Abstract
The OTOA gene (Locus: DFNB22) is reported to be one of the causative genes for non-syndromic autosomal recessive hearing loss. The copy number variations (CNVs) identified in this gene are also known to cause hearing loss, but have not been identified in Japanese patients with hearing loss. Furthermore, the clinical features of OTOA-associated hearing loss have not yet been clarified. In this study, we performed CNV analyses of a large Japanese hearing loss cohort, and identified CNVs in 234 of 2262 (10.3%, 234/2262) patients with autosomal recessive hearing loss. Among the identified CNVs, OTOA gene-related CNVs were the second most frequent (0.6%, 14/2262). Among the 14 cases, 2 individuals carried OTOA homozygous deletions, 4 carried heterozygous deletions with single nucleotide variants (SNVs) in another allele. Additionally, 1 individual with homozygous SNVs in the OTOA gene was also identified. Finally, we identified 7 probands with OTOA-associated hearing loss, so that its prevalence in Japanese patients with autosomal recessive hearing loss was calculated to be 0.3% (7/2262). As novel clinical features identified in this study, the audiometric configurations of patients with OTOA-associated hearing loss were found to be mid-frequency. This is the first study focused on the detailed clinical features of hearing loss caused by this gene mutation and/or gene deletion.
Keywords: OTOA, DFNB22, hearing loss, copy number variations
1. Introduction
Hereditary hearing loss affects approximately one in 500–600 infants in developed countries, and genetic causes account for at least 50% of all childhood hearing loss [1]. Approximately 100 genes have been recognized as causative for sensorineural hearing loss (SNHL) [2]. Next-generation sequencing (NGS) analysis has become a powerful tool for finding variants in many rare genes, and has allowed genetic epidemiology to be clarified [3,4]. We have recently reported a series of studies on various relatively rare genes in the Japanese population, including POU4F3 [5], WFS1 [6], OTOF [7], and STRC [8]. The study was performed as one in a series of findings on specific genes that were published based on the same cohort.
In general, most of the causal mutations in these genes are small insertions/deletions (indels) or single nucleotide variants (SNVs). Recently, copy number variations (CNVs), that is, the alteration through deletion, insertion and/or duplication of more than 1 kbp, involving the genes associated with hearing loss have been observed in several patients with hearing loss (HL) [8,9]. Shearer et al. reported that 143 CNVs were identified in 16 of 89 deafness-associated genes from 686 patients, with the greatest number of CNVs identified in the STRC and OTOA genes, comprising 73% and 13% of all identified CNVs, respectively [9].
The OTOA gene (Locus: DNFB22) was first reported as one of the responsible genes for non-syndromic autosomal recessive hearing loss by Zwaenepoel et al. in 2002 [10]. OTOA is located on chromosome 16p12.2, and encodes otoancorin, a protein required for limbal attachment of the tectorial membrane, which is important for conditioning proper stimulation of the inner hair cells [11,12].
To date, 27 different variants [9,10,12,13,14,15,16,17,18,19,20,21,22] and 24 long or whole gene deletions [9,13,15,16,19,20,23,24,25] in the OTOA gene have been reported to cause SNHL in various ethnic groups, mainly in the Middle-Eastern countries. Although previous papers reported on the SNVs, indels, splicing variants, or CNVs, the detailed clinical characteristics of patients with OTOA variants still remain unclear.
In the present study, we aimed to clarify the prevalence and the clinical characteristics of OTOA-associated SNHL by using the NGS platform to identify small variants and CNVs in the OTOA gene, and confirmed their existence via direct sequencing or high-resolution array genomic hybridization (aCGH) analysis.
2. Materials and Methods
2.1. Subjects
This study was undertaken using data from a total of 2262 Japanese autosomal recessive sensorineural hearing loss (ARSNHL) probands (including sporadic cases) registered from 67 otorhinolaryngology departments in Japan between May 2013 and November 2018. The ages of the probands ranged from 0 to 86 years (mean 21.3 years). To participate in this study, written informed consent was obtained from all patients or the family members of the proband. All procedures were approved by the Shinshu University Ethical Committee as well as the respective ethical committees of the other participating institutions. All methods were in accordance with the Shinshu University Ethical Committee for Human Genetic Research guidelines and regulations.
This study was conducted in accordance with the Declaration of Helsinki, with the protocol approved by the Ethics Committee of Shinshu University School of Medicine No. 387-4 September 2012 and No. 576-2 May 2017.
2.2. Short Variant Analysis Including SNVs, Indels, and Splicing Variants
We developed amplicon libraries, using an Ion AmpliSeq™ Custom Panel (ThermoFisher Scientific, Waltham, MA, USA), for 68 genes previously reported as genetic causes of non-syndromic hearing loss (Supplementary Table S1), and performed emulsion PCR and sequencing, in line with the manufacturer’s instructions. The detailed procedures have been described in our published paper [26]. NGS was performed with an Ion Torrent Personal Genome Machine (PGM) system using an Ion PGM 200 Sequencing Kit and an Ion 318 Chip (ThermoFisher Scientific) or Ion Proton™ system using the Ion PI™ HiQ™ Sequencing 200 Kit and Ion PI™ Chip (ThermoFisher Scientific). We mapped the sequence data against the human genome sequence (build GRCh37/hg19) with a Torrent Mapping Alignment Program. After sequence mapping, the DNA variant regions were stacked with Torrent Variant Caller plug-in software. After variant detection, we analyzed their effects using ANNOVAR software [27]. The variants (missense, nonsense, insertion/deletion and splicing variants) affecting the amino acid sequence were selected from among the identified variants. Variants were further selected as less than 1% of (1) the ExAC [28], (2) gnomAD [29], (3) 3.5KJPN [30] databases, and (4) the 333 in-house Japanese normal hearing controls. We employed direct sequencing to confirm the selected variants.
The pathogenicity of a variant was evaluated based on the criteria of the ACMG (American College of Medical Genetics) standards and guidelines [31]. For missense variants, in particular, functional prediction software, including Sorting Intolerant from Tolerant (SIFT) [32], Polymorphism Phenotyping (PolyPhen2) [33], Likelihood Ratio Test (LRT) [34], Mutation Taster [35], Mutation Assessor [36], Rare Exome Variant Ensemble Learner (REVEL) [37], and Combined Annotation Dependent Depletion (CADD) [38] were used on the ANNOVAR software. We also evaluated the conservation of the variant site in 170 vertebrates from HGMD Professional. [39]. Segregation analysis was performed for each proband and family members (if samples were obtained or available) by direct sequencing.
2.3. Copy Number Variations (CNVs) Analysis
We performed a CNV detection method with Ion AmpliSeq sequencing and multiplex PCR-based targeted genome enrichment. The detailed protocol has been described elsewhere [40]. The read depth data was used for copy number analysis. From the results of the CNVs analysis of the 2262 probands, we picked up 14 patients with OTOA gene CNVs.
