Abstract
WAX INDUCER1/SHINE1 (WIN1) belongs to the AP2/EREBP transcription factor family and plays an important role in wax and cutin accumulation in plants. Here we show that BnWIN1 from Brassica napus (Bn) has dual functions in wax accumulation and oil synthesis. Overexpression (OE) of BnWIN1 led to enhanced wax accumulation and promoted growth without adverse effects on oil synthesis under salt stress conditions. Lipid profiling revealed that BnWIN1-OE plants accumulated more waxes with elevated C29-alkanes, C31-alkanes, C28-alcohol, and C29-alcohol relative to wild type (WT) under salt stress. Moreover, overexpression of BnWIN1 also increased seed oil content under normal growth conditions. BnWIN1 directly bound to the promoter region of genes encoding biotin carboxyl carrier protein 1 (BCCP1), glycerol-3-phosphate acyltransferase 9 (GPAT9), lysophosphatidic acid acyltransferase 5 (LPAT5), and diacylglycerol acyltransferase 2 (DGAT2) involved in the lipid anabolic process. Overexpression of BnWIN1 resulted in upregulated expression of numerous genes involved in de novo fatty acid synthesis, wax accumulation, and oil production. The results suggest that BnWIN1 is a transcriptional activator to regulate the biosynthesis of both extracellular and intracellular lipids.
Keywords: WAX INDUCER1/SHINE1 (WIN1), wax accumulation; salt stress; oil synthesis; transcriptional regulation; Brassica napus
1. Introduction
Brassica napus (B. napus) is the second largest oilseed crop widely grown in the world behind only soybean. It provides high-quality edible oil for human health, renewable industrial feedstock, and bioenergy [1,2]. The production of B. napus suffers frequently environmental stresses during growth seasons. High salinity, featured with increased sodium concentration in cells, is one of the major adverse factors inhibiting plant growth and development [3]. Plants are sessile and have evolved to cope with different stresses via diverse changes including morphological modification and physiological, biochemical, or molecular responses [4]. One of such changes is wax accumulation in terrestrial plants in response to osmotic stress [5,6]. Waxes, located either outside of the cuticle or within the cuticular matrix, function as a hydrophobic barrier in preventing water loss from non-stomatal transpiration in plants and are important for plant tolerance to drought [7,8,9]. Waxes also have sophisticated roles in the protection against pathogen infection, insect attack, and ultraviolet radiation [10,11]. In addition, epicuticular waxes prevent organs from fusion and restrict the deposition of dust and other air pollutants [5,6,12].
Epicuticular waxes are mostly composed of very-long-chain fatty acids (VLCFAs), alkanes, aldehydes, alcohols, ketones, and esters, which are derived from fatty acids [13,14,15]. Fatty acids are synthesized by FA synthase (FAS) complex in plastids [16]. The acetyl-CoA carboxylase (ACCase) complex, consisting of biotin carboxyl carrier protein (BCCP), biotin carboxylase (BC), and carboxyltransferase (CT), uses acetyl-CoA and CO2 to produce malonyl-CoA, the first committed step in de novo FA synthesis [16,17]. After being released from FAS and exported from plastids, free FAs are activated to acyl-coenzyme A (CoA) by long-chain acyl-CoA synthetases (LACSs) for further elongation by the fatty acyl-coA elongase (FAE) complex in the endoplasmic reticulum (ER) to produce VLCFAs, with chain lengths ranging from 20 to 34 carbons [18]. VLCFA derivatives, the major constituents of waxes, are synthesized via two independent pathways using VLCFAs as substrates in ER: the alcohol pathway that produces very-long-chain VLC-fatty primary alcohols and their esters, and the alkane-forming pathway by decarbonylation to generate VLC-alkanes, secondary n-alcohols and ketones [13,14,19,20]. Wax constituents synthesized in the ER are transported to the plasma membrane, exported to the epidermal cell wall, and ultimately loaded in the outer surface of aerial plant tissues [21].
