Abstract
The onset of leaf senescence is triggered by external cues and internal factors such as phytohormones and signaling pathways involving transcription factors (TFs). Abscisic acid (ABA) strongly induces senescence and endogenous ABA levels are finely tuned by many senescence-associated TFs. Here, we report on the regulatory function of the senescence-induced TF OsWRKY5 TF in rice (Oryza sativa). OsWRKY5 expression was rapidly upregulated in senescing leaves, especially in yellowing sectors initiated by aging or dark treatment. A T-DNA insertion activation-tagged OsWRKY5-overexpressing mutant (termed oswrky5-D) promoted leaf senescence under natural and dark-induced senescence (DIS) conditions. By contrast, a T-DNA insertion oswrky5-knockdown mutant (termed oswrky5) retained leaf greenness during DIS. Reverse-transcription quantitative PCR (RT-qPCR) showed that OsWRKY5 upregulates the expression of genes controlling chlorophyll degradation and leaf senescence. Furthermore, RT-qPCR and yeast one-hybrid analysis demonstrated that OsWRKY5 indirectly upregulates the expression of senescence-associated NAM/ATAF1/2/CUC2 (NAC) genes including OsNAP and OsNAC2. Precocious leaf yellowing in the oswrky5-D mutant might be caused by elevated endogenous ABA concentrations resulting from upregulated expression of ABA biosynthesis genes OsNCED3, OsNCED4, and OsNCED5, indicating that OsWRKY is a positive regulator of ABA biosynthesis during leaf senescence. Furthermore, OsWRKY5 expression was suppressed by ABA treatment. Taken together, OsWRKY5 is a positive regulator of leaf senescence that upregulates senescence-induced NAC, ABA biosynthesis, and chlorophyll degradation genes.
Keywords: rice, leaf senescence, abscisic acid (ABA), OsWRKY, NAC
1. Introduction
Leaf senescence is the final stage of plant development and involves diverse molecular and cellular processes such as degradation of chlorophylls and macromolecules, and remobilization of nutrients into newly developing or storage organs through expression of senescence-associated genes (SAGs). The onset of leaf senescence begins with chlorophyll degradation and proceeds to hydrolysis of macromolecules (proteins, lipids, and nucleic acids); this is followed by cell death [1,2,3].
Genetic studies have revealed the contribution of chlorophyll catabolic enzymes to sequential reactions of chlorophyll degradation. The STAY-GREEN (SGR) protein is a magnesium (Mg)-dechelatase, which produces pheophytin a by removing Mg from chlorophyll a [4,5]. Thus, functional deficiency of SGR orthologs leads to a strong stay-green phenotype in diverse plant species including Arabidopsis thaliana [6], rice (Oryza sativa) [4], pea (Pisum sativum) [7], tomato (Solanum lycopersicum), bell pepper (Capsicum annuum) [8], and soybean (Glycine max) [9]. Failure to convert pheophytin a to pheophorbide a due to mutation of NON-YELLOW COLORING3 (NYC3), encoding an α/β hydrolase-fold family protein, delays leaf senescence during dark-induced senescence (DIS) [10]. Knockdown of rice pheophorbide a oxygenase (OsPAO) leads to accumulation of pheide a and prolongs leaf greenness during dark incubation [11]. SAGs identified during leaf senescence in rice encode putative proteins involved in metabolic programing [12]; Osh36 and Osl85 encode an aminotransferase and isocitrate lyase, which participate in amino acid and fatty acid metabolism, respectively.
Abscisic acid (ABA) participates in multiple aspects of plant development including leaf senescence, seed germination, stomatal closure, and root development [13,14,15,16,17,18]. Specifically, expression of ABA biosynthetic genes such as those encoding 9′-cis-epoxycarotenoid dioxygenases (NCEDs) is induced by leaf senescence, elevating endogenous ABA levels in Arabidopsis leaves [19,20]. Increased levels of endogenous ABA can activate chlorophyll degradation pathways mediated by senescence-associated transcription factors (TFs) [21,22,23,24]. For instance, ABA induces the expression of the genes encoding ABA-RESPONSIVE ELEMENT (ABRE)-BINDING TRANSCRIPTION FACTOR 2 (ABF2), ABF3, and ABF4, which directly bind to the SGR1 promoter, accelerating chlorophyll degradation in Arabidopsis leaves [21]. In rice, ABA-promoted expression of OsNAP directly upregulates chlorophyll degradation genes (CDGs) such as SGR, NYC1, NYC3, and RCCR1, leading to early leaf senescence [25].
The WRKY TFs participate in various biological processes such as biotic and abiotic stress, seed development, seed dormancy, and germination [26]. Genome-wide analyses have revealed that many WRKY genes are strongly induced by leaf senescence [27,28], suggesting that WRKY TFs are involved in regulating leaf senescence. Following identification of AtWRKY6 as a regulator of leaf senescence [29], other WRKY TFs regulating leaf senescence have been functionally characterized. For example, mutation of Arabidopsis AtWRKY53 confers a delayed leaf senescence phenotype by specifically altering regulation of its target genes [30]. Overexpression of AtWRKY22, a target gene of AtWRKY53, accelerates leaf senescence [31]. AtWRKY54 and AtWRKY70 act as negative regulators of leaf senescence by interacting independently with WRKY30 [32]. AtWRKY45 mediates gibberellic acid (GA)-induced leaf senescence by interacting with a DELLA protein, RGL1 [33]. AtWRKY75 increases salicylic acid (SA) and H2O2 levels by activating SID2 and repressing CAT2, respectively, resulting in early leaf senescence [34]. Heterologous expression of rice OsWRKY23 promotes leaf senescence in Arabidopsis [35]. Rice OsWRKY42 induces reactive oxygen species (ROS) by directly downregulating the expression of OsMT1d encoding metallothionein protein and thereby promoting leaf senescence [36]. Unlike Arabidopsis WRKY TFs involved in the regulation of leaf senescence; however, few OsWRKY TFs have been identified as functioning in the execution of leaf senescence.
In this study, we found that OsWRKY5 expression is upregulated at the onset of leaf senescence. The OsWRKY5-overexpressing oswrky5-D mutation promoted leaf yellowing under aging and dark treatment, while an oswrky5-knockdown mutant exhibited a delayed senescence phenotype during DIS. Reverse-transcription quantitative PCR (RT-qPCR) analysis suggested that CDGs and SAGs were upregulated by senescence-induced OsWRKY5. Furthermore, OsWRKY5 seemed to indirectly regulate the expression of senescence-associated NAC (senNAC) genes such as OsNAP and OsNAC2, which are upstream regulators of CDGs and SAGs. OsWRKY5 elevated endogenous ABA levels by upregulating the expression of ABA biosynthetic genes. Our results thus provide evidence that OsWRKY5 acts as a positive regulator of leaf senescence in rice.
2. Results
2.1. Characterization of OsWRKY5
OsWRKY5 (Os05g04640), a member of rice WRKY TF family, consists of six exons with 1509 bp of open reading frame in 5379 bp of genomic DNA. OsWRKY5 is predicted to encode a 502 amino acid protein with a molecular mass of 52.3 kDa (http://rice.plantbiology.msu.edu/index.shtml). The WRKY domain of OsWRKY5 has a single consensus motif (WRKYGQK) and a zinc-finger C2H2 motif (Cx5Cx23HxH), indicating that OsWRKY5 belongs to the group II WRKY TF family [37]. From sequence alignment of WRKY domains between OsWRKY5 and group II Arabidopsis thaliana WRKY (AtWRKY) proteins, we found that the domain sequences of OsWRKY5 are quite similar to those of AtWRKY6 and AtWRKY47, members of the subgroup IIb AtWRKY TF family (Figure S1). To examine the subcellular localization of OsWRKY5, we transiently expressed the 35S::YFP-OsWRKY5 construct in onion epidermal cells. The fluorescent signal of YFP-OsWRKY5 fusions was observed exclusively in nuclei (Figure S2), indicating that OsWRKY5 is a nuclear-localized protein.
