Abstract
An imbalance in our microbiota may contribute to many human diseases, but the mechanistic underpinnings of dysbiosis remain poorly understood. We argue that dysbiosis is secondary to a defect in microbiota-nourishing immunity, a part of our immune system that balances the microbiota to attain colonization resistance against environmental exposure to microorganisms. We discuss this new hypothesis and its implications for ulcerative colitis, an inflammatory bowel disease of the large intestine.
Keywords: microbiota, dysbiosis, ulcerative colitis, epithelial cell metabolism
INTRODUCTION
Anti-infective immunity has been studied extensively for over a century and is considered to be our body’s main tactic to deal with microbes. Numerous seminal concepts were established by studying how anti-infective immunity detects microbial intruders and removes them from our body. However, in the wake of recent advances in high-throughput sequencing, it is becoming increasingly clear that interactions between our immune system and our resident microbial communities, the microbiota, represent a distinct type of immune response that differs in its goals but is complementary in its function to anti-infective immunity (Fig. 1).1 Initial attempts to describe these interactions proposed the term “acceptive immunity” to convey the concept that our immune system detects commensal microbes with the goal of developing immunological tolerance, thereby giving rise to a peaceful coexistence.2
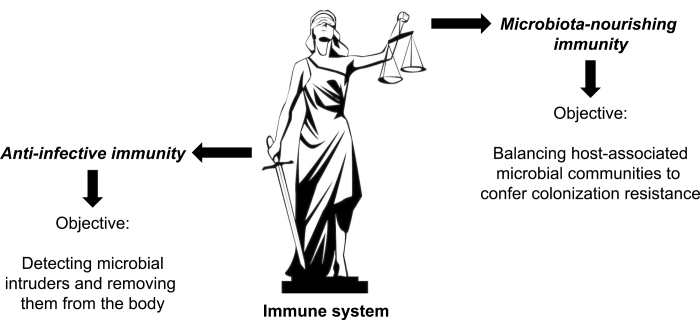
FIGURE 1.
Our immune system employs 2 different tactics for interacting with microbes. The schematic illustrates that our immune system bears a resemblance to Lady Justice in that it protects our body from microbial invasion by fighting infection with one arm, while balancing host-associated microbial communities with the other. Anti-infective immunity represents the arm of our immune system that functions in detecting intruding microbes, such as infection with pathogens or entry of commensal microbes during injury, and generating immune responses aiding in the removal of the intruder from the body. Microbiota-nourishing immunity refers to a separate arm of our immune system that functions in balancing the microbiota composition to establish colonization resistance, which in turn protects the host against constant environmental exposure to microbes.
Yet an alternative view holds that the microbiota should be viewed as an additional organ that fulfills important functions for human health.3 Viewing our interaction with commensal microbes from this perspective suggests that host cell types and mechanisms involved in nourishing and shaping the microbiota serve an important function in maintaining our microbial organ.4 In turn, maintaining a balanced microbial community confers niche protection against pathogens or pathobionts, a property known as colonization resistance.5 We propose the term “microbiota-nourishing immunity” to describe the part of our immune system that balances the microbiota to generate colonization resistance (Fig. 1). In addition to host immune mechanisms that sculpt our microbiota,6 microbiota-nourishing immunity includes immune mechanisms conferring colonization resistance, an effector function mediated by our microbial organ. Thus, microbiota-nourishing immunity contains components of both host and microbial origin, which distinguishes this concept from acceptive immunity. Whereas the main goal of anti-infective immunity is to detect and eliminate microbial intruders from our body, the principle objective of microbiota-nourishing immunity is to balance our microbial self, thereby protecting the host from continuous environmental exposure to microbes that represent potential intruders (Fig. 2). Thus, anti-infective immunity and microbiota-nourishing immunity fulfill complementary functions to shield us from possible harm from microbial intruders. Here, we discuss the implications of this theory for diseases associated with an imbalance in the microbiota composition (dysbiosis) with a special emphasis on ulcerative colitis.
