Abstract
Aims
The Brugada syndrome (BrS) is an inherited cardiac disorder predisposing to ventricular arrhythmias. Despite considerable efforts, its genetic basis and cellular mechanisms remain largely unknown. The objective of this study was to identify a new susceptibility gene for BrS through familial investigation.
Methods and results
Whole-exome sequencing performed in a three-generation pedigree with five affected members allowed the identification of one rare non-synonymous substitution (p.R211H) in RRAD, the gene encoding the RAD GTPase, carried by all affected members of the family. Three additional rare missense variants were found in 3/186 unrelated index cases. We detected higher levels of RRAD transcripts in subepicardium than in subendocardium in human heart, and in the right ventricle outflow tract compared to the other cardiac compartments in mice. The p.R211H variant was then subjected to electrophysiological and structural investigations in human cardiomyocytes derived from induced pluripotent stem cells (iPSC-CMs). Cardiomyocytes derived from induced pluripotent stem cells from two affected family members exhibited reduced action potential upstroke velocity, prolonged action potentials and increased incidence of early afterdepolarizations, with decreased Na+ peak current amplitude and increased Na+ persistent current amplitude, as well as abnormal distribution of actin and less focal adhesions, compared with intra-familial control iPSC-CMs Insertion of p.R211H-RRAD variant in control iPSCs by genome editing confirmed these results. In addition, iPSC-CMs from affected patients exhibited a decreased L-type Ca2+ current amplitude.
Conclusion
This study identified a potential new BrS-susceptibility gene, RRAD. Cardiomyocytes derived from induced pluripotent stem cells expressing RRAD variant recapitulated single-cell electrophysiological features of BrS, including altered Na+ current, as well as cytoskeleton disturbances.
Keywords: Brugada syndrome, RAD GTPase, Induced pluripotent stem cells, Sodium current, L-type calcium current, Actin cytoskeleton
Translational perspective
The Brugada syndrome (BrS) is an inherited cardiac disorder predisposing to ventricular arrhythmias and sudden cardiac death. In a large family affected by BrS, we identified a rare missense mutation in RRAD, the gene encoding the RAD GTPase. The mutation leads to electrical and structural defects consistent with BrS in cardiomyocytes differentiated from induced pluripotent stem cells of the family proband. A small but significant excess burden of rare missense variants in RRAD was found in unrelated BrS patients, suggesting that RRAD may be a new susceptibility gene for BrS.
Introduction
The Brugada syndrome (BrS) is a rare inherited disorder characterized by a specific but labile pattern at the electrocardiogram (ECG), i.e. a coved ST-segment elevation followed by a negative T wave in the right precordial leads, revealing electrical dysfunctions that predispose to ventricular arrhythmias and sudden cardiac death (SCD).1,2 Despite numerous studies over the last 25 years, the molecular basis for the ECG abnormalities and the mechanism underlying ventricular arrhythmias and SCD in the context of BrS are still incompletely understood.
Mutations in the SCN5A gene, which encodes the main cardiac Na+ channel α-subunit Nav1.5, are found in 20–25% of BrS probands.3,4 These mutations result in a reduction of the cardiac Na+ current (INa) and are thought to decrease rapid cardiomyocyte depolarization and cardiac conduction, a mechanism that may play a key role in BrS pathogenesis.5 Other rare variants have been reported as predisposing to BrS, affecting more than 20 genes encoding proteins either involved in Nav1.5 regulation, or encoding L-type Ca2+ channel subunits and K+ channels subunits mainly involved in IK,ATP and Ito generation.6,7 However, these additional variants account for <5% of cases, bringing the molecular diagnosis success rate to only 25–30% of cases.
Apart from SCN5A, the genes associated with BrS susceptibility have been identified only in few unrelated patients or in small families and often through candidate gene approaches: there is increasing evidence that they may play only a minor role, if any, in the phenotype of the patients.8–10 Moreover, extended familial genetic investigations revealed numerous occurrences of phenocopies and non-penetrance for the identified familial SCN5A variants in families affected by BrS, questioning the autosomal dominant transmission pattern initially proposed.11 Recently, the identification of three risk loci for BrS by genome-wide association study confirmed a more complex pattern of disease transmission.12 Despite this genetic complexity, studies on familial cases remain useful to discover new genes involved in disease susceptibility and understand better the pathogenesis of BrS.
In the present study, by applying a whole-exome sequencing approach to a large pedigree with BrS and documented history of SCD, we identified a rare non-synonymous variant (p.R211H) in the RRAD gene, which encodes the RAD (Ras Associated with Diabetes) GTPase.13 We then demonstrated RAD involvement in BrS pathogenesis based on functional studies in cardiomyocytes differentiated from (i) induced pluripotent stem cells (iPSC-CMs) obtained from four family members, two affected members, including the proband, and two unaffected members, and (ii) an isogenic pair of wild type and p.R211H-RRAD iPSC-CMs generated from one control individual with independent genetic background. The results were further confirmed in neonatal mouse cardiomyocytes with adenoviral-mediated expression of wild type and p.R211H RAD transcripts.
Methods
The study was conducted according to the principles set forth under the Declaration of Helsinki (1989) and European guidelines for clinical and genetic research. Institutional review board approvals of the study were obtained before the initiation of patient enrolment. Informed written consent was obtained from each patient who agreed to participate in the clinical and genetic study. H9 human embryonic stem cells were used as positive controls for human iPSC characterization under agreement n°RE13-004 from the Agence de la Biomédecine.
All the experimental procedures performed in the context of this study are available in detail in the Supplementary material online.
Results
Clinical report: a familial case of Brugada syndrome
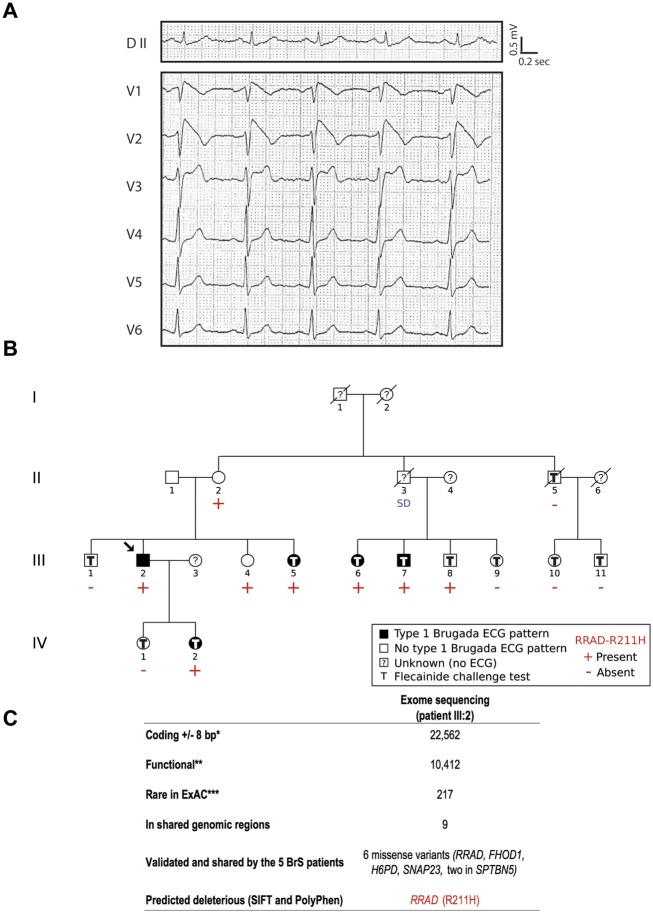
The index case (individual III:2; Figure 1), a 41-year-old man, was first diagnosed after a systematic ECG. He presented a typical but labile BrS Type I ECG pattern (Figure 1A). He mentioned that he had previously experienced palpitations associated with near syncope and nocturnal agonal respiration. After implantable cardioverter-defibrillator implantation, neither recurrent symptoms nor ventricular arrhythmias have been observed. During familial screening, four relatives were identified as affected after flecainide challenge (Figure 1B). In addition, individual III:8 exhibited a BrS Type II ECG pattern under flecainide (Supplementary material online, Figure S1). None of these individuals had presented relevant symptoms. However, an unexplained sudden death occurred at rest in a 41-year-old proband's uncle (individual II:3). Electrocardiogram and clinical parameters for this French family are provided in Table 1.
