Abstract
Limb-girdle muscular dystrophy recessive 1 (LGMDR1), previously known as LGMD2A, is a rare disease caused by mutations in the CAPN3 gene. It is characterized by progressive weakness of shoulder, pelvic, and proximal limb muscles that usually appears in children and young adults and results in loss of ambulation within 20 years after disease onset in most patients. The pathophysiological mechanisms involved in LGMDR1 remain mostly unknown, and to date, there is no effective treatment for this disease. Here, we review clinical and experimental evidence suggesting that dysregulation of Ca2+ homeostasis in the skeletal muscle is a significant underlying event in this muscular dystrophy. We also review and discuss specific clinical features of LGMDR1, CAPN3 functions, novel putative targets for therapeutic strategies, and current approaches aiming to treat LGMDR1. These novel approaches may be clinically relevant not only for LGMDR1 but also for other muscular dystrophies with secondary calpainopathy or with abnormal Ca2+ homeostasis, such as LGMD2B/LGMDR2 or sporadic inclusion body myositis.
Keywords: calpain 3, calcium, LGMD2A, LGMDR1, muscular dystrophies, calpainopathy
1. Overview of Calcium Homeostasis in the Skeletal Muscle
Ca2+ plays a vital role in a wide range of cellular processes such as gene transcription, membrane resealing, secretion, neurotransmission, as well as cell differentiation, proliferation, or survival [1,2]. In skeletal muscle fibers, Ca2+ is crucial for both electric activation along the motor endplate and skeletal muscle contraction. In addition, Ca2+ is involved in many other functions such as protein synthesis, protein degradation, fiber type shifting, Ca2+-regulated proteolysis, transcription factor modulation, mitochondrial adaptation, cell plasticity, and respiration [3]. Therefore, tight regulation of Ca2+ levels is essential for the proper function of skeletal muscle (Figure 1).
Figure 1.
Representation of Ca2+ fluxes in the muscle fiber. Upon sarcolemmal depolarization reaching T-tubules (1), DHPRs undergo a conformational change that activates RyR1 channels and results in Ca2+ release from the SR (2). Ca2+ diffuses to the sarcomere where it initiates muscle contraction (3). Muscle relaxation takes place when Ca2+ is sequestered into the SR by SERCAs (4) or pumped out of the fiber by membrane channels (NCX, PMCA) (5). Cytosolic Ca2+ also binds CaM, which activates the Ca2+-dependent signaling pathways resulting in muscle gene regulation (6). Cytosolic Ca2+ also reaches mitochondria (7), where it stimulates metabolism and ATP synthesis required for muscle contraction and relaxation.
1.1. Ca2+ in Excitation-Contraction Coupling
Muscle contraction initiates by depolarization of the sarcolemma in response to acetylcholine release from motoneurons. The action potential propagates into the triads, which are anatomical structures formed by the association of sarcolemmal transverse tubules (T-tubules) and sarcoplasmic reticulum (SR) terminal cisternae [4]. Triads play an essential role in excitation-contraction coupling (ECC) since they allow close contact and synchronization between crucial receptors in the sarcolemma and the SR. See [3,5] for review. Membrane depolarization activates dihydropyridine receptors (DHPRs) in the T-tubules [6], and this results in activation of the closely apposed ryanodine receptors (RyRs), the main Ca2+ release channels in the SR [7]. RyR1, the predominant RyR isoform in skeletal muscle, releases Ca2+ from the SR into the cytosol either after activation by DHPRs or increased cytosolic Ca2+ levels [8]. RyR1 function is modulated by post-translational modifications, such as S-nitrosylation, S-glutathionylation, and phosphorylation by both Protein Kinase A (PKA) and Ca2+/calmodulin-dependent protein kinase II (CaMKII) [9]. RyRs operate in coordination with other proteins in order to maintain the balance between Ca2+ release, Ca2+ storage, and Ca2+ reuptake [10]. Indeed, a variety of proteins and small molecules, both in the SR lumen and cytosol are needed for this tight coordination [9]. On the cytosolic side, calmodulin (CaM), a Ca2+ sensor, has a dual effect on RyR1, functioning as an activator at low cytosolic Ca2+ levels and as an inhibitor at high cytosolic Ca2+ levels [11]. On the SR luminal side, calsequestrin (CSQ) forms a complex with RyRs, junctin, and triadin. RyR function is inhibited by the binding of CSQ, which essentially depends on luminal Ca2+ concentration [12]. CSQ is the main Ca2+-binding protein in the SR lumen, and it functions as an endogenous regulator of Ca2+ fluxes and as a Ca2+ reservoir with a moderate affinity but high capacity to bind Ca2+ [13]. After Ca2+ release into the cytosol through RyR1, Ca2+ binds to various cytosolic Ca2+ buffers, such as ATP and CaM, and it can also be sequestered by mitochondria. At the sarcomere, the contractile unit of the skeletal muscle, troponin C undergoes a Ca2+-dependent conformational change that ultimately results in myosin and actin cross-bridge cycling and muscle contraction [14]. Upon muscle excitation, cytosolic Ca2+ levels raise from ~100 nM (resting levels) to ~10µM in slow fibers, and ~18 µM in fast fibers [15].
Muscle relaxation initiates by lowering of cytosolic Ca2+ back to resting levels, which mainly relies on sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) pumps [16], and sarcolemmal Ca2+ transporters such as Na+/Ca2+ exchangers (NCX1-3) and the plasma membrane Ca2+-ATPase (PMCA) [3]. SERCA actively transports Ca2+ from the cytosol into the SR against a large concentration gradient at the expense of ATP hydrolysis. Fast muscle fibers express SERCA1a isoform, whereas slow fibers and cardiac muscle express SERCA2a isoform [16]. Since SERCA pumps are major ATP consumers, they are strongly affected by changes in cell energetics and ATP supply [17]. Similarly to RyRs, SERCAs are also modulated by several cytosolic and SR luminal proteins, as well as by post-translational modifications, including N-glycosylation, S-glutathionylation, and phosphorylation [18], although there is some controversy over SERCA modulation through phosphorylation. Also, the short integral membrane proteins phospholamban (PLN) and sarcolipin (SLN) inhibit SERCA activity [19]. Phosphorylation of PLN by PKA or CaMKII results in SERCA activation through dissociation of PLN from the Ca2+ pump [18].
1.2. Ca2+-Mediated Signaling Pathways
In addition to its role in ECC, Ca2+ is also a key regulator of gene transcription triggered by different stimuli. For instance, intracellular Ca2+ signals mediate transcriptional changes necessary for skeletal muscle adaptation in response to changes in activation patterns [20]. CaMK pathway and Calcineurin (Cn), a Ca2+/CaM-dependent serine/threonine protein phosphatase, have crucial roles in many of these Ca2+-mediated signaling processes. Cn stimulates the transcription of both NFAT and NF-κB targeted genes [21,22]. While the Cn/NFAT pathway responds preferentially to sustained and low-amplitude elevations of intracellular Ca2+, high amplitude oscillations activate NF-κB [23]. NFAT signaling induces the slow gene program during muscle regeneration and maintains the slow fiber phenotype in the adult muscle tissue, whereas NF-κB regulates the gene program for myoblast proliferation and differentiation [24,25]. Interestingly, depending on the stimulus nature, NF-κB can behave either as a promoter or antagonist of apoptosis [26].
CaMK pathway is involved in the regulation of contraction-induced Ca2+ handling, and mitochondrial biogenesis. It also regulates gene expression in skeletal muscle, promoting the slow to fast fiber shift [27]. Also, during muscle development and adaptation, CaMK activates critical transcription factors such as MEF2 (myocyte enhancer factor 2) via phosphorylation of class II histone deacetylases (HDACs) [28]. CaMKII, the main CaMK in the skeletal muscle, is involved in the maintenance of myofiber phenotype and muscle growth [29,30]. In addition, CaMKII is one of the upstream activators of AMP-activated protein kinase (AMPK), an energy sensor that coordinates cell growth and autophagy, as well as regulates mitochondrial function and biogenesis. Under low intracellular ATP levels, AMPK promotes catabolic pathways to generate more ATP and suppresses anabolic pathways such as the high ATP consuming mTORC1 pathway, to inhibit cell growth [31].
1.3. Ca2+ in Mitochondria
Mitochondria are, together with SR, main Ca2+ storages, and they also function as a source of intracellular Ca2+ [2]. They account for about 15% of the cytosolic volume in oxidative fibers [32], and they are located close to the SR in the skeletal muscle. There is a significant interplay between mitochondria and SR through the mitochondria-associated SR membrane (MAM), which is essential for cell physiology and Ca2+ homeostasis [33]. Ca2+ regulates mitochondrial metabolism, biogenesis, motility, distribution, and plasticity [34,35,36]. In turn, mitochondria can also affect intracellular Ca2+ levels. Thus, ECC triggers transient Ca2+ increases in the mitochondrial matrix, which are essential to promote mitochondrial metabolism and ATP synthesis. This is required to balance the ATP consumption of actomyosin cross-bridge cycling and SERCA pumps during contraction and relaxation, respectively [37]. Alternatively, Ca2+ overload in the mitochondria may induce the opening of permeability transient pores (PTP) that results in a massive release of Ca2+ and pro-apoptotic factors such as cytochrome C (Cyt-c) into the cytosol, which, in turn, activate caspase 9 and initiate the apoptotic program [38,39].
Ca2+ is also involved in mitochondrial biogenesis through regulation of the CaMKII pathway. Indeed, the expression of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC1α), a key regulator of mitochondrial biogenesis, depends on the Ca2+-mediated activation of CaMKII [40]. Finally, mitochondrial motility also relies on cytosolic [Ca2+]. Indeed, myosin-Va, a Ca2+ sensor molecule, may regulate mitochondria-bound molecular motors allowing mitochondrial movements along cytoskeletal fibers and controlling the distribution of mitochondria to enhance Ca2+ buffering and ATP production in regions with high cytosolic [Ca2+] [36].
2. Limb-Girdle Muscular Dystrophy-Recessive 1
LGMDR1, also known as calpainopathy and previously symbolized LGMD2A [41], is caused by homozygous or compound heterozygous mutation in the CAPN3 gene encoding for the proteolytic enzyme calpain 3 (CAPN3) [42]. Recently, heterozygous mutations in the CAPN3 gene have been reported to cause autosomal dominant limb-girdle muscular dystrophy-4 (LGMDD4), with a later onset and milder phenotype [43,44,45], although the mechanisms underlying these cases still need some clarification [46]. The prevalence of LGMDR1 ranges from 1 to 9 cases per 100,000 people, and it represents almost 30% of all LGMD cases in open populations [47,48,49,50,51,52,53] with some ancestral mutations responsible for specific ethnic or geographic clusters [54,55,56,57,58]. By September 2019, there are more than 480 pathogenic variants of CAPN3 reported in the Leiden Open Variation database [59]. The molecular spectrum covers all CAPN3 exons with some hot regions related to severe or benign phenotypes, as well as intronic variants [51,60].
LGMDR1 is characterized by progressive muscle weakness and degeneration, with a predominant effect on shoulder, pelvic, and proximal limb muscles [61]. There is no affection of cardiac and facial muscles, and no cognitive defects have been reported in this disease [47]. Age of onset is highly variable, although initial symptoms usually appear between eight and 15 years [62] and patients loss ambulation around 10 to 20 years after the onset [54,55]. However, lately, benign forms are being increasingly reported with preserved ambulation even after reaching 60 or more years old. In general, these benign forms have metabolic symptoms at onset (myalgia, cramps, and exercise intolerance) or even asymptomatic hyper-creatine kinase-emia that may carry on for years before muscle weakness. Symptoms of the classical LGMDR1 phenotype fit with the criteria described by Erb in 1884 to define juvenile muscular dystrophy [54]. However, there is certain variability regarding disease progression and severity related to gender, as well as the type and localization of mutations [51]. Moreover, a phenomenon known as de novo intermolecular complementation (iMOC) of CAPN3 may also lead to a milder phenotype in compound heterozygotes [63]. Additionally, in some families, there is considerable phenotypic variability among patients with identical mutations [64], which makes prognosis in LGMDR1 very challenging [47].
