Abstract
Biodiversity in plant shape is mainly attributable to the diversity of leaf shape, which is largely determined by the transient morphogenetic activity of the leaf margin that creates leaf serrations. However, the precise mechanism underlying the establishment of this morphogenetic capacity remains poorly understood. We report here that INDOLE-3-BUTYRIC ACID RESPONSE 5 (IBR5), a dual-specificity phosphatase, is a key component of leaf-serration regulatory machinery. Loss-of-function mutants of IBR5 exhibited pronounced serrations due to increased cell area. IBR5 was localized in the nucleus of leaf epidermis and petiole cells. Introducing a C129S mutation within the highly conserved VxVHCx2GxSRSx5AYLM motif of IBR5 rendered it unable to rescue the leaf-serration defects of the ibr5-3 mutant. In addition, auxin reporters revealed that the distribution of auxin maxima was expanded ectopically in ibr5-3. Furthermore, we found that the distribution of PIN1 on the plasma membrane of the epidermal and cells around the leaf vein was compromised in ibr5-3. We concluded that IBR5 is essential for the establishment of PIN-FORMED 1 (PIN1)-directed auxin maxima at the tips of leaf serration, which is vital for the elaborated regulation during its formation.
Keywords: Arabidopsis, leaf serrations, IBR5, auxin maxima, PIN1
1. Introduction
Leaf serrations determine the morphology of leaf shape, which mainly contributes to the biodiversity of plant shape [1,2,3]. On this basis, leaves with slight serrations are classified as simple leaf, while leaves with strong dissections are termed compound leaves [4]. Hence, the development of leaf serration is an intriguing process, attracting much attention from many scientists.
Recent molecular studies demonstrated that the formation of leaf serration is a sophisticated process that is fine-tuned by various environmental and developmental factors, such as light, phytohormone, and temperature [3,5,6,7,8]. Among these factors, auxin is critical in determining the morphology of leaf serrations [9]. As an essential hormone, auxin is involved in almost every biological activity, ranging from gravitropism [10] to the abiotic stress response [11], and from cell expansion [12] to the formation of leaf margin [9,13]. The morphogenesis of leaf serrations depends on the guidance of auxin maxima at their tips [9], and that is regulated by many factors, such as four auxin biosynthesis proteins, YUCCA1/2/4/6 (YUC1/2/4/6), Indole-3-Acetic Acid Carboxyl Methyltransferase (IAMT1), auxin influx carrier AUXIN-RESISTANT 1 (AUX1), and auxin efflux protein PIN-FORMED 1 (PIN1) [14,15,16,17,18,19,20,21].
In addition to the reported factors in auxin signaling pathways, CUP-SHAPED COTYLEDON2 (CUC2) plays vital roles in the generation of auxin maxima through promotion of the polar distribution of PIN1 [13]. Conversely, auxin also represses the expression of CUC2 during the morphogenesis of leaf serrations [9,13]. Moreover, auxin and ASYMMETRIC LEAVES1 (AS1) signaling pathways converge to suppress the expression of the KNOTTED1-like homeobox (KNOX) gene BREVIPEDICELLUS (BP), which plays a key role in generating the auxin maxima at the tips of leaf serrations [14]. Despite the known importance of auxin maxima in determining the formation of leaf serrations, the factors affecting the establishment of auxin maxima in this process remain mostly unknown.
Mitogen-activated protein kinases (MAPKs) form a highly conserved family of proteins that are dephosphorylated by MAPKs phosphatases (MKPs) in a large number of physiological processes [22]. IBR5 encodes one of the five dual-specificity phosphatases (MKP1, MKP2, DUAL-SPECIFICITY PROTEIN TYROSINE PHOSPHATASE 1 (DsPTP1), PROPYZAMIDE HYPERSENSITIVE 1 (PHS1), and INDOLE-3-BUTYRIC ACID RESPONSE 5 (IBR5) and that all of them contain a highly conserved dual-specificity motif VxVHCx2GxSRSx5AYLM in Arabidopsis [22,23]. MKP2 and DsPTP1 have been reported to possess phosphatase activity against MPK3/6 and MPK4, respectively [24,25]. Dephosphorylation of MPK12 by IBR5 was confirmed in auxin signaling pathways in the root [26]. The expression of IBR5 has been detected in almost all tissues throughout developmental stages, including roots, petals, sepals, anther filaments, carpels of flowers, green siliques, and leaf serrations [23], and loss-of-function mutants of IBR5 displayed pleiotropic phenotypes, such as longer roots, shorter hypocotyls, smaller petals, and pronounced serrations [23,26,27]. However, the molecular mechanism of IBR5-mediated leaf serration formation remains unknown.
In this study, we report the functional analysis of IBR5 in the development of leaf serrations. IBR5 was constitutively expressed and localized in the nucleus of leaf epidermal cells. Loss-of-function mutants of IBR5 exhibited pronounced leaf serrations due to enhanced cell expansion but not normal cell proliferation. Moreover, IBR5 was necessary for the establishment of auxin maxima that is restricted at the tips of leaf serrations. The abundance of PIN1 on the plasma membrane of the epidermal cells and cells around the leaf vein were compromised in ibr5-3. Also, IBR5C129S lost the ability to rescue the defective leaf serrations of ibr5-3. Our results demonstrated that IBR5 is a critical factor in the regulation of leaf serration development by altering auxin maxima distribution.
2. Results
2.1. Identification and Phenotypic Characterization of IBR5 Loss-of-Function Mutants
The development of leaf serrations is an intriguing process, which has triggered our interests. Using a reverse genetic approach to understand the molecular mechanism underlying the development of leaf serrations, we analyzed two T-DNA insertion mutants exhibiting defective leaf serration development. We found that the T-DNA fragments of the two mutants were inserted into the sequence of the IBR5 locus. In agreement with a previous report [23,26,27], one T-DNA fragment was inserted at the 5′ untranslated region at 161 bp upstream of the ATG and the other was inserted in the and second exon of the IBR5 genomic sequence. These two mutants were termed ibr5-2 (SALK_032185) and ibr5-3 (SALK_039359C), respectively (Figure 1A) [23,26,27]. The upstream and downstream sequences of the inserted sites were intact in these two mutants as revealed by PCR analysis. To verify whether the T-DNA insertion disrupted the expression of IBR5, we conducted qRT-PCR analysis using total RNAs isolated from the leaves of WT and ibr5 mutants with three paired primers designed to amplify three different regions of the transcript. Decreased expression of IBR5 was detected in ibr5-2, which suggested this mutant was a weak allele with the full-length transcript at a reduced level (Supplementary Figure S1A–C). However, only the portion of the transcript that was upstream of the T-DNA insertion site could be detected in ibr5-3, implying that this mutant produced a truncated IBR5 with the first 100 amino acids. Hence, ibr5-3 was used for further analysis unless stated otherwise.
