Abstract
The central oscillator generating cyanobacterial circadian rhythms comprises KaiA, KaiB, and KaiC proteins. Their interactions cause KaiC phosphorylation and dephosphorylation cycles over approximately 24 h. KaiB interacts with phosphorylated KaiC in competition with SasA, an output protein harboring a KaiB-homologous domain. Structural data have identified KaiB–KaiC interaction sites; however, KaiB mutations distal from the binding surfaces can impair KaiB–KaiC interaction and the circadian rhythm. Reportedly, KaiB and KaiC exclusively form a complex in a 6:6 stoichiometry, indicating that KaiB–KaiC hexamer binding shows strong positive cooperativity. Here, mutational analysis was used to investigate the functional significance of this cooperative interaction. Results demonstrate that electrostatic complementarity between KaiB protomers promotes their cooperative assembly, which is indispensable for accurate rhythm generation. SasA does not exhibit such electrostatic complementarity and noncooperatively binds to KaiC. Thus, the findings explain KaiB distal mutation effects, providing mechanistic insights into clock protein interplay.
Keywords: cooperative interaction, circadian rhythm, clock protein, native mass spectrometry, cooperativity
1. Introduction
Circadian rhythms enable organisms, including mammals and plants, to coordinate their biochemistry and physiology with daily environmental alterations that are caused by Earth’s light and dark cycles. Cyanobacteria are photoautotrophic organisms that use circadian rhythms to regulate their photosynthetic activity. A similar regulatory mechanism has been observed in some eukaryotic algae and plants; accordingly, cyanobacteria are considered to be one of the most useful model organisms to study circadian biology [1]. Cyanobacteria utilize a central oscillator to maintain their circadian rhythms. It comprises three proteins—KaiA, KaiB, and KaiC [2]—which interact in the presence of adenosine 5′-triphosphate (ATP), resulting in periodic KaiC phosphorylation and dephosphorylation cycles over 24 h. The KaiABC circadian clock has been successfully reconstituted in vitro without daylight oscillation, suggesting that the clock functions autonomously, irrespective of transcriptional and translational feedback systems [3].
KaiC forms a homohexamer with a double-ring structure composed of the CI and CII rings (Figure 1a) [4,5]. Two specific residues, Ser431 and Thr432 (denoted as S and T, respectively), that are positioned in the CII ring are periodically phosphorylated and dephosphorylated over 24 h as follows: KaiC-S/T → KaiC-S/pT → KaiC-pS/pT → KaiC-pS/T → KaiC-S/T, where “p” represents the phosphorylated residue [6]. KaiA stimulates KaiC phosphorylation through its interaction with the C-terminal region of the CII ring [5,7,8], whereas KaiB preferentially binds to the CI ring of phosphorylated KaiC (p-KaiC), which accelerates dephosphorylation [6,9,10,11].
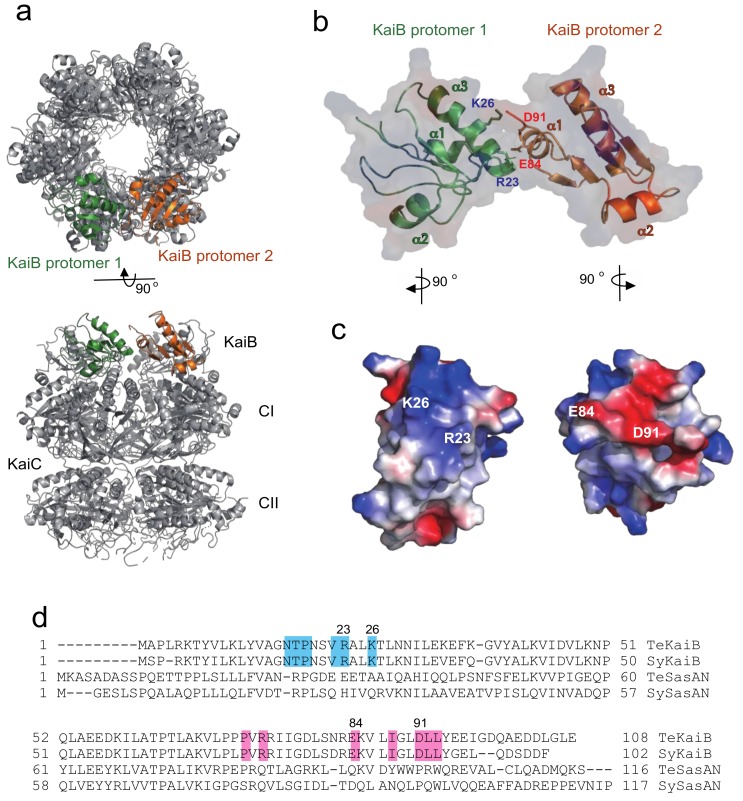
Figure 1.