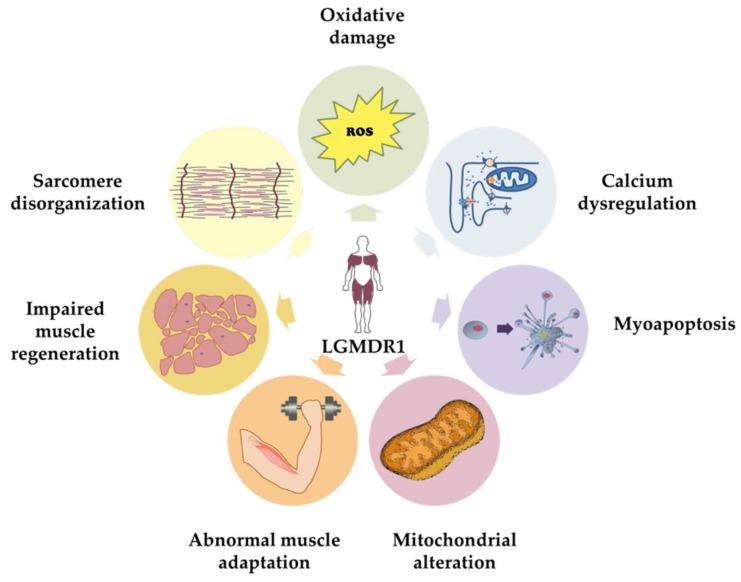
Since the discovery of CAPN3 as the gene responsible for LGMDR1, several groups have been trying to identify the pathogenic mechanisms that may give rise to the clinical and histological features of LGMDR1. Although to date, these mechanisms are not entirely understood, there is solid evidence indicating that CAPN3 is a multifunctional protein. Different studies performed in animal models and human samples have shown that CAPN3 deficiency is associated with different features in the skeletal muscle such as oxidative damage [65,66], Ca2+ dysregulation [67,68], sarcomere disorganization [69], mitochondrial abnormalities [66,70,71,72], abnormal muscle adaptation [73,74], and impaired muscle regeneration [71], which together would lead to inflammation, necrosis, fibrosis, atrophy, and progressive muscle degeneration, characteristic of LGMDR1 (Figure 2 and Figure 3). Indeed, patients in the early stages of the disease present an increased concentration of serum creatine kinase (CK), which is an unspecific hallmark of muscle damage [55,75]. Some patients at this stage present eosinophilic infiltrations associated with peripheral blood eosinophilia that have an unclear pathogenic significance [50,54,76]. Fibrosis is often present, and it tends to increase with disease progression [75].
Figure 2.
Illustration of the pathological features of CAPN3 deficiency in the skeletal muscle.
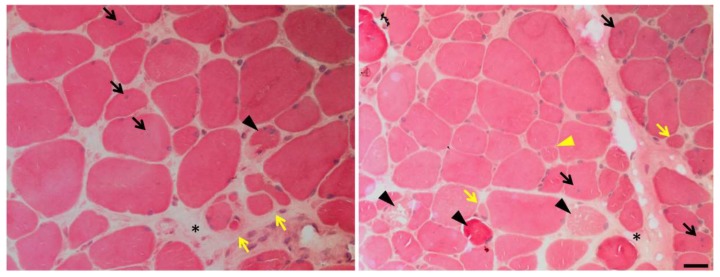
Figure 3.
Muscle biopsy of a LGMDR1 patient. Hematoxylin and eosin staining shows endomysial fibrosis (black asterisks), central nuclei (black arrows), fiber splitting (yellow triangle), necrosis (black triangles), atrophic fibers (yellow arrows) and increased variation in fiber size and shape. Scale bar: 25 μm.
Muscle biopsies from LGMDR1 patients also present general dystrophic features, such as necrotic areas with regenerative regions, fiber size variability, central nuclei, and disorganized myofibrils (Figure 3). Finally, muscles from LGMDR1 patients may present myonuclear apoptosis and a peculiar pattern of focal degeneration, as well as fibers with a lobulated pattern, commonly attributed to sarcomere and mitochondria disorganization [50].
3. CAPN3 Localization and Function
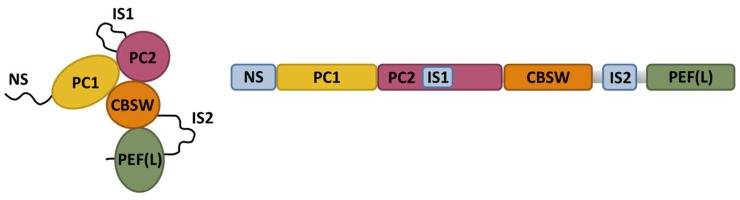
CAPN3 belongs to the calpain superfamily of Ca2+-dependent non-lysosomal cysteine proteases. Calpains have relevant functions in many cellular processes, including cell motility, apoptosis, cell differentiation, and cell-cycle regulation. In these processes, intracellular Ca2+ activates calpains, which then cleave specific substrates [77,78,79,80]. In mammals, the ubiquitous CAPN1 (µ-calpain) and CAPN2 (m-calpain) are the most extensively studied calpains. CAPN3 shares catalytic domain structure with CAPN1 and CAPN2 but CAPN3 has three unique regions, namely NS, IS1, and IS2, (Figure 4) that confer this protein some unusual features, such as an extreme instability and fast autodegradation rate, and a Na+-dependent activation [81].
Figure 4.
Schematic representation of CAPN3 structure. CAPN3 is comprised of two protease core domains (PC1 and PC2), a calpain-type β-sandwich domain (CBSW), and a penta E-F hand domain (PEF) that binds four calcium ions and may contribute to CAPN3 dimerization. The three specific regions of CAPN3 (NS, IS1, and IS2) are shown in blue. Schematics have been modified from Ye et al., 2018 [82]. The right panel depicts the estimated tertiary structure of CAPN3.
One of the most remarkable features of CAPN3 is its extreme autodegradation rate [83], which has hindered conventional biochemical analysis. Indeed, the native structure of CAPN3 remains unresolved, as well as the accurate interactions of CAPN3 with other proteins. Another extremely unusual characteristic of CAPN3 is its ability to regain proteolytic function after its autolytic dissociation. This occurs through iMOC, a process where two autolytic fragments of CAPN3 reconstitute an active core protease domain [84]. With regards to its proteolytic function, CAPN3 can be activated at physiological intracellular concentrations of Ca2+ (100 nM) and Na+ (15 mM) [85], and thus, Na+ is responsible for the low Ca2+ levels required to activate CAPN3. In fact, in absence of Na+, the Ca2+ concentration required for CAPN3 autolysis would be around 0.1 mM. Hence, in the skeletal muscle, CAPN3 autolytic activity is suppressed in vivo, through its binding to connectin/titin [86]. CAPN3 presents both proteolytic as well as non-proteolytic functions [81]. Different studies have identified several mechanisms dependent on CAPN3 proteolytic function, such as mechanosensory transduction and sarcomere remodeling after exercise [81,87]. Non-proteolytic features of CAPN3, independent of its protease activity, have been identified through comparative studies in CAPN3 knockout and knock-in mice. These studies indicate that CAPN3 contributes to the maintenance of Ca2+ homeostasis through the stabilization of critical Ca2+-handling proteins, which relies on non-proteolytic functions of CAPN3 [88,89,90]. These functions will be further discussed in the following section.
CAPN3 presents a broad distribution within the muscle fiber, having been found at the sarcomere, membrane fraction, sarcoplasmic reticulum (SR), cytosol, and even the nucleus [88,91,92]. CAPN3 has been mainly implicated in the regulation of muscle contraction and sarcomere stability [85,92,93,94]. Within the sarcomere, CAPN3 is localized in several regions, where it interacts with different proteins. CAPN3 interacts at the Z-line with α-actinin-3, tropomyosin, and LIM-domain binding protein 3. CAPN3 also interacts with titin, a large scaffold protein that plays a vital role in sarcomere assembly and passive tension of myofibrils, as well as in mechanosensory transduction pathways [92,95]. Interestingly, CAPN3 binds to titin at the N2A and M-line regions, with different affinity depending on the sarcomere length. Thus, the presence of CAPN3 at the N2A region is increased compared to the M-line region when the sarcomere stretches. This location shift along different titin regions is facilitated by CAPN3 proteolytic activity [93], and it is crucial for the dissociation of Muscle Ankyrin Repeat Protein-2 (MARP-2) from titin, and its translocation to the nucleus to transmit signals of mechanical perturbation [96], which suggests that CAPN3 may function as a sensor of sarcomere integrity.
Titin stabilizes CAPN3 by preventing its autodegradation [69], and therefore, CAPN3 activity may be regulated by titin binding. On the other hand, since titin is also a CAPN3 substrate, CAPN3 may be responsible for the fast turnover of titin [97], and likely other sarcomere proteins, an aspect that is necessary for appropriate maintenance of the sarcomere structure [98,99]. In this line, C3KO mice present abnormal A-bands at the sarcomere and delayed myofibrillogenesis [69], together with an accumulation of high molecular weight ubiquitin-protein conjugates [87]. Altogether these findings suggest that proper interaction between CAPN3 and titin is essential for sarcomere maintenance and remodeling [100].
4. Ca2+-Mediated Pathogenic Mechanisms Involved in CAPN3 Deficiency
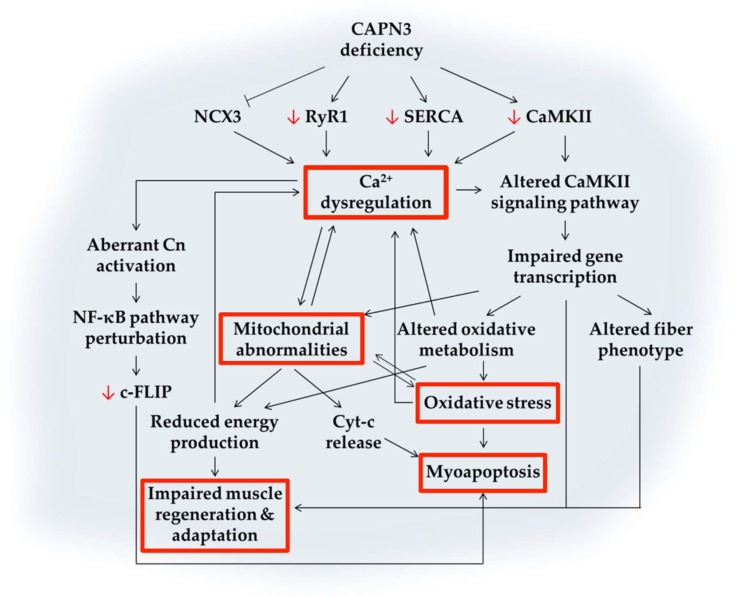
Evidence obtained from animal models and human patients implicates Ca2+ homeostasis as a pathophysiological mechanism underlying different muscular dystrophies, including LGMDR1 [5]. Several mouse models of LGMDR1 have been used to understand the pathogenic mechanisms resulting from CAPN3 deficiency. Among these models, the CAPN3 knockout mouse lines Capn3−/− and C3KO [69,101], and the CAPN3 knock-in mice Capn3CS/CS, expressing a structurally intact but inactive CAPN3 [96,102] have enabled to identify specifically non-proteolytic and proteolytic functions of CAPN3. Noteworthy, the phenotype of these mouse models does not fully recapitulate the severity of LGMDR1 in human patients, likely due to the higher regenerative potential of the murine muscle [103] and/or the increased Ca2+ buffering capacity of murine fibers [67]. Therefore, data from cellular models and muscle samples of patients with LGMDR1 need to be considered in order to unravel the molecular pathways involved in LGMDR1. During the last years, our group has been working on understanding these mechanisms using human biopsies as well as primary and immortalized human myotubes [67,68,104]. Below, we will review recent findings on pathogenic mechanisms associated with CAPN3 deficiency, with a particular focus on the involvement of Ca2+ dysregulation, as summarized in Figure 5.
Figure 5.
Schematic representation of putative Ca2+-mediated pathogenic mechanisms triggered by CAPN3 deficiency. CAPN3 deficiency results in reduced levels of RyR1, SERCA, and CaMKII. In addition, NCX3 activity may also be reduced. The decreased function of major Ca2+-handling proteins results in Ca2+ dysregulation and increased intracellular [Ca2+]. Reduced CaMKII levels together with Ca2+ dysregulation compromise CaMK downstream signaling pathways, which may lead to impaired gene transcription, mitochondrial abnormalities, oxidative stress, altered fiber phenotype, and impaired muscle regeneration. Mitochondrial abnormalities aggravate Ca2+ dysregulation and oxidative damage. They may also impact energy production and promote apoptosis through Cyt-c release and activation of caspases. Among these mechanisms, multiple feedback loops lead to altered Ca2+ levels and may result in myoapoptosis and muscle waste. Black arrows or blunt ends indicate enhancing or inhibitory effects, respectively. Red arrows indicate decreased protein expression. Text in red boxes represent several pathological features of CAPN3, as described in Figure 2.