Figure 1.
ibr5-3 mutants showing defective leaf shape. (A) The schematic illustration of the T-DNA insertion mutants for IBR5 used in this study [23,26,27]. (B) Phenotypic characterization of WT, ibr5-2 and ibr5-3. The white arrows indicate the leaf serrations. Bar, 1 cm. (C) The length analysis of the third leaf in WT, ibr5-2, and ibr5-3. Error bars indicate ±SD; 12 leaves from 12 individual plants were analyzed. (D) The width analysis of the third leaf in WT, ibr5-2, and ibr5-3. Error bars are ±SD; 12 leaves from 12 individual plants were analyzed. (E) The maximum height analysis of the most proximal serrations of the third leaf in WT, ibr5-2, and ibr5-3. Error bars are ± SD; 11 leaves from 11 individual plants were analyzed. Two asterisks mean significant differences (p < 0.01 from Student’s t-test). (F) The maximum width analysis of the most proximal serrations of the third leaf in WT, ibr5-2, and ibr5-3. Error bars are ±SD; 11 leaves from 11 individual plants were analyzed. Two asterisks mean significant differences (p < 0.01 from Student’s t-test).
Homozygous ibr5 mutants showed developmental defects in leaf serrations as compared to the wild type (WT) when grown in soil (Figure 1B and Supplementary Figure S2), but the length and width of the first to fourth leaves were normal in these mutants (Figure 1C,D). However, the shape of the leaf margin in ibr5 mutants was extremely irregular due to the increased height and width of the leaf serrations (Figure 1E,F and Supplementary Figure S3A–F). To investigate the underlying reasons for these defects, the cell size and number were monitored in the most proximal leaf serrations of the third leaves in WT and ibr5-3. We observed a larger cell size and normal cell number in ibr5-3 (Supplementary Figure S4A–D). Here, we conclude that the pronounced serrations of ibr5-3 were due to a larger cell size, which suggested that IBR5 is involved in cell expansion during the formation of leaf serrations.
2.2. IBR5 but Not IBR5C129S Rescues the Leaf-Serration Defects of ibr5-3
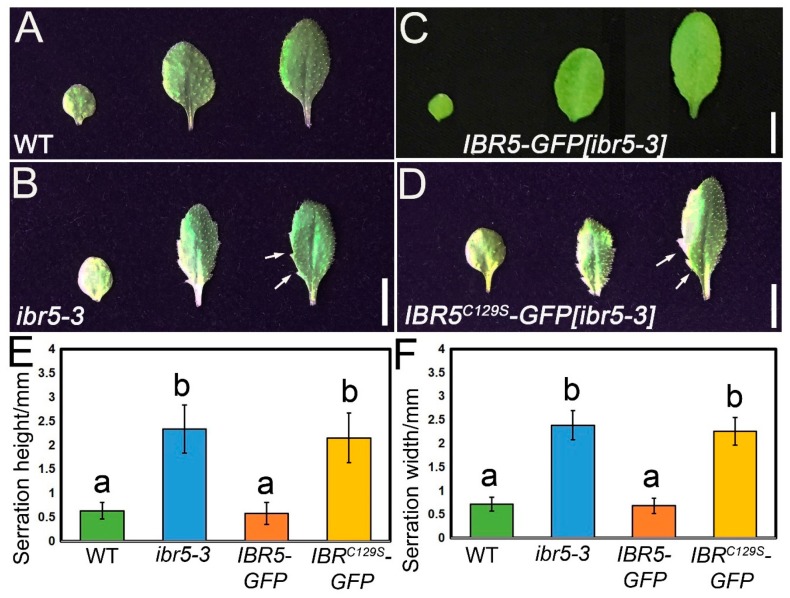
Previous studies showed that ibr5 mutants exhibit pleiotropic defects, such as slight dwarfing, fewer lateral roots, and smaller petals [23,28,29]. Here, we showed that a pronounced leaf serration phenotype is associated with this mutant (Figure 1B,E,F). IBR5 was expressed in almost all tissues analyzed by microarrays [30]. To further validate its predicted expression pattern, we introduced an IBR5 genomic fragment fused with an in-frame GFP (IBR5-GFP) into the ibr5-3 background. The IBR5 genomic fragment fully rescued the defects of leaf serrations in ibr5-3 (Figure 2A–C,E,F), demonstrating that GFP translational fusion did not affect the function of IBR5. Therefore, complementary lines could be employed to study IBR5 subcellular localization in different tissues.
Figure 2.
The functionality of IBR5 relies on its dual-specificity phosphatase activity. (A) Representative images of the first, second, and third leaves in the WT. Bar, 5 mm. (B) Representative images of the first, second, and third leaves in the WT. Bar, 5 mm. (C) Representative images of the first, second, and third leaves in IBR5pro::IBR5 genomic fragment–GFP rescued lines. Bar, 5 mm. (D) Representative images of the first, second, and third leaves in IBR5pro::IBR5 genomic fragment C126S–GFP rescued lines. Bar, 5 mm. (E) The maximum height analysis of the most proximal serrations of the third leaf in WT, ibr5-3, and rescued lines. Error bars are ± SD, with 13 leaves analyzed. Different characters mean significant differences (p < 0.01 from Student’s t-test). (F) The maximum width analysis of the most proximal serrations of the third leaf in the WT and ibr5-3. Error bars are ± SD, with 11 leaves analyzed. Different characters mean significant differences (p < 0.01 from Student’s t-test).
Given that IBR5 is a dual-specificity phosphatase, we wonder whether its regulation on the development of leaf serrations relied on its catalytic activity. Hence, we introduced a point mutation into the sequence of IBR5 and in which the conserved VxVHCx2GxSRSx5AYLM motif was changed to VxVHSx2GxSRSx5AYLM (IBR5C129S). This mutation has been shown to abolish the ability of IBR5 to trigger the dephosphorylation process with a nucleophilic attack on the phosphorus atom of the phosphotyrosine or phosphothreonine substrate [25,31,32]. We introduced an IBR5C129S–GFP fusion fragment into ibr5-3 and found that the exogenous IBR5C129S–GFP failed to compensate for the defects of its leaf serrations (Figure 2A,B,D–F), suggesting that IBR5-mediated development of leaf serrations is dependent on its phosphatase activity.
