Abstract
Head and neck squamous cell carcinoma (HNSCC) is a highly aggressive tumor and the sixth most common cancer worldwide. Current treatment strategies for HNSCC are surgery, radiotherapy, chemotherapy, immunotherapy or combinatorial therapies. However, the overall 5-year survival rate of HNSCC patients remains at about 50%. Cancer stem cells (CSCs), a small population among tumor cells, are able to self-renew and differentiate into different tumor cell types in a hierarchical manner, similar to normal tissue. In HNSCC, CSCs are proposed to be responsible for tumor initiation, progression, metastasis, drug resistance, and recurrence. In this review, we discuss the molecular and cellular characteristics of CSCs in HNSCC. We summarize current approaches used in the literature for identification of HNSCC CSCs, and mechanisms required for CSC regulation. We also highlight the role of CSCs in treatment failure and therapeutic targeting options for eliminating CSCs in HNSCC.
Keywords: cancer stem cells, head and neck squamous cell carcinoma, lymph node metastasis, resistance
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common type of cancer worldwide, with about 200 000 newly diagnosed cases and approximately 128 000 deaths per year.1,2 Arising from the epithelium lining, the oral cavity, tongue, pharynx, larynx, and sinonasal tract, HNSCC is more likely to metastasize than other cancers, with around 50% lymph node metastasis at diagnosis.3 Currently, the standard treatments for HNSCC include surgery with adjuvant radiotherapy and chemotherapy. However, the 5-year survival rates of HNSCC patients remain below 50% and these have only marginally improved in the past few decades.
In recent years, intensive research on molecular mechanism regulating HNSCC has revolutionized the treatment for patients with this metastatic disease. Targeted therapy and immune checkpoint inhibition have been developed and continue to be applied in HNSCC patients.4 Despite these advancements, eradication or control of HNSCC has not been achieved, and the majority of patients will develop treatment resistance. The main root of this resistance and subsequent treatment failure is intratumoral heterogeneity, a key feature of HNSCC, comprising of a mixture of cells displaying differential degrees of sensitivity to cancer treatment. The observed heterogeneity could be the consequence of genetic alterations, epigenetic modification, environmental differences, and changes in cell properties.5 Two models have been proposed that may explain heterogeneity, the clonal evolution model, and the cancer stem cell model. In this review, we will briefly discuss these two models of tumor heterogeneity generation and focus on characterization of HNSCC cancer stem cells (CSCs), their cellular origin, molecular regulation, and prospective therapeutic options.
Clonal evolution versus CSCs
The heterogeneity of tumors is mainly a result of genetic or epigenetic differences between different cell types associated with tumors and tumor cells. Two models of cancer progression and metastasis progression have been proposed: the clonal evolution model and the CSC model.
The clonal evolution model
The clonal evolution model was initially proposed by Peter Nowell in 1976.6 Similar to Darwin’s natural selection, Nowell believed that cancer is an evolutionary process driven by expansion of adapted subclones that carry selectively advantageous mutations. Accumulation of advantageous mutations in cancer cells, as a result of high genetic instability, allows them to outcompete other clones in the tissue ecosystems. Therapeutic intervention can provide potent selective pressure and allow expansion of the resistant clones. The clonal evolution model posits that all tumor cells have equal potential to form tumors, which might help to explain the progression and treatment resistance in certain cancers, such as chronic lymphocytic leukemia and acute myeloid leukemia.7,8 And indeed, there is evidence supporting the genetic instability of solid cancer and its contribution to the genetic heterogeneity of solid tumors.9 Using xenotransplantation, cell surface marker, and clonal cell analyses, previous studies have found that HNSCC progression follows the clonal evolution model.10 Furthermore, another study described two different patterns of clonal dynamics in advanced head and neck cancer by assessing paired primary tumors and distant metastasis from 26 HNSCC patients and sequencing a panel of recurrently mutated genes.11 However, more and more scientific evidence supports the hierarchical model of most solid tumors.
The CSC model
The cancer stem cell model was proposed nearly a half century ago. According to the CSC model, CSCs are at the top of the hierarchy, symmetrically splitting to complement the CSC pool, and one-way asymmetric division produces low tumorigenic daughter cells. CSCs share similar features to normal tissue stem cells, including the ability to self-renew, proliferate, and differentiate. The main difference between the CSC model and the clonal evolution model is that it proposes a hierarchical organization of tumors. In 1994, Dick and colleagues first isolated acute myeloid leukemia stem cells based on the expression cell surface markers.12 They found that CD34+CD38− cells can give rise to a large number of colony-forming progenitor cells in transplanted SCID mice, whereas CD34+CD38+ and CD34− cells do not have these characteristics.12 After identification of leukemia stem cells, the presence of CSCs was also identified in various solid tumors,13–15 further confirming the CSC model.
Note that the clonal evolution and CSC models are not mutually exclusive in cancers. Given the randomness of obtaining additional genetic mutations, the clonal evolution model predicts that each cell can acquire the characteristics of CSCs, whereas CSCs would be expected to evolve by clonal evolution.
Identification of HNSCC CSCs
Currently, the gold standard to define a CSC population is whether they demonstrate long-term clonal growth capabilities in functional repopulation assays, including serial transplantation into recipients or in vivo lineage tracing. Traditionally, putative CSCs in HNSCC are isolated from primary patient samples or cell lines by their unique marker expression pattern, followed by a functional limiting dilution transplantation assay. So far, multiple markers have been described in literature as putative CSC markers in HNSCC in vitro. Below we briefly discuss some commonly used markers. Table 1 provides a list of markers published for HNSCC CSCs isolation. Recently, in vivo lineage tracing assays have been used to make great contributions to identification of HNSCC CSCs, and we will summarize application of this technique in SCC CSCs.
Table 1 .
CSC markers for HNSCC CSCs isolation.
| Genes | Assay | Functions |
|---|---|---|
| CD24 | Flow cytometry | CD24+ cells promote tumor initiation, growth and angiogenesis in HNSCC43,44 |
| CD29 | Flow cytometry | CD29+ cells are related to tumor invasion ability, migration, and lymph node metastasis of HNSCC34,173–175 |
| CD44 | Flow cytometry | CD44+ cells display the characteristics of CSCs in HNSCC73,74,76,176–178 |
| CD98 | Flow cytometry | CD98+ cells are capable of generating tumors in nude mice and express high levels of cell cycle and DNA repair genes179 |
| CD133 | Flow cytometry | CD133+ cells demonstrate CSC properties in HNSCC36–38 |
| ALDH | ALDEFLUOR assay | ALDHhigh population display EMT characteristics, enhanced colony forming and metastasis abilities57,59,61 |
| c-Met | Flow cytometry | c-Met+ cells demonstrate chemoresistance, metastasis and CSC properties in HNSCC49,50,180 |
| Side population | DNA dye exclusion assay | SP cells exist in HNSCC cell lines show higher metastatic potential71 |
| Sphere | Sphere-formation assay | Spherical cells are able to survive detachment from their native microenvironment and to form cellular aggregates in an anchorage-independent manner73 |
CSC isolation in vitro
There are several approaches to isolation of HNSCC CSCs in vitro: including fluorescence-activated cell sorting (FACS) approaches based on cell surface marker expression (Fig. 1), aldehyde dehydrogenase (ALDH) activity, or different efflux ability (side population); and sphere-forming assays using nonattached culture conditions.
Figure 1 .
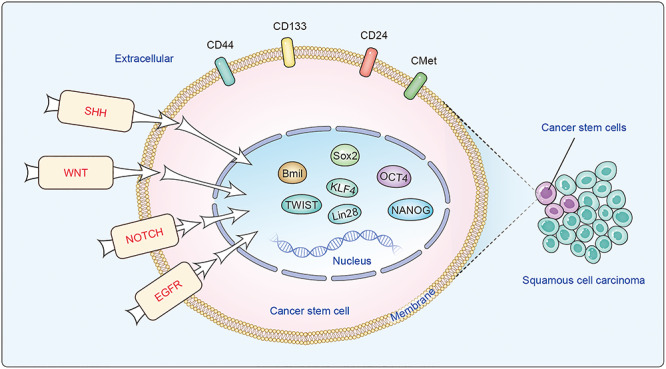
Properties of cancer stem cells (CSCs) in head and neck squamous cell carcinoma (HNSCC).
CD44. Prince et al. first published a study showing that a HNSCC CD44+ cell population displayed higher tumor-initiating ability than CD44− counterparts in xenografts, proving the existence of CSCs in HNSCC.14 Since then, CD44 has become the most well-established and commonly used CSC marker in HNSCCs.16–19 CD44 is a type I transmembrane glycoprotein involved in intercellular interactions, cell adhesion, and cell migration.20 There are multiple isoforms of CD44 produced by alternative RNA splicing, including the standard CD44 isoform with no variable exons (CD44s), and isoform variants with different exons of CD44 (CD44v).21 The extracellular domain of CD44 can bind to various ligands, including hyaluronan, growth factors, cytokines, and matrix metalloproteinases.22 CD44 is involved in activation of a variety of receptor tyrosine kinases-induced cascades, including HGF/c-Met, Src/FAK, and PI3K/AKT, which are responsible for increased proliferation and survival of cells.23–25 Its expression is associated with locoregional recurrence, histopathological grade of malignancy, lymph node metastasis, resistance to therapy, and clinical outcome of HNSCC patients.26 However, recent studies have also raised concern in using CD44 as a CSC marker in HNSCC. For example, systemic examination of CD44s and CD44v6 demonstrates that there is a comparable level of CD44s and CD44v6 expression between normal and benign or malignant epithelia of the head and neck.27,28 In addition, CD44 displays constitutive expression patterns in all HNSCC cell lines, thereby reinforcing it as a reliable marker for CSCs in HNSCC cell lines.29,30 Therefore, the value of CD44 as a marker for the HNSCC CSCs may need to be further re-evaluated.