KaiB–KaiB interface in the KaiB–KaiC complex. (a) Overall structure of the KaiB–KaiC complex. This structural model is based on the amino acid sequence of wild-type TeKaiB and the crystal structure of the complex in which TeKaiB has three mutation sites (PDB code: 5JWQ). The KaiC hexamer forms a double-ring structure composed of CI and CII rings and KaiB forms a hexameric ring on the CI ring. The two KaiB protomers are colored green and orange. (b,c) The KaiB–KaiB lateral interaction and surface electrostatic complementarity between two KaiB protomers. (d) Sequence alignment of KaiB and SasA originating from Synechococcus elongatus PCC 7942 and Thermosynechococcus elongatus. Sequences were aligned using the Dali server. The residues involved in the KaiB–KaiB lateral interactions are highlighted in cyan (protomer 1) and pink (protomer 2).
SasA is a circadian clock-output protein with an N-terminal domain that is homologous to KaiB and a C-terminal EnvZ-like histidine kinase domain [12,13]. It interacts with the CI ring of the phosphorylated KaiC hexamer using its N-terminal domain [12], which triggers the autophosphorylation of SasA at its catalytic histidine residue (e.g., His160 in Thermosynechococcus elongatus SasA). The phosphorylated SasA is released from KaiC, and it subsequently interacts with and transfers the phosphate group to its cognate response regulator, RpaA [14]. The phosphorylated RpaA then regulates the transcription of its target genes that are associated with circadian rhythms. KaiB and SasA have a common interaction surface on the CI ring of the phosphorylated KaiC and therefore compete with each other to bind to KaiC [15,16,17].
Previous structural studies regarding the KaiB–KaiC interaction using crystallography, electron microscopy, solution scattering, and native mass spectrometry (MS) have revealed that the KaiC hexamer can accommodate a maximum of six KaiB protomers (Figure 1a). The crystallographic data revealed that the KaiB–KaiC interaction sites are mainly composed of a KaiC B-loop and KaiB α2 helix. Intriguingly, mutations in KaiB that are distal from these binding surfaces, e.g., R22A and D90G in Synechococcus elongatus KaiB, can impair its interaction with the KaiC hexamer and the consequent circadian rhythm [18,19]. These mutations are located at the lateral KaiB–KaiB interaction surfaces in the KaiB–KaiC complex [20]. In a previous study, native MS titration analysis revealed that KaiB and KaiC form a complex exclusively in a 6:6 stoichiometry, indicating that KaiB and the KaiC hexamer bind with strong positive cooperativity [21]. However, the biological relevance of this cooperative interaction remains to be elucidated.
Here, the molecular mechanisms and functional significance of the cooperative binding of KaiB to the KaiC hexamer are addressed by mutational analysis using native MS. In addition, the KaiC-binding property of SasA compared with that of KaiB is characterized. Finally, based on the findings, the importance of the cooperative binding of KaiB to the KaiC hexamer for regulating circadian rhythms is discussed.
2. Results and Discussion
2.1. Complex Formation of KaiC with KaiB Mutants
First, the identification of structural factors contributing to the mechanism of cooperative binding of KaiB to the KaiC hexamer was attempted. The crystal structure of the KaiB–KaiC complex (PDB code: 5JWQ) revealed that six KaiB molecules are arranged into a ring structure on the KaiC CI ring, with each KaiB protomer in contact with its neighboring KaiB protomers (Figure 1a) [20]. In the lateral KaiB–KaiB interface, interactions between each KaiB molecule in the ring involve the binding of positively charged residues (Arg23 and Lys26) of the α3 helix of one KaiB protomer with negatively charged residues (Glu84 and Asp91) of the α1 helix of its clockwise neighbor (Figure 1b,c) [20]. The amino acid residues involved in interaction surfaces are conserved among cyanobacteria (Figure 1d and Figure S1). The importance of electrostatic complementarity in the KaiB–KaiC interaction was examined by mutational experiments in conjunction with native MS analysis. For this purpose, the basic amino acid residues, Arg23 and Lys26, were both replaced with aspartate in an N-terminal truncated mutant of T. elongatus KaiB (termed TeKaiB10–108), which has been reported to be monomeric and a higher-affinity binder to its cognate KaiC than the wild-type protein [16,22]. Therefore, we deleted the N-terminal segment of KaiB to facilitate native MS detection of the KaiB–KaiC complex. The mutant thus constructed was designated as TeKaiB10–108/DD. We confirmed that the DD mutation affected neither the secondary structure nor tetramer formation of TeKaiB in the absence of KaiC (Figure S2). Further, we used a T. elongatus KaiC mutant with its Ser431 and Thr432 substituted by aspartate residues (TeKaiCDD) because this KaiC mutant mimics the fully phosphorylated form and exhibits a higher binding affinity to TeKaiB and its monomeric mutants than those that mimic the nonphosphorylated form [22].