2.3. IBR5 Is Expressed and Localized in the Nucleus of Cells Forming the Leaf
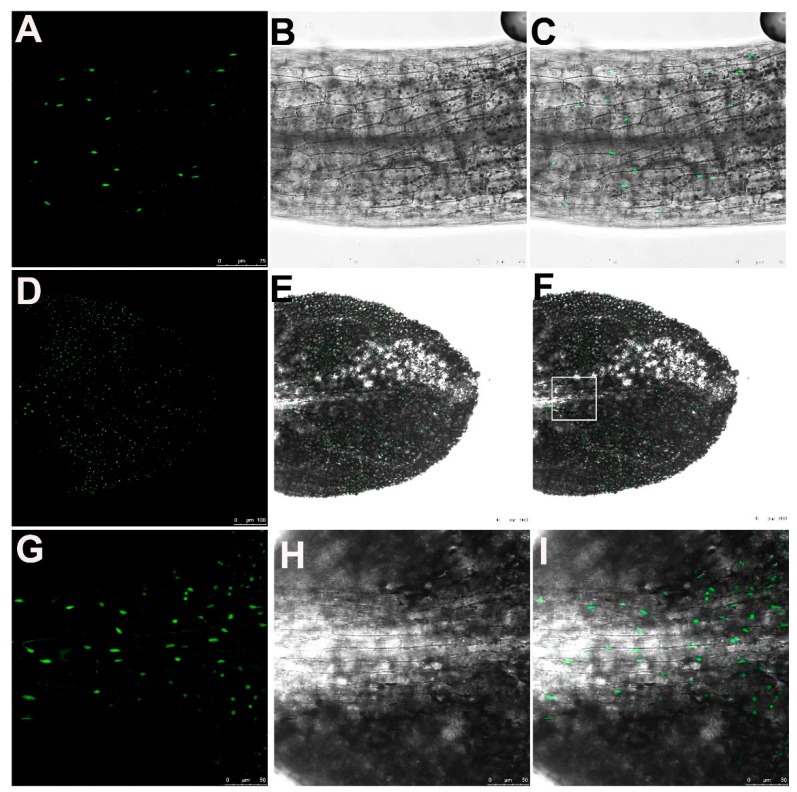
To better understand the roles of IBR5 in the development of leaf serrations, it is necessary to define its subcellular localization. Recent studies conducted by transient expression in tobacco leaf epidermal cells showed that IBR5 generated two spliced transcripts IBR5.1 and IBR5.3, which would be translated into two products localized in both the nucleus and cytoplasm or only the nucleus, respectively [27]. As a fast and convenient approach, transient heterozygous expression could not always reveal the real subcellular localization, especially in some tissues, such as petiole and leaf vascular bundles. Therefore, we used stable complementary Arabidopsis lines in the ibr5 mutant background (IBR5–GFP). Confocal imaging showed that clear and intense GFP signals were accumulated in the nucleus of the petiole and cells around the leaf vein (Figure 3). Furthermore, only IBR5.1–GFP was detected in the leaves of fully rescued IBR5–GFP plants (Supplementary Figure S5), which indicates that IBR5.1 but not IBR5.3 functions in leaf serration development.
Figure 3.
Expression pattern and subcellular localization of IBR5. (A–C) Representative images of IBR5 expressed and localized in the nucleus of petiole cells. Bars, 75 µm. (D–F) Representative images of IBR5 expressed and localized in the nucleus of epidermal cells of 2-day-old leaves. Bars, 100 µm. (G–I) Representative images of IBR5 expressed and localized in the nucleus in the boxed region in the figure (F). Bars, 100 µm.
2.4. Auxin Distribution Is Perturbed in ibr5-3
Next, we attempted to investigate the roles of auxin in the development of leaf serrations. Auxin maxima were demonstrated to act as a critical factor for determining the size of leaf serrations [9,13]. Given the enhanced cell expansion in ibr5 mutants (Supplementary Figure S4A–C), we hypothesized that auxin maxima were perturbed in these mutants.
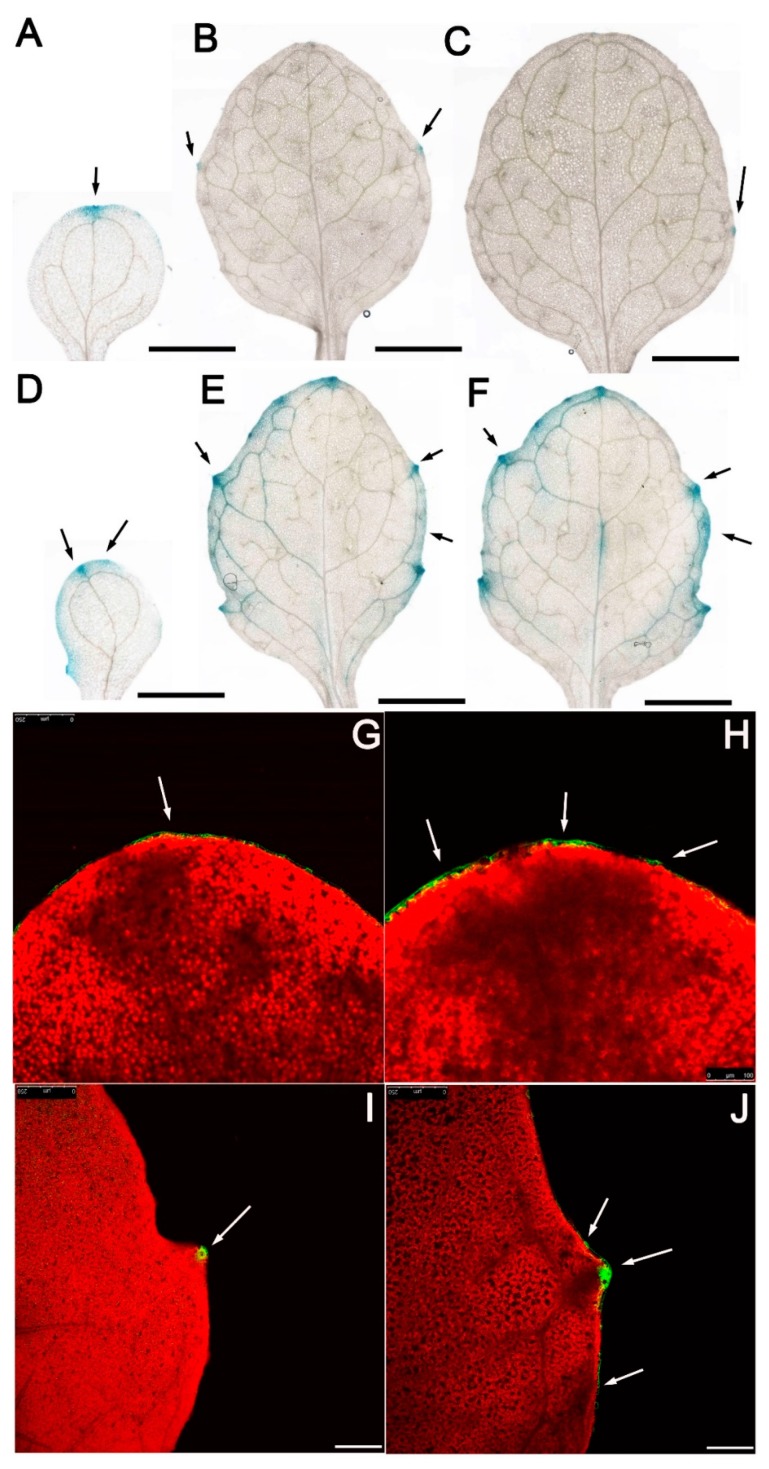
To determine if auxin distribution was perturbed in ibr5-3, we crossed an auxin reporter line DR5:GUS [33] into ibr5-3 and analyzed the GUS signals in vivo. The punctuated GUS signals were spatially restricted at the tips of 10-day-old leaf serrations and leaf apex in the WT (Supplementary Figure S6), consistent with previous work [9]. By contrast, strong and ectopic GUS signals expanded from the leaf apex and the tips of leaf serrations to nearly the whole leaf margin in ibr5-3 (Supplementary Figure S6), demonstrating that enhanced auxin response and perturbed auxin maxima existed in ibr5 mutants. To obtain high-quality images for the distributed sites of GUS signals, we analyzed the first, third, and fourth leaves by light transmission microscopy. We found that GUS signals were not only distributed at the leaf apex and the tips of leaf serrations in ibr5-3 (Figure 4A–C) but expanded to most if not all the sinus and covered the whole leaf margin in these three leaves (Figure 4E,F). To confirm these results, we then introduced another auxin reporter line DR5-GFP into ibr5-3 and WT plants. Similarly, the third leaves of WT and ibr5-3 were selected as targets for further analysis. Confocal imaging showed that GFP signals were spatially restricted and accumulated at the leaf apex and the tips of leaf serrations (Figure 4G,I), demonstrating that the auxin maxima were strictly restricted to the tips of the leaf serrations and leaf apex. While, more GFP signals were accumulated and ectopically expanded from the leaf apex and tips of leaf serrations to the adjacent regions in ibr5-3 (Figure 4H,J), a similar result that was also observed in DR5:GUS lines. Since auxin maxima at the tips of leaf serrations are responsible for determining the formation of leaf serrations [34], the pronounced serrations observed in ibr5-3 can be explained reasonably by the enhanced auxin response and ectopic auxin maxima (Figure 4D–F). Collectively, we propose that the pronounced serrations were due to the ectopic auxin maxima and more active auxin response in ibr5-3.
