Abstract
Tamoxifen displays wide inter-individual variability (IIV) in its pharmacokinetics and treatment outcome. Data on tamoxifen pharmacokinetics and pharmacogenetics from black African breast cancer patient populations is lacking. We investigated the pharmacokinetic and pharmacogenetic profile of tamoxifen and its major active metabolite, endoxifen, in Ethiopian breast cancer patients. A total of 81 female breast cancer patients on adjuvant tamoxifen therapy were enrolled. Tamoxifen (Tam) and its major metabolites, N-desmethyltamoxifen (NDM), 4-hydroxy-tamoxifen (4-HT), and (Z)-endoxifen (E) were quantified using LC-MS/MS. Genotyping for CYP2D6, CYP2C9, CYP2C19, CYP3A5, POR, and ABCB1 and UGT2B15 and copy number variation for CYP2D6 were done. The proportion of patients with low endoxifen level (<5.9 ng/mL) was 35.8% (median concentration 7.94 ng/mL). The allele frequency of CYP2D6 gene deletion (*5) and duplication (*1×N or *2×N) was 4.3% and 14.8%, respectively. Twenty-six percent of the patients carried duplicated or multiplicated CYP2D6 gene. An increase in CYP2D6 activity score was associated with increased endoxifen concentration and MRE/NDM (p < 0.001). The IIV in endoxifen concentration and MRE/NDM was 74.6% and 59%, respectively. CYP2D6 diplotype explained 28.2% and 44% of the variability in absolute endoxifen concentration and MRE/NDM, respectively. The explanatory power of CYP2D6 diplotype was improved among ABCB1c.4036G carriers (43% and 65.2%, respectively for endoxifen concentration and MRE/NDM) compared to A/A genotype. CYP2C9, CYP2C19, and CYP3A5 genotypes had no significant influence on endoxifen concentration or MRE/NDM. In conclusion, we report a high rate of low endoxifen level as well as large IIV in tamoxifen and its metabolite concentrations. CYP2D6 is significant predictor of plasma endoxifen level in a gene-dose dependent manner.
Keywords: tamoxifen, endoxifen, pharmacokinetics, pharmacogenetics, CYP2D6, ABCB1, breast cancer
1. Introduction
Tamoxifen is widely used as adjuvant therapy for women with estrogen receptor-positive breast cancer [1]. Its use for 5 years has been shown to be highly effective in reducing breast cancer recurrence and cancer-specific mortality [2]. Despite its proven clinical effectiveness, not all breast cancer patients gain benefit from tamoxifen treatment; approximately 30–50% of patients relapse or eventually die from the disease [2]. Some individuals also experience treatment associated side effects [3].
The causes for breast cancer relapse or occurrence of side effects after tamoxifen treatment are attributed to a variety of factors, including pharmacogenetic variations in relevant drug-metabolizing enzymes involved in the metabolic activation of tamoxifen into endoxifen [1,4]. The bioactivation of tamoxifen involves several CYP450 drug metabolizing enzymes (DMEs) including, CYP2D6, CYP3A4, CYP3A5, CYP2C9, and CYP2C19, with key roles for CYP2D6 and CYP3A [5]. The wide interindividual variability in the concentration of tamoxifen and its metabolites was mainly attributed to genetic polymorphism in CYP2D6 [3,6,7,8]. Previous studies showed that patients with low CYP2D6 enzyme activity or taking medication which inhibit CYP2D6 activity were associated with lower endoxifen concentration [7,9]. Other studies also demonstrated that patients exhibiting endoxifen concentration below a proposed therapeutic threshold (5.9 ng/mL), were significantly associated with a higher risk of recurrence or death [3,10].
The link between patients’ CYP2D6 genotype and plasma levels of tamoxifen and its metabolites has been established in population of European and Asian descent [10,11,12], but not in sub-Saharan populations. Studies also showed that CYP2D6 explains about 39–58% of the interpatient variability in endoxifen concentration [8] and residual variability is largely unexplained [7,12]. The extent to which CYP2D6 determines the wide interpatient variability of endoxifen under standard tamoxifen treatment is not well characterized in black African populations. Although CYP2D6 is the predominant enzyme in the metabolic conversion of tamoxifen into endoxifen, previous studies reported inconsistent findings about the association between CYP2D6 polymorphism and clinical outcome of tamoxifen therapy.
The polymorphic CYP2D6 gene exhibits pronounced interethnic differences in allele frequency distribution. Ethnic differences in tamoxifen metabolism and treatment outcome are also well recognized [13,14]. Reports on the association of CYP2D6 genotype with clinical outcome in different population is not consistent, and hence lack of consensus to recommend genotype-based personalized tamoxifen treatment. About 29% of Ethiopians carry functionally active CYP2D6 gene duplications and multiplications associated with increased enzyme activity [15]. Given this unique CYP2D6 genotype constitution, the pharmacokinetic profile of tamoxifen in this population may have important implications for tamoxifen dosing recommendations to improve clinical effectiveness of the treatment. CYP2C9, CYP2C19, and CYP3A5 are also involved in tamoxifen metabolism, but the impact of their genotype on the pharmacokinetics (PK) of tamoxifen are conflicting [4,6]. Furthermore, data on the impact of other genetic variations such as the role of POR and non-genetic determinants on endoxifen plasma concentration is limited. There is growing evidence that altered ABCB1 activity may affect tamoxifen pharmacokinetics and possibly tamoxifen efficacy in breast cancer patients. The primary active tamoxifen metabolites, endoxifen and 4-hydroxytamoxifen, are substrates for ABCB1 [16]. The three common SNPs in the protein coding region of ABCB1 gene (rs1128503 (1236T>C), rs2032582 (2677T>G/A), and rs1045642 (3435T>C) have been the focus of many pharmacokinetic and disease association studies [17] with inconsistent and controversial results [18]. Other studies have also investigated the relationship between the C3435T polymorphism of ABCB1 gene and risk of breast cancer; whose results are also conflicting [19,20]. On the other hand, the relevance of ABCB1c.4036A>G (rs3842A>G) has, however, been consistently demonstrated in African population in relation to the PK of antiretroviral drugs. Several studies have showed that ABCB1c.4036A>G (rs3842A>G) is a significant predictor of plasma and intracellular efavirenz concentration in Uganda [21,22], South Africa [23], Ethiopia and Tanzania [24] as well as in other populations [25]. We hypothesize that this SNP may also influence cellular transport of tamoxifen metabolites in our study population. In the present study, we investigated the pharmacokinetics profile and the influence of CYP2D6, CYP2C9, CYP2C19, CYP3A5, POR, ABCB1 and UGT2B15 polymorphisms on the plasma level of tamoxifen and its active metabolites in Ethiopian breast cancer patients receiving adjuvant tamoxifen.
2. Results
2.1. Sociodemographic Characterstics
A total of 89 patients (mean age = 39 ± 8.5) who were on tamoxifen treatment were enrolled. Of these, eight patients were excluded as the plasma level of tamoxifen and endoxifen were below the limit of quantification, possibly related to noncompliance to therapy. The remaining 81 patients, all with tamoxifen concentration >150 nM were included in the analysis. All the patients completed full course of chemotherapy after primary surgical treatment (modified radical mastectomy). The median duration of tamoxifen use was 11 months (IQR 6–18 months). The socio-demographic and tumor characteristics are displayed in Table 1.
Table 1.
Socio-demographic and tumor characteristics of tamoxifen treated breast cancer patients at the radiotherapy center, Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia.
| Parameters | Value | |
|---|---|---|
| Socio-demographics | ||
| Age (years, mean ± SD *) | 39 ± 8.5 | |
| BMI (kg/m2, mean ± SD) | 24.6 (±3.86) | |
| Chemotherapy regimen used | N (%) | |
| FAC ∝ | 37 (45.7) | |
| AC | 11 (11.1) | |
| AC - T | 35 (43.2) | |
| Duration of tamoxifen use (months, median + IQR ♣) | 11 (6–18) | |
| Menopausal status | N (%) | |
| Premenopausal | 56 (69.1) | |
| Postmenopausal | 25 (30.9) | |
| Tumor characteristics | N (%) | |
| Histologic type of tumor | Ductal | 79 (97.5) |
| Lobular | 2 (2.5) | |
| Degree of differentiation | Well differentiated | 21 (25.9) |
| Moderately differentiated | 44(54.3) | |
| Poorly differentiated | 16 (19.8) | |
| Lymph node involvement | Negative | 15 (18.5) |
| Positive | 66 (81.5) | |
| Distant metastatic site | No known distant metastasis | 10 (12.3) |
| Bone, lymph node, or lung only | 63 (77.8) | |
| Liver, CNS, lung + other organs | 8 (9.9) |
♣ IQR inter quartile range; * SD standard deviation; ∝ FAC (5-Flourouracil 500 mg/m2, Adriamycin [Doxorubicin] 50 mg/m2, and Cyclophosphamide 500 mg/m2), AC (Adriamycin 50 mg/m2 and Cyclophosphamide 600 mg/m2), and AC-T (four cycles of Adriamycin 60 mg/m2 and Cyclophosphamide 600 mg/m2 followed by another 4 cycles of Taxol 175 mg/m2).