2.2. OsWRKY5 Is Upregulated during Leaf Senescence
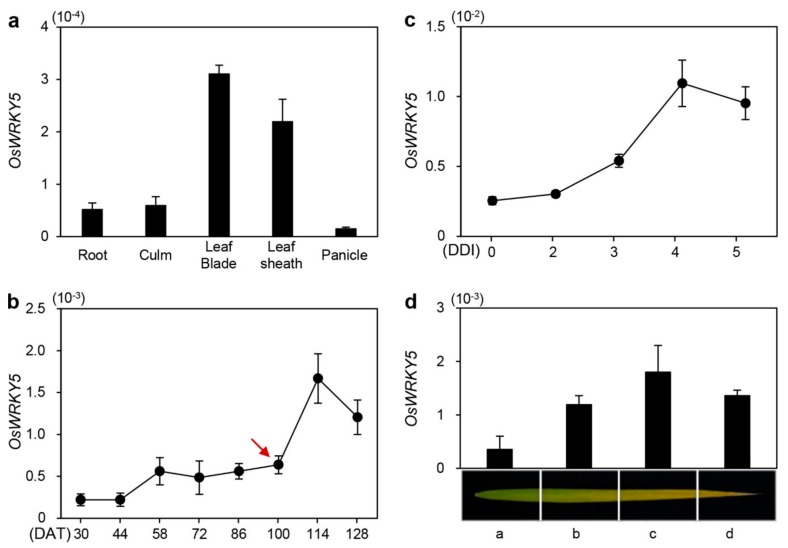
To examine the spatial expression of OsWRKY5, we investigated transcript levels of OsWRKY5 in rice organs including root, culm, leaf blade, leaf sheath, and panicle at the reproductive stage (Figure 1a). OsWRKY5 was preferentially expressed in the leaf blade and leaf sheath. Previous transcriptome analysis [28] showed upregulation of 47 rice WRKY TFs including OsWRKY5 in flag leaves during natural senescence (NS). Therefore, we determined age-dependent changes in OsWRKY5 expression in flag leaves of wild-type (WT; japonica cultivar ‘Dongjin’) rice grown in a paddy field under natural long-day (NLD) conditions (>14 h light/day). While OsWRKY5 was constitutively expressed in developing leaves at the vegetative stage, OsWRKY5 expression was dramatically upregulated in senescing leaves at the reproductive stage (Figure 1b). In addition, OsWRKY5 expression gradually increased in detached leaves of four-week-old WT leaves during DIS (Figure 1c). We further found that OsWRKY5 transcripts accumulated in the yellowing sector (region c) more than in the green sector (region a) of senescing flag leaves (Figure 1d). These results suggest that OsWRKY5 is involved in the onset and progression of leaf senescence in rice.
Figure 1.
Expression profiles of OsWRKY5 in rice. (a) OsWRKY5 mRNA levels in detached organs from the japonica cultivar ‘Dongjin’ (hereafter wild type; WT) at the heading stage. OsWRKY5 was mainly expressed in leaf blade and leaf sheath. (b,c) Changes in OsWRKY5 expression level in leaf blades of WT rice grown in a paddy field (b) or in the greenhouse (c) under natural long day conditions (≥14 h light/day). (c) Detached leaves of three-week-old WT were incubated in 3 Mm 2-(N-morpholino)ethanesulfonic (MES) buffer (pH 5.8) at 28 °C in complete darkness. Red arrow indicates heading date. (d) Expression of OsWRKY5 measured in flag leaves divided into four regions from the green sector (a) to the yellow sector (d) at 128 days after transplanting (DAT). OsWRKY5 mRNA levels were determined by RT-qPCR analysis and normalized to that of OsUBQ5 (Os01g22490). Mean and SD values were obtained from at least three biological samples. Experiments were repeated twice with similar results. DDI, day(s) of dark incubation.
2.3. OsWRKY5 Positively Regulates the Progression of Leaf Senescence
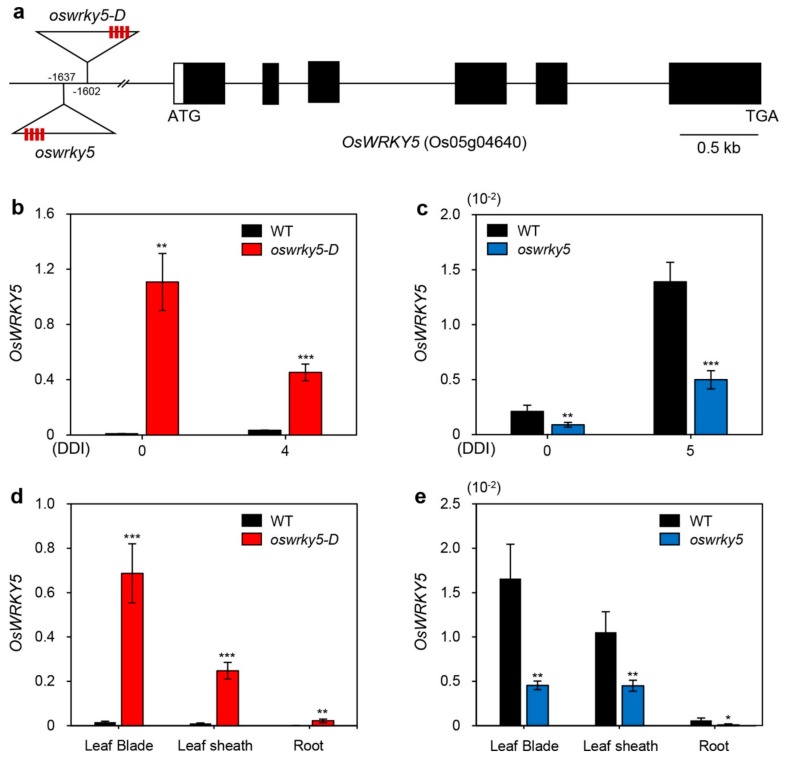
To examine the function of OsWRKY5 in leaf senescence, we identified activation-tagged and loss-of-function mutants. To this end, we obtained two independent T-DNA insertion lines (PFG_3A-15928 and PFG_3A-06060) from the RiceGE database (http://signal.salk.edu/cgi-bin/RiceGE) in which each T-DNA fragment with an activation tag (4 × 35S promoter) was integrated into the promoter region of OsWRKY5 (Figure 2a). Nucleotide sequences used for determining the locations of T-DNA insertions have been posted on the RiceGE database. Based on the comparison with OsWRKY5 promoter sequence, we predicted that T-DNA fragments of oswrky5-D and oswrky5 are inserted in the 1602-bp and 1637-bp upstream of the start codon of OsWRKY5 gene, respectively. They were confirmed by our re-sequencing the regions of T-DNA insertion regions of two mutant lines. To verify the expression levels of OsWRKY5 in these mutant lines, we measured OsWRKY5 expression levels in detached leaves of four-week-old mutant plants during DIS. RT-qPCR showed that OsWRKY5 transcripts accumulated to high levels in PFG_3A-15928 compared to the WT due to the activation-tagged T-DNA insertion; by contrast, in PFG_3A-06060, the T-DNA insertion in the promoter region of OsWRKY5 reduced expression of OsWRKY5 (Figure 2b,c). These results indicate that PFG_3A-15928 and PFG_3A-06060 are dominant activation and recessive knockdown mutants, respectively (hereafter termed oswrky5-D and oswrky5, respectively). To confirm this, we further investigated the OsWRKY5 expression in leaf blade, leaf sheath, and root separated from WT and mutant lines grown in paddy soil for three weeks. Similar to expression patterns of OsWRKY5 shown in detached leaves during DIS (Figure 2b,c), OsWRKY5 transcripts highly accumulated in all tissues of oswrky5-D compared with the WT, while they were significantly lower in oswrky5 than in the WT (Figure 2d,e).
Figure 2.
Mutation of OsWRKY5 by T-DNA insertion. (a) Schematic diagram depicting the positions of T-DNA insertions in the promoter region of OsWRKY5 (LOC_Os05g04640). Black and white bars represent exons and 5′-untranslated region, respectively. Open triangles indicate the location of the OsWRKY5 T-DNA insertions (oswrky5-D, PFG_3A-15928; oswrky5, PFG_3A-06060). Red boxes on triangles represent tetramerized 35S enhancers (4 × 35S). (b,c) Total RNA was isolated from detached leaves of WT and mutant lines (oswrky5-D and oswrky5) under DIS as shown in Figure 3a,b. (d,e) OsWRKY5 mRNA levels were measured in rice tissues separated from three-week-old WT and mutant lines. Transcript levels of OsWRKY5 in oswrky5-D (b,d) and oswrky5 (c,e) were determined by RT-qPCR and normalized to the transcript levels of OsUBQ5. Mean and SD values were obtained from more than three biological replicates. Asterisks indicate a statistically significant difference from WT, as determined by Student’s t-test (* p < 0.05, ** p < 0.01, *** p < 0.001). DDI, day(s) of dark incubation.
