/mL) have been reported to co-isolate with EVs in blood samples (d = 1.13–1.19) subjected to ultracentrifugation.30,40 Since EVs may themselves carry apolipoproteins,63–65 it is not possible to definitively conclude if apolipopoteins are present or absent from the EV fraction using this isolation method. This may explain the high content of neutral lipids such as CE and glycerolipids we observed in the pellet of fractionated serum, or it is possible that these particular lipid classes are enriched in EVs isolated from serum. This method is also not ideal for high throughput isolation of EVs from clinical samples. Ultracentrifugation takes hours to complete and the number of samples that can be isolated is limited by the rotor capacity (typically 4–6 samples at a time can be isolated). Lastly, although we defined a method to enhance the reproducibility of data produced by the MS/MSALL approach, this method is not an absolute quantitation of lipid concentration. Although between group comparisons are accurate, it is not possible to compare lipids within a given class, as the ionization efficiency varies with chain lengths.
This study reports an optimized workflow for untargeted lipidomic analysis of serum EVs. A simple ultracentrifugation technique isolates at least three clear populations of EVs. The upper three fractions contain more buoyant EVs that are phospholipid rich, but lack CD63, TSG101, or flotillin. Fractions 5 and 6 are of medium buoyancy, and contain CD63, TSG101, but are negative for flotillin. The pellet is dense, ceramide rich, and contains CD63, TSG101, and flotillin. These results provided new insights into the classical ultracentrifugation method used to isolate serum EVs. Future studies will determine if the lipid composition from subfractions of EVs can be used for diagnostic or prognostic measures of disease activity.
Supplemental material
Supplementary_Information-0801-njh-final_resub for Lipidomic characterization of extracellular vesicles in human serum by Suming Chen, Amrita Datta-Chaudhuri, Pragney Deme, Alex Dickens, Raha Dastgheyb, Pavan Bhargava, Honghao Bi and Norman J Haughey in Journal of Circulating Biomarkers
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institutes of Health awards (AG057420, DA040390, MH077542, MH075673, MH110246, MH105280, and MH096636).
ORCID iDs: Pragney Deme  https://orcid.org/0000-0001-9117-891X
https://orcid.org/0000-0001-9117-891X
Norman J Haughey  https://orcid.org/0000-0001-5194-4122
https://orcid.org/0000-0001-5194-4122
Supplemental material: Supplemental material for this article is available online.
References
- 1. Yanez-Mo M, Siljander PRM, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 2015; 4: 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Bi 2014; 30: 255–289. [DOI] [PubMed] [Google Scholar]
- 3. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013; 200(4): 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Verkhratsky A, Matteoli M, Parpura V, et al. Astrocytes as secretory cells of the central nervous system: idiosyncrasies of vesicular secretion. Embo J 2016; 35(3): 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tucher C, Bode K, Schiller P, et al. Extracellular vesicle subtypes released from activated or apoptotic T-lymphocytes carry a specific and stimulus-dependent protein cargo. Front Immunol 2018; 9: 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keller S, Sanderson MP, Stoeck A, et al. Exosomes: from biogenesis and secretion to biological function. Immunol Lett 2006; 107(2): 102–108. [DOI] [PubMed] [Google Scholar]
- 7. Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Bio 2010; 11(8): 593–598. [DOI] [PubMed] [Google Scholar]
- 8. van der Pol E, Boing AN, Harrison P, et al. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev 2012; 64(3): 676–705. [DOI] [PubMed] [Google Scholar]
- 9. Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015; 523(7559): 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hogan MC, Bakeberg JL, Gainullin VG, et al. Identification of biomarkers for PKD1 using urinary exosomes. J Am Soc Nephrol 2015; 26(7): 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015; 527(7578): 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo JH, Cui YZ, Yan ZQ, et al. Phosphoproteome characterization of human colorectal cancer SW620 cell-derived exosomes and new phosphosite discovery for C-HPP. J Proteome Res 2016; 15(11): 4060–4072. [DOI] [PubMed] [Google Scholar]
- 13. Goetzl EJ, Schwartz JB, Abner EL, et al. High complement levels in astrocyte-derived exosomes of Alzheimer disease. Ann Neurol 2018; 83(3): 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goetzl L, Merabova N, Darbinian N, et al. Diagnostic potential of neural exosome cargo as biomarkers for acute brain injury. Ann Clin Transl Neurol 2018; 5(1): 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuwano N, Kato TA, Mitsuhashi M, et al. Neuron-related blood inflammatory markers as an objective evaluation tool for major depressive disorder: an exploratory pilot case-control study. J Affect Disord 2018; 240: 88–98. [DOI] [PubMed] [Google Scholar]
