Abstract
Autophagy and inflammasomes are shown to interact in various situations including infectious disease, cancer, diabetes and neurodegeneration. Since multiple layers of molecular regulators contribute to the interplay between autophagy and inflammasome activation, the detail of such interplay remains largely unknown. Non-coding RNAs (ncRNAs), which have been implicated in regulating an expanding list of cellular processes including immune defense against pathogens and inflammatory response in cancer and metabolic diseases, may join in the crosstalk between inflammasomes and autophagy in physiological or disease conditions. In this review, we summarize the latest research on the interlink among ncRNAs, inflammasomes and autophagy and discuss the emerging role of these three in multiple signaling transduction pathways involved in clinical conditions. By analyzing these intriguing interconnections, we hope to unveil the mechanism inter-regulating these multiple processes and ultimately discover potential drug targets for some refractory diseases.
Keywords: ncRNAs, innate and adaptive immunity, inflammasome, autophagy, precision medicine
Introduction
Autophagy (self-eating), first described by Ashford and Porter in 19621, is a ubiquitous eukaryotic cellular process to provide the metabolic needs and the renewal of some organelles. The important step of autophagy (macroautophagy) process is the formation of autophagophore, which needs the preinitiation complex to activate the Class III PI3K complex (Bclin1, ATG14L, VPS34 and VPS15), and generate PI3P at the site of nucleation of the isolation membrane, finally forming the phagophore2. Then an elongation reaction enlarges with phosphatidylethanolamine-conjugated LC3 until forming an autophagosome. Lysosome then fuses with autophagosome and degrades cargoes. Chaperone-mediated autophagy can select cargoes with specific target sequences to lysosome. Thus, autophagy can be classified as selective and nonselective ones. The selective process can specifically remove protein aggregates (aggrephagy), organelles such as peroxisomes (pexophagy), mitochondria (mitophagy), endoplasmic reticulum (reticulophagy), ribosomes (ribophagy), lipids (lipophagy) and bacteria (xenophagy). Growing evidence indicates that autophagy plays a key part in a broad scope of cellular functions including metabolism, inflammation, and immunity3,4, but the actual role and underlying mechanism of autophagy in the physiological and pathological processes remains incompletely understood.
As a barrier to bacterial infection, the autophagic pathway has multiple functions, such as the degradation of macromolecules5, an alternative source of anabolic nutrients under starvation6, and the degradation of intracellular pathogens as a means against infection7. On the other hand, certain bacteria, such as Pseudotuberculosis8, Yersinia pestis9 and Chlamydia trachomatis,10 can survive and thrive by taking advantage of the autophagy machinery to evade the host defense11. But it is evident that autophagy can be modulated to enable an effective immune response. Therefore, autophagy may serve as an auxiliary control measure for some infectious diseases but may not work for infections of other microorganisms. Moreover, autophagy has a similar complex role in cancer and metabolic diseases12, thus of great impact on human life. We strongly believe that further dissecting the mechanisms of the survival or death of pathogens in autophagic conditions should lead to better strategies for effective treatment of different diseases especially infectious diseases.
By crosstalk with autophagy, inflammasomes influence the effector cells of innate and adaptive immunity during inflammatory response13. Inflammasome recognizes pathogens like bacteria and damage-associated molecular pattern components via specific receptors, such as NOD-like receptors (NLRs) (Fig. 1)14. Classic inflammasomes including NLRP1, NLRP3, NLRC4 and AIM2 inflammasomes directly or indirectly activate caspase-1, resulting in a highly inflammatory form of programmed cell death (pyroptosis) and facilitating maturation of IL-1β and IL-183,15–17. Among them, NLRP1 is relatively unique in the family of NLRs containing FIIND (function-to-find domain) domain and CARD (caspase activation and recruitment domain) domain at the C-terminus of the protein. NLRP1 was earlier reported to be activated by hydrolysis of the lethal factor (LF) of Bacillus anthracis and the mechanisms were recently characterized15. Vance et al. uncovered that LF could hydrolyze the N-terminal peptide of NLRP1 to produce a new N-terminal. This new N-terminus promotes the degradation of NLRP1 via the N-end rule proteasomal degradation pathway, while the FIIND domain is hydrolyzed by proteasomal degradation and forms a CARD domain at the C-terminus (984aa-1233aa), thereby completing inflammasome assembly and activation processes16. Up to date, NLRP3 appears to be the most well studied inflammasome. As an important component of innate immunity, NLRP3 plays an important role in both immune response and a variety of disease processes17, including Type 2 diabetes18, Alzheimer's Disease19, and atherosclerosis20. Therefore, as the core of the inflammatory response, NLRP3 inflammasome studies may indicate new targets for the treatment of various inflammatory diseases. Due to the diversity of NLRP3 agonists, the specific mechanism of action and signal axis of agonist-receptor proteins remain to be fully understood. Different from NLRP3 and other NLRs, the NLRC4 inflammasome does not contain PYD domain, whereas its CARD domain directly binds with Caspase 1 to coordinate complex assembly. The function of NLRC4 is closely associated with the NAIP protein (NLR family apoptosis inhibitory protein), which can bind to flagellin and type III secretion apparatus and other bacterial structural components20,21. Mechanistically, the NAIP protein uses a catalytic surface that is highly similar to the NLRC4 protein to initiate a “self-replication” activation process of the NLRC4 protein, and avoiding recognition of the NAIP’s own surface ensures the specific activation for NLRC4 inflammasome21. AIM2 inflammasome is characterized as a DNA inflammasome by its ability to detect foreign dsDNA including viral dsDNA, bacterial dsDNA and even broken host dsDNA and mitochondria DNA22. Hence, AIM2 inflammasome is a very important intracytoplasmic sensor, which is involved in infection and autoimmune disease. Although there is evidence indicating that NLRP2, 6, 7, 12 and IFI16 inflammasomes can also form inflammasome complexes, their specific physiological functions require further details3. Unlike classic inflammasome, non-classic inflammasome caspase-11 senses bacterial lipopolysaccharide (LPS) and also regulates IL-1β and IL-18 expression17. The overexpressed IL-1β and IL-18 then accelerate the deterioration of inflammation factors. On the contrary, some pathogen-associated proteins produced after infection can inhibit the formation and activation of inflammasome leading to the decay of IL-1β and IL-18, which may hamper pathogen eradication and trigger chronic infection or result in cell death23. Therefore, an alternative approach to clearing the pathogens, such as autophagy, may serve as a compensatory mechanism to strengthen host defense by modulating inflammasome-driven responses.
Figure 1 .
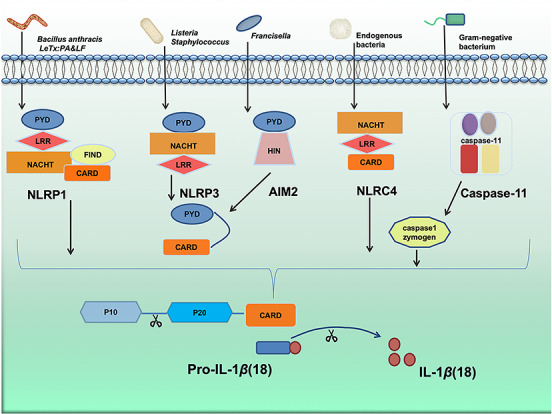
The process of bacteria activating inflammasomes. Different bacteria activate different inflammasomes. Bacillus anthracis LeTx activate NLRP1 by PA & LF. Listeria and Staphylococcus aureus activate NLRP3 by bacterial pore-forming toxins, bacterial RNA, and bacterial cell wall components LPS. The endogenous bacteria and Francisella active NLRC4 and bacterial DNA sensor including AIM2, NLRP3, and NLRP1 inducing the cleavage of caspase-1 to release IL-1β and IL-18. Gram-negative bacterial LPS activate the non-classical inflammasome caspase-11. PA, protective antigen; LF: lethal factor; LPS, lipopolysaccharide; NLRP1, NACHT, LRR and PYD domains-containing protein 1; NLRC4, NLR family CARD domain-containing protein 4; NLRP3, NACHT, LRR and PYD domains-containing protein 3; AIM2, absent in melanoma 2; IL-1β, interleukin 1 beta; IL-18, interleukin 18.
A non-coding RNA (ncRNA) refers to an RNA that does not encode a protein, which are often classified into different categories based on the length: transfer RNAs and ribosomal RNA, small RNA [microRNA (miRNA) and siRNA (small interfering RNA)]24, piRNA (Piwi-interacting RNA), small nucleolar RNAs (snoRNA), nuclear RNAs (snRNAs), repeat associated small interfering RNAs (scaRNAs), circular RNA (circRNA), long intergenic noncoding RNA (lincRNA) and long noncoding RNA (lncRNA)25–28. A regulatory RNA primarily exercises its biological function at post transcription levels. Approximately 75% of the human genome can be transcribed into RNAs, but 74% of which are not translated into proteins29. ncRNAs play an important role in many biological processes, such as in embryonic formation and development, gene expression, and metabolism30. ncRNAs are also found to be associated with tumor triggering, development, progression and metastasis31, as well as diseases including diabetes, metabolic syndrome32, colitis33, autoimmunity34 and neurodegeneration35. With the development of genomics and bioinformatics tools, especially the extensive application of high-throughput sequencing technology and single cell RNA sequencing (scRNA-Seq), increasing regulatory mechanisms of non-coding RNAs have been unfolded. Recently, ncRNAs, especially microRNAs, have been found implicated in the development of inflammatory responses and autophagy36, suggesting that the ncRNA is a potential precise target for treating autophagy-driven and inflammasome-associated diseases. For determining such a target, it is necessary to better explore the effects of ncRNAs on the interplay between autophagy and inflammasomes. Herein we are aimed at systematically analyzing the current understanding about the intersection of the role and mechanism among autophagy, inflammasome and ncRNAs in infection and inflammatory diseases.
Interaction between autophagy and inflammasome-mediated process
Researches in the crosstalk between autophagy and inflammasome activation are mostly known in pathological progression of bacterial infection (Fig. 2). Activation of the inflammasome and autophagy is critically important for host defenses against microbial invasion37. These two entities coordinate to enhance host defense and mammals are fortunate to have such a powerful pair of regulatory mechanisms to keep various diseases in check. However, the fact is that autophagy and inflammasome not only function in their own ways but also form complex interrelationship to exert an even stronger and critical impact in physiological and pathophysiological conditions. Although both inflammasome activation and autophagy play beneficial roles in host defense against infection, they do have distinct and sometimes contrary effects. Inflammasome activation leads to inflammatory responses38 and even results in cell death (pyroptosis), while autophagy tends to inhibit inflammation4. When autophagy is impaired in monocytes, IL-1β would be increased along with retention of dysfunctional mitochondria39. Importantly, autophagy down-regulates inflammasome activation by removing defective mitochondria (mitophagy)40. However, certain pathogens (e.g., Salmonella) can subvert host immune responses through hiding in autophagic vacuoles to avoid phagocytic clearance41. On the other hand, inflammasome components are also shown to regulate autophagy42. Furthermore, bacterial virulence factors or other induction factors may impact host response. The latest research in our lab also indicates that the CRISPR-Cas adaptive immune system may regulate host defense and alter inflammasome activation by augmenting autophagy43. Taken together, there is apparent complicate interaction between autophagy and the inflammasome-mediated process. Understanding the underlying mechanisms may help create better weapons against bacterial infection.
Figure 2 .