Figure 4.
Auxin accumulation monitored by GUS staining and confocal microscopy in ibr5-3. (A–C) Representative images of GUS staining for 10-day-old DR5-GUS transgenic plants in WT. Bars, 5 mm. From left to right: cotyledon, the third true leaf, and the fifth true leaf. The black arrows indicate the auxin maxima sites. (D–F) The representative images of GUS staining for 10-day-old DR5-GUS transgenic plants in ibr5-3. Bars, 5 mm. From left to right: cotyledon, the third true leaf, and the fifth true leaf. The black arrows indicate the auxin maxima sites. (G,H) Representative images of confocal imaging for the third leaf of 10-day-old DR5-GFP transgenic plants in WT. Bars, 100 µm. The white arrows indicate the auxin maxima sites. (I,J) Representative images of confocal imaging for the third leaf of 10-day-old DR5-GFP transgenic plants in ibr5-3. Bars, 100 µm. The white arrows indicate the auxin maxima sites.
2.5. The Distribution of PIN1 Is Compromised in ibr5-3
We have shown that the auxin response was enhanced in the leaf apex and the tips of leaf serrations. We next investigated why an enhanced auxin response was characterized in ibr5 mutants. Leaves serve as the source to produce auxin that is necessary for the development of other organs [35,36]. Firstly, we determined the transcriptional level of auxin biosynthetic genes expressed in leaves, YUC1, YUC2, YUC4, and YUC6 [19,20,28], and our qRT-PCR results showed that the expression levels of these four genes were not affected in the ibr5 mutant (Supplementary Figure S7). These results suggested that the process of auxin biosynthesis was normal in ibr5-3.
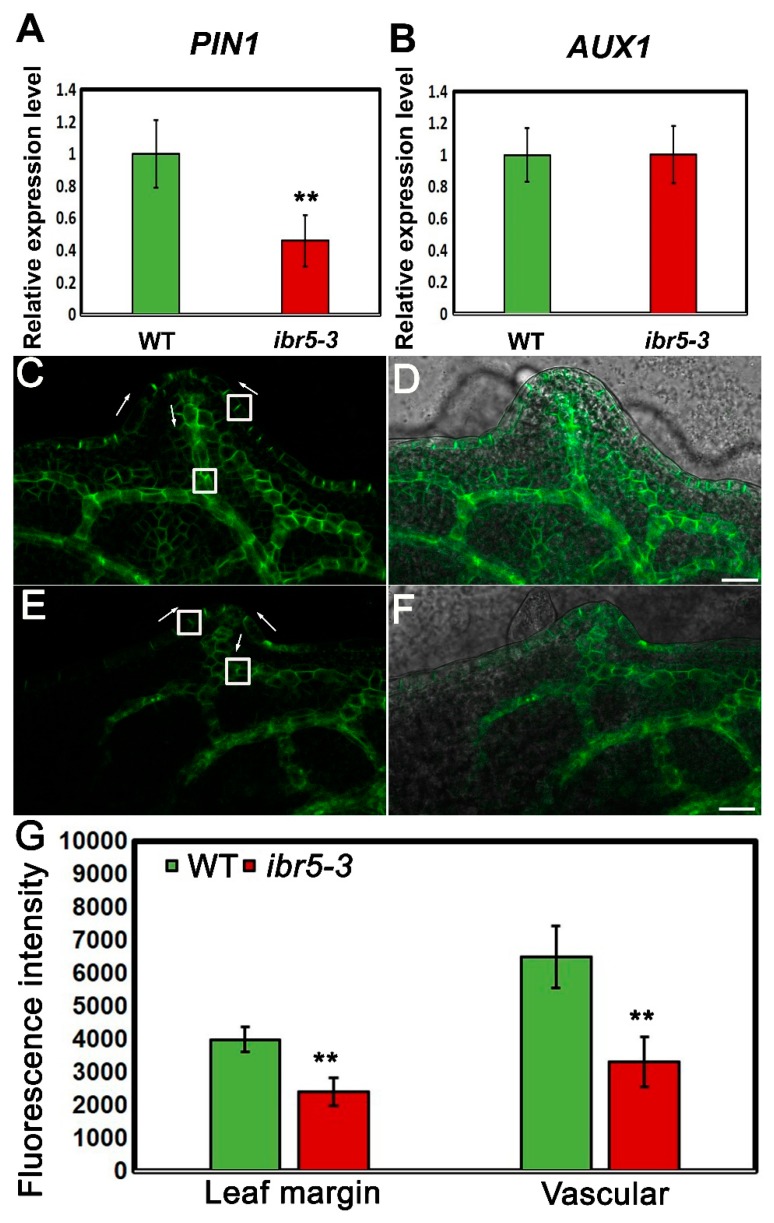
PIN1 is an auxin efflux carrier and primarily responsible for the polar transport of auxin, and its convergence points mark the auxin maxima guiding the formation of leaf serrations [4,14,17,34]. We wondered whether the expression and localization of PIN1 were affected in ibr5-3. qRT-PCR results showed that the transcriptional level of PIN1 but not AUX1 was decreased in ibr5-3 (Figure 5A,B), implying the abundance of PIN1 was related to IBR5. We then employed a PIN1-GFP transgenic line [36] to mark the localization and accumulation of PIN1 and crossed it into ibr5-3. GFP signals were distributed asymmetrically on the plasma membrane of epidermal and cells around the leaf vein of leaf serrations in the WT background (Figure 5C,D), which indicated the auxin transport pathway (i.e., from the sinus to the tip of serrations and finally into vascular tissue) as reported [9]. GFP signals were also found to be localized on the plasma membrane of cells around the leaf vein and epidermal cells in ibr5-3 (Figure 5E,F), demonstrating that the polar localization of PIN1 was not affected and the direction of auxin transport was normal in the absence of IBR5. However, the intensity of the GFP signal was much weaker on the plasma membrane of both epidermal cells and cells around the leaf vein in ibr5-3 versus WT (Figure 5E), indicating that the abundance of PIN1 on the plasma membrane was compromised in ibr5-3. Given that PIN1 acts as an auxin efflux carrier, the reduced abundance of PIN1 on the plasma membrane likely led to the reduced rate of auxin transport. Hence, the transport pathway of auxin produced in the leaf was likely to be partially limited, which may cause an enhanced auxin response and auxin maxima expanded from the tips of leaf serration to the adjacent regions in ibr5-3.