Wax content and composition differ depending on plant species, tissues, environmental stresses, and development stages, which are modulated by multiple metabolic processes [14,22]. Numerous enzymes responsible for wax synthesis have been identified through mutant screening, and mutations in these genes exhibited reduced wax accumulation [7,8,9,23,24,25]. For example, ECERIFERUM3 (CER3) catalyzes the reduction of VLCFA to produce fatty aldehydes, whereas CER1 catalyzes the decarbonylation of fatty aldehydes to n-alkanes. Loss of function in the CER1 and CER3 leads to a substantial decrease in alkanes [25,26]. CER2 plays a role in fatty acid elongation with chain length from C28 to C34 [27,28]. LACS1 and LACS2 are responsible for activation of VLC fatty acids for wax components accumulation. The cuticular wax in Arabidopsis lacs1 and lacs2 mutants was decreased [29]. Diacylgycerol acyltransferase 2 (DGAT2) catalyzes the ester formation between fatty acids and alcohols, and loss of DGAT2 leads to reduced wax accumulation [19]. Arabidopsis CER5 is responsible for wax transport, and mutation in CER5 leads to reduced epicuticular wax and increased intracellular wax content [30,31]. These results suggest that wax accumulation is modulated by diverse regulatory processes.
Due to the complicated networks of wax anabolism, transcription factors may play important roles through regulating coordinately the induction of multiple metabolic pathways [7,32]. WAX INDUCER1/SHINE1 (WIN1/SHN1), containing AP2/ERF domain binding to the GCC (AGCCGCC) box and dehydration responsive element (DRE) of target genes, was first identified to be important for wax accumulation and was involved in drought tolerance in Arabidopsis [7,8]. Subsequently, WIN1-related members in the AP2/ERF family, such as rice OsWR1 and Medicago WXP1, were found to play a positive role in wax accumulation and drought tolerance [33,34]. Arabidopsis WRINKLED4 (WRI4), another AP2/ERF transcription factor, also increases the expression of genes involved in wax accumulation [35]. These results suggest that WIN1 and its related transcription factors are important for extracellular lipids synthesis. However, the role of WIN1 in intracellular lipids remains to be elucidated. Here, we show that BnWIN1 from B. napus has effects on both wax and storage lipid accumulation. Overexpression (OE) of BnWIN1 results in enhanced wax accumulation and increased tolerance to salt stress whereas it enhances oil synthesis under normal growth conditions.
2. Results
2.1. BnWIN1 Is Expressed in Various Tissues in B. Napus
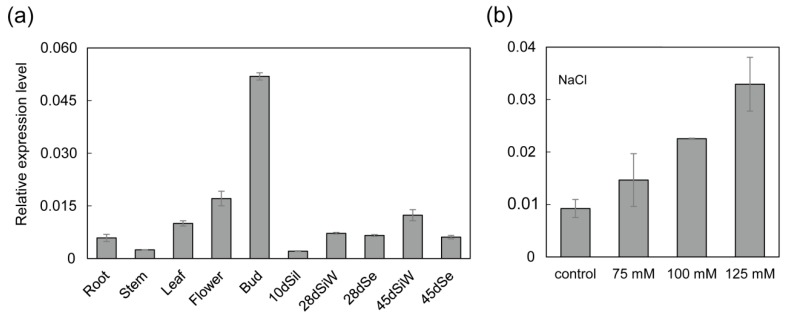
To investigate temporal and spatial distribution of BnWIN1 mRNA in B. napus (Supplementary Figure S1), total RNA was extracted from various tissues of cultivar (cv.) Westar (B. napus) plants, and the expression pattern of BnWIN1 was analyzed by quantitative reverse transcription PCR (qRT-PCR). The BnWIN1 mRNA was detected in various tissues. The BnWIN1 transcript level was highest in flower bud relative to other tissues. The BnWIN1 expression levels in flower, leaf, developing seed, silique wall, and root were higher than in stem and 10-day silique (Figure 1a).
Figure 1.
BnWIN1 is expressed in various tissues and is induced by salt stress in Brassica napus. (a) The expression pattern of BnWIN1 in B. napus detected by qRT-PCR normalized to BnActin. Total RNA was extracted from various tissues of B. napus. Values are means ± SD (n = 3 separated samples). Bud, flower bud; 10dSil, silique at 10 days after flowering; 28dSiW and 45dSiW, silique wall at 28 and 45 days after flowering, respectively; 28dSe and 45dSe, seed at 28 and 45 days after flowering, respectively. (b) The transcript level of BnWIN1 was induced by NaCl stress in B. napus. Total RNA was extracted from 21-day-old plants treated with 0 (control), 75, 100, and 125 mM NaCl for two weeks. Actin was used as an internal standard. Values are mean ± SD (n = 3 separate samples).