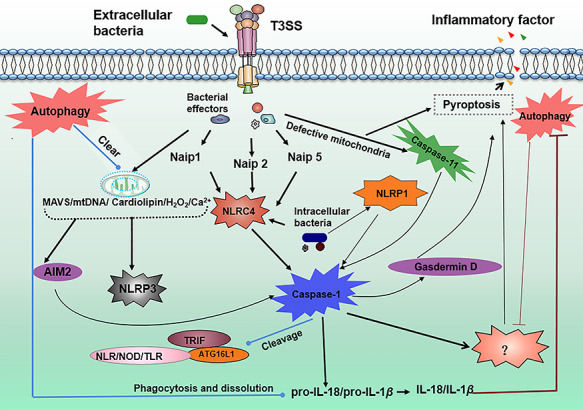
Interplay between autophagy and inflammasome activation during bacterial infection. Extracellular inject virulence factors into host cells via T3SS. Some of the virulence factors activate NLRC4 mediated by NAIP proteins, and other virulence factors have strong destructive power to mitochondria. The damaged mitochondria release mtDNA, cardiolipin, ROS, and other activators of AIM2 and NLRP3. Then, AIM2, NLRP3, Caspase-11 and NLPC4 combine with caspase-1 to form inflammasomes. Activated inflammasomes cleave proteins that are important for autophagy progress, such as ATG16L1, TRIF, and other receptor proteins, and trigger pyroptosis and liberate a number of inflammatory factors. However, activated autophagy during bacterial infection will attenuate pyroptosis and clear defective mitochondria which leads to the downregulation of such activators as mtDNA, cardiolipin, ROS, and Ca2+ of inflammasomes. Autophagy can also swallow proIL-18 and IL-1β which are necessary for mature inflammatory factors. NLRC4, NLR family CARD domain-containing protein 4; NLRP3, NACHT, LRR and PYD domains-containing protein 3; AIM2, absent in melanoma 2; mtDNA, mitochondria DNA; ROS, Reactive oxygen species.
A question follows naturally: which of these processes is more important, or the dominant, in the interaction between autophagy and inflammasome? Recent studies have uncovered that deletion of LC3B and Atg16L1, or depletion of Beclin1 along with introduction of an Atg4B dominant negative construct could apparently increase IL-1β secretion, showing that autophagy is an integral regulator of the inflammasome via regulation of ROS and autophagosomal degradation44,45. ROS, produced by NADPH oxidases in mitochondria in response to bacterial infection, requires pattern recognition receptors (PRR) such as TLRs, NLRs or lectins to activate inflammasomes46,47. One explanation is that inflammasomes are inhibited under autophagic condition48. In the deficiency of autophagy-related genes, such as atg5, atg7, atg12, and atg16L1 (which are implicated in regulating endotoxin-induced inflammasome)49, autophagy does not function normally, resulting in accumulation of dysfunctional mitochondria and increased ROS release. Inflammasomes in innate immune cells recognize ROS and produce proinflammatory cytokines50, while autophagy can reduce the accumulation of mitochondria by clearing the dysfunctional ones and peroxisomes, to slow down inflammatory responses. Collectively, these reports demonstrate that while ROS may enhance inflammation, it also activates autophagy to serve as a negative feedback during bacterial infection.
Moreover, inflammasomes are also regulated by autophagy through augmenting or decreasing IL-1β secretion, a determinant for inflammatory response51. Other evidence supports that ubiquitination of ASC is detectable upon absent in melanoma 2 (AIM2) stimulation regulatory proteins, which are able to stimulate autophagy in response to the invasion of bacteria52. A portion of ASC-containing inflammasomes are found to be redirected towards autophagosomes and autophagolysosomes upon NLRP3 or AIM2 activation53, indicating that autophagy often accompanies inflammasome activation. As discussed above, NLRs are cytoplasmic receptors that play a crucial role in the innate immune response by recognizing bacteria and initiating a fierce inflammatory reaction. These findings together indicate that balanced inflammatory responses are resulted from the NLRs controlling or coordinating autophagy47. Some bacteria are reported to involve interaction between inflammasome and autophagy (Table 1).
Table 1.
Microbes activate inflammasome and autophagy.
| Microbe | Adaptation in inflammasome | Adaptation in autophagy. | Ref |
| Mycobacterium tuberculosis | Mycobacterium tuberculosis can inhibit the expression of IL-1β through metalloproteinase being encoded by zmp1 gene. | IL-4 and IL-13 inhibit IFNγ or starvation-induced autophagic elimination of M. tuberculosis. | [54] [55] |
| P. aeruginosa | P. aeruginosa import effector protein ExoU and restrain the activation of caspase-1 and IL-1β. | Caspase-1 cleavage of TRIF thus inhibits autophagy and β-interferon production during P. aeruginosa infection. | [56] [57] |
| Bacillus anthracis | Bacillus anthracis lethal toxin induced activation of caspase-1 was able to attenuate proIL-1β due to the cell death and the activation of the caspase-1. | Autophagy induction may protect cells against anthrax lethal toxin. | [58] [59] |
| Mycobacterium marinum | M. marinum secretion system ESX-1 plays an important role in the process of escaping from the host cell’s phagocytosis. | M. marinum are escorted by a distinct polar autophagocytic vacuole. Cell-to-cell transmission is inhibited once autophagy is impaired and the host cells die. | [60] [61] |
| Yersinia pseudotuberculosis | Effector molecules YopK and T3SS interact and inhibit host recognition of T3SS and activation of inflammasome. YopK defects can enhance the activity of inflammasome and host bacterial clearance. | Cells showed significant cytoplasmic localization of p53 and reduced LC3-I to LC3-II conversion responding to Yersinia pseudotuberculosis infection | [62] [63] |
| Staphylococcus aureus | IL-1β produced by the activation of inflammatory cells can promote chemotaxis of neutrophils and bacterial clearance. | Atg5 deficiency inhibits bacterial degradation in autolysosomes. | [64] [65] |
| Shigella flexneri | Shigella flexneri concerts modulation of pro-death and pro-survival signaling pathways to circumvent the innate immune response. | IcsB and VirA act synergistically at vicinity of the vacuole membrane to allow Shigella flexneri to escape from LC3-positive vacuoles. | [66] [67] |
| Listeria monocytogenes | Inflammasome-mediated inhibition of Listeria monocytogenes-stimulated immunity is important for bacterial accumulation in CD8α (+) DCs. | L. monocytogenes prevents phagosomes forming. | [68] [69] |
| Porphyromonas gingivalis (Pg) | P. gingivalis reduces IL-1β secretion and escapes host immune response, owing to P. gingivalis and Pg-LPS differentially controlled the NLRP3 inflammasome pathway in endothelial cells. | The bacteria evade endocytic trafficking to lysosomes by trafficking to autophagosomes. | [70] [71] |
|
Legionella
pneumophila |
L. pneumophil may have encountered little selective pressure to evade PRRs recognition and immune responses. | L. pneumophilal replication vacuoles have autophagy markers. It is postulated that the bacterium delays autophagosome maturation early after infection. | [72] [73] |
| Burkholderia pseudomallei | The specific mutants of B. pseudomallei eg. sifA and sdhA, aberrantly enter the cytosol and trigger caspase-11 protects against bacteria that escape the vacuole. | B. pseudomalle prevents bacterial colocalization with LC3 autophagosome marker. | [74] [75] |
Crosstalk between autophagy and inflammasome activation is not limited to infections. Both autophagy and inflammasome are related to protein secretion, such as end-binding protein 1 (EB1)76, which seems to be a reciprocal mechanism. Furthermore, the dectin-1/Syk pathway activates unconventional, vesicle-mediated protein secretion that is dependent on both inflammasome complex assembly and autophagy activity in human macrophages77. TRIM20 (a subset of tripartite motif proteins) and TRIM21 were reported to directly bind with their respective cargo, leading to autophagic degradation. Further studies suggest that TRIM20 targets inflammasome components, such as NLRP3, NLRP1, and pro-caspase-1 for autophagic degradation, whereas TRIM21 targets IRF378. In addition, autophagy is shown to regulate the secretion of proinflammatory cytokines through deletion of Atg16L1, an Atg5-Atg12 binding protein, which triggers the production and release of large amounts of inflammatory cytokines in response to LPS and other pathogen associated molecular patterns (PAMPs) of bacterial pathogens in mice. It will be interesting to understand more details of the interaction between autophagy and inflammasome modulation, as our knowledge in this interplay is only a tip of the iceberg.
ncRNAs intermediating between inflammasomes and autophagy
miRNAs are small RNAs about 18-22 nucleotides and function at RNA silencing through base-pairing with complementary sequences of three prime untranslated region (3’-UTR) or 5’-UTR of mRNA to regulate transcriptional gene expression79. For example, miRNA-223 was reported to target NLRP3 3’-UTR to suppress NLRP3 protein expression80,81, indicating that inflammasomes can be repressed by ncRNA (miRNA). Similarly, miRNAs are able to target the autophagy associated genes82, which highlights the double role of miRNAs in regulating both inflammasomes and autophagy. Tables 2 and 3 list the miRNAs and lncRNAs that are known to be involved in inflammasomes or autophagy machineries in different biological processes and their functional mechanisms. Except for miRNA103, all other miRNAs appear to regulate inflammasome and autophagy by targeting mRNA 3’-UTR of different genes. Among them, miR-301b and miR-302b were reportedly involved in Pseudomonas aeruginosa and Klebsiella pneumoniae infection-induced inflammation, which may have potential link with inflammasome and require further investigation83. Likewise, a recent study reported that P. aeruginosa infection may be linked to inflammasomes84 but its link with other miRNAs needs to be examined. Therefore, further studies are needed to find out the role of miRNAs and inflammasomes in other situations.
Table 2.
Crosstalk of ncRNAs and inflammasomes.
| ncRNAs | Function | Interaction | Ref |
| microRNA7 | Controlling mRNA expression | NLRP3 as a target gene of miR-7 | [132] |
| microRNA155 | Roles in physiological and pathological processes | Increasing inflammasome-associated gene expression | [133] |
| microRNA377 | A biomarker of oxidative stress | Activating O2(−)/p38 MAPK/TXNIP/NLRP3 inflammasome pathway when overexpressed | [134] |
| microRNA223 | Regulating expression levels of other genes | Binding to NLRP3 3’-UTR sites | [80] |
| microRNA143 | Regulating expression levels of other genes | miR-143 increasing the expression of AIM2 and ASC mRNAs | [135] |
| microRNA20a | Controlling mRNA expression | Regulating expression of NLRP3 by targeting TXNIP | [136] |
| microRNA133a-1 | Regulating expression levels of other genes | Suppressing inflammasome activation trough UCP2 | [137] |
| microRNABART15 | Epstein-Barr virus miRNA | Targeting NLRP3 3′-UTR | [138] |
| microRNA9 | Inhibiting target mRNAs by binding to their 3′-UTRs | Targeting ELAVL1 to inhibit pyroptosis | [139] |
| microRNA21 | Inhibiting phosphatases | Activating NLRP3 in liver fibrosis | [140] |
| microRNA92a | Inhibiting endothelial cell angiogenesis | SREBP2-miR-92a-inflammasome responding to oxidative stress | [141] |
| microRNA146a | Anti-inflammatory microRNA | Upregulating inflammasome gene activation in diabetic nephropathy | [142] |
| microRNA-30c-5 | Regulating expression levels of other genes | Inhibiting NLRP3 inflammasome in atherosclerosis | [143] |
| microRNA-495 | A tumor-suppressor | Suppressing NLRP3 signaling pathway in endothelial cell injury | [144] |
| miR-186 | Controlling mRNA expression | Suppressing NLRP3 signaling to control neuropathic pain | [75] |
| lncRNA XIST | Involved in cell proliferation migration, inflammation process and apoptosis | Inhibiting bacteria induced production of NLRP3 inflammasome | [76] |
| lncRNA ANRIL | Participating in NF-κB-mediated signaling | Increasing NLRP3 activation in nephropathy | [77] |
| lncRNA SNHG1 | A competing endogenous RNA | Regulating NLRP3 Pathway in parkinson’s disease | [78] |
| lncRNA Neat1 | Regulating biological processes | Increasing activation of inflammasomes in in innate immunity | [145] |
Table 3.