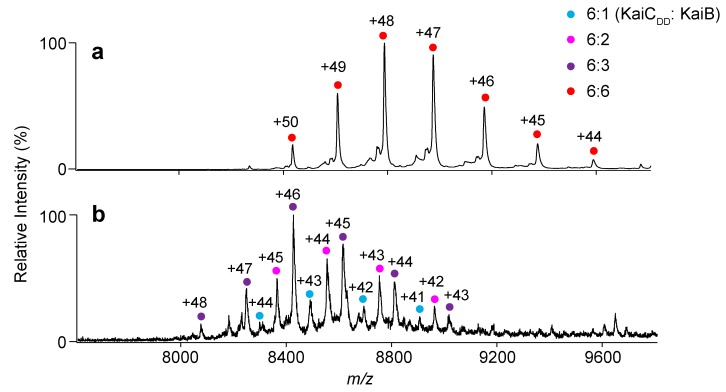
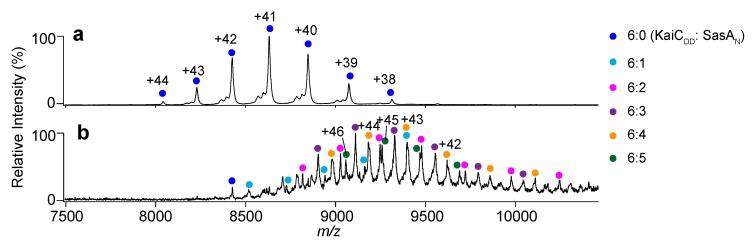
The MS titration data confirmed that TeKaiB10–108 and TeKaiCDD formed a complex exclusively in a 6:6 stoichiometry (Figure 2a and Figure S3a). This indicates that six TeKaiB protomers bind to the TeKaiC hexamer with strong positive cooperativity, which is consistent with previous observations obtained using synechococcal Kai (SyKai) proteins [21,23]. In contrast, TeKaiB10–108/DD formed complexes with TeKaiCDD in varying stoichiometries from 1:6 to 3:6 (Figure 2b and Figure S3b). However, in comparison with TeKaiB10–108, which has a native lateral interface, TeKaiB10–108/DD showed virtually no difference in its binding affinity to monomeric KaiC (Figure S4). These results indicated that the electrostatic complementarity at the lateral KaiB–KaiB interface promotes the cooperative assembly of KaiB protomers on the KaiC hexamer.
Figure 2.
Native MS analysis of KaiB–KaiC complex formation. Mass spectra of mixtures of TeKaiCDD and (a) TeKaiB10–108 or (b) TeKaiB10–108/DD at a 1:3.5 molar ratio (TeKaiCDD to TeKaiB). The cyan, magenta, purple, and red circles show the ion series of 6:1, 6:2, 6:3, and 6:6 complexes of TeKaiCDD and TeKaiB mutants, respectively.