SNP genotyping results for CYP2D6, CYP2C9, CYP2C19, CYP3A5, POR, ABCB1 and UGT2B15 are presented in Table 2. All the genotype frequencies were in Hardy-Weinberg Equilibrium (p > 0.05). A total of 20 (25.9%) patients had CYP2D6 copy number of 3 or more. The allele frequencies of CYP2D6 *5 (gene deletion), and *1 or *2 gene duplication/multiplication were 4.3 and 14.8%, respectively. The frequencies of CYP2D6 activity score (AS) and phenotype categories are presented in Table 3. Based on the CPIC CYP2D6 phenotype assignment, 61.7 and 22.2% of the patients are predicted to be normal metabolizers (NMs) and ultra-rapid metabolizers (UMs), respectively. The prevalence of poor (PMs) and intermediate metabolizers (IMs) was low, accounting for 1.2 and 3.7%, respectively.
Table 2.
The genotype and allele frequency distribution of tamoxifen metabolizing enzymes’ and transporter genes.
| Gene | Variant allele | Allele frequency (%) |
|---|---|---|
| CYP2D6 | *2 | 33.3 |
| *4 | 4.9 | |
| *5 | 4.3 | |
| *10 | 1.9 | |
| *17 | 10.5 | |
| *1×N or *2×N † | 14.8 | |
| CYP2C9 | *2 | 4.3 |
| *3 | 7.4 | |
| CYP2C19 | *2 | 11.7 |
| *3 | 1.2 | |
| CYP3A5 | *3 | 67 |
| POR | *28 | 12.4 |
| ABCB1 c.4036A>G | G | 14.8 |
| ABCB1 c.3435C>T | T | 16.9 |
| UGT2B15 *2 | T | 20.2 |
| UGT2B15 *4 | C | 40.3 |
† Number of CYP2D6 gene copies.
Table 3.
CYP2D6 activity score and plasma concentrations of tamoxifen and its metabolite and respective metabolic ratios.
| CYP2D6 Activity Score | Phenotype Group † | N (%) |
|---|---|---|
| >2 | UM ¥ | 18 (22.2) |
| 2 | NM | 37 (45.7) |
| 1.5 | NM | 12 (14.8) |
| 1 | NM or IM | 10 (12.4) |
| 0.5 | IM | 3 (3.7) |
| 0 | PM | 1 (1.2) |
| Plasma concentrations | values | CV (%) ‡ |
| Tamoxifen (ng/mL, median + IQR #) | 138 (105–200.5) | 56.2 |
| N-Desmethyl-tamoxifen (ng/mL, median + IQR) | 257 (185.5–343) | 53.1 |
| 4-Hydroxy-tamoxifen (ng/mL, median + IQR) | 3.04 (2.15–4.0) | 61 |
| Z-Endoxifen (ng/mL, median + IQR) | 7.94 (4.68–13.75) | 74.7 |
| Metabolic ratio | values | CV (%) ‡ |
| MRεE/NDM (median + IQR) | 0.038 (0.021–0.06) | 59 |
| MRγNDM/Tam (median + IQR) | 1.81 (1.51–2.26) | 28.6 |
| MRλ4-HT/Tam (median + IQR) | 0.022 (0.016–0.029) | 38.8 |
| MR♠E/4-HT (median + IQR) | 2.85 (2.10–3.73) | 43.9 |
† Phenotype translated based on Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and tamoxifen therapy; ¥ UM ultrarapid metabolizer; NM normal metabolizer; IM intermediate metabolizer; PM poor metabolizer; ‡ coefficient of variation; # IQR interquartile range; ε metabolic ratio of endoxifen to N-desmethyl-tamoxifen; γ metabolic ratio of N-desmethyl-tamoxifen to tamoxifen; λ metabolic ratio of 4-Hydroxy-tamoxifen to tamoxifen; ♠ metabolic ratio of endoxifen to 4-hydroxy-tamoxifen.
2.2. Plasma Tamoxifen and Its Metabolite Concentration Profile
The plasma concentrations of tamoxifen and its metabolites are depicted in Table 3. The median concentration of endoxifen (7.94 ng/mL), was 2.6 times higher than 4-HT (3.04 ng/mL). The proportion of patients with low endoxifen concentration (<5.9 ng/mL) was 35.8%. This low level of endoxifen was detected in patients classified as PM/PM (1/1), IM/PM (3/3), IM/IM (1/1), NM/PM (6/9, 66.7%), NM/IM (5/12, 41.7%), and NM/NM (11/35, 31.4%) diplotypes. There was pronounced interindividual variability in endoxifen concentration (coefficient of variation 74.7%) and in metabolic ratio of endoxifen to NDM (coefficient of variation 59%) (Table 3).
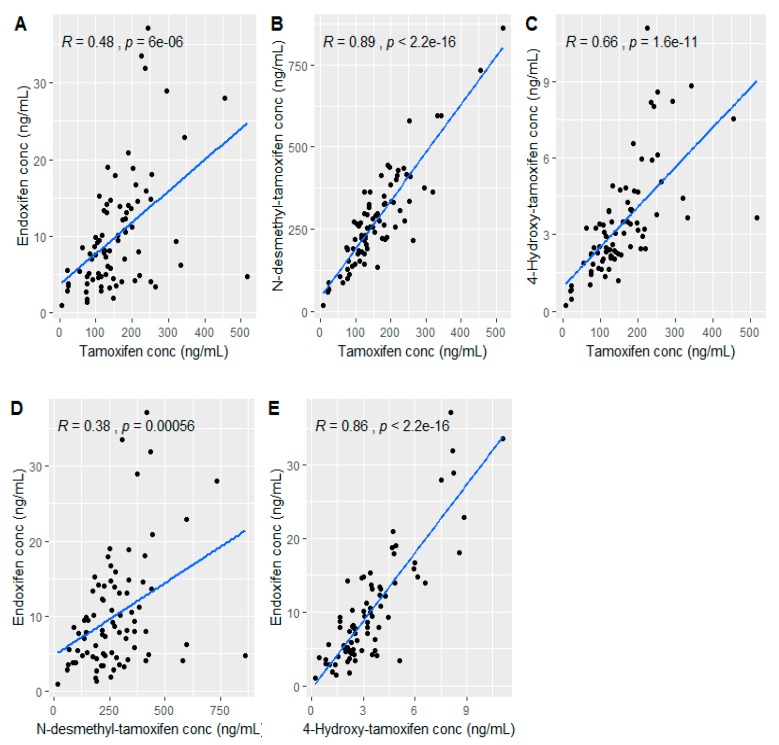
Pearson correlation analysis between tamoxifen and its metabolites revealed a strong positive association between tamoxifen and formation of NDM (Pearson correlation coefficient (R) = 0.89), and tamoxifen and formation of 4-HT (R = 0.66) (Figure 1). Thus, an increase in the concentration of tamoxifen was associated with a corresponding increase in NDM and 4HT levels. Formation of endoxifen is also strongly correlated with 4-HT level (R = 0.86). However, the correlation between NDM and formation of endoxifen (R = 0.38) as well as tamoxifen and formation of endoxifen (R = 0.48) was moderate.
Figure 1.
Scatter plot showing the Pearson’s correlation between endoxifen versus tamoxifen (A), N-desmethyl-tamoxifen versus tamoxifen (B) and 4-hydroxy-tamoxifen versus tamoxifen (C), endoxifen versus N-desmethyl-tamoxifen (D) and endoxifen versus 4-hydroxy-tamoxifen (E).