Crosstalk of ncRNAs and autophagy.
| ncRNAs | Function | Interaction | Ref |
| microRNA24-3p | Regulating gene expression | Suppressing DEDD transcription | [146] |
| microRNA539 | Regulating gene expression | Targeting the 3’-UTR of mek which suppressed autophagy in H9C2 cells | [147] |
| antimicroRNA30b | Targeting 3′-UTR of genes | miR-30b represses autophagy then promotes TNF-α-induced apoptosis | [148] |
| microRNA221 | Targeting tumor suppressors | Targeting the autophagy gene beclin-1 in tumor cell death | [149] |
| microRNA27a | A brain-specific miRNA | Binding to FoxO3a mRNA which regulates autophagy | [150] |
| microRNA199a-5p | Regulating gene expression | Inhibiting cisplatin-induced drug resistance by inhibiting of autophagy process | [151] |
| microRNA128 | A brain-enriched miRNA involved in gene expression regulating | Repressing mTOR signaling that regulates cytotoxicity | [152] |
| microRNA103/107 | One of the only known miRNA which can target 5’-UTR | Preserving end-stage of autophagy via regulating diacylglycerol kinase C signal pathway | [153] |
| microRNA99 | Antiviral miRNA | Increasing autophagy during hepatitis B virus infection | [154] |
| microRNA146a | A mediator of inflammation | Repressing Bcl-2 and promoting autophagy in Hypoxia condition | [155] |
| microRNA20a | Regulating gene expression | Targeting ATG7 and ATG16L1 in macrophage cells | [156] |
| microRNA21 | Regulating gene expression | Inhibiting autophagy via PTEN/Akt/HIF-1α and Akt-mTOR pathways | [157] |
| microRNA34a | Regulating gene expression | Mediating SIRT1/mTOR autophagy pathway | [158] |
| microRNA23a | Regulating the expressions of other genes | Repressing autophagy in premature senescence | [159] |
| microRNA144* | Regulating the expression of genes involved in erythropoiesis and other process | Targeting the autophagy protein DRAM2 during mycobacterium tuberculosis infection. | [160] |
| microRNA195 | Affecting stability and translation of mRNAs | Targeting GABARAPL1 which is a upstream regulator of autophagy under hypoxic conditions | [161] |
| microRNA222 | A known extracellular RNA | Inhibiting autophagy after cardiac-specific overexpression | [162] |
| microRNA140-5p | A miRNA signature of kinds tumors | Regulating IP3k2 induced drug resistance by inducing autophagy in osteosarcoma cells | [163] |
| microRNA22 | Binding to the 3’-UTR of other mRNAs | Targeting p38α to regulate regulates starvation-induced | [164] |
| microRNA124-3p | Neuronal cell associated miR | Inhibiting autophagy gene beclin-1 in breast cancer cell | [165] |
| microRNA30a-3p | Highly expressed in heart cells | Targeting BECN1 to regulate autophagy during L. donovani infection | [166] |
| microRNAlet-7 g | Regulating gene expression | Regulating autophagy by rapamycin signaling in granulosa cells | [167] |
| microRNA153 | Regulating gene expression | Regulating autophagy through targeting Mcl-1 in cardiomyocytes | [168] |
| microRNA384-5p | Mainly functioning in neural system. | Targeting Beclin-1 to regulate autophagy in macrophage cell | [169] |
| microRNA965 | Shrimp miR | Targeting the autophagy gene ATG5 against virus infection | [170] |
| microRNA126 | Regulating gene expression | Alerting cell metabolism to induce autophagy in malignant mesothelioma | [171] |
| microRNA19a-39/19b-3p | Oxidative stress associated miR | Targeting TGF-beta1 to inhibit autophagy | [172] |
| microRNA33 | Strongly associated with lipid metabolism | Reprograming autophagy induced by Mycobacterium tuberculosis | [173] |
| microRNA299-5p | Regulating gene expression | Targeting autophagy gene atg5 to control apoptosis | [174] |
| microRNA141 | Regulating gene expression | Targeting HMGB1 to regulate autophagy | [175] |
| microRNA181a-5p | A miR involved in multiple processes | Repressing autophagy in during detachment induction | [176] |
| microRNA376 | Important for cancer formation | Regulating macroautophagy | [177] |
| microRNA26b | Playing roles in hypoxia, neuronal differentiation, hepatocellular carcinoma, etc. | Targeting ULK2 to inhibit autophagy in cancer cells | [178] |
| microRNA495 | Regulating gene expression | Targeting autophagy gene atg3 during starvation | [179] |
| microRNA1273g-3p | Mainly involved in HIF-1 signaling pathway and the nervous system | Regulating glucose fluctuation-induced autophagy | [180] |
| microRNA301a/b | Regulating gene expression | Increasing cell autophagy in hypoxia condition | [181] |
| microRNA142a-5p | Targeting 3′-UTR of mRNAs | Regulating beclin-1-mediated autophagy after LPS-Induced | [182] |
| microRNA1303 | Regulating gene expression | Targeting autophagy gene atg2b during bacterial infection | [183] |
| microRNA129-5p | Regulating gene expression | Targeting HMGB1 in breast cancer | [184] |
| microRNA183 | Regulating gene expression | Targeting UVRAG, a regulator of autophagy and apoptosis in colorectal cancer | [185] |
| microRNA96 | Targeting 3’-UTR of genes | Targeting MTOR or ATG7 respectively at different expression levels | [186] |
| microRNALET7I | Regulating gene expression | Increasing autophagy via targeting IGF1R to protect T cell from death | [187] |
| microRNA451 | Regulating gene expression | Targeting TSC1 to regulate autophagy | [188] |
| microRNA204-5P | Cancer associated miR | Inhibiting activity of LC3B-II | [189] |
| microRNA200C | Mainly associated with cancer | Targeting UBQLN1 to inhibit autophagy in breast cancer cells | [190] |
| microRNA17-5p | Regulating gene expression | Targets Mcl-1 and STAT3 to active autophagy in macrophages after mycobacterium tuberculosis infection | [191] |
| microRNA212 | Regulating gene expression | Inhibiting starvation induced autophagy in cancer cells by targeting SIRT1 | [192] |
| microRNA14-3p | Mainly involved in cardiac morphogenesis and cancer | Targeting GABARAPL1 which inhibits autophagy in gastric cancer cells | [193] |
| microRNA497 | Regulating gene expression | Repressing autophagy in reoxygenation injury in cardiomyocytes | [194] |
| microRNA23b-3p | Mainly involved in cancer | Targeting autophagy genes atg12 and hmgb2 in gastric cancer cells | [195] |
| microRNA125a | Mainly involved in monocytes during mycobacterium infection | Blocking M. tuberculosis-induced autophagy | [196] |
| microRNA188-3p | Inducing autophagic cell death in cancer cells | Targeted by lncRNA APF and targeting atg7 | [197] |
| microRNA423-5p | Regulating gene expression. | Increasing autophagy activity in hepatocellular carcinoma | [198] |
| microRNA638 | Regulating gene expression | Targeting TFAP2A/AP-2α and regulating autophagy in melanoma cells | [199] |
| microRNA214 | Associated with various cancers with functions varying in different tissues | Targets UCP2 to promote autophagy in breast cancers | [200] |
| microRNA20a | Hypoxia sensitive miR | Targeting autophagy gens ATG5/FIP200 in colorectal cancer | [164] |
| lncRNAHotair | Playing roles in myeloid transcriptional regulation | HOTAIRM1 acting as a miR-20a/106b sponge to regulate autophagy pathway | [201] |
| lncRNAH19 | Involved in diabetic cardiomyopathy | H19 inhibiting autophagy activation by promoting mTOR phosphorylation in cardiomyocytes | [202] |
| lncRNA HNF1A-AS1 | Involved in carcinogenesis and cancer | Regulating autophagy by miR-30b which can target Bcl-2 | [203] |
| lncRNAMALAT1 | Transcript 1 of metastasis-associated lung adenocarcinoma | Promoting autophagy activation in aggressive pancreatic cancer | [204] |
| lncRNANBR2 | Regulating diverse biological processes | LKB1-AMPK- NBR2 feedback loop under energy stress | [205] |
| lncRNA TINCR | Mainly sponging miRs | Sp3 binding to TINCR promoters and promoting autophagy in cutaneous squamous cell carcinoma | [206] |
| lncRNA CA7-4 | Mainly sponging miRs | Promoting autophagy by sponging MIR877-3P and MIR5680 in endothelial cells | [207] |
| lncRNA17A | Involoved in neurodegenerative disorders | Promoting autophagy in Alzheimer’s disease model | [208] |
| lncRNA OGFRP1 | Regulating diverse biological processes | Inhibiting autophagy though AKT/mTOR signaling in coronary artery endothelial cells | [209] |
| lncRNA BLACAT1 | Mainly sponging miRs | Promoting autophagy gene atg7 expression via miR-17 in lung cancer | [210] |
| lncRNA MEG3 | Involved in cancer and bacterial infection | Promoting autophagy in glioma and during bacterial infection | [211] [99] |
Similar to miRNAs, siRNA, a double-stranded RNA approximately 20-25 nt in length, interferes with the expression of genes by base-pairing with complementary sequences to degrade mRNA after transcription85. siRNA has been an indispensable tool for manipulating gene expression in wide-spread scopes of experimental studies including research involving inflammasome and autophagy.
Different from miRNAs and siRNAs, circRNAs do not have 5′ or 3′ ends and usually function as a sponge of miRNAs86. Thus far, although only a small portion of circRNAs, such as circRNA mm9, circRNA CBT15, circ005915 and circR1011, are reportedly associated with NLRP3 inflammasome activation after PM2.5 treatment87, circRNAs are speculated to contribute to inflammasome activation in other potential biological processes. Also notable is that circPAN388, circHIPK389, circ10407590, circHECTD191, circRNA.283792, circ000194693, ciR-01209194, circNRIP195 and circACR96 were found to regulate autophagy by targeting autophagy-associated miRNAs, implying that autophagy is strongly impacted by the circRNA-miRNA axis. However, there might be other mechanisms with small ncRNAs in coordinating inflammasome assembly and autophagy machinery, requiring further investigations.
As a sponge of miRNAs, lncRNAs have been increasingly recognized for important function in a variety of cellular activities in recent years. Though lncRNAs are acknowledged to be linked to the autophagy process, only limited literature showed their direct association with inflammasomes. However, studies did indicate that lncRNAs were involved in inflammation and immune diseases97. We summarized recent researches that reported the association of lncRNAs with inflammasome (Table 2) and autophagy (Table 3). Among them, lncRNA-MEG3 is the only lncRNA confirmed to regulate autophagy during bacterium (P. aeruginosa and Mycobacteria) infection98,99, while others predominantly linked to cancers (Table 3). These studies provide a novel perspective to further explore ncRNAs in regulating inflammasomes and autophagy. Nevertheless, it is currently unclear whether lncRNAs can link with inflammasome activation and autophagy processes at the same time. Until 2017 when Meng et al. found the shRNA disturbance of NLRP3 inflammasome through autophagy activation in alleviating ischemia reperfusion-induced damage100, which is a critical example to link the ncRNA, inflammasome, and autophagy together. Then in 2018, Xue et al. confirmed that Cox2 (lincRNA) interferes with NLRP3 inflammasome- and autophagy-mediated inflammation. Mechanistically, these authors delineated that Cox2 promotes the NF-κB p65 nuclear translocation by directly binding to p65, leading to increased IL-1β secretion. Meanwhile, Cox2 regulates the expression of NLRP3 and ASC, which further promotes the IL-1β secretion. The increased IL-1β then enhances the TIR-domain-containing adapter-inducing interferon-β (TRIF) cleavage. Hence, the TRIF-mediated autophagy is inhibited101. These studies elucidate the links between ncRNAs and inflammasome-autophagy crosstalk and highlight the necessity to further delve into more links and the underlying mechanisms.