Figure 1.
Electrocardiographic pattern and genetic investigation in a family with Brugada syndrome. (A) DII peripheral lead and V1–V6 precordial leads of the proband (III:2) under baseline condition. (B) Family pedigree. The arrow indicates the proband. Electrocardiograms were recorded in 14 individuals, under baseline conditions only for three individuals (II:1, II:2, and III:4), or under baseline conditions and during flecainide challenge for 11 individuals. DNA was collected for 14 individuals. (C) Exome filters. *SO terms (VEP): stop_gained, stop_lost, frameshift_variant, missense_variant, splice_donor_variant, splice_acceptor_variant, initiator_codon_variant, inframe_insertion, inframe_deletion, splice_region_variant, synonymous_variant, and stop_retained_variant. **SO terms (VEP): stop_gained, stop_lost, frameshift_variant, missense_variant, splice_donor_variant, splice_acceptor_variant, initiator_codon_variant, inframe_insertion, and inframe_deletion. ***MAF (Minor Allele Frequency) <0.1% in all ExAC populations. SD, sudden death.
Table 1.
Electrocardiogram and clinical parameters of the family members
| Family members | Chr16 haplotype | Type I ECG pattern | Baseline |
Flecainide |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RR (ms) | P (ms) | PR (ms) | QRS (ms) | QTc (ms) | TPE (ms) | RR (ms) | P (ms) | PR (ms) | QRS (ms) | QTc (ms) | TPE (ms) | |||
| III:2 | Yes | Spontaneous | 828 | 98 | 165 | 100 | 410 | 72 | — | — | — | — | — | — |
| III:5 | Yes | Drug induced | 942 | 90 | 139 | 97 | 410 | 59 | 955 | 87 | 160 | 104 | 438 | 67 |
| III:6 | Yes | Drug induced | 1015 | 76 | 145 | 133 | 426 | 67 | 949 | 72 | 144 | 149 | 447 | 72 |
| III:7 | Yes | Drug induced | 711 | 108 | 152 | 118 | 395 | 67 | 872 | 106 | 196 | 127 | 402 | 66 |
| III:8 | Yes | Noa | 724 | 81 | 152 | 91 | 375 | 22 | 755 | 95 | 196 | 95 | 385 | 51 |
| IV:2 | Yes | Drug induced | 617 | 83 | 122 | 88 | 409 | 64 | 649 | 87 | 157 | 116 | 464 | 52 |
| II:5 | No | No | 882 | 117 | 200 | 101 | 424 | 69 | 829 | 109 | 217 | 113 | 427 | 68 |
| III:1 | No | No | 1262 | 112 | 204 | 89 | 339 | 55 | 986 | 115 | 206 | 119 | 407 | 63 |
| III:9 | No | No | 832 | 82 | 158 | 89 | 394 | 61 | 849 | 103 | 206 | 99 | 439 | 100 |
| III:10 | No | No | 938 | 93 | 166 | 91 | 358 | 52 | 942 | 100 | 204 | 90 | 385 | 59 |
| III:11 | No | No | 928 | 97 | 177 | 104 | 397 | 73 | 867 | 111 | 221 | 107 | 405 | 70 |
| IV:1 | No | No | 897 | 93 | 148 | 87 | 375 | 62 | 679 | 103 | 162 | 103 | 446 | 118 |
P, P wave; PR, PR interval; QRS, QRS complex, QTc, corrected QT interval; TPE, time interval between the peak and the end of the T wave.
Type II ECG pattern under flecainide (Supplementary material online, Figure S1).
Identification of a rare RRAD variant in a familial case of Brugada syndrome
Whole-exome sequencing on the index case unveiled 217 rare coding variants (Supplementary material online, Methods). After analysing variants located in genomic regions shared by all affected family members and co-segregation analyses, six missense variants were detected in all five affected patients (Figure 1C). They were located in RRAD and FHOD1 on chromosome 16, H6PD on chromosome 1, and SNAP23 and SPTBN5 (two variants) on chromosome 15 (Supplementary material online, Table S1). While there were six non-affected carriers for each variant on chromosomes 1 and 15, both variants located on chromosome 16 (in RRAD and FHOD1) were carried by only three non-affected relatives (Supplementary material online, Table S1; Figure 1B): the individual III:8 presenting with a BrS Type II ECG pattern, and two women (II:2 and III:4) for whom drug challenge could not be performed. Furthermore, among the six missense variants, the RRAD variant (p.Arg211His) was the only one predicted to be damaging by SIFT and PolyPhen-2 tools (Supplementary material online, Tables S1 and S2). This variant is also associated to the highest CADD score (CADD PHRED score: 33) and is currently reported as the least frequent in the gnomAD database (1/242 446 alleles; Supplementary material online, Table S1). By screening the four coding exons of RRAD in 186 unrelated BrS patients using Sanger sequencing, we identified three additional rare missense mutations in isolated cases: p.Asp46Tyr, p.Gln186Arg, and p.Val215Met (Supplementary material online, Table S2). Overall, we detected a trend for enrichment (P = 0.042, odds ratio = 6.98, 95% confidence interval: 0.79–84.33) in rare RRAD non-synonymous variants (with a MAF below 0.1% in gnomAD) among the BrS cases (three carriers out of 186 cases) compared to a population of 856 reference individuals of French origin, for which RRAD genotype status was available in-house (2 carriers out of 856 individuals; Supplementary material online, Table S2).