3.3. Association of Pharmacogenetics and Pharmacokinetics of Tamoxifen Its Metabolites
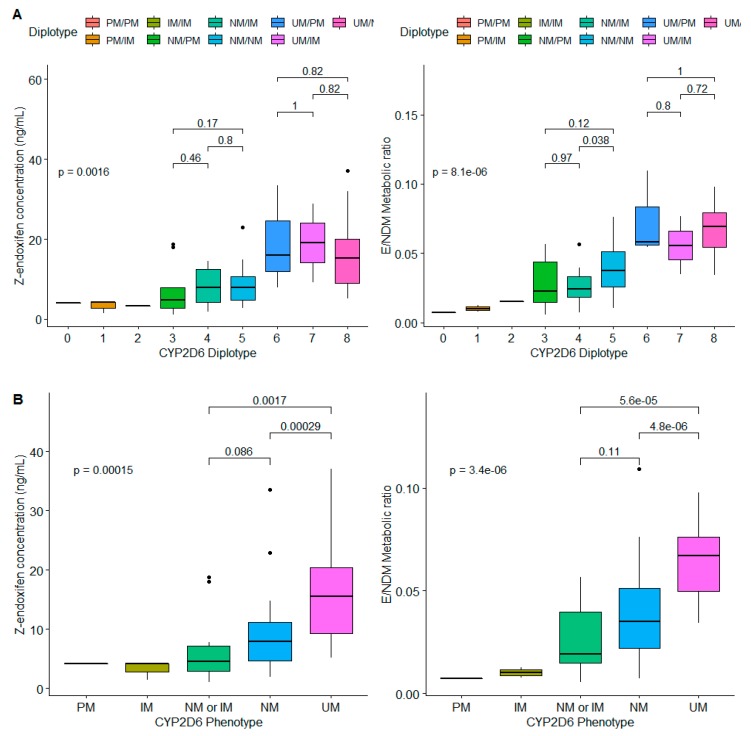
Analysis of variance showed that the median endoxifen level, MRE/NDM and MR4HT/Tam were significantly different among CYP2D6 diplotype (p < 0.001) or genotype-predicted phenotype groups (p < 0.001) (Figure 2). Patients with UM/EM, UM/IM and UM/PM diplotypes were associated with higher median endoxifen concentrations or metabolic ratios (MRE/NDM and MR4-HT/Tam) (Figure 2A). Similarly, a corresponding increase in absolute endoxifen concentration and metabolic ratio (MRE/NDM) was observed with an increase in the activity score of CYP2D6 phenotype (p < 0.001) (Figure 2B).
Figure 2.
Plasma concentration of endoxifen and metabolic ratio (MRE/NDM) by CYP2D6 diplotype (A) and phenotype (B).
In patients categorized into CYP2D6 NM/IM diplotype group, significant difference in MRE/NDM (p = 0.036) and MRE/4-HT (p = 0.022) was observed between genotypes *1/*10, *1/*17 and *2/*17. Among patients classified under NM/NM diplotypes, significant differences were also found between *1/*1, *1/*2, and *2/*2 genotypes in tamoxifen (p < 0.001), NDM (p < 0.001), and 4-HT concentrations (p < 0.001) as well as corresponding metabolic ratios; MRE/NDM (p < 0.001), MRE/4-HT (p = 0.025) and MR4-HT/Tam (p = 0.007).
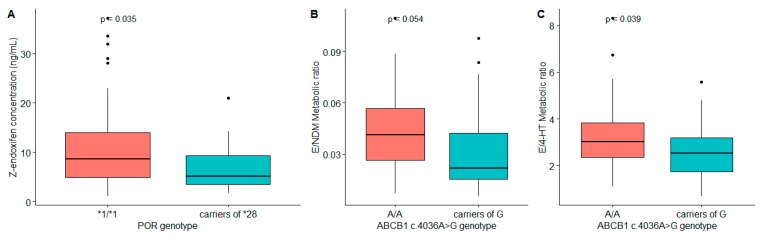
In addition to CYP2D6 genotype, a moderate association POR genotype with endoxifen level was observed (p = 0.035) (Figure 3A). On the other hand, ABCB1 (rs3842) genotype showed a trend of association with MRE/NDM (p = 0.054) and moderate association with MRE/4-HT (p = 0.039). Carriers of the G allele for ABCB1 (rs3842) had a lower MRE/NDM (0.022 vs. 0.041; p = 0.054) and MRE/4-HT (2.52 vs. 3.02, p = 0.039) ratios compared to the wild type genotype (Figure 3B,C).
Figure 3.
Plasma endoxifen concentration and metabolic ratios (MRE/NDM and MRE/4-HT) plotted by POR (A) or ABCB1 c.4036A>G genotype (B,C).
Patients with at least one variant allele for UGT2B15 (c.1568A>C, rs4148269) showed lower median plasma concentrations of tamoxifen (125.9 vs. 173.8 ng/mL, p = 0.045). The median NDM/Tam (1.86 vs. 1.54, p = 0.004) and E/4-HT (2.96 vs. 2.06, p = 0.029) ratios were also higher in carriers of C allele for UGT2B15 (c.1568A>C, rs4148269) compared to the wild type genotype. However, there was no significant difference in median plasma tamoxifen metabolite concentrations or metabolic ratios between CYP2C9, CYP2C19 CYP3A5, UGT2B15*2 (c.253G>T) or ABCB1 c.3435C>T genotypes. No difference was also detected in the median plasma tamoxifen concentrations and its metabolites (p > 0.05), between pre- and postmenopausal patients, and different BMI groups as well as with age.
The impact of CYP2D6 diplotype or phenotype on the interpatient variability of endoxifen or MRE/NDM was modeled using linear regression. The results indicated that CYP2D6 diplotype explained 28.2% of the variability in endoxifen concentration (coefficient of determination (R2) = 0.282, p < 0.001). Similarly, CYP2D6 phenotype explained 26.3% of the variability in endoxifen concentration (R2 = 0.263, p < 0.001). POR contributed 3.4% of the variability in absolute endoxifen concentration (R2 = 0.034, p = 0.054). CYP2D6 diplotype explained 44% of variability in MRE/NDM and combined with ABCB1c.4036A/G genotype, it explained 46.7% (R2 = 0.467, p < 0.001) of the variability in MRE/NDM. Phenotype and ABCB1c.4036A>G explained 40.9% of the variability in MRE/NDM (R2 = 0.409, p < 0.001). ABCB1c.4036A>G contributed 4% the variability in MRE/NDM (R2 = 0.040, p = 0.042).
The impact of CYP2D6 on endoxifen variability and MRE/NDM (a marker for CYP2D6 metabolic activity) was assessed stratifying patients by ABCB1 genotype groups. Interestingly, CYP2D6 diplotype explained 43% of the variability in endoxifen concentration among ABCB1c.4036G carriers (R2 = 0.43, p = 0.004) (explanatory power increased from 28.2 to 43%), compared to A/A genotype (R2 = 0.231, p < 0.001). Similarly, CYP2D6 diplotype explained 65.2% of the variability in MRE/NDM (improved from 44 to 65.2%) among ABCB1c.4036G carriers (R2 = 0.652, p < 0.001), compared to A/A genotype (R2 = 0.356, p < 0.001).
3. Discussion
In the present study, we investigated the pharmacokinetic profile of tamoxifen and its active metabolites in breast cancer patients, and explored the impact of CYP2D6, CYP2C9, CYP2C19, CYP3A5, POR, ABCB1 and UGT2B15 genotypes on endoxifen and MRE/NDM in Ethiopians, a population with high frequency distribution of active CYP2D6 gene duplication or multiplication. Our major findings include; (i) a wide between-patient variability in plasma concentrations of tamoxifen and its metabolites, (ii) significant proportion of patients (35.8%) with subtherapeutic plasma endoxifen concentrations (<5.9 ng/mL), (iii) CYP2D6 diplotype is a significant predictor of both endoxifen concentration and MRE/NDM, (iv) and a moderate effect of POR*28 on plasma endoxifen concentration and ABCB1c.4036A>G genotype on MRE/NDM. To our knowledge, this is the first study to investigate the pharmacokinetics and pharmacogenetics of tamoxifen in breast cancer patients from subSaharan African population and to explore the impact of POR and ABCB1c.4036A>G pharmacogenetics on tamoxifen PK.