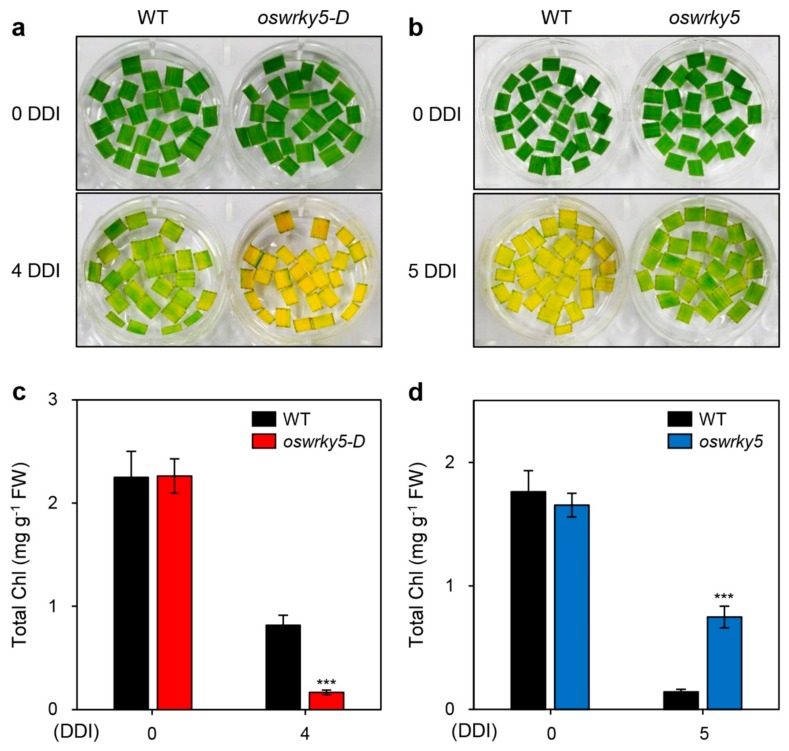
To determine the phenotypic difference between WT and mutant lines during DIS, we next incubated detached leaves of three-week-old WT, oswrky5-D, and oswrky5 plants in 3 mM MES buffer (pH 5.8) at 28 °C under complete darkness. While oswrky5-D showed accelerated leaf yellowing compared with the WT, the oswrky5 leaves retained their green color longer than the WT leaves (Figure 3a,b). Consistent with the leaf color, the total chlorophyll content of oswrky5-D was less than that of the WT after DIS, whereas oswrky5 maintained higher total chlorophyll levels during DIS compared with the WT (Figure 3c,d).
Figure 3.
OsWRKY5 promotes leaf yellowing under dark-induced senescence (DIS) conditions. WT and mutant lines (oswrky5-D and oswrky5) were grown in paddy soil for four weeks under natural long day conditions (≥14 h light/day). (a,b) Yellowing of detached leaves induced in 3 mM MES buffer (pH 5.8) at 28 °C under complete darkness. Changes in leaf color (a,b) and total chlorophyll (Chl) contents (c,d) of oswrky5-D or oswrky5 mutants compared with the WT after 4 or 5 days of dark incubation (DDI), respectively. Mean and SD values were obtained from more than three biological replicates. Asterisks indicate a statistically significant difference from WT, as determined by Student’s t-test (*** p < 0.001). Experiments were repeated twice with similar results. FW, fresh weight.
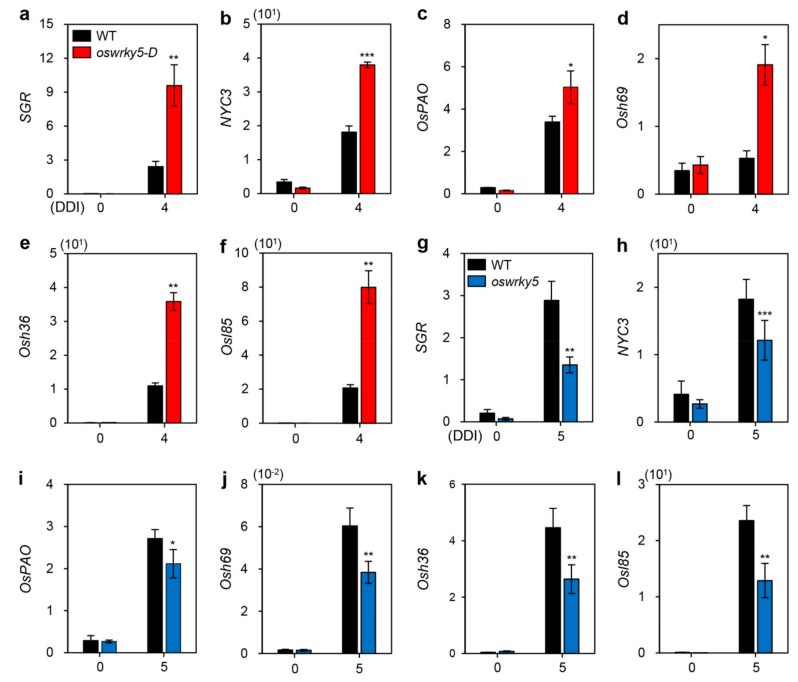
In senescing leaves, chlorophylls are sequentially degraded by chlorophyll-degrading enzymes associated with upregulation of CDGs, including SGR [4], NYC3 [10], and OsPAO [11]. Many other SAGs are also upregulated during DIS in rice, with products identified as seed imbibition protein (Osh69), glyoxylate aminotransferase (Osh36), and isocitrate lyase (Osl85) [12]. We therefore measured transcript levels of CDGs and SAGs in detached leaves of three-week-old WT, oswrky5-D, and oswrky5 plants under DIS conditions as shown in Figure 2. RT-qPCR analysis revealed that expression of CDGs and SAGs was upregulated in oswrky5-D after 4 days of dark incubation (DDI) (Figure 4a–f) but downregulated in oswrky5 after 5 DDI when compared with the WT (Figure 4g–l). These results demonstrate that OsWRKY5 promotes the onset and progression of leaf senescence by upregulating expression of CDGs and SAGs.
Figure 4.
Altered expression of CDGs and SAGs in oswrky5-D and oswrky5 during DIS. Total RNA was isolated from detached leaves of WT and mutant lines (oswrky5-D and oswrky5) under DIS as shown in Figure 3 (3a,b). Expression of CDGs and SAGs in oswrky5-D (a–f) or oswrky5 (g–l) was compared with that in the WT after 4 or 5 DDI, respectively. Transcript levels of CDGs (a–c and g–i) and SAGs (d–f and j–l) were determined by RT-qPCR analysis and normalized to that of OsUBQ5. Mean and SD values were obtained from more than three biological replicates. Asterisks indicate a statistically significant difference from WT, as determined by Student’s t-test (* p < 0.05, ** p < 0.01, *** p < 0.001). Experiments were repeated twice with similar results. CDGs, Chl degradation genes; DDI, day(s) of dark incubation; SAGs, senescence-associated genes.