CD133. CD133, a transmembrane glycoprotein, is a well-known cell surface marker for isolation of a panel of human normal and malignant tissue stem cells.31,32 Although CD133 is often used to isolate HNSCC CSCs, the reproducibility of using it as a marker for HNSCC CSCs is still under debate. Some studies detected no CD133 expression in freshly prepared HNSCC patient samples,20,33,34 whereas other studies showed that cells sorted for high expression of CD133 have similar patterns of clonogenicity compared to CD133− cells.35 In contrast, investigators reported high expression of CD133 is a CD44+ cell population.36 In addition, CD133+ cells were found to have increased clonality, migratory ability, stemness, and drug resistance when compared with CD133− cells in some HNSCC cell lines.37–40 The expression of CD133 in HNSCC prognosis also remains controversial.41,42
CD24. Another commonly used marker CD24, a cell surface glycoprotein involved in cell adhesion and metastasis, is often expressed in tumorigenic CSCs in HNSCC.43–45 CD24 expression level is linked to cisplatin sensitivity and affects expression of critical apoptotic, stem, and drug resistance genes in HNSCC.46 A CD24+ cell population demonstrated a greater ability to self-renew and a greater resistance to chemotherapy in HNSCC.46 Furthermore, CD24+ cells can promote angiogenesis of HNSCC using a mouse model.44 However, CD44high/CD24low or CD44v3+/CD24− cells show higher tumor-initiating ability, clonogenic capacity, and higher drug resistance, suggesting a distinct role of CD24 in different CSC populations in HNSCC.47,48
c-Met. c-Met, the tyrosine kinase receptor for hepatocyte growth factor (HGF), also serves as a cell surface marker for CSCs in HNSCC.49,50 Expression of c-Met is associated with progression, invasion, angiogenesis, and metastasis of HNSCC.51–53 The c-Met pathway also participates in cross-talk of other signaling pathways, including cellular Src kinase (c-Src), phosphotidylinsitol-3-OH kinase (PI3K), α serine/threonine-protein kinase (Akt), and mitogen-activated protein kinase (MAPK).50,54 Sun et al. showed that c-Met can be used as a single marker for HNSCC CSCs and a c-Met+ cell population was responsible for cisplatin-resistance and metastasis.49 However, in retrospective studies, no consensus has been reached regarding whether expression of c-Met has an impact on overall survival or progression-free survival in HNSCC patients or not.55,56
ALDH activity. HNSCC CSCs have demonstrated elevated ALDH activity, which can allow for detoxification of aldehydes and oxidization of retinoic acid.57–59 Thanks to the emergence of ALDEFLUOR flow cytometry assays, researchers have been able to sort live cells with high ALDH activity (ALDHhigh) and characterize the function of ALDHhigh cells in HNSCC progression.60 ALDHhigh subpopulations in HNSCC display a more tumorigenic phenotype and resistance to radiotherapy and chemotherapy.57,59,61 Interestingly, studies have shown that ALDHhigh HNSCC cells can sensitize autologous lymphocytes, whereas the ALDHlow counterparts have limited ability to activate lymphocytes, suggesting the existence of unique CSC antigens in ALDHhigh CSCs.62 To date, 19 ALDH genes have been identified within the human genome. In HNSCC, ALDH1 expression is often increased in primary isolated tumors or cell lines.63,64 However, inconsistent results fosters uncertainty on whether ALDH1 can serve as a predictor of HNSCC prognosis.45,65
Side population. CSCs can also be obtained by isolating the side population (SP) cells based on the ability to efflux Hoechst 33342 dye. SP cells have been successfully used to identify CSC populations in a variety of solid tumors, including HNSCC.66–69 The ability of SP cells to expel the dye lies in expression of a group of transmembrane transporters, which are involved in efflux of the chemotherapeutic drug and resistance to chemotherapy.70 Previous reports have also shown that more SP cells exist in HNSCC cell lines with high metastatic potential than those with low metastatic potential, indicating that SP cells might be responsible for metastatic spreading of HNSCC.71
Sphere-forming ability. Sphere-forming assays have been widely used to assess the self-renewal and differentiation capability of CSCs in vitro. The concept of using spherical cultures to isolate CSC is based on the capacity of CSCs to survive detachment from their native microenvironment and to form cellular aggregates in an anchorage-independent manner. In HNSCC, the expression of known CSCs markers, including CD44, CD133, and ABCG2, can be dramatically enriched under serum-free medium culture conditions.36,72 However, the ability to form spheres varies between different HNSCC samples.73 In comparison with isolation of CSCs using cellular markers, sphere-forming assays are less specific and are often used as a validation assay for isolated CSCs.
It has become clear that a single marker is not sufficient to isolate a pure CSC subpopulation from a given tumor. Therefore, combination of multiple markers is needed for identification and characterization of CSCs. For example, ALDHhighCD44high has become popular as a marker for HNSCC CSCs.59,74,75 In addition, CD44+SSEA4+ CSCs show greater tumorigenic capacity compared with the CD44+ subpopulation, SSEA4+ subpopulation, and parental cells.76
Because tumor initiation is one of the central characteristics of CSCs, xenografts of putative CSC populations in immunodeficient mice remain the gold standard for verification of CSC properties. In fact, application of this approach has successfully guided researchers to identify CSCs in HNSCC.14,72,75 However, there are several drawbacks in using this xenograft model. First, the current isolation approach inevitably causes damage of surface markers during the enzyme digestion process. Second, even orthotopic transplantation may not truly reflect the tumor microenvironment, such as stromal component and cellular architecture, of that patient’s original tumor. Third, the immunodeficient mice used in the xenograft assay lack natural immunosurveillance and cell cytokines, which could impart selective forces on tumor cells and inhibit the growth of the real CSCs within a patient’s tumor.
In vivo identification of CSCs
Determining the cancer cells that are critical for tumor development in their native niche is important for understanding their regulation. In recent years, genetic lineage tools have been deployed in study of CSCs in vivo. The most widely used lineage tracing approach is the Cre-lox system, in which expression of Cre recombinase is driven by a cell-specific promoter.77 With this approach, Cre activity can be transiently induced by administration of small molecules, allowing expression of a reporter gene. Lineage tracing studies are extremely useful to study the cellular hierarchies in cancer homeostasis.
Using genetic lineage tracing, Chen et al. clonally traced tumor cells in vivo in an unperturbed HNSCC induced by carcinogen. They found that Bmi1+ CSCs were responsible for initiation, development, and metastasis of HNSCC.75 Interestingly, cisplatin could effectively kill proliferating cells, but it could not kill Bmi1+ CSCs, which may be the cause of HNSCC recurrence.75 Moreover, Bmi1CreER;Rosa26DTA mice carrying HNSCC had a significantly reduced number of SCC relapses after Bmi1+ cell ablation.75 Furthermore, in a murine cutaneous SCC model, Oshimori et al. showed that in the genetic lineage tracing of TGF-β the expressing cells multiplied faster and were responsible for acceleration of SCC tumor growth.78 Boumahdi showed that Sox2+ cells and their progeny cells are the driving force of skin squamous cell carcinoma induced by DMBA/TPA.79 Interestingly, results from in vivo microscopy of mouse breast tumors show undisputedly the existence of CSCs in unperturbed mammary tumors and demonstrate CSC plasticity.80
Although these murine models help to characterize the CSC component of SCCs in vivo, we must bear in mind that differences exist between mice and humans. In addition, some of the cytokines in mouse and human are not functionally equivalent, indicating that regulation of CSCs might follow different regimens in these two species.
Regulation of HNSCC CSCs
Stem cell factors and HNSCC CSCs
Comparison of molecular signatures between CSCs and normal stem cells uncovered a great deal of overlap between these two different kinds of cell populations.81 Interestingly, genes associated with self-renewal, angiogenesis, migration, and anti-apoptosis were largely shared between these two stem cell populations. In particular, factors that are highly enriched in embryonic stem cells (ESCs), such as OCT4 (Octamer-binding transcription factor 4), SOX2 (Sex determining region Y-box 2), NANOG (Nanog Homeobox), KLF4 (Krüppel-like factor4), and Lin28/Let-7, are often linked to the stem-cell like feature of HNSCC CSCs (Fig. 1).