Although CYP2D6 is involved in various tamoxifen metabolic pathways, it is the sole enzyme responsible for the formation of endoxifen from NDM [1]. Previous studies have consistently demonstrated a significant gene-dose effect of CYP2D6 polymorphism for tamoxifen metabolism [3,6,7,8]. Our study also showed an increase in endoxifen concentration and MRE/NDM with increase in CYP2D6 activity score, indicating a gene-dose effect of endoxifen formation. However, CYP2D6 gene is highly variable among different ethnic groups, resulting in variations in concentrations of tamoxifen and its metabolites, mainly endoxifen [26,27].
In the present study, CYP2D6 diplotype and ABCB1c.4036A/G genotype explained 46.7% the variability in MRE/NDM. With slightly different genotype interpretation applied for CYP2D6 AS, previous study demonstrated that the contribution of CYP2D6 to the IIV of endoxifen formation via NDM, as revealed from MRE/NDM, was 46% (Asians), 55% (Lebansese) and 55% (Caucasians) [3]. Our result is comparable to Asians but lower than Lebanese or Caucasians. The explained variability of absolute endoxifen concentration by CYP2D6 diplotype in our study (28.2%) was also lower than that reported in Asians and Caucasians (39–58%) [7], suggesting that other factors could contribute for the bioavailability of endoxifen in our study population.
Previous studies reported that variations in the concentration of endoxifen can be related to variations in tamoxifen concentrations itself [28,29]. A high between-patient variations in plasma tamoxifen (CV = 56.2%) and NDM concentrations (CV = 53.1%) was also observed in our study. The Pearson’s correlation coefficient between endoxifen and tamoxifen (R = 0.48) as well as endoxifen and NDM (R = 0.38) also showed that, endoxifen level would only moderately be predicted by plasma levels of tamoxifen and NDM. This could mean that the extent of absorption of tamoxifen from the gut or the quality of the formulation or food-drug interaction could also be potential factors influencing the bioavailability of tamoxifen thereby impacting the plasma level of endoxifen.
Our study also showed significant differences in MRE/NDM between patients with genotype CYP2D6 *1/*10, *1/*17 and *2/*17, which belong to EM/IM dipotype. Similarly, differences were also observed in endoxifen level between *1/*1, *1/*2, and *2/*2 genotypes which are grouped under same NM/NM diplotype groups (Figure 2). In agreement with our finding, recent study demonstrated that the concentrations of tamoxifen, endoxifen 4-HT, and NDM were significantly different between genotypes classed under same NMs [8]. The range of activity between CYP2D6*1/*1, *1/*2, and *2/*2 genotypes could be explained by the differences in the level of expression of mRNA, associated with the presence or absence of enhancer SNP in the enhancer regions distant to the CYP2D6 gene [30].
Previous studies showed that tamoxifen efficacy is dependent on attaining a certain threshold of endoxifen level. According to Madlensky et al., women in the upper quintiles of endoxifen concentration, but not of tamoxifen and other metabolite levels, had a lower recurrence rate compared to those in the lowest quintile. Moreover, endoxifen concentration >5.97 ng/mL was associated with a 30% lower risk of additional breast cancer events and proposed as the therapeutic threshold for endoxifen [10]. A similar endoxifen level (13 nM) was also noted to occupy 90 % of the estrogen receptor (ER) in vitro and 93% of CYP2D6 PMs fell below this concentration [12]. Subsequent study also demonstrated that, low endoxifen concentration (<5.97 ng/mL) was associated with distant relapse free survival [3], indicating the significance of this therapeutic threshold. The results of our study showed that significant proportion of Ethiopian patients (35.8%) were unable to attain this proposed therapeutic threshold for endoxifen concentrations. This is in agreement with previous study reported in Sweden (30%) [29], but higher compared to that reported in the U.S. (20%) [10].
In our study, the low level of endoxifen was detected in patients classified to PM/PM, IM/PM, IM/IM, NM/PM, NM/IM, as well as NM/NM diplotypes. Poor/intermediate metabolizer genotype was identified as a predictors of low endoxifen concentration in earlier study [10]. Moreover, it was also reported that 99% of PMs can be predicted by CYP2D6 null alleles in homozygous variant or heterozygous genotype [31]. Our result is in agreement with this notion, given that all the null or lower activity diplotypes (PM/PM, IM/PM, and IM/IM) were associated with low endoxifen level. In such patients, the clinical outcome of the cancer could be less favorable, though its association was not investigated in the present study. Thus, in line with the recent CPIC therapeutic recommendations, CYP2D6 PMs in our study are recommended alternate hormonal therapy such as aromatase inhibitors (AIs) for post-menopausal women or AIs along with ovarian function suppression in premenopausal women. Escalation of tamoxifen dose from 20–40 mg/day can also be considered for PMs if there is contraindications to AIs [32].
In contrast, other studies demonstrated that not all PMs have endoxifen concentrations below the proposed threshold concentration (5.9 ng/mL) [10,33]. For example, it was observed that 24 % of the PMs were still able to generate therapeutic concentrations of endoxifen despite the lack of metabolic activity of CYP2D6 [33]. This could partly be attributed to environmental factors. A previous study demonstrated that black Africans were associated with lower rates of CYP2D6-dependent drug metabolism compared to Caucasians of the same apparent genotype [34]. Moreover, among individuals of the same CYP2D6 genotype, it was revealed that Ethiopians in living in Sweden had higher CYP2D6 metabolic activity than Ethiopians in Ethiopia [34], implying the profound impact of environmental factors in CYP2D6 catalyzed drug metabolism. The frequency of CYP2D6 PM (1.2%) and CYP2D6 duplication (26%) identified in this study is lower as compared to previous report [34].
In the present study, patients with NM/IM (41.7%) and NM/NM diplotypes (31.4%), all belonging to NM phenotypes had lower endoxifen concentrations (<5.9 ng/mL). Recently, it was reported that the activity of CYP2D6 is impacted not only by genetic variations within the CYP2D6 gene, but also by regulatory mechanisms including genetic polymorphisms in the enhancer regions or differential expression of transcription factors, which can modulate the rate of translation of mRNA into protein [35]. Genotype panels typically interrogate a limited number of SNPs identifying more commonly observed CYP2D6 alleles and provide limited information on gene copy number and structural variants [35]. It is therefore possible that a patient carries allele(s) that was not tested for and that may contribute to the variability seen in these groups. In addition, the impact environmental factors on CYP2D6 catalyzed drug metabolism cannot also be underestimated [34]. Moreover, other individual-specific and biologic factors may also modulate CYP2D6 expression levels or enzyme activity and contribute to the variability CYP2D6 enzymatic activity. Emerging evidence showed the contribution of the microbiome on overall health and its role in drug metabolism and response [36]. A recent study also provided evidence that nutritional status, specifically fasting, alters P450-mediated drug metabolism in animal studies [37]. However, the extent a person’s microbiome, nutritional and fasting status influences CYP activity and CYP2D6 metabolizer status, intra- and interindividual variability, in particular, remains to be investigated in future studies. Notably, the low level of endoxifen detected in large proportion of patients with predicted NM phenotypes in our study may suggest the need for therapeutic drug monitoring (TDM) of endoxifen in addition to genotyping for the common functional CYP2D6 variant alleles.
Cytochrome P450 oxidoreductase (POR) is the only electron donor for all microsomal cytochrome P450 monooxygenases (CYP) responsible for oxidation of more than 80% of drugs [38]. In our study, we observed that POR*28 carriers were associated with low plasma endoxifen level, although the effect is moderate (p = 0.035). Previous reports indicated that genetic variants of POR (e.g., POR*28 (A503V)) were implicated in the impaired activity of CYP2D6 and CYP3A4 [39,40], possibly contributing to genetic variability in drug metabolism.