We designed a custom aCGH for 68 genes previously reported as genetic causes of non-syndromic hearing loss using the Agilent web software (Agilent SureDesign, Agilent Technologies, Santa Clara, CA, USA), with the probes covering specific chromosomal regions of those genes at 150–200 bp intervals as a design-setting on the Agilent 8 × 60 K platform (Agilent Technologies, Santa Clara, CA, USA) [41]. There were 235 probes laid across the OTOA region (chr16:21,740,000–21,772,500). We used the same DNA samples as used for the amplicon resequencing, with quality assessment also performed. Five micrograms of genomic DNA were fragmented, and labeled with cyanine-3 for reference DNA samples and cyanine-5 for subjects, and then hybridized. We performed scanning of the array with a G2600D SureScan Microarray Scanner (Agilent Technologies) according to the manufacturer’s recommended protocols, and analyzed scanned aCGH data using CytoGenomics software version 3.0.6.6 (Agilent Technologies).
2.4. Clinical Evaluations
Clinical information including the age of onset of SNHL, the result of newborn hearing screening (NHS), pedigree, the presence of subjective progression in SNHL, and episodes of vertigo/dizziness were collected from each proband from a review of the medical charts.
Hearing loss was evaluated using pure-tone audiometry and severity of SNHL was classified by a pure-tone average (PTA) over 500, 1000, 2000 and 4000 Hz. If an individual did not respond to the maximum hearing level at a frequency, 5 dB was added to the maximum hearing level. The severity of HL was classified as follows: mild (PTA: 20–40 dB HL), moderate (41–70 dB HL), severe (71–95 dB HL), and profound (>95 dB HL). Audiometric configuration was categorized into low-frequency, mid-frequency (U-shaped), high-frequency (gently or steeply sloping), or flat based on a previous report [42].
3. Results
3.1. Identified OTOA Variants and Their Prevalence in Japanese ARSNHL Patients
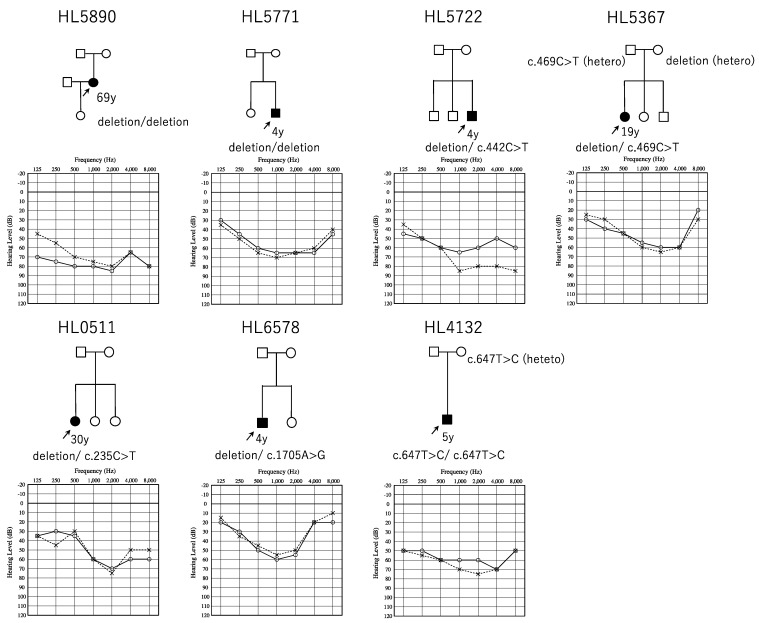
Of 2262 cases, CNVs in the 68 target genes were detected in 234 cases (10.3%, 234/2262). The most frequent gene with CNVs was the STRC gene (8.4%, 190/2262), followed by the OTOA gene (0.6%, 14/2262). Among the 14 cases with OTOA gene CNVs, two carried homozygous deletions, nine carried heterozygous deletions, and three carried three copies (one-copy gain). Among the nine cases with heterozygous deletions in the OTOA gene, four cases have possibly disease-causing small variants of the OTOA gene in the other allele. Additionally, we identified one case with OTOA gene homozygous SNVs. Finally, we identified seven probands with OTOA-associated HL in this study (Table 1). Thus, the prevalence of OTOA-associated HL in Japanese ARSNHL patients was calculated to be 0.3% (7/2262). All were sporadic cases, and there were no affected family members (Figure 1). No candidate pathogenic variants in the other 67 deafness genes were detected in these seven individuals. Unfortunately, we could not obtain un-affected sibling samples as shown in Figure 1. Thus, the segregation analysis was not performed for these families.
Table 1.
Summary of the clinical features and identified variants of individuals with OTOA variants in this study.
| Newborn | Average | Audiometric | Hearing | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hearing | Hearing Level | Age at | Configuration | Loss | Vertigo/ | |||||||
| ID | Zygosity | Allele #1 | Allele #2 | Onset | Screening R/L | R/L (dB) | Audiogram | R/L | Progression | Dizziness | ||
| HL5771 | homo | whole gene deletion | whole gene deletion | 3y | N/A | 58.75/62.5 | 4y | MF/MF | - | |||
| HL5890 | homo | whole gene deletion | whole gene deletion | childhood | N/A | 77.5/72.5 | 69y | Flat/MF | progressive | + | ||
| HL0511 | compound hetero | whole gene deletion | c.235C>T | p.(Arg79Trp) | 7y | N/A | 56.25/55 | 30y | HF/MF | progressive | - | |
| HL5722 | compound hetero | whole gene deletion | c.442C>T | p.(Arg148*) | 0m | refer/refer | 58.75/76.25 | 7y | Flat/HF | - | ||
| HL5367 | compound hetero | whole gene deletion | c.469C>T | p.(Arg157Cys) | 5y | N/A | 55/57.5 | 19y | MF/MF | progressive | - | |
| HL6578 | compound hetero | whole gene deletion | c.1705A>G | p.(Lys569Glu) | 0m | refer/refer | 46.25/42.5 | 4y | MF/MF | - | ||
| HL4132 | homo | c.647T>C | p.(Phe216Ser) | c.647T>C | p.(Phe216Ser) | 0m | refer/refer | 62.5/68.75 | 5y | Flat/MF | - |
All variants were indicated in NM_144672. y: year(s), m: month(s), N/A: not applicable (not received NHS), HF: high-frequency sensorineural hearing loss, MF: mid-frequency sensorineural hearing loss.
Figure 1.
Pedigree and audiograms for each family with OTOA variants. Arrows show the probands in each family. The ages indicated in the pedigree represent the time at which the audiogram was obtained. Genetic findings for each individual tested are also noted in the pedigree.