2.2. Overexpression of BnWIN1 Enhanced Salt Stress Tolerance
The BnWIN1 transcript level was markedly induced by salt stress when plants were subjected to 125 mM NaCl treatment for two weeks, as compared to control conditions without salt stress treatment (Figure 1b). The result suggests that BnWIN1 is involved in salt stress. To test this hypothesis, the full length of BnWIN1 complementary DNA (cDNA) was cloned from cv. Westar by reverse transcript (polymerase chain reaction) PCR using mRNA extracted from leaves as template and was overexpressed in cv. Westar plants under the control of the 35S promoter. The expression level in BnWIN1-OE plants was 21-fold increased as compared to wild type (WT) plants (Figure 2c).
Figure 2.
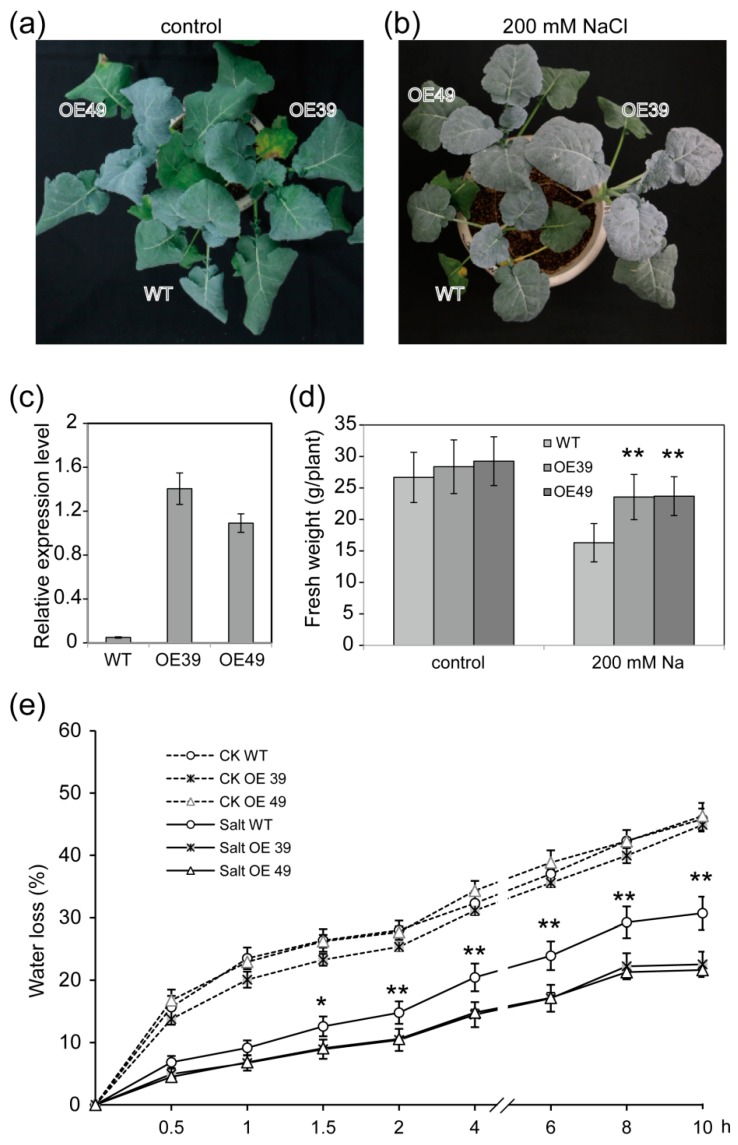
Overexpression of BnWIN1 enhanced plants growth and reduced water loss under salt stress. (a,b) The phenotype of BnWIN1-overexpression (OE) plants under control and salt stress conditions. Forty-day-old plants grown in soil in pots were treated with 200 mM NaCl for 30 days. (c) The BnWIN1 expression level of BnWIN1-OE plants confirmed by real-time PCR with β-Actin as an internal standard. Values are means ± SD (n = 3 separated samples). (d) Enhanced growth of BnWIN1-OE plants under salt stress. Forty-day-old plants grown in soil in pots were treated with 200 mM NaCl for 30 days, and fresh weight was measured. Values are means ± SD (n = 12 plants). (e) Water loss from detached leaves. Values are mean ± SD (n = 3 independent experiments). * and ** indicate significant difference at p < 0.05 and p < 0.01, respectively, compared with wild type (WT) plants based on Student’s t test.