Apart from tamoxifen activating enzymes, genetic variants in drug transporters have been reported to influence pharmacokinetics and play a role to a variable degree in the clinical outcome of tamoxifen treatment and to confer treatment resistance for many anticancer drugs [41]. Results of the present study did not show significant effect of ABCB1c.3435C>T on tamoxifen and its metabolites level. However, it was observed that carriers of G allele for ABCB1c.4036A>G had a marginally lower MRE/NDM (0.025 vs. 0.036; p = 0.054) compared to the wild type allele (ABCB1, allele A) indicating that polymorphism of ABCB1 ABCB1c.4036A>G could influence exposure to endoxifen. Interestingly, among ABCB1c.4036G carriers, improved predictive effect of the variability in endoxifen concentration (43%) and endoxifen formation from NDM (65.2%) was observed by CYP2D6 diplotype. A previous study demonstrated that ABCB1 influences the systemic exposure of tamoxifen and its active metabolites, 4-HT and endoxifen [16]. Moreover, several other studies also substantiated the impact of ABCB1 in relation to clinical outcome of tamoxifen. ABCB1 polymorphism was associated with risk of recurrence or disease free survival (DFS), in breast cancer patients on tamoxifen treatment [16,42,43].
Variant alleles of CYP2C9, CYP2C19 and CYP3A5 could alter tamoxifen metabolism and exposure to its metabolites especially formation of 4-hydroxy-tamoxifen [1]. However, no evidence of association was observed between CYP2C9, CYP2C19 and CYP3A5 genotypes and plasma concentrations of tamoxifen or its metabolites. Our finding is substantiated with previous studies [6,11,33]. On the other hand, other studies have reported lower plasma concentrations of 4-HT and endoxifen among carriers of CYP2C9*2 or *3 alleles [3,12]. The disparity in the results could be attributed to the difference in sample size or the variation in allele distributions.
In addition of the significant effect of CYP2D6 genotype, variations in glucuronidation activity of the uridine 5’-diphosphate-glucuronosyltransferase (UGT) also seem to play a role in maintaining optimal levels of both 4-hydroxy-tamoxifen and endoxifen. Modified glucouronidation could affect half-lives of circulating tamoxifen and its metabolites thereby influencing the effectiveness of active metabolites in the treatment of breast cancer [44]. In the present study, patients with C allele for UGT2B15c.1568A>C (rs4148269) showed a lower plasma concentrations of tamoxifen, compared to those with wild type genotype. Although we could not determine plasma levels of tamoxifen-glucouronide, our finding may suggest increased glucouronidation of tamoxifen. Higher glucouronidation activity of UGT2B15c.1568A>C (rs4148269) genotype was also reported against endoxifen and 4-hydroxytamoxifen [44] and against oxazepam [45].
On the other hand, our result did not show significant variations in tamoxifen and its metabolites concentration with UGT2B15*2 (c.253G>T, rs1902023). Similarly, no correlation was detected between this polymorphism and both hydroxylated tamoxifen metabolites and their corresponding glucouronide product [12,44]. However, UGT2B15*2 (c.253G>T, rs1902023) was demonstrated to be a determinant of glucouronidation of oxazepam [45,46] and lorazepam [47] in the human liver, suggesting a substrate-dependent effect of this polymorphism.
Our study has some limitations. We present a significant impact of CYP2D6 and a marginal effect of POR and ABCB1 on the pharmacokinetics of tamoxifen and its metabolites. However, the sample size included in our study was not powered enough and might be a reason for lack of significant association of POR and ABCB1 genotypes with tamoxifen and its metabolites PK. In addition, association of plasma concentration of the active tamoxifen metabolites to clinical outcome was not assessed. The treatment of cancer in patients with comorbidities can be challenging as these individuals are underrepresented in clinical studies [48]. In addition to the CYP2D6 genetics that much of the controversy around tamoxifen therapy has focused, drug-drug interactions (DDI) involving CYP2D6 in polypharmacy for co-morbidities markedly increases the complexity of tamoxifen metabolism and efficacy. The concurrent use of CYP2D6 inhibitors and tamoxifen would be expected to substantially reduce endoxifen concentrations, and there is clinical evidence to support this expectation [6,9,49]. For example, NMs who were also taking CYP2D6 inhibitors had endoxifen plasma concentrations that were 58% lower than NMs not on CYP2D6 inhibitors. Similarly, IMs on CYP2D6 inhibitors had endoxifen concentrations that were 38% lower than IMs not on CYP2D6 inhibitors [6]. As tamoxifen has complex pharmacokinetics, with more than a dozen drug-metabolizing enzymes and transporters involved in its disposition, enzyme inducers may increase the activity of several of these pathways, including phase II enzymes, ABC transporters, and various CYP enzymes other than CYP2D6 [50]. There is a growing evidence that enzyme inducers can substantially alter the disposition of endoxifen, reducing tamoxifen efficacy [51].
4. Materials and Methods
4.1. Patients
This study was conducted at the radiotherapy center of Tikur Anbessa specialized hospital, Addis Ababa University, Addis Ababa, Ethiopia. A total of 81 female breast cancer patients who were on adjuvant tamoxifen were enrolled. These patients had completed a full course of chemotherapy and were on tamoxifen 20 mg/day for at least three months. None of the study participants were taking any known CYP2D6 enzyme inhibitor as co-medication. The study protocol was approved by the Institutional Review Board (IRB) of the College of Health Sciences, Addis Ababa University (AAU) (Ref No: 011/16/2016), the Armauer Hansen Research Institute Ethical Review Committee (AAERC) (Ref No: PO26/16) and the Ethiopian National Research Ethics Review Committee (NRERC) (Ref No: 3.10/235/2017). Signed informed consent was obtained from individual study participants prior to study enrolment. Patients’ demographic, clinical and tumor characteristics including age, menopausal status, performance status, weight, height, body mass index (BMI), histologic type of the tumor, degree of differentiation, tumor size, lymph node involvement, hormone receptor status and co-morbidity status were collected.
4.2. Genotyping of CYP2D6, CYP2C9, CYP2C19, CYP3A5, POR, ABCB1 and UGT2B15
Whole blood sample was collected in EDTA containing vacutainer tube and genomic DNA was isolated from peripheral leukocytes using QIAamp DNA Midi Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer’s instruction. SNP genotyping was performed using TaqManTM drug metabolism genotyping assay reagents (Applied Biosystems, USA) for allelic discrimination as described previously [52] with the following ID numbers for each SNP: C_27102425_10 for CYP2D6*2 (rs16947), C_27102431_D0 for CYP2D6*4 (rs3892097), C_11484460_40 for CYP2D6*10 (rs1065852), C_2222771_A0 for CYP2D6*17 (rs28371706), C_26201809_30 for CYP3A5*3 (rs776746), C_25625805_10 for CYP2C9*2 (rs1799853), C_27104892_10 for CYP2C9*3 (rs1057910), C_25986767_70 for CYP2C19*2 (rs4244285), C_27861809_10 for CYP2C19*3 (rs4986893), C_8890131_30 for POR*28 (rs1057868) and C_11711730_20 for ABCB1c.4036A>G (rs3842), C_7586657_20 for ABCB1c.3435C>T (rs1045642), C_27028164_10 for UGT2B15*2 (rs1902023), C_9440184_20 for UGT2B15*4 (rs4148269). The genomic DNA samples were amplified in 96-well plates on QuantStudioTM 12K Flex Real-Time PCR system (Applied Biosystems, Life Technologies Holding, Singapore). The final volume for each reaction was 10 μL, consisting of TaqManTM fast advanced master mix (Applied Biosystems, Waltham, MA, USA), TaqManTM 20× drug metabolism genotyping assays mix (Applied Biosystems) and genomic DNA. The PCR conditions consisted of an initial step at 60 °C for 30 s, hold stage at 95 °C for 10 min and PCR stage for 40 cycles step 1 with 95 °C for 15 and step 2 with 60 °C for 1 min and after read stage with 60 °C for 30 s. The characterized SNPs were selected on the basis of their potential to influence the functionality of enzymes to affect the disposition and transport of tamoxifen or its metabolite.