3.2. Confirmation of CNVs and Short Variants, and The Pathogenic Interpretation of These Variants
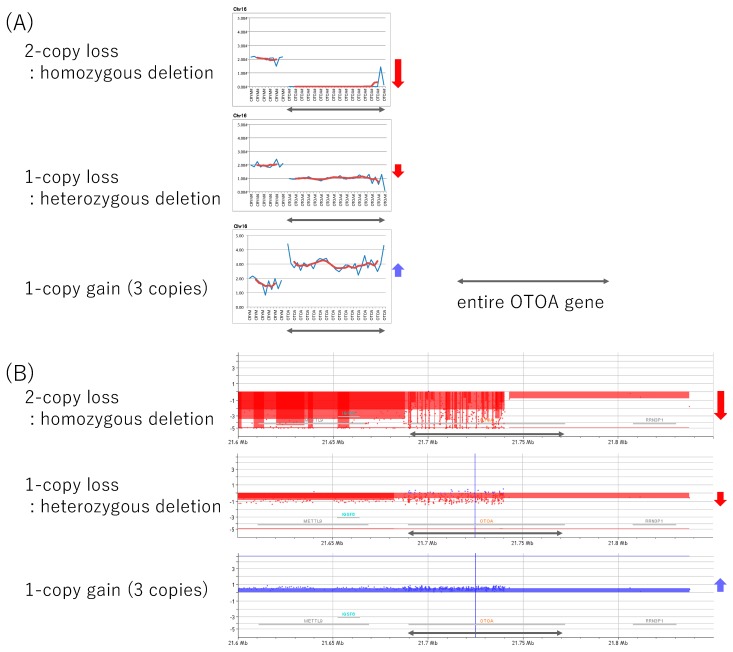
In this study, we detected CNVs by using NGS read data as a first screening step followed by confirmation with aCGH. We performed aCGH analysis to confirm the CNVs for five individuals. Two of them (HL5890 and HL5771) carried homozygous deletions in the OTOA gene and three (HL5722, HL5367, and HL6578) carried heterozygous deletions. All cases had entire OTOA gene deletions, and the aCGH results were consistent with the NGS-based analysis results. Furthermore, deletions in all cases included the METTL9 and IGSF6 genes, which are located upstream of the OTOA gene. Figure 2 shows the results of NGS analysis and aCGH analysis in these cases. We also performed aCGH analysis for a case with three copies as a technical confirmation, and the results were consistent with the NGS analysis results. Therefore, we believe that CNV analysis using NGS data is reliable, even for heterozygous deletions, homozygous deletions, and one-copy gains in the OTOA gene. Unfortunately, the total amount of DNA available for HL0511 was not sufficient for aCGH analysis, so we did not perform aCGH analysis for this patient.
Figure 2.
The results of copy number variation (CNV) analysis. (A) The results of CNV analysis based on next-generation sequencing (NGS) read depth data for patients with two-copy loss (homozygous deletion), one-copy loss (heterozygous deletion), or one-copy gain (three copies) in the OTOA gene identified in the present study. (B) The results of aCGH analysis for the same patients. Black arrows indicate the OTOA region. Red arrows indicate deletions, and blue arrows indicate duplications.
All five single nucleotide variants (c.235C>T, c.442C>T, c.469C>T, c.1705A>G, and c.647T>C) identified in this study were evaluated according to the ACMG standards and guidelines [31]. All variants were novel, and were not observed or observed in very low frequency in the control population database (PM2) (Table 2). One mutation (c.442C>T) was categorized as a “likely pathogenic” variant as this variant is a nonsense variant (p.(Asp148*)) leading to the stop codon (PVS1). Three missense variants (c.235C>T, c.469C>T, and c.1705A>G) detected in trans with a pathogenic (whole gene deletion) variant (PM3) were categorized as being of “uncertain significance”. The remaining missense variant identified as homozygous (c.647T>C) was also categorized as of “uncertain significance”. All four missense variants were predicted as deleterious and have high CADD scores.
Table 2.
Possible causative variant identified in this study.
| Prediction Score | Allele Frequency in Controls | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino | |||||||||||||
| Nucleotide | Acid | PolyPhen | Mut_ | Mut_ | ACMG | ||||||||
| Changes | Change | SIFT * | 2_HVAR * | LRT * | Taster * | Assessor * | REVEL * | Cadd | Exac | Gnomad | 3.5kJPN | Guidelines | |
| c.235C>T | p.(Arg79Trp) | D(0.4) | B(0.166) | N(0.132) | N(0.09) | M(0.552) | 0.21 | 23.6 | 0.00000824 | 0.00000812 | N/A | Uncertain Significance | PM2,PM3 |
| c.442C>T | p.(Arg148*) | - | - | N(0.225) | A(0.81) | - | - | 35 | 0.0000247 | 0.0000163 | N/A | Likely Pathogenic | PVS1, PM2 |
| c.469C>T | p.(Arg157Cys) | D(0.912) | D(0.916) | D(0.629) | D(0.548) | M(0.752) | 0.285 | 34 | 0.0000165 | 0.0000203 | N/A | Uncertain Significance | PM2,PM3 |
| c.1705A>G | p.(Lys569Glu) | D(0.427) | D(0.875) | D(0.629) | D(0.441) | M(0.567) | 0.598 | 31 | N/A | N/A | N/A | Uncertain Significance | PM2,PM3 |
| c.647T>C | p.(Phe 216Ser) | D(0.721) | D(0.764) | D(0.629) | D(0.412) | M(0.741) | 0.326 | 24.3 | N/A | N/A | N/A | Uncertain Significance | PM2 |
* The Prediction Score of each algorithm included in the ANNOVAR software was converted from the original scoring system. A score closer to 1.0 indicated the variant was predicted to be more damaging. A, disease causing automatic (Mutation Taster); B, benign (PolyPhen2_HVAR); D, deleterious (SIFT, LRT), probably damaging (PolyPhen2), or disease causing (Mutation Taster); M, medium (Mutation Assessor); N, Neutral (LRT). PVS: evidence of Pathogenicity—Very Strong, PM: evidence of Pathogenicity—Moderate.
3.3. Clinical Features of OTOA-Associated SNHL Patients
Table 1 summarizes the clinical findings of the seven affected individuals identified in this study. The age of onset of HL ranged from congenital to childhood. All congenital cases were identified through NHS, but the other childhood onset cases did not receive NHS. Most of the cases have bilateral symmetrical SNHL (Figure 1), and the severity of HL ranged from moderate to severe. Interestingly, most cases showed mid-frequency HL. Based on the audiometric configuration classification criteria previously reported, mid-frequency HL was observed in nine ears, flat type in three ears, and high-frequency HL in two ears. Progression of HL was noticed, based on the medical charts, for three (all adults: HL5890, HL0511, and HL5367) of the seven individuals. Serial audiograms could be obtained from one individual (HL5367), and the averaged hearing threshold (PTA) was observed to have slowly deteriorated from 41.25 dB at 4 years old to 55 dB at 19 years old. Vertigo/dizziness is rare among patients with OTOA-associated HL, and only one individual (HL5890) was found to have episodes of vertigo.