Figure 5.
The distribution of PIN1 in the leaf serrations of WT and ibr5-3. (A) The relative expression level of PIN1 in WT and ibr5-3. Error bars are ± SE, n = 3 independent replicates and each with 3 biological replicates analyzed in each assay. (B) The relative expression level of AUX1 in WT and ibr5-3. Error bars are ± SE, n = 3 independent replicates and each with 3 biological replicates analyzed in each assay. (C,D) The representative image of PIN1-GFP in the 10-day-old leaf serrations of WT. Bar, 20 µm. The white boxed regions were used for analysis. The white arrows indicate the direction of auxin transport. (E,F) The representative image of PIN1-GFP in the 10-day-old leaf serrations of ibr5-3. Bar, 20 µm. The white boxed regions were used for analysis. The white arrows indicate the direction of auxin transport. (G) The quantified analysis of the fluorescence intensity of PIN1-GFP in 10-day-old leaf serrations in WT and ibr5-3. Error bars are ± SE, n = 3 independent replicates and each with 12 leaves analyzed in each assay. Two asterisks mean significant differences (p < 0.01 from Student’s t-test).
Here, we conclude that IBR5 is a critical factor in the formation of leaf serrations through modulation of the abundance of PIN1 on the plasma membrane of leaf epidermal and cells around the leaf vein, which is responsible for transporting auxin produced by leaves to other organs. The blockage of this pathway may lead to an enhanced auxin response in the leaf margins and pronounced leaf serrations.
3. Discussion
Plant leaves are essential for light harvest, gas exchange, starch accumulation, and phytohormone production [1,4,36,37,38]. These processes are significantly influenced by the morphology of leaf shape, which is primarily determined by the size of leaf serrations [13]. Studies on the regulatory mechanism underlying the development of leaf serrations have revealed a role for the auxin signaling network in this process [9,13]. Moreover, the expression of IBR5 was induced by external auxin [29]. Here we demonstrated that IBR5 is a key regulator in the development of leaf serrations through modulation of the accumulation of PIN1 on the plasma membrane of the epidermal and cells around the leaf vein. PIN1 is critical for regulating the rate of auxin transport and restricting the auxin maxima to the tips of leaf serrations (Figure 6). IBR5 was highly expressed in almost every tissue, and consequently, its mutants exhibited pleiotropic defects, such as slight dwarfism, smaller petals, and defective leaf vasculature patterns [23,27,28]. Here, we showed that an enhanced auxin response was characterized in leaf serrations of the ibr5 mutant (Figure 4). Our studies systematically investigated the molecular mechanisms of IBR5 in the development of leaf serrations. Moreover, we provided mutually verifiable evidence to support our conclusion that excessive and ectopic auxin distribution exists in the leaf serrations of ibr5-3 mutants.
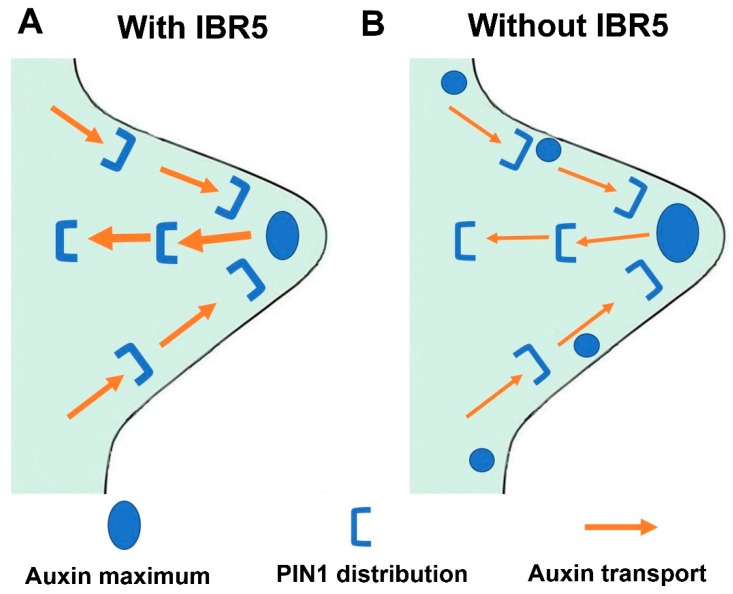
Figure 6.
The schematic model for this work. (A) With IBR5, auxin maxima were restricted at the tips of leaf serrations. (B) Without IBR5, the distribution of PIN1 on the plasma membrane of the epidermal and cells around the leaf vein was compromised, the rate of auxin transport was reduced, and auxin maxima were enlarged and expended from the tip to the adjacent regions.
Auxin is an important hormone that plays a critical role in determining the shape of leaf serrations [9,13]. ibr5 mutants exhibited defective leaf shapes due to pronounced leaf serrations (Figure 1B), which was similar to the loss-of-function mutants of AS1 and AUXIN RESISTANT1 (AXR1) caused by the abnormal distribution of auxin maxima [14]. We initially speculated that this phenotype was caused by perturbed auxin maxima as implied by the previous study [9]. Indeed, auxin maxima were expanded ectopically in the tips of leaf serrations in ibr5-3 (Figure 4B) due to blocked auxin transport caused by the compromised PIN1 abundance on the plasma membrane of the epidermal and cells around the leaf vein of leaf serrations (Figure 4 and Figure 5). This demonstrated once again that PIN1 is a critical factor, and its expression level is of paramount importance in regulating the development of leaf serrations [13]. IBR5 and PIN1 are two differently localized proteins in the nucleus and plasma membrane, respectively (Figure 2 and Figure 5). However, the direct link between them remains elusive. Combined with the decreased expression level of PIN1 at the transcriptional and translational level in ibr5-3 (Figure 5), we propose that IBR5 could act upstream of PIN1 to activate its expression in leaf serrations via dephosphorylating and activating another transcription factor. Moreover, the phosphorylation of PIN1 is essential for its polar distribution, which determines the auxin transport direction and patterns [39,40]. Hence, this suggests another possible link between IBR5 and PIN1. Because the functionality of IBR5 was achieved by modulating the auxin transport in a PIN1-dependent manner, further efforts should be dedicated to investigating the direct link between these two components.