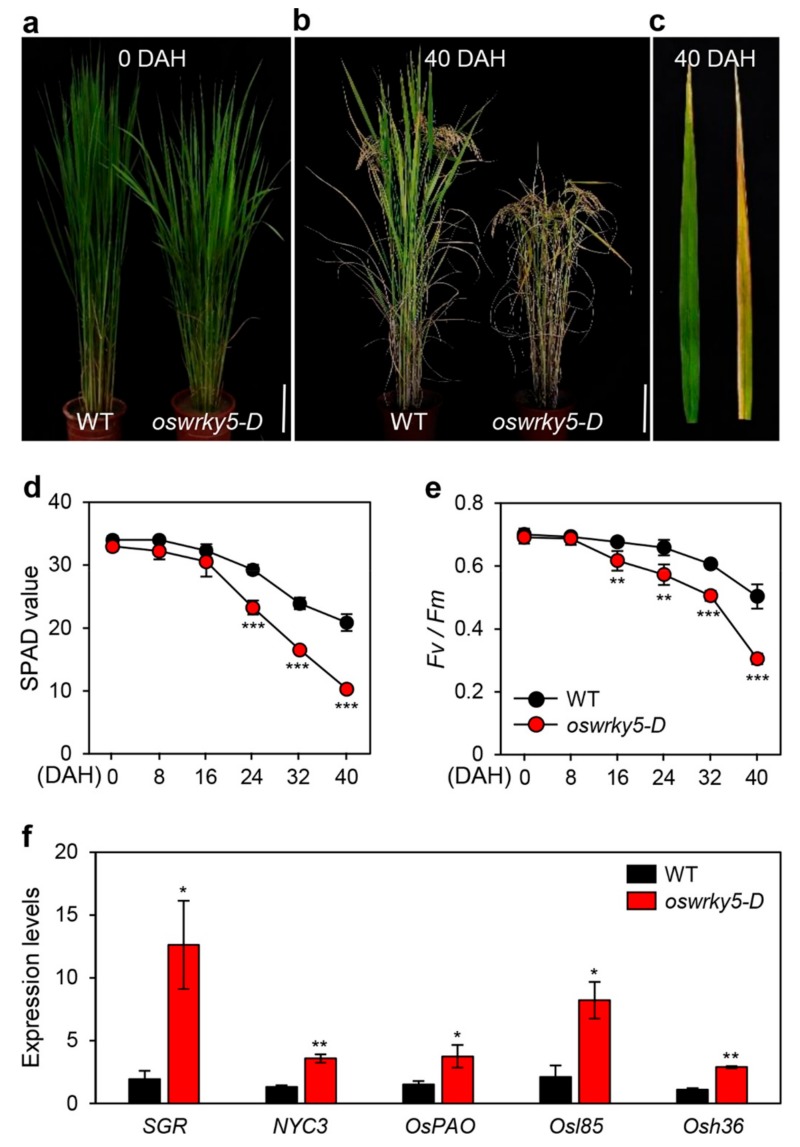
To further examine how OsWRKY5 overexpression affects leaf senescence during vegetative and reproductive stages, we monitored age-dependent leaf yellowing in WT and oswrky5-D plants grown under NLD conditions (>14 h daylight) in the field (37° N latitude, Suwon, South Korea). While there was no significant difference in leaf color between the WT and oswrky5-D until heading (Figure 5a), the leaves of oswrky5-D showed a precocious leaf senescence phenotype at 40 days after heading (DAH) (Figure 5b,c). The SPAD value, a parameter for leaf greenness, indicated lower levels of green pigments in flag leaves of oswrky5-D compared with the WT at 24 DAH (Figure 5d). Leaf greenness is closely linked to photosynthetic capacity [38,39]. Thus, reduced SPAD value led to a relatively lower Fv/Fm ratio (efficiency of photosystem II) in oswrky5-D than in WT at 24 DAH (Figure 5e). Similar to expression patterns of CDGs and SAGs during DIS, CDG and SAG transcripts were more abundant in the senescing flag leaves of oswrky5-D than in those of WT at 40 DAH (Figure 5f). These results indicate that OsWRKY5 acts as a positive regulator of leaf senescence during both NS and DIS.
Figure 5.
oswrky5-D promotes leaf senescence during NS. WT and oswrky5-D plants were grown in a paddy field under natural long-day conditions (≥14 h light/day). (a,b) Phenotypes of WT and oswrky5-D plants at heading (0 DAH) (a) and 40 days after heading (DAH) (b). White scale bars = 20 cm. (c) Senescing flag leaves of WT (left) and oswrky5-D (right) at 40 DAH. Photos shown are representative of five independent plants. (d–e) Changes in SPAD value (d) and photosystem II (PSII) activity (Fv/Fm) (e) in flag leaves at heading. (f) Expression of CDGs and SAGs measured in senescing flag leaves (c). Transcript levels were determined by RT-qPCR analysis and normalized to that of OsUBQ5. Mean and SD values were obtained from more than three biological replicates. Asterisks indicate a statistically significant difference from WT, as determined by Student’s t-test (* p < 0.05, ** p < 0.01, *** p < 0.001).
2.4. OsWRKY5 Upregulates SenNAC Genes
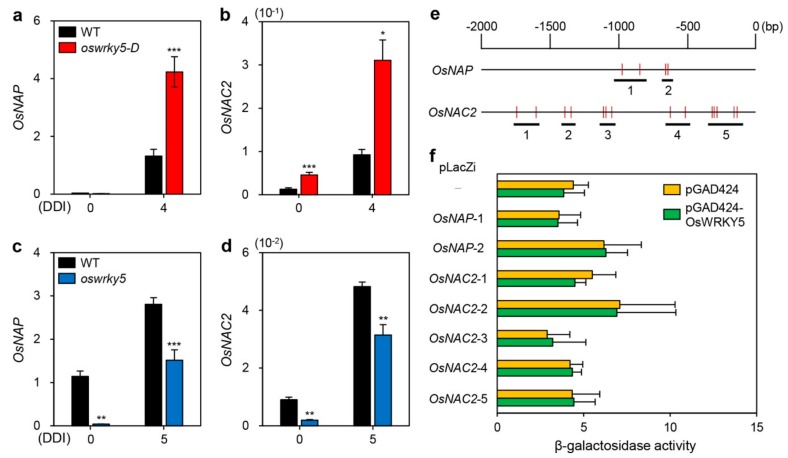
Previous studies have shown that senNACs including OsNAP and OsNAC2 promote leaf senescence by upregulating expression of CDGs and SAGs [25,40]. To determine whether OsWRKY5 participates in NAC TF-mediated senescence pathways, we examined the expression levels of OsNAP and OsNAC2 in detached leaves of WT, oswrky5-D, and oswrky5 under DIS conditions. RT-qPCR showed that compared with WT, the expression levels of OsNAP and OsNAC2 were higher in oswrky5-D during dark incubation (Figure 6a,b), while they were downregulated at 0 and 5 DDI in oswrky5 compared with WT (Figure 6c,d). These results suggest that OsWRKY5 promote leaf senescence by upregulating the expression of OsNAP and OsNAC2.
Figure 6.
OsWRKY5 indirectly regulates expression of senescence-induced NAC TFs. (a–d) Total RNA was isolated from detached leaves of WT and mutant lines (oswrky5-D and oswrky5), as shown in Figure 3 (a,b). Transcript levels of OsNAP (a,c) and OsNAC2 (b,d) were determined by RT-qPCR analysis and normalized to the transcript levels of OsUBQ5. Mean and SD values were obtained from more than three biological replicates. Asterisks indicate a statistically significant difference from WT, as determined by Student’s t-test (* p < 0.05, ** p < 0.01, *** p < 0.001). (e,f) Interaction of OsWRKY5 with the promoters of OsNAP and OsNAC2 by yeast one-hybrid assays. (e) Numbers represent upstream base pairs from the transcriptional initiation sites of OsNAP and OsNAC2. Vertical red lines represent the W-box core sequence (TGAC). Horizontal black bars represent regions containing repetitive TGAC sequences. (f) β-Galactosidase activity of bait plasmids (pGAD424 and pGAD424-OsWRKY5) evaluated by the absorbance of chloramphenicol red, a hydrolysis product of chlorophenol red-β-D-galactopyranoside (CPRG). Empty bait (pGAD424) and prey plasmids (-) were used for negative controls. Experiments were repeated twice with similar results. DDI, day(s) of dark incubation.
WRKY TFs regulate the transcription of their target genes by recognizing a consensus cis-element, the so-called W-box [26]. The W-box has been generally defined as 5′-TTGAC(C/T)-3′ with an invariant TGAC core sequence essential for WRKY binding [37,41]. Since repetitive TGAC sequences enhance WRKY binding efficiency, we searched for the TGAC core sequence within 2 kb upstream of the transcriptional initiation sites of OsNAP and OsNAC2, and found two regions (‒1001 ~ ‒765 and ‒657 ~ ‒575) in the promoter of OsNAP and five regions (‒1760 ~ ‒1574, ‒1411 ~ ‒1309, ‒1135 ~ ‒1021, ‒660 ~ ‒480, and ‒352 ~ ‒98) in the promoter of OsNAC2 (Figure 6e). To investigate whether the OsWRKY5 TF directly binds to the promoters of OsNAP and OsNAC2, we performed yeast one-hybrid assays. However, we could not find any difference between GAL4AD and GAL4AD-OsWRKY5 by measuring β-galactosidase activity of lacZ reporter genes, indicating that OsWRKY5 seems not to interact with the promoter of OsNAP or OsNAC2 (Figure 6f).