OCT4. OCT4 is highly expressed in ESCs and has an essential role in self-renewal and differentiation by regulating the pluripotent potential of these cells.82,83 It has been shown that high expression of OCT4 is associated with poor survival and strongly independent prognostic effects on HNSCC progression.72,84,85 In HNSCC, OCT4 and its target gene CIP2A were co-expressed in a CD24 positive side-population and were responsible for increased aggressiveness and radioresistancy.86 Inhibition of OCT4 expression results in reduced tumorigenic ability of HNSCC CSCs, whereas enforced expression of OCT4 in HNSCC leads to increased tumorigenicity and epithelial-mesenchymal transition (EMT) transformation of HNSCC CSCs.87
SOX2. SOX2 has emerged as a factor in maintaining self-renewal of ESC and is often enriched in HNSCC through amplification on chromosome 3q26.82,88,89 Several studies have shown that SOX2 nuclear expression is closely associated with tumor recurrence and poor prognosis in patients with HNSCC.90–94 The SOX2-mediated pathway is critical in HNSCC initiation and progression by regulating acquisition of CSC-like and radiochemoresistant properties in HNSCC.89,94 Overexpression of SOX2 promotes cell proliferation via cyclin B1 expression and CSC features, including self-renewal and chemoresistance.92 Moreover, silence of SOX2 in HNSCC CSCs substantially inhibits their self-renewal capacity, chemoresistance, invasion capacity, and in vivo tumorigenicity.92
NANOG. Similar to OCT4, a high level of NANOG is linked to poor survival and independent prognostic effects on HNSCC progression.72,84 Expression of NANOG is elevated in both the side population and in the tumorsphere of HNSCC cell lines, implying that NANOG plays a role in regulation of HNSCC CSCs.72,84 Indeed, knockdown of NANOG can effectively block CSC-like properties and increase drug sensitivity and apoptosis of HNSCC CSCs.95 Interestingly, NANOG protein stability can be regulated by human protein kinase Cε via phosphorylation at T200 and T280 residues.96 Inhibition of T200A or T280A phosphorylation in NANOG can lead to decreased cell proliferation, colony formation, invasion, migration of the CSC population in HNSCC cells.96
KLF4. KLF4 is a relatively large family of zinc finger transcription factors belonging to sp1-like transcription factors. The role of KLF4 remains controversial in HNSCC. It has been shown that expression of KLF4 displays an inconsistent pattern in HNSCC.97,98 Although KLF4 protein expression is decreased in the majority of HNSCC patient samples, there is still persistent KLF4 expression detected in some HNSCC.97 HNSCC with high KLF4 expression is often associated with a lower disease-specific survival, whereas ectopic KLF4 expression promotes HNSCC progression.88 However, using a genetic mouse model, a study has demonstrated that conditional knockout of Klf4 expression in the oral epithelium promotes development of malignant oral SCC lesions, suggesting a potential tumor suppressor role of Klf4 in HNSCC.99
LIN28/Let7. As a well-known RNA-binding protein, LIN28 plays a critical role in regulating the balance between stemness and differentiation in ESCs via regulation of the microRNA Let-7.100 Lin28 is highly expressed during embryogenesis and is critical for the determination of stemness state in multiple tissue lineages.101 In contrast, Let-7 often serves as a tumor suppressor in a variety of cancers, most likely through targeting oncogenes, such as RAS or HMGA2.102,103 In HNSCC, dysregulation of Let-7 is associated with patient clinical outcomes.104 Furthermore, Let-7 expression is dramatically decreased in ALDHhigh putative CSCs compared to ALDHlow population.105 Functional assays further confirm that Lin28B-Let7 is required for Oct4 and Sox2 expression, and for the self-renewal properties of HNSCC CSCs.103 Specifically, targets of Let7, including ARID3B and HMGA2, can directly regulate Oct4 and Sox2 expression via binding to their promoter.103
BMI1. BMI1 (B lymphoma Mo-MLV insertion region 1 homolog) is a polycomb group (PcG) protein that plays an important role in the self-renewal and epigenetic regulation of normal and cancer stem cells. BMI1 serves as a key component of the polycomb repressive complex 1 (PRC1), which represses gene expression through monoubiquitination of histone H2A.106,107 High BMI1 often correlates with advanced stages, aggressive clinicopathological behaviors, stem cell-like properties, drug resistance, and poor prognosis in HNSCC.14,59,75,108 Down-regulation of BMI1 can reduce the cell sphere and colony formation.109 Pharmacological targeting of BMI1, using a small molecule inhibitor, dramatically impaired tumorigenesis in HNSCC.75,110,111
Signaling pathway and HNSCC CSCs
Signaling pathways, including Sonic Hedgehog (SHH), Wnt, epidermal growth factor receptor (EGFR), and Notch, that control normal stem cell self-renewal and differentiation are often aberrantly activated in HNSCC CSCs (Fig. 1). Therefore, identification of the crucial pathways necessary for CSC maintenance may provide important therapeutic targets for HNSCC.
SHH pathway. The SHH signaling pathway is an important mechanism for embryonic development and homeostasis of mature tissues.112 The pathway is activated on binding of SHH to the PTCH receptor, which in turn derepresses the Smoothened (SMO) transmembrane receptor.113 Activation of SMO then triggers the SHH signaling cascade via recruitment and activation of GLI family transcription factors, including GLI1, GLI2, and GLI3.113 Previous studies have shown that SHH signaling is upregulated in various CSCs, including breast cancer, liver cancer, brain tumors, and gastric cancer.114–117 In HNSCC, elevated levels of GLI, PTCH1, SMO, and SHH can be detected in tumor compared with normal oral mucosa.118 In addition, expression of GLI1 is associated with lymph node metastasis, recurrence, clinical stages, and poor prognosis. Consistently, blockage of the SHH signaling pathway leads to inhibition of tumor growth and angiogenesis in HNSCC.119 Moreover, inhibition of GLI3 in HNSCC cell lines leads to a decrease of CD44high CSCs and sphere-forming ability by inhibition of OCT4 and BMI1 expression.17
WNT pathway. The WNT signaling pathway is an evolutionarily conserved pathway critical for stem cell self-renewal and fate determination.120 Based on the dependency of β-catenin, the WNT signaling pathway can be divided into two, namely canonical and non-canonical pathways. Activation of the canonical WNT/β-catenin signaling pathway requires binding of WNT ligands to Frizzled and its co-receptor, lipoprotein receptor-related protein 5 or 6 (LRP 5/6), which then recruits the β-catenin degradation complex and allows accumulation of β-catenin in the cytoplasm.121 Consequently, β-catenin will be translocated into the cell nucleus to promote transcription of downstream targets via binding to the N-terminal of T-cell factor/lymphoid enhancing factor (TCF/LEF) transcription factors.121 Previous studies have demonstrated an important role of the WNT/β-catenin signaling pathway in maintenance of HNSCC CSCs. For example, treatment of WNT activator or WNT inhibitor can alter the CSC proliferation rate in HNSCC, while this treatment shows minimal effect on the parental HNSCC cells, suggesting a role for the WNT/β-catenin signaling pathway in HNSCC CSCs.122–124 In addition, a recent study showed that Wnt activation in HNSCC increased the properties of CSCs, such as sphere formation and invasiveness.125
EGFR pathway. The EGFR is a transmembrane protein that is activated via binding of its specific ligands, including epidermal growth factor (EGF) and transforming growth factor α (TGFα), which triggers activation of an intracellular signaling cascade to control cell growth, differentiation, and survival.126 Elevated levels of EGFR are often observed in HNSCC, and increased activity of the EGF pathway has been associated with resistance to treatment and poor clinical outcome.127 Results have shown that high expression of EGFR promotes a higher functional proportion of ALDHhigh CSCs in a high aggressive human papillomavirus-16 (HPV-16) + cell line UM-SCC-104.128 In addition, the CSC cell surface marker CD44 can interact with EGFR to enhance progression of HNSCC.129
NOTCH pathway. Notch signaling plays an essential role in a variety of stem cell processes, such as cell proliferation, differentiation, and self-renewal.130 Four Notch receptors (Notch 1–4) and five Notch ligands (Jagged-1, Jagged-2, Delta-1, Delta-3, and Delta-4) have been reported in mammals.131 When the Notch receptor is activated, the Notch intracellular domain (NICD) will be released from the plasma membrane and translocates into the nucleus.131 Together with the CSL transcription factors, NICD will then induce the expression of its target genes, such as Hes-1 and Hey-1.131 Inhibition of NOTCH1 by antibody or inhibitor is able to repress tumor growth and CSC function in multiple tumors.132,133 In addition, NOTCH1 inhibition can impair the tumorigenesis and CSC self-renewal of HNSCC using a xenograft model.134 However, studies have also reported that Notch1 might serve as a tumor-suppressor gene in HNSCC.135,136
EMT and HNSCC CSCs
EMT is a process in which epithelial cells change their morphology, lose their polarity and acquire the migratory properties of mesenchymal cells. During EMT, epithelial cells gradually lose expression of intercellular adhesion molecules, such as E-cadherin and keratin, and gain expression of vimentin, N-cadherin, and fibronectin.137 EMT is mainly mediated by a core set of transcription factors, including SNAIL, SLUG, TWIST1, ZEB1, and ZEB2.138 Aberrant regulation of these EMT transcription factors can induce cancer cell plasticity and promote tumor initiation and metastatic spread. In addition, EMT transcription factors are able to induce many other traits, such as cell survival and stem cell-like features.139 Mani et al. first provided experimental evidence that induction of EMT leads to upregulation of stem cell markers in breast cancer cells.140 Since then, accumulating evidence in various solid tumors supports that EMT induction not only promotes tumor cell invasion and metastasis, but also enhances drug resistance and enriches CSCs.141–143
In HNSCC cell lines, studies have shown that ALDHhigh putative CSCs display EMT characteristics and enhanced colony forming abilities.144 In addition, two different types of CSCs displaying distinct phenotypes exist in HNSCC, including CD44+ ESAlow EMT CSCs expressing EMT markers and CD44+ ESAhigh non-EMT CSCs displaying epithelial characteristics.145 However, only the ALDH1+ EMT CSCs in HNSCC are able to metastasize and form new tumors.145 Hence, some researchers suggested that high expression of EMT transcription factors, stemness-related factors, and ALDH1+ cells should be used as conditions for identifying CSC of HNSCC.144,146 Studies have found simultaneous high expression of EMT-related genes (SNAIL, SLUG, TWIST, Vimentin, and Fibronectin) and stemness-related genes (OCT4, NANOG and BMI1) in metastatic cell lines of HNSCC.144,147 BMI1 can be directly regulated by TWIST, and synergistically promotes EMT and the tumor-initiating capability of HNSCC with TWIST.147 In addition, overexpression of Twist results in upregulation of HNSCC CSC markers, such as ALDH and CD133 in HNSCC cell lines.111
Recently, studies indicated that EMT is not a two-level process from epithelial to complete mesenchymal state, but gradual events passing through different intermediate hybrid states. Different stages of EMT show different cellular plasticity, invasiveness, and metastasis potential,148 which further supports the heterogeneous nature of EMT. Furthermore, different sources of cells may affect whether or not cells undergo EMT. For example, in murine cutaneous SCC, hair follicle-derived cancer cells are more prone to undergo EMT than interfollicular epidermis-derived cancer cells. And genes of EMT in hair follicle lineages are regulated by transcription factors related to hair follicle stemness and differentiation.149 These findings may provide an explanation why some types of cancer cells retain an epithelial organization rather than mesenchymal traits when metastasis occurs.143 Metastatic cancer cells may be located in a different type of intermediate hybrid state of EMT or these cancer cells may not easily undergo the EMT process. Therefore the findings do not dispute the hypothesis that EMT is necessary to maintain the CSC phenotype.