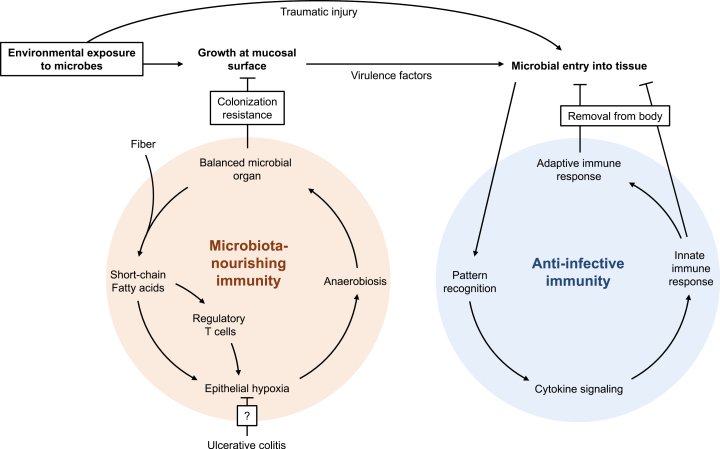
FIGURE 2.
Anti-infective immunity and microbiota-nourishing immunity in the large intestine. A balanced microbial organ converts dietary fiber into short-chain fatty acids, a signal processed by regulatory T cells and epithelial cells to generate epithelial hypoxia. In turn, epithelial hypoxia helps maintain anaerobiosis, thereby balancing the microbial organ to attain colonization resistance. The first line of defense against environmental exposure to microbes is colonization resistance conferred by microbiota-nourishing immunity, which prevents incoming microorganisms from gaining a foothold. When microbes are introduced into tissue—either because of traumatic injury or because virulence factors enable a pathogen to overcome barrier functions—the intruders are recognized by anti-infective immunity through pattern recognition. The consequent cytokine storm orchestrates innate and adaptive immune responses designed to remove the microbe from our body.
Epithelial Metabolism Balances the Microbiota
One of the reasons why functions contributing to microbiota-nourishing immunity remain poorly studied is the remarkable diversity in microbial taxa carriage between individuals,7 which has hampered progress in defining balance. In turn, in the absence of an obvious definition for microbiota balance, it is difficult to identify immune mechanisms maintaining this trait in microbial communities.6 The germ-organ theory overcomes this impasse by proposing that the metabolism of epithelial cells balances microbial communities in the large intestine by maintaining anaerobiosis to ensure a dominance of obligate anaerobic bacteria belonging to the classes Clostridia (phylum Firmicutes) and Bacteroidia (phylum Bacteroidetes).4 The concept of balance applied by the germ-organ theory is based on the ecological view that the immune system maintains homeostasis by shaping the microbiota to be beneficial.8 A microbial community dominated by Clostridia and Bacteroidia provides benefit because these microbes encode a broad spectrum of enzymes for hydrolyzing different complex carbohydrates (fiber) that cannot be processed by host enzymes in the upper gastrointestinal tract.9 Thus, by maintaining a dominance of obligate anaerobic bacteria, the colonic epithelium balances the microbiota to ensure the microbial community provides nutritional benefit through the generation of fermentation products, such as short-chain fatty acids, from fiber.4
Mechanistically, the host limits the amount of oxygen emanating from the colonic surface by polarizing the epithelial metabolism toward oxidative phosphorylation, which is characterized by high oxygen consumption, thereby producing epithelial hypoxia.10 Microbiota-nourishing immunity monitors the presence of a balanced microbial community by detecting microbiota-derived short-chain fatty acids, which activate epithelial PPARγ (peroxisome proliferator-activated receptor gamma) signaling11 and maintain the colonic regulatory T cell pool.12, 13 In turn, epithelial PPARγ signaling and regulatory T cells cooperate to polarize epithelial metabolism toward oxidative phosphorylation and mitochondrial β-oxidation of fatty acids to preserve epithelial hypoxia in the colon (Fig. 2).11 The picture emerging from these studies is that epithelial cell metabolism plays a key role in microbiota-nourishing immunity by prominently influencing the nutritional environment of resident microbes to balance the microbiota composition.