Previous studies have reported that the microRNA osa-miR164b is closely associated with the post-transcriptional regulation of OsNAC2, resulting in reduction of OsNAC2 mRNA levels [42,43]. To examine whether OsWRKY5 regulates endogenous levels of osa-miR164b during DIS, we determined the expression of osa-miR164b in detached leaves of three-week-old WT, oswrky5-D, and oswrky5 plants using stem-loop RT-PCR analysis. It revealed no difference in the levels of osa-miR164b among genotypes (Figure S3), indicating that OsWRKY5 seems not to participate in osa-miR164b-mediated senescence pathway.
2.5. OsWRKY5 Is Involved in ABA Biosynthesis Pathway
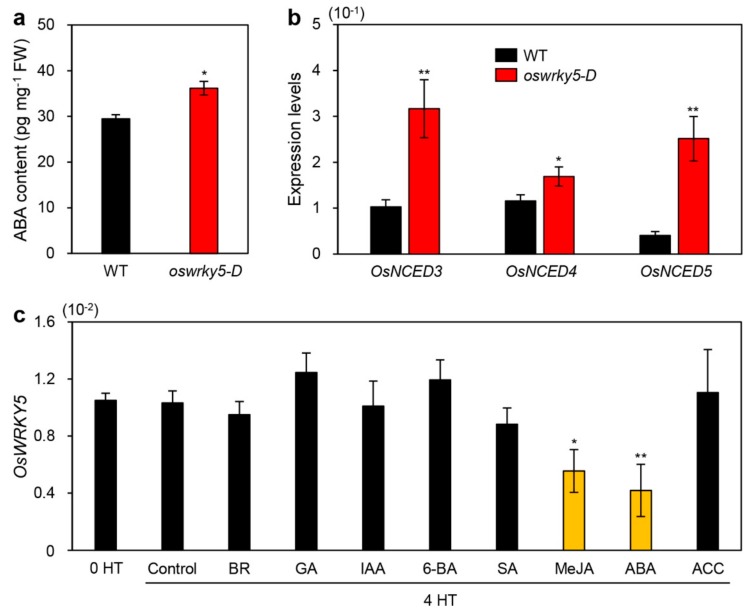
Among phytohormones affecting the onset and progression of leaf senescence [2], ABA activates senescence-associated regulatory pathways, leading to acceleration of leaf senescence [44]. Genetic studies have revealed that the endogenous ABA concentration is delicately controlled by senNACs such as OsNAP and OsNAC2 [25,40]. Considering that OsWRKY5 upregulated the expression of OsNAP and OsNAC2, we speculated that OsWRKY5 is mainly involved in regulating ABA biosynthesis. Indeed, the endogenous ABA concentration was significantly higher in leaves of three-week-old oswrky5-D plants than in those of the WT (Figure 7a). RT-qPCR analysis showed that ABA biosynthesis genes including OsNCED3, OsNCED4, and OsNCED5 were significantly upregulated in oswrky5-D leaves compared with the WT (Figure 7b). This strongly suggests that the early leaf senescence of oswrky5-D is mainly due to an increase in ABA biosynthesis after heading.
Figure 7.
OsWRKY5 participates in ABA-mediated senescence pathways. (a) Endogenous ABA contents measured in leaves of WT and oswrky5-D plants grown in paddy soil for 3 weeks under LD conditions. FW, fresh weight. (b) Total RNA was extracted from leaves of the same WT and oswrky5-D plants used for the analysis shown in Figure 6A. Transcript levels of ABA biosynthetic genes including OsNCED3, OsNCED4, and OsNCED5 were determined by RT-qPCR analysis and normalized to transcript levels of OsUBQ5. Mean and SD values were obtained from more than three biological replicates. Asterisks indicate a statistically significant difference from WT, as determined by Student’s t-test (* p < 0.05, ** p < 0.01). (c) Ten-day-old WT seedlings grown on 0.5X MS phytoagar medium at 28 °C under continuous light conditions were transferred to 0.5X MS liquid medium only (control) or 0.5X MS liquid medium containing 50 μM epibrassinolide (BR), 50 μM gibberellic acid (GA), 50 μM 3-indoleacetic acid (IAA), 50 μM 6-benzylaminopurine (6-BA), 100 μM salicylic acid (SA), 50 μM methyl jasmonic acid (MeJA), 50 μM abscisic acid (ABA), or 50 μM 1-aminocyclopropane-1-carboxylic acid (ACC). Total RNA was isolated from leaves after 4 h of treatment. OsWRKY5 mRNA levels were determined by RT-qPCR analysis and normalized to transcript levels of OsUBQ5. Mean and SD values were obtained from more than three biological replicates. Asterisks on orange bars indicate a statistically significant difference from the control, as determined by Student’s t-test (*p < 0.05, ** p < 0.01). Experiments were repeated twice with similar results.
To investigate whether phytohormones affect the expression of OsWRKY5, we next measured the expression of OsWRKY5 in ten-day-old WT seedlings exogenously treated with epibrassinolide (BR), gibberellic acid (GA), indole-3-acetic acid (IAA), 6-benzylaminopurine (6-BA), salicylic acid (SA), methyl jasmonic acid (MeJA), ABA, or 1-aminocyclopropane-1-carboxylic acid (ACC). RT-qPCR showed that OsWRKY5 expression was significantly reduced by MeJA and ABA treatments (Figure 7c), indicating that excessive levels of ABA decrease the expression of OsWRKY5.
3. Discussion
3.1. OsWRKY5 Promotes Leaf Yellowing during NS and DIS
We found that OsWRKY5 participates in the ABA-mediated regulatory pathways of leaf senescence. OsWRKY5 was expressed in leaves and its transcription was activated by aging and dark treatment (Figure 1b,c). In addition, we found four W-boxes and a single G-box in the 1500-bp upstream of transcription initiation site of OsWRKY5 (Figure S4). These cis-elements are recognized by WRKY, bZIP, bHLH, and NAC transcription factors which are involved in the regulation of leaf senescence [28], implying that expression of OsWRKY5 could be regulated by senescence-induced transcription factors. The WRKY domain of OsWRKY5 has the highest amino acid similarity to that of AtWRKY6 (Figure S1). Similar to the early leaf senescence phenotype of AtWRKY6-OX in Arabidopsis [29], the progression of leaf senescence was much faster in the oswrky5-D mutant than in WT plants under NS and DIS conditions. (Figure 3 and Figure 5), and the oswrky5 knockdown mutant showed markedly delayed leaf senescence (Figure 3).
Many senescence-induced TFs directly or indirectly regulate expression of their target genes, including CDGs, SAGs, and other senescence-associated TFs. OsNAP directly binds to the promoters of SGR, NYC1, NYC3, RCCR1, and Osl57 (encoding a putative 3-ketoacyl-CoA thiolase). OsNAP also indirectly regulates the expression of Osh36 and Osh69, whose amino acid sequences are quite similar to those of Arabidopsis thaliana aminotransferase and Brassica oleracea seed imbibition protein, respectively [25]. OsNAC2 enhances chlorophyll degradation by directly interacting with the promoters of SGR and NYC3 [40]. We therefore speculate that OsWRKY5 upregulates the expression of CDGs and SAGs by regulating senNACs. Upregulation of OsNAP and OsNAC2 was observed in oswrky5-D, resulting in early leaf yellowing (Figure 4 and Figure 5). However, OsWRKY5 does not bind to the promoter regions of OsNAP and OsNAC2 despite the presence of repetitive TGAC core sequences (Figure 6e,f), suggesting that it indirectly regulates expression of these genes.
WRKY TFs can physically interact with other TFs involved in leaf senescence. For example, Besseau et al. (2012) showed that expression of Arabidopsis WRKY30, WRKY53, WRKY54, and WRKY70 is induced during leaf senescence and WRKY53, WRKY54, and WRKY70 interact independently with WRKY30 in yeast two-hybrid assays [32]. In Arabidopsis, WRKY45 functions in GA-mediated leaf senescence by interacting with the DELLA protein RGA-LIKE1 (RGL1) characterized as a repressor of GA signaling [33]. Recently, TT2, a MYB family member, was identified as an interacting partner of WRKY27 in upland cotton (Gossypium hirsutum L.) [45]. Therefore, exploring the possible interaction networks of the OsWRKY5 TF in the regulation of senNACs should provide more insight into the mechanism of leaf senescence.