2.2. Complex Formation of KaiC with SasA
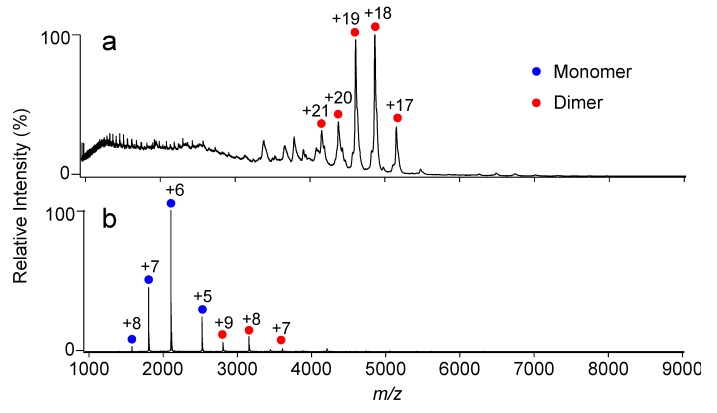
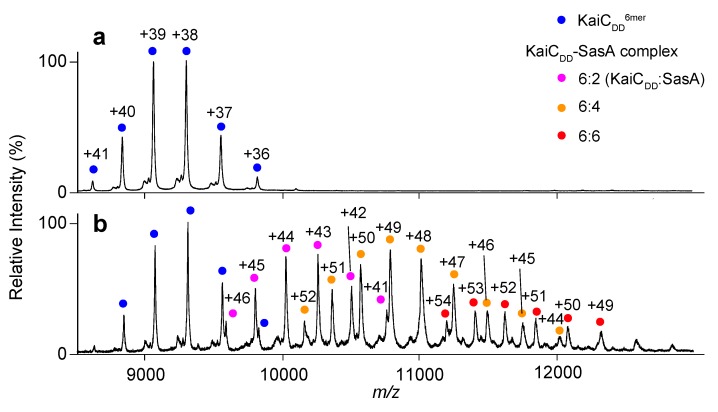
The N-terminal domain of SasA adopts a thioredoxin fold that is homologous to the KaiC-bound form of KaiB (Figure S5). This suggests that KaiB and SasA share a KaiC binding site [15,24]. Intriguingly, most amino acid residues involved in the lateral KaiB–KaiB interactions, including the complementary charged residues, are not conserved in SasA (Figure 1d). This prompted the investigation of whether SasA can bind to KaiC hexamers in a cooperative manner. The KaiC mutant TeKaiCDD has the highest affinity for SasA among mutants mimicking the phosphorylated state [16,17]. Therefore, TeKaiCDD was used for analyzing its interaction with TeSasA. Native MS analysis indicated TeSasA formed a dimer (Figure 3a), which is in contradiction to the previous report describing that the full-length SasA forms a trimer based on size exclusion chromatography and sedimentation equilibrium analyses [17]. This discrepancy might be attributed to the elongated, non-globular structure of full-length SasA and co-existence of higher oligomers that were detected in the chromatographic analysis in the previous solution condition, which hinder interpretation of those hydrodynamic data. Native MS-based titration of TeKaiCDD with TeSasA detected TeSasA/TeKaiCDD complexes in 2:6, 4:6, and 6:6 stoichiometries, suggesting that TeSasA interacts with KaiC as dimer (Figure 4 and Figure S6a). To eliminate potential ambiguities in data interpretation caused by the dimerization property of TeSasA, we used the N-terminal domain of SasA (TeSasAN), which was confirmed to be monomeric (Figure 3b), consistent with the previous report [17].
Figure 3.
Native MS analysis of SasA and its mutant. Mass spectra of (a) TeSasA and (b) TeSasAN. The blue and red circles show the ion series of the SasA monomer and dimer, respectively.
Figure 4.
Native MS analysis of TeKaiCDD–TeSasA complex formation. Mass spectra of mixtures of TeKaiCDD and TeSasA at (a) 1:0 and (b) 1:1 molar ratios (TeKaiCDD to TeSasA). The blue, magenta, orange, and red circles show the ion series of the TeKaiCDD homohexamer and 6:2, 6:4, and 6:6 complexes of TeKaiCDD and TeSasA, respectively.
Titration of TeKaiCDD with TeSasAN gave rise to their complexes in varying stoichiometries from 6:1 to 6:5 (Figure 5 and Figure S6b). This indicated that, in marked contrast to the KaiB–KaiC interaction, the SasA N-terminal domain exhibited no obvious binding cooperativity to the KaiC hexamer. The cooperative binding of KaiB to the KaiC hexamer may cause a synergistic release of SasA from the oscillator complex for strong downstream signaling to regulate the circadian rhythm.
Figure 5.