Balanced Microbial Communities Confer Colonization Resistance
Whereas the nutritional benefit of converting fiber into fermentation products only concerns the colonic microbiota, the ability to confer colonization resistance is a universal property of balanced microbial communities that applies to all host surfaces. Colonization resistance is acquired after birth through the incorporation of maternal and environmental microbes into the developing microbiota. Microorganisms continue to accumulate over time until all available nutrient-niches are occupied by a suitable tenant, and the microbiota reaches a balanced equilibrium state.14 Stable coexistence requires that each member within the microbial community can utilize at least one of the available nutrients better than any other member, a concept known as the nutrient-niche hypothesis.15 Importantly, priority effects make it difficult for an incoming microbe to unseat the occupant of a particular nutrient-niche,16 which explains why the vast majority of strains in the microbiota of healthy volunteers are resident for years.17 Therefore, a balanced microbiota creates a challenge for newly arriving microbes to establish permanent residency, as the best seats in the house are already taken.18 As a result of colonization resistance, a balanced microbiota generates resilience against the stress imposed by a constant environmental exposure to microbes, which constitutes the main effector function of microbiota-nourishing immunity (Fig. 2).
Microbiota-nourishing Immunity and Communicable Diseases
Microbial pathogens are defined as microorganisms that can cause communicable diseases (also known as infectious diseases) because they can overcome host defenses in individuals with an intact immune system. Conventional wisdom holds pathogenic microbes overcome host defenses using their virulence factors, a view classically applied to components of anti-infective immunity (Fig. 1). Because one of the major defenses initially encountered by mucosal pathogens is colonization resistance, it stands to reason that some virulence factors of mucosal pathogens are designed to overcome microbiota-nourishing immunity. Consistent with this idea, recent work shows that virulence factors of enteric pathogens target host components of microbiota-nourishing immunity to create a new nutrient-niche that enables the pathogenic microbe to triumph over colonization resistance.19 As an example of this strategy, we will briefly discuss the pathogenesis of the murine pathogen Citrobacter rodentium (phylum Proteobacteria).
Citrobacter rodentium is a member of the attaching and effacing (AE) pathogens, a group of luminal enteric pathogens that includes enteropathogenic Escherichia coli (EPEC) and enterohaemorrhagic E. coli (EHEC) (phylum Proteobacteria).20 Attaching and effacing pathogens share a virulence factor, a type III secretion system (T3SS) that injects bacterial proteins into the cytosol of epithelial cells to mediate intimate attachment to the mucosal surface.21 Epithelial injury inflicted by C. rodentium triggers epithelial repair responses characterized by elongation of the crypts of Lieberkühn, a pathological change termed colonic crypt hyperplasia.22 Crypt elongation results from excessive division of transit-amplifying cells, an early intermediate epithelial cell type that divides a finite number of times before terminally differentiating into mature colonic epithelial cells (colonocytes) or goblet cells.23 Dividing transit-amplifying cells obtain energy through anaerobic glycolysis, which is characterized by conversion of glucose into lactate even in the presence of oxygen,24 a process known as the Warburg metabolism.25 An accumulation of transit-amplifying cells during colonic crypt hyperplasia thus increases the amount of oxygen emanating from the mucosal surface, which impairs microbiota-nourishing immunity by disrupting anaerobiosis in the colon. By elevating oxygen availability, colonic crypt hyperplasia generates a new luminal nutrient-niche in which C. rodentium can expand by using aerobic respiration to consume microbiota-derived fermentation products, such as formate.26 Through this series of events, C. rodentium uses its virulence factors to create a new nutrient-niche to overcome colonization resistance,19 the main effector function of microbiota-nourishing immunity (Fig. 2).