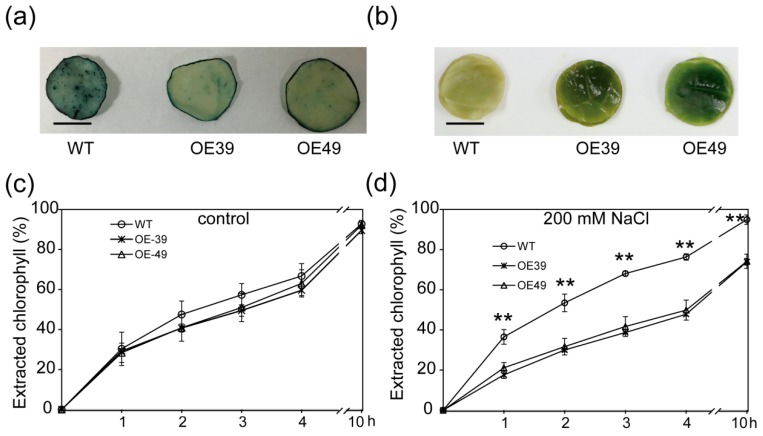
To investigate whether BnWIN1 is involved in osmotic stress, BnWIN1-OE and WT plants were subjected to salt stress. Under control conditions, BnWIN1-OE plants were not visibly different from WT plants (Figure 2a,d). However, BnWIN1-OE plants grew better than WT when plants were treated with 200 mM NaCl. The fresh weight of BnWIN1-OE plants was increased by 45% compared to WT (Figure 2b,d). BnWIN1-OE plants exhibited more glaucous appearance in the leaf surface than that of WT plants under salt stress conditions (Figure 2b). Water loss from salt-stressed detached leaves of BnWIN1-OE plants was much less than that of WT and, in particular, non-stomatal evaporation from OE leaves was 70% of WT after detachment for 10 h (Figure 2e). Leaf tissues in BnWIN1-OE were less stained with toluidine blue (TB) than in WT (Figure 3a), suggesting that OE leaves were less damaged than those of WT under salt stress. Moreover, the chlorophyll in salt-stressed BnWIN1-OE leaves was leached much slower than in WT leaves; the chlorophyll extracted rate in OE leaves was ~40% of the total amount, whereas that of WT leaves was 68% after incubation in ethanol for 4 h (Figure 3b–d). The results suggest that BnWIN1-OE plants had reduced permeability of leaf surface as compared to WT under salt stress. Thus, BnWIN1-OE plants display more tolerance to salt stress.
Figure 3.
Overexpression of BnWIN1 reduced cuticular permeability under salt stress. (a) The reduced tubulin staining in BnWIN1-OE plants under salt stress conditions. Scale bar = 1 cm. (b–d) The chlorophyll extraction rate of BnWIN1-OE and WT plants under control and salt stress conditions. Forty-day-old plants grown in soil in pots were treated without (control) or with 200 mM NaCl for 30 days. Values are mean ± SD (n = 3 independent experiments). Scale bar = 1 cm. ** indicate significant difference at p < 0.01 compared to WT based on Student’s t test.