4.1. Calcium Dysregulation
CAPN3 has been found to interact with several key Ca2+-handling proteins, such as RyR1, CSQ, and SERCA [88]. In this line, several studies have shown that CAPN3 deficiency results in abnormal Ca2+ handling in the skeletal muscle. Indeed, previous studies performed on the CAPN3 knockout mice C3KO and Capn3−/−, indicate that these mice present reduced RyR1 expression together with a decrease in SR Ca2+ release [89,90]. Also, Capn3−/− knockout myotubes display reduced SR Ca2+ levels together with a lower response to SERCA inhibitors compared to wild-type myotubes [90]. Moreover, we have also contributed to a study showing reduced RyR1 expression and CaMKII signaling in muscles from LGMDR1 patients and C3KO mice [73]. At the triads, CAPN3 is part of a complex comprised of RyR1, AldoA, and CaMKII. Interestingly, in the absence of CAPN3, in C3KO mice, both RyR1 and CaMKII protein levels are decreased while AldoA is mislocalized. Thus, CAPN3 has been proposed as a structural stabilizer of RyR1 complexes at the triads [73,89]. This structural function of CAPN3 may depend on specific genetic regions since a recent study with LGMDR1 patients has reported a new CAPN3 mutation that does not result in diminished RyR1 in the skeletal muscle [70].
Our group has recently found that mouse and human myotubes deficient for CAPN3 display decreased SERCA protein levels as well as impaired Ca2+ reuptake into the SR [67,68]. Moreover, we also found reduced SERCA expression in muscle samples from LGMDR1 patients. In line with these findings, a recent study has shown that in sporadic inclusion body myositis, a common acquired muscle disease associated with aging, there is a secondary calpainopathy and a concomitant reduction of SERCA proteins that leads to Ca2+ dyshomeostasis [105]. Interestingly, we have found that SERCA deficiency in CAPN3 knockdown myotubes resulted in increased resting intracellular [Ca2+] in human myotubes, but not in mouse myotubes [67]. This is likely due to the fact that mouse muscle fibers have a higher Ca2+ buffer capacity, since parvalbumin, a major cytosolic Ca2+ buffer, is highly expressed in the mouse muscle [68]. In any case, SERCA mRNA levels were found unchanged in CAPN3 deficient samples, and therefore, we propose that CAPN3 is necessary to stabilize SERCA proteins and prevent their degradation, similarly to RyR1 and CaMKII. Interestingly, impaired SERCA function leads to SR Ca2+ depletion and ultimately to SR stress that activates the unfolded protein response (UPR) [106,107]. Indeed, in CAPN3 deficient myotubes, we found that several makers of SR stress and UPR are upregulated such as GRP78, CHOP, HERP, and the XBP1 spliced variant [68].
Finally, CAPN3 may also regulate Ca2+ homeostasis by increasing the activity of NCX3. In HEK293T Michel et al. describe an increased activity of the NCX3 muscle isoform, NCX3-AC, following CAPN3 cleavage [108]. This Na+/Ca2+ exchanger is found at the triads and extrudes Ca2+ across the sarcolemma to lower intracellular [Ca2+] during relaxation [108]. Nevertheless, more studies need to be performed in order to verify the regulation of NCX3 by CAPN3 in myotubes as well as in LGMDR1 models.
4.2. Abnormal Muscle Adaptation
The skeletal muscle is a remarkably adaptable tissue that responds to physiological and environmental challenges by altering its size and composition [109]. Ca2+ plays a fundamental role in muscle adaptation to changes in functional demand by activating specific Ca2+-dependent transcriptional pathways, such as CaMK pathways, in order to control muscle growth, fiber type transition, or mitochondrial biogenesis. In particular, CaMKII has been proposed as a major sensor of muscle activity that translates it into phenotypic adaptations by regulating the transcription of specific genes [30]. CaMKII signaling has been found severely compromised in the CAPN3 knockout mice, C3KO, and thus, abnormal muscle adaptation has been proposed as a major instrumental mechanism in LGMDR1 [73,74]. Indeed, in C3KO mice, reduced CaMKII expression results in an impaired slow myogenic program [73]. This may explain why muscles highly enriched in slow-twitch fibers, such as soleus and diaphragm are the most severely affected in C3KO mice, and also why there seems to be a preferential involvement of slow fibers in LGMDR1 muscle biopsies [73]. Moreover, after endurance exercise, CAPN3 deficient muscles fail to upregulate several groups of genes associated with muscle adaptation, such as myofibrillar, mitochondrial, and metabolic genes [74]. In this line, we have observed that human myotubes deficient for CAPN3 show reduced protein levels of CaMKII (unpublished data).
4.3. Mitochondrial Abnormalities
Several studies support the notion that CAPN3 is an important modulator of mitochondrial function, and its absence seems to have major consequences over this organelle [72,110]. LGMDR1 patients and CAPN3 knockdown mice present abundant mitochondria with abnormal spatial distribution [50,72,111], and a recent study points toward mutation-specific patterns of mitochondrial dysfunction in different LGMDR1 patients [70]. Moreover, in C3KO mice, mitochondria display a swollen appearance with disrupted membranes [72], while in LGMDR1 muscle biopsies, several mitochondrial genes have been found to be deregulated [112]. Mitochondrial abnormalities would have a direct impact on several pathomechanisms in LGMDR1, including Ca2+ dysregulation, energy deficits, oxidative stress, and ultimately, they could lead to cell death through the release of different pro-apoptotic factors into the cytosol (Figure 3). Indeed, C3KO muscles present decreased ATP production and increased oxidative stress [72], while LGMDR1 muscles show Cyt-c mislocalization to the cytosol [91], which may promote activation of caspases and apoptosis [113]. Likewise, mitochondria biogenesis and function are severely affected by Ca2+ dyshomeostasis [5]. Thus, while Ca2+ promotes mitochondrial metabolism and ATP synthesis, sustained high intracellular Ca2+ levels, such as the ones found in CAPN3 deficient myotubes [67,68], may result in mitochondrial Ca2+ overload and eventually lead to mitochondrial dysfunction and muscle cell degeneration. On the other hand, mitochondria biogenesis is modulated by PGC1α, which is in turn regulated by Cn and CaMK pathway, and therefore it is highly susceptible to Ca2+ dysregulation [40,114]. In this line, during muscle regeneration, CAPN3 knockout mice have proved unable to increase mitochondrial DNA content, as well as PGC1α and ATP5D transcripts, likely due to diminished CaMKII signaling [71].
4.4. Oxidative Stress
Oxidative and nitrosative stress have been associated with muscle wasting in several muscular dystrophies, including LGMDR1, where NAPDH oxidase appears to be one potential source of oxidative stress in LGMDR1 muscle biopsies [65]. In LGMDR1 biopsies, elevated reactive oxygen species (ROS), increased oxidized proteins, and lipid peroxidation have been reported [65,66]. Oxidative stress has also been associated with C3KO mouse muscles [72], where defective mitochondria may be a contributing factor since they are considered to generate the majority of cellular free radicals. Reciprocally, oxidative stress may also lead to mitochondrial damage [115], as well as Ca2+ dysregulation [116]. Therefore, identification of the cause-effect relationship among these features is very challenging. In any case, oxidative stress can cause cell death through necrotic or apoptotic pathways.
In response to oxidative stress, several protective mechanisms are activated to buffer extra ROS, such as superoxide dismutases (SOD). An increment in their concentration is indicative of oxidative stress. Accordingly, in CAPN3 knockout mouse increased levels of SOD have been reported, indicating a suitable protective response against oxidative stress [66]. In contrast, a reduction of antioxidant defense mechanisms has been described in LGMDR1 biopsies [66], pointing toward a higher vulnerability of human LGMDR1 muscles compared to mouse C3KO muscles.
4.5. Impaired Muscle Regeneration
Impaired muscle regeneration is another main pathological feature of LGMDR1 [71]. Muscle regeneration is a complex process that is initiated upon injury of muscle cells and can be divided into several stages: activation and proliferation of satellite cells, which are the muscle stem cells, differentiation or fusion of the muscle stem cells, maturation of the newly formed muscle fibers and the remodeling of muscle fibers. This process requires high levels of protein synthesis, proper mRNA translation, and energy consumption. Different signaling pathways underlie these processes, such as AMP-activated protein kinase (AMPK) signaling, which is regulated by CaMKII and liver kinase B1 (LKB1) [117,118]. In C3KO mice, during muscle regeneration after CTX injection, muscle fiber growth is arrested due to increased AMPK phosphorylation, inhibition of mTORC1, and energy shortage [71]. Regeneration is one of the most energy-consuming cellular processes, and therefore, C3KO mice fibers are not able to activate genes necessary to adapt to a new situation [119,120]. Moreover, abnormal sarcomere organization in C3KO mice may also contribute to impaired muscle regeneration [69].
Interestingly, in regenerating C3KO muscles [71], as well as in LGMDR1 biopsies [121], a similar miRNA pattern has been reported to the one described in muscles with impaired myofiber repair/regeneration and subsequent fibrosis, showing increased Pax7 expression and downregulation of the muscle specific miRNAs miR-1, miR-133a, and miR-206. These miRNAs, also known as dystromirs, are involved in myogenesis by promoting muscle proliferation (miR-133a) and differentiation (miR-1 and miR-206) [122]. Downregulation or inhibition of miR-1 and miR-206 is associated to increase in the proliferation of satellite cells and Pax7 expression in vivo [123,124]. On the other hand, miR-133a is involved in the maintenance of adult skeletal muscle structure, function, bioenergetics and myofiber identity [125]. Remarkably, mir-1, miR-206, and miR-133a have been proposed as disease biomarkers for Duchenne muscular dystrophy [122].
Finally, MyoD modulation by CAPN3 has been demonstrated in murine cell cultures, which signals CAPN3 as a potential player during muscle regeneration [126]. Moreover, a defective fusion of C3KO myoblast has been described in vitro, due to an accumulation of the β-catenin-M-cadherin complex at the membrane [127]. CAPN3 regulates the localization of β-catenin [127]. At the same time, FRZB, through the inhibition of the Wnt pathway, may prevent the translocation of β-catenin to the nucleus [104]. Interestingly, upregulated FRZB expression has been found in LGMDR1 muscle samples [104]. Therefore, CAPN3 seems to be involved in the fusion and maturation processes of myogenic cells.
4.6. Myoapoptosis
Several studies have reported apoptotic myonuclei in muscle samples from LGMDR1 patients [91] and Capn3−/− CAPN3 knockout mice [101]. Conversely, similar studies performed in the C3KO mice fail to detect apoptotic nuclei in muscle fibers [69]. Differences in Ca2+ buffer capacity or protective response against oxidative stress may account for the lack of apoptotic nuclei in this mouse model of LGMDR1.
In LGMDR1, apoptosis may be triggered through several pathways (see Figure 5). First, CAPN3 activates NFκB in a Ca2+ dependent manner, through degradation of the NF-κB inhibitor IκBα [101]. Activation of NF-κB in the skeletal muscle may lead to protein degradation, inflammation, and fibrosis [128], but it also may promote muscle cell survival under certain conditions [129,130]. This seems to be the case in LGMDR1 since previous works have shown that CAPN3 regulates the expression of NF-κB-dependent survival genes to prevent apoptosis in skeletal muscle, such as c-FLIP [129]. In particular, the expression of c-FLIP, a master anti-apoptotic regulator downstream NF-κB signaling pathway, is downregulated in human myotubes and mouse muscle deficient for CAPN3 [68]. Deregulations in the NF-κB pathway could be part of the mechanism responsible for the muscle wasting resulting from CAPN3 deficiency.
On the other hand, Cn also plays a significant role in apoptosis through the dephosphorylation of proteins involved in the apoptotic pathway, such as caspase 9. Phosphorylation of caspase 9 by Akt pathway results in caspase 9 inhibition, while Cn triggers caspase activity via dephosphorylation [131]. Third, Cyt-c leakage from mitochondria promotes apoptosome generation in the cytosol, which activates caspase 9 and the downstream caspase cascade, and may result in myoapoptosis [113]. Interestingly, a cytosolic Cyt-c localization has been reported in LGMD2A muscle biopsies [91].