Links of autophagy, inflammasomes, and ncRNAs to human diseases
Inflammasome activation are critical for host immunity against pathogens, so their disturbance is closely related to the occurrence and development of various human diseases. In addition to bacterial infection and immunity, Type 2 diabetes is also found to be associated with the NLRP3 inflammasome102,103. The primary causes of Type 2 diabetes are insulin resistance and functional deficiency of pancreatic beta cells. Long-term high glucose concentration stimulates the islet cells to activate NLPR3 and trigger inflammation and compound the dysfunction of pancreatic beta cells, ultimately leading to development of Type 2 diabetes104. The damage of mitochondria apparatus is also closely associated with insulin resistance and functional damage of pancreatic beta cells, due to its critical energy generating function105. It is not surprising that autophagy process as a cell self-protection mechanism plays a vital role in maintaining the structure and function of pancreatic beta cells and in improving insulin resistance. However, the present understanding about the link between autophagy and diabetes is still preliminary, and further investigation is needed to provide insight into the mechanisms of the disease etiology and to offer improved treatments.
The NLRP3 inflammasome is also found to be linked with the occurrence of nonalcoholic fatty liver, by its high expression in the model of nonalcoholic fatty liver in mice106. Another study found that the long-time high fat intake caused the decline of mitochondrial autophagy level and mitochondrial breakdown, which in turn were associated with an impaired mitophagy to aggravate liver NLRP3 inflammasome activation in a murine nonalcoholic steatohepatitis model107. Again, these studies confirmed that normal level autophagy is vital to maintain cell metabolism in the liver and keep the liver functioning properly. Numerous studies have also demonstrated dysfunctional autophagy levels in a variety of liver diseases caused by alcohol, drugs, viral hepatitis, and ischemia-reperfusion injury108. These results show the important effect of autophagy on the complex assembly of inflammasomes in liver disorders. With further understanding of the role of autophagy in liver diseases, it may be possible to treat liver diseases by rescuing autophagy.
Inflammasome activation also triggers cardiovascular and cerebrovascular diseases and acute interstitial renal injury109. A major factor of hypertension pathophysiology is the NLRP3-associated inflammation110, while NF-κB, the effective activator of NLRP3 was reported to be suppressed by the activation of AMPK in the hypertension process111. Peng et al. found that P2X7R contributes to the progression of atherosclerosis by promoting NLRP3 inflammasome activation112, whereas Sirt6 was reported to stabilize atherosclerosis plaques by promoting autophagy113. These studies illustrate that autophagy may be involved in the energy consumption mechanisms to adjust metabolism and maintain homeostasis, and that altering the delicate regulation of these processes will impose tissue injury and cause serious organ dysfunction and disease development.
In inflammatory diseases, inflammasome was found to have a beneficial role in inflammatory bowel disease (IBD), showing that NLRP3 inflammasome activation helps maintain the intestinal microbial balance of flora and suppresses colitis-associated tumors. Furthermore, other NLR family proteins including NLRC4, NLRP6, and NLRP3 may also play a role in colitis114.
As the central link in inflammation, inflammasomes may be involved in genetic diseases, such as Webster’s syndrome, whose gene mutation may over-activate NLRP3 inflammasomes, resulting in excessive production of pro-inflammatory factors115. The existing data showed that one of the factors in the development of acute lung injury is the dysregulated inflammatory responses116. Moreover, studies have shown that the serum levels of IL-1β and IL-18 in cancer patients are positively correlated with the malignancy of tumors and negatively correlated with the survival rate of patients, revealing the close association between inflammasomes and tumors117. lncRNA GAS5 was found to suppress ovarian cancer by inducing inflammasome formation118. In summary, inflammasome activation is involved in genetic diseases, chronic inflammatory diseases, cancer, besides others.
Like inflammasome, autophagy is also involved with nervous system diseases and cardiovascular diseases (cardiomyopathy, cardiac hypertrophy, ischemic heart disease, and heart failure). In the brain of patients with Alzheimer’s disease (AD), accelerated accumulation of autophagosomes and Alzheimer’s amyloid plaques activate the NLRP3 inflammasome119. Undoubtedly, regulatory ncRNAs are also tightly associated with diseases, such as cancer, nervous system diseases, and heart disease. Many lncRNAs are shown to be associated with breast cancer. As an example, the aberrant expression of lncRNAs that regulate the telomerase TERT gene in breast cancer cells (including precancerous cells) may increase telomerase activity, thereby enhancing cancer cell growth and inhibiting apoptosis120. The cardiovascular system has a particularly rich source of miRNAs, which play diverse roles in cardiovascular diseases. Certain miRNAs may be attributed to the potential susceptibility of the heart and blood vessels to injury, while other miRNAs may help sustain cardiovascular function and homeostasis121. The roles of ncRNA in inflammasome activation- or autophagy-related diseases only began to be unfolded and are worth further studying. For example, miR-30A was the first small noncoding RNA identified as an autophagy regulator by targeting the beclin1 gene in a spectrum of cancer cells122. Altogether, we have just briefly touched the base of these complex intertwined regulatory pathways, and it is almost impossible to build a complete hierarchy map. Nevertheless, typical examples have already demonstrated the important interplay between inflammasome, autophagy, and ncRNA in human diseases. We here provide some online resource links (http://www.cuilab.cn/lncrnadisease for lncRNA and http://www.cuilab.cn/hmdd for microRNA).
Targeting ncRNAs to treat diseases involving autophagy and inflammasomes
Although RNA drugs have been extensively designed and developed for 30 years, only a handful of RNA-based small molecule drugs are currently on the market worldwide, such as Exondys 51 (for Duchenne Muscular Dystrophy)123, Macugen (for vascular disease)124, Vitravene (for cytomegalovirus disease)125, Kynamro (for familial hypercholesterolemia)126, Defitelio (for veno-occlusive disease)127, and Spinraza (for muscular atrophy)128. Recently, Dr. Matt Disney of the Scripps Research Institute in Florida, has discovered a small molecule called targaprimir-96 that binds to a precursor of miRNA-96, pri-miR-96. Pri-miR-96 generates mir-96 through RNA mutation, which is a carcinogenic miRNA, and can reduce the activity of FOXO1 to perform an important role in triggering breast cancer129. However, there is still a long way before making it a clinical drug. Chemist Amanda Hargrove’s team at Duke University analyzed the chemical informatics of ligands for 100 targeted RNAs and found that the same library of compounds used to target proteins could also be used for targeting RNAs, meaning lower risk for humans due to their solubility, permeability and toxicity130. We believe that as screening technology fast advances, RNAs targeted drug research would be more feasible and eventually enter the clinic. Another way to target ncRNAs is interrupting the regulation between them. For example, lncRNA TGFB2-OT1 can regulate miRNAs in autophagy and inflammation of vascular endothelial cells131. If the interactions between lncRNAs and miRNAs were prevented, the subsequent cascade of reactions would be terminated.
Summary and perspective
The fundamental understanding of the crosstalk between inflammasome activation and autophagy will add to the design of therapeutics for various diseases, such as chronic inflammatory and drug resistance in infection and cancer. Recent research sheds light on the molecular complexity of autophagy and inflammasome activation under bacterial infection, which may be harnessed to control drug resistance to infection. With the development of new technologies like CRISPR-Cas enabling the easy editing of autophagy factors and inflammasome machineries for improving knowledge in this field, more insight will be illuminated with the continued research.
In the three-party interplay, ncRNAs will have a big impact on disease through intermediating between autophagy and inflammasome. We know that more than half of the eukaryote DNA are transcribed into RNA, most of which are ncRNAs. Accumulating evidence suggests that ncRNAs play important roles in biological processes including development, physiologic functions, metabolism, and disease. However, unlike the mechanism about autophagy and inflammasomes, much less is known about how ncRNAs regulate these processes in each of the many conditions: where the ncRNAs are from; where they go to; what/how they function; or even whether each or most of them have an important biological significance. Thanks to the high-throughput screening technologies, for example next-generation sequencing (NGS) and scRNA-Seq, characterization of ncRNA would be unprecedentedly faster. It is clear that ncRNAs have a wide range of functions. The main task in the near future is to discover their functional roles and potential links to diseases, which will be truly time-consuming and full of difficulty. It is hoped that a thorough understanding of the regulatory network of ncRNAs, inflammasomes and autophagy will provide the breakthrough in the demystification of the function of the interactome. ncRNAs-based therapeutics or regulators for inflammasome or autophagy may add to the treatments to enhance efficacy and reduce side effects, representing the exciting, revolutionary feature of precision medicine.
Acknowledgements
This work was supported by National Institutes of Health (Grants No. AI101973-01, AI109317-01A1, and AI097532-01A1 as well as P20 GM103442 and GM113123 for the UND COREs). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare no conflict of interest.