4. Discussion
In our cohort of 2262 Japanese ARSNHL patients, we identified seven probands with OTOA-associated HL, including two cases with homozygous deletions, four cases with heterozygous deletions in trans to a SNVs, and one case with homozygous SNVs. The frequency of OTOA-associated HL in Japanese ARSNHL patients was calculated to be 0.3% (7/2262). In a previous report analyzing a larger number of patients, Shearer et al. identified five probands with OTOA-associated HL among 686 SNHL patients from American probands, so that the frequency of OTOA-associated HL was calculated to be 0.7% among all SNHL patients (5/686) [9]. Sloan-Heggen et al. identified eight probands with OTOA-associated HL among 1119 unrelated SNHL patients from various ethnic populations (0.7%) [16] and also identified six OTOA-associated HL cases among 302 Iranian patients (2.0%) [13]. Our results were comparable with the studies on both the American patients and various ethnic populations, but noticeably lower than that on the Iranian patients. These differences may reflect differences in the ratio of consanguineous patients among each cohort.
To elucidate the prevalence of OTOA CNVs in the normal hearing population, we also performed NGS analysis for 152 normal hearing controls (data not shown). The controls were aged from 20–30 years, and pure-tone audiometry was performed for each control, showing normal hearing. Among the 152 controls, none carried a copy number loss of the OTOA gene, but one case carried three copies of the OTOA gene. It was unclear whether the one-copy gain of the OTOA gene was pathogenic or neutral (no impact on phenotype). However, the identification of a one-copy gain of the OTOA gene from a control case, suggests that this one-copy gain of the OTOA gene was not associated with any phenotypes. Therefore, the CNVs of OTOA were rare in Japanese control population.
For all OTOA gene CNVs identified in this study, the aCGH results showed that the whole OTOA gene as well as whole METTL9 and IGSF6 genes were deleted or duplicated. In the previous three reports analyzing the deletion region in detail [23,24,43], all cases carried a whole OTOA, METTL9 and IGSF6 gene deletion as in this study. One plausible reason for relatively large number of CNVs observed in this area and same types of deletion including OTOA, METTL9, and IGSF6 were observed even in different ethnic population, is the segmental duplications of the region in chromosome 16p12.2. There is a highly homologous sequences before and after chr16p12.2, including the OTOA, METTL9, and IGSF6 genes [23,43,44,45]. Further, these segmental duplication increased mis-homologous recombination in this region, and may act as a hotspot for CNVs. As a result of this mis-homologous recombination, the similar CNVs in this area (including the OTOA, METTL9, and IGSF6 genes) may be commonly observed in many ethnic populations.
The OTOA gene has a pseudogene located 820Kb downstream, which has a high sequence similarity and 99% or more homology in the exon 20–28 region of the OTOA gene [23]. Therefore, the mapping quality of this region was degraded and SNV detection in this region is challenging when using short-read NGS [46]. Except for one variant (c.2960_2961delAT), all variants identified in this study and previous studies were located in exon 3-19 (summarized in Table 3).
Table 3.
Summary of variants identified in this and previous studies (NM_144672).
| Allele Frequency | Prediction Score | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino Acid | GnomAD | Polyphen2 | Mut | Mut | |||||||
| Change | Change | Exon | Exac03 | Exome | Sift * | _Hvar * | LRT * | Taster * | Assessor * | Revel * | CADD | Reference |
| missense/nonsense variant | ||||||||||||
| c.131T>C | p.(Ile44Thr) | 3 | 0.0000494 | 0.0000731 | D | P | D | D | M | N/A | 23.8 | Christina M. Sloan-Heggen, 2016 [16] |
| c.235C>T | p.(Arg79Trp) | 5 | 0.00000824 | 0.00000812 | D(0.4) | B(0.166) | N(0.132) | N(0.09) | M(0.552) | 0.21 | 23.6 | this study |
| c.313A>T | p.(Lys105*) | 6 | N/A | N/A | - | - | - | - | - | - | - | Christina M. Sloan-Heggen, 2016 [16] |
| c.442C>T | p.(Arg148*) | 7 | 0.0000247 | 0.0000163 | - | - | N(0.225) | A(0.81) | - | - | 35 | this study |
| c.446C>A | p.(Ala149Asp) | 7 | 0.000016 | N/A | D | D | N | P | - | - | 28.8 | Shearer, 2014 [9] |
| c.469C>T | p.(Arg157Cys) | 7 | 0.0000165 | 0.0000203 | D(0.912) | D(0.916) | D(0.629) | D(0.548) | M(0.752) | 0.285 | 34 | this study |
| c.647T>C | p.(Phe216Ser) | 8 | N/A | N/A | D(0.721) | D(0.764) | D(0.629) | D(0.412) | M(0.741) | 0.326 | 24.3 | this study |
| c.878A>G | p.(Gln293Arg) | 10 | N/A | N/A | D | P | D | D | M | - | 24.2 | L. He, 2018 [17] |
| c.1025A>T | p.(Asp342Val) | 11 | N/A | N/A | D(0.784) | D(0.719) | N(0.388) | D(0.81) | M(0.552) | 0.453 | 26.7 | Walsh, 2006 [18] |
| c.1249C>T | p.(Leu417Phe) | 12 | 0.0000165 | 0.0000163 | D | P | D | D | M | - | 28.6 | Tsai, 2013 [19] |
| c.1282G>T | p.(Val428Phe) | 12 | N/A | N/A | D | P | N | P | L | - | 24.7 | Cabanillas, 2018 [20] |
| c.1352G>A | p.(Gly451Asp) | 13 | 0.00000824 | 0.00000407 | D(0.912) | D(0.971) | D(0.439) | D(0.524) | M(0.567) | 0.768 | 24.8 | K Lee, 2013 [21] |
| c.1705A>G | p.(Lys569Glu) | 16 | N/A | N/A | D(0.427) | D(0.875) | D(0.629) | D(0.441) | M(0.567) | 0.598 | 31 | this study |