Native MS analysis of TeKaiCDD–TeSasAN complex formation. Mass spectra of mixtures of TeKaiCDD and TeSasAN at (a) 1:0 and (b) 1:3.5 molar ratios (TeKaiCDD to TeSasAN). The blue, cyan, magenta, purple, orange, and green circles show the ion series of the TeKaiCDD homohexamer and 6:1, 6:2, 6:3, 6:4, and 6:5 complexes of TeKaiCDD and TeSasAN, respectively.
2.3. Effects of KaiB Mutantation on Circadian Oscillations
To address the functional relevance of the cooperative KaiB–KaiC interaction, an in vitro clock system of SyKai proteins [25] of wild-type and mutant KaiB was used to examine if the disruption of charge complementarity at the lateral KaiB–KaiB interface showed any effects on circadian oscillation. We confirmed the DD mutation of SyKaiB impaired its cooperative interaction with KaiCDT (Figure S7).
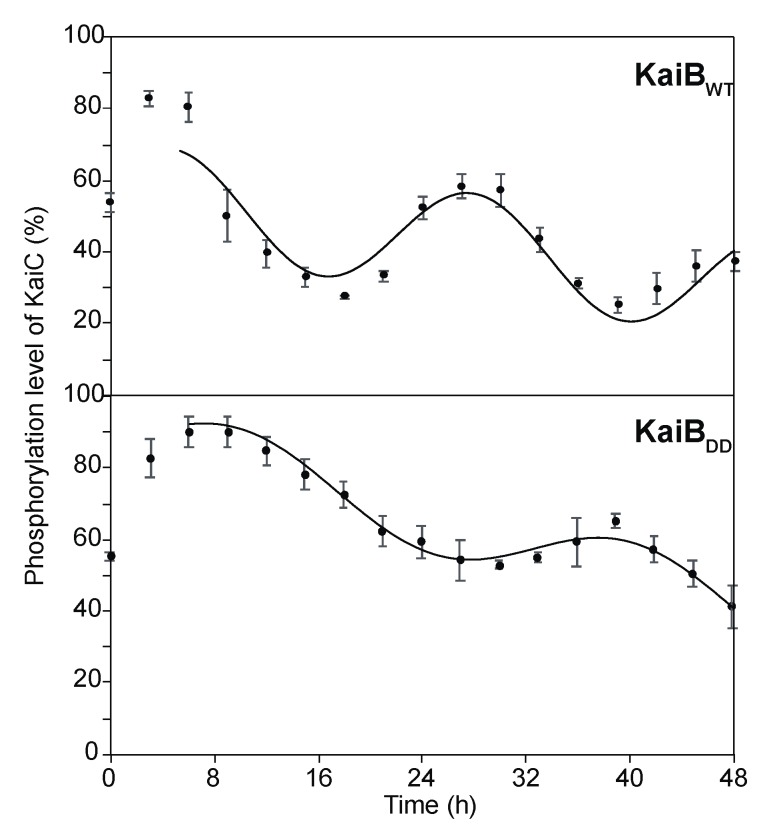
As shown in Figure 6 and Figure S8, the SyKaiBDD mutant perturbed the periodic oscillation of the phosphorylated KaiC level, which resulted in prolonged time periods of the circadian rhythm, i.e., 30.4 h, in comparison with the wild type (23.2 h). Furthermore, the mutation caused considerable reduction in the amplitude of the circadian oscillation. These results indicate that the electrostatic complementarity at the KaiB–KaiB interaction surface is a determining factor for an accurate circadian clock oscillation. Previous studies have reported that a KaiB R22A mutation decreased its affinity for the KaiC hexamer and a KaiB D90G mutation severely impaired the S. elongatus PCC 7942 circadian rhythm [19]. Because these mutations can compromise the charge complementarity between the two KaiB protomers, the observed effects can be explained by loss or attenuation of the positive cooperativity of KaiB–KaiC hexamer binding.
Figure 6.
Circadian oscillations in vitro. Phosphorylated KaiC levels in the presence of wild-type SyKaiB and its mutant SyKaiBDD. Plots show the percentage ratios of phosphorylated KaiC to total KaiC in the presence of KaiBWT and KaiBDD. The relative amounts of unphosphorylated and phosphorylated KaiC were estimated using densitometry. The data represent the means ± SD from three independent experiments. The simulated time course showing circadian oscillations in the phosphorylated KaiC level were generated using an integrated rhythm-analyzing program [27].