HNSCC CSC and drug resistance
There are several mechanisms of drug resistance in cancer, including reduced drug aggregation, changes in drug target, reduced apoptosis, and increased DNA damage repair. According to the CSC hypothesis, the stem cell-like properties of CSCs help tumors to resist chemotherapy agents. However, studies have also shown that treatment with chemotherapy drugs can increase stemness in multiple solid tumors.150–152 To study the chemoresistance of HNSCC CSCs, researchers need to take into account the specific markers that are used for isolation of CSCs, because CSC markers may possess the molecular features to antagonize chemotherapy.
For example, CD44 can interact with hyaluronan to promote phospholipase C-mediated intracellular Ca2+ mobilization in order to prevent apoptotic effects caused by cisplatin.126 In addition, hyaluronan triggers an interaction between CD44v3 and OCT4-SOX2-NANOG, which in turn induces expression of miR-302 and survival proteins (cIAP-1, cIAP-2, and XIAP), thereby allowing cells to become tolerant to cisplatin.153 Another marker, CD133, is critical in chemoresistance by positively regulating expression of CSC-related genes.154 Chemotherapeutic drugs, such as cisplatin and erlotinib, can increase the level of reactive oxygen species (ROS) and lipid peroxidation-derived aldehydes.155 It is reasonable to suspect that high activity of ALDHs, which catalyze oxidation of aldehydes, can help in metabolizing these reactive aldehydes and reducing oxidative stress in HNSCC CSCs. Indeed, targeting the activity of ALDH using small molecule inhibitors, greatly impairs the cisplatin resistance in HNSCC.156 In HNSCC, c-Met-expression cells exhibit cisplatin-resistant capacity.49 Blockage of c-Met with ARQ 197 and crizotinib, two small molecule inhibitors of c-Met, results in reduced tumorigenesis in HNSCC both in vivo and in vitro.157 These studies indicate that markers for isolating CSCs are also responsible for drug resistance in CSCs.
Stem cell factors also take part in chemoresistance of HNSCC CSCs. For instance, SOX2 can promote cisplatin resistance of HNSCC CSCs through ABCG2 (ATP-binding cassette super-family G member 2) expression.92 Upregulation of OCT4 and NANOG has also been associated with chemoresistance in HNSCC.158 Drug resistance mediated by a HNSCC CSC marker is often involved in activation of the PI-3 kinase/AKT signaling pathway, in which increased phosphorylation of AKT inhibits activation of the apoptotic effect.63
A recent study suggests the existence of plasticity between CSC and non-CSC populations.23 This has led researchers to wonder whether targeting both CSC and non-CSC populations will be needed for better treatment. A study by Chen et al. provided experimental evidence demonstrating reduced primary tumor volume and lymph node metastasis through the means of targeting mouse HNSCC with PTC-209 (Bmi1 inhibitor) and cisplatin.75
Clinical and therapeutic implications of HNSCC CSCs
Despite the advances in immune-checkpoint inhibitors in HNSCC, early studies from clinical trials show limited efficacy with monotherapy in HNSCC compared to traditional chemotherapy. Thus, more effective approaches are required for HNSCC. Targeting CSCs could improve the efficacy of cancer therapy because CSCs can resist chemotherapy and radiotherapy, leading to a population of cancer cells left behind to continue to grow and spread. Although intensive efforts have been made towards targeting CSCs in different tumor types, most of them showed limited effect, probably because of the failure to identify the bona fide CSCs population. Besides, researchers also need to be aware that CSCs are heterogeneous based on the genetically unstable nature of cancer cells. Another point of note is that CSC-targeted therapy is possibly toxic toward normal stem cells. Here, we discuss some recent preclinical and clinical progress in targeting HNSCC CSCs.
Targeting cell surface markers
Cell surface markers have been explored previously as possible pharmaceutical targets of CSCs in HNSCC. Targeting CD44v6 using chimeric monoclonal antibody U36 in patient or monoclonal antibody BIWA in xenograft results showed that this approach can reduce tumor size or can be useful for fluorescence-guided surgery.159,160 Furthermore, Yu et al. showed that targeting CD133 can overcome the chemoresistance in HNSCC-derived SP cells, suggesting that cell surface makers are promising targets for CSC inhibition.39
Targeting tyrosine kinases
Receptor tyrosine kinases promote tumor progression in HNSCC and are an attractive therapeutic target. Cetuximab is a US Food and Drug Administration (FDA)-approved monoclonal antibody targeting EGFR in HNSCC. Treatment with cetuximab also promotes differentiation of CSCs and impairs resistance of CSC to chemo/radiation.161 Also, a newly discovered gamboge derivative compound 2 showed effective inhibition of HNSCC by targeting a CSC population through EGFR tyrosine phosphorylation.162 However, another study has demonstrated that the EGFR-targeted agents, such as cetuximab and erlotinib, show only modest effects on tumor control of HNSCC.161
Targeting stem cell factors
Progression of HNSCC is associated with Bmi1-positive CSCs, which are responsible for tumor invasion, drug resistance, and lymph node metastasis. Hence, Bmi1 serves as a potential CSC therapeutic target for HNSCC. Inhibition of BMI1 by the small molecule inhibitor PTC-209, enhances the therapeutic effect of cisplatin and inhibits lymph node metastasis in HNSCC.75 In addition, a study has shown that the flavonoid derivative 2-(3-hydroxyphenyl)-5-methylnaphthyridin-4-one (CSC-3436) can effectively inhibit EMT, cancer stemness, and migration/invasion abilities via downregulation of the Twist/Bmi1-Akt/β-catenin pathway.111 Heat shock protein 90 inhibitors, KU711 and Ku757, are able to effectively target CSCs in HNSCC and inhibit the expression of BMI1 and EMT.163
Targeting immune checkpoint inhibitors
To date, various immunotherapy approaches have been developed to target HNSCC, including vaccines, T cell infusion, immune checkpoint inhibitors, and tumor-specific monoclonal antibodies.164 In 2016, anti-PD1 antibodies (nivolumab and pembrolizumab) were approved by the FDA for treatment of HNSCC in the second-line setting. In these pilot studies, results have demonstrated that nivolumab and pembrolizumab can improve overall survival of HNSCC compared with traditional chemotherapy.4,165,166 However, response rates to nivolumab and pembrolizumab in HNSCC remain low, ranging only from 13% to 20%, whereas survival has improved in one of 10 patients who received these therapies. The heterogeneous nature of HNSCC might be the key to the unresponsiveness and resistance to immunotherapy. A recent study using a mouse model has shown that TGF-β responsive CSCs are responsible for resistance of immunotherapy and cancer recurrence through selective expression of CD80, a well-known immune checkpoint protein,167 supporting the notion that targeting CSCs might improve the current immunotherapy of HNSCC.
Targeting HNSCC CSCs by natural compounds
In recent decades, natural compounds have been recognized more and more as useful drugs to target CSCs in multiple cancer types, including HNSCC.168–172,174,175 For example, BE-43547A2, which belongs to the naturally occurring cyclic depsipeptides, can selectively target pancreatic CSCs.178 Researcher have also found several naturally derived compounds that can ablate CSCs in melanoma more effectively.177 In addition, curcumin, a natural product extracted from Curcuma longa plants, can suppress stemness of HNSCC CSCs through inhibition of RXRα.178
Concluding remarks and future perspectives
CSCs constitute a small portion of tumor cells and display the ability to renovate new tumors. Hence, targeting the CSC population has become an effective method of cancer treatment, although CSC-targeting therapeutics are still in their infancy. Although intensive studies have been performed to investigate the characteristics and regulation of CSCs in HNSCC, many of these were based on in vitro data and none have gone through the early clinical stages. As previously mentioned, for development of new strategies for targeting CSCs in HNSCC, we need a better understanding of CSC properties and their druggable targets. Furthermore, it is important to realize that precision medicine might provide great value for treatment of different patients who show different CSC regulation.
Acknowledges
This work was supported by the National Institutes of Health (Grant No.: R01 DE15964) (C.Y.W).