References
- 1. Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thoresen SB, Pedersen NM, Liestøl K, et al. . A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res 2010;316(20):3368–78. doi: 10.1016/j.yexcr.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 3. Castrejón-Jiménez NS, Leyva-Paredes K, Hernández-González JC, et al. The role of autophagy in bacterial infections. Biosci Trends 2015;9(3):149–59. doi: 10.5582/bst.2015.01035. [DOI] [PubMed] [Google Scholar]
- 4. Joven J, Guirro M, Mariné-Casadó R, et al. . Autophagy is an inflammation-related defensive mechanism against disease. In: Oxidative Stress and Inflammation in Non-communicable Diseases-Molecular Mechanisms and Perspectives in Therapeutics. Springer, 2014, 43–59. [DOI] [PubMed] [Google Scholar]
- 5. Finkbeiner S. The autophagy lysosomal pathway and neurodegeneration. Cold Spring Harb Perspect Biol 2019. doi: 10.1101/cshperspect.a033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling .Cell Res 2014;24(1):42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nozawa T, Minowa-Nozawa A, Aikawa C, et al. . The STX6-VTI1B-VAMP3 complex facilitates xenophagy by regulating the fusion between recycling endosomes and autophagosomes. Autophagy 2017;13(1):57–69. doi: 10.1080/15548627.2016.1241924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moreau K, Lacas-Gervais S, Fujita N, et al. . Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. Cell Microbiol 2010;12(8):1108–23. doi: 10.1111/j.1462-5822.2010.01456.x. [DOI] [PubMed] [Google Scholar]
- 9. Pujol C, Klein KA, Romanov G A, et al. . Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect Immun 2009;77(6):251–61. doi: 10.1128/IAI.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yasir M, Pachikara ND, Bao X, et al. . Regulation of chlamydial infection by host autophagy and vacuolar ATPase-bearing organelles. Infect Immun 2011;79(10):4019–28. doi: 10.1128/IAI.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bah A, Vergne I. Macrophage autophagy and bacterial infections. Front Immunol 2017;8:1483. doi: 10.3389/fimmu.2017.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldsmith J, Levine B, Debnath J. Chapter two - autophagy and cancer metabolism. Methods Enzymol 2014;542:25–57. doi: 10.1016/B978-0-12-416618-9.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rao L, Eissa NT. Autophagy in pulmonary innate immunity. J Innate Immun 2019. doi: 10.1159/000497414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen GY, Nunez G. Inflammasomes in intestinal inflammation and cancer. Gastroenterology 2011;141(6):1986–99. doi: 10.1053/j.gastro.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet 2006;38(2):240–4. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 16. Sandstrom A, Mitchell PS, Goers L, et al. . Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 2019;364(6435). doi: 10.1126/science.aau1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karagianni P, Nezos A, Ioakeim F, et al. . Analysis of NLRP3, MVK and TNFRSF1A variants in adult Greek patients with autoinflammatory symptoms. Clin Exp Rheumatol 2018;36(6 Suppl 115):86–89. [PubMed] [Google Scholar]
- 18. Murphy AM, Smith CE, Murphy LM, et al. . Potential interplay between dietary saturated fats and genetic variants of the NLRP3 Inflammasome, to modulate insulin resistance and diabetes risk: Insights from a meta-analysis of 19,005 individuals. Mol Nutr Food Res 2019;e1900226. doi: 10.1002/mnfr.201900226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tejera D, Mercan D, Sanchez-Caro JM, et al. . Systemic inflammation impairs microglial Abeta clearance through NLRP3 inflammasome. EMBO J 2019;e101064. doi: 10.15252/embj.2018101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma P, Zha S, Shen X, et al. NFAT5 mediates hypertonic stress-induced atherosclerosis via activating NLRP3 inflammasome in endothelium. Cell Commun Signal 2019;17(1):102. doi: 10.1186/s12964-019-0406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu Z, Zhou Q, Zhang C, et al. . Structural and biochemical basis for induced self-propagation of NLRC4. Science 2015;350(6259):399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 22. Roberts TL, Idris A, Dunn JA, et al. . HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 2009;323(5917):1057–60. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 23. Zoete MR, Palm NW, Zhu S, et al. . Inflammasomes. Cold Spring Harb Perspect Biol 2014;6(12):a016287. doi: 10.1101/cshperspect.a016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li S, Xu Z, Sheng J. tRNA-derived small RNA: A novel regulatory small non-coding RNA. Genes (Basel) 2018;9(5):246. doi: 10.3390/genes9050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng J, Kapranov P, Drenkow J, et al. . Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005;308(5725):1149–54. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 26. The ENCODE Project Consortium . Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murayama S, Ikezoe J, Godwin JD, et al. . Pulmonary infection in patients with cyclosporine, azathioprine, and corticosteroids after cardiac transplantation. Clinical and Radiographic Assessment. Nihon Igaku Hoshasen Gakkai Zasshi 1991;51(7):780–9. [PubMed] [Google Scholar]
- 28. Hadjicharalambous MR, Lindsay MA. Long non-coding RNAs and the innate immune response. Noncoding RNA 2019;5(2):34. doi: 10.3390/ncrna5020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet 2015;31(5):239–51. doi: 10.1016/j.tig.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu Y, Wu W, Han Q, et al. . New insights into the interplay between non-coding RNAs and RNA-binding protein HnRNPK in regulating cellular functions. Cells 2019;8(1):62. doi: 10.3390/cells8010062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McMullen JR, Drew BG. Long non-coding RNAs (lncRNAs) in skeletal and cardiac muscle: Potential therapeutic and diagnostic targets? Clin Sci (Lond) 2016;130(24):2245–56. doi: 10.1042/CS20160244. [DOI] [PubMed] [Google Scholar]
- 32. Zaiou M. Circular RNAs as potential biomarkers and therapeutic targets for metabolic diseases. Adv Exp Med Biol 2019;1134:177–91. doi: 10.1007/978-3-030-12668-1_10. [DOI] [PubMed] [Google Scholar]
- 33. Yang F, Li XF, Cheng LN, et al. . Long non-coding RNA CRNDE promotes cell apoptosis by suppressing miR-495 in inflammatory bowel disease. Exp Cell Res 2019. doi: 10.1016/j.yexcr.2019.06.029. [DOI] [PubMed] [Google Scholar]
- 34. Sterbova M, Pazourkova E, Santorova-Pospisilova S, et al. . The use of human inflammatory response and autoimmunity RT2 lncRNA PCR Array for plasma examination in breast cancer patients prior to therapy. Neoplasma 2019;2019:641–6. doi: 10.4149/neo_2018_180907N679. [DOI] [PubMed] [Google Scholar]
- 35. Simchovitz A, Hanan M, Niederhoffer N, et al. . NEAT1 is overexpressed in Parkinson's disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. FASEB J 2019. doi: 10.1096/fj.201900830R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim JK, Kim TS, Basu J, et al. . MicroRNA in innate immunity and autophagy during mycobacterial infection. Cell Microbiol 2017;19(1):e12687. doi: 10.1111/cmi.12687. [DOI] [PubMed] [Google Scholar]
- 37. Krakauer T. Inflammasomes, autophagy, and cell death: The trinity of innate host defense against intracellular bacteria. Mediators Inflamm 2019;2019:2471215. doi: 10.1155/2019/2471215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: Host cell death and inflammation. Nat Rev Microbiol 2009;7(2):99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy 2005;1(2):66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- 40. Burgh R, Nijhuis L, Pervolaraki K, et al. . Defects in mitochondrial clearance predispose human monocytes to interleukin-1β hypersecretion. J Biological Chem 2014;289(8):5000–12. doi: 10.1074/jbc.M113.536920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang LD, Yan J, Niu H, et al. . Autophagy and ubiquitination in Salmonella infection and the related inflammatory responses. Front Cell Infect Microbiol 2018;8:78. doi: 10.3389/fcimb.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saitoh T, Fujita N, Yoshimori T, et al. . Autophagy and innate immunity. Tanpakushitsu Kakusan Koso 2008;53(16 Suppl):2279–85. [PubMed] [Google Scholar]
- 43. Wu Q, Wang B, Zhou C, et al. . Bacterial type I CRISPR-Cas systems influence inflammasome activation in mammalian host by promoting autophagy. Immunology 2019. doi: 10.1111/imm.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beare PA, Gilk SD, Larson CL, et al. . Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. MBio 2011;2(4):e00175–11. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodgers MA, Bowman JW, Liang QM, et al. . Regulation where autophagy intersects the inflammasome. Antioxid Redox Signal 2014;20(3):495–506. doi: 10.1089/ars.2013.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim JY, Paton JC, Briles DE, et al. . Streptococcus pneumoniae induces pyroptosis through the regulation of autophagy in murine microglia. Oncotarget 2015;6(42):44161–78. doi: 10.18632/oncotarget.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sauer JD, Pereyre S, Archer KA, et al. . Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc Natl Acad Sci USA 2011;108(30):12419–24. doi: 10.1073/pnas.1019041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. West AP, Brodsky IE, Rahner C, et al. . TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 2011;472(7344):476–80. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tal MC, Sasai, Lee HK, et al. . Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA 2009;106(8):2770–5. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lara-Tejero M, Sutterwala FS, Ogura Y, et al. . Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med 2006;203(6):1407–12. doi: 10.1084/jem.20060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol 2013;13(6):397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abdelaziz DHA, Khalil H, Cormet-Boyaka E, et al. . The cooperation between the autophagy machinery and the inflammasome to implement an appropriate innate immune response: Do they regulate each other? Immunol Rev 2015;265(1):194–204. doi: 10.1111/imr.12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shi CS, Shenderov K, Huang NN, et al. . Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 2012;13(3):255–63. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Master SS, Rampini SK, Davis AS, et al. . Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe 2008;3(4):224–32. doi: 10.1016/j.chom.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Biswas T, Tsodikov OV. Hexameric ring structure of the N-terminal domain of Mycobacterium tuberculosis DnaB helicase. FEBS J 2008;275(12):3064–71. doi: 10.1111/j.1742-4658.2008.06460.x. [DOI] [PubMed] [Google Scholar]
- 56. Sutterwala FS, Ogura Y, Flavell RA. The inflammasome in pathogen recognition and inflammation. J Leukoc Biol 2007;82(2):259–64. doi: 10.1189/jlb.1206755. [DOI] [PubMed] [Google Scholar]
- 57. Li D, Guabiraba R, Besnard AG, et al. . IL-33 promotes ST2-dependent lung fibrosis by the induction of alternatively activated macrophages and innate lymphoid cells in mice. J Allergy Clin Immunol 2014;134(6):1422–32. doi: 10.1016/j.jaci.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li XL, Ezelle HJ, Kang TJ, et al. . An essential role for the antiviral endoribonuclease, RNase-L, in antibacterial immunity. Proc Natl Acad Sci USA 2008;105(52):20816–21. doi: 10.1073/pnas.0807265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tan ZX, Xiao BJ, Liao YH. Effects of acute hypoxia on microvessels response and anti-oxidation enzyme in rats. Zhongguo Ying Yong Sheng Li Xue Za Zhi 2009;25(4):438–9. [PubMed] [Google Scholar]