The KaiB hexameric ring formed on the KaiC hexamer can bind KaiA molecules [20,26], resulting in a rapid depletion of free KaiA molecules in the clock system. Therefore, cooperative binding of the KaiB–KaiC hexamer may facilitate a robust clock oscillation by enabling the efficient sequestration of KaiA molecules.
The findings suggest that the cooperative binding of the KaiB protomers to the KaiC hexamer enables sharp phase switching of the circadian oscillator complex through the synergistic release of the SasA clock-output protein and the coincidental sequestration of the KaiA phosphorylation enhancer. These mechanisms contribute to the synchronous nature of the cyanobacterial circadian system.
3. Materials and Methods
3.1. Protein Expression and Purification
KaiA, KaiB, KaiC, and SasA from Synechococcus elongatus PCC 7942 and T. elongatus were expressed in Escherichia coli. SyKaiA and SyKaiC were expressed and purified as Strep-tagged recombinant proteins [21,28]. TeKaiA, TeKaiB, TeKaiC, TeSasA, and SyKaiB were expressed as glutathione S-transferase (GST)-tagged recombinant proteins [4] and purified after the cleavage of the GST-tag, as described previously [16,21]. The mutated proteins, TeKaiB10–108, TeKaiB10–108/DD, SyKaiBDD, SyKaiCDT, TeKaiCDD, and SasAN were expressed and purified by the same method used for their wild-type counterparts [16,22,29,30]. For the KaiB mutant, the basic amino acid residues, Arg23 and Lys26, in TeKaiB10–108 and the corresponding Arg22 and Lys25 in SyKaiB were substituted with aspartate residues.
3.2. Size Exclusion Chormatography
Size exclusion chromatography analysis of TeKaiB or TeKaiBDD was performed with a Superdex 200 increase column (GE Healthcare Japan, Tokyo, Japan) equilibrated with 20 mM Tris-HCl (pH 8.0) containing 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA and 1 mM DTT at 0.75 min/mL flow rate. The elution profiles of proteins were monitored by absorbance at 280 nm.
3.3. Circular Dichroism Spectra
TeKaiB10–108 or TeKaiB10–108/DD (0.2 mg/mL) was dissolved in 20 mM phosphate buffer (pH 7.8) containing 150 mM NaCl. Measurements of circular dichroism spectra were performed in a 1-mm quartz cuvette at a room temperature using a spectropolarimeter (J-725, JASCO, Tokyo, Janan). After subtraction of the spectrum of the buffer alone, data were represented as mean residue ellipticities.
3.4. Native MS Analysis
The purified TeKaiCDD (20 µM) was titrated with either KaiB proteins (TeKaiB10–108 and TeKaiB10–108/DD) or SasA proteins (TeSasAWT and TeSasAN). The mixed protein solutions were incubated at 37 °C for 6 h, and were subsequently buffer-exchanged into 150 mM ammonium acetate, pH 6.8, using a Bio-Spin 6 column (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. In analyses of the Kai proteins from Synechococcus elongatus PCC 7942, the purified SyKaiCDT (20 µM) was mixed with SyKaiB or SyKaiBDD (30 µM), followed by incubation at 30 °C for 6 h. Approximately 2–5 µL of the buffer-exchanged protein solutions were immediately analyzed by nanoflow electrospray ionization MS using gold-coated glass capillaries prepared inhouse. Spectra were recorded on a SYNAPT G2-Si HDMS (Waters, Wilmslow, UK) in the positive ionization mode at 1.33 kV with a 150 V sampling cone voltage and source offset voltage, 0-V trap and transfer collision energy, and 5-mL/min trap gas flow. The spectra were calibrated using 1 mg/mL cesium iodide and analyzed using MassLynx software (Waters). Spectral measurements were performed at least in triplicate.
3.5. Measurement of Time-dependent Phosphorylated KaiC Levels
Reaction mixtures of SyKaiA (1.5 µM) and SyKaiC (3.4 µM) were incubated in the presence of 4.2 µM SyKaiBWT, or SyKaiBDD at 30 °C, and 3 μL aliquots of the reaction mixtures were removed to stop the reaction at specific time intervals, as described previously [28]. The aliquots were analyzed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie Brilliant Blue staining. The intensities of the bands were measured by densitometry using ImageJ 1.41 software (National Institutes of Health). The upper bands corresponded to phosphorylated KaiC, and the lower band corresponded to nonphosphorylated KaiC [25]. The relative proportions of phosphorylated KaiC to total KaiC (phosphorylated KaiC level) from the band intensities were calculated and the circadian oscillations associated with each phosphorylated KaiC level were analyzed using the rhythm-analyzing program [27].