4.3. Copy Number Variation
The CYP2D6 gene copy number for the patient DNA samples was determined by TaqManTM Copy Number Assay (Life Technologies, California, USA) according to the manufacturer’s recommendations. The genomic DNA samples were amplified in 96-well plates on QuantStudioTM 12K Flex Real-Time PCR system (Applied Biosystems, Life Technologies Holding, Singapore) using comparative ΔΔCT method. CYP2D6 exon 9 (TaqMan™ Copy Number Assay ID: Hs00010001_cn), and intron 6 (TaqMan™ Copy Number Assay ID: Hs04502391_cn) were used as primers. RNase P (ID: 431683) was used as the internal control for copy number analysis (Life Technologies). All the samples were run in triplicate. The final volume for each reaction was 10 μL, consisting of TaqMan™ Genotyping Master Mix (Applied Biosystems), TaqMan™ Copy Number Assay mix (20×, Applied Biosystems), TaqMan™ Copy Number Reference Assay mix (20×, Applied Biosystems). The PCR conditions consisted of an initial step at 60 °C for 30 s, hold stage at 95 °C for 10 min and PCR stage for 40 cycles step 1 with 95 °C for 15 and step 2 with 60 °C for 1 min and after read stage with 60 °C for 30 s. Relative quantification of CYP2D6 gene copy number was performed using CopyCaller™ Software (Life Technologies). Genomic DNA samples with known CYP2D6 gene copy number (0, 1, 2, 3 and 4 CYP2D6 gene copies) obtained from a previous study [15] were used as controls.
4.4. Quantification of Plasma Tamoxifen and Its Metabolites
Blood samples were collected in EDTA tube, centrifuged at 2500 g for 10 minutes, and the separated plasma was stored at −80 °C until analysis. Tamoxifen (Tam) and its major metabolites N-desmethyltamoxifen (NDM), (Z)-4-hydroxytamoxifen (4-HT), and (Z)-endoxifen (E) were quantified in the TDM Laboratory, Department of Clinical Pharmacology, Karolinska Institutet (Stockholm, Sweden), by ultra-high-performance liquid chromatography (UHPLC) followed by electrospray tandem mass spectrometry (LC-MS/MS). Briefly, 400 µL internal standard solution (containing 2 ng/mL of Z/E-endoxifen-d5, and 4-OH-tamoxifen-d5, and 20 ng/mL of N-desmethyl-tamoxifen-d5, and tamoxifen-d5) was added to 200 µL of thawed plasma samples and mixed for 10 minutes in a micro plate shaker (Vx-2400 Multitube mixer, Troemner, Philadelphia, USA), followed by centrifugation at 3400 rpm at 4 °C for 10 min. The supernatant was then transferred to new glass vial and evaporated to dryness using vacuum centrifuge. The residue was dissolved in 50 µL methanol over a microplate shaker for 10 min and the resulting solution was then centrifuged at 3400 rpm for 10 min at 4 °C. Ten microliter (10 µL) of the solution was injected into the LC system (Dionex Ultimate 3000, UHPLC focused, Thermo Scientific, California, USA) coupled with a mass spectrometer (TSQ Quantiva, Thermo Scientific). Separation occurred on an Accucore C-18 column (150 × 2.1 mm, Thermo Scientific). The mobile phase for gradient elution consisted of solution A (10 mmol/L ammonium formate with 0.005% formic acid in water) and solution B (10 mmol/L ammonium formate with 0.005% formic acid in 99% methanol). The LC conditions were: column oven temperature of 40 °C, flow rate of 0.50 mL/min, autosampler temperature of 10 °C. Detection was in positive ion mode using electrospray ionization and monitored using Trace Finder software (Thermo Scientific). Under this condition, the retention times were 3.25, 3.26, 3.38, 4.03, and 4.11 min for E-endoxifen, Z-endoxifen, 4-OH-tamoxifen, N-desmethyltamoxifen, and tamoxifen, respectively. Calibration curves were constructed by plotting the ratio of the area of the compound and the internal standard against the standard analyte concentration. Three quality control (QC) samples involving, (i) low-level QC [QCL] (0.3 ng/mL of E and 4-HT and 3 ng/mL of NDM and Tam, (ii) mid-level QC [QCM] (8 ng/mL of E and 4-HT and 80 ng/mL of NDM and Tam), and (iii) high level QC [QCH] (80 ng/mL of E and 4-HT and 800 ng/mL of NDM and Tam) were also included at each run.
4.5. Genotype Based Phenotype Assignment
Phenotype classification from genotype was based on the recent Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and tamoxifen therapy [32]. Functional status of each allele was assigned an activity score (AS) value ranging from 0 to 1 [0 for no function (*4, *5), 0.5 for decreased function (*10, and *17) and 1.0 for normal function alleles (*1, and *2)]. The combination of any two alleles represents the patient’s diplotype. CYP2D6 activity score (AS) was then determined as the sum of the values assigned to each allele of the diplotype. Accordingly, CYP2D6 AS was translated into patient’s phenotype status as poor metabolizer, PM (AS = 0), intermediate metabolizer, IM (AS = 0.5), normal metabolizer, NM (AS = 1.5–2) and ultra-rapid metabolizer, UM (AS > 2). Patients with AS of 1 were classified as a CYP2D6 normal metabolizer or intermediate metabolizers (NM or IM).
4.6. Statistical Analysis
Descriptive statistics were computed to explore the demographic characteristics, clinical profiles, genotype frequencies of participants. Chi-square test was used to compare the observed and expected genotype frequency according to Hardy–Weinberg equilibrium. The plasma concentrations of tamoxifen and its active metabolites were described as median with interquartile range (IQR). Interpatient variability (IIV) of plasma endoxifen and endoxifen/N-desmethyltamoxifen metabolic ratio (MRE/NDM), a marker for CYP2D6 metabolic activity, was described by coefficient of variation (%COV). ANOVA or independent t-test was used to determine the association of log transformed concentrations of tamoxifen and its metabolites and MRE/NDM across CYP2D6 diplotypes, phenotypes, non CYP2D6 genotypes (CYP2C9, CYP2C19, CYP3A5, POR, or ABCB1c.4036A>G), and other patient specific factors (i.e., age, BMI, menopausal status, etc). Pearson correlation coefficient was determined to assess the magnitude of association between tamoxifen and its metabolites. The impact of CYP2D6 diplotype and phenotype to explain the absolute concentration of endoxifen or MRE/NDM was assessed using linear regression. Adjusted coefficient of determination (adjusted R2) was used to define the proportion of the variance in endoxifen concentration and MRE/NDM that could be predictable from CYP2D6 diplotype and phenotype. The data were analyzed using SPSS for Windows (version 21.0, IBM Corporation, New York, USA). Graphs were prepared using ggplot2 package in R (version 3.5.1) [53].
5. Conclusions
In conclusion, we report a high rate of low plasma endoxifen level and wide between-patient variability in plasma concentration of endoxifen, the main therapeutically active metabolites of tamoxifen. CYP2D6 genotype and its activity score is a significant predictor of plasma endoxifen exposure. Tamoxifen therapy guided by CYP2D6 genotyping in clinical practice may assist treatment decision-making for optimal patient benefit. Therapeutic drug monitoring is also recommended for those with normal metabolizer phenotypes. The marginal effect of POR and ABCB1 on tamoxifen and its metabolites PK found in this study needs further investigation in a larger sample size. Despite the simplified AS system recommended by CPIC for genotype-phenotype translation, our study signals further refinement of the approach, probably considering other sources of variability such as POR and ABCB1. Future studies are also recommended to investigate the impact of the CYP2D6 genetic variations on the clinical outcome of breast cancer in Ethiopian patients.
Acknowledgments
The authors would like to extend their sincere appreciation to Armauer Hansen Research Institute (AHRI) for the financial assistance required for the study.
Author Contributions
Conceptualization, J.H.A., E.M. and E.A.; methodology, J.H.A., E.M. and E.A.; software, J.H.A.; validation, E.A.; formal analysis, J.H.A. and E.A.; investigation, J.H.A. and E.A.; resources, A.F., A.A., R.H. and E.A.; data curation, J.H.A.; writing—original draft preparation, J.H.A.; writing—review and editing, J.H.A., E.M., A.F., A.A., R.H. and E.A.; visualization, J.H.A.; supervision, E.M., A.A., R.H. and E.A.; project administration, A.A., R.H. and E.A.; funding acquisition, J.H.A., A.A. and R.H.