Conflict of interest
None declared.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3. Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 4. Ferris RL, BlumenscheinG, Jr., Fayette J, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 2016;375:1856–67. doi: 10.1056/NEJMoa1602252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 6. Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 7. Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015;526:525–30. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012;481:506–10. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017;168:613–28. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 10. Cameron SR, Dahler AL, Endo-Munoz LB, et al. Tumor-initiating activity and tumor morphology of HNSCC is modulated by interactions between clonal variants within the tumor. Lab Invest 2010;90:1594–603. doi: 10.1038/labinvest.2010.131. [DOI] [PubMed] [Google Scholar]
- 11. Melchardt T, Magnes T, Hufnagl C, et al. Clonal evolution and heterogeneity in metastatic head and neck cancer—an analysis of the Austrian study Group of Medical Tumour Therapy study group. Eur J Cancer 2018;93:69–78. doi: 10.1016/j.ejca.2018.01.064. [DOI] [PubMed] [Google Scholar]
- 12. Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 13. Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A 2007;104:973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 16. Kuo SZ, Honda CO, Li WT, et al. Metformin results in diametrically opposed effects by targeting non-stem cancer cells but protecting cancer stem cells in head and neck squamous cell carcinoma. Int J Mol Sci 2019;20. doi: 10.3390/ijms20010193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodrigues M, Miguita L, De Andrade NP, et al. GLI3 knockdown decreases stemness, cell proliferation and invasion in oral squamous cell carcinoma. Int J Oncol 2018;53:2458–72. doi: 10.3892/ijo.2018.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bae WJ, Koo BS, Lee SH, et al. Inhibitor of DNA binding 2 is a novel therapeutic target for stemness of head and neck squamous cell carcinoma. Br J Cancer 2017;117:1810–8.doi: 10.1038/bjc.2017.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kerk SA, Finkel KA, Pearson AT, et al. 5T4-targeted therapy ablates cancer stem cells and prevents recurrence of head and neck squamous cell carcinoma. Clin Cancer Res 2017;23:2516–27.doi: 10.1158/1078-0432.CCR-16-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Margaritescu C, Pirici D, Simionescu C, et al. The utility of CD44, CD117 and CD133 in identification of cancer stem cells (CSC) in oral squamous cell carcinomas (OSCC). Rom J Morphol Embryol 2011;52:985–93. PMID: 22119814. [PubMed] [Google Scholar]
- 21. Negi LM, Talegaonkar S, Jaggi M, et al. Role of CD44 in tumour progression and strategies for targeting. J Drug Target 2012;20:561–73. doi: 10.3109/1061186X.2012.702767. [DOI] [PubMed] [Google Scholar]
- 22. Dzwonek J, Wilczynski GM. CD44: Molecular interactions, signaling and functions in the nervous system. Front Cell Neurosci 2015;9:175. doi: 10.3389/fncel.2015.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marjanovic ND, Weinberg RA, Chaffer CL. Cell plasticity and heterogeneity in cancer. Clin Chem 2013;59:168–79. doi: 10.1373/clinchem.2012.184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matzke A, Sargsyan V, Holtmann B, et al. Haploinsufficiency of c-met in cd44−/− mice identifies a collaboration of CD44 and c-met in vivo. Mol Cell Biol 2007;27:8797–806. doi: 10.1128/MCB.01355-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Skupien A, Konopka A, Trzaskoma P, et al. CD44 regulates dendrite morphogenesis through Src tyrosine kinase-dependent positioning of the Golgi. J Cell Sci 2014;127:5038–51. doi: 10.1242/jcs.154542. [DOI] [PubMed] [Google Scholar]
- 26. Garcia AS, Assao A, Carvalho AL, et al. The stem cell markers expression CD44v6 and podoplanin in lip cancer: Clinical significance. Virchows Arch 2019. 2019;474(6):745‑54. doi: 10.1007/s00428-019-02539-3. [DOI] [PubMed] [Google Scholar]
- 27. Mack B, Gires O. CD44s and CD44v6 expression in head and neck epithelia. PLoS One 2008;3:e3360. doi: 10.1371/journal.pone.0003360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J, Zhou J, Lu J, et al. Significance of CD44 expression in head and neck cancer: A systemic review and meta-analysis. BMC Cancer 2014;14:15. doi: 10.1186/1471-2407-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pries R, Wittkopf N, Hasselbacher K, et al. Constitutive expression of the potential stem cell marker CD44 in permanent HNSCC cell lines. HNO 2008;56:461–6. doi: 10.1007/s00106-008-1707-0. [DOI] [PubMed] [Google Scholar]
- 30. Pries R, Witrkopf N, Trenkle T, et al. Potential stem cell marker CD44 is constitutively expressed in permanent cell lines of head and neck cancer. In Vivo 2008;22:89–92. PMID:18396788. [PubMed] [Google Scholar]
- 31. Miraglia S, Godfrey W, Yin AH, et al. A novel five-transmembrane hematopoietic stem cell antigen: Isolation, characterization, and molecular cloning. Blood 1997;90:5013–21. PMID:9389721. [PubMed] [Google Scholar]
- 32. Glumac PM, LeBeau AM. The role of CD133 in cancer: A concise review. Clin Transl Med 2018;7:18. doi: 10.1186/s40169-018-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okamoto H, Fujishima F, Nakamura Y, et al. Significance of CD133 expression in esophageal squamous cell carcinoma. World J Surg Oncol 2013;11:51. doi: 10.1186/1477-7819-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Moraes FP, Lourenco SV, Ianez RC, et al. Expression of stem cell markers in oral cavity and oropharynx squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol 2017;123:113–22. doi: 10.1016/j.oooo.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 35. Harper LJ, Piper K, Common J, et al. Stem cell patterns in cell lines derived from head and neck squamous cell carcinoma. J Oral Pathol Med 2007;36:594–603. doi: 10.1111/j.1600-0714.2007.00617.x [DOI] [PubMed] [Google Scholar]
- 36. Okamoto A, Chikamatsu K, Sakakura K, et al. Expansion and characterization of cancer stem-like cells in squamous cell carcinoma of the head and neck. Oral Oncol 2009;45:633–9. doi: 10.1016/j.oraloncology.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 37. Zhang Q, Shi S, Yen Y, et al. A subpopulation of CD133(+) cancer stem-like cells characterized in human oral squamous cell carcinoma confer resistance to chemotherapy. Cancer Lett 2010;289:151–60. doi: 10.1016/j.canlet.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 38. Zhou L, Wei X, Cheng L, et al. CD133, one of the markers of cancer stem cells in Hep-2 cell line. Laryngoscope 2007;117:455–60. doi: 10.1097/01.mlg.0000251586.15299.35. [DOI] [PubMed] [Google Scholar]
- 39. Yu CC, Hu FW, Yu CH, et al. Targeting CD133 in the enhancement of chemosensitivity in oral squamous cell carcinoma-derived side population cancer stem cells. Head Neck 2016;38:E231–8. doi: 10.1002/hed.23975. [DOI] [PubMed] [Google Scholar]
- 40. Lu BC, Li J, Yu WF, et al. Elevated expression of Nrf2 mediates multidrug resistance in CD133(+) head and neck squamous cell carcinoma stem cells. Oncol Lett 2016;12:4333–8. doi: 10.3892/ol.2016.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hang D, Dong HC, Ning T, et al. Prognostic value of the stem cell markers CD133 and ABCG2 expression in esophageal squamous cell carcinoma. Dis Esophagus 2012;25:638–44. doi: 10.1111/j.1442-2050.2011.01298.x. [DOI] [PubMed] [Google Scholar]
- 42. Fan Z, Li M, Chen X, et al. Prognostic value of cancer stem cell markers in head and neck squamous cell carcinoma: A meta-analysis. Sci Rep 2017;7:43008. doi: 10.1038/srep43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Satpute PS, Hazarey V, Ahmed R, et al. Cancer stem cells in head and neck squamous cell carcinoma: A review. Asian Pac J Cancer Prev 2013;14:5579–87. doi: 10.7314/apjcp.2013.14.10.5579. [DOI] [PubMed] [Google Scholar]
- 44. Zimmerer RM, Ludwig N, Kampmann A, et al. CD24+ tumor-initiating cells from oral squamous cell carcinoma induce initial angiogenesis in vivo. Microvasc Res 2017;112:101–8. doi: 10.1016/j.mvr.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 45. Koukourakis MI, Giatromanolaki A, Tsakmaki V, et al. Cancer stem cell phenotype relates to radio-chemotherapy outcome in locally advanced squamous cell head-neck cancer. Br J Cancer 2012;106:846–53. doi: 10.1038/bjc.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Modur V, Joshi P, Nie D, et al. CD24 expression may play a role as a predictive indicator and a modulator of Cisplatin treatment response in head and neck squamous cellular carcinoma. PLoS One 2016;11:e0156651. doi: 10.1371/journal.pone.0156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ghuwalewala S, Ghatak D, Das P, et al. CD44(high)CD24(low) molecular signature determines the cancer stem cell and EMT phenotype in oral squamous cell carcinoma. Stem Cell Res 2016;16:405–17. doi: 10.1016/j.scr.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 48. Todoroki K, Ogasawara S, Akiba J, et al. CD44v3+/CD24- cells possess cancer stem cell-like properties in human oral squamous cell carcinoma. Int J Oncol 2016;48:99–109. doi: 10.3892/ijo.2015.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun S, Wang Z. Head neck squamous cell carcinoma c-met(+) cells display cancer stem cell properties and are responsible for cisplatin-resistance and metastasis. Int J Cancer 2011;129:2337–48. doi: 10.1002/ijc.25927. [DOI] [PubMed] [Google Scholar]