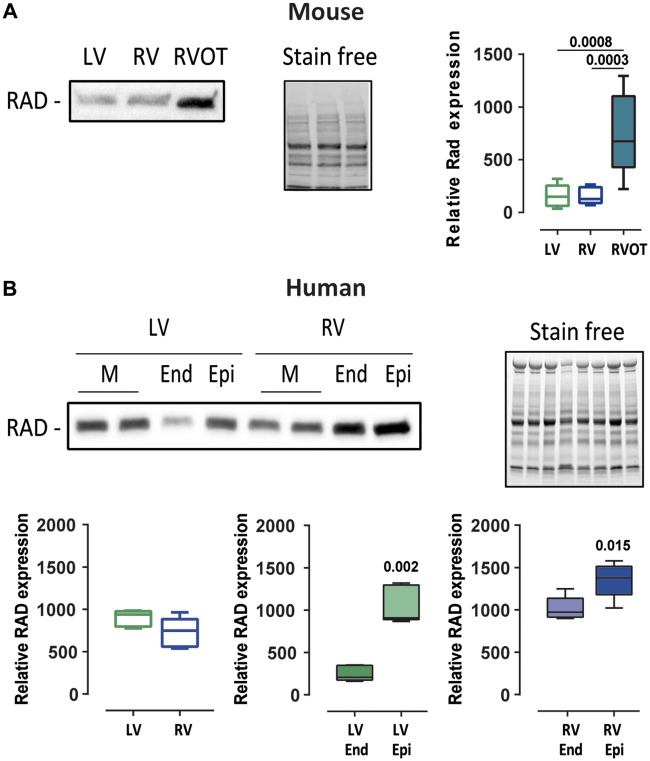
Since BrS is characterized by abnormal electrical activity in the right ventricle outflow track (RVOT) region, we compared Rad expression levels in mouse RVOT to its expression in the rest of the right ventricular and the left ventricular free walls. Rad was found four-fold more expressed in the RVOT than in the other ventricular parts (Figure 2A). We also investigated the expression of RAD in human left and right ventricles and observed higher expression in the subepicardium than in the subendocardium for both compartments (Figure 2B).
Figure 2.
Rad protein expression pattern in mouse and human hearts. (A) Left: representative western blot of mouse Rad expression in the left ventricular free wall, the right ventricular free wall (without outflow tract), and the right ventricular outflow tract from one mouse. Right: box plot of Rad expression normalized to stain free for nine mice. Statistical test: Kruskal–Wallis test. (B) Top: representative western blot of RAD expression in left ventricular and right ventricular transmural (M), subendocardial (End), and subepicardial (Epi) samples from one individual. Bottom: box plots of RAD expression normalized to stain free for three individuals. Mann–Whitney test.
In order to decipher the electrophysiological consequences of the missense variant in RRAD (and/or possibly in FHOD1) within the family, we differentiated cardiomyocytes from iPSCs obtained from the index case (BrS1; individual III:2; three iPSC clones: BrS101, BrS102, and BrS103) and his unaffected brother (Ctl1; individual III:1; two clones: Ctl103 and Ctl104) who carries the rare variants on chromosomes 1 and 15 but not on chromosome 16.
Slow rate and reduced action potential upstroke velocity linked to INa and ICa,L disruption in Brugada syndrome cardiomyocytes derived from induced pluripotent stem cells
We first investigated the properties of the ventricular-like action potentials. BrS1 iPSC-CMs displayed slower spontaneous rhythms than Ctl1 iPSC-CMs and lower action potential upstroke velocity (Figure 3A–C). Based on these results, we investigated the biophysical properties of the Na+ and L-type Ca2+ currents. Membrane capacitance in BrS1 iPSC-CMs was slightly, though not significantly, higher (55.5 ± 25.5 pF, n = 68) than in Ctl1 iPSC-CMs (47.5 ± 22.8 pF; n = 51; P = 0.07). INa density was significantly lower (by ∼40% at −25 mV) in BrS1 iPSC-CMs (36.8 ± 16.7 pA/pF) than in Ctl1 iPSC-CMs (58.8 ± 16.5 pA/pF; Figure 3D and E), a result consistent with a lower NaV1.5 protein level in BrS1 iPSC-CMs than in Ctl1 iPSC-CMs (Supplementary material online, Figure S3) while NaV1.5 transcript level was not significantly changed (BrS1: 0.7 ± 0.4, 3 clones; Ctl1: 1.0 ± 0.5, 2 clones). Steady-state activation and inactivation gating properties did not differ between BrS1 and Ctl1 iPSC-CMs but recovery from inactivation was slightly faster in BrS1 iPSC-CMs than in Ctl1 iPSC-CMs (Supplementary material online, Table S4). ICaL density was also lower (by ∼30% at −5 mV) in BrS1 iPSC-CMs (18.9 ± 6.0 pA/pF) compared to Ctl1 iPSC-CMs (26.3 ± 7.1 pA/pF) without any disturbance of activation gating properties (Figure 3F and G; Supplementary material online, Table S4). Similar results have been obtained on iPSC-CMs derived from the proband's daughter carrying the p.R211H RRAD variant and affected by BrS (BrS2; individual IV:2; Supplementary material online, Figure S4 and Table S4); her unaffected sister was used as control (Ctl2; individual IV:1). No difference in INa and ICaL densities between the three BrS1 iPSC-CM clones and between the two Ctl1 iPSC-CM clones was observed (Supplementary material online, Figure S5).
Figure 3.
Spontaneous action potentials, INa and ICaL properties in induced pluripotent stem cell-derived cardiomyocytes from the proband (BrS1) and his unaffected brother (Ctl1). (A) Representative action potential recordings of spontaneously beating Ctl1 and BrS1 cardiomyocytes derived from induced pluripotent stem cells. (B and C) Box plots of cycle length (B; spontaneous CL) and maximum upstroke velocity (C; dV/dTmax) of spontaneous action potentials. Statistical test: Mann–Whitney test. (D and F) Superimposed representative INa (D) and ICaL (F) traces recorded in Ctl1 and BrS1 cardiomyocytes derived from induced pluripotent stem cells (voltage-clamp protocols in insets; stimulation frequency: 0.5 and 0.125 Hz for INa and ICaL, respectively). (E and G) Mean (±SEM) current density-voltage relationship for peak INa (E) and ICa,L (G). **P < 0.01 and ***P < 0.001 (two-way analysis of variance with Bonferroni post hoc test for multiple comparisons).
Prolonged action potential duration, early afterdepolarizations, and calcium flux disturbances in Brugada syndrome cardiomyocytes derived from induced pluripotent stem cells
When paced at a cycle length of 700 ms, BrS1 iPSC-CMs showed longer action potentials than Ctl1 iPSC-CMs (Figure 4A and B). No significant difference was observed between the three BrS1 iPSC-CM clones and between the two Ctl1 iPSC-CM clones (Supplementary material online, Figure S6). Interestingly, BrS1 iPSC-CMs also exhibited a larger persistent Na+ current than Ctl1 iPSC-CMs Figure 4C and D). Early afterdepolarizations (EADs) were observed in 80% and 50% of BrS1 iPSC-CMs derived from BrS101 and BrS103 clones, respectively (Figure 4E and F) and in BrS2 iPSC-CMs (Supplementary material online, Figure S4). No EAD was observed in Ctl1 iPSC-CMs and in iPSC-CMs derived from clone BrS102.
Figure 4.