Microbiota-nourishing Immunity and Noncommunicable Diseases
Medical conditions triggered by an underlying, nontransmissible host defect are termed noncommunicable diseases. Noncommunicable diseases caused by a defect in anti-infective immunity increase the risk for contracting opportunistic infections, and these diseases fit the classic definition of an immunodeficiency.27 However, there is evidence that a weakening of microbiota-nourishing immunity can also produce immunodeficiency. For example, antibiotic therapy impairs colonization resistance by disrupting the microbial component of microbiota-nourishing immunity, thereby rendering patients more susceptible to infection with the opportunistic pathogen Clostridium difficile (class Clostridia).28 Fecal microbiota transplants can restore a balanced composition of our microbial organ, thereby restoring colonization resistance after antibiotic therapy.29 Alternatively, immunodeficiencies might be caused by impairing one of the host components of microbiota-nourishing immunity. The hypothesis that the host balances the microbiota composition implies that dysbiosis is an expected consequence of disrupting the underlying host control mechanisms.6 Thus, dysbiosis might be a generic microbial signature of impaired microbiota-nourishing immunity. We will explore the validity of this hypothesis for ulcerative colitis, a noncommunicable disease associated with dysbiosis.30
Impairment of Microbiota-nourishing Immunity During Ulcerative Colitis
Ulcerative colitis (UC) is an inflammatory bowel disease (IBD) that causes chronic inflammation with lesions closely resembling those triggered by infection with an AE pathogen in a primate model.31 As discussed previously, the T3SS shared by AE pathogens triggers colonic crypt hyperplasia to increase epithelial oxygenation in a mouse model, which drives a luminal expansion of the facultative anaerobic C. rodentium (family Enterobacteriaceae).26 Similarly, ulcerative colitis is associated with a dysbiotic expansion of facultative anaerobic Enterobacteriaceae (phylum Proteobacteria).30, 32 The oxygen hypothesis proposes that this change in the microbiota composition reflects a disruption of anaerobiosis in the lumen of the large intestine during ulcerative colitis (Fig. 2).33, 34 Experimental support for this idea is provided by the observation that microbial respiration is a metabolic signature of dysbiosis in a mouse model of chemically induced colitis.35 The mechanism by which colonic crypt hyperplasia increases epithelial oxygenation in a mouse model of C. rodentium infection is an accumulation of undifferentiated transit-amplifying cells and a concomitant decline in the numbers of colonocytes and goblet cells, which causes a shift in epithelial metabolism from mitochondrial β-oxidation of fatty acids to anaerobic glycolysis.26 Consistent with the idea that a similar shift in epithelial metabolism occurs in humans, colonic epithelial cells from ulcerative colitis patients exhibit reduced mitochondrial β-oxidation of butyrate to carbon dioxide.36 Furthermore, ulcerative colitis is associated with a reduction in the number of goblet cells and a concomitant thinning of the mucus layer.37, 38 Collectively, these data support the idea that dysbiosis during ulcerative colitis is triggered by a shift in epithelial metabolism that impairs microbiota-nourishing immunity by disrupting anaerobiosis in the colon (Fig. 2). However, additional work is needed to establish causality by demonstrating that epithelial dysfunction is the main driver of dysbiosis during ulcerative colitis in humans.