- 50. Lim YC, Kang HJ, Moon JH. C-met pathway promotes self-renewal and tumorigenecity of head and neck squamous cell carcinoma stem-like cell. Oral Oncol 2014;50:633–9. doi: 10.1016/j.oraloncology.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 51. Zeng Q, Chen S, You Z, et al. Hepatocyte growth factor inhibits anoikis in head and neck squamous cell carcinoma cells by activation of ERK and Akt signaling independent of NFkappa B. J Biol Chem 2002;277:25203–8. doi: 10.1074/jbc.M201598200. [DOI] [PubMed] [Google Scholar]
- 52. De Herdt MJ, Baatenburg de Jong RJ. HGF and c-MET as potential orchestrators of invasive growth in head and neck squamous cell carcinoma. Front Biosci 2008;13:2516–26. PMID:17981731. [DOI] [PubMed] [Google Scholar]
- 53. Di Renzo MF, Olivero M, Martone T, et al. Somatic mutations of the MET oncogene are selected during metastatic spread of human HNSC carcinomas. Oncogene 2000;19:1547–55. doi: 10.1038/sj.onc.1203455. [DOI] [PubMed] [Google Scholar]
- 54. Arnold L, Enders J, Thomas SM. Activated HGF-c-met Axis in head and neck cancer. Cancers (Basel) 2017;9. doi: 10.3390/cancers9120169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. da Costa A, Costa FD, Araujo DV, et al. The roles of PTEN, cMET, and p16 in resistance to cetuximab in head and neck squamous cell carcinoma. Med Oncol 2018;36:8. doi: 10.1007/s12032-018-1234-0. [DOI] [PubMed] [Google Scholar]
- 56. Vsiansky V, Gumulec J, Raudenska M, et al. Prognostic role of c-met in head and neck squamous cell cancer tissues: A meta-analysis. Sci Rep 2018;8:10370. doi: 10.1038/s41598-018-28672-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Clay MR, Tabor M, Owen JH, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck 2010;32:1195–201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Krishnamurthy S, Dong Z, Vodopyanov D, et al. Endothelial cell-initiated signaling promotes the survival and self-renewal of cancer stem cells. Cancer Res 2010;70:9969–78. doi: 10.1158/0008-5472.CAN-10-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nor C, Zhang Z, Warner KA, et al. Cisplatin induces Bmi-1 and enhances the stem cell fraction in head and neck cancer. Neoplasia 2014;16:137–46. doi: 10.1593/neo.131744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Storms RW, Trujillo AP, Springer JB, et al. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci U S A 1999;96:9118–23. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bertrand G, Maalouf M, Boivin A, et al. Targeting head and neck cancer stem cells to overcome resistance to photon and carbon ion radiation. Stem Cell Rev 2014;10:114–26. doi: 10.1007/s12015-013-9467-y. [DOI] [PubMed] [Google Scholar]
- 62. Prince MEP, Zhou L, Moyer JS, et al. Evaluation of the immunogenicity of ALDH (high) human head and neck squamous cell carcinoma cancer stem cells in vitro. Oral Oncol 2016;59:30–42. doi: 10.1016/j.oraloncology.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen YC, Chen YW, Hsu HS, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun 2009;385:307–13. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 64. Yu CC, Lo WL, Chen YW, et al. Bmi-1 regulates Snail expression and promotes metastasis ability in head and neck squamous cancer-derived ALDH1 positive cells. J Oncol 2011;2011. doi: 10.1155/2011/609259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang Y, Zhe H, Gao P, et al. Cancer stem cell marker ALDH1 expression is associated with lymph node metastasis and poor survival in esophageal squamous cell carcinoma: A study from high incidence area of northern China. Dis Esophagus 2012;25:560–5. doi: 10.1111/j.1442-2050.2011.01279.x. [DOI] [PubMed] [Google Scholar]
- 66. Chiba T, Kita K, Zheng YW, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology 2006;44:240–51. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 67. Luo Y, Ellis LZ, Dallaglio K, et al. Side population cells from human melanoma tumors reveal diverse mechanisms for chemoresistance. J Invest Dermatol 2012;132:2440–50. doi: 10.1038/jid.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wu C, Wei Q, Utomo V, et al. Side population cells isolated from mesenchymal neoplasms have tumor initiating potential. Cancer Res 2007;67:8216–22. doi: 10.1158/0008-5472.CAN-07-0999 [DOI] [PubMed] [Google Scholar]
- 69. Zhang P, Zhang Y, Mao L, et al. Side population in oral squamous cell carcinoma possesses tumor stem cell phenotypes. Cancer Lett 2009;277:227–34. doi: 10.1016/j.canlet.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 70. Tabor MH, Clay MR, Owen JH, et al. Head and neck cancer stem cells: The side population. Laryngoscope 2011;121:527–33. doi: 10.1002/lary.21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Song J, Chang I, Chen Z, et al. Characterization of side populations in HNSCC: Highly invasive, chemoresistant and abnormal Wnt signaling. PLoS One 2010;5:e11456. doi: 10.1371/journal.pone.0011456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chiou SH, Yu CC, Huang CY, et al. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res 2008;14:4085–95. doi: 10.1158/1078-0432.CCR-07-4404. [DOI] [PubMed] [Google Scholar]
- 73. Lim YC, Oh SY, Cha YY, et al. Cancer stem cell traits in squamospheres derived from primary head and neck squamous cell carcinomas. Oral Oncol 2011;47:83–91. doi: 10.1016/j.oraloncology.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 74. Seino S, Shigeishi H, Hashikata M, et al. CD44(high)/ALDH1(high) head and neck squamous cell carcinoma cells exhibit mesenchymal characteristics and GSK3beta-dependent cancer stem cell properties. J Oral Pathol Med 2016;45:180–8. doi: 10.1111/jop.12348. [DOI] [PubMed] [Google Scholar]
- 75. Chen D, Wu M, Li Y, et al. Targeting BMI1(+) cancer stem cells overcomes Chemoresistance and inhibits metastases in squamous cell carcinoma. Cell Stem Cell 2017;20:621–34e6. doi: 10.1016/j.stem.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Noto Z, Yoshida T, Okabe M, et al. CD44 and SSEA-4 positive cells in an oral cancer cell line HSC-4 possess cancer stem-like cell characteristics. Oral Oncol 2013;49:787–95. doi: 10.1016/j.oraloncology.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 77. Kim H, Kim M, Im SK, et al. Mouse Cre-LoxP system: General principles to determine tissue-specific roles of target genes. Lab Anim Res 2018;34:147–59. doi: 10.5625/lar.2018.34.4.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Oshimori N, Oristian D, Fuchs E. TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 2015;160:963–76. doi: 10.1016/j.cell.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Boumahdi S, Driessens G, Lapouge G, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 2014;511:246–50. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 80. Zomer A, Ellenbroek SI, Ritsma L, et al. Intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells 2013;31:602–6. doi: 10.1002/stem.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schepers AG, Snippert HJ, Stange DE, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 2012;337:730–5. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 82. Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol 2005;6:872–84. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 83. Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998;95:379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 84. Habu N, Imanishi Y, Kameyama K, et al. Expression of Oct3/4 and Nanog in the head and neck squamous carcinoma cells and its clinical implications for delayed neck metastasis in stage I/II oral tongue squamous cell carcinoma. BMC Cancer 2015;15:730. doi: 10.1186/s12885-015-1732-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sinha N, Mukhopadhyay S, Das DN, et al. Relevance of cancer initiating/stem cells in carcinogenesis and therapy resistance in oral cancer. Oral Oncol 2013;49:854–62. doi: 10.1016/j.oraloncology.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 86. Ventela S, Sittig E, Mannermaa L, et al. CIP2A is an Oct4 target gene involved in head and neck squamous cell cancer oncogenicity and radioresistance. Oncotarget 2015;6:144–58. doi: 10.18632/oncotarget.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tsai LL, Hu FW, Lee SS, et al. Oct4 mediates tumor initiating properties in oral squamous cell carcinomas through the regulation of epithelial-mesenchymal transition. PLoS One 2014;9:e87207. doi: 10.1371/journal.pone.0087207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bayo P, Jou A, Stenzinger A, et al. Loss of SOX2 expression induces cell motility via vimentin up-regulation and is an unfavorable risk factor for survival of head and neck squamous cell carcinoma. Mol Oncol 2015;9:1704–19. doi: 10.1016/j.molonc.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Freier K, Knoepfle K, Flechtenmacher C, et al. Recurrent copy number gain of transcription factor SOX2 and corresponding high protein expression in oral squamous cell carcinoma. Genes Chromosomes Cancer 2010;49:9–16. doi: 10.1002/gcc.20714. [DOI] [PubMed] [Google Scholar]
- 90. Du L, Yang Y, Xiao X, et al.Sox2 nuclear expression is closely associated with poor prognosis in patients with histologically node-negative oral tongue squamous cell carcinoma. Oral Oncol 2011;47:709–13. doi: 10.1016/j.oraloncology.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 91. Schrock A, Bode M, Goke FJ, et al. Expression and role of the embryonic protein SOX2 in head and neck squamous cell carcinoma. Carcinogenesis 2014;35:1636–42. doi: 10.1093/carcin/bgu094. [DOI] [PubMed] [Google Scholar]