| c.1728T>G | p.(Ile576Met) | 16 | 0.000033 | 0.0000284 | D | P | D | D | M | - | 23.8 | Christina M. Sloan-Heggen, 2016 [16] |
| c.1865T>A | p.(Leu622His) | 17 | 0.000008 | N/A | D | P | D | D | - | - | 29.1 | P Fontana, 2017 [15] |
| c.1807G>T | p.(Val603Phe) | 16 | N/A | 0.00000406 | T | P | N | D | M | - | 26.6 | Ammar-Khodja, 2015 [22]; Christina M. Sloan-Heggen, 2016 [16] |
| c.1814G>C | p.(Cys605Ser) | 17 | N/A | N/A | T | P | D | D | M | - | 26.8 | Christina M. Sloan-Heggen, 2016 [16] |
| c.1879C>T | p.(Pro627Ser) | 17 | 0.000033 | 0.0000366 | D(0.496) | D(0.916) | D(0.629) | D(0.548) | M(0.567) | 0.446 | 31 | K Lee, 2013 [21]; Christina M. Sloan-Heggen, 2015 [13] |
| c.1939G > C | p.(Gly647Arg) | 18 | N/A | 0.0000122 | T(0.363) | P(0.604) | D(0.629) | D(0.478) | M(0.567) | 0.813 | 23.6 | Christina M. Sloan-Heggen, 2015 [13] |
| c.2201A>G | p.(Gln734Arg) | 19 | 0.00000824 | 0.00000407 | T(0.330) | B(0.339) | N(0.229) | D(0.330) | M(0.723) | 0.079 | 8.163 | Christina M. Sloan-Heggen, 2015 [13] |
| splicing variant | ||||||||||||
| c.151+1G>A | N/A | N/A | - | - | - | D(0.81) | - | - | 26.3 | Christina M. Sloan-Heggen, 2015 [13] | ||
| c.1320+2T>C | N/A | N/A | - | - | - | D(0.81) | - | - | 24.2 | Zwaenepoel, 2002 [10] | ||
| c.1320+5G>C | N/A | 0.00001 | - | - | - | D | - | - | 21.7 | Bong Jik Kim, 2019 [12] | ||
| c.2208−1G>A | 0.000036 | N/A | - | - | - | D(0.81) | - | - | 22.4 | Christina M. Sloan-Heggen, 2015 [13] | ||
| small deletion | ||||||||||||
| c.827delT | p.(Ile276fs) | 9 | 0.000025 | N/A | - | - | - | N/A | - | - | 35 | Shearer, 2014 [9]; Christina M. Sloan-Heggen, 2016 [16]; Sommen, 2016 [14] |
| c.1765delC | p.(Gln589fs) | 17 | 0.000025 | N/A | - | - | - | D | - | - | 28.5 | Bong Jik Kim, 2019 [12] |
| c.2960_2961delAT | p.(His987fs) | 25 | 0.000094 | N/A | - | - | - | N/A | - | - | 25.3 | Sommen, 2016 [14] |
All variants were indicated in NM_144672. * The Prediction Score of each algorithm included in the ANNOVAR software was converted from the original scoring system. A score closer to 1.0 indicated the mutation was more damaging, and that closer to 0 indicated it was more tolerant. A disease causing automatic (Mutation Taster); B, benign (PolyPhen2); D, deleterious (SIFT, LRT), probably damaging (PolyPhen2), or disease causing (Mutation Taster); L, low (Mutation Assessor); M, medium (Mutation Assessor); N, Neutral (LRT), polymorphism (Mutation Taster); P, possibly damaging (PolyPhen2), polymorphism automatic (Mutation Taster); T, Tolerated (SIFT).
In this study, we identified nine cases with one-copy loss of the OTOA gene. Among these nine cases, four cases carried one-copy loss of the OTOA gene with candidate SNVs in the trans allele; however, five cases carried only one-copy loss of the OTOA gene. Shearer et al. also reported five cases among 686 cases that carried one-copy loss of the OTOA gene without any other SNVs in the OTOA gene [4]. Among these cases, there might be some cases with SNVs in the exon 20–28 region that cause OTOA-associated HL. To confirm these cases, newer technologies such as long-read NGS are required.
In this study, we used aCGH to confirm the CNVs identified from NGS results. Array CGH has been the gold standard for copy number analysis, but it is time-consuming and costly. Thus, now we employ NGS as the standard CNVs analysis method as it is possible to detect the SNVs and CNVs in one experiment [15]. In addition, we are currently trying to establish a social health insurance-based platform using NGS as standard CNV detection method as it is possible to detect SNVs and CNVs at the same time and it is more cost- and time-effective.
The severity of the OTOA-associated HL varied from moderate to severe, but most of the cases showed moderate HL (86%, 6/7 individuals) in this study. Also in previous reports, the severity of HL varied significantly from mild to profound (summarized in Table 4). Even in cases of homozygous OTOA gene deletions, significant differences were observed in the severity of HL. These differences in the severity of HL may be due to other environmental or genetic factors including aging. The progress of HL in patients with OTOA-associated HL has not been specifically described in previous reports. In the present study, three adult cases noticed progression of HL, and the progression was confirmed by serial audiograms in one patient in whom the averaged hearing threshold (PTA) was slowly deteriorated from 41.25 dB at 4 years old to 55 dB at 19 years old. From these observations, progressive HL appears to be a common trend in OTOA-associated HL. With regard to the age of onset, three cases showed congenital HL and others showed prelingual to childhood onset in this study. In previous reports, the age of onset was pre-childhood in most cases, but two cases of adult onset were reported [9,16]. All three cases with congenital HL identified in this study were identified through NHS screening. Thus, we estimated that most cases of OTOA-associated HL may be congenital and could be identified through NHS screening. However, in cases not undergoing such screening, the HL was mild to moderate and progressed slowly, and was identified in childhood or later.
Table 4.
Summary of clinical features associated with OTOA variants from this and previous studies.