Funding
This research was supported by Armauer Hansen Research Institute (AHRI) through the BSPP Program, a grant obtained from Sida-Ethiopia Bilateral Program.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Saladores P.H., Precht J.C., Schroth W., Brauch H., Schwab M. Impact of metabolizing enzymes on drug response of endocrine therapy in breast cancer. Expert Rev. Mol. Diagn. 2013;13:349–365. doi: 10.1586/erm.13.26. [DOI] [PubMed] [Google Scholar]
- 2.EBCTCG Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 3.Saladores P., Mürdter T., Eccles D., Chowbay B., Zgheib N.K., Winter S., Ganchev B., Eccles B., Gerty S., Tfayli A., et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharm. J. 2015;15:84–94. doi: 10.1038/tpj.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Vries Schultink A.H.M., Zwart W., Linn S.C., Beijnen J.H., Huitema A.D.R. Effects of Pharmacogenetics on the Pharmacokinetics and Pharmacodynamics of Tamoxifen. Clin. Pharm. 2015;54:797–810. doi: 10.1007/s40262-015-0273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein D.J., Thorn C.F., Desta Z., Flockhart D.A., Altman R.B., Klein T.E. PharmGKB summary: Tamoxifen pathway, pharmacokinetics. Pharm. Genom. 2013;23:643–647. doi: 10.1097/FPC.0b013e3283656bc1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Y., Desta Z., Stearns V., Ward B., Ho H., Lee K.-H., Skaar T., Storniolo A.M., Li L., Araba A., et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J. Natl. Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 7.Schroth W., Winter S., Mürdter T., Schaeffeler E., Eccles D., Eccles B., Chowbay B., Khor C.C., Tfayli A., Zgheib N.K., et al. Improved Prediction of Endoxifen Metabolism by CYP2D6 Genotype in Breast Cancer Patients Treated with Tamoxifen. Front. Pharmacol. 2017;8:582. doi: 10.3389/fphar.2017.00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puszkiel A., Arellano C., Vachoux C., Evrard A., Le Morvan V., Boyer J.-C., Robert J., Delmas C., Dalenc F., Debled M., et al. Factors Affecting Tamoxifen Metabolism in Patients With Breast Cancer: Preliminary Results of the French PHACS Study. Clin. Pharmacol. Ther. 2019;106:585–595. doi: 10.1002/cpt.1404. [DOI] [PubMed] [Google Scholar]
- 9.Stearns V., Johnson M.D., Rae J.M., Morocho A., Novielli A., Bhargava P., Hayes D.F., Desta Z., Flockhart D.A. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J. Natl. Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 10.Madlensky L., Natarajan L., Tchu S., Pu M., Mortimer J., Flatt S.W., Nikoloff D.M., Hillman G., Fontecha M.R., Lawrence H.J., et al. Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clin. Pharmacol. Ther. 2011;89:718–725. doi: 10.1038/clpt.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim J.S.L., Chen X.A., Singh O., Yap Y.S., Ng R.C.H., Wong N.S., Wong M., Lee E.J.D., Chowbay B. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br. J. Clin. Pharmacol. 2011;71:737–750. doi: 10.1111/j.1365-2125.2011.03905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mürdter T.E., Schroth W., Bacchus-Gerybadze L., Winter S., Heinkele G., Simon W., Fasching P.A., Fehm T., German Tamoxifen and AI Clinicians Group. Eichelbaum M., et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin. Pharmacol. Ther. 2011;89:708–717. doi: 10.1038/clpt.2011.27. [DOI] [PubMed] [Google Scholar]
- 13.Livaudais J.C., Hershman D.L., Habel L., Kushi L., Gomez S.L., Li C.I., Neugut A.I., Fehrenbacher L., Thompson B., Coronado G.D. Racial/ethnic differences in initiation of adjuvant hormonal therapy among women with hormone receptor-positive breast cancer. Breast Cancer Res. Treat. 2012;131:607–617. doi: 10.1007/s10549-011-1762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farias A.J., Du X.L. Ethnic differences in initiation and timing of adjuvant endocrine therapy among older women with hormone receptor-positive breast cancer enrolled in Medicare Part D. Med. Oncol. 2016;33:19. doi: 10.1007/s12032-016-0732-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aklillu E., Persson I., Bertilsson L., Johansson I., Rodrigues F., Ingelman-Sundberg M. Frequent distribution of ultrarapid metabolizers of debrisoquine in an ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J. Pharmacol. Exp. Ther. 1996;278:441–446. [PubMed] [Google Scholar]
- 16.Iusuf D., Teunissen S.F., Wagenaar E., Rosing H., Beijnen J.H., Schinkel A.H. P-glycoprotein (ABCB1) transports the primary active tamoxifen metabolites endoxifen and 4-hydroxytamoxifen and restricts their brain penetration. J. Pharmacol. Exp. Ther. 2011;337:710–717. doi: 10.1124/jpet.110.178301. [DOI] [PubMed] [Google Scholar]
- 17.Wang D., Johnson A.D., Papp A.C., Kroetz D.L., Sadée W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharm. Genom. 2005;15:693–704. doi: 10.1097/01.fpc.0000178311.02878.83. [DOI] [PubMed] [Google Scholar]
- 18.Leschziner G.D., Andrew T., Pirmohamed M., Johnson M.R. ABCB1 genotype and PGP expression, function and therapeutic drug response: A critical review and recommendations for future research. Pharm. J. 2007;7:154–179. doi: 10.1038/sj.tpj.6500413. [DOI] [PubMed] [Google Scholar]
- 19.Tazzite A., Kassogue Y., Diakité B., Jouhadi H., Dehbi H., Benider A., Nadifi S. Association between ABCB1 C3435T polymorphism and breast cancer risk: A Moroccan case-control study and meta-analysis. BMC Genet. 2016;17:126. doi: 10.1186/s12863-016-0434-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amjadi O., Hedayatizadeh-Omran A., Alizadeh-navaei R. Association between MDR1 (C3435T) gene polymorphism and risk of breast cancer: An Iranian case-control study—WCRJ. WCRJ. 2018;5:e1126. [Google Scholar]
- 21.Mukonzo J.K., Okwera A., Nakasujja N., Luzze H., Sebuwufu D., Ogwal-Okeng J., Waako P., Gustafsson L.L., Aklillu E. Influence of efavirenz pharmacokinetics and pharmacogenetics on neuropsychological disorders in Ugandan HIV-positive patients with or without tuberculosis: A prospective cohort study. BMC Infect. Dis. 2013;13:261. doi: 10.1186/1471-2334-13-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukonzo J.K., Röshammar D., Waako P., Andersson M., Fukasawa T., Milani L., Svensson J.O., Ogwal-Okeng J., Gustafsson L.L., Aklillu E. A novel polymorphism in ABCB1 gene, CYP2B6*6 and sex predict single-dose efavirenz population pharmacokinetics in Ugandans. Br. J. Clin. Pharmacol. 2009;68:690–699. doi: 10.1111/j.1365-2125.2009.03516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swart M., Ren Y., Smith P., Dandara C. ABCB1 4036A>G and 1236C>T Polymorphisms Affect Plasma Efavirenz Levels in South African HIV/AIDS Patients. Front. Genet. 2012;3:236. doi: 10.3389/fgene.2012.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ngaimisi E., Habtewold A., Minzi O., Makonnen E., Mugusi S., Amogne W., Yimer G., Riedel K.-D., Janabi M., Aderaye G., et al. Importance of Ethnicity, CYP2B6 and ABCB1 Genotype for Efavirenz Pharmacokinetics and Treatment Outcomes: A Parallel-Group Prospective Cohort Study in Two Sub-Saharan Africa Populations. PLoS ONE. 2013;8:e67946. doi: 10.1371/journal.pone.0067946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elens L., Vandercam B., Yombi J.-C., Lison D., Wallemacq P., Haufroid V. Influence of host genetic factors on efavirenz plasma and intracellular pharmacokinetics in HIV-1-infected patients. Pharmacogenomics. 2010;11:1223–1234. doi: 10.2217/pgs.10.94. [DOI] [PubMed] [Google Scholar]