- 92. Lee SH, Oh SY, Do SI, et al. SOX2 regulates self-renewal and tumorigenicity of stem-like cells of head and neck squamous cell carcinoma. Br J Cancer 2014;111:2122–30. doi: 10.1038/bjc.2014.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Deng P, Wang J, Zhang X, et al. AFF4 promotes tumorigenesis and tumor-initiation capacity of head and neck squamous cell carcinoma cells by regulating SOX2. Carcinogenesis 2018;39:937–47. doi: 10.1093/carcin/bgy046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Keysar SB, Le PN, Miller B, et al. Regulation of head and neck squamous cancer stem cells by PI3K and SOX2. J Natl Cancer Inst 2017;109. doi: 10.1093/jnci/djw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Huang CE, Yu CC, Hu FW, et al. Enhanced chemosensitivity by targeting Nanog in head and neck squamous cell carcinomas. Int J Mol Sci 2014;15:14935–48. doi: 10.3390/ijms150914935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xie X, Piao L, Cavey GS, et al. Phosphorylation of Nanog is essential to regulate Bmi1 and promote tumorigenesis. Oncogene 2014;33:2040–52. doi: 10.1038/onc.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tai SK, Yang MH, Chang SY, et al. Persistent Kruppel-like factor 4 expression predicts progression and poor prognosis of head and neck squamous cell carcinoma. Cancer Sci 2011;102:895–902. doi: 10.1111/j.1349-7006.2011.01859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Foster KW, Ren S, Louro ID, et al.Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: Transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ 1999;10:423–34. PMID:10392904. [PubMed] [Google Scholar]
- 99. Abrigo M, Alvarez R, Paparella ML, et al. Impairing squamous differentiation by Klf4 deletion is sufficient to initiate tongue carcinoma development upon K-Ras activation in mice. Carcinogenesis 2014;35:662–9. doi: 10.1093/carcin/bgt349. [DOI] [PubMed] [Google Scholar]
- 100. Tsialikas J, Romer-Seibert J. LIN28: Roles and regulation in development and beyond. Development 2015;142:2397–404. doi: 10.1242/dev.117580. [DOI] [PubMed] [Google Scholar]
- 101. Shyh-Chang N, Zhu H, Yvanka de Soysa T, et al. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 2013;155:778–92. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 103. Chien CS, Wang ML, Chu PY, et al. Lin28B/Let-7 regulates expression of Oct4 and Sox2 and reprograms oral squamous cell carcinoma cells to a stem-like state. Cancer Res 2015;75:2553–65. doi: 10.1158/0008-5472.CAN-14-2215. [DOI] [PubMed] [Google Scholar]
- 104. Wald AI, Hoskins EE, Wells SI, et al. Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck 2011;33:504–12. doi: 10.1002/hed.21475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yu CC, Chen YW, Chiou GY, et al. MicroRNA let-7a represses chemoresistance and tumourigenicity in head and neck cancer via stem-like properties ablation. Oral Oncol 2011;47:202–10. doi: 10.1016/j.oraloncology.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 106. Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 107. Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 2003;423:302–5. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 108. Kaseb HO, Fohrer-Ting H, Lewis DW, et al. Identification, expansion and characterization of cancer cells with stem cell properties from head and neck squamous cell carcinomas. Exp Cell Res 2016;348:75–86. doi: 10.1016/j.yexcr.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. He Q, Liu Z, Zhao T, et al. Bmi1 drives stem-like properties and is associated with migration, invasion, and poor prognosis in tongue squamous cell carcinoma. Int J Biol Sci 2015;11:1–10. doi: 10.7150/ijbs.10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wang Q, Li Z, Wu Y, et al. Pharmacological inhibition of Bmi1 by PTC-209 impaired tumor growth in head neck squamous cell carcinoma. Cancer Cell Int 2017;17:107. doi: 10.1186/s12935-017-0481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lai YJ, Yu WN, Kuo SC, et al. CSC-3436 inhibits TWIST-induced epithelial-mesenchymal transition via the suppression of Twist/Bmi1/Akt pathway in head and neck squamous cell carcinoma. J Cell Physiol 2019;234:9118–29. doi: 10.1002/jcp.27589. [DOI] [PubMed] [Google Scholar]
- 112. Milla LA, Gonzalez-Ramirez CN, Palma V. Sonic hedgehog in cancer stem cells: A novel link with autophagy. Biol Res 2012;45:223–30. doi: 10.4067/S0716-97602012000300004. [DOI] [PubMed] [Google Scholar]
- 113. Tickle C, Towers M. Sonic hedgehog Signaling in limb development. Front Cell Dev Biol 2017;5:14. doi: 10.3389/fcell.2017.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Xu Q, Yuan X, Liu G, et al. Hedgehog signaling regulates brain tumor-initiating cell proliferation and portends shorter survival for patients with PTEN-coexpressing glioblastomas. Stem Cells 2008;26:3018–26. doi: 10.1634/stemcells.2008-0459. [DOI] [PubMed] [Google Scholar]
- 115. Zhou M, Hou Y, Yang G, et al. LncRNA-Hh strengthen cancer stem cells generation in Twist-positive breast cancer via activation of hedgehog Signaling pathway. Stem Cells 2016;34:55–66. doi: 10.1002/stem.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhang C, Li C, He F, et al. Identification of CD44+CD24+ gastric cancer stem cells. J Cancer Res Clin Oncol 2011;137:1679–86. doi: 10.1007/s00432-011-1038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhang K, Che S, Pan C, et al. The SHH/Gli axis regulates CD90-mediated liver cancer stem cell function by activating the IL6/JAK2 pathway. J Cell Mol Med 2018;22:3679–90. doi: 10.1111/jcmm.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Cavicchioli Buim ME, Gurgel CA, Goncalves Ramos EA, et al. Activation of sonic hedgehog signaling in oral squamous cell carcinomas: A preliminary study. Hum Pathol 2011;42:1484–90. doi: 10.1016/j.humpath.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 119. Fan HX, Wang S, Zhao H, et al.Sonic hedgehog signaling may promote invasion and metastasis of oral squamous cell carcinoma by activating MMP-9 and E-cadherin expression. Med Oncol 2014;31:41. doi: 10.1007/s12032-014-0041-5. [DOI] [PubMed] [Google Scholar]
- 120. Nusse R, Clevers H. Wnt/beta-catenin Signaling, disease, and emerging therapeutic modalities. Cell 2017;169:985–99. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 121. Clevers H, Loh KM, Nusse R.. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 122. Felthaus O, Ettl T, Gosau M, et al. Cancer stem cell-like cells from a single cell of oral squamous carcinoma cell lines. Biochem Biophys Res Commun 2011;407:28–33. doi: 10.1016/j.bbrc.2011.02.084. [DOI] [PubMed] [Google Scholar]
- 123. Lim YC, Kang HJ, Kim YS, et al. All-trans-retinoic acid inhibits growth of head and neck cancer stem cells by suppression of Wnt/beta-catenin pathway. Eur J Cancer 2012;48:3310–8. doi: 10.1016/j.ejca.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 124. Warrier S, Bhuvanalakshmi G, Arfuso F, et al. Cancer stem-like cells from head and neck cancers are chemosensitized by the Wnt antagonist, sFRP4, by inducing apoptosis, decreasing stemness, drug resistance and epithelial to mesenchymal transition. Cancer Gene Ther 2014;21:381–8. doi: 10.1038/cgt.2014.42. [DOI] [PubMed] [Google Scholar]
- 125. Le PN, Keysar SB, Miller B, et al. Wnt signaling dynamics in head and neck squamous cell cancer tumor-stroma interactions. Mol Carcinog 2019;58:398–410. doi: 10.1002/mc.22937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wang SJ, Bourguignon LY. Hyaluronan and the interaction between CD44 and epidermal growth factor receptor in oncogenic signaling and chemotherapy resistance in head and neck cancer. Arch Otolaryngol Head Neck Surg 2006;132:771–8. doi: 10.1001/archotol.132.7.771. [DOI] [PubMed] [Google Scholar]
- 127. Argiris A, Harrington KJ, Tahara M, et al. Evidence-based treatment options in recurrent and/or metastatic squamous cell carcinoma of the head and neck. Front Oncol 2017;7:72. doi: 10.3389/fonc.2017.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Tang AL, Hauff SJ, Owen JH, et al. UM-SCC-104: A new human papillomavirus-16-positive cancer stem cell-containing head and neck squamous cell carcinoma cell line. Head Neck 2012;34:1480–91. doi: 10.1002/hed.21962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Perez A, Neskey DM, Wen J, et al. CD44 interacts with EGFR and promotes head and neck squamous cell carcinoma initiation and progression. Oral Oncol 2013;49:306–13. doi: 10.1016/j.oraloncology.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Chatterjee S, Sil PC. Targeting the crosstalks of Wnt pathway with hedgehog and Notch for cancer therapy. Pharmacol Res 2019;142:251–61. doi: 10.1016/j.phrs.2019.02.027. [DOI] [PubMed] [Google Scholar]
- 131. Meurette O, Mehlen P. Notch Signaling in the tumor microenvironment. Cancer Cell 2018;34:536–48. doi: 10.1016/j.ccell.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 132. Chu Q, Orr BA, Semenkow S, et al. Prolonged inhibition of glioblastoma xenograft initiation and clonogenic growth following in vivo Notch blockade. Clin Cancer Res 2013;19:3224–33. doi: 10.1158/1078-0432.CCR-12-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Pannuti A, Foreman K, Rizzo P, et al. Targeting Notch to target cancer stem cells. Clin Cancer Res 2010;16:3141–52. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhao ZL, Zhang L, Huang CF, et al. NOTCH1 inhibition enhances the efficacy of conventional chemotherapeutic agents by targeting head neck cancer stem cell. Sci Rep 2016;6:24704. doi: 10.1038/srep24704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006;7:131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 138. Stemmler MP, Eccles RL, Brabletz S, et al. Non-redundant functions of EMT transcription factors. Nat Cell Biol 2019;21:102–12. doi: 10.1038/s41556-018-0196-y. [DOI] [PubMed] [Google Scholar]