Another mechanism contributing to a dysbiotic expansion of Enterobacteriaceae during colitis is the generation of nitrate in the gut lumen,39 which depends on elevated synthesis of the host enzyme inducible nitric oxide synthase (iNOS).40 The reaction catalyzed by iNOS is the production of nitric oxide from L-arginine,41 which significantly increases nitric oxide concentrations in colonic luminal gas of individuals with ulcerative colitis.42 Nitric oxide can react to form peroxynitrite, which can be further converted to nitrate in a reaction catalyzed by carbon dioxide.43 In mouse models of genetically or chemically induced colitis, an expansion of facultative anaerobic Enterobactericeae, such as E. coli, can be explained mechanistically by an increased luminal availability of oxygen and nitrate.35, 40Enterobactericeae coli uses respiration to consume microbiota-derived formate, which is present at elevated luminal concentrations during murine colitis.35
The observation that ulcerative colitis responds to antibiotic therapy44 suggests that dysbiosis caused by disrupting microbiota-nourishing immunity can exacerbate intestinal inflammation. Consistent with this idea, intestinal inflammation can be moderated in mouse models by blunting an expansion of facultative anaerobic Enterobacteriaceae through selective inhibition of microbial pathways for respiration and formate oxidation, which are operational only during episodes of inflammation.45 Yet, the hypothesis that dysbiosis is caused by a disruption of microbiota-nourishing immunity suggests that an alternative strategy for rebalancing the microbiota would be to reverse the shift in epithelial metabolism observed in ulcerative colitis patients,36 thereby restoring host control over the ecosystem.6 Activation of epithelial PPARγ signaling promotes a differentiation of transit-amplifying cells into colonocytes,46 thus reducing crypt hyperplasia.47 The idea that epithelial differentiation rebalances the microbiota is supported by the observation that treatment of ulcerative colitis patients with the PPARγ agonist mesalazine (5-aminosalicylic acid)48 lowers the abundance of facultative anaerobic Proteobacteria in the colonic microbiota,49 which is indicative of restoring anaerobiosis in the colon.19 These data raise the intriguing possibility that topical treatment of the epithelial surface with mesalazine, a first-line therapy for bringing mild to moderate cases of ulcerative colitis into remission,48, 50–52 restores microbiota-nourishing immunity to rebalance the microbiota. However, additional work is needed to provide direct evidence that mesalazine restores epithelial hypoxia to rebalance the microbiota. Targeting of epithelial metabolism would explain why mesalazine is no longer effective in patients with severe acute ulcerative colitis53 because severe epithelial erosion, which accompanies this disease manifestation, eliminates the presumed treatment target.48, 52
CONCLUSIONS
The picture emerging from these studies is that ulcerative colitis might be an acquired immunodeficiency that is caused, at least in part, by an impairment in microbiota-nourishing immunity (Fig. 2). Dysbiosis observed during ulcerative colitis is likely to be a microbial signature of epithelial dysfunction,54 which emerges as a promising therapeutic target for restoring microbiota-nourishing immunity. However, the concept of microbiota-nourishing immunity might hold relevance beyond ulcerative colitis for a wide range of noncommunicable diseases associated with gut dysbiosis such as colorectal cancer,55 cancer cachexia,56 irritable bowel syndrome,57, 58 inflammaging,59 or necrotizing enterocolitis.60 A better understanding of immune mechanisms underlying microbiota-nourishing immunity promises to provide insights into the etiology of a broad spectrum of human diseases.
Supported by: YL.was supported by Vaadia-BARD Postdoctoral Fellowship FI-505–2014. Work in AJB’s laboratory was supported by USDA/NIFA award 2015-67015-22930 and by Public Health Service Grants AI044170, AI096528, AI112445 and AI112949.
REFERENCES
- 1. Kisseleva EP. Innate immunity underlies symbiotic relationships. Biochemistry (Mosc). 2014;79:1273–1285. [DOI] [PubMed] [Google Scholar]
- 2. Klimovich VB. Actual problems of evolutional immunology. Zh Evol Biokhim Fiziol. 2002;38:442–451. [PubMed] [Google Scholar]
- 3. O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Byndloss MX, Bäumler AJ. The germ-organ theory of non-communicable diseases. Nat Rev Microbiol. 2018;16:103–110. [DOI] [PubMed] [Google Scholar]