| Hereditary | Onset | Average hearing level | Zygosity | Allele #1 | Allele #2 | Reference | ||
|---|---|---|---|---|---|---|---|---|
| AR/Spo | 3y | moderate | homo | whole gene deletion | whole gene deletion | this study | ||
| AR/Spo | childhood | severe | homo | whole gene deletion | whole gene deletion | this study | ||
| AR | prelingual | N/A | homo | whole gene deletion | whole gene deletion | Shahin, 2010 [23] | ||
| AR | N/A | mild to moderate | homo | whole gene deletion | whole gene deletion | Bademci, 2014 [24] | ||
| AR | 0−10y | moderate to severe | homo | Whole gene deletion | whole gene deletion | Shearer, 2014 [9] | ||
| N/A | 21−30y | N/A | homo | whole gene deletion | whole gene deletion | Shearer, 2014 [9] | ||
| AR | prelingual | moderate to severe | homo | whole gene deletion | whole gene deletion | Christina M. Sloan-Heggen, 2015 [13] | ||
| N/A | N/A | N/A | homo | whole gene deletion | whole gene deletion | Christina M. Sloan-Heggen, 2016 [16] | ||
| AD | adult | severe to profound | homo | whole gene deletion | whole gene deletion | Christina M. Sloan-Heggen, 2016 [16] | ||
| Spo | congenital | severe to profound | homo | whole gene deletion | whole gene deletion | Christina M. Sloan-Heggen, 2016 [16] | ||
| AR | 1−13y | severe | homo | 58000bp deletion | 58000bp deletion | Alkowari, 2017 [25] | ||
| AR | prelingual | severe | homo | c.151+1G>A | c.151+1G>A | Christina M. Sloan-Heggen, 2015 [13] | ||
| AR/Spo | 0m | moderate | homo | c.647T>C | p.(Phe216Ser) | c.647T>C | p.(Phe216Ser) | this study |
| AR | prelingual | moderate to severe | homo | c.1025A>T | p.(Asp342val) | c.1025A>T | p.(Asp342val) | Walsh, 2006 [18] |
| AR | prelingual | moderate to severe | homo | c1320+2T>C | c.1320+2T>C | Zwaenepoel, 2002 [10] | ||
| AR | prelingual | severe | homo | c.1352G>A | p.(Gly451Asp) | c.1352G>A | p.(Gly451Asp) | K Lee, 2013 [21] |
| AR | prelingual | severe to profound | homo | c.1807G>T | p.(Val603Phe) | c.1807G>T | p.(Val603Phe) | Ammar-Khodja, 2015 [22] |
| AR | prelingual | severe | homo | c.1879C>T | p.(Pro627Ser) | c.1879C>T | p.(Pro627Ser) | K Lee, 2013 [21] |
| AR | prelingual | moderate to severe | homo | c.1879C>T | p.(Pro627Ser) | c.1879C>T | p.(Pro627Ser) | Christina M. Sloan-Heggen, 2015 [13] |
| AR | prelingual | moderate to severe | homo | c.1939G C | p.(Gly647Arg) | c.1939G>C | p.(Gly647Arg) | Christina M. Sloan-Heggen, 2015 [13] |
| AR | prelingual | moderately severe to profound | homo | c.2201A>G | p.(Gln734Arg) | c.2201A>G | p.(Gln734Arg) | Christina M. Sloan-Heggen, 2015 [13] |
| AR/Spo | 7y | moderate | compound hetero | whole gene deletion | c.235C>T | p.(Arg79Trp) | this study | |
| N/A | 0−10y | N/A | compound hetero | whole gene deletion | c.446C>A | p.(Ala149Asp) | Shearer, 2014 [9] | |
| AR/Spo | 5y | moderate | compound hetero | whole gene deletion | c.469C>T | p.(Arg157Cys) | this study | |
| N/A | 0−10y | N/A | compound hetero | whole gene deletion | c.827delT | p.(Ile276fs) | Shearer, 2014 [9] | |
| Spo | congenital | N/A | compound hetero | whole gene deletion | c.827delT | p.(Ile276fs) | Christina M. Sloan-Heggen, 2016 [16] | |
| AR | childhood | N/A | compound hetero | whole gene deletion | c.1282G>T | p.(Val428Phe) | Cabanillas, 2018 [20] | |
| AD | congenital | N/A | compound hetero | whole gene deletion | c.1728T>G | p.(Ile576Met) | Christina M. Sloan-Heggen, 2016 [16] | |
| AR/Spo | congenital | moderate | compound hetero | whole gene deletion | c.1705A>G | p.(Lys569Glu) | this study | |
| Spo | childhood | severe to profound | compound hetero | whole gene deletion | c.1807G>T | p.(Val603Phe) | Christina M. Sloan-Heggen, 2016 [16] | |
| Spo | congenital | mild to moderate | compound hetero | whole gene deletion | c.1814G>C | p.(Cys605Ser) | Christina M. Sloan-Heggen, 2016 [16] | |
| AR | prelingual | severe | compound hetero | whole gene deletion | c.1865T>A | p.(Leu622His) | P Fontana, 2017 [15] | |
| N/A | N/A | N/A | compound hetero | multi exon deletion | c.1249C>T | p.(Leu417Phe) | Tsai, 2013 [19]t | |
| AR/Spo | 0m | moderate | compound hetero | deletion | c.442C>T | p.(Arg148*) | this study | |
| AR | prelingual | N/A | compound hetero | deletion | c.2960_2961delAT | p.His987fs | Sommen, 2016 [14] | |
| Spo | before 6 years | moderate | compound hetero | micro deletion | c.878A>G | p.(Gln293Arg) | L. He, 2018 [17] | |
| Spo | congenital | mild to moderate | compound hetero | c.131T>C | p.(Ile44Thr) | c.313A>T | p.(Lys105*) | Christina M. Sloan-Heggen, 2016 [16] |
| AR | prelingual | N/A | compound hetero | c.827delT | p.(Ile276fs) | c.2960_2961delAT | p.(His987fs) | Sommen, 2016 [14] |
| AR | congenital | moderate | compound hetero | c.1320+5G>C | c.1765delC | p.(Gln589fs) | Bong Jik Kim, 2019 [12] |
AD: autosomal dominant. AR: autosomal recessive. Spo: sporadic. N/A: not available. y: year(s), m: month(s).
It is noteworthy that mid-frequency HL was most commonly observed in individuals with OTOA variants in this study. In addition, flat HL and high-frequency HL were also observed in some cases. In previous reports, only Alkowari et al. have provided detailed audiograms of the three cases from one family with homozygous OTOA deletions, and the audiometric configurations of these patients were mid-frequency HL [25]. Interestingly, otoancorin, encoded by the OTOA gene, is a protein that acts as a glycosylphosphatidylinositol (GPI) anchorage, and is important for limbal attachment of the tectorial membrane (TM) [11,12]. The TECTA gene (Locus: DFNA8/12) encoding α-tectorin, a major non-collagenous glycoprotein of TM, which is expressed in the spiral limbus during TM development [10,11], is also known as a genetic cause of mid-frequency HL [47,48,49]. The similarities between the clinical characteristics of HL in patients with OTOA and TECTA gene mutations reflect the mechanism of deafness caused by TM impairment. The results of this study will be useful for the selection of more appropriate treatment for patients as well as the further understanding of the disease-causing mechanisms of OTOA-associated HL.
5. Conclusions
Here, we presented the detailed clinical characteristics of the seven patients with OTOA-associated HL identified from 2262 unrelated Japanese ARNSHL patients. The prevalence of OTOA-associated HL in Japanese ARSNHL patients was calculated to be 0.3%. This is the first report of HL caused by this gene mutation in Japanese patients with HL. The remarkable clinical characteristics of the patients with OTOA variants was congenital or early onset, progressive, mid-frequency HL.
Acknowledgments
We thank the participants of the Deafness Gene Study Consortium [4] for providing samples and clinical information. We also thank Sachiko Matsuda and Fumiko Tomioka for their technical assistance with this research.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/9/715/s1, Table S1: 68 deafness-causative genes.
Author Contributions
Conceptualization, S.N. and S.U.; methodology, S.N.; software, S.N.; validation, K.S., T.K., T.Y., K.W., H.M., and T.K.; formal analysis, K.S. and S.N.; investigation, S.U.; resources, S.A., A.O., H.M., R.M., M.T., and Y.K.; data curation, K.S., H.M., and S.N.; writing—original draft preparation, K.S. and S.K.; writing—review and editing, K.S., S.K., and S.N.; visualization, K.S.; supervision, S.U.; project administration, S.U.; funding acquisition, S.U.