Action potential repolarization, persistent Na+ current (INaP) and intracellular Ca2+ handling in cardiomyocytes derived from induced pluripotent stem cells from the proband (BrS1) and his unaffected brother (Ctl1). (A) Representative action potentials recorded at a pacing cycle length of 700 ms in Ctl1 and BrS1 cardiomyocytes derived from induced pluripotent stem cells. (B) Box plot of action potential duration (APD) at 30% (APD30), 50% (APD50), and 90% (APD90) of full repolarization. Statistical test: two-way analysis of variance with Bonferroni test for multiple comparisons. (C) Representative currents recorded during 1-s voltage-clamp ramps from −120 mV to +40 mV (inset; stimulation frequency: 0.2 Hz) before (Control) and after tetrodotoxin (TTX) perfusion (top traces) in Ctl1 and BrS1 cardiomyocytes derived from induced pluripotent stem cells Bottom traces show the corresponding TTX-sensitive currents. (D) Box plot of the TTX-sensitive INaP at −20 mV. Mann–Whitney test. (E) Representative examples of early afterdepolarizations recorded in spontaneously beating cardiomyocytes differentiated from BrS101 and BrS103 iPSC clones (BrS1 patient). (F) Early afterdepolarization incidence in the different cardiomyocytes derived from induced pluripotent stem cell lines. χ2 test; P values vs. Ctl1. (G) Representative [Ca2+]i transients from Ctl1 and BrS1 cardiomyocytes derived from induced pluripotent stem cells with an example of abnormal Ca2+ oscillations following peak [Ca2+]i transient in BrS cardiomyocytes derived from induced pluripotent stem cells. (H) Box plot of [Ca2+]i transient 75% decay time (Ca2+ transient 75%) in Ctl1 and BrS1 cardiomyocytes derived from induced pluripotent stem cells Mann–Whitney test.
Given these results, we investigated intracellular Ca2+ flux properties. BrS1 iPSC-CMs exhibited longer [Ca2+]i transient decay times than Ctl1 iPSC-CMs (Figure 4G and H), suggesting an impairment of the Ca2+ recycling. Moreover, we observed abnormal [Ca2+]i oscillations in 40% of the BrS1 iPSC-CMs (Figure 4G). These events are consistent with action potential prolongation and EAD appearance observed in these cells.
Cytoskeleton defects in Brugada syndrome cardiomyocytes derived from induced pluripotent stem cells
The BrS1 iPSC-CMs displayed impaired F-actin organization and cortical distribution of troponin I, while Ctl1 iPSC-CMs displayed a striated distribution of both proteins. This cytoskeleton defect was observed in 70% of BrS1 iPSC-CMs but in only 8% of Ctl1 iPSC-CMs (Figure 5A). Three-dimensional views using confocal microscopy showed that Ctl1 iPSC-CMs had flat cell bodies, whereas BrS1 iPSC-CMs exhibited round cell shape making the cell thickness larger than in Ctl1 iPSC-CMs (Figure 5B). In addition, the density of vinculin-containing adhesion complexes was lower in BrS1 iPSC-CMs than in Ctl1 iPSC-CMs, with impaired localization of vinculin (Supplementary material online, Figure S7). Similar results have been obtained in BrS2 iPSC-CMs (Supplementary material online, Figure S8).
Figure 5.
Cytoskeleton in cardiomyocytes derived from induced pluripotent stem cells obtained from the proband (BrS1) and his unaffected brother (Ctl1). (A) Left: representative immunostainings of filamentous actin (F-actin; stained with phalloidin) and troponin I and merged acquisitions with nucleus staining with DAPI in Ctl1 and BrS1 cardiomyocytes derived from induced pluripotent stem cells (top panels) and corresponding fluorescence distribution measured at the level of the yellow dotted lines (fluorescence-distance relationships; bottom panels). Scale bars: 20 µm. Right panel: percentage of Ctl1 (n = 10) and BrS1 (n = 18) cardiomyocytes derived from induced pluripotent stem cells with striated (in white) and cortical (in black) F-actin staining. Statistical test: Fisher’s exact test. (B) Left: representative three-dimensional acquisition illustrations of a Ctl1 and a BrS1 cardiomyocytes derived from induced pluripotent stem cell after immunostaining of F-actin and Troponin I (left panels). Width: 107.12 µm; height: 12 µm; and depth: 107.12 µm. Right panel: box plots of thickness and cell circularity (cell width/cell length ratio) in Ctl1 (n = 10) and BrS1 (n = 12) cardiomyocytes derived from induced pluripotent stem cells Mann–Whitney test.
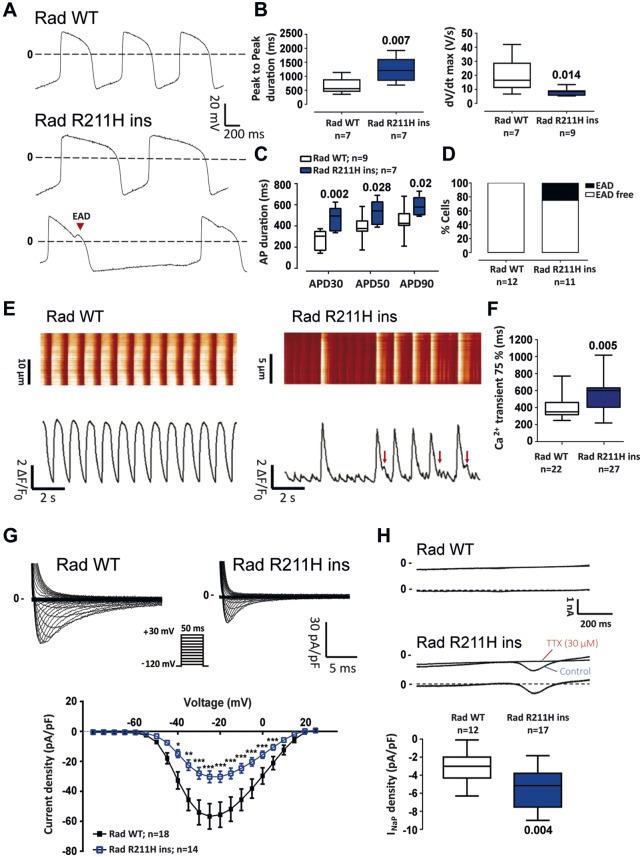
RRAD p.R211H knock in isogenic line recapitulates the Brugada syndrome phenotype observed in the Brugada syndrome family members cell lines
RRAD p.R211H variant was inserted into an extra-familial control iPSC line by CRISPR/Cas9 technology (Supplementary material online, Figure S9A). Action potential recordings showed that genome edited (Rad R211H ins) iPSC-CMs had slower spontaneous rhythms, lower action potential upstroke velocity and longer action potential duration than control (Rad WT) iPSC-CMs (Figure6A–C). Early afterdepolarizations were observed in 20% of Rad R211H ins iPSC-CMs and in none of the Rad WT iPSC-CMs (Figure 6D). Intracellular calcium recordings showed that Rad R211H ins iPSC-CMs exhibited uneven beating rate when compared with Rad WT iPSC-CMs as well as a slowing in the calcium reuptake (Figure 6E and F). Rad R211H ins iPSC-CMs exhibited a significantly lower peak INa density than Rad WT iPSC-CMs (30.3 ± 14.2 pA/pF vs. 56.8 ± 35.0 pA/pF, respectively, at −25 mV), with no change in steady-state activation and inactivation properties (Supplementary material online, Table S4). Rad R211H ins iPSC-CMs also exhibited a larger persistent Na+ current than RAD WT iPSC-CMs (Figure 6G and H). No appreciable difference between Rad R211H ins iPSC-CMs and Rad WT iPSC-CMs in ICaL density was observed (Supplementary material online, Figure S9). Finally, as in iPSC-CMs of the family members affected by BrS, the insertion of the p.R211H variant in the extra-familial control line triggered cytoskeleton defects in ∼40% of the cardiomyocytes (Supplementary material online, Figure S9C).