- 60. Gårdhagen R, Lantz J, Carlsson F, et al. . Quantifying turbulent wall shear stress in a stenosed pipe using large eddy simulation. J Biomech Eng 2010;132(6):061002. doi: 10.1115/1.4001075. [DOI] [PubMed] [Google Scholar]
- 61. Gerstenmaier L, Pilla R, Herrmann L, et al. . The autophagic machinery ensures nonlytic transmission of mycobacteria. Proc Natl Acad Sci USA 2015;112(7):E687–92. doi: 10.1073/pnas.1423318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brodsky IE, Palm NW, Sadanand S, et al. . A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe 2010;7(5):376–87. doi: 10.1016/j.chom.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chiang CY, Uzoma I, Lane DJ, et al. . A reverse-phase protein microarray-based screen identifies host signaling dynamics upon Burkholderia spp. infection. Front Microbiol 2015;6:683. doi: 10.3389/fmicb.2015.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Amano A, Nakagawa I, Yoshimori T. Autophagy in innate immunity against intracellular bacteria. J Biochem 2006;140(2):161–6. doi: 10.1093/jb/mvj162. [DOI] [PubMed] [Google Scholar]
- 65. Miller LS, Pietras EM, Uricchio LH, et al. . Inflammasome-mediated production of IL-1β is required for neutrophil recruitment against Staphylococcus aureus in vivo. J Immunol 2007;179(10):6933–42. doi: 10.4049/jimmunol.179.10.6933. [DOI] [PubMed] [Google Scholar]
- 66. Campbell-Valois FX, Sachse M, Sansonetti PJ, et al. . Escape of actively secreting Shigella flexneri from ATG8/LC3-positive vacuoles formed during cell-to-cell spread is facilitated by IcsB and VirA. MBio. 2015, 6(3):e02567–14. doi: 10.1128/mBio.02567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ashida H, Ogawa M, Kim M, et al. . Bacteria and host interactions in the gut epithelial barrier. Nat Chem Biol 2011;8(1):36–45. doi: 10.1038/nchembio.741. [DOI] [PubMed] [Google Scholar]
- 68. Birmingham CL, Canadien V, Kaniuk NA, et al. . Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature 2008;451(7176):350–4. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 69. Williams CR, Sorrentino F, Murphy TE, et al. . Synchronization states and multistability in a ring of periodic oscillators: Experimentally variable coupling delays. Chaos 2013;23(4):043117. doi: 10.1063/1.4829626. [DOI] [PubMed] [Google Scholar]
- 70. Andriankaja O, Trevisan M, Falkner K, et al. . Association between periodontal pathogens and risk of nonfatal myocardial infarction. Community Dent Oral Epidemiol 2011;39(2):177–85. doi: 10.1111/j.1600-0528.2010.00582.x. [DOI] [PubMed] [Google Scholar]
- 71. Huck O, Elkaim R, Davideau JL, et al. . Porphyromonas gingivalis-impaired innate immune response via NLRP3 proteolysis in endothelial cells. Innate Immun 2015;21(1):65–72. doi: 10.1177/1753425914523459. [DOI] [PubMed] [Google Scholar]
- 72. Amer AO, Swanson MS. Autophagy is an immediate macrophage response to legionella pneumophila. Cell Microbiol 2005;7(6):765–78. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pereira MS, Morgantetti GF, Massis LM, et al. . Activation of NLRC4 by flagellated bacteria triggers caspase-1-dependent and -independent responses to restrict Legionella pneumophila replication in macrophages and in vivo. J Immunol 2011;187(12):6447–55. doi: 10.4049/jimmunol.1003784. [DOI] [PubMed] [Google Scholar]
- 74. Cullinane M, Gong L, Li XL, et al. . Stimulation of autophagy suppresses the intracellular survival of Burkholderia pseudomallei in mammalian cell lines. Autophagy 2008;4(6):744–53. doi: 10.4161/auto.6246. [DOI] [PubMed] [Google Scholar]
- 75. Aachoui Y, Leaf IA, Hagar JA, et al. . Caspase-11 protects against bacteria that escape the vacuole. Science 2013;339(6122):975–8. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang LJ, Huang HY, Huang MP, et al. . The microtubule-associated protein EB1 links AIM2 inflammasomes with autophagy-dependent secretion. J Biol Chem 2014;289(42):29322–33. doi: 10.1074/jbc.M114.559153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Öhman T, Teirilä L, Lahesmaa-Korpinen AM, et al. . Dectin-1 pathway activates robust autophagy-dependent unconventional protein secretion in human macrophages. J Immunol 2014;192(12):5952–62. doi: 10.4049/jimmunol.1303213. [DOI] [PubMed] [Google Scholar]
- 78. Kimura T, Jain A, Choi SW, et al. . TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol 2015;210(6):973–89. doi: 10.1083/jcb.201503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ambros V. The functions of animal microRNAs. Nature 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 80. Yang Z, Zhong LN, Xian RH, et al. . MicroRNA-223 regulates inflammation and brain injury via feedback to NLRP3 inflammasome after intracerebral hemorrhage. Mol Immunol 2015;65(2):267–76. doi: 10.1016/j.molimm.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 81. Yang F, Lou GH, Zhou XT, et al. . MicroRNA-223 acts as an important regulator to Kupffer cells activation at the early stage of con A-induced acute liver failure via AIM2 signaling pathway. Cell Physiol Biochem 2014;34(6):2137–52. doi: 10.1159/000369658. [DOI] [PubMed] [Google Scholar]
- 82. Chen S, Wu J, Jiao K, et al. . MicroRNA-495-3p inhibits multidrug resistance by modulating autophagy through GRP78/mTOR axis in gastric cancer. Cell Death Dis 2018;9(11):1070. doi: 10.1038/s41419-018-0950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhou XK, Li XF, Wu M. MiRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct Target Ther 2018;3:14. doi: 10.1038/s41392-018-0006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pu QQ, Gan CP, Li RP, et al. . Atg7 deficiency intensifies inflammasome activation and pyroptosis in Pseudomonas Sepsis. J Immunol 2017;198(8):3205–13. doi: 10.4049/jimmunol.1601196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Subhan MA, Torchilin VP. Efficient nanocarriers of siRNA therapeutics for cancer treatment. Transl Res 2019. doi: 10.1016/j.trsl.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 86. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32(5):453–61. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhong YJ, Wang YX, Zhang C, et al. . Identification of long non-coding RNA and circular RNA in mice after intra-tracheal instillation with fine particulate matter. Chemosphere 2019;235:519–26. doi: 10.1016/j.chemosphere.2019.06.122. [DOI] [PubMed] [Google Scholar]
- 88. Shang J, Chen WM, Liu S, et al. . CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res 2019;85:106198. doi: 10.1016/j.leukres.2019.106198. [DOI] [PubMed] [Google Scholar]
- 89. Chen XY, Mao R, Su WM, et al. . Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy 2019. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chi GN, Xu DH, Zhang BY, et al. . Matrine induces apoptosis and autophagy of glioma cell line U251 by regulation of circRNA-104075/BCL-9. Chem Biol Interact 2019;308:198–205. doi: 10.1016/j.cbi.2019.05.030. [DOI] [PubMed] [Google Scholar]
- 91. Han B, Zhang Y, Zhang YH, et al. . Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: Implications for cerebral ischemic stroke. Autophagy 2018;14(7):1164–84. doi: 10.1080/15548627.2018.1458173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhou ZB, Niu YL, Huang GX, et al. . Silencing of circRNA.2837 plays a protective role in sciatic nerve injury by sponging the miR-34 family via regulating neuronal autophagy. Mol Ther - Nucleic Acids 2018;12:718–29. doi: 10.1016/j.omtn.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li XX, Diao HY. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J Cell Physiol 2019;234(8):13807–19. doi: 10.1002/jcp.28061. [DOI] [PubMed] [Google Scholar]
- 94. Cheng YS, Luo W, Li Z, et al. . CircRNA-012091/PPP1R13B-mediated lung fibrotic response in silicosis via ER stress and autophagy. Am J Respir Cell Mol Biol 2019. doi: 10.1165/rcmb.2019-0017OC. [DOI] [PubMed] [Google Scholar]
- 95. Zhang X, Wang S, Wang HX, et al. . Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer 2019;18(1):20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhou LY, Zhai M, Huang Y, et al. . The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/FAM65B pathway. Cell Death Differ 2019;26(7):1299–15. doi: 10.1038/s41418-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Pawar K, Hanisch C, Palma Vera SE, et al. . Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci Rep 2016;6:19416. doi: 10.1038/srep19416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li RP, Fang LZ, Pu QQ, et al. . MEG3-4 is a miRNA decoy that regulates IL-1β abundance to initiate and then limit inflammation to prevent Sepsis during lung infection. Sci Signal 2018;11(536):eaao2387. doi: 10.1126/scisignal.aao2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sharbati S, Ravon F, Einspanier R, et al. . Mycobacterium smegmatis but not Mycobacterium avium subsp. hominissuis causes increased expression of the long non-coding RNA MEG3 in THP-1-derived human macrophages and associated decrease of TGF-Β. Microorganisms 2019;7(3):E63. doi: 10.3390/microorganisms7030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Meng Z, Song MY, Li CF, et al. . ShRNA interference of NLRP3 inflammasome alleviate ischemia reperfusion-induced myocardial damage through autophagy activation. Biochem Biophys Res Commun 2017;494(3/4):728–35. doi: 10.1016/j.bbrc.2017.10.111. [DOI] [PubMed] [Google Scholar]
- 101. Xue ZY, Zhang ZM, Liu HK, et al. . LincRNA-Cox2 regulates NLRP3 inflammasome and autophagy mediated neuroinflammation. Cell Death Differ 2019;26(1):130–45. doi: 10.1038/s41418-018-0105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Dixit VD. Nlrp3 inflammasome activation in type 2 diabetes: Is it clinically relevant? Diabetes 2013;62(1):22–4. doi: 10.2337/db12-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Luo BB, Huang F, Liu YL, et al. . NLRP3 inflammasome as a molecular marker in diabetic cardiomyopathy. Front Physiol 2017;8:519. doi: 10.3389/fphys.2017.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lamkanfi M, Mueller JL, Vitari AC, et al. . Glyburide inhibits the cryopyrin/Nalp3 inflammasome. J Cell Biol 2009;187(1):61–70. doi: 10.1083/jcb.200903124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sergi D, Naumovski N, Heilbronn LK, et al. . Mitochondrial (dys) function and insulin resistance: From pathophysiological molecular mechanisms to the impact of diet. Front Physiol 2019;10:532. doi: 10.3389/fphys.2019.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kamari Y, Shaish A, Vax E, et al. . Lack of interleukin-1α or interleukin-1β inhibits transformation of steatosis to steatohepatitis and liver fibrosis in hypercholesterolemic mice. J Hepatol 2011;55(5):1086–94. doi: 10.1016/j.jhep.2011.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhang NP, Liu XJ, Xie L, et al. . Impaired mitophagy triggers NLRP3 inflammasome activation during the progression from nonalcoholic fatty liver to nonalcoholic steatohepatitis. Lab Invest 2019;99(6):749–63. doi: 10.1038/s41374-018-0177-6. [DOI] [PubMed] [Google Scholar]
- 108. Zhao YM, Cai HB, Zhou PM, et al. . Protective effect of ulinastatin on hepatic ischemia reperfusion injury through autophagy activation in Chang liver cells. J Cell Biochem 2019;120(9):14960–70. doi: 10.1002/jcb.28758. [DOI] [PubMed] [Google Scholar]
- 109. Hansson GK, Klareskog L. Pulling down the plug on atherosclerosis: Cooling down the inflammasome. Nat Med 2011;17(7):790–1. doi: 10.1038/nm0711-790. [DOI] [PubMed] [Google Scholar]