Previously reported results have demonstrated that the catalytic activity of IBR5 was critical for its function in petal size and root development [26,27]. These results were also supported by our findings that IBR5-GFPC129S failed to rescue the defective leaf serrations of ibr5-3 (Figure 2). Therefore, the functionality of IBR5 was conferred by its dual-specificity phosphatase activity in the development of leaf serrations, similar to its roles in roots [26]. Hence, the substrate of IBR5 is necessary for a better understanding of its roles in many developmental processes. For instance, MPK12 was identified as one substrate that was efficiently dephosphorylated and inactivated by IBR5, and acted as a negative regulator of auxin signaling in root development [26]. Nevertheless, MPK12 mutants showed normal leaf serrations as reported [41], which suggested that MPK12 plays no role in IBR5-mediated leaf-serration development. Because of the absence of the consensus sequence recognized by IBR5, it is impossible to evaluate more likely substrates via in silico prediction. Based on the subcellular localization, as well as the phenotypes of IBR5 loss-of-function mutants, we considered that some transcription factors expressed in leaf serrations may be the putative substrates of IBR5, such as Arabidopsis CUC2 and CUC3 [9,13]. Overexpression of CUC2 and CUC3 exhibited pronounced and dissected leaf serrations [13], similar to those of ibr5-3. CUC2 acts earlier and possibly through limiting growth of the sinus and/or promoting growth of the teeth, whereas CUC3 appears to act later to sustain teeth growth [42]. Moreover, CUC2 activity is required for building up discrete maxima of auxin via a modification of auxin transport [43]. Hence, CUC2 is likely to be a substrate of IBR5 in leaf serration development. Auxin response factors (ARFs) were provided to be phosphorylated and dephosphorylated, and also involved in cell expansion [44,45], which suggests that they may act as substrates of IBR5 in leaf development.
In summary, IBR5 is critical for the development of leaf serrations as phosphatase enzymatic activity is required. Moreover, IBR5 acted through regulating the distribution of auxin maxima at the tips of leaf serrations via modulation of the abundance of PIN1, an observation that provides further support for the importance of auxin dynamics in this process [13,26]. Collectively, this work sheds light on the roles of IBR5 in the development of leaf serrations, which is intriguing and deserves to be studied in the future.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The T-DNA insertion lines, SALK_032185 (ibr5-2) and SALK_039359C (ibr5-3), were obtained from the European Arabidopsis Stock Center (NASC, http://arabidopsis.info). The upstream and downstream sequences of the inserted sites of ibr5-2 and ibr5-3 were amplified via ZP2/ZP12 and ZP3/ZP4, respectively. Then, the PCR product was sequenced and aligned with the sequence of the IBR5 genomic sequence. The primers are listed in Supplementary Table S1.
All the experiments were conducted in Col-0 background. The plants were grown in a 4:1:1 mix of Fafard 4P:perlite:vermiculite under an 18 h lightness/6 h darkness cycle at 22 °C in the greenhouse of Shanghai Jiao Tong University. For the seedlings’ culture on plates, surface-sterilized Arabidopsis seeds were plated on Murashige and Skoog basal medium with vitamins (MS) (Phytotechlab, http://www.phytotechlab.com/). All plates were kept at 4 °C in darkness for 3 days before being transferred to the growth chamber with a 16 h lightness/8 h darkness cycle at 22 °C. Stable Arabidopsis transformations were done using the floral-dip method as described [46].
4.2. PCR, RT-PCR, qRT-PCR and Vectors Construct
Total leaf RNAs was isolated using TIANGEN RNAprep pure Plant Kit (DP432, http://www.tiangen.com/) according to the manufacturer’s instructions. In total, 1 μg RNA was used to synthesize the first-strand cDNA using the Rever Tra Ace-a-First strand cDNA synthesis kit (TOYOBO, http://www.toyobo.cn/). Primers used for qRT-PCR are as follows: ZP1/ZP2 for the F1/R1 of IBR5; ZP3/ZP4 for the F2/R2 of IBR5, ZP5/ZP6 for the F3/R3 of IBR5, ZP206/ZP207 for YUC1, ZP208/ZP209 for YUC2, ZP210/ZP211 for YUC4, ZP212/ZP213 for YUC6, ZP214/ZP215 for PIN1, ZP216/ZP217 for AUX1, ZP200/ZP210 for GAPDH, ZP202/ZP203 for ACTIN2, and ZP204/ZP205 for TUB2. Primers for RT-PCR are as follows: ZP5/ZP218 for the F3/GFPR of IBR5-GFP and ZP204/ZP205 for TUB2. Detailed information for these primers is listed in Supplementary Table S1.
The qRT-PCR was performed with the Bio-Rad CFX96™ Real-Time System (Bio-Rad, CA, USA) using SYBR® Green Real-time PCR master mix (TOYOBO CO., LTD, Japan). Each 40-µL reaction system contained 20 µL SYBR® Green Realtime PCR master mix, 2 µL cDNA, and 2 µL primers (10 µM). The program was as follows: 95 °C for 1 min and 95 °C for 15 s, 58 °C for 15 s, and 72 °C for 15 s repeated for 46 cycles. Fluorescence data were collected during the 72-°C step and analyzed with the BioRad CFX Manager (Bio-Rad, USA). Total RNA extraction was extracted from the third leaves of 10-day-old plants and with three independent biological replicates. Quantification of relative expression values was performed, and the reference genes were selected as described [47,48].
The vectors mentioned in this work were modified from pCAMBIA1301. The functional IBR5-GFP reporter vector driven by its own promotor was constructed using In-Fusion technology (http://www.clontech.com) to insert the IBR5 genomic fragment without a stop codon into pCAMBIA1301. The IBR5C129S-GFP reporter vector was generated by replacing the 429-bp sequence with a manually synthesized sequence including the point mutation. The pointed sequence was inserted into sequence of IBR5-GFP through two restriction enzyme sites: XbaI and PstI. The point mutated sequence was synthesized in Sangon Biotech Company (Shanghai, www.sangon.com/). The vectors of IBR5-GFP and IBR5C129S-GFP were transformed into Arabidopsis via the floral dipping method. For these vectors, more than 15 independent transgenic lines (stable F3 homozygous lines) were obtained and analyzed for each, and only one representative line was exhibited: ZP11/ZP12 for IBR-GFP. All primers used in this study are listed in Supplemental Table S1.