Lastly, sustained SR stress, elevated intracellular calcium, and oxidative stress, all of which underlie LGMDR1 pathology, may also lead to apoptosis through caspase activation. Therefore, further studies are needed to elucidate the molecular mechanisms that trigger the apoptotic pathway and muscle waste in LGMDR1. These findings may be highly valuable for identification of novel therapeutic candidates.
5. Therapeutic Approaches for LGMDR1
Similar to other hereditary disorders, there are currently no effective therapies to treat LGMD. Moreover, treatments for these diseases have mostly been symptomatic so far. However, new treatments, both non-disease- and disease-specific, are emerging for LGMD [132]. Among non-disease-specific treatments, both strength and aerobic exercise training are beneficial for LGMD. In particular, strength training improves strength in LGMDR1, although improvement generally seems smaller than with endurance training [133,134]. One proposed pharmacological approach is to increase muscle mass by enhancing positive regulators of muscle growth, or by inhibiting negative regulators, such as myostatin. In this regard, in the Capn3−/− mouse model of LGMDR1, treatment with AAV-carrying a mutant myostatin propeptide with an inactive C-terminal domain has shown increased muscle mass and improved force generation [135]. On the other hand, a Phase I/II trial with MYO-029, a recombinant human antibody that inhibits myostatin activity has shown good tolerability but minimal improvement in the muscle strength and the pathology of patients with LGMD [136].
Although a meaningful effort has been made in all these areas to address the challenge of developing therapies for LGMDR1, to date, most groups are focused on correcting the primary genetic defect through the use of adeno-associated virus (AAV) gene therapy, transcriptional modification approaches (exon skipping and induction of stop codon read-through) or cell therapy (stem cells and induced pluripotent stem cells) [141]. In the particular case of LGMDR1, there are currently few ongoing therapeutic approaches, and so far, they are only showing a moderate efficacy. Therefore, there is a need for developing new therapeutic strategies directed toward alternative targets of the disease, such as the ones summarized in Figure 2. To date, in the LGMDR1 mouse model, transfer of the AAV-mediated CAPN3 gene has shown safety and efficacy, resulting in significant myopathology amelioration [137]. Moreover, gene correction in LGMDR1 patient-specific iPS cells has been successfully achieved. Those genetically corrected iPS cells might be evaluated in vitro and in vivo once differentiated into skeletal muscle progenitors, in order to address the efficient restoration of full-length CAPN3 [140]. Finally, Sarepta, in association with Nationwide Children’s Hospital, has recently announced a gene therapy program using the AAVrh74 vector, designed to replace CAPN3 in the skeletal muscle via systemic administration. AAVrh74 has a robust affinity for muscle cells, with a relatively low level of pre-existing immunity [142]. This vector has been previously used in other gene therapy programs targeting Duchenne Muscular Dystrophy and five other LGMDs [143,144]. All the therapeutic approaches for LGMDR1 described above are summarized in Table 1.
Table 1.
Therapeutic strategies for limb-girdle muscular dystrophy recessive 1 (LGMDR1).
| Therapy | Clinical-Pharmacological Use | State | Comments | Ref. |
|---|---|---|---|---|
| Pharmacological Therapy | ||||
| MYO-029 | Myostatin human recombinant neutralizing antibody | Competed I/II trial | Minimal improvement in muscle strength | [136] |
| Gene therapy | ||||
| AAV-delivered mutant myostatin propeptide | Prevention of the cleavage of myostatin propeptide | Preclinical | Increased muscle mass and force generation in mice | [135] |
| AAV-mediated transfer of calpain 3 | Increase of calpain 3 expression and function | Preclinical | Rescue of the contractile force deficits in mice | [137] |
| Plasmid DNA | Increase of calpain 3 expression and function | Active project | [138] | |
| AAVrh74 vector | Increase of calpain 3 expression and function | Active project | Systemic delivery to muscle | [139] |
| Cell therapy | ||||
| iPSC | Increase of calpain 3 expression and function | Active project | [140] |
6. Future Directions and Conclusions
The role of Ca2+ dysregulation as a pathological mechanism underlying LGMDR1 is increasingly becoming accepted. Here, we have reviewed different participants disrupted in LGMDR1 that regulate Ca2+ homeostasis. Several of these Ca2+-handling proteins may be interesting candidates for therapeutic approaches for calpainopathies. In this line, treatment with RyR stabilizers has shown an improvement of muscle function in mouse models of Duchenne and LGMDR4 muscular dystrophies [116,145]. Thus, it would be interesting to test these compounds as well as other therapeutic approaches targeting Ca2+-handling proteins compromised in LGMDR1 such as CaMKII and SERCAs. SERCA overexpression could be achieved through AAV-mediated gene therapy [146,147], but several drugs may also prove useful in increasing SERCA expression or function, including adrenoreceptor blockers, adrenergic agonists, hormones, glucocorticoids, natural antioxidants, and small molecule SERCA activators such as CDN1163 [58,148,149]. Alternatively, reduction or down-regulation of SERCA inhibitors SLN [150] and PLN [151] could be considered. Finally, inhibition of SERCA degradation through the ubiquitin-proteasome system has been shown to rescue SERCA1 function in a cellular model of Brody disease [152], and therefore, it might also have therapeutic potential for LGMDR1.
Several therapeutic approaches targeting mitochondrial dysfunction and oxidative stress have been shown to improve muscle function in different dystrophic mouse models and could therefore also be effective in ameliorating LGMDR1 phenotype. These therapies include cyclosporine A and cyclophilin D inhibitors (Debio 025) targeting mitochondrial PTP opening, pargyline for the excessive accumulation of reactive oxygen species and overexpression of PGC1α for mitochondrial dysfunction in post-necrotic dystrophic muscles [153].
While genetic correction of CAPN3 is so far one of the most promising therapeutic approaches aiming to cure LGMDR1, there is still much work to be done before gene therapy is available for these patients. Indeed, these therapies need to overcome several hindrances in LGMDR1 patients, including potential immune response resulting from AAV or CAPN3 introduction, as well as efficient delivery of viral vectors to affected muscles without off-target effects. Moreover, it should be taken into account that not every LGMDR1 patient may qualify for this kind of therapy. In this context, therapeutic approaches targeting key Ca2+-handling proteins in LGMDR1 may help ameliorate dystrophic progression in LGMDR1, since, as discussed in this review, Ca2+ dysregulation seems to play a central pathogenic role in this disease. In any case, further studies of the different pathogenic mechanisms underlying LGMDR1 are essential for helping us understand the many elusive functions of CAPN3 in the skeletal muscle.
Acknowledgments
We thank Inma Cobos for helpful comments on muscle histopathology. We dedicate this work to all patients with CAPN3 mutations and their families.
Abbreviations
| AAV | Adeno-associated virus |
| Akt | Protein kinase B |
| AldoA | Aldolase isoform A |
| AMP | Adenosine monophosphate |
| AMPK | AMP-activated protein kinase |
| ATP | Adenosine triphosphate |
| ATP5D | ATP synthase subunit delta |
| C3KO | Calpain 3 knockout mouse |
| CaM | Calmodulin |
| CAMK | Ca2+/calmodulin-dependent protein kinase family |
| CAMKII | Ca2+/calmodulin-dependent protein kinase type II |
| CAPN1 | Calpain 1 |
| CAPN2 | Calpain 2 |
| CAPN3 | Human calpain 3 gene |
| CAPN3 | Human calpain 3 protein |
| Capn3−/− | Calpain 3 knockout mouse |
| CBSW | Calpain-type beta-sandwich |
| c-FLIP | Cellular FLICE inhibitory protein |
| CK | Creatine Kinase |
| Cn | Calcineurin |
| CSQ | Calsequestrin |
| CTX | Cardiotoxin |
| Cyt-c | Cytochrome C |
| DHPR | Dihydropyridine receptor |
| ECC | Excitation-contraction coupling |
| FRZB | Frizzled Related Protein |
| HDAC | Class II histone deacetylases |
| IκBα | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, isoform alpha |
| iMOC | Intermolecular complementation |
| iPSC | Induced pluripotent stem cell |
| IS1 | Insertion sequence 1 |
| IS2 | Insertion sequence 2 |
| LGMD | Limb girdle muscular dystrophy |
| LGMD2A | Limb girdle muscular dystrophy type 2A, renamed LGMDR1 |
| LGMD2B | Limb girdle muscular dystrophy type 2B, renamed LGMDR2 |
| LGMDD4 | Limb girdle muscular dystrophy dominant 4 |
| LGMDR1 | Limb girdle muscular dystrophy recessive 1, caused by mutations in CAPN3 |
| LGMDR2 | Limb girdle muscular dystrophy recessive 2, caused by mutations in dysferlin |
| LGMDR4 | Limb girdle muscular dystrophy recessive 4, caused by mutations in β-sarcoglycan |
| LKB1 | Liver kinase B1 |
| MAM | Mitochondria-associated sarcoplasmic reticulum membrane |
| MARP-2 | Muscle Ankyrin Repeat Protein-2 |
| MEF2 | Myocyte enhancer factor-2 |
| miRNA | Micro ribonucleic acid |
| mRNA | Messenger ribonucleic acid |
| mTOR | Mammalian target of rapamycin |
| mTORC1 | Mammalian target of rapamycin complex 1 |
| MyoD | Myoblast determination protein 1 |
| NCX | Na+/Ca2+ exchanger |
| NCX3 | Na+/Ca2+ exchanger isoform 3 |
| NFAT | Nuclear factor of activated T-cells |
| NF-κB | Nuclear factor kappa-ligjht-chain-enhancer of activated B cells |
| NS | N-terminal addition sequence |
| Pax7 | Paired box protein 7 |
| PC1 | Protease core subdomain 1 |
| PC2 | Protease core subdomain 2 |
| PEF | Penta E-F hand |
| PGC1α | Peroxisome proliferator activated receptor gamma coactivator 1 alpha |
| PKA | Protein kinase A |
| PLN | Phospholamban |
| PMCA | Plasma membrane calcium ATPase |
| PTP | Permeability Transient Pore |
| ROS | Reactive oxygen species |
| RyR | Ryanodine receptor |
| RyR1 | Ryanodine receptor isoform 1 |
| SERCA | Sarco/endoplasmic reticulum Ca2+-ATPase |
| SERCA1 | Sarco/endoplasmic reticulum Ca2+-ATPase isoform 1 |
| SERCA2a | Sarco/endoplasmic reticulum Ca2+-ATPase isoform 2a |
| SLN | Sarcolipin |
| SOD | Superoxide dismutase |
| SR | Sarcoplasmic reticulum |
| T-tubule | Transverse tubule |
| UPR | Unfolded protein rsponse |
| Wnt | Wingless-related integration site |
Author Contributions
Investigation, J.L.-E., L.M.-M., N.N.-G., A.S., A.L.d.M., and A.V.-I.; writing—original draft preparation, J.L.-E., L.M.-M., and A.V.-I.; writing—review and editing, J.L.-E., L.M.-M., N.N.-G., A.S., A.L.d.M., and A.V.-I.; visualization, J.L.-E, L.M.-M., and N.N.-G.; supervision, A.L.d.M. and A.V.-I.; project administration, A.V.-I.; funding acquisition, A.L.d.M. and A.V.-I.