- 16. Karnati HK, Garcia JH, Tweedie D, et al. Neuronal enriched extracellular vesicle proteins as biomarkers for brain traumatic injury. J Neurotrauma 2019; 36(7): 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Srivastava A, Moxley K, Ruskin R, et al. A non-invasive liquid biopsy screening of urine-derived exosomes for miRNAs as biomarkers in endometrial cancer patients. AAPS J 2018; 20(5): 82. [DOI] [PubMed] [Google Scholar]
- 18. Roy S, Hochberg FH, Jones PS. Extracellular vesicles: the growth as diagnostics and therapeutics; a survey. J Extracell Vesicles 2018; 7(1): 1438720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Llorente A, Skotland T, Sylvanne T, et al. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. BBA-Mol Cell Biol L 2013; 1831(7): 1302–1309. [DOI] [PubMed] [Google Scholar]
- 20. Kreimer S, Belov AM, Ghiran I, et al. Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J Proteome Res 2015; 14(6): 2367–2384. [DOI] [PubMed] [Google Scholar]
- 21. Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res 2017; 66: 30–41. [DOI] [PubMed] [Google Scholar]
- 22. Buzas EI, Gyorgy B, Nagy G, et al. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol 2014; 10(6): 356–364. [DOI] [PubMed] [Google Scholar]
- 23. Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 2009; 9(8): 581–593. [DOI] [PubMed] [Google Scholar]
- 24. Muller L, Hong CS, Stolz DB, et al. Isolation of biologically-active exosomes from human plasma. J Immunol Methods 2014; 411: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Joshi P, Benussi L, Furlan R, et al. Extracellular vesicles in Alzheimer’s disease: friends or foes? Focus on aβ-vesicle interaction. Int J Mol Sci 2015; 16(3): 4800–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu G, Yang L, Cai Y, et al. Emerging roles of extracellular vesicles in neurodegenerative disorders: focus on HIV-associated neurological complications. Cell Death Dis 2016; 7(11): e2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chistiakov DA, Chistiakov AA. α-Synuclein-carrying extracellular vesicles in Parkinson’s disease: deadly transmitters. Acta Neurol Belg 2017; 117(1): 43–51. [DOI] [PubMed] [Google Scholar]
- 28. An TX, Qin SH, Xu Y, et al. Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J Extracell Vesicles 2015; 4: 27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou H, Pisitkun T, Aponte A, et al. Exosomal Fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int 2006; 70(10): 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han XL, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res 2003; 44(6): 1071–1079. [DOI] [PubMed] [Google Scholar]
- 31. Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov 2005; 4(7): 594–610. [DOI] [PubMed] [Google Scholar]
- 32. Wang M, Han XL. Advanced shotgun lipidomics for characterization of altered lipid patterns in neurodegenerative diseases and brain injury. Methods Mol Biol 2016; 1303: 405–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cajka T, Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal Chem 2016; 88(1): 524–545. [DOI] [PubMed] [Google Scholar]
- 34. Han XL, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev 2005; 24(3): 367–412. [DOI] [PubMed] [Google Scholar]
- 35. Simons B, Kauhanen D, Sylvanne T, et al. Shotgun lipidomics by sequential precursor ion fragmentation on a hybrid quadrupole time-of-flight mass spectrometer. Metabolites 2012; 2(1): 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu XC, Chen YP, Subramanian R. Comparison of information-dependent acquisition, SWATH, and MSAll techniques in metabolite identification study employing ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. Anal Chem 2014; 86(2): 1202–1209. [DOI] [PubMed] [Google Scholar]
- 37. Rost HL, Rosenberger G, Navarro P, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol 2014; 32(3): 219–223. [DOI] [PubMed] [Google Scholar]
- 38. Tsugawa H, Cajka T, Kind T, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods 2015; 12(6): 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA 2016; 113(8): E968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yuana Y, Levels J, Grootemaat A, et al. Co-isolation of extracellular vesicles and highdensity lipoproteins using density gradient ultracentrifugation. J Extracell Vesicles 2014; 3(1): 23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sampaio JL, Gerl MJ, Klose C, et al. Membrane lipidome of an epithelial cell line. P Natl Acad Sci USA 2011; 108(5): 1903–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han X. Lipid alterations in the earliest clinically recognizable stage of Alzheimer’s disease: implication of the role of lipids in the pathogenesis of Alzheimer’s disease. Curr Alzheimer Res 2005; 2(1): 65–77. [DOI] [PubMed] [Google Scholar]