Interestingly, despite of constitutive expression of OsWRKY5 in oswrky5-D, there were no significant difference in senescence phenotype and expression of CDGs and SAGs between WT and oswrky5-D before leaf senescence (Figure 3a and Figure 5a). Leaf senescence is a complex process involving numerous regulators [27,46]. These regulators are mostly induced by the onset of senescence. OsWRKY5 may require other cofactors to upregulate the expression levels of CDGs and SAGs during leaf senescence. Thus, even though OsWRKY5 transcripts are highly accumulated in oswrky5-D during vegetative growth, it seems that OsWRKY5 overexpression is not much as effective as a senescence promoter, possibly due to a lack of senescence-induced cofactors.
3.2. OsWRKY5 Mediates ABA-Induced Leaf Senescence
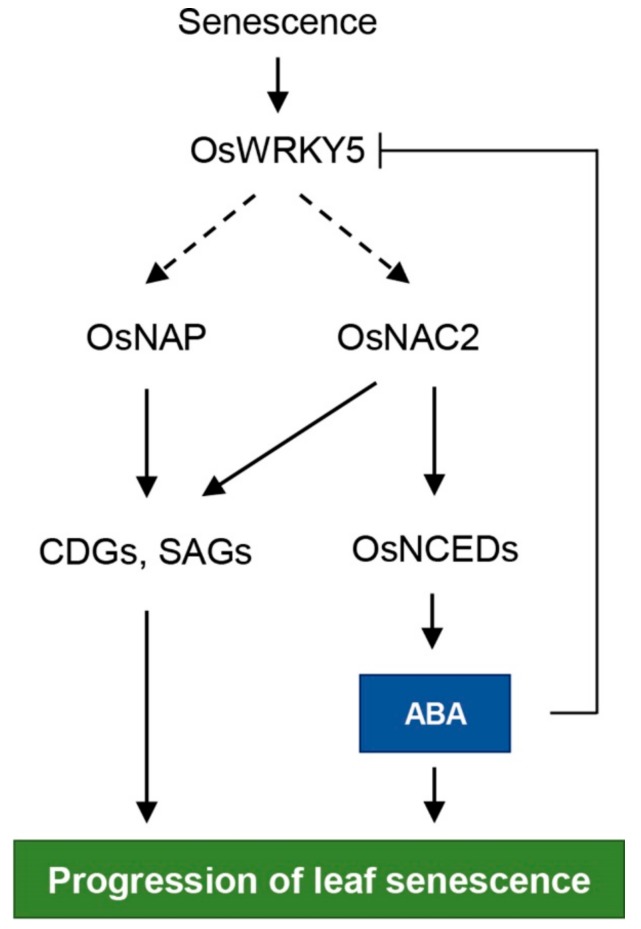
ABA promotes the onset and progression of leaf senescence [47]. Thus, endogenous ABA levels are elevated by upregulation of ABA biosynthesis genes during leaf senescence, promoting further ABA-induced leaf senescence [44,48]. Arabidopsis 9-CIS-EPOXYCAROTENOID DIOXYGENASE (NCED) genes, encoding a rate-limiting enzyme in ABA biosynthesis, are upregulated during NS [19,49]. Dark incubation induces expression of OsNCED3 in rice leaves [50], and overexpression of OsNCED3 accelerates leaf yellowing in rice during dark incubation. NAP increases ABA biosynthesis by directly upregulating transcription of ABSCISIC ALDEHYDE OXIDASE3 (AAO3), leading to chlorophyll degradation during dark incubation in Arabidopsis [20]. In rice, although transcription of ABA biosynthesis genes, such as OsNCED1, OsNCED3, OsNCED4, and OsZEP, is inhibited by OsNAP, the functional ortholog of Arabidopsis NAP, overexpression of OsNAP leads to precocious leaf senescence by directly regulating CDGs and SAGs [25]. OsNAC2 elevates endogenous ABA content by directly binding to the promoters of OsNCED3 and OsZEP, thereby promoting leaf senescence [40]. Transgenic Arabidopsis plants heterologously expressing foxtail millet (Setaria italica) NAC1 (SiNAC1) show enhanced transcription of ABA biosynthesis genes, NCED2 and NCED3, resulting in early leaf senescence [51]. Although ABA signaling pathways mediated by WRKY TFs are involved in multiple aspects of plant development including leaf senescence [52], molecular evidence for WRKY TF involvement in ABA biosynthesis is limited. In this study, we found that OsWRKY5 upregulates transcription of ABA biosynthesis genes, OsNCED3, OsNCED4, and OsNCED5 (Figure 7b), suggesting that OsWRKY5 functions in the promotion of leaf senescence by increasing ABA biosynthesis (Figure 7a). Furthermore, based on the involvement of OsWRKY5 in OsNAC2 expression (Figure 6b,d), OsWRKY5 probably activates an OsNAC2-mediated ABA biosynthetic pathway (Figure 8).
Figure 8.
Proposed model for the role of OsWRKY5 in leaf senescence. Arrows indicate activation and bar-ended line represents inhibition. Solid and dashed arrows represent direct and indirect regulation, respectively.
Plants have developed several regulatory mechanisms to restore ABA homeostasis during leaf senescence. For example, in tomato, NAP2 directly regulates expression of genes regulating ABA biosynthesis (NCED1) and ABA degradation (CYP707A2) to establish ABA homeostasis during leaf senescence [53]. ABA-induced OsNAP represses the accumulation of endogenous ABA in rice, indicating that OsNAP participates in a negative feedback mechanism on ABA biosynthesis [25]. Expression of OsNAC2 is differentially regulated by ABA concentration; OsNAC2 expression is upregulated by 20 μM ABA, but inhibited by ABA concentrations over 40 μM [40]. Because the expression of OsWRKY5 is reduced by excessive ABA treatment (Figure 7c), it is highly possible that OsWRKY5 transcription is repressed by excessive concentrations of ABA via a negative feedback regulatory mechanism.
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Experimental Treatments
The Oryza sativa japonica cultivar ‘Dongjin’ (parental line), and the oswrky5-D and oswrky5 mutants were grown in a growth chamber under LD conditions (14 h light at 28 °C/10 h dark at 25 °C) and in a rice paddy field under NLD conditions (≥14 h sunlight/day, 37° N latitude) in Seoul, Republic of Korea. The T-DNA insertion activation-tagged oswrky5-D and knockdown oswrky5 mutants were obtained from the Crop Biotech Institute at Kyung Hee University, Republic of Korea [54,55].
For dark treatment, detached leaves of rice plants grown in the growth chamber for 3 weeks were incubated in 3 mM 2-(N-morpholino)ethanesulfonic (MES) buffer (pH 5.8) with the abaxial side up at 28 °C in complete darkness. To detect OsWRKY5 transcript levels under various hormone treatments, WT seeds were sterilized with 70% ethanol and 2% NaClO, and then germinated and grown on half-strength Murashige and Skoog (0.5X MS, Duchefa, The Netherlands) solid medium under LD conditions for 10 days in a growth chamber. Ten-day-old plants were transferred to 0.5X MS liquid medium containing 50 μM epibrassinolide (BR), 50 μM GA, 50 μM IAA, 50 μM 6-BA, 100 μM SA, 50 μM MeJA, 50 μM ABA, or 50 μM ACC. Total RNA was extracted from leaves harvested at 0 and 6 h of treatment.
4.2. Subcellular Localization
Full-length cDNA of OsWRKY5 was amplified using gene-specific primers (Table S1), cloned into pCR8/GW/TOPO vector (Invitrogen, Carlsbad, CA, USA), and then transferred into pEarleyGate104 (pEG104) gateway binary vector using Gateway LR clonase II enzyme mix (Invitrogen), resulting in a 35S::YFP-OsWRKY5 construct. The pEG104 vector and recombinant constructs were introduced into onion (Allium cepa) epidermal cells using a DNA particle delivery system (Biolistic PDS-1000/He, Bio-Rad, Hercules, CA, USA). After incubation at 25 °C for 16 h, green fluorescence was detected using a confocal laser scanning microscope (SP8X, Leica, Wetzlar, Germany). To visualize nuclei, samples were stained with 10 mL of 1 μg·mL−1 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) dissolved in water for 10 min then viewed using a fluorescence microscope under ultraviolet light irradiation with appropriate filters.