Funding
This research was funded by Instituto de Salud Carlos III, co-funded by European Regional Development Fund/European Social Fund, “Investing in your future” (A.V.-I., PI17/00676; A.L.d.M., PI17/01841); the Basque Government (A.V.-I., 2016111091) and Diputación Foral de Gipuzkoa (A.V.-I., 2018-000117-01-B and 2019-00362-01-B). A.V.-I. holds a Ramón y Cajal contract funded by the Spanish Ministry of Economy and Competitiveness, and J.L.-E. holds a PhD fellowship from the Basque Government.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Berridge M.J., Lipp P., Bootman M.D. The Versatility and Universality of Calcium Signalling. Nat. Rev. Mol. Cell. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Cheng X., Zhang X., Yu L., Xu H. Calcium Signaling in Membrane Repair. Semin. Cell Dev. Biol. 2015;45:24–31. doi: 10.1016/j.semcdb.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gehlert S., Bloch W., Suhr F. Ca2+-Dependent Regulations and Signaling in Skeletal Muscle: From Electro-Mechanical Coupling to Adaptation. Int. J. Mol. Sci. 2015;16:1066–1095. doi: 10.3390/ijms16011066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Qusairi L., Laporte J. T-Tubule Biogenesis and Triad Formation in Skeletal Muscle and Implication in Human Diseases. Skelet. Muscle. 2011;1:26. doi: 10.1186/2044-5040-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallejo-Illarramendi A., Toral-Ojeda I., Aldanondo G., López de Munain A. Dysregulation of Calcium Homeostasis in Muscular Dystrophies. Expert Rev. Mol. Med. 2014;16:e16. doi: 10.1017/erm.2014.17. [DOI] [PubMed] [Google Scholar]
- 6.Protasi F. Structural Interaction between RyRs and DHPRs in Calcium Release Units of Cardiac and Skeletal Muscle Cells. Front. Biosci. 2002;7:650–658. doi: 10.2741/A801. [DOI] [PubMed] [Google Scholar]
- 7.Santulli G., Lewis D.R., Marks A.R. Physiology and Pathophysiology of Excitation-Contraction Coupling: The Functional Role of Ryanodine Receptor. J. Muscle Res. Cell Motil. 2017;38:37–45. doi: 10.1007/s10974-017-9470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanner J.T., Georgiou D.K., Joshi A.D., Hamilton S.L. Ryanodine Receptors: Structure, Expression, Molecular Details, and Function in Calcium Release. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capes E.M., Loaiza R., Valdivia H.H. Ryanodine Receptors. Skelet. Muscle. 2011;1:18. doi: 10.1186/2044-5040-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi A.E., Dirksen R.T. Sarcoplasmic Reticulum: The Dynamic Calcium Governor of Muscle. Muscle Nerve. 2006;33:715–731. doi: 10.1002/mus.20512. [DOI] [PubMed] [Google Scholar]
- 11.Rodney G.G., Williams B.Y., Strasburg G.M., Beckingham K., Hamilton S.L. Regulation of RYR1 Activity by Ca2+ and Calmodulin. Biochemistry. 2000;39:7807–7812. doi: 10.1021/bi0005660. [DOI] [PubMed] [Google Scholar]
- 12.Beard N.A., Wei L., Dulhunty A.F. Ca2+ Signaling in Striated Muscle: The Elusive Roles of Triadin, Junctin, and Calsequestrin. Eur. Biophys. J. 2009;39:27–36. doi: 10.1007/s00249-009-0449-6. [DOI] [PubMed] [Google Scholar]
- 13.Novák P., Soukup T. Calsequestrin Distribution, Structure and Function, Its Role in Normal and Pathological Situations and the Effect of Thyroid Hormones. Physiol. Res. 2011;60:439–452. doi: 10.33549/physiolres.931989. [DOI] [PubMed] [Google Scholar]
- 14.Calderón J.C., Bolaños P., Caputo C. The Excitation-Contraction Coupling Mechanism in Skeletal Muscle. Biophys. Rev. 2014;6:133–160. doi: 10.1007/s12551-013-0135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo I.Y., Ehrlich B.E. Signaling in Muscle Contraction. Cold Spring Harb. Perspect. Biol. 2015;7:a006023. doi: 10.1101/cshperspect.a006023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Periasamy M., Kalyanasundaram A. SERCA Pump Isoforms: Their Role in Calcium Transport and Disease. Muscle Nerve. 2007;35:430–442. doi: 10.1002/mus.20745. [DOI] [PubMed] [Google Scholar]
- 17.Murphy R.M., Larkins N.T., Mollica J.P., Beard N.A., Lamb G.D. Calsequestrin Content and SERCA Determine Normal and Maximal Ca2+ Storage Levels in Sarcoplasmic Reticulum of Fast- and Slow-Twitch Fibres of Rat. J. Physiol. 2009;587:443–460. doi: 10.1113/jphysiol.2008.163162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vangheluwe P., Raeymaekers L., Dode L., Wuytack F. Modulating Sarco (Endo) Plasmic Reticulum Ca2+ ATPase2 (SERCA2) Activity: Cell Biological Implications. Cell Calcium. 2005;38:291–302. doi: 10.1016/j.ceca.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 19.Shaikh S.A., Sahoo S.K., Periasamy M. Phospholamban and Sarcolipin: Are They Functionally Redundant or Distinct Regulators of the Sarco(Endo)Plasmic Reticulum Calcium ATPase? J. Mol. Cell. Cardiol. 2016;91:81–91. doi: 10.1016/j.yjmcc.2015.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tavi P., Westerblad H. The Role of in Vivo Ca2+ Signals Acting on Ca2+-Calmodulin-Dependent Proteins for Skeletal Muscle Plasticity. J. Physiol. 2011;589:5021–5031. doi: 10.1113/jphysiol.2011.212860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chin E.R., Olson E.N., Richardson J.A., Yang Q., Humphries C., Shelton J.M., Wu H., Zhu W., Bassel-Duby R., Williams R.S. A Calcineurin-Dependent Transcriptional Pathway Controls Skeletal Muscle Fiber Type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alzuherri H., Chang K.C. Calcineurin Activates NF-κB in Skeletal Muscle C2C12 Cells. Cell. Signal. 2003;15:471–478. doi: 10.1016/S0898-6568(02)00120-1. [DOI] [PubMed] [Google Scholar]
- 23.Valdés J.A., Gaggero E., Hidalgo J., Leal N., Jaimovich E., Carrasco M.A. NFAT Activation by Membrane Potential Follows a Calcium Pathway Distinct from Other Activity-Related Transcription Factors in Skeletal Muscle Cells. Am. J. Physiol. Physiol. 2008;294:C715–C725. doi: 10.1152/ajpcell.00195.2007. [DOI] [PubMed] [Google Scholar]
- 24.McCullagh K.J.A., Calabria E., Pallafacchina G., Ciciliot S., Serrano A.L., Argentini C., Kalhovde J.M., Lomo T., Schiaffino S. NFAT Is a Nerve Activity Sensor in Skeletal Muscle and Controls Activity-Dependent Myosin Switching. Proc. Natl. Acad. Sci. USA. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson J.M., Bakkar N., Guttridge D.C. NF-κB Signaling in Skeletal Muscle Health and Disease. In: Pavlath G.K., editor. Myogenesis. Elsevier Inc.; Cambridge, MA, USA: 2011. [DOI] [PubMed] [Google Scholar]
- 26.Kaltschmidt B., Kaltschmidt C., Hofmann T.G., Hehner S.P., Dröge W., Schmitz M.L. The Pro- or Anti-Apoptotic Function of NF-κB Is Determined by the Nature of the Apoptotic Stimulus. Eur. J. Biochem. 2000;267:3828–3835. doi: 10.1046/j.1432-1327.2000.01421.x. [DOI] [PubMed] [Google Scholar]
- 27.Eilers W., Jaspers R.T., De Haan A., Ferri C., Valdivieso P., Flück M. CaMKII Content Affects Contractile, but Not Mitochondrial, Characteristics in Regenerating Skeletal Muscle. BMC Physiol. 2014;14 doi: 10.1186/s12899-014-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potthoff M.J., Wu H., Arnold M.A., Shelton J.M., Backs J., Mcanally J., Richardson J.A., Bassel-duby R., Olson E.N. Histone Deacetylase Degradation and MEF2 Activation Promote the Formation of Slow-Twitch Myofibers. J. Clin. Investig. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramerova I., Torres J.A., Eskin A., Nelson S.F., Spencer M.J. Calpain 3 and CaMKIIβ Signaling Are Required to Induce HSP70 Necessary for Adaptive Muscle Growth after Atrophy. Hum. Mol. Genet. 2018;27:1642–1653. doi: 10.1093/hmg/ddy071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin E.R. Role of Ca2+/Calmodulin-Dependent Kinases in Skeletal Muscle Plasticity. J. Appl. Physiol. 2005;99:414–423. doi: 10.1152/japplphysiol.00015.2005. [DOI] [PubMed] [Google Scholar]
- 31.Mihaylova M.M., Shaw R.J. The AMPK Signalling Pathway Coordinates Cell Growth, Autophagy and Metabolism. Nat. Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J., Dhakal K., Yi J. Mitochondrial Ca2+ Uptake in Skeletal Muscle Health and Disease. Sci. China Life Sci. 2016;59:770–776. doi: 10.1007/s11427-016-5089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisner V., Csordas G., Hajnoczky G. Interactions between Sarco-Endoplasmic Reticulum and Mitochondria in Cardiac and Skeletal Muscle—Pivotal Roles in Ca2+ and Reactive Oxygen Species Signaling. J. Cell Sci. 2013;126:2965–2978. doi: 10.1242/jcs.093609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraysse B., Nagi S.M., Boher B., Ragot H., Lainé J., Salmon A., Fiszman M.Y., Toussaint M., Fromes Y. Ca2+ Overload and Mitochondrial Permeability Transition Pore Activation in Living δ-Sarcoglycan-Deficient Cardiomyocytes. Am. J. Physiol. Physiol. 2010;299:C706–C713. doi: 10.1152/ajpcell.00545.2009. [DOI] [PubMed] [Google Scholar]
- 35.Celsi F., Pizzo P., Brini M., Leo S., Fotino C., Pinton P., Rizzuto R. Mitochondria, Calcium and Cell Death: A Deadly Triad in Neurodegeneration. Biochim. Biophys. Acta-Bioenerg. 2009;1787:335–344. doi: 10.1016/j.bbabio.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi M., Weaver D., Hajnóczky G. Control of Mitochondrial Motility and Distribution by the Calcium Signal: A Homeostatic Circuit. J. Cell Biol. 2004;167:661–672. doi: 10.1083/jcb.200406038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi A.E., Boncompagni S., Dirksen R.T. Sarcoplasmic Reticulum-Mitochondrial Symbiosis: Bidirectional Signaling in Skeletal Muscle. Exerc. Sport Sci. Rev. 2009;37:29–35. doi: 10.1097/JES.0b013e3181911fa4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giorgi C., Baldassari F., Bononi A., Bonora M., De Marchi E., Marchi S., Missiroli S., Patergnani S., Rimessi A., Suski J.M., et al. Mitochondrial Ca2+ and Apoptosis. Cell Calcium. 2012;52:36–43. doi: 10.1016/j.ceca.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernardi P., von Stockum S. The Permeability Transition Pore as a Ca2+ Release Channel: New Answers to an Old Question. Cell Calcium. 2012;52:22–27. doi: 10.1016/j.ceca.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright D.C. Mechanisms of Calcium-Induced Mitochondrial Biogenesis and GLUT4 Synthesis. Appl. Physiol. Nutr. Metab. 2008;32:840–845. doi: 10.1139/H07-062. [DOI] [PubMed] [Google Scholar]
- 41.Straub V., Murphy A., Udd B. 229th ENMC International Workshop: Limb Girdle Muscular Dystrophies—Nomenclature and Reformed Classification Naarden, the Netherlands, 17–19 March 2017. Neuromuscul. Disord. 2018;28:702–710. doi: 10.1016/j.nmd.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., et al. Mutations in the Proteolytic Enzyme Calpain 3 Cause Limb-Girdle Muscular Dystrophy Type 2A. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 43.Vissing J., Barresi R., Witting N., Van Ghelue M., Gammelgaard L., Bindoff L.A., Straub V., Lochmüller H., Hudson J., Wahl C.M., et al. A Heterozygous 21-Bp Deletion in CAPN3 Causes Dominantly Inherited Limb Girdle Muscular Dystrophy. Brain. 2016;139:2154–2163. doi: 10.1093/brain/aww133. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Thompson J.M., Niu Z., Tracy J.A., Moore S.A., Swenson A., Wieben E.D., Milone M. Autosomal Dominant Calpainopathy Due to Heterozygous CAPN3 c.643_663del21. Muscle Nerve. 2018;57:657–683. doi: 10.1002/mus.25970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Thompson J.M., Moore S.A., Liewluck T. A Novel CAPN3 Mutation in Late-Onset Limb-Girdle Muscular Dystrophy with Early Respiratory Insufficiency. J. Clin. Neurosci. 2018;53:229–231. doi: 10.1016/j.jocn.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sáenz A., López de Munain A. Dominant LGMD2A: Alternative Diagnosis or Hidden Digenism? Brain. 2017;140:e7. doi: 10.1093/brain/aww281. [DOI] [PubMed] [Google Scholar]
- 47.Dinçer P., Leturcq F., Richard I., Piccolo F., Yalnizoǧlu D., De Toma C., Akçören Z., Broux O., Deburgrave N., Brenguier L., et al. A Biochemical, Genetic, and Clinical Survey of Autosomal Recessive Limb Girdle Muscular Dystrophies in Turkey. Ann. Neurol. 1997;42:222–229. doi: 10.1002/ana.410420214. [DOI] [PubMed] [Google Scholar]
- 48.Topaloğlu H., Dincer P., Richard I., Akçören Z., Alehan D., Ozme S., Caglar M., Karaduman A., Urtizberea J.A., Beckmann J. Calpain-3 Deficiency Causes a Mild Muscular Dystrophy in Childhood. Neuropediatrics. 1997;28:212–216. doi: 10.1055/s-2007-973702. [DOI] [PubMed] [Google Scholar]
- 49.Richard I., Brenguier L., Dincer P., Roudaut C., Bady B., Burgunder J., Chemaly R., Garcia C., Halaby G., Jackson C., et al. Multiple Independent Molecular Etiology for Limb-Girdle Muscular Dystrophy Type 2A Patients from Various Geographical Origins. Am. J. Hum. Genet. 1997;60:1128–1138. [PMC free article] [PubMed] [Google Scholar]
- 50.Chae J., Minami N., Jin Y., Nakagawa M., Murayama K., Igarashi F., Nonaka I. Calpain 3 Gene Mutations: Genetic and Clinico-Pathologic Findings in Limb-Girdle Muscular Dystrophy. Neuromuscul. Disord. 2001;11:547–555. doi: 10.1016/S0960-8966(01)00197-3. [DOI] [PubMed] [Google Scholar]
- 51.De Paula F., Vainzof M., Passos-Bueno M.R., Rita de Cássia M.P., Matioli S.R., Anderson L.V.B., Nigro V., Zatz M. Clinical Variability in Calpainopathy: What Makes the Difference? Eur. J. Hum. Genet. 2002;10:825–832. doi: 10.1038/sj.ejhg.5200888. [DOI] [PubMed] [Google Scholar]
- 52.Zatz M., Starling A. Calpains and Disease. N. Engl. J. Med. 2005;352:2413–2423. doi: 10.1056/NEJMra043361. [DOI] [PubMed] [Google Scholar]
- 53.Orphanet. [(accessed on 2 September 2019)]; Available online: https://www.orpha.net/consor/cgi-bin/Disease_Search.php?lng=ES&data_id=870&Disease_Disease_Search_diseaseGroup=LGMD2A&Disease_Disease_Search_diseaseType=Pat&Enfermedad(es)/grupo de enfermedades=Distrofia-muscular-de-cinturas-autos-mica-recesiva-tipo-2.