- 43. Griffiths WJ, Hornshaw M, Woffendin G, et al. Discovering oxysterols in plasma: a window on the metabolome. J Proteome Res 2008; 7(8): 3602–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pallebage-Gamarallage MM, Takechi R, Lam V, et al. Post-prandial lipid metabolism, lipid-modulating agents and cerebrovascular integrity: implications for dementia risk. Atheroscler Suppl 2010; 11(1): 49–54. [DOI] [PubMed] [Google Scholar]
- 45. Mielke MM, Haughey NJ, Bandaru VV, et al. Plasma sphingomyelins are associated with cognitive progression in Alzheimer’s disease. J Alzheimer’s Dis: JAD 2011; 27(2): 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gonzalez CE, Venkatraman VK, An Y, et al. Peripheral sphingolipids are associated with variation in white matter microstructure in older adults. Neurobiol Aging 2016; 43: 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mielke MM, Haughey NJ, Han D, et al. The association between plasma ceramides and sphingomyelins and risk of Alzheimer’s disease differs by sex and APOE in the Baltimore Longitudinal Study of Aging. J Alzheimer’s Dis: JAD 2017; 60(3): 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sacktor N, Haughey N, Cutler R, et al. Novel markers of oxidative stress in actively progressive HIV dementia. J Neuroimmunol 2004; 157(1–2): 176–184. [DOI] [PubMed] [Google Scholar]
- 49. Bandaru VV, McArthur JC, Sacktor N, et al. Associative and predictive biomarkers of dementia in HIV-1-infected patients. Neurology 2007; 68(18): 1481–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haughey NJ, Steiner J, Nath A, et al. Converging roles for sphingolipids and cell stress in the progression of neuro-AIDS. Front Biosci 2008; 13: 5120–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haughey NJ, Bandaru VV, Bae M, et al. Roles for dysfunctional sphingolipid metabolism in Alzheimer’s disease neuropathogenesis. Biochimt Biophys Acta 2010; 1801(8): 878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bandaru VV, Mielke MM, Sacktor N, et al. A lipid storage-like disorder contributes to cognitive decline in HIV-infected subjects. Neurology 2013; 81(17): 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Walkley SU, Suzuki K. Consequences of NPC1 and NPC2 loss of function in mammalian neurons. Biochim Biophys Acta 2004; 1685(1–3): 48–62. [DOI] [PubMed] [Google Scholar]
- 54. Tallaksen CM, Berg JE. Miglustat therapy in juvenile Sandhoff disease. J Inherit Metab Dis 2009; 32(Suppl 1): S289–293. [DOI] [PubMed] [Google Scholar]
- 55. Lipina C, Hundal HS. Sphingolipids: agents provocateurs in the pathogenesis of insulin resistance. Diabetologia 2011; 54(7): 1596–1607. [DOI] [PubMed] [Google Scholar]
- 56. Crescenzo R, Bianco F, Coppola P, et al. Increased skeletal muscle mitochondrial efficiency in rats with fructose-induced alteration in glucose tolerance. Br J Nutr 2013; 110(11): 1996–2003. [DOI] [PubMed] [Google Scholar]
- 57. Bae M, Bandaru VV, Patel N, et al. Ceramide metabolism analysis in a model of binge drinking reveals both neuroprotective and toxic effects of ethanol. J Neurochem 2014; 131(5): 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mathews AT, Famodu OA, Olfert MD, et al. Efficacy of nutritional interventions to lower circulating ceramides in young adults: FRUVEDomic pilot study. Physiol Rep 2017; 5(13): e13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang H, Ralle M, Wolfgang MJ, et al. Copper-dependent amino oxidase 3 governs selection of metabolic fuels in adipocytes. PLoS Biol 2018; 16(9): e2006519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bijlsma S, Bobeldijk L, Verheij ER, et al. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal Chem 2006; 78(2): 567–574. [DOI] [PubMed] [Google Scholar]
- 61. Li J, Hoene M, Zhao X, et al. Stable isotope-assisted lipidomics combined with nontargeted isotopomer filtering, a tool to unravel the complex dynamics of lipid metabolism. Anal Chem 2013; 85(9): 4651–4657. [DOI] [PubMed] [Google Scholar]
- 62. Osteikoetxea X, Balogh A, Szabo-Taylor K, et al. Improved characterization of EV preparations based on protein to lipid ratio and lipid properties. PloS One 2015; 10(3): e0121184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nikitidou E, Khoonsari PE, Shevchenko G, et al. Increased release of apolipoprotein E in extracellular vesicles following amyloid-β protofibril exposure of neuroglial co-cultures. J Alzheimer’s Dis: JAD 2017; 60(1): 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pienimaeki-Roemer A, Kuhlmann K, Bottcher A, et al. Lipidomic and proteomic characterization of platelet extracellular vesicle subfractions from senescent platelets. Transfusion 2015; 55(3): 507–521. [DOI] [PubMed] [Google Scholar]
- 65. van Niel G, Bergam P, Di Cicco A, et al. Apolipoprotein E regulates amyloid formation within endosomes of pigment cells. Cell Rep 2015; 13(1): 43–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Information-0801-njh-final_resub for Lipidomic characterization of extracellular vesicles in human serum by Suming Chen, Amrita Datta-Chaudhuri, Pragney Deme, Alex Dickens, Raha Dastgheyb, Pavan Bhargava, Honghao Bi and Norman J Haughey in Journal of Circulating Biomarkers