Figure 6.
Rad p.R211H insertion in an extra-familial control line by genome editing: electrophysiological characterization. (A) Representative action potential recordings from control (Rad WT) and mutated (Rad WT) conditions. (B) Box plots of peak to peak duration and action potential upstroke velocity (dV/dt max) of Rad WT and Rad WT cardiomyocytes derived from induced pluripotent stem cells Mann–Whitney test. (C) Mean action potential duration (APD) at 30% (APD30), 50% (APD50), and 90% (APD90) of full repolarization at a pacing cycle length of 1 s. Two-way ANOVA with Bonferroni test for multiple comparisons. (D) Early afterdepolarization incidence in Rad WT and Rad R211H ins cardiomyocytes derived from induced pluripotent stem cells. (E) Representative [Ca2+]i transients obtained in Rad WT and Rad R211H ins cardiomyocytes derived from induced pluripotent stem cells and their corresponding fluorescence map. (F) [Ca2+]i transient 75% decay time (Ca2+ transient 75%) in Rad WT and Rad R211H ins cardiomyocytes derived from induced pluripotent stem cells Mann–Whitney test. (G) Superimposed representative INa traces recorded in Rad WT and Rad R211H ins cardiomyocytes derived from induced pluripotent stem cells (upper panel; voltage-clamp protocol in inset) and mean (±SEM) current density-membrane potential relationships (bottom). *P < 0.05, **P < 0.01, and ***P < 0.001 (two-way analysis of variance with Bonferroni post hoc test for multiple comparisons). (H) Top: representative currents recorded during 1-s voltage-clamp ramps from −120 mV to +40 mV (stimulation frequency: 0.2 Hz) before (Control) and after tetrodotoxin (TTX) perfusion (top traces) in Rad WT and Rad R211H ins cardiomyocytes derived from induced pluripotent stem cells and corresponding TTX-sensitive persistent sodium current (INaP, bottom traces). Bottom: box plots of INaP density. Mann–Whitney test.
In addition, the effects of RRAD p.R211H variant were also investigated in neonatal mouse ventricular myocytes infected with adenoviruses encoding Green Fluorescent Protein (GFP) alone, wild type human RAD plus GFP, or p.R211H-RAD plus GFP. These experiments confirmed the results obtained with iPSC-CMs (Supplementary material online, Results).
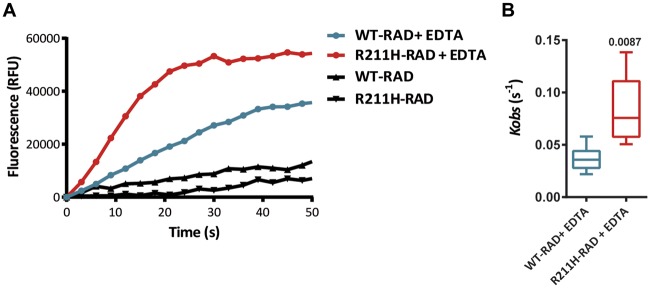
The p.R211H variant triggers a gain of function effect on RAD GTPase activity
To directly assess the impact of the p.R211H variant on RAD activity, we performed nucleotide exchange kinetics measurements on WT-RAD and R211H-RAD proteins. As no guanine exchange factor is described for RAD GTPase, kinetics were initiated by Mg2+ chelation with ethylenediaminetetraacetic acid (EDTA) R211H-RAD protein showed an increased activity compared to WT-RAD (Figure 7A and B) with Kobs values that were significantly higher (respectively 0.083 ± 0.025 s−1 and 0.03 ± 0.008 s−1). These results suggest that the variant has a direct positive effect on RAD GTPase activity, consistent with a gain of function of the protein.
Figure 7.
RAD protein GTPase activity analysis. (A) Representative fluorescence kinetics traces of WT-RAD and R211H-RAD GDP/GTP-mant exchange in the presence or absence of EDTA. (B) Box plot of Kobs values of EDTA-induced GDP/GTP-mant exchanges in WT-RAD (n = 6) and R211H-Rad (n = 5) cardiomyocytes. Statistical test: Mann–Whitney test.
Take home figure.
The p.R211H mutation in the gene encoding RAD GTPase is involved in Brugada syndrome pathogenesis, leading to both electrical and structural defects in cardiomyocytes differentiated from induced pluripotent stem cells of affected patients.
Discussion
In this study, we identified a novel BrS-susceptibility gene, RRAD, and showed that the RRAD variant identified in a familial form of the disease leads to a BrS-typical electrical defect, i.e. reduced amplitude of INa, coupled with cardiomyocyte cytoskeletal abnormalities. At the cellular level, such dual effect has never been reported in the context of this hereditary arrhythmia.
In the past two decades, BrS has been associated to more than 20 susceptibility genes.3,4 Most of those genes, including SCN5A, have been identified using functional candidate approaches, with only few mutations co-segregating with BrS ECG anomalies in familial forms. In the present study, we applied a hypothesis‐free approach, based on whole‐exome sequencing and identity-by-descent analysis, to a familial case with genetically unexplained BrS. Following this strategy, we identified six missense variants shared by the five family members exhibiting the BrS phenotype. Among these variants, the p.R211H substitution in the RRAD gene was (i) the least frequent in public databases (reported in only one case), (ii) the only variant predicted as deleterious by both SIFT and Polyphen-2, and (iii) the variant with the highest CADD score. These results indicate that p.R211H is the most likely causal variant in this familial case, independently of the biological function of RRAD gene product.
We then tested the relative contribution of RRAD variants in BrS susceptibility by screening for rare non-synonymous variants within its sequence among unrelated index cases compared to reference individuals, all of French origin. We observed a trend for enrichment in rare non-synonymous variants among cases (P < 0.05)—with 3/186 affected individuals carrying rare missense mutations vs. 2/856 reference individuals—thus strengthening the likelihood of RRAD involvement in BrS pathophysiology.