- 54.Fardeau M., Hillaire D., Mignard C., Feingold N., Feingold J., Mignard D., De Ubeda B., Collin H., Tomé F.M.S., Richard I., et al. Juvenile Limb-Girdle Muscular Dystrophy Clinical, Histopathological and Genetic Data from a Small Community Living in the Reunion Island. Brain. 1996;119:295–308. doi: 10.1093/brain/119.1.295. [DOI] [PubMed] [Google Scholar]
- 55.Urtasun M., Sáenz A., Roudaut C., Poza J.J., Urtizberea J.A., Cobo A.M., Richard I., García Bragado F., Leturcq F., Kaplan J.C., et al. Limb-Girdle Muscular Dystrophy in Guipúzcoa (Basque Country, Spain) Brain. 1998;121:1735–1747. doi: 10.1093/brain/121.9.1735. [DOI] [PubMed] [Google Scholar]
- 56.Allamand V., Broux O., Bourg N., Richard I., Tischfield J.A., Hodes M.E., Conneally P.M., Fardeau M., Jackson C.E., Beckmann J.S. Genetic Heterogeneity of Autosomal Recessive Limb-Girdle Muscular Dystrophy in a Genetic Isolate (Amish) and Evidence for a New Locus. Hum. Mol. Genet. 1995;4:459–463. doi: 10.1093/hmg/4.3.459. [DOI] [PubMed] [Google Scholar]
- 57.Pantoja-Melendez C.A., Miranda-Duarte A., Roque-Ramirez B., Zenteno J.C. Epidemiological and Molecular Characterization of a Mexican Population Isolate with High Prevalence of Limb-Girdle Muscular Dystrophy Type 2A Due to a Novel Calpain-3 Mutation. PLoS ONE. 2017;12:1–13. doi: 10.1371/journal.pone.0170280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fanin M., Benedicenti F., Fritegotto C., Nascimbeni A., Peterle E., Stanzial F., Cristofoletti A., Castellan C., Angelini C. An Intronic Mutation Causes Severe LGMD2A in a Large Inbred Family Belonging to a Genetic Isolate in the Alps. Clin. Genet. 2012;82:601–602. doi: 10.1111/j.1399-0004.2012.01873.x. [DOI] [PubMed] [Google Scholar]
- 59.Leiden Database. [(accessed on 2 September 2019)]; Available online: https://databases.lovd.nl/shared/genes/CAPN3.
- 60.Blázquez L., Azpitarte M., Sáenz A., Goicoechea M., Otaegui D., Ferrer X., Illa I., Gutierrez-Rivas E., Vilchez J.J., López De Munain A. Characterization of Novel CAPN3 Isoforms in White Blood Cells: An Alternative Approach for Limb-Girdle Muscular Dystrophy 2A Diagnosis. Neurogenetics. 2008;9:173–182. doi: 10.1007/s10048-008-0129-1. [DOI] [PubMed] [Google Scholar]
- 61.Richard I., Hogrel J.-Y., Stockholm D., Payan C.A.M., Fougerousse F., Calpainopathy Study Group. Eymard B., Mignard C., López de Munain A., Fardeau M., et al. Natural History of LGMD2A for Delineating Outcome Measures in Clinical Trials. Ann. Clin. Transl. Neurol. 2016;3:248–265. doi: 10.1002/acn3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gallardo E., Saenz A., Illa I. Limb-Girdle Muscular Dystrophy 2A. In: Aminoff M.J., Boller F., Swaab D.F., editors. Muscular Dystrophies. Elsevier, B.V.; Amsterdam, The Netherlands: 2011. [DOI] [Google Scholar]
- 63.Sáenz A., Ono Y., Sorimachi H., Goicoechea M., Leturcq F., Blázquez L., García-Bragado F., Marina A., Poza J.J., Azpitarte M., et al. Does the Severity of the LGMD2A Phenotype in Compound Heterozygotes Depend on the Combination of Mutations? Muscle Nerve. 2011;44:710–714. doi: 10.1002/mus.22194. [DOI] [PubMed] [Google Scholar]
- 64.Schessl J., Walter M.C., Schreiber G., Schara U., Müller C.R., Lochmüller H., Bönnemann C.G., Korinthenberg R., Kirschner J. Phenotypic Variability in Siblings with Calpainopathy (LGMD2A) Acta Myol. 2008;27:54–58. [PMC free article] [PubMed] [Google Scholar]
- 65.Rajakumar D., Alexander M., Oommen A. Oxidative Stress, NF-KB and the Ubiquitin Proteasomal Pathway in the Pathology of Calpainopathy. Neurochem. Res. 2013;38:2009–2018. doi: 10.1007/s11064-013-1107-z. [DOI] [PubMed] [Google Scholar]
- 66.Nilsson M.I., Macneil L.G., Kitaoka Y., Alqarni F., Suri R., Akhtar M., Haikalis M.E., Dhaliwal P., Saeed M., Tarnopolsky M.A. Redox State and Mitochondrial Respiratory Chain Function in Skeletal Muscle of LGMD2A Patients. PLoS ONE. 2014;9:e102549. doi: 10.1371/journal.pone.0102549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toral-Ojeda I., Aldanondo G., Lasa-Elgarresta J., Lasa-Fernández H., Fernández-Torrón R., López de Munain A., Vallejo-Illarramendi A., Lasa-Fernandez H., Fernandez-Torron R., Lopez de Munain A., et al. Calpain 3 Deficiency Affects SERCA Expression and Function in the Skeletal Muscle. Expert Rev. Mol. Med. 2016;18:e7. doi: 10.1017/erm.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toral-Ojeda I., Aldanondo G., Lasa-Elgarresta J., Lasa-Fernandez H., Vesga-Castro C., Mouly V., de Munain A.L., Vallejo-Illarramendi A. Muscle Cell and Tissue-Current Status of Research Field. IntechOpen; London, UK: 2018. A novel functional in vitro model that recapitulates human muscle disorders; pp. 133–153. [DOI] [Google Scholar]
- 69.Kramerova I., Kudryashova E., Tidball J.G., Spencer M.J. Null Mutation of Calpain 3 (P94) in Mice Causes Abnormal Sarcomere Formation in Vivo and in Vitro. Hum. Mol. Genet. 2004;13:1373–1388. doi: 10.1093/hmg/ddh153. [DOI] [PubMed] [Google Scholar]
- 70.El-Khoury R., Traboulsi S., Hamad T., Lamaa M., Sawaya R., Ahdab-Barmada M. Divergent Features of Mitochondrial Deficiencies in LGMD2A Associated With Novel Calpain-3 Mutations. J. Neuropathol. Exp. Neurol. 2019;78:88–98. doi: 10.1093/jnen/nly113. [DOI] [PubMed] [Google Scholar]
- 71.Yalvac M.E., Amornvit J., Braganza C., Chen L., Hussain S.R.A., Shontz K.M., Montgomery C.L., Flanigan K.M., Lewis S., Sahenk Z. Impaired Regeneration in Calpain-3 Null Muscle Is Associated with Perturbations in MTORC1 Signaling and Defective Mitochondrial Biogenesis. Skelet. Muscle. 2017;7:1–18. doi: 10.1186/s13395-017-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kramerova I., Kudryashova E., Wu B., Germain S., Vandenborne K., Romain N., Haller R.G., Verity M.A., Spencer M.J. Mitochondrial Abnormalities, Energy Deficit and Oxidative Stress Are Features of Calpain 3 Deficiency in Skeletal Muscle. Hum. Mol. Genet. 2009;18:3194–3205. doi: 10.1093/hmg/ddp257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kramerova I., Kudryashova E., Ermolova N., Saenz A., Jaka O., López de munain A., Spencer M.J. Impaired Calcium Calmodulin Kinase Signaling and Muscle Adaptation Response in the Absence of Calpain 3. Hum. Mol. Genet. 2012;21:3193–3204. doi: 10.1093/hmg/dds144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kramerova I., Ermolova N., Eskin A., Hevener A., Quehenberger O., Armando A.M., Haller R., Romain N., Nelson S.F., Spencer M.J. Failure to Up-Regulate Transcription of Genes Necessary for Muscle Adaptation Underlies Limb Girdle Muscular Dystrophy 2A (Calpainopathy) Hum. Mol. Genet. 2016;25:2194–2207. doi: 10.1093/hmg/ddw086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fanin M., Angelini C. Protein and Genetic Diagnosis of Limb Girdle Muscular Dystrophy Type 2A: The Yield and the Pitfalls. Muscle Nerve. 2015;52:163–173. doi: 10.1002/mus.24682. [DOI] [PubMed] [Google Scholar]
- 76.Krahn M., Goicoechea M., Hanisch F., Groen E., Bartoli M., Pécheux C., Garcia-Bragado F., Leturcq F., Jeannet P.Y., Lobrinus J.A., et al. Eosinophilic Infiltration Related to CAPN3 Mutations: A Pathophysiological Component of Primary Calpainopathy? Clin. Genet. 2011;80:398–402. doi: 10.1111/j.1399-0004.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 77.Carafoli E., Molinari M. Calpain: A Protease in Search of a Function? Biochem. Biophys. Res. Commun. 1998;247:193–203. doi: 10.1006/bbrc.1998.8378. [DOI] [PubMed] [Google Scholar]
- 78.Santella L., Kyozuka K., De Riso L., Carafoli E. Calcium, Protease Action, and the Regulation of the Cell Cycle. Cell Calcium. 1998;23:123–130. doi: 10.1016/S0143-4160(98)90110-5. [DOI] [PubMed] [Google Scholar]
- 79.Wang K.K.W. Calpain and Caspase: Can You Tell the Difference? Trends Neurosci. 2000;23:20–26. doi: 10.1016/S0166-2236(99)01479-4. [DOI] [PubMed] [Google Scholar]
- 80.Glading A., Lauffenburger D.A., Wells A. Cutting to the Chase: Calpain Proteases in Cell Motility. Trends Cell Biol. 2002;12:46–54. doi: 10.1016/S0962-8924(01)02179-1. [DOI] [PubMed] [Google Scholar]
- 81.Ono Y., Ojima K., Shinkai-Ouchi F., Hata S., Sorimachi H. An Eccentric Calpain, CAPN3/P94/Calpain-3. Biochimie. 2016;122:169–187. doi: 10.1016/j.biochi.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 82.Ye Q., Campbell R.L., Davies P.L. Structures of Human Calpain-3 Protease Core with and without Bound Inhibitor Reveal Mechanisms of Calpain Activation. J. Biol. Chem. 2018;293:4056–4070. doi: 10.1074/jbc.RA117.001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sorimachi H., Toyama-Sorimachi N., Saido T.C., Kawasaki H., Sugita H., Miyasaka M., Arahata K.I., Ishiura S., Suzuki K. Muscle-Specific Calpain, P94, Is Degraded by Autolysis Immediately after Translation, Resulting in Disappearance from Muscle. J. Biol. Chem. 1993;268:10593–10605. [PubMed] [Google Scholar]
- 84.Ono Y., Shindo M., Doi N., Kitamura F., Gregorio C.C., Sorimachi H. The N- and C-Terminal Autolytic Fragments of CAPN3/P94/Calpain-3 Restore Proteolytic Activity by Intermolecular Complementation. Proc. Natl. Acad. Sci. USA. 2014;111:E5527–E5536. doi: 10.1073/pnas.1411959111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ono Y., Ojima K., Torii F., Takaya E., Doi N., Nakagawa K., Hata S., Abe K., Sorimachi H. Skeletal Muscle-Specific Calpain Is an Intracellular Na+- Dependent Protease. J. Biol. Chem. 2010;285:22986–22998. doi: 10.1074/jbc.M110.126946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ono Y., Torii F., Ojima K., Doi N., Yoshioka K., Kawabata Y., Labeit D., Labeit S., Suzuki K., Abe K., et al. Suppressed Disassembly of Autolyzing P94/CAPN3 by N2A Connectin/Titin in a Genetic Reporter System. J. Biol. Chem. 2006;281:18519–18531. doi: 10.1074/jbc.M601029200. [DOI] [PubMed] [Google Scholar]