- 110. Yu XJ, Zhang DM, Jia LL, et al. . Inhibition of NF-κB activity in the hypothalamic paraventricular nucleus attenuates hypertension and cardiac hypertrophy by modulating cytokines and attenuating oxidative stress. Toxicol Appl Pharmacol 2015;284(3):315–22. doi: 10.1016/j.taap.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 111. Zhai C, Shi WH, Feng W, et al. . Activation of AMPK prevents monocrotaline-induced pulmonary arterial hypertension by suppression of NF-κB-mediated autophagy activation. Life Sci 2018;208:87–95. doi: 10.1016/j.lfs.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 112. Peng K, Liu LS, Wei DH, et al. . P2X7R is involved in the progression of atherosclerosis by promoting NLRP3 inflammasome activation. Int J Mol Med 2015;35(5):1179–88. doi: 10.3892/ijmm.2015.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang TT, Sun C, Hu L, et al. . Sirt6 stabilizes atherosclerosis plaques by promoting macrophage autophagy and reducing contact with endothelial cells. Biochimie Et Biol Cell 2019. doi: 10.1139/bcb-2019-0057. [DOI] [PubMed] [Google Scholar]
- 114. Chen GY, Liu MC, Wang FY, et al. . A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol 2011;186(12):7187–94. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Agostini L, Martinon F, Burns K, et al. . NALP3 forms an IL-1beta-processing inflammasome with increased activity in muckle-Wells autoinflammatory disorder. Immunity 2004;20(3):319–25. doi: 10.1016/S1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 116. Dostert C, Pétrilli V, Van Bruggen R, et al. . Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 2008;320(5876):674–7. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wang HX, Lu XL, Huang WJ, et al. . Pyroptosis is involved in cryopreservation and auto-transplantation of mouse ovarian tissues and pyroptosis inhibition improves ovarian graft function. Res Vet Sci 2019;124:52–6. doi: 10.1016/j.rvsc.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 118. Li J, Yang C, Li YG, et al. . LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome formation. Biosci Rep 2017;2017:BSR20171150. doi: 10.1042/BSR20171150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Halle A, Hornung V, Petzold GC, et al. . The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol 2008;9(8):857–65. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Yan R, Wang KJ, Peng R, et al. . Genetic variants in lncRNA SRA and risk of breast cancer. Oncotarget 2016;7(16):22486–96. doi: 10.18632/oncotarget.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kriegel AJ, Gartz M, Afzal MZ, et al. . Molecular approaches in HFpEF: MicroRNAs and iPSC-derived cardiomyocytes. J Cardiovasc Transl Res 2017;10(3):295–304. doi: 10.1007/s12265-016-9723-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhang J, Wang PY, Wan L, et al. . The emergence of noncoding RNAs as Heracles in autophagy. Autophagy 2017;13(6):1004–24. doi: 10.1080/15548627.2017.1312041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Khan N, Eliopoulos H, Han LX, et al. . Eteplirsen treatment attenuates respiratory decline in ambulatory and non-ambulatory patients with Duchenne muscular dystrophy. J Neuromuscul Dis 2019;6(2):213–25. doi: 10.3233/JND-180351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Battaglia Parodi M, Bartolo E, Brue C, et al. . Pegaptanib: Choroidal neovascularization in patients with age-related macular degeneration and previous arterial thromboembolic events. Eur J Ophthalmol 2018;28(1):58–62. doi: 10.5301/ejo.5001060. [DOI] [PubMed] [Google Scholar]
- 125. Steininger C. Novel therapies for cytomegalovirus disease. Recent Pat Antiinfect Drug Discov 2007;2(1):53–72. doi: 10.2174/157489107779561634. [DOI] [PubMed] [Google Scholar]
- 126. Parham JS, Goldberg AC. Mipomersen and its use in familial hypercholesterolemia. Expert Opin Pharmacother 2019;20(2):127–31. doi: 10.1080/14656566.2018.1550071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Richardson P, Aggarwal S, Topaloglu O, et al. . Systematic review of defibrotide studies in the treatment of veno-occlusive disease/sinusoidal obstruction syndrome (VOD/SOS). Bone Marrow Transplant 2019. doi: 10.1038/s41409-019-0474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Canadian Agency for Drugs and Technologies in Health . Pharmacoeconomic Review Report. Nusinersen (Spinraza) (Biogen Canada Inc.). Ottawa (ON): Canadian agency for drugs and technologies in Health, 2018. [PubMed]
- 129. Velagapudi SP, Cameron MD, Haga CL, et al. . Design of a small molecule against an oncogenic noncoding RNA . Proc Natl Acad Sci U S A 2016;113(21):5898–903. doi: 10.1073/pnas.1523975113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mullard A. Small molecules against RNA targets attract big backers .Nat Rev Drug Discov 2017;16(12):813–815. doi: 10.1038/nrd.2017.239. [DOI] [PubMed] [Google Scholar]
- 131. Huang S, Lu W, Ge D, et al. . A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells . Autophagy 2015;11(12):2172–83. doi: 10.1080/15548627.2015.1106663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhou Y, Lu M, Du RH, et al. . MicroRNA-7 targets nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson's disease. Mol Neurodegener 2016;11:28. doi: 10.1186/s13024-016-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Chen S, Smith BA, Iype J, et al. . MicroRNA-155-deficient dendritic cells cause less severe GVHD through reduced migration and defective inflammasome activation. Blood 2015;126(1):103–12. doi: 10.1182/blood-2014-12-617258. [DOI] [PubMed] [Google Scholar]
- 134. Wang W, Ding XQ, Gu TT, et al. . Pterostilbene and allopurinol reduce fructose-induced podocyte oxidative stress and inflammation via microRNA-377. Free Radic Biol Med 2015;83:214–26. doi: 10.1016/j.freeradbiomed.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 135. Momeni M, Reza Mirzaei M, Zainodini N, et al. MiR-143 induces expression of AIM2 and ASC in Jurkat cell line .Iran J Immunol 2013;10(2):103–9. doi: IJIv10i2A5. [PubMed] [Google Scholar]
- 136. Li XF, Shen WW, Sun YY, et al. . MicroRNA-20a negatively regulates expression of NLRP3-inflammasome by targeting TXNIP in adjuvant-induced arthritis fibroblast-like synoviocytes . Joint Bone Spine 2016;83(6):695–700. doi: 10.1016/j.jbspin.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 137. Bandyopadhyay S, Lane T, Venugopal R, et al. . MicroRNA-133a-1 regulates inflammasome activation through uncoupling protein-2 . Biochem Biophys Res Commun 2013;439(3):407–12. doi: 10.1016/j.bbrc.2013.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Haneklaus MGerlic M, Kurowska-Stolarska M, et al. . Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol 2012;189(8):3795–9. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 139. Jeyabal P, Thandavarayan RA, Joladarashi D, et al. . MicroRNA-9 inhibits hyperglycemia-induced pyroptosis in human ventricular cardiomyocytes by targeting ELAVL1 . Biochem Biophys Res Commun 2016;471(4):423–9. doi: 10.1016/j.bbrc.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Ning ZW, Luo XY, Wang GZ, et al. . MicroRNA-21 mediates angiotensin II-induced liver fibrosis by activating NLRP3 Inflammasome/IL-1beta Axis via targeting Smad7 and Spry1 . Antioxid Redox Signal 2017;27(1):1–20. doi: 10.1089/ars.2016.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Chen Z, Wen L, Martin M, et al. . Oxidative stress activates endothelial innate immunity via sterol regulatory element binding protein 2 (SREBP2) transactivation of microRNA-92a. Circulation 2015;131(9):805–14. doi: 10.1161/CIRCULATIONAHA.114.013675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Bhatt K, Lanting LL, Jia Y, et al. . Anti-inflammatory role of MicroRNA-146a in the pathogenesis of diabetic nephropathy . J Am Soc Nephrol 2016;27(8):2277–88. doi: 10.1681/ASN.2015010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Li P, Zhong X, Li J, et al. . MicroRNA-30c-5p inhibits NLRP3 inflammasome-mediated endothelial cell pyroptosis through FOXO3 down-regulation in atherosclerosis. Biochem Biophys Res Commun 2018;503(4):2833–40. doi: 10.1016/j.bbrc.2018.08.049. [DOI] [PubMed] [Google Scholar]
- 144. Zhou T, Xiang DK, Li SN, et al. . MicroRNA-495 ameliorates cardiac microvascular endothelial cell injury and inflammatory reaction by suppressing the NLRP3 Inflammasome Signaling pathway . Cell Physiol Biochem 2018;49(2):798–815. doi: 10.1159/000493042. [DOI] [PubMed] [Google Scholar]
- 145. Zhang P, Cao L, Zhou R, et al. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun 2019;10(1):1495. doi: 10.1038/s41467-019-09482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Yu G, Jia Z, Dou Z. miR-24-3p regulates bladder cancer cell proliferation, migration, invasion and autophagy by targeting DEDD .Oncol Rep 2017;37(2):1123–31. doi: 10.3892/or.2016.5326. [DOI] [PubMed] [Google Scholar]
- 147. Hui J, Huishan W, Tao L, et al. . miR-539 as a key negative regulator of the MEK pathway in myocardial infarction . Herz 2016;42(8):781–9. doi: 10.1007/s00059-016-4517-2. [DOI] [PubMed] [Google Scholar]
- 148. Chen Z, Jin T, Lu Y. AntimiR-30b inhibits TNF-alpha mediated apoptosis and attenuated cartilage degradation through enhancing autophagy .Cell Physiol Biochem 2016;40(5):883–94. [DOI] [PubMed] [Google Scholar]
- 149. Pradhan AK, Talukdar S, Bhoopathi P, et al. . Mda-7/IL-24 mediates cancer cell-specific death via regulation of miR-221 and the beclin-1 axis. Cancer Res 2017;77(4):949–59. doi: 10.1158/0008-5472.CAN-16-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Sun L, Zhao M, Wang Y, et al. . Neuroprotective effects of miR-27a against traumatic brain injury via suppressing FoxO3a-mediated neuronal autophagy . Biochem Biophys Res Commun 2016;482(4):1141–7. doi: 10.1016/j.bbrc.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 151. Li Y, Jiang W, Hu Y, et al. . MicroRNA-199a-5p inhibits cisplatin-induced drug resistance via inhibition of autophagy in osteosarcoma cells . Oncol Lett 2016;12(5):4203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Chen PH, Cheng CH, Shih CM, et al. . The inhibition of microRNA-128 on IGF-1-activating mTOR Signaling involves in Temozolomide-induced Glioma cell apoptotic death . PLoS One 2016;11(11):e0167096. doi: 10.1371/journal.pone.0167096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Park JK, Peng H, Katsnelson J, et al. . MicroRNAs-103/107 coordinately regulate macropinocytosis and autophagy . J Cell Biol 2016;215(5):667–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Lin Y, Deng W, Pang J, et al. . The microRNA-99 family modulates hepatitis B virus replication by promoting IGF-1R/PI3K/Akt/mTOR/ULK1 signaling-induced autophagy . Cell Microbiol 2017;19(5):e12709. doi: 10.1111/cmi.12709. [DOI] [PubMed] [Google Scholar]
- 155. Chen G, Gao X, Wang J, et al. . Hypoxia-induced microRNA-146a represses Bcl-2 through Traf6/IRAK1 but not Smad4 to promote chondrocyte autophagy . Biol Chem 2017;398(4):499–507. doi: 10.1515/hsz-2016-0211. [DOI] [PubMed] [Google Scholar]
- 156. Guo L, Zhao J, Qu Y, et al. . microRNA-20a inhibits Autophagic process by targeting ATG7 and ATG16L1 and Favors mycobacterial survival in macrophage cells. Front Cell Infect Microbiol 2016;6:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Song L, Liu S, Zhang L, et al. . MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1alpha feedback loop and the Akt-mTOR signaling pathway . Tumour Biol 2016;37(9):12161–8. [DOI] [PubMed] [Google Scholar]
- 158. Kou X, Liu X, Chen X, et al. . Ampelopsin attenuates brain aging of D-gal-induced rats through miR-34a-mediated SIRT1/mTOR signal pathway . Oncotarget 2016;7(46):74484–95. doi: 10.18632/oncotarget.12811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Zhang JA, Zhou BR, Xu Y, et al. . MiR-23a-depressed autophagy is a participant in PUVA- and UVB-induced premature senescence . Oncotarget 2016;7(25):37420–35. doi: 10.18632/oncotarget.9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kim JK, Lee HM, Park KS, et al. . MIR144* inhibits antimicrobial responses against mycobacterium tuberculosis in human monocytes and macrophages by targeting the autophagy protein DRAM2 . Autophagy 2016;13(2):423–41. doi: 10.1080/15548627.2016.1241922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Mo J, Zhang D, Yang R.. MicroRNA-195 regulates proliferation, migration, angiogenesis and autophagy of endothelial progenitor cells by targeting GABARAPL1. Biosci Rep 2016;36(5):e00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Su M, Chen Z, Wang C, et al. . Cardiac-specific overexpression of miR-222 induces heart failure and inhibits autophagy in mice . Cell Physiol Biochem 2016;39(4):1503–11. doi: 10.1159/000447853. [DOI] [PubMed] [Google Scholar]