The RAD protein is a member of the RGK subfamily of Ras GTPases that has previously been associated with ventricular arrhythmias in mice.14 Here, we showed higher levels of RRAD protein in subepicardium than in subendocardium in human heart, as well as predominant expression of Rad in the right ventricle outflow tract compared to the other cardiac compartments in mice: both expression patterns are fully concordant with a role in BrS pathogenesis.6
Results obtained with iPSC-CMs demonstrate that the RRAD p.R211H variant reduces INa by about 40%. To our knowledge, this is the first report of an effect of RAD on the cardiac Na+ channel. The involvement of the RRAD p.R211H variant in INa down-regulation was confirmed by inserting the mutation by genome editing in a control human iPSC line obtained from a healthy individual unrelated to the family and by expressing the mutated protein in neonatal mouse ventricular cardiomyocytes in primary culture. The studies performed on this second model suggest that the p.R211H variant leads to a gain of function of RAD. Indeed, its effect on INa is mimicked by a 10-fold overexpression of wild-type RAD and at this level of expression, both the WT and the p.R211H RAD have the same effect on INa. This gain-of-function hypothesis is supported by the two-fold faster GDP/GTP exchange activity of RAD that we found in the mutant protein compared to the WT RAD. Interestingly, the arginine-211 residue is highly conserved among species and among the RGK protein members. It is localized between the G4 and the G5 sites of the RAD guanine recognition area located in the G domain and may play an important role on RAD affinity for GTP.15 Our results suggest that at least part of p.R211H RAD effects on INa involves a down-regulation of Nav1.5 expression at the protein level, without any alteration of SCN5A transcription. Further studies will be needed to elucidate the mechanisms of this down-regulation and whether RAD also interacts with Nav1.5 or one of its regulatory subunits to directly modulate its function.
The iPSC-CMs of the mutated patients and the genome edited Rad R211H ins iPSC-CMs were also characterized by the presence of a large persistent Na+ current and the occurrence of EADs. The effects of RRAD p.R211H variant on the Na+ current, i.e. a decrease in Na+ peak current amplitude and the occurrence of a persistent Na+ current, are similar to those produced by the SCN5A mutation p.1795insD, which causes an overlap syndrome combining the phenotypic traits of BrS and Type 3 long QT syndrome.16 However, mouse studies suggest that RAD is preferentially expressed in the RVOT which could limit the effects of the mutation to this specific area, which is insufficient to trigger appreciable QT prolongation. This would explain why the RRAD p.R211H carriers exhibit normal QT intervals.
At first glance, EADs are not expected to be involved in arrhythmias in the context of BrS. However, they have also been observed in iPSC-CMs from a BrS patient with a TBX5 gene variant,17 as well as in iPSC-CMs of BrS patients carrying SCN5A mutations.18 In this latter study, those arrhythmic events were linked to abnormal Ca2+ handling, which also led to delayed afterdepolarizations. The investigation of Ca2+ handling in our BrS1 and Rad R211H ins cell lines showed delayed Ca2+ recapture and abnormal Ca2+ release during recapture in both lines. Combined with the persistent Na+ current, these results are consistent with the APD prolongation and EAD appearance. This phenomenon could be linked to the immaturity of the cells: on the one hand, a recent study showed that until day 30 of differentiation, iPS-CMs express a different isoform of NaV1.5 protein,19 and on the other hand CaV1.2 is not properly localized close to the ryanodine receptor RYR2 due to the absence of T-tubule structures.20 These phenotypic traits seem to be characteristic of BrS in iPSC-CMs but their involvement in triggering arrhythmias in the patient remains uncertain.
The iPSC-CMs of the family members carrying the RRAD p.R211H variant were also characterized by a reduction of the L-type Ca2+ current compared to intra-familial control iPSC-CMs. A reduction of this current has already been involved in some forms of BrS.6,21 Interestingly, RAD has previously been shown to regulate the cardiac L-type Ca2+ current by controlling the Cav1.2 channel trafficking to the sarcolemma.14,22 Altogether, these data suggested that the pathophysiological mechanisms of RRAD-related BrS included a combined decrease of both Na+ and Ca2+ current amplitude, which had never been described before. However, the investigation of the isogenic Rad R211H ins iPSC-CMs only confirmed the loss of function of the Na+ current. This discrepancy could be explained by the presence of other variant(s) limiting this dual electrical modulation to this particular family. Nonetheless, the isogenic model validates that RRAD p.R211H variant is sufficient to provoke a pro-arrhythmic phenotype and suggests that the main BrS causal defects induced by this variant are a dysregulation of the Na+ channel and an alteration of the cytoskeleton.
Indeed, in addition to the electrical defects, RRAD p.R211H disorganizes the striated architecture of the cardiomyocyte cytoskeleton, and disturbs the localization of focal adhesions, which leads to cell rounding. Again here, the gain-of-function effect of the mutation RAD GTPase activity is proven, since the overexpression of WT RAD induces similar effects in neonatal mouse cardiomyocytes, and more severely with the overexpression of p.R211H-RAD. RAD is known to interact with the cytoskeleton.23,24 In the present study, RAD p.R211H acts like a down-regulator of the F-actin cytoskeleton consolidation - most likely through the inhibition of Rho Kinase activity (Supplementary material online, Figure S12), leading to a decrease in cell contractility and focal adhesion formation and maturation.25 These structural defects, which might be limited to the RVOT given the preferred localization of RAD protein in this cardiac region, could lead to decreased cell-to-cell connection and abnormal cardiac conduction26 and thus play a role in BrS. There are indeed accumulating evidences that structural anomalies in the myocardium, such as fatty infiltrations and fibrosis, may trigger electrical anomalies in relation to BrS.27–29 Furthermore, several susceptibility genes or loci, such as MOG130,31 or HEY2, have been shown to regulate not only electrical activity but also cardiac morphogenesis.32,33 Taken together, these data indicate that structural ventricular anomalies may contribute significantly to the expression of the electrical features typical of BrS.
Study limitations
One of the limitations of the present study relates to the immature state of the iPSC-CMs. At the electrical level, the lack of IK1 and the low Ito density prevent the cardiomyocytes to display proper action potential shape and resting membrane potential. At the structural level, the absence of cell polarity and T-tubule structures makes it difficult to study calcium handling and cytoskeleton integrity due to lack of proper localization of the proteins involved. Moreover, Brugada syndrome being related to RVOT, these cells fail to capture the complexity of the phenotype due to lack of cardiac layer specification (endocardium, myocardium, and epicardium) and chamber specification (right or left ventricles/auricles). Despite these limitations, the fact that iPSC-CMs express the main ion currents and display spontaneous contractile activity makes it an accurate model to study the impact of a specific variant at the single cell level in a patient-specific manner.
In conclusion, p.R211H RRAD variant induces a gain of function of RAD, which reduces the amplitude of the Na+ current, a mechanism already associated with BrS, and induces a persistent Na+ current. In addition, this variant leads to cytoskeleton anomalies and defects in cell morphology. This impaired structural integrity of cardiomyocytes, which had never been related to BrS, might alter conduction by destabilizing myocardial structure. Furthermore, the fact that the insertion of the mutation in an external control cell line recapitulates the overall phenotype confirms the involvement of the p.R211H RRAD variant in the BrS phenotype observed in the other models. The relative contribution of myocardial structural abnormalities vs. electrical alterations remains uncertain and should be subjected to further investigations.
Supplementary Material
Acknowledgements
The authors thank Virginie Forest and Agnès Carcouet (l'institut du thorax) for their expert technical assistance, as well as the members of the Centre d'Investigation Clinique and the Centre de Référence des troubles du rythme héréditaires ou rares de l'Ouest and the Centre de Ressources Biologiques at the CHU of Nantes. Genomic analysis, confocal imaging experiments, and neonatal mouse cardiomyocyte isolation and culture were performed with the support of the GenoBIRD, MicroPiCell, and Therassay core facilities (Biogenouest), respectively. Finally, the authors are grateful to the patients and families who agreed to participate in the research.