- 87.Kramerova I., Kudryashova E., Venkatraman G., Spencer M.J. Calpain 3 Participates in Sarcomere Remodeling by Acting Upstream of the Ubiquitin-Proteasome Pathway. Hum. Mol. Genet. 2005;14:2125–2134. doi: 10.1093/hmg/ddi217. [DOI] [PubMed] [Google Scholar]
- 88.Ojima K., Ono Y., Ottenheijm C., Hata S., Suzuki H., Granzier H., Sorimachi H. Non-Proteolytic Functions of Calpain-3 in Sarcoplasmic Reticulum in Skeletal Muscles. J. Mol. Biol. 2011;407:439–449. doi: 10.1016/j.jmb.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kramerova I., Kudryashova E., Wu B., Ottenheijm C., Granzier H., Spencer M.J. Novel Role of Calpain-3 in the Triad-Associated Protein Complex Regulating Calcium Release in Skeletal Muscle. Hum. Mol. Genet. 2008;17:3271–3280. doi: 10.1093/hmg/ddn223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dayanithi G., Richard I., Viero C., Mazuc E., Mallie S., Valmier J., Bourg N., Herasse M., Marty I., Lefranc G., et al. Alteration of Sarcoplasmic Reticulum Ca2+ Release in Skeletal Muscle from Calpain 3-Deficient Mice. Int. J. Cell Biol. 2009;2009:340346. doi: 10.1155/2009/340346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baghdiguian S., Martin M., Richard I., Pons F., Astier C., Bourg N., Hay R.T., Chemaly R., Halaby G., Loiselet J., et al. Calpain 3 Deficiency Is Associated with Myonuclear Apoptosis and Profound Perturbation of the IκBα/NF-κB Pathway in Limb-Girdle Muscular Dystrophy Type 2A. Nat. Med. 1999;5:503–511. doi: 10.1038/8385. [DOI] [PubMed] [Google Scholar]
- 92.Sorimachi H., Kinbara K., Kimura S., Takahashi M., Ishiura S., Sasagawa N., Sorimachi N., Shimada H., Tagawa K., Maruyama K., et al. Muscle-Specific Calpain, P94, Responsible for Limb Girdle Muscular Dystrophy Type 2A, Associates with Connectin through IS2, a P94-Specific Sequence. J. Biol. Chem. 1995;270:31158–31162. doi: 10.1074/jbc.270.52.31158. [DOI] [PubMed] [Google Scholar]
- 93.Ojima K., Ono Y., Doi N., Yoshioka K., Kawabata Y., Labeit S., Sorimachi H. Myogenic Stage, Sarcomere Length, and Protease Activity Modulate Localization of Muscle-Specific Calpain. J. Biol. Chem. 2007;282:14493–14504. doi: 10.1074/jbc.M610806200. [DOI] [PubMed] [Google Scholar]
- 94.Gunning P.W., Hardeman E.C., Lappalainen P., Mulvihill D.P. Tropomyosin—Master Regulator of Actin Filament Function in the Cytoskeleton. J. Cell Sci. 2015;128:2965–2974. doi: 10.1242/jcs.172502. [DOI] [PubMed] [Google Scholar]
- 95.Escolar D.M., O’Carroll P., Leshner R. Neuromuscular Disorders. Elsevier Inc.; Philadelphia, PA, USA: 2011. Treatment and Management of Muscular Dystrophies; pp. 343–372. [DOI] [Google Scholar]
- 96.Ojima K., Kawabata Y., Nakao H., Nakao K., Doi N., Kitamura F., Ono Y., Hata S., Suzuki H., Kawahara H., et al. Dynamic Distribution of Muscle-Specific Calpain in Mice Has a Key Role in Physical-Stress Adaptation and Is Impaired in Muscular Dystrophy. J. Clin. Investig. 2010;120:2672–2683. doi: 10.1172/JCI40658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Taveau M., Bourg N., Sillon G., Roudaut C., Bartoli M., Richard I. Calpain 3 Is Activated through Autolysis within the Active Site and Lyses Sarcomeric and Sarcolemmal Components. Mol. Cell. Biol. 2003;23:9127–9135. doi: 10.1128/MCB.23.24.9127-9135.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zak R., Martin A.F., Prior G., Rabinowitz M. Comparison of Turnover of Several Myofibrillar Proteins and Critical Evaluation of Double Isotope Method. J. Biol. Chem. 1977;252:3430–3435. [PubMed] [Google Scholar]
- 99.Isaacs W.B., Kim I.S., Struve A., Fulton A.B. Biosynthesis of Titin in Cultured Skeletal Muscle Cells. J. Cell Biol. 1989;109:2189–2195. doi: 10.1083/jcb.109.5.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beckmann J.S., Spencer M. Calpain 3, the “Gatekeeper” of Proper Sarcomere Assembly, Turnover and Maintenance. Neuromuscul. Disord. 2008;18:913–921. doi: 10.1016/j.nmd.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Richard I., Roudaut C., Marchand S., Baghdiguian S., Herasse M., Stockholm D., Ono Y., Suel L., Bourg N., Sorimachi H., et al. Loss of Calpain 3 Proteolytic Activity Leads to Muscular Dystrophy and to Apoptosis-Associated IκBα/Nuclear Factor ΚB Pathway Perturbation in Mice. J. Cell Biol. 2000;151:1583–1590. doi: 10.1083/jcb.151.7.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spencer M.J., Guyon J.R., Sorimachi H., Potts A., Richard I., Herasse M., Chamberlain J., Dalkilic I., Kunkel L.M., Beckmann J.S. Stable Expression of Calpain 3 from a Muscle Transgene in Vivo: Immature Muscle in Transgenic Mice Suggests a Role for Calpain 3 in Muscle Maturation. Proc. Natl. Acad. Sci. USA. 2002;99:8874–8879. doi: 10.1073/pnas.132269299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sacco A., Mourkioti F., Tran R., Choi J., Llewellyn M., Kraft P., Shkreli M., Delp S., Pomerantz J.H., Artandi S.E., et al. Short Telomeres and Stem Cell Exhaustion Model Duchenne Muscular Dystrophy in Mdx/MTR Mice. Cell. 2010;143:1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jaka O., Casas-Fraile L., Azpitarte M., Aiastui A., López de Munain A., Sáenz A. FRZB and Melusin, Overexpressed in LGMD2A, Regulate Integrin Β1D Isoform Replacement Altering Myoblast Fusion and the Integrin-Signalling Pathway. Expert Rev. Mol. Med. 2017;19:1–16. doi: 10.1017/erm.2017.3. [DOI] [PubMed] [Google Scholar]
- 105.Amici D.R., Pinal-Fernandez I., Mázala D.A.G., Lloyd T.E., Corse A.M., Christopher-Stine L., Mammen A.L., Chin E.R. Calcium Dysregulation, Functional Calpainopathy, and Endoplasmic Reticulum Stress in Sporadic Inclusion Body Myositis. Acta Neuropathol. Commun. 2017;5:24. doi: 10.1186/s40478-017-0427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sehgal P., Szalai P., Olesen C., Praetorius H.A., Nissen P., Christensen S.B., Engedal N., Møller J.V. Inhibition of the Sarco/Endoplasmic Reticulum (ER) Ca2-ATPase by Thapsigargin Analogs Induces Cell Death via ER Ca2 Depletion and the Unfolded Protein Response. J. Biol. Chem. 2017;292:19656–19673. doi: 10.1074/jbc.M117.796920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mekahli D., Bultynck G., Parys J.B., De Smedt H., Missiaen L. Endoplasmic-Reticulum Calcium Depletion and Disease. Cold Spring Harb. Perspect. Biol. 2011;3:131–154. doi: 10.1101/cshperspect.a004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Michel L.Y.M., Hoenderop J.G.J., Bindels R.J.M. Calpain-3-Mediated Regulation of the Na+-Ca2+ exchanger Isoform 3. Pflugers Arch. Eur. J. Physiol. 2016;468:243–255. doi: 10.1007/s00424-015-1747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Matsakas A., Patel K. Intracellular Signalling Pathways Regulating the Adaptation of Skeletal Muscle to Exercise and Nutritional Changes. Histol. Histopathol. 2009;24:209–222. doi: 10.14670/HH-24.209. [DOI] [PubMed] [Google Scholar]
- 110.Cohen N., Kudryashova E., Kramerova I., Anderson L.V.B., Beckmann J.S., Bushby K., Spencer M.J. Identification of Putative in Vivo Substrates of Calpain 3 by Comparative Proteomics of Overexpressing Transgenic and Nontransgenic Mice. Proteomics. 2006;6:6075–6084. doi: 10.1002/pmic.200600199. [DOI] [PubMed] [Google Scholar]
- 111.Kramerova I., Beckmann J.S., Spencer M.J. Molecular and Cellular Basis of Calpainopathy (Limb Girdle Muscular Dystrophy Type 2A) Biochim. Biophys. Acta. 2007;1772:128–144. doi: 10.1016/j.bbadis.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 112.Sáenz A., Azpitarte M., Armañanzas R., Leturcq F., Alzualde A., Inza I., García-Bragado F., De la Herran G., Corcuera J., Cabello A., et al. Gene Expression Profiling in Limb-Girdle Muscular Dystrophy 2A. PLoS ONE. 2008;3:e3750. doi: 10.1371/journal.pone.0003750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kroemer G., Galluzzi L., Brenner C. Mitochondrial Membrane Permeabilization in Cell Death. Physiol. Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 114.Handschin C., Rhee J., Lin J., Tarr P.T., Spiegelman B.M. An Autoregulatory Loop Controls Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1alpha Expression in Muscle. Proc. Natl. Acad. Sci. USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guo C., Sun L., Chen X., Zhang D. Oxidative Stress, Mitochondrial Damage and Neurodegenerative Diseases. Neural Regen. Res. 2013;8:2003–2014. doi: 10.3969/j.issn.1673-5374.2013.21.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bellinger A.M., Reiken S., Carlson C., Mongillo M., Liu X., Rothman L., Matecki S., Lacampagne A., Marks A.R. Hypernitrosylated Ryanodine Receptor/Calcium Release Channels Are Leaky in Dystrophic Muscle Andrew. Nat. Med. 2009;15:325–330. doi: 10.1038/nm.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Raney M.A., Turcotte L.P. Evidence for the Involvement of CaMKII and AMPK in Ca2+-Dependent Signaling Pathways Regulating FA Uptake and Oxidation in Contracting Rodent Muscle. J. Appl. Physiol. 2008;104:1366–1373. doi: 10.1152/japplphysiol.01282.2007. [DOI] [PubMed] [Google Scholar]