4.3. Leaf Serration Height and Width, Cell Size, and Cell Number Analysis
Leaf serration height and width analysis was referred to the reported study [9]. The third leaf of 10-day-old plants was fixed in 2.5% glutaraldehyde for 20 min within vacuum. The samples of adaxial cell length measurement were selected from the basal region of the first most proximal serration except the epidermis of the leaf. Images were acquired by the light microscope (E200, Nikon, Japan) and analyzed by the ImageJ (http://rsb.info.nih.gov/ij/).
4.4. GUS Staining and Vascular Pattern Analysis
The 10-day-old plants of ibr5-3 and WT were immersed into GUS solution (50 mM Na3PO4 (pH 7.0), 50 mM NaH2PO4, 10 mg/mL X-Gluc and 0.02% (v/v) TritonX-100, 10 mM Na2EDTA, 0.5 mM K3[Fe(CN)6], 0.5 mM K4[Fe(CN)6]) under dark at 37 °C for 12 h. After that, the samples (stable T3 homozygous lines) were washed with 70% ethanol for 36 h twice until the leaves become transparent. The GUS staining images were taken via a Leica light microscope (M205A) with a CCD camera.
4.5. Fluorescence Observation and Analysis
The leaves from10-day-old IBR5-GFP transgenic plants (stable T3 homozygous lines) were captured by epifluorescence using an inverted laser scanning confocal microscope (SP5, Leica, German). DR5-GFP images were captured by an inverted laser scanning confocal microscope (LSM780, Zeiss, German). Next, 15% of Argon laser power was used to excite the sample, the pinhole was set as 86 µm, and the detector ranged from the 505- to the 550-nm band pass filter for GFP and from the 600- to the 650-nm band pass filter for chlorophyll fluorescence, which were placed in front of PMT2. The fluorescence intensity was analyzed via ImageJ (http://rsb.info.nih.gov/ij/) as the subsequent steps: 1) Open the image and draw the regions of interest (ROI) (boxed region containing the PIN1 fluorescence of plasma membrane) via the “rectangle tool”. 2) Select “Analyze–Tools-ROI Manager” and open a dialog box. 3) Select “Analyze–Set Measurements” to select the “Area, Min and max gray value, Integrated density and Mean gray value” to measure. 4) Click on “Measure” in the ROI Manager window. The detailed results were exhibited in a results window and could be saved as Excel files.
Acknowledgments
We thank the NASC for kindly providing the plant materials described in this article. We thank Hongwei Xue for providing DR5-GUS, DR5-GFP, and PIN1-GFP transgenic seeds.
Abbreviations
| AS1 | ASYMMETRIC LEAVES1 |
| ARFs | auxin response factors |
| AUX1 | auxin influx carrier |
| AXR1 | AUXIN RESISTANT1 |
| BP | BREVIPEDICELLUS |
| CUC2 | CUP-SHAPED COTYLEDON2 |
| DsPTP1 | DUAL-SPECIFICITY PROTEIN TYROSINE PHOSPHATASE 1 |
| IAMT1 | Indole-3-Acetic Acid Carboxyl Methyltransferase |
| IBR5 | INDOLE-3-BUTYRIC ACID RESPONSE 5 |
| KNOX | KNOTTED1-like homeobox |
| MAPKs | Mitogen-activated protein kinases |
| MKPs | MAPKs phosphatases |
| PHS1 | PROPYZAMIDE HYPERSENSITIVE 1 |
| PIN1 | PIN-FORMED 1 |
| WT | wild-type |
| YUC1 | YUCCA1 |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/18/4429/s1.
Author Contributions
Conceptualization, X.K. and K.Z.; Data curation, G.H. and C.Z.; Formal analysis, Y.X.; Funding acquisition, K.Z.; Investigation, X.K., G.H. and C.Z.; Project administration, X.K.; Software, G.H.; Supervision, K.Z.; Validation, X.S.; Visualization, Y.X. and J.W.; Writing—original draft, X.K., G.H. and K.Z.; Writing—review and editing, X.K. and J.G.
Funding
This work was supported by the China Transgenic Program (2016zx08005-003-003).
Conflicts of Interest
The authors declare no competing financial or non-financial interests.
References
- 1.Tsukaya H. Leaf development. Arab. Book. 2013;11:e0163. doi: 10.1199/tab.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar M., Ori N. Compound leaf development in model plant species. Curr. Opin. Plant Biol. 2015;23:61–69. doi: 10.1016/j.pbi.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Bar M., Ori N. Leaf development and morphogenesis. Development. 2014;141:4219–4230. doi: 10.1242/dev.106195. [DOI] [PubMed] [Google Scholar]
- 4.Tsukaya H. Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 2006;57:477–496. doi: 10.1146/annurev.arplant.57.032905.105320. [DOI] [PubMed] [Google Scholar]
- 5.Kozuka T., Kobayashi J., Horiguchi G., Demura T., Sakakibara H., Tsukaya H., Nagatani A. Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 2010;153:1608–1618. doi: 10.1104/pp.110.156802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozuka T., Kong S.G., Doi M., Shimazaki K., Nagatani A. Tissue-autonomous promotion of palisade cell development by phototropin 2 in Arabidopsis. Plant Cell. 2011;23:3684–3695. doi: 10.1105/tpc.111.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar R., Kushalappa K., Godt D., Pidkowich M.S., Pastorelli S., Hepworth S.R., Haughn G.W. The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell. 2007;19:2719–2735. doi: 10.1105/tpc.106.048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maugarny-Calès A., Laufs P. Getting leaves into shape: A molecular, cellular, environmental and evolutionary view. Development. 2018;145:dev161646. doi: 10.1242/dev.161646. [DOI] [PubMed] [Google Scholar]
- 9.Kawamura E., Horiguchi G., Tsukaya H. Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 2010;62:429–441. doi: 10.1111/j.1365-313X.2010.04156.x. [DOI] [PubMed] [Google Scholar]
- 10.Huang G., Liang W., Sturrock C.J., Pandey B.K., Giri J., Mairhofer S., Wang D., Muller L., Tan H., York L.M., et al. Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat. Commun. 2018;9:2346. doi: 10.1038/s41467-018-04710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giri J., Bhosale R., Huang G., Pandey B.K., Parker H., Zappala S., Yang J., Dievart A., Bureau C., Ljung K., et al. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat. Commun. 2018;9:1408. doi: 10.1038/s41467-018-03850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones A.M., Im K.H., Savka M.A., Wu M.J., Dewitt N.G., Shillito R., Binns A.N. Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science. 1998;282:1114–1117. doi: 10.1126/science.282.5391.1114. [DOI] [PubMed] [Google Scholar]
- 13.Bilsborough G.D., Runions A., Barkoulas M., Jenkins H.W., Hasson A., Galinha C., Laufs P., Hay A., Prusinkiewicz P., Tsiantis M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA. 2011;108:3424–3429. doi: 10.1073/pnas.1015162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay A., Barkoulas M., Tsiantis M. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development. 2006;133:3955–3961. doi: 10.1242/dev.02545. [DOI] [PubMed] [Google Scholar]