Funding
This study was funded by the Health and Labor Sciences Research Grant for Research on Rare and Intractable diseases and Comprehensive Research on Disability Health and Welfare from the Ministry of Health, Labor and Welfare of Japan (S.U. H29-Nanchitou(Nan)-Ippan-031), the Grant-in-Aid from Japan Agency for Medical Research and Development (AMED) (S.U. 16kk0205010h0001, 18ek0109363h0001), Grant-in-Aid for Young Scientists (T.K. 19K18765) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (S.U. 15H02565).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Data Availability Statement
The sequencing data are available in the DDBJ databank of Japan (Accession number: JGAS00000000200).
References
- 1.Morton C.C., Nance W.E. Newborn Hearing Screening—A Silent Revolution. N. Engl. J. Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Hereditary Hearing Loss Homepage. [(accessed on 13 May 2019)]; Available online: https://hereditaryhearingloss.org.
- 3.Miyagawa M., Naito T., Nishio S.Y., Kamatani N., Usami S.I. Targeted Exon Sequencing Successfully Discovers Rare Causative Genes and Clarifies the Molecular Epidemiology of Japanese Deafness Patients. PLoS ONE. 2013;8:e71381. doi: 10.1371/journal.pone.0071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishio S.Y., Usami S.I. Deafness Gene Variations in a 1120 Nonsyndromic Hearing Loss Cohort: Molecular Epidemiology and Deafness Mutation Spectrum of Patients in Japan. Ann. Otol. Rhinol. Laryngol. 2015;124:49S–60S. doi: 10.1177/0003489415575059. [DOI] [PubMed] [Google Scholar]
- 5.Kitano T., Miyagawa M., Nishio S.-Y., Moteki H., Oda K., Ohyama K., Miyazaki H., Hidaka H., Nakamura K.-I., Murata T., et al. POU4F3 mutation screening in Japanese hearing loss patients: Massively parallel DNA sequencing-based analysis identified novel variants associated with autosomal dominant hearing loss. PLoS ONE. 2017;12:e0177636. doi: 10.1371/journal.pone.0177636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi M., Miyagawa M., Nishio S.Y., Moteki H., Fujikawa T., Ohyama K., Sakaguchi H., Miyanohara I., Sugaya A., Naito Y., et al. WFS1 mutation screening in a large series of Japanese hearing loss patients: Massively parallel DNA sequencing-based analysis. PLoS ONE. 2018;13:e0193359. doi: 10.1371/journal.pone.0193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasa Y.I., Nishio S.Y., Sugaya A., Kataoka Y., Kanda Y., Taniguchi M., Nagai K., Naito Y., Ikezono T., Horie R., et al. OTOF mutation analysis with massively parallel DNA sequencing in 2265 Japanese sensorineural hearing loss patients. PLoS ONE. 2019;14:e0215932. doi: 10.1371/journal.pone.0215932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokota Y., Moteki H., Nishio S.-Y., Yamaguchi T., Wakui K., Kobayashi Y., Ohyama K., Miyazaki H., Matsuoka R., Abe S., et al. Frequency and clinical features of hearing loss caused by STRC deletions. Sci. Rep. 2019;9:4408. doi: 10.1038/s41598-019-40586-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shearer A.E., Kolbe D.L., Azaiez H., Sloan C.M., Frees K.L., Weaver A.E., Clark E.T., Nishimura C.J., Black-Ziegelbein E.A., Smith R.J.H. Copy number variants are a common cause of non-syndromic hearing loss. Genome Med. 2014;6:37. doi: 10.1186/gm554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zwaenepoel I., Mustapha M., Leibovici M., Verpy E., Goodyear R., Liu X.Z., Nouaille S., Nance W.E., Kanaan M., Avraham K.B., et al. Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc. Natl. Acad. Sci. USA. 2002;99:6240–6245. doi: 10.1073/pnas.082515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukashkin A.N., Legan P.K., Weddell T.D., Lukashkina V.A., Goodyear R.J., Welstead L.J., Petit C., Russell I.J., Richardson G.P. A mouse model for human deafness DFNB22 reveals that hearing impairment is due to a loss of inner hair cell stimulation. Proc. Natl. Acad. Sci. USA. 2012;109:19351–19356. doi: 10.1073/pnas.1210159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim B.J., Kim D.-K., Han J.H., Oh J., Kim A.R., Lee C., Kim N.K., Park H.-R., Kim M.Y., Lee S., et al. Clarification of glycosylphosphatidylinositol anchorage of OTOANCORIN and human OTOA variants associated with deafness. Hum. Mutat. 2019;40:525–531. doi: 10.1002/humu.23719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sloan-Heggen C.M., Babanejad M., Beheshtian M., Simpson A.C., Booth K.T., Ardalani F., Frees K.L., Mohseni M., Mozafari R., Mehrjoo Z., et al. Characterising the spectrum of autosomal recessive hereditary hearing loss in Iran. J. Med. Genet. 2015;52:823–829. doi: 10.1136/jmedgenet-2015-103389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommen M., Schrauwen I., Vandeweyer G., Boeckx N., Corneveaux J.J., Ende J.V.D., Boudewyns A., De Leenheer E., Janssens S., Claes K., et al. DNA Diagnostics of Hereditary Hearing Loss: A Targeted Resequencing Approach Combined with a Mutation Classification System. Hum. Mutat. 2016;37:812–819. doi: 10.1002/humu.22999. [DOI] [PubMed] [Google Scholar]
- 15.Fontana P., Morgutti M., Pecile V., Lenarduzzi S., Cappellani S., Falco M., Scarano F., Lonardo F. A novel OTOA mutation in an Italian family with hearing loss. Gene Rep. 2017;9:111–114. doi: 10.1016/j.genrep.2017.10.002. [DOI] [Google Scholar]
- 16.Sloan-Heggen C.M., Bierer A.O., Shearer A.E., Kolbe D.L., Nishimura C.J., Frees K.L., Ephraim S.S., Shibata S.B., Booth K.T., Campbell C.A., et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Qual. Life Res. 2016;135:441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He L., Pang X., Liu H., Chai Y., Wu H., Yang T. Targeted next-generation sequencing and parental genotyping in sporadic Chinese Han deaf patients. Clin. Genet. 2018;93:899–904. doi: 10.1111/cge.13182. [DOI] [PubMed] [Google Scholar]