Funding
This work was supported by the Fondation pour la Recherche Médicale (Equipe FRM DEQ20140329545 to J.J.S.), the Agence Nationale de la Recherche (ANR-12-BSC1-0013-01 to F.C., ANR-14-CE10-0001-01 to R.R., ANR-15-CE14-0019-01 to N.G., ANR-15-CE14-0006-01 to C.M.), the National Institutes of Health (R01 HL128170 to J.C.W.), the Leducq Foundation (18CVD05 to J.C.W.), the Fédération Française de Cardiologie (FFC R15070NN to N.G.; subvention FFC 2017 to V.P.), and the Pays-de-la-Loire regional council (VaCaRMe programme to R.R.).
Conflict of interest: J.C.W. is a co-founder of Khloris Biosciences. All other authors have nothing to disclose.
See page 3094 for the editorial comment on this article (doi: 10.1093/eurheartj/ehz448)
References
- 1. Brugada P, Brugada J.. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol 1992;20:1391–1396. [DOI] [PubMed] [Google Scholar]
- 2. Priori SG, Blomström-Lundqvist C, Mazzanti A, Blom N, Borggrefe M, Camm J, Elliott PM, Fitzsimons D, Hatala R, Hindricks G, Kirchhof P, Kjeldsen K, Kuck KH, Hernandez-Madrid A, Nikolaou N, Norekvål TM, Spaulding C, Van Veldhuisen DJ; ESC Scientific Document Group. 2015 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: the Task Force for the Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death of the European Society of Cardiology (ESC). Endorsed by: association for European Paediatric and Congenital Cardiology (AEPC). Eur Heart J 2015;36:2793–2867. [DOI] [PubMed] [Google Scholar]
- 3. Kapplinger JD, Tester DJ, Alders M, Benito B, Berthet M, Brugada J, Brugada P, Fressart V, Guerchicoff A, Harris-Kerr C, Kamakura S, Kyndt F, Koopmann TT, Miyamoto Y, Pfeiffer R, Pollevick GD, Probst V, Zumhagen S, Vatta M, Towbin JA, Shimizu W, Schulze-Bahr E, Antzelevitch C, Salisbury BA, Guicheney P, Wilde AA, Brugada R, Schott JJ, Ackerman MJ.. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm 2010;7:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andorin A, Behr ER, Denjoy I, Crotti L, Dagradi F, Jesel L, Sacher F, Petit B, Mabo P, Maltret A, Wong LC, Degand B, Bertaux G, Maury P, Dulac Y, Delasalle B, Gourraud JB, Babuty D, Blom NA, Schwartz PJ, Wilde AA, Probst V.. Impact of clinical and genetic findings on the management of young patients with Brugada syndrome. Heart Rhythm 2016;13:1274–1282. [DOI] [PubMed] [Google Scholar]
- 5. Postema PG, van Dessel PF, Kors JA, Linnenbank AC, van Herpen G, Ritsema van Eck HJ, van Geloven N, de Bakker JM, Wilde AA, Tan HL.. Local depolarization abnormalities are the dominant pathophysiologic mechanism for type 1 electrocardiogram in Brugada syndrome. A study of electrocardiograms, vectorcardiograms, and body surface potential maps during ajmaline provocation. J Am Coll Cardiol 2010;55:789–797. [DOI] [PubMed] [Google Scholar]
- 6. Gourraud J-B, Barc J, Thollet A, Le Scouarnec S, Le Marec H, Schott JJ, Redon R, Probst V.. The Brugada syndrome: a rare arrhythmia disorder with complex inheritance. Front Cardiovasc Med 2016;3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Portero V, Le Scouarnec S, Es-Salah-Lamoureux Z, Burel S, Gourraud JB, Bonnaud S, Lindenbaum P, Simonet F, Violleau J, Baron E, Moreau E, Scott C, Chatel S, Loussouarn G, O'Hara T, Mabo P, Dina C, Le Marec H, Schott JJ, Probst V, Baró I, Marionneau C, Charpentier F, Redon R.. Dysfunction of the voltage-gated K+ channel β2 subunit in a familial case of Brugada syndrome. J Am Heart Assoc 2016;5:e003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Risgaard B, Jabbari R, Refsgaard L, Holst AG, Haunsø S, Sadjadieh A, Winkel BG, Olesen MS, Tfelt-Hansen J.. High prevalence of genetic variants previously associated with Brugada syndrome in new exome data. Clin Genet 2013;84:489–495. [DOI] [PubMed] [Google Scholar]
- 9. Le Scouarnec S, Karakachoff M, Gourraud JB, Lindenbaum P, Bonnaud S, Portero V, Duboscq-Bidot L, Daumy X, Simonet F, Teusan R, Baron E, Violleau J, Persyn E, Bellanger L, Barc J, Chatel S, Martins R, Mabo P, Sacher F, Haïssaguerre M, Kyndt F, Schmitt S, Bézieau S, Le Marec H, Dina C, Schott JJ, Probst V, Redon R.. Testing the burden of rare variation in arrhythmia-susceptibility genes provides new insights into molecular diagnosis for Brugada syndrome. Hum Mol Genet 2015;24:2757–2763. [DOI] [PubMed] [Google Scholar]
- 10. Ackerman MJ. Genetic purgatory and the cardiac channelopathies: exposing the variants of uncertain/unknown significance issue. Heart Rhythm 2015;12:2325–2331. [DOI] [PubMed] [Google Scholar]
- 11. Probst V, Wilde AAM, Barc J, Sacher F, Babuty D, Mabo P, Mansourati J, Le Scouarnec S, Kyndt F, Le Caignec C, Guicheney P, Gouas L, Albuisson J, Meregalli PG, Le Marec H, Tan HL, Schott JJ.. SCN5A mutations and the role of genetic background in the pathophysiology of Brugada syndrome. Circ Cardiovasc Genet 2009;2:552–557. [DOI] [PubMed] [Google Scholar]
- 12. Bezzina CR, Barc J, Mizusawa Y, Remme CA, Gourraud JB, Simonet F, Verkerk AO, Schwartz PJ, Crotti L, Dagradi F, Guicheney P, Fressart V, Leenhardt A, Antzelevitch C, Bartkowiak S, Borggrefe M, Schimpf R, Schulze-Bahr E, Zumhagen S, Behr ER, Bastiaenen R, Tfelt-Hansen J, Olesen MS, Kääb S, Beckmann BM, Weeke P, Watanabe H, Endo N, Minamino T, Horie M, Ohno S, Hasegawa K, Makita N, Nogami A, Shimizu W, Aiba T, Froguel P, Balkau B, Lantieri O, Torchio M, Wiese C, Weber D, Wolswinkel R, Coronel R, Boukens BJ, Bézieau S, Charpentier E, Chatel S, Despres A, Gros F, Kyndt F, Lecointe S, Lindenbaum P, Portero V, Violleau J, Gessler M, Tan HL, Roden DM, Christoffels VM, Le Marec H, Wilde AA, Probst V, Schott JJ, Dina C, Redon R.. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet 2013;45:1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reynet C, Kahn CR.. Rad: a member of the Ras family overexpressed in muscle of type II diabetic humans. Science 1993;262:1441–1444. [DOI] [PubMed] [Google Scholar]