- 118.Shan T., Zhang P., Liang X., Bi P., Yue F., Kuang S. Lkb1 Is Indispensable for Skeletal Muscle Development, Regeneration, and Satellite Cell Homeostasis. Stem Cells. 2014;32:2893–2907. doi: 10.1002/stem.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Morita M., Gravel S.P., Hulea L., Larsson O., Pollak M., St-Pierre J., Topisirovic I. MTOR Coordinates Protein Synthesis, Mitochondrial Activity. Cell Cycle. 2015;14:473–480. doi: 10.4161/15384101.2014.991572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Morita M., Gravel S.P., Chénard V., Sikström K., Zheng L., Alain T., Gandin V., Avizonis D., Arguello M., Zakaria C., et al. MTORC1 Controls Mitochondrial Activity and Biogenesis through 4E-BP-Dependent Translational Regulation. Cell Metab. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 121.Rosales X.Q., Malik V., Sneh A., Chen L., Lewis S., Kota J., Gastier-Foster J.M., Astbury C., Pyatt R., Reshmi S., et al. Impaired Regeneration in LGMD2A Supported by Increased PAX7-Positive Satellite Cell Content and Muscle-Specific Microrna Dysregulation. Muscle Nerve. 2013;47:731–739. doi: 10.1002/mus.23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roberts T.C., Blomberg K.E.M., McClorey G., El Andaloussi S., Godfrey C., Betts C., Coursindel T., Gait M.J., Smith C.E., Wood M.J. Expression Analysis in Multiple Muscle Groups and Serum Reveals Complexity in the MicroRNA Transcriptome of the Mdx Mouse with Implications for Therapy. Mol. Ther. Nucleic Acids. 2012;1:e39. doi: 10.1038/mtna.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen J.F., Tao Y., Li J., Deng Z., Yan Z., Xiao X., Wang D.Z. MicroRNA-1 and MicroRNA-206 Regulate Skeletal Muscle Satellite Cell Proliferation and Differentiation by Repressing Pax7. J. Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dey B.K., Gagan J., Dutta A. MiR-206 and -486 Induce Myoblast Differentiation by Downregulating Pax7. Mol. Cell. Biol. 2011;31:1329. doi: 10.1128/MCB.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu N., Bezprozvannaya S., Shelton J.M., Frisard M.I., Hulver M.W., McMillan R.P., Wu Y., Voelker K.A., Grange R.W., Richardson J.A., et al. Mice Lacking MicroRNA 133a Develop Dynamin 2-Dependent Centronuclear Myopathy. J. Clin. Investig. 2011;121:3258–3268. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stuelsatz P., Pouzoulet F., Lamarre Y., Dargelos E., Poussard S., Leibovitch S., Cottin P., Veschambre P. Down-Regulation of MyoD by Calpain 3 Promotes Generation of Reserve Cells in C2C12 Myoblasts. J. Biol. Chem. 2010;285:12670–12683. doi: 10.1074/jbc.M109.063966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kramerova I., Kudryashova E., Wu B., Spencer M.J. Regulation of the M-Cadherin-b-Catenin Complex by Calpain 3 during Terminal Stages of Myogenic Differentiation. Mol. Cell. Biol. 2006;26:8437–8447. doi: 10.1128/MCB.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li H., Malhotra S., Kumar A. Nuclear Factor-Kappa B Signaling in Skeletal Muscle Atrophy. J. Mol. Med. 2008;86:1113–1126. doi: 10.1007/s00109-008-0373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Benayoun B., Baghdiguian S., Lajmanovich A., Bartoli M., Daniele N., Gicquel E., Bourg N., Raynaud F., Pasquier M., Suel L., et al. NFkB-Dependent Expression of the Antiapoptotic Factor c-FLIP Is Regulated by Calpain 3, the Protein Involved in Limb-Girdle Muscular Dystrophy Type 2A Be. FASEB J. 2008;22:1521–1529. doi: 10.1096/fj.07-8701com. [DOI] [PubMed] [Google Scholar]
- 130.Elbaz M., Yanay N., Laban S., Rabie M., Mitrani-Rosenbaum S., Nevo Y. Life or Death by NFκB, Losartan Promotes Survival in Dy2J/Dy2J Mouse of MDC1A. Cell Death Dis. 2015;6:e1690. doi: 10.1038/cddis.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Groenendyk J., Lynch J., Michalak M. Calreticulin, Ca2+, and Calcineurin-Signaling from the Endoplasmic Reticulum. Mol. Cells. 2004;17:383–389. [PubMed] [Google Scholar]
- 132.Vissing J. Limb Girdle Muscular Dystrophies: Classification, Clinical Spectrum and Emerging Therapies. Curr. Opin. Neurol. 2016;29:635–641. doi: 10.1097/WCO.0000000000000375. [DOI] [PubMed] [Google Scholar]
- 133.Sveen M.L., Andersen S.P., Ingelsrud L.H., Blichter S., Olsen N.E., Jønck S., Krag T.O., Vissing J. Resistance Training in Patients with Limb-Girdle and Becker Muscular Dystrophies. Muscle Nerve. 2013;47:163–169. doi: 10.1002/mus.23491. [DOI] [PubMed] [Google Scholar]
- 134.Sczesny-Kaiser M., Kowalewski R., Schildhauer T.A., Aach M., Jansen O., Grasmücke D., Güttsches A.K., Vorgerd M., Tegenthoff M. Treadmill Training with HAL Exoskeleton-A Novel Approach for Symptomatic Therapy in Patients with Limb-Girdle Muscular Dystrophy-Preliminary Study. Front. Neurosci. 2017;11:1–9. doi: 10.3389/fnins.2017.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bartoli M., Poupiot J., Vulin A., Fougerousse F., Arandel L., Daniele N., Roudaut C., Noulet F., Garcia L., Danos O., et al. AAV-Mediated Delivery of a Mutated Myostatin Propeptide Ameliorates Calpain 3 but Not α-Sarcoglycan Deficiency. Gene Ther. 2007;14:733–740. doi: 10.1038/sj.gt.3302928. [DOI] [PubMed] [Google Scholar]
- 136.Wagner K.R., Fleckenstein J.L., Amato A.A., Barohn R.J., Bushby K., Escolar D.M., Flanigan K.M., Pestronk A., Tawil R., Wolfe G.I., et al. A Phase I/II Trial of MYO-029 in Adult Subjects with Muscular Dystrophy. Ann. Neurol. 2008;63:561–571. doi: 10.1002/ana.21338. [DOI] [PubMed] [Google Scholar]
- 137.Roudaut C., Le Roy F., Suel L., Poupiot J., Charton K., Bartoli M., Richard I. Restriction of Calpain3 Expression to the Skeletal Muscle Prevents Cardiac Toxicity and Corrects Pathology in a Murine Model of Limb-Girdle Muscular Dystrophy. Circulation. 2013;128:1094–1104. doi: 10.1161/CIRCULATIONAHA.113.001340. [DOI] [PubMed] [Google Scholar]
- 138.Coalition to Cure Calpain 3. [(accessed on 15 May 2019)]; Available online: http://www.curecalpain3.org.
- 139.Sarepta Therapeutics. [(accessed on 16 May 2019)]; Available online: https://www.sarepta.com/
- 140.Selvaraj S., Filareto A., Kiley J., Voytas D., Kyba M., Perlingeiro R. Gene Correction of LGMD2A Patient-Specific IPS Cells for Targeted Autologous Cell Therapy. Mol. Ther. 2016;24:S125–S126. doi: 10.1016/S1525-0016(16)33121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Straub V., Bertoli M. Where Do We Stand in Trial Readiness for Autosomal Recessive Limb Girdle Muscular Dystrophies? Neuromuscul. Disord. 2015;26:111–125. doi: 10.1016/j.nmd.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 142.Rodino-Klapac L.R., Janssen P.M.L., Shontz K.M., Canan B., Montgomery C.L., Griffin D., Heller K., Schmelzer L., Handy C., Clark K.R., et al. Micro-Dystrophin and Follistatin Co-Delivery Restores Muscle Function in Aged DMD Model. Hum. Mol. Genet. 2013;22:4929–4937. doi: 10.1093/hmg/ddt342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sondergaard P.C., Griffin D.A., Pozsgai E.R., Johnson R.W., Grose W.E., Heller K.N., Shontz K.M., Montgomery C.L., Liu J., Clark K.R., et al. AAV.Dysferlin Overlap Vectors Restore Function in Dysferlinopathy Animal Models. Ann. Clin. Transl. Neurol. 2015;2:256–270. doi: 10.1002/acn3.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gruntman A.M., Flotte T.R. Delivery of Adeno-Associated Virus Gene Therapy by Intravascular Limb Infusion Methods. Hum. Gene Ther. Clin. Dev. 2015;26:159–164. doi: 10.1089/humc.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Andersson D.C., Meli A.C., Reiken S., Betzenhauser M.J., Umanskaya A., Shiomi T., D’Armiento J., Marks A.R. Leaky Ryanodine Receptors in β-Sarcoglycan Deficient Mice: A Potential Common Defect in Muscular Dystrophy. Skelet. Muscle. 2012;2:1–9. doi: 10.1186/2044-5040-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mázala D.A.G., Pratt S.J.P., Chen D., Molkentin J.D., Lovering R.M., Chin E.R. SERCA1 Overexpression Minimizes Skeletal Muscle Damage in Dystrophic Mouse Models. Am. J. Physiol. Physiol. 2015;308:C699–C709. doi: 10.1152/ajpcell.00341.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zsebo K., Yaroshinsky A., Rudy J.J., Wagner K., Greenberg B., Jessup M., Hajjar R.J. Long-Term Effects of AAV1/SERCA2a Gene Transfer in Patients with Severe Heart Failure: Analysis of Recurrent Cardiovascular Events and Mortality. Circ. Res. 2014;114:101–108. doi: 10.1161/CIRCRESAHA.113.302421. [DOI] [PubMed] [Google Scholar]
- 148.Yi J., Ma C., Li Y., Weisleder N., Ríos E., Ma J., Zhou J. Mitochondrial Calcium Uptake Regulates Rapid Calcium Transients in Skeletal Muscle during Excitation-Contraction (E-C) Coupling. J. Biol. Chem. 2011;286:32436–32443. doi: 10.1074/jbc.M110.217711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Qaisar R., Bhaskaran S., Ranjit R., Sataranatarajan K., Premkumar P., Huseman K., Van Remmen H. Restoration of SERCA ATPase Prevents Oxidative Stress-Related Muscle Atrophy and Weakness. Redox Biol. 2019;20:68–74. doi: 10.1016/j.redox.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Voit A., Patel V., Pachon R., Shah V., Bakhutma M., Kohlbrenner E., McArdle J.J., Dell’Italia L.J., Mendell J.R., Xie L.H., et al. Reducing Sarcolipin Expression Mitigates Duchenne Muscular Dystrophy and Associated Cardiomyopathy in Mice. Nat. Commun. 2017;8:1068. doi: 10.1038/s41467-017-01146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tsuji T., del Monte F., Yoshikawa Y., Abe T., Shimizu J., Nakajima-Takenaka C., Taniguchi S., Hajjar R.J., Takaki M. Rescue of Ca2+ Overload-Induced Left Ventriclur Dysfunction by Targeted Ablation of Phospholamban. Am. J. Physiol. Circ. Physiol. 2008;296:H310–H317. doi: 10.1152/ajpheart.00975.2008. [DOI] [PubMed] [Google Scholar]
- 152.Bianchini E., Testoni S., Gentile A., Calì T., Ottolini D., Villa A., Brini M., Betto R., Mascarello F., Nissen P., et al. Inhibition of Ubiquitin Proteasome System Rescues the Defective Sarco(Endo)Plasmic Reticulum Ca2+-ATPase (SERCA1) Protein Causing Chianina Cattle Pseudomyotonia. J. Biol. Chem. 2014;289:33073–33082. doi: 10.1074/jbc.M114.576157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Katsetos C.D., Koutzaki S., Melvin J.J. Mitochondrial Dysfunction in Neuromuscular Disorders. Semin. Pediatr. Neurol. 2013;20:202–215. doi: 10.1016/j.spen.2013.10.010. [DOI] [PubMed] [Google Scholar]