- 26.Zhou S.-F. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin. Pharm. 2009;48:689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 27.Woo H.I., Lee S.K., Kim J., Kim S.W., Yu J., Bae S.Y., Lee J.E., Nam S.J., Lee S.-Y. Variations in plasma concentrations of tamoxifen metabolites and the effects of genetic polymorphisms on tamoxifen metabolism in Korean patients with breast cancer. Oncotarget. 2017;8:100296–100311. doi: 10.18632/oncotarget.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balkenende E.M.E., Dahhan T., Linn S.C., Jager N.G.L., Beijnen J.H., Goddijn M. A prospective case series of women with estrogen receptor-positive breast cancer: Levels of tamoxifen metabolites in controlled ovarian stimulation with high-dose tamoxifen. Hum. Reprod. 2013;28:953–959. doi: 10.1093/humrep/des445. [DOI] [PubMed] [Google Scholar]
- 29.Fotoohi A.K., Karim H., Lafolie P., Pohanka A., Östervall J., Hatschek T., Vitols S. Pronounced Interindividual But Not Intraindividual Variation in Tamoxifen and Metabolite Levels in Plasma During Adjuvant Treatment of Women With Early Breast Cancer. Ther. Drug Monit. 2016;38:239–245. doi: 10.1097/FTD.0000000000000257. [DOI] [PubMed] [Google Scholar]
- 30.Wang D., Poi M.J., Sun X., Gaedigk A., Leeder J.S., Sadee W. Common CYP2D6 polymorphisms affecting alternative splicing and transcription: Long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum. Mol. Genet. 2014;23:268–278. doi: 10.1093/hmg/ddt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Goetz M.P., Sangkuhl K., Guchelaar H.-J., Schwab M., Province M., Whirl-Carrillo M., Symmans W.F., McLeod H.L., Ratain M.J., Zembutsu H., et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin. Pharmacol. Ther. 2018;103:770–777. doi: 10.1002/cpt.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teft W.A., Gong I.Y., Dingle B., Potvin K., Younus J., Vandenberg T.A., Brackstone M., Perera F.E., Choi Y.-H., Zou G., et al. CYP3A4 and seasonal variation in vitamin D status in addition to CYP2D6 contribute to therapeutic endoxifen level during tamoxifen therapy. Breast Cancer Res. Treat. 2013;139:95–105. doi: 10.1007/s10549-013-2511-4. [DOI] [PubMed] [Google Scholar]
- 34.Aklillu E., Herrlin K., Gustafsson L.L., Bertilsson L., Ingelman-Sundberg M. Evidence for environmental influence on CYP2D6-catalysed debrisoquine hydroxylation as demonstrated by phenotyping and genotyping of Ethiopians living in Ethiopia or in Sweden. Pharmacogenetics. 2002;12:375–383. doi: 10.1097/00008571-200207000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Gaedigk A., Dinh J.C., Jeong H., Prasad B., Leeder J.S. Ten Years’ Experience with the CYP2D6 Activity Score: A Perspective on Future Investigations to Improve Clinical Predictions for Precision Therapeutics. J. Pers. Med. 2018;8:15. doi: 10.3390/jpm8020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koppel N., Rekdal V.M., Balskus E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science. 2017:356. doi: 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Vries E.M., Lammers L.A., Achterbergh R., Klümpen H.-J., Mathot R.A.A., Boelen A., Romijn J.A. Fasting-Induced Changes in Hepatic P450 Mediated Drug Metabolism Are Largely Independent of the Constitutive Androstane Receptor CAR. PLoS ONE. 2016;11:e0159552. doi: 10.1371/journal.pone.0159552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hart S.N., Zhong X.-B. P450 oxidoreductase: Genetic polymorphisms and implications for drug metabolism and toxicity. Expert Opin. Drug Metab. Toxicol. 2008;4:439–452. doi: 10.1517/17425255.4.4.439. [DOI] [PubMed] [Google Scholar]
- 39.Hart S.N., Wang S., Nakamoto K., Wesselman C., Li Y., Zhong X. Genetic polymorphisms in cytochrome P450 oxidoreductase influence microsomal P450-catalyzed drug metabolism. Pharmacol. Genom. 2008;18:11–24. doi: 10.1097/FPC.0b013e3282f2f121. [DOI] [PubMed] [Google Scholar]
- 40.Sandee D., Morrissey K., Agrawal V., Tam H.K., Kramer M.A., Tracy T.S., Giacomini K.M., Miller W.L. Effects of genetic variants of human P450 oxidoreductase on catalysis by CYP2D6 in vitro. Pharmacol. Genom. 2010;20:677–686. doi: 10.1097/FPC.0b013e32833f4f9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tulsyan S., Mittal R.D., Mittal B. The effect of ABCB1 polymorphisms on the outcome of breast cancer treatment. Pharmgenom. Pers. Med. 2016;9:47–58. doi: 10.2147/PGPM.S86672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teh L.K., Mohamed N.I., Salleh M.Z., Rohaizak M., Shahrun N.S., Saladina J.J., Shia J.K.S., Roslan H., Sood S., Rajoo T.S., et al. The risk of recurrence in breast cancer patients treated with tamoxifen: Polymorphisms of CYP2D6 and ABCB1. AAPS J. 2012;14:52–59. doi: 10.1208/s12248-011-9313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sensorn I., Sukasem C., Sirachainan E., Chamnanphon M., Pasomsub E., Trachu N., Supavilai P., Pinthong D., Wongwaisayawan S. ABCB1 and ABCC2 and the risk of distant metastasis in Thai breast cancer patients treated with tamoxifen. Onco Targets Ther. 2016;9:2121–2129. doi: 10.2147/OTT.S100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romero-Lorca A., Novillo A., Gaibar M., Bandrés F., Fernández-Santander A. Impacts of the Glucuronidase Genotypes UGT1A4, UGT2B7, UGT2B15 and UGT2B17 on Tamoxifen Metabolism in Breast Cancer Patients. PLoS ONE. 2015;10:e0132269. doi: 10.1371/journal.pone.0132269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Court M., Hao Q., Krishnaswamy S., Bekaii-Saab T., Al-Rohaimi A., Von Moltke L. UDPglucuronosyltransferase (UGT) 2B15 pharmacogenetics: UGT2B15 D85Y genotype and gender are major determinants of oxazepam glucuronidation by human liver. J. Pharmacol. Exp. Ther. 2004;310:656–665. doi: 10.1124/jpet.104.067660. [DOI] [PubMed] [Google Scholar]
- 46.He X., Hesse L.M., Hazarika S., Masse G., Harmatz J.S., Greenblatt D.J., Court M.H. Evidence for oxazepam as an in vivo probe of UGT2B15: Oxazepam clearance is reduced by UGT2B15 D85Y polymorphism but unaffected by UGT2B17 deletion. Br. J. Clin. Pharmacol. 2009;68:721–730. doi: 10.1111/j.1365-2125.2009.03519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung J.-Y., Cho J.-Y., Yu K.-S., Kim J.-R., Jung H.-R., Lim K.-S., Jang I.-J., Shin S.-G. Effect of the UGT2B15 genotype on the pharmacokinetics, pharmacodynamics, and drug interactions of intravenous lorazepam in healthy volunteers. Clin. Pharmacol. Ther. 2005;77:486–494. doi: 10.1016/j.clpt.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Lee L., Cheung W.Y., Atkinson E., Krzyzanowska M.K. Impact of comorbidity on chemotherapy use and outcomes in solid tumors: A systematic review. J. Clin. Oncol. 2011;29:106–117. doi: 10.1200/JCO.2010.31.3049. [DOI] [PubMed] [Google Scholar]
- 49.Borges S., Desta Z., Li L., Skaar T.C., Ward B.A., Nguyen A., Jin Y., Storniolo A.M., Nikoloff D.M., Wu L., et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: Implication for optimization of breast cancer treatment. Clin. Pharmacol. Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 50.Powers J.L., Buys S.S., Fletcher D., Melis R., Johnson-Davis K.L., Lyon E., Malmberg E.M., McMillin G.A. Multigene and Drug Interaction Approach for Tamoxifen Metabolite Patterns Reveals Possible Involvement of CYP2C9, CYP2C19, and ABCB1. J. Clin. Pharm. 2016;56:1570–1581. doi: 10.1002/jcph.771. [DOI] [PubMed] [Google Scholar]
- 51.Hansten P.D. The Underrated Risks of Tamoxifen Drug Interactions. Eur. J. Drug Metab. Pharm. 2018;43:495–508. doi: 10.1007/s13318-018-0475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmed J.H., Makonnen E., Yimer G., Seifu D., Bekele A., Assefa M., Aseffa A., Howe R., Fotoohi A., Hassan M., et al. CYP2J2∗7 Genotype Predicts Risk of Chemotherapy-Induced Hematologic Toxicity and Reduced Relative Dose Intensity in Ethiopian Breast Cancer Patients. Front. Pharmacol. 2019;10 doi: 10.3389/fphar.2019.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2018. [Google Scholar]