Acknowledgments
We thank Junko Moriwaki (Ritsumeikan Univ.) for her helps of the phosphorylation oscillation experiments.
Abbreviations
| ATP | adenosine 5′-triphosphate |
| p-KaiC | phosphorylated KaiC |
| MS | mass spectrometry |
| Te | Thermosynechococcus elongatus (strain BP-I) |
| Sy | Synechococcus elongatus (strain PCC 7942) |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
Supplementary Materials
The Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/18/4550/s1.
Author Contributions
Conceptualization, R.M., H.Y. and K.K.; data curation, R.M., Y.Y., K.I. and K.T.; formal analysis, R.M., Y.Y., K.I. and H.Y.; funding acquisition, R.M., Y.Y., K.I. and K.K.; investigation, R.M., Y.Y., K.I. and K.T.; project administration, K.T., S.U., H.Y. and K.K.; methodology, R.M., Y.Y., K.I. and K.T.; resources, K.K.; supervision, K.T., S.U., H.Y. and K.K.; validation, Y.Y. and K.I.; visualization, R.M., Y.Y., K.I. and H.Y.; writing-original draft, R.M. and H.Y.; writing-review and editing, H.Y. and K.K.
Funding
This work was supported by grants (JP15K18492 to R.M., JP18J21063 to Y.Y., JP16H00784 and 17K19247 to K.T, JP18K14671 to K.I., and JP25102001 and JP25102008 to K.K.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, by the Okazaki ORION project, and by the Joint Research by Exploratory Research Center on Life and Living Systems (ExCELLS).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Bünning E. The Physiological Clock. 3rd ed. English Universities Press; New York, NY, USA: Springer; London, UK: 1973. [Google Scholar]
- 2.Ishiura M., Kutsuna S., Aoki S., Iwasaki H., Andersson C.R., Tanabe A., Golden S.S., Johnson C.H., Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 3.Iwasaki H., Taniguchi Y., Ishiura M., Kondo T. Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J. 1999;18:1137–1145. doi: 10.1093/emboj/18.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashi F., Suzuki H., Iwase R., Uzumaki T., Miyake A., Shen J.R., Imada K., Furukawa Y., Yonekura K., Namba K., et al. ATP-Induced hexameric ring structure of the cyanobacterial circadian clock protein KaiC. Genes Cells. 2003;8:287–296. doi: 10.1046/j.1365-2443.2003.00633.x. [DOI] [PubMed] [Google Scholar]
- 5.Pattanayek R., Wang J., Mori T., Xu Y., Johnson C.H., Egli M. Visualizing a circadian clock protein: Crystal structure of KaiC and functional insights. Mol. Cell. 2004;15:375–388. doi: 10.1016/j.molcel.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Nishiwaki T., Satomi Y., Kitayama Y., Terauchi K., Kiyohara R., Takao T., Kondo T. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim Y.I., Dong G., Carruthers C.W., Jr., Golden S.S., LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. USA. 2008;105:12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwasaki H., Nishiwaki T., Kitayama Y., Nakajima M., Kondo T. KaiA-Stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kageyama H., Nishiwaki T., Nakajima M., Iwasaki H., Oyama T., Kondo T. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol. Cell. 2006;23:161–171. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 10.Williams S.B., Vakonakis I., Golden S.S., LiWang A.C. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: A potential clock input mechanism. Proc. Natl. Acad. Sci. USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitayama Y., Iwasaki H., Nishiwaki T., Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwasaki H., Williams S.B., Kitayama Y., Ishiura M., Golden S.S., Kondo T. A kaiC-Interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 13.Stock A.M., Robinson V.L., Goudreau P.N. Two-Component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 14.Takai N., Nakajima M., Oyama T., Kito R., Sugita C., Sugita M., Kondo T., Iwasaki H. A KaiC-Associating SasA-RpaA two-Component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang Y.G., Cohen S.E., Phong C., Myers W.K., Kim Y.I., Tseng R., Lin J., Zhang L., Boyd J.S., Lee Y., et al. Circadian rhythms. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science. 