4.3. Determination of Photosynthetic Activity, Total Chlorophyll, and SPAD Value
To evaluate photosynthetic activity, the middle section of the flag leaf of plants grown in a paddy field under NLD conditions was adapted in the dark for 10 min. The Fv/Fm ratio was then measured using an OS-30p+ instrument (Opti-Sciences, Hudson, NH, USA). Total chlorophyll content was measured in rice leaves grown in the growth chamber for 4 weeks. Pigment was extracted from detached leaves incubated in complete darkness using 80% ice-cold acetone. After centrifugation at 10,000× g for 15 min at 10 °C, the absorbance of supernatants was measured at 647 and 663 nm using a UV/VIS spectrophotometer (BioTek Instruments, Winooski, VT, USA). The concentration of chlorophyll was calculated as previously described [56]. The change of SPAD value was determined in the flag leaf of plants grown in a paddy field under NLD conditions using a SPAD-502 instrument (KONICA MINOLTA, Tokyo, Japan).
4.4. RT-qPCR and Stem-Loop RT-qPCR Analysis
Total RNA was extracted from rice tissues using an RNA Extraction kit (MG Med, Seoul, Korea), according to the manufacturer’s instructions. For synthesis of first-strand cDNA, 2 μg of total RNA was used for reverse transcription (RT) in 20 μL volume with oligo(dT)15 primer and M-MLV reverse transcriptase (Promega, Madison, WI, USA). For quantification of miR164b, stem-loop pulsed RT was conducted from 2 μg of total RNA in 20 μL volume using a miR164b-specific stem-loop primer and M-MLV reverse transcriptase (Promega) with the following conditions: 16 °C for 30 min followed by pulsed RT of 40 cycles at 16 °C for 2 min, 42 °C for 1 min, and 50 °C for 1 s, and then inactivation of reverse transcription at 70 °C for 5 min [57]. All RT products were diluted with 80 µL distilled water.
qPCR was performed with gene-specific primers and normalized to UBIQUITIN5 (UBQ5) (Os01g22490) or rice U6 snRNA (Table S1) according to the 2−ΔΔCt method [58]. The 20 µL reaction mixture included 2 µL cDNA from RT or stem-loop pulsed RT, 1 µL 0.5 µM primer, and 10 μL 2X GoTaq master mix (Promega). qPCR amplifications were conducted with a LightCycler 480 (Roche, Basel, Switzerland) using the following program: 94 °C for 2 min followed by 40 cycles of 94 °C for 15 s and 60 °C for 1 min.
4.5. Yeast One-Hybrid Assays
The coding sequence of OsWRKY5 was amplified by PCR. The PCR product was subcloned into the EcoRI and PstI sites of pGAD424 vector (Clontech, Kusatsu, Japan). Fragments of OsNAP and OsNAC2 promoters containing the repetitive W-box core sequence (TGAC) were amplified by PCR and then cloned into pLacZi vector using EcoRI-XbaI, EcoRI-XbaI, SalI-XhoI, SalI-XhoI, SalI-XhoI, SalI-XhoI, and SalI-XhoI sites, generating OsNAP-1::LacZi, OsNAP-2::LacZi, OsNAC2-1::LacZi, OsNAC2-2::LacZi, OsNAC2-3::LacZi, OsNAC2-4::LacZi, and OsNAC2-5::LacZi reporter constructs, respectively (Clontech, USA). These vectors and empty vector were transformed into yeast strain YM4271 using the PEG/LiAc method, and yeast cells were incubated in SD/-His/-Leu liquid medium. β-Galactosidase activity was determined by absorbance of chloramphenicol red, a hydrolysis product of chlorophenol red-β-D-galactopyranoside (CPRG), at 595 nm using a UV/VIS spectrophotometer (BioTek Instruments, Winooski, VT, USA) according to the Yeast Protocol Handbook (Clontech).
4.6. Determination of ABA Content
Four-week-old leaves of WT and oswrky5-D were pulverized in liquid nitrogen and then homogenized in 80% methanol containing 1 mM butylated hydroxytoluene as an antioxidant. Extracts incubated for 12 h at 4 °C were centrifuged at 4000× g for 20 min. The supernatant was passed through a Sep-Pak C18 cartridge (Waters, Milford, MA, USA) as described previously [59], and the eluate was subjected to an enzyme-linked immunosorbent assay (ELISA) using an ABA ELISA kit (MyBioSource, San Diego, CA, USA) according to the manufacturer’s instructions.
5. Conclusions
We investigated a WRKY TF family member in rice, OsWRKY5, which acts as a positive regulator of leaf senescence. OsWRKY5 upregulates both ABA biosynthesis and transcription of CDGs and SAGs during leaf senescence, thereby promoting leaf yellowing. OsWRKY5 transcription contributes to regulating leaf senescence in rice.
Abbreviations
| ABA | abscisic acid |
| DDI | day(s) of dark-induced senescence |
| DAH | day(s) after heading |
| NS | natural senescence |
| LD | long day |
| NLD | natural long day |
| DIS | dark-induced senescence |
| RT-qPCR | reverse transcription-quantitative PCR |
| CDG | chlorophyll degradation gene |
| SAG | senescence-associated gene |
| TF | transcription factor |
| senNAC | senescence-associated NAC gene |
| WT | wild-type |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/18/4437/s1.
Author Contributions
T.K. and K.K. performed experiments and analyzed data. K.K. and N.-C.P. conceived the study, designed and supervised the project. T.K., K.K., and N.-C.P. wrote and edited the manuscript. S.-H.K. assisted in analyzing the data. G.A. developed plant materials and provided advice about the manuscript. All authors read and approved the final manuscript.