- 139. Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol 2014;16:488–94. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 140. Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wang Z, Li Y, Ahmad A, et al. Targeting miRNAs involved in cancer stem cell and EMT regulation: An emerging concept in overcoming drug resistance. Drug Resist Updat 2010;13:109–18. doi: 10.1016/j.drup.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Santisteban M, Reiman JM, Asiedu MK, et al. Immune-induced epithelial to mesenchymal transition in vivo generates breast cancer stem cells. Cancer Res 2009;69:2887–95. doi: 10.1158/0008-5472.CAN-08-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med 2017;23:1124–34. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 144. Chen C, Wei Y, Hummel M, et al. Evidence for epithelial-mesenchymal transition in cancer stem cells of head and neck squamous cell carcinoma. PLoS One 2011;6:e16466. doi: 10.1371/journal.pone.0016466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Biddle A, Liang X, Gammon L, et al. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res 2011;71:5317–26. doi: 10.1158/0008-5472.CAN-11-1059. [DOI] [PubMed] [Google Scholar]
- 146. Chen YS, Huang WL, Chang SH, et al. Enhanced filopodium formation and stem-like phenotypes in a novel metastatic head and neck cancer cell model. Oncol Rep 2013;30:2829–37. doi: 10.3892/or.2013.2772. [DOI] [PubMed] [Google Scholar]
- 147. Yang MH, Hsu DS, Wang HW, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol 2010;12:982–92. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 148. Pastushenko I, Brisebarre A, Sifrim A, et al. Identification of the tumour transition states occurring during EMT. Nature 2018;556:463–8. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 149. Latil M, Nassar D, Beck B, et al. Cell-type-specific chromatin states differentially prime squamous cell carcinoma tumor-initiating cells for epithelial to Mesenchymal transition. Cell Stem Cell 2017;20:191–204e5. doi: 10.1016/j.stem.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zhang F, Duan S, Tsai Y, et al. Cisplatin treatment increases stemness through upregulation of hypoxia-inducible factors by interleukin-6 in non-small cell lung cancer. Cancer Sci 2016;107:746–54. doi: 10.1111/cas.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Korkaya H, Kim GI, Davis A, et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol Cell 2012;47:570–84. doi: 10.1016/j.molcel.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. El Khoury F, Corcos L, Durand S, et al. Acquisition of anticancer drug resistance is partially associated with cancer stemness in human colon cancer cells. Int J Oncol 2016;49:2558–68. doi: 10.3892/ijo.2016.3725. [DOI] [PubMed] [Google Scholar]
- 153. Bourguignon LY, Wong G, Earle C, et al. Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem 2012;287:32800–24. doi: 10.1074/jbc.M111.308528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Lee J, Park M, Ko Y, et al. Ectopic overexpression of CD133 in HNSCC makes it resistant to commonly used chemotherapeutics. Tumour Biol 2017;39:1010428317695534. doi: 10.1177/1010428317695534. [DOI] [PubMed] [Google Scholar]
- 155. Raha D, Wilson TR, Peng J, et al. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res 2014;74:3579–90. doi: 10.1158/0008-5472.CAN-13-3456. [DOI] [PubMed] [Google Scholar]
- 156. Kim J, Shin JH, Chen CH, et al. Targeting aldehyde dehydrogenase activity in head and neck squamous cell carcinoma with a novel small molecule inhibitor. Oncotarget 2017;8:52345–56. doi: 10.18632/oncotarget.17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Saintigny P, WilliamWN, Jr., Foy JP, et al. Met receptor tyrosine kinase and chemoprevention of Oral cancer. J Natl Cancer Inst 2018;110. doi: 10.1093/jnci/djx186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Tsai LL, Yu CC, Chang YC, et al. Markedly increased Oct4 and Nanog expression correlates with cisplatin resistance in oral squamous cell carcinoma. J Oral Pathol Med 2011;40:621–8. doi: 10.1111/j.1600-0714.2011.01015.x. [DOI] [PubMed] [Google Scholar]
- 159. Colnot DR, Quak JJ, Roos JC, et al. Phase I therapy study of 186Re-labeled chimeric monoclonal antibody U36 in patients with squamous cell carcinoma of the head and neck. J Nucl Med 2000;41:1999–2010. PMID:11138685. [PubMed] [Google Scholar]
- 160. Odenthal J, Rijpkema M, Bos D, et al. Targeting CD44v6 for fluorescence-guided surgery in head and neck squamous cell carcinoma. Sci Rep 2018;8:10467. doi: 10.1038/s41598-018-28059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Setubal Destro Rodrigues MF, Gammon L, Rahman MM, et al. Effects of cetuximab and erlotinib on the behaviour of cancer stem cells in head and neck squamous cell carcinoma. Oncotarget 2018;9:13488–500. doi: 10.18632/oncotarget.24416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Deng R, Wang X, Liu Y, et al. A new gamboge derivative compound 2 inhibits cancer stem-like cells via suppressing EGFR tyrosine phosphorylation in head and neck squamous cell carcinoma. J Cell Mol Med 2013;17:1422–33. doi: 10.1111/jcmm.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Subramanian C, Kovatch KJ, Sim MW, et al. Novel C-terminal heat shock protein 90 inhibitors (KU711 and Ku757) are effective in targeting head and neck squamous cell carcinoma cancer stem cells. Neoplasia 2017;19:1003–11. doi: 10.1016/j.neo.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Moy JD, Moskovitz JM, Ferris RL. Biological mechanisms of immune escape and implications for immunotherapy in head and neck squamous cell carcinoma. Eur J Cancer 2017;76:152–66. doi: 10.1016/j.ejca.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): An open-label, multicentre, phase 1b trial. Lancet Oncol 2016;17:956–65. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 166. Harrington KJ, Ferris RL, BlumenscheinG, Jr.,et al. Nivolumab versus standard, single-agent therapy of investigator’s choice in recurrent or metastatic squamous cell carcinoma of the head and neck (CheckMate 141): Health-related quality-of-life results from a randomised, phase 3 trial. Lancet Oncol 2017;18:1104–15. doi: 10.1016/S1470-2045(17)30421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Miao Y, Yang H, Levorse J, et al. Adaptive immune resistance emerges from tumor-initiating stem cells. Cell 2019;177(5):1172‑1186.e14. doi: 10.1016/j.cell.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Tsai CF, Hsieh TH, Lee JN, et al. Curcumin Suppresses Phthalate-Induced Metastasis and the Proportion of Cancer Stem Cell (CSC)-like Cells via the Inhibition of AhR/ERK/SK1 Signaling in Hepatocellular Carcinoma. J Agric Food Chem. 2015;63(48):10388–98. [DOI] [PubMed] [Google Scholar]
- 169. Baharuddin P, Satar N, Fakiruddin KS, et al. Curcumin improves the efficacy of cisplatin by targeting cancer stem-like cells through p21 and cyclin D1-mediated tumour cell inhibition in non-small cell lung cancer cell lines. Oncol Rep. 2016;35(1):13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Sun Y, Ding Y, Li D, et al. Cyclic Depsipeptide BE-43547A2 : Synthesis and Activity against Pancreatic Cancer Stem Cells. Angew Chem Int Ed Engl. 2017;56(46):14627–31. [DOI] [PubMed] [Google Scholar]
- 171. Sztiller-Sikorska M, Koprowska K, Majchrzak K, et al. Natural compounds' activity against cancer stem-like or fast-cycling melanoma cells. PLoS One. 2014;9(3):e90783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Jiang P, Xu C, Zhou M, et al. RXRalpha-enriched cancer stem cell-like properties triggered by CDDP in head and neck squamous cell carcinoma (HNSCC). Carcinogenesis. 2018;39(2):252–62. [DOI] [PubMed] [Google Scholar]
- 173. Lin HC, Wu CL, Chen YL, et al. High-level beta1-integrin expression in a subpopulation of highly tumorigenic oral cancer cells. Clin Oral Investig 2014;18:1277–84. doi: 10.1007/s00784-013-1088-y. [DOI] [PubMed] [Google Scholar]
- 174. Ahsan MS, Yamazaki M, Maruyama S, et al. Differential expression of perlecan receptors, alpha-dystroglycan and integrin beta1, before and after invasion of oral squamous cell carcinoma. J Oral Pathol Med 2011;40:552–9. doi: 10.1111/j.1600-0714.2010.00990.x. [DOI] [PubMed] [Google Scholar]
- 175. Yen YC, Hsiao JR, Jiang SS, et al. Insulin-like growth factor-independent insulin-like growth factor binding protein 3 promotes cell migration and lymph node metastasis of oral squamous cell carcinoma cells by requirement of integrin beta1. Oncotarget 2015;6:41837–55. doi: 10.18632/oncotarget.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Mohanta S, Siddappa G, Valiyaveedan SG, et al. Cancer stem cell markers in patterning differentiation and in prognosis of oral squamous cell carcinoma. Tumour Biol 2017;39:1010428317703656. doi: 10.1177/1010428317703656. [DOI] [PubMed] [Google Scholar]
- 177. Mishra A, Sriram H, Chandarana P, et al. Decreased expression of cell adhesion genes in cancer stem-like cells isolated from primary oral squamous cell carcinomas. Tumour Biol 2018;40:1010428318780859. doi: 10.1177/1010428318780859. [DOI] [PubMed] [Google Scholar]
- 178. Athanassiou-Papaefthymiou M, Shkeir O, Kim D, et al. Evaluation of CD44 variant expression in oral, head and neck squamous cell carcinomas using a triple approach and its clinical significance. Int J Immunopathol Pharmacol 2014;27:337–49. doi: 10.1177/039463201402700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Martens-de Kemp SR, Brink A, Stigter-van Walsum M, et al. CD98 marks a subpopulation of head and neck squamous cell carcinoma cells with stem cell properties. Stem Cell Res 2013;10:477–88. doi: 10.1016/j.scr.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 180. Yap TA, Olmos D, Brunetto AT, et al. Phase I trial of a selective c-MET inhibitor ARQ 197 incorporating proof of mechanism pharmacodynamic studies. J Clin Oncol 2011;29:1271–9. doi: 10.1200/JCO.2010.31.0367. [DOI] [PubMed] [Google Scholar]