- 15.Barkoulas M., Hay A., Kougioumoutzi E., Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 2008;40:1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- 16.Kim J.I., Murphy A.S., Baek D., Lee S.W., Yun D.J., Bressan R.A., Narasimhan M.L. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2011;62:3981–3992. doi: 10.1093/jxb/err094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wenzel C.L., Schuetz M., Yu Q., Mattsson J. Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J. 2007;49:387–398. doi: 10.1111/j.1365-313X.2006.02977.x. [DOI] [PubMed] [Google Scholar]
- 18.Qin G., Gu H., Zhao Y., Ma Z., Shi G., Yang Y., Pichersky E., Chen H., Liu M., Chen Z., et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell. 2005;17:2693–2704. doi: 10.1105/tpc.105.034959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Y., Dai X., Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W., Xu B., Wang H., Li J., Huang H., Xu L. YUCCA genes are expressed in response to leaf adaxial-abaxial juxtaposition and are required for leaf margin development. Plant Physiol. 2011;157:1805–1819. doi: 10.1104/pp.111.186395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasprzewska A., Carter R., Swarup R., Bennett M., Monk N., Hobbs J.K., Fleming A. Auxin influx importers modulate serration along the leaf margin. Plant J. 2015;83:705–718. doi: 10.1111/tpj.12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartels S., Gonzalez Besteiro M.A., Lang D., Ulm R. Emerging functions for plant MAP kinase phosphatases. Trends Plant Sci. 2010;15:322–329. doi: 10.1016/j.tplants.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Monroe-Augustus M., Zolman B.K., Bartel B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell. 2003;15:2979–2991. doi: 10.1105/tpc.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J.S., Ellis B.E. Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J. Biol. Chem. 2007;282:25020–25029. doi: 10.1074/jbc.M701888200. [DOI] [PubMed] [Google Scholar]
- 25.Gupta R., Huang Y., Kieber J., Luan S. Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J. 1998;16:581–589. doi: 10.1046/j.1365-313x.1998.00327.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee J.S., Wang S., Sritubtim S., Chen J.G., Ellis B.E. Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant J. 2009;57:975–985. doi: 10.1111/j.1365-313X.2008.03741.x. [DOI] [PubMed] [Google Scholar]
- 27.Jayaweera T., Siriwardana C., Dharmasiri S., Quint M., Gray W.M., Dharmasiri N. Alternative splicing of Arabidopsis IBR5 pre-mRNA generates two IBR5 isoforms with distinct and overlapping functions. PLoS ONE. 2014;9:e102301. doi: 10.1371/journal.pone.0102301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson K.L., Ramm S., Kappel C., Ward S., Leyser O., Sakamoto T., Kurata T., Bevan M.W., Lenhard M. The Tinkerbell (Tink) Mutation Identifies the Dual-Specificity MAPK Phosphatase INDOLE-3-BUTYRIC ACID-RESPONSE5 (IBR5) as a Novel Regulator of Organ Size in Arabidopsis. PLoS ONE. 2015;10:e0131103. doi: 10.1371/journal.pone.0131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strader L.C., Monroe-Augustus M., Bartel B. The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 2008;8:41. doi: 10.1186/1471-2229-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolboxv. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farooq A., Zhou M.M. Structure and regulation of MAPK phosphatases. Cell. Signal. 2004;16:769–779. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Neel B.G., Tonks N.K. Protein tyrosine phosphatases in signal transduction. Curr. Opin. Cell Biol. 1997;9:193–204. doi: 10.1016/S0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 33.Sassi M., Lu Y., Zhang Y., Wang J., Dhonukshe P., Blilou I., Dai M., Li J., Gong X., Jaillais Y., et al. COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development. 2012;139:3402–3412. doi: 10.1242/dev.078212. [DOI] [PubMed] [Google Scholar]
- 34.Scarpella E., Marcos D., Friml J., Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;20:1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ljung K., Bhalerao R.P., Sandberg G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001;28:465–474. doi: 10.1046/j.1365-313X.2001.01173.x. [DOI] [PubMed] [Google Scholar]
- 36.Scarpella E., Barkoulas M., Tsiantis M. Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. 2010;2:a001511. doi: 10.1101/cshperspect.a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichihashi Y., Aguilar-Martinez J.A., Farhi M., Chitwood D.H., Kumar R., Millon L.V., Peng J., Maloof J.N., Sinha N.R. Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc. Natl. Acad. Sci. USA. 2014;111:E2616–E2621. doi: 10.1073/pnas.1402835111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beerling D.J., Osborne C.P., Chaloner W.G. Evolution of leaf-form in land plants linked to atmospheric CO2 decline in the Late Palaeozoic era. Nature. 2001;410:352–354. doi: 10.1038/35066546. [DOI] [PubMed] [Google Scholar]
- 39.Weller B., Zourelidou M., Frank L., Barbosa I.C., Fastner A., Richter S., Jürgens G., Hammes U.Z., Schwechheimer C. Dynamic PIN-FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux-dependent growth. Proc. Natl. Acad. Sci. USA. 2017;114:E887–E896. doi: 10.1073/pnas.1614380114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barbosa I.C., Hammes U.Z., Schwechheimer C. Activation and polarity control of PIN-FORMED auxin transporters by phosphorylation. Trends Plant Sci. 2018;23:523–538. doi: 10.1016/j.tplants.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Jakobson L., Vaahtera L., Toldsepp K., Nuhkat M., Wang C., Wang Y.S., Horak H., Valk E., Pechter P., Sindarovska Y., et al. Natural Variation in Arabidopsis Cvi-0 Accession Reveals an Important Role of MPK12 in Guard Cell CO2 Signaling. PLoS Biol. 2016;14:e2000322. doi: 10.1371/journal.pbio.2000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasson A., Plessis A., Blein T., Adroher B., Grigg S., Tsiantis M., Boudaoud A., Damerval C., Laufs P. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell. 2011;23:54–68. doi: 10.1105/tpc.110.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maugarny-Cales A., Cortizo M., Adroher B., Borrega N., Goncalves B., Brunoud G., Vernoux T., Arnaud N., Laufs P. Dissecting the pathways coordinating patterning and growth by plant boundary domains. PLoS Genet. 2019;15:e1007913. doi: 10.1371/journal.pgen.1007913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boure N., Kumar S.V., Arnaud N. The BAP Module: A Multisignal Integrator Orchestrating Growth. Trends Plant Sci. 2019;24:602–610. doi: 10.1016/j.tplants.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Tabata R., Ikezaki M., Fujibe T., Aida M., Tian C.E., Ueno Y., Yamamoto K.T., Machida Y., Nakamura K., Ishiguro S. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 2010;51:164–175. doi: 10.1093/pcp/pcp176. [DOI] [PubMed] [Google Scholar]
- 46.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 47.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 48.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034-1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.