- 18.Walsh T., Abu Rayan A., Abu Sa’Ed J., Shahin H., Shepshelovich J., Lee M.K., Hirschberg K., Tekin M., Salhab W., Avraham K.B., et al. Genomic analysis of a heterogeneous Mendelian phenotype: Multiple novel alleles for inherited hearing loss in the Palestinian population. Hum. Genom. 2006;2:203–211. doi: 10.1186/1479-7364-2-4-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai E.A., Berman M.A., Conlin L.K., Rehm H.L., Francey L.J., Deardorff M.A., Holst J., Kaur M., Gallant E., Clark D.M., et al. PECONPI: A Novel Software for Uncovering Pathogenic Copy Number Variations in Non-Syndromic Sensorineural Hearing Loss and Other Genetically Heterogeneous Disorders. Am. J. Med. Genet. Part A. 2013;161:2134–2147. doi: 10.1002/ajmg.a.36038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabanillas R., Diñeiro M., Cifuentes G.A., Castillo D., Pruneda P.C., Álvarez R., Sánchez-Durán N., Capín R., Plasencia A., Viejo-Díaz M., et al. Comprehensive genomic diagnosis of non-syndromic and syndromic hereditary hearing loss in Spanish patients. BMC Med. Genomics. 2018;11:58. doi: 10.1186/s12920-018-0375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee K., Chiu I., Santos-Cortez R., Basit S., Khan S., Azeem Z., Andrade P., Kim S., Ahmad W., Leal S. Novel OTOA mutations cause autosomal recessive non-syndromic hearing impairment in Pakistani families. Clin. Genet. 2013;84:294. doi: 10.1111/cge.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ammar-Khodja F., Bonnet C., Dahmani M., Ouhab S., Lefevre G.M., Ibrahim H., Hardelin J.-P., Weil D., Louha M., Petit C. Diversity of the causal genes in hearing impaired Algerian individuals identified by whole exome sequencing. Mol. Genet. Genom. Med. 2015;3:189–196. doi: 10.1002/mgg3.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahin H., Walsh T., Rayyan A.A., Lee M.K., Higgins J., Dickel D., Lewis K., Thompson J., Baker C., Nord A.S., et al. Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur. J. Hum. Genet. 2010;18:407. doi: 10.1038/ejhg.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bademci G., Diaz-Horta O., Guo S., Duman D., Van Booven D., Foster J., II, Cengiz F.B., Blanton S., Tekin M. Identification of Copy Number Variants Through Whole-Exome Sequencing in Autosomal Recessive Nonsyndromic Hearing Loss. Genet. Test. Mol. Biomarkers. 2014;18:658–661. doi: 10.1089/gtmb.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alkowari M.K., Vozzi D., Bhagat S., Krishnamoorthy N., Morgan A., Hayder Y., Logendra B., Najjar N., Gandin I., Gasparini P., et al. Targeted sequencing identifies novel variants involved in autosomal recessive hereditary hearing loss in Qatari families. Mutat. Res. 2017 doi: 10.1016/j.mrfmmm.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Miyagawa M., Nishio S.Y., Ikeda T., Fukushima K., Usami S.I. Massively Parallel DNA Sequencing Successfully Identifies New Causative Mutations in Deafness Genes in Patients with Cochlear Implantation and EAS. PLoS ONE. 2013;8:e75793. doi: 10.1371/journal.pone.0075793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Exome Aggregation Consortium Database (ExAC) [(accessed on 13 May 2019)]; Available online: http://exac.broadinstitute.org/
- 29.The Genome Aggregation Database (gnomAD) [(accessed on 13 May 2019)]; Available online: https://gnomad.broadinstitute.org/
- 30.Integrative Japanese Genome Variation Database (3.5KJPN) [(accessed on 13 May 2019)]; Available online: https://ijgvd.megabank.tohoku.ac.jp/statistics/statistics-3.5kjpn-all.
- 31.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 33.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 36.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The Human Gene Mutation Database professional (HGMD) [(accessed on 13 May 2019)]; Available online: http://www.hgmd.cf.ac.uk/
- 40.Nishio S.Y., Moteki H., Usami S.I. Simple and efficient germline copy number variant visualization method for the Ion AmpliSeqTM custom panel. Mol. Genet. Genomic Med. 2018;6:678–686. doi: 10.1002/mgg3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moteki H., Azaiez H., Sloan-Heggen C.M., Booth K., Nishio S.-Y., Wakui K., Yamaguchi T., Kolbe D.L., Iwasa Y.-I., Shearer A.E., et al. Detection and confirmation of deafness-causing copy number variations in the STRC gene by massively parallel sequencing and comparative genomic hybridization. Ann. Otol. Rhinol. Laryngol. 2016;125:918–923. doi: 10.1177/0003489416661345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazzoli M.G.V.C., Van Camp G.U.Y., Newton V., Giarbini N., Declau F., Parving A. Recommendations for the Description of Genetic and Audiological Data for Families with Nonsyndromic Hereditary Hearing Impairment. Audiol. Med. 2003;1:148–150. [Google Scholar]
- 43.Tassano E., Ronchetto P., Calcagno A., Fiorio P., Gimelli G., Capra V., Scala M. ‘Distal 16p12.2 microdeletion’ in a patient with autosomal recessive deafness-22. J. Genet. 2019;98:56. doi: 10.1007/s12041-019-1107-0. [DOI] [PubMed] [Google Scholar]
- 44.Itsara A., Cooper G.M., Baker C., Girirajan S., Li J., Absher D., Krauss R.M., Myers R.M., Ridker P.M., Chasman D.I., et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 2008;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin J., Han C., Gordon L.A., Terry A., Prabhakar S., She X., Xie G., Hellsten U., Chan Y.M., Altherr M., et al. The sequence and analysis of duplication-rich human chromosome 16. Nature. 2004;432:988–994. doi: 10.1038/nature03187. [DOI] [PubMed] [Google Scholar]
- 46.Mandelker D., Amr S.S., Pugh T., Gowrisankar S., Shakhbatyan R., Duffy E., Bowser M., Harrison B., Lafferty K., Mahanta L., et al. Comprehensive diagnostic testing for stereocilin: An approach for analyzing medically important genes with high homology. J. Mol. Diagnostics. 2014;16:639–647. doi: 10.1016/j.jmoldx.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Moteki H., Nishio S.-Y., Hashimoto S., Takumi Y., Iwasaki S., Takeichi N., Fukuda S., Usami S.-I. TECTA mutations in Japanese with mid-frequency hearing loss affected by zona pellucida domain protein secretion. J. Hum. Genet. 2012;57:587–592. doi: 10.1038/jhg.2012.73. [DOI] [PubMed] [Google Scholar]
- 48.De Heer A.R., Pauw R.J., Huygen P.L.M., Collin R.W.J., Kremer H., Cremers C.W.R.J. Flat threshold and mid-frequency hearing impairment in a dutch DFNA8/12 family with a novel mutation in TECTA: Some evidence for protection of the inner ear. Audiol. Neurotol. 2009;14:153–162. doi: 10.1159/000171477. [DOI] [PubMed] [Google Scholar]
- 49.Moreno-Pelayo M.A., Del Castillo I., Villamar M., Romero L., Hernández-Calvín F.J., Herraiz C., Barberá R., Navas C., Moreno F. A cysteine substitution in the zona pellucida domain of α-tectorin results in autosomal dominant, postlingual, progressive, mid frequency hearing loss in a Spanish family. J. Med Genet. 2001;38:e13. doi: 10.1136/jmg.38.5.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data are available in the DDBJ databank of Japan (Accession number: JGAS00000000200).