- 163. Wei R, Cao G, Deng Z, et al. . miR-140-5p attenuates chemotherapeutic drug-induced cell death by regulating autophagy through inositol 1,4,5-trisphosphate kinase 2 (IP3k2) in human osteosarcoma cells . Biosci Rep 2016;36(5):e00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Li G, Wang G, Ma L, et al. . miR-22 regulates starvation-induced autophagy and apoptosis in cardiomyocytes by targeting p38alpha . Biochem Biophys Res Commun 2016;478(3):1165–72. doi: 10.1016/j.bbrc.2016.08.086. [DOI] [PubMed] [Google Scholar]
- 165. Zhang F, Wang B, Long H, et al. Decreased miR-124-3p expression prompted breast cancer cell progression mainly by targeting Beclin-1 .Clin Lab 2016;62(6):1139–45. [DOI] [PubMed] [Google Scholar]
- 166. Singh AK, Pandey RK, Shaha C, et al. . MicroRNA expression profiling of Leishmania donovani-infected host cells uncovers the regulatory role of MIR30A-3p in host autophagy . Autophagy 2016;12(10):1817–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Zhou J, Yao W, Liu K, et al. . MicroRNA let-7g regulates mouse granulosa cell autophagy by targeting insulin-like growth factor 1 receptor. Int J Biochem Cell Biol 2016;78:130–40. doi: 10.1016/j.biocel.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 168. Zou Y, Liu W, Zhang J, et al. . miR-153 regulates apoptosis and autophagy of cardiomyocytes by targeting Mcl-1 . Mol Med Rep 2016;14(1):1033–9. doi: 10.3892/mmr.2016.5309. [DOI] [PubMed] [Google Scholar]
- 169. Wang B, Zhong Y, Huang D, et al. . Macrophage autophagy regulated by miR-384-5p-mediated control of Beclin-1 plays a role in the development of atherosclerosis . Am J Transl Res 2016;8(2):606–14. [PMC free article] [PubMed] [Google Scholar]
- 170. Shu L, Li C, Zhang X. The role of shrimp miR-965 in virus infection. Fish Shellfish Immunol 2016;54:427–34. doi: 10.1016/j.fsi.2016.04.129. [DOI] [PubMed] [Google Scholar]
- 171. Tomasetti M, Monaco F, Manzella N, et al. . MicroRNA-126 induces autophagy by altering cell metabolism in malignant mesothelioma . Oncotarget 2016;7(24):36338–52. doi: 10.18632/oncotarget.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Zou M, Wang F, Gao R, et al. . Autophagy inhibition of hsa-miR-19a-3p/19b-3p by targeting TGF-beta R II during TGF-beta1-induced fibrogenesis in human cardiac fibroblasts. Sci Rep 2016;6:24747. doi: 10.1038/srep24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Ouimet M, Koster S, Sakowski E, et al. . Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism . Nat Immunol 2016;17(6):677–86. doi: 10.1038/ni.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Zhang M, Xie Y, Yan R, et al. . Curcumin ameliorates alveolar epithelial injury in a rat model of chronic obstructive pulmonary disease. Life Sci 2016;164:1–8. doi: 10.1016/j.lfs.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 175. Zhu H, Huang L, Zhu S, et al. . Regulation of autophagy by systemic admission of microRNA-141 to target HMGB1 in l-arginine-induced acute pancreatitis in vivo . Pancreatology 2016;16(3):337–46. doi: 10.1016/j.pan.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 176. Wei JL, Li YC, Ma ZL, et al. . MiR-181a-5p promotes anoikis by suppressing autophagy during detachment induction in the mammary epithelial cell line MCF10A . Protein Cell 2016;7(4):305–9. doi: 10.1007/s13238-016-0255-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Tekirdag KA, Akkoc Y, Kosar A, et al. . MIR376 family and cancer . Histol Histopathol, 2016;31(8):841–55. doi: 10.14670/HH-11-752. [DOI] [PubMed] [Google Scholar]
- 178. John Clotaire DZ, Zhang B, Wei N, et al. . MiR-26b inhibits autophagy by targeting ULK2 in prostate cancer cells. Biochem Biophys Res Commun 2016;472(1):194–200. doi: 10.1016/j.bbrc.2016.02.093. [DOI] [PubMed] [Google Scholar]
- 179. Li W, Yang Y, Hou X, et al. . MicroRNA-495 regulates starvation-induced autophagy by targeting ATG3. FEBS Lett 2016;590(6):726–38. doi: 10.1002/1873-3468.12108. [DOI] [PubMed] [Google Scholar]
- 180. Guo J, Sang Y, Yin T, et al. . miR-1273g-3p participates in acute glucose fluctuation-induced autophagy, dysfunction, and proliferation attenuation in human umbilical vein endothelial cells. Am J Physiol Endocrinol Metab 2016;310(9):E734–43. doi: 10.1152/ajpendo.00444.2015. [DOI] [PubMed] [Google Scholar]
- 181. Guo YJ, Liu JX, Guan YW. Hypoxia induced upregulation of miR-301a/b contributes to increased cell autophagy and viability of prostate cancer cells by targeting NDRG2. Eur Rev Med Pharmacol Sci 2016;20(1):101–8. [PubMed] [Google Scholar]
- 182. Zhou Z, You Z. Mesenchymal stem cells alleviate LPS-induced acute lung injury in mice by MiR-142a-5p-controlled pulmonary endothelial cell autophagy .Cell Physiol Biochem 2016;38(1):258–66. doi: 10.1159/000438627. [DOI] [PubMed] [Google Scholar]
- 183. Au KY, Pong JC, Ling WL, et al. . MiR-1303 regulates mycobacteria induced autophagy by targeting Atg2B. PLoS One 2016;11(1):e0146770. doi: 10.1371/journal.pone.0146770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Luo J, Chen J, He L. Mir-129-5p attenuates irradiation-induced autophagy and decreases Radioresistance of breast cancer cells by targeting HMGB1. Med Sci Monit 2015;21:4122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Huangfu L, Liang H, Wang G, et al. . miR-183 regulates autophagy and apoptosis in colorectal cancer through targeting of UVRAG. Oncotarget 2016;7(4):4735–45. doi: 10.18632/oncotarget.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Ma Y, Yang HZ, Dong BJ, et al. . Biphasic regulation of autophagy by miR-96 in prostate cancer cells under hypoxia . Oncotarget 2014;5(19):9169–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Hou C, Zhu M, Sun M, et al. . MicroRNA let-7i induced autophagy to protect T cell from apoptosis by targeting IGF1R . Biochem Biophys Res Commun 2014;453(4):728–34. doi: 10.1016/j.bbrc.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 188. Song L, Su M, Wang S, et al. . MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1 . J Cell Mol Med 2014;18(11):2266–74. doi: 10.1111/jcmm.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Sümbül AT, Göğebakan B, Ergün S, et al. . miR-204-5p expression in colorectal cancer: An autophagy-associated gene . Tumour Biol 2014;35(12):12713–9. doi: 10.1007/s13277-014-2596-3. [DOI] [PubMed] [Google Scholar]
- 190. Sun Q, Liu T, Yuan Y, et al. . MiR-200c inhibits autophagy and enhances radiosensitivity in breast cancer cells by targeting UBQLN1 . Int J Cancer 2015;136(5):1003–12. doi: 10.1002/ijc.29065. [DOI] [PubMed] [Google Scholar]
- 191. Kumar R, Sahu SK, Kumar M, et al. . MicroRNA 17-5p regulates autophagy in mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3 . Cell Microbiol 2016;18(5):679–91. doi: 10.1111/cmi.12540. [DOI] [PubMed] [Google Scholar]
- 192. Ramalinga M, Roy A, Srivastava A, et al. . MicroRNA-212 negatively regulates starvation induced autophagy in prostate cancer cells by inhibiting SIRT1 and is a modulator of angiogenesis and cellular senescence. Oncotarget 2015;6(33):34446–57. doi: 10.18632/oncotarget.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Du F, Feng Y, Fang J, et al. . MicroRNA-143 enhances chemosensitivity of Quercetin through autophagy inhibition via target GABARAPL1 in gastric cancer cells. Biomed Pharmacother 2015;74:169–77. doi: 10.1016/j.biopha.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 194. Li X, Zeng Z, Li Q, et al. . Inhibition of microRNA-497 ameliorates anoxia/reoxygenation injury in cardiomyocytes by suppressing cell apoptosis and enhancing autophagy. Oncotarget 2015;6(22):18829–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. An Y, Zhang Z, Shang Y, et al. . miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis 2015;6:e1766. doi: 10.1038/cddis.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Kim JK, Yuk JM, Kim SY, et al. . MicroRNA-125a inhibits autophagy activation and antimicrobial responses during mycobacterial infection. J Immunol 2015;194(11):5355–65. doi: 10.4049/jimmunol.1402557. [DOI] [PubMed] [Google Scholar]
- 197. Wang K, Liu CY, Zhou LY, et al. . APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 198. Stiuso P, Potenza N, Lombardi A, et al. . MicroRNA-423-5p promotes autophagy in cancer cells and is increased in serum from Hepatocarcinoma patients treated with Sorafenib. Mol Ther Nucleic Acids 2015;4:e233. doi: 10.1038/mtna.2015.8. [DOI] [PubMed] [Google Scholar]
- 199. Bhattacharya A, Schmitz U, Raatz Y, et al. . miR-638 promotes melanoma metastasis and protects melanoma cells from apoptosis and autophagy. Oncotarget 2015;6(5):2966–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Yu X, Luo A, Liu Y, et al. . MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol Cancer 2015;14:208. doi: 10.1186/s12943-015-0480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Chen ZH, Wang WT, Huang W, et al. . The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway . Cell Death Differ 2017;24(2):212–24. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Zhuo C, Jiang R, Liu X, et al. . LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy . Oncotarget 2017;8(1):1429–37. doi: 10.18632/oncotarget.13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Liu Z, Wei X, Zhang A, et al. . Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p . Biochem Biophys Res Commun 2016;473(4):1268–75. doi: 10.1016/j.bbrc.2016.04.054. [DOI] [PubMed] [Google Scholar]
- 204. Li L, Chen H, Gao Y, et al. . Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther 2016;15(9):2232–43. doi: 10.1158/1535-7163.MCT-16-0008. [DOI] [PubMed] [Google Scholar]
- 205. Liu X, Xiao ZD, Han L, et al. . LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress . Nat Cell Biol 2016;18(4):431–42. doi: 10.1038/ncb3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Zhou W, Zhang S, Li J, et al. . lncRNA TINCR participates in ALA-PDT-induced apoptosis and autophagy in cutaneous squamous cell carcinoma . J Cell Biochem 2019;120(8):13893–902. doi: 10.1002/jcb.28662. [DOI] [PubMed] [Google Scholar]
- 207. Zhao X, Su L, He X, et al. . Long noncoding RNA CA7-4 promotes autophagy and apoptosis via sponging MIR877-3P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy 2019;1–16. doi: 10.1080/15548627.2019.1598750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Wang X, Zhang M, Liu H. LncRNA17A regulates autophagy and apoptosis of SH-SY5Y cell line as an in vitro model for Alzheimer's disease .Biosci Biotechnol Biochem 2019;83(4):609–21. doi: 10.1080/09168451.2018.1562874. [DOI] [PubMed] [Google Scholar]
- 209. Zhang X, Liu J, Gu Y, et al. Down-regulation of lncRNA OGFRP1 induces autophagy and growth inhibition by AKT/mTOR signaling pathway in HCAECs .Cell Biol Int 2019;43(2):158–66. doi: 10.1002/cbin.11081. [DOI] [PubMed] [Google Scholar]
- 210. Huang FX, Chen HJ, Zheng FX, et al. LncRNA BLACAT1 is involved in chemoresistance of nonsmall cell lung cancer cells by regulating autophagy .Int J Oncol 2019;54(1):339–47. doi: 10.3892/ijo.2018.4614. [DOI] [PubMed] [Google Scholar]
- 211. Zhao H, Wang X, Feng X, et al. . Long non-coding RNA MEG3 regulates proliferation, apoptosis, and autophagy and is associated with prognosis in glioma . J Neurooncol 2018;140(2):281–8. doi: 10.1007/s11060-018-2874-9. [DOI] [PubMed] [Google Scholar]