Funding
This work was carried out with the support of the Cooperative Research Program for Agricultural Science & Technology Development (PJ013146), Rural Development Administration, South Korea, and the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (NRF-2017R1A2B3003310).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Guo Y., Gan S. Leaf senescence: Signals, execution, and regulation. Curr. Top. Dev. Biol. 2005;71:83–112. doi: 10.1016/S0070-2153(05)71003-6. [DOI] [PubMed] [Google Scholar]
- 2.Lim P.O., Kim H.J., Nam H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 3.Liu L., Zhou Y., Zhou G., Ye R., Zhao L., Li X., Lin Y. Identification of early senescence-associated genes in rice flag leaves. Plant Mol. Biol. 2008;67:37–55. doi: 10.1007/s11103-008-9300-1. [DOI] [PubMed] [Google Scholar]
- 4.Park S.-Y., Yu J.-W., Park J.-S., Li J., Yoo S.-C., Lee N.-Y., Lee S.-K., Jeong S.-W., Seo H.S., Koh H.-J., et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell. 2007;19:1649–1664. doi: 10.1105/tpc.106.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimoda Y., Ito H., Tanaka A. Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell. 2016;28:2147–2160. doi: 10.1105/tpc.16.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren G., An K., Liao Y., Zhou X., Cao Y., Zhao H., Ge X., Kuai B. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 2007;144:1429–1441. doi: 10.1104/pp.107.100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato Y., Morita R., Nishimura M., Yamaguchi H., Kusaba M. Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc. Natl. Acad. Sci. USA. 2007;104:14169–14174. doi: 10.1073/pnas.0705521104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barry C.S., McQuinn R.P., Chung M.-Y., Besuden A., Giovannoni J.J. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008;147:179–187. doi: 10.1104/pp.108.118430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang C., Li C., Li W., Wang Z., Zhou Z., Shen Y., Wu M., Wu Y., Li G., Kong L.A., et al. Concerted evolution of D1 and D2 to regulate chlorophyll degradation in soybean. Plant J. 2014;77:700–712. doi: 10.1111/tpj.12419. [DOI] [PubMed] [Google Scholar]
- 10.Morita R., Sato Y., Masuda Y., Nishimura M., Kusaba M. Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009;59:940–952. doi: 10.1111/j.1365-313X.2009.03919.x. [DOI] [PubMed] [Google Scholar]
- 11.Tang Y., Li M., Chen Y., Wu P., Wu G., Jiang H. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J. Plant Physiol. 2011;168:1952–1959. doi: 10.1016/j.jplph.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 12.Lee R.-H., Wang C.-H., Huang L.-T., Chen S.-C.G. Leaf senescence in rice plants: Cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 2001;52:1117–1121. doi: 10.1093/jexbot/52.358.1117. [DOI] [PubMed] [Google Scholar]
- 13.Häffner E., Konietzki S., Diederichsen E. Keeping Control: The role of senescence and development in plant pathogenesis and defense. Plants. 2015;4:449–488. doi: 10.3390/plants4030449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piao W., Kim E.-Y., Han S.-H., Sakuraba Y., Paek N.-C. Rice phytochrome B (OsPhyB) negatively regulates dark- and starvation-induced leaf senescence. Plants. 2015;4:644–663. doi: 10.3390/plants4030644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivero R.M., Kojima M., Gepstein A., Sakakibara H., Mittler R., Gepstein S., Blumwald E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. USA. 2007;104:19631–19636. doi: 10.1073/pnas.0709453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandler P.M., Robertson M. Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu. Rev. Plant Biol. 1994;45:113–141. doi: 10.1146/annurev.pp.45.060194.000553. [DOI] [Google Scholar]
- 17.Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 18.Hauser F., Li Z., Waadt R., Schroeder J.I. SnapShot: Abscisic acid signaling. Cell. 2017;171:1708.e1. doi: 10.1016/j.cell.2017.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finkelstein R. Abscisic acid synthesis and response. Arabdopsis Book. 2013;11:e0166. doi: 10.1199/tab.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J., Worley E., Udvardi M.A. NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell. 2014;26:4862–4874. doi: 10.1105/tpc.114.133769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao S., Gao J., Zhu X., Song Y., Li Z., Ren G., Zhou X., Kuai B. ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant. 2016;9:1272–1285. doi: 10.1016/j.molp.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Park D.-Y., Shim Y., Gi E., Lee B.-D., An G., Kang K., Paek N.-C. The MYB-related transcription factor RADIALIS-LIKE3 (OsRL3) functions in ABA-induced leaf senescence and salt sensitivity in rice. Environ. Exp. Bot. 2018;156:86–95. doi: 10.1016/j.envexpbot.2018.08.033. [DOI] [Google Scholar]
- 23.Robatzek S., Somssich I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence-and defense-related processes. Plant J. 2001;28:123–133. doi: 10.1046/j.1365-313X.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang K., Gan S.-S. An Abscisic Acid-AtNAP Transcription factor SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol. 2012;158:961–969. doi: 10.1104/pp.111.190876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang C., Wang Y., Zhu Y., Tang J., Hu B., Liu L., Ou S., Wu H., Sun X., Chu J., et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA. 2014;111:10013–10018. doi: 10.1073/pnas.1321568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Guo Y., Cai Z., Gan S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004;27:521–549. doi: 10.1111/j.1365-3040.2003.01158.x. [DOI] [Google Scholar]
- 28.Liu L., Xu W., Hu X., Liu H., Lin Y. W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 2016;6:20881. doi: 10.1038/srep20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robatzek S., Somssich I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao Y., Laun T., Zimmermann P., Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004;55:853–867. doi: 10.1007/s11103-005-2142-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X., Jiang Y., Yu D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells. 2011;31:303–313. doi: 10.1007/s10059-011-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Besseau S., Li J., Palva E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012;63:2667–2679. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L., Xiang S., Chen Y., Li D., Yu D. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant. 2017;10:1174–1189. doi: 10.1016/j.molp.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Guo P., Li Z., Huang P., Li B., Fang S., Chu J., Guo H. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell. 2017;29:2854–2870. doi: 10.1105/tpc.17.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jing S., Zhou X., Song Y., Yu D. Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul. 2009;58:181–190. doi: 10.1007/s10725-009-9366-z. [DOI] [Google Scholar]
- 36.Han M., Kim C.-Y., Lee J., Lee S.-K., Jeon J.-S. OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in Rice. Mol. Cells. 2014;37:532–539. doi: 10.14348/molcells.2014.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 38.Netto A.T., Campostrini E., de Oliveira J.G., Bressan-Smith R.E. Photosynthetic pigments, nitrogen, chlorophyll a fluorescence and SPAD-502 readings in coffee leaves. Sci. Hort. 2005;104:199–209. doi: 10.1016/j.scienta.2004.08.013. [DOI] [Google Scholar]
- 39.Sim C.C., Zaharah A.R., Tan M.S., Goh K.J. Rapid determination of leaf chlorophyll concentration, photosynthetic activity and NK concentration of Elaies guineensis via correlated SPAD-502 chlorophyll index. Asian J. Agric. Res. 2015;9:132–138. [Google Scholar]
- 40.Mao C., Lu S., Lv B., Zhang B., Shen J., He J., Luo L., Xi D., Chen X., Ming F. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017;174:1747–1763. doi: 10.1104/pp.17.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciolkowski I., Wanke D., Birkenbihi R.P., Somssich I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang Y., Xie K., Xiong L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014;65:2119–2135. doi: 10.1093/jxb/eru072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang D., Chen W., Dong J., Li J., Yang F., Wu Z., Zhou H., Wang W., Zhuang C. Overexpression of miR164b-resistant OsNAC2 improves plant architecture and grain yield in rice. J. Exp. Bot. 2018;69:1533–1543. doi: 10.1093/jxb/ery017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H., Zhou C. Signal transduction in leaf senescence. Plant Mol. Biol. 2013;82:539–545. doi: 10.1007/s11103-012-9980-4. [DOI] [PubMed] [Google Scholar]
- 45.Gu L., Dou L., Guo Y., Wang H., Li L., Wang C., Ma L., Wei H., Yu S. The WRKY transcription factor GhWRKY27 coordinates the senescence regulatory pathway in upland cotton (Gossypium hirsutum L.) BMC Plant Biol. 2019;19:116. doi: 10.1186/s12870-019-1688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.-F., Wu S.-H., Swidzinski J., Ishizaki K., et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- 47.Noodén L.D. Abscisic acid, auxin, and other regulators of senescence. In: Noodén L.D., Leopold A.C., editors. Senescence and Aging in Plants. Academic Press; San Diego, CA, USA: 1988. pp. 329–355. [Google Scholar]
- 48.Breeze E., Harrison E., McHattie S., Hughes L., Hickman R., Hill C., Kiddle S., Kim Y.-S., Penfold C.A., Jenkins D., et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 2011;23:873–894. doi: 10.1105/tpc.111.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Graaff E., Schwacke R., Schneider A., Desimone M., Flügge U.-I., Kunze R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006;141:776–792. doi: 10.1104/pp.106.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Y., Guo Y., Liu Y., Zhang F., Wang Z., Wang H., Wang F., Li D., Mao D., Luan S., et al. 9-cis-Epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 2018;9:162. doi: 10.3389/fpls.2018.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren T., Wang J., Zhao M., Gong X., Wang S., Wang G., Zou C. Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in foxtail millet. Planta. 2018;247:53–68. doi: 10.1007/s00425-017-2770-0. [DOI] [PubMed] [Google Scholar]
- 52.Rushton D.L., Tripathi P., Rabara R.C., Lin J., Ringler P., Boken A.K., Langum T.J., Smidt L., Boomsma D.D., Emme N.J., et al. WRKY transcription factors: Key components in abscisic acid signaling. Plant Biotechnol. J. 2012;10:2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- 53.Ma X., Zhang Y., Turečková V., Xue G.-P., Fernie A.R., Mueller-Roeber B., Balazadeh S. The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol. 2018;177:1286–1302. doi: 10.1104/pp.18.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeon J.-S., Lee S., Jung K.-H., Jun S.-H., Jeong D.-H., Lee J., Kim C., Jang S., Yang K., Nam J., et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- 55.Jeong D.-H., An S., Park S., Kang H.-G., Park G.-G., Kim S.-R., Sim J., Kim Y.O., Kim M.K., Kim S.R., et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- 56.Porra R.J. Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta BBA Bioenerg. 1989;975:384–394. [Google Scholar]
- 57.Varkonyi-Gasic E., Wu R., Wood M., Walton E.F., Hellens R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 59.Yang J., Zhang J., Wang Z., Zhu Q., Wang W. Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 2001;127:315–323. doi: 10.1104/pp.127.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.