- 5. Olsan EE, Byndloss MX, Faber F, et al. . Colonization resistance: the deconvolution of a complex trait. J Biol Chem. 2017;292:8577–8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Byndloss MX, Pernitzsch SR, Bäumler AJ. Healthy hosts rule within: ecological forces shaping the gut microbiota. Mucosal Immunol. 2018;11:1299–1305. [DOI] [PubMed] [Google Scholar]
- 7. Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Foster KR, Schluter J, Coyte KZ, et al. . The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El Kaoutari A, Armougom F, Gordon JI, et al. . The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. [DOI] [PubMed] [Google Scholar]
- 10. Litvak Y, Byndloss MX, Bäumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018;362:eaat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Byndloss MX, Olsan EE, Rivera-Chávez F, et al. . Microbiota-activated PPAR-γ signaling inhibits dysbiotic enterobacteriaceae expansion. Science. 2017;357:570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith PM, Howitt MR, Panikov N, et al. . The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science. 2013;341:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Atarashi K, Tanoue T, Shima T, et al. . Induction of colonic regulatory T cells by indigenous clostridium species. Science. 2011;331:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sommer F, Anderson JM, Bharti R, et al. . The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–638. [DOI] [PubMed] [Google Scholar]
- 15. Freter R, Brickner H, Fekete J, et al. . Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun. 1983;39:686–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2018;15:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faith JJ, Guruge JL, Charbonneau M, et al. . The long-term stability of the human gut microbiota. Science. 2013;341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sassone-Corsi M, Raffatellu M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol. 2015;194:4081–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rivera-Chávez F, Lopez CA, Bäumler AJ. Oxygen as a driver of gut dysbiosis. Free Radic Biol Med. 2017;105:93–101. [DOI] [PubMed] [Google Scholar]
- 20. Luperchio SA, Newman JV, Dangler CA, et al. . Citrobacter rodentium, the causative agent of transmissible murine colonic hyperplasia, exhibits clonality: synonymy of C. rodentium and mouse-pathogenic Escherichia coli. J Clin Microbiol. 2000;38:4343–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wong AR, Pearson JS, Bright MD, et al. . Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol. 2011;80:1420–1438. [DOI] [PubMed] [Google Scholar]
- 22. Collins JW, Keeney KM, Crepin VF, et al. . Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol. 2014;12:612–623. [DOI] [PubMed] [Google Scholar]
- 23. Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan YY, Davidson LA, Callaway ES, et al. . A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am J Physiol Gastrointest Liver Physiol. 2015;309:G1–G9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lopez CA, Miller BM, Rivera-Chávez F, et al. . Virulence factors enhance citrobacter rodentium expansion through aerobic respiration. Science. 2016;353:1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kovanen PE, Leonard WJ. Cytokines and immunodeficiency diseases: critical roles of the gamma©-dependent cytokines interleukins 2, 4, 7, 9, 15, and 21, and their signaling pathways. Immunol Rev. 2004;202:67–83. [DOI] [PubMed] [Google Scholar]
- 28. Kuijper EJ, Coignard B, Tüll P; ESCMID Study Group for Clostridium difficile; EU Member States; European Centre for Disease Prevention and Control Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;12(Suppl 6):2–18. [DOI] [PubMed] [Google Scholar]
- 29. Bakken JS, Borody T, Brandt LJ, et al. ; Fecal Microbiota Transplantation Workgroup Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol. 2011;9:1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morgan XC, Tickle TL, Sokol H, et al. . Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mansfield KG, Lin KC, Xia D, et al. . Enteropathogenic Escherichia coli and ulcerative colitis in cotton-top tamarins (Saguinus oedipus). J Infect Dis. 2001;184:803–807. [DOI] [PubMed] [Google Scholar]
- 32. Lepage P, Häsler R, Spehlmann ME, et al. . Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. [DOI] [PubMed] [Google Scholar]
- 33. Rigottier-Gois L. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. Isme J. 2013;7:1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henson MA, Phalak P. Microbiota dysbiosis in inflammatory bowel diseases: in silico investigation of the oxygen hypothesis. BMC Syst Biol. 2017;11:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hughes ER, Winter MG, Duerkop BA, et al. . Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe. 2017;21:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roediger WE. The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet. 1980;2:712–715. [DOI] [PubMed] [Google Scholar]