- 14. Yada H, Murata M, Shimoda K, Yuasa S, Kawaguchi H, Ieda M, Adachi T, Murata M, Ogawa S, Fukuda K.. Dominant negative suppression of Rad leads to QT prolongation and causes ventricular arrhythmias via modulation of L-type Ca2+ channels in the heart. Circ Res 2007;101:69–77. [DOI] [PubMed] [Google Scholar]
- 15. Rojas AM, Fuentes G, Rausell A, Valencia A.. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J Cell Biol 2012;196:189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Portero V, Casini S, Hoekstra M, Verkerk AO, Mengarelli I, Belardinelli L, Rajamani S, Wilde AAM, Bezzina CR, Veldkamp MW, Remme CA.. Anti-arrhythmic potential of the late sodium current inhibitor GS-458967 in murine Scn5a-1798insD+/- and human SCN5A-1795insD+/- iPSC-derived cardiomyocytes. Cardiovasc Res 2017;113:829–838. [DOI] [PubMed] [Google Scholar]
- 17. Bersell KR, Yang T, Hong CC, Wells QS, Kannankeril PJ, Roden DM.. Genomic editing in iPSCs establishes that a rare TBX5 variant causes Brugada syndrome. Heart Rhythm 2016;13(Suppl):S9. [Google Scholar]
- 18. Liang P, Sallam K, Wu H, Li Y, Itzhaki I, Garg P, Zhang Y, Vermglinchan V, Lan F, Gu M, Gong T, Zhuge Y, He C, Ebert AD, Sanchez-Freire V, Churko J, Hu S, Sharma A, Lam CK, Scheinman MM, Bers DM, Wu JC.. Patient-specific and genome-edited induced pluripotent stem cell-derived cardiomyocytes elucidate single cell phenotype of Brugada syndrome. J Am Coll Cardiol 2016;68:2086–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Veerman CC, Mengarelli I, Lodder EM, Kosmidis G, Bellin M, Zhang M, Dittmann S, Guan K, Wilde AAM, Schulze-Bahr E, Greber B, Bezzina CR, Verkerk AO.. Switch from fetal to adult SCN5A isoform in human induced pluripotent stem cell-derived cardiomyocytes unmasks the cellular phenotype of a conduction disease-causing mutation. J Am Heart Assoc 2017;6:e005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Satin J, Itzhaki I, Rapoport S, Schroder EA, Izu L, Arbel G, Beyar R, Balke CW, Schiller J, Gepstein L.. Calcium handling in human embryonic stem cell‐derived cardiomyocytes. Stem Cells 2008;26:1961. [DOI] [PubMed] [Google Scholar]
- 21. Antzelevitch C, Pollevick GD, Cordeiro JM, Casis O, Sanguinetti MC, Aizawa Y, Guerchicoff A, Pfeiffer R, Oliva A, Wollnik B, Gelber P, Bonaros EP Jr, Burashnikov E, Wu Y, Sargent JD, Schickel S, Oberheiden R, Bhatia A, Hsu LF, Haïssaguerre M, Schimpf R, Borggrefe M, Wolpert C.. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation 2007;115:442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang G, Zhu X, Xie W, Han P, Li K, Sun Z, Wang Y, Chen C, Song R, Cao C, Zhang J, Wu C, Liu J, Cheng H.. Rad as a novel regulator of excitation-contraction coupling and β-adrenergic signaling in heart. Circ Res 2010;106:317–327. [DOI] [PubMed] [Google Scholar]
- 23. Zhu J, Bilan PJ, Moyers JS, Antonetti DA, Kahn CR.. Rad, a novel Ras-related GTPase, interacts with skeletal muscle beta-tropomyosin. J Biol Chem 1996;271:768–773. [DOI] [PubMed] [Google Scholar]
- 24. Bilan PJ, Moyers JS, Kahn CR.. The Ras-related protein rad associates with the cytoskeleton in a non-lipid-dependent manner. Exp Cell Res 1998;242:391–400. [DOI] [PubMed] [Google Scholar]
- 25. Ward Y, Yap SF, Ravichandran V, Matsumura F, Ito M, Spinelli B, Kelly K.. The GTP binding proteins Gem and Rad are negative regulators of the Rho-Rho kinase pathway. J Cell Biol 2002;157:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kant S, Holthöfer B, Magin TM, Krusche CA, Leube RE.. Desmoglein 2-dependent arrhythmogenic cardiomyopathy is caused by a loss of adhesive function. Circ Cardiovasc Genet 2015;8:553–563. [DOI] [PubMed] [Google Scholar]
- 27. Catalano O, Antonaci S, Moro G, Mussida M, Frascaroli M, Baldi M, Cobelli F, Baiardi P, Nastoli J, Bloise R, Monteforte N, Napolitano C, Priori SG.. Magnetic resonance investigations in Brugada syndrome reveal unexpectedly high rate of structural abnormalities. Eur Heart J 2009;30:2241–2248. [DOI] [PubMed] [Google Scholar]
- 28. Nademanee K, Raju H, de Noronha SV, Papadakis M, Robinson L, Rothery S, Makita N, Kowase S, Boonmee N, Vitayakritsirikul V, Ratanarapee S, Sharma S, van der Wal AC, Christiansen M, Tan HL, Wilde AA, Nogami A, Sheppard MN, Veerakul G, Behr E.. Fibrosis, connexin-43, and conduction abnormalities in the Brugada syndrome. J Am Coll Cardiol 2015;66:1976–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sieira J, Dendramis G, Brugada P.. Pathogenesis and management of Brugada syndrome. Nat Rev Cardiol 2016;13:744–756. [DOI] [PubMed] [Google Scholar]
- 30. Kattygnarath D, Maugenre S, Neyroud N, Balse E, Ichai C, Denjoy I, Dilanian G, Martins RP, Fressart V, Berthet M, Schott JJ, Leenhardt A, Probst V, Le Marec H, Hainque B, Coulombe A, Hatem SN, Guicheney P.. MOG1: a new susceptibility gene for Brugada syndrome . Circ Cardiovasc Genet 2011;4:261–268. [DOI] [PubMed] [Google Scholar]
- 31. Zhou J, Wang L, Zuo M, Wang X, Ahmed AS, Chen Q, Wang QK.. Cardiac sodium channel regulator MOG1 regulates cardiac morphogenesis and rhythm. Sci Rep 2016;6:21538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sakata Y, Kamei CN, Nakagami H, Bronson R, Liao JK, Chin MT.. Ventricular septal defect and cardiomyopathy in mice lacking the transcription factor CHF1/Hey2. Proc Natl Acad Sci USA 2002;99:16197–16202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Veerman CC, Podliesna S, Tadros R, Lodder EM, Mengarelli I, de Jonge B, Beekman L, Barc J, Wilders R, Wilde AAM, Boukens BJ, Coronel R, Verkerk AO, Remme CA, Bezzina CR.. The Brugada syndrome susceptibility gene HEY2 modulates cardiac transmural ion channel patterning and electrical heterogeneity. Circ Res 2017;121:537–548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.