2015;349:324–328. doi: 10.1126/science.1260031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami R., Mutoh R., Iwase R., Furukawa Y., Imada K., Onai K., Morishita M., Yasui S., Ishii K., Valencia Swain J.O., et al. The roles of the dimeric and tetrameric structures of the clock protein KaiB in the generation of circadian oscillations in cyanobacteria. J. Biol. Chem. 2012;287:29506–29515. doi: 10.1074/jbc.M112.349092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valencia S.J., Bitou K., Ishii K., Murakami R., Morishita M., Onai K., Furukawa Y., Imada K., Namba K., Ishiura M. Phase-Dependent generation and transmission of time information by the KaiABC circadian clock oscillator through SasA-KaiC interaction in cyanobacteria. Genes Cells. 2012;17:398–419. doi: 10.1111/j.1365-2443.2012.01597.x. [DOI] [PubMed] [Google Scholar]
- 18.Garces R.G., Wu N., Gillon W., Pai E.F. Anabaena circadian clock proteins KaiA and KaiB reveal a potential common binding site to their partner KaiC. EMBO J. 2004;23:1688–1698. doi: 10.1038/sj.emboj.7600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hitomi K., Oyama T., Han S., Arvai A.S., Getzoff E.D. Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm. J. Biol. Chem. 2005;280:19127–19135. doi: 10.1074/jbc.M411284200. [DOI] [PubMed] [Google Scholar]
- 20.Tseng R., Goularte N.F., Chavan A., Luu J., Cohen S.E., Chang Y.G., Heisler J., Li S., Michael A.K., Tripathi S., et al. Structural basis of the day-Night transition in a bacterial circadian clock. Science. 2017;355:1174–1180. doi: 10.1126/science.aag2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugiyama M., Yagi H., Ishii K., Porcar L., Martel A., Oyama K., Noda M., Yunoki Y., Murakami R., Inoue R., et al. Structural characterization of the circadian clock protein complex composed of KaiB and KaiC by inverse contrast-matching small-Angle neutron scattering. Sci. Rep. 2016;6:35567. doi: 10.1038/srep35567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iida T., Mutoh R., Onai K., Morishita M., Furukawa Y., Namba K., Ishiura M. Importance of the monomer-Dimer-Tetramer interconversion of the clock protein KaiB in the generation of circadian oscillations in cyanobacteria. Genes Cells. 2015;20:173–190. doi: 10.1111/gtc.12211. [DOI] [PubMed] [Google Scholar]
- 23.Snijder J., Burnley R.J., Wiegard A., Melquiond A.S., Bonvin A.M., Axmann I.M., Heck A.J. Insight into cyanobacterial circadian timing from structural details of the KaiB-KaiC interaction. Proc. Natl. Acad. Sci. USA. 2014;111:1379–1384. doi: 10.1073/pnas.1314326111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vakonakis I., Klewer D.A., Williams S.B., Golden S.S., LiWang A.C. Structure of the N-Terminal domain of the circadian clock-Associated histidine kinase SasA. J. Mol. Biol. 2004;342:9–17. doi: 10.1016/j.jmb.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., Iwasaki H., Oyama T., Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 26.Snijder J., Schuller J.M., Wiegard A., Lossl P., Schmelling N., Axmann I.M., Plitzko J.M., Forster F., Heck A.J. Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Science. 2017;355:1181–1184. doi: 10.1126/science.aag3218. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto K., Onai K., Ishiura M. RAP, an integrated program for monitoring bioluminescence and analyzing circadian rhythms in real time. Anal. Biochem. 2005;340:193–200. doi: 10.1016/j.ab.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Oyama K., Azai C., Nakamura K., Tanaka S., Terauchi K. Conversion between two conformational states of KaiC is induced by ATP hydrolysis as a trigger for cyanobacterial circadian oscillation. Sci. Rep. 2016;6:32443. doi: 10.1038/srep32443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayashi F., Itoh N., Uzumaki T., Iwase R., Tsuchiya Y., Yamakawa H., Morishita M., Onai K., Itoh S., Ishiura M. Roles of two ATPase-Motif-Containing domains in cyanobacterial circadian clock protein KaiC. J. Biol. Chem. 2004;279:52331–52337. doi: 10.1074/jbc.M406604200. [DOI] [PubMed] [Google Scholar]
- 30.Murakami R., Miyake A., Iwase R., Hayashi F., Uzumaki T., Ishiura M. ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes Cells. 2008;13:387–395. doi: 10.1111/j.1365-2443.2008.01174.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.