- 37. Strugala V, Dettmar PW, Pearson JP. Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn’s disease. Int J Clin Pract. 2008;62:762–769. [DOI] [PubMed] [Google Scholar]
- 38. McCormick DA, Horton LW, Mee AS. Mucin depletion in inflammatory bowel disease. J Clin Pathol. 1990;43:143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dudhgaonkar SP, Tandan SK, Kumar D, et al. . Influence of simultaneous inhibition of cyclooxygenase-2 and inducible nitric oxide synthase in experimental colitis in rats. Inflammopharmacology. 2007;15:188–195. [DOI] [PubMed] [Google Scholar]
- 40. Winter SE, Winter MG, Xavier MN, et al. . Host-derived nitrate boosts growth of E. Coli in the inflamed gut. Science. 2013;339:708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Palmer RM, Ashton DS, Moncada S. Vascular endothelial cells synthesize nitric oxide from L-arginine. Nature. 1988;333:664–666. [DOI] [PubMed] [Google Scholar]
- 42. Lundberg JO, Hellström PM, Lundberg JM, et al. . Greatly increased luminal nitric oxide in ulcerative colitis. Lancet. 1994;344:1673–1674. [DOI] [PubMed] [Google Scholar]
- 43. Szabó C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6:662–680. [DOI] [PubMed] [Google Scholar]
- 44. Khan KJ, Ullman TA, Ford AC, et al. . Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011;106:661–673. [DOI] [PubMed] [Google Scholar]
- 45. Zhu W, Winter MG, Byndloss MX, et al. . Precision editing of the gut microbiota ameliorates colitis. Nature. 2018;553:208–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tylichová Z, Straková N, Vondráček J, et al. . Activation of autophagy and pparγ protect colon cancer cells against apoptosis induced by interactive effects of butyrate and DHA in a cell type-dependent manner: the role of cell differentiation. J Nutr Biochem. 2017;39:145–155. [DOI] [PubMed] [Google Scholar]
- 47. Rousseaux C, El-Jamal N, Fumery M, et al. . The 5-aminosalicylic acid antineoplastic effect in the intestine is mediated by pparγ. Carcinogenesis. 2013;34:2580–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rousseaux C, Lefebvre B, Dubuquoy L, et al. . Intestinal antiinflammatory effect of 5-aminosalicylic acid is dependent on peroxisome proliferator-activated receptor-gamma. J Exp Med. 2005;201:1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu J, Chen N, Wu Z, et al. . 5-aminosalicylic acid alters the gut bacterial microbiota in patients with ulcerative colitis. Front Microbiol. 2018;9:1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brogden RN, Sorkin EM. Mesalazine. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in chronic inflammatory bowel disease. Drugs. 1989;38:500–523. [DOI] [PubMed] [Google Scholar]
- 51. Greenfield SM, Punchard NA, Teare JP, et al. . Review article: the mode of action of the aminosalicylates in inflammatory bowel disease. Aliment Pharmacol Ther. 1993;7:369–383. [DOI] [PubMed] [Google Scholar]
- 52. Zhou SY, Fleisher D, Pao LH, et al. . Intestinal metabolism and transport of 5-aminosalicylate. Drug Metab Dispos. 1999;27:479–485. [PubMed] [Google Scholar]
- 53. Kedia S, Ahuja V, Tandon R. Management of acute severe ulcerative colitis. World J Gastrointest Pathophysiol. 2014;5:579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Litvak Y, Byndloss MX, Tsolis RM, et al. . Dysbiotic proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol. 2017;39:1–6. [DOI] [PubMed] [Google Scholar]
- 55. Arthur JC, Perez-Chanona E, Mühlbauer M, et al. . Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338: 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pötgens SA, Brossel H, Sboarina M, et al. . Klebsiella oxytoca expands in cancer cachexia and acts as a gut pathobiont contributing to intestinal dysfunction. Sci Rep. 2018;8:12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Carroll IM, Ringel-Kulka T, Siddle JP, et al. . Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:521–30, e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Krogius-Kurikka L, Lyra A, Malinen E, et al. . Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Conley MN, Wong CP, Duyck KM, et al. . Aging and serum MCP-1 are associated with gut microbiome composition in a murine model. PeerJ. 2016;4:e1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Normann E, Fahlén A, Engstrand L, et al. . Intestinal microbial profiles in extremely preterm infants with and without necrotizing enterocolitis. Acta Paediatr. 2013;102:129–136. [DOI] [PubMed] [Google Scholar]