2.3. BnWIN1 Enhanced Wax Accumulation under Salt Stress While It Enhanced Oil Accumulation under Normal Growth Conditions
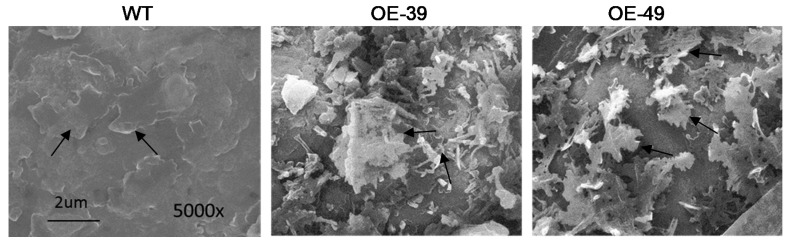
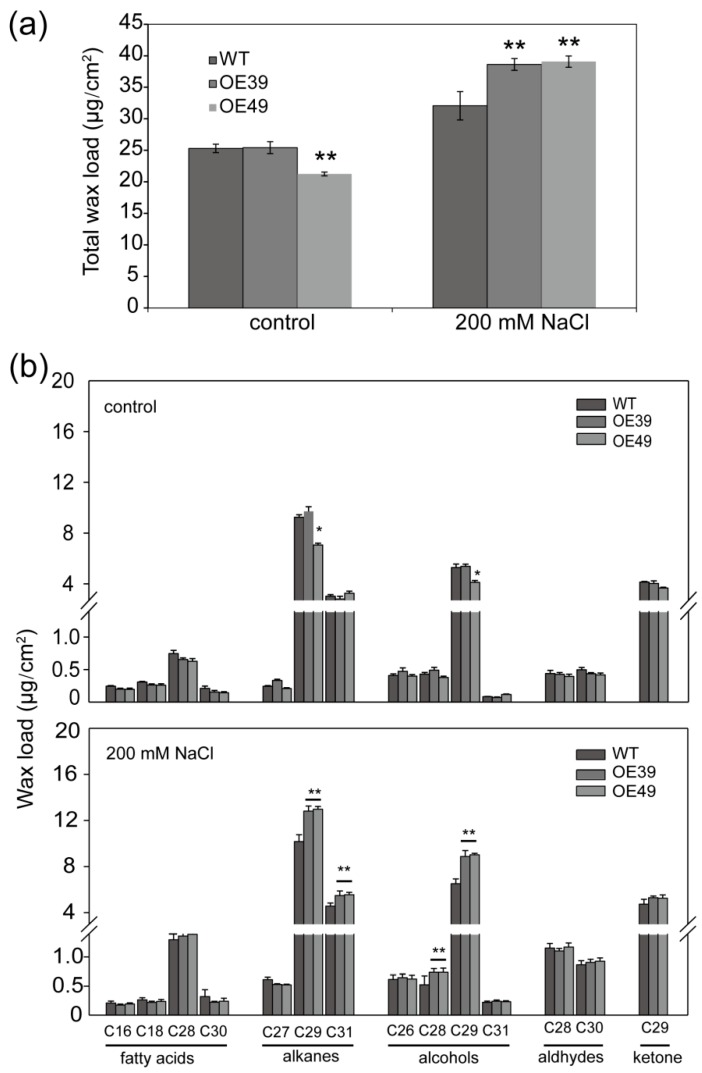
To further investigate the effect of BnWIN1 on wax accumulation, the structure of leaf surface was observed under microscopy. BnWIN1-OE leaf surface displayed a heavy deposition of epicuticular wax crystals, exhibiting an enriched typical mixture of platelets, joint-plates, and polygonal rodlets protruding, as compared to WT (Figure 4). To quantify wax accumulation, chloroform extracts from the leaf epicuticular wax were analyzed by GC/MS. Under normal growth conditions, total wax content in BnWIN1-OE39 leaf surface was not significantly different from that of WT plants, whereas total wax in BnWIN1-OE49 was reduced as compared to WT. When plants were subjected to 200 mM NaCl treatment for 30 days, the total wax content deposited on per unit area of BnWIN1-OE leaf surface was significantly higher, with a 20%~22% increase, as compared with WT (Figure 5a). C29-alkane, C31-alkane, C29-alcohol, and C29-ketone are major wax components in B. napus. The increased wax accumulation in BnWIN1-OE plants resulted from an increase in C29-alkane, C31-alkane, C28-alcohol, and C29-alcohol species. The C29-alkane and C29-alcohol were elevated by 27% and 38%, respectively, relative to WT plants (Figure 5b). The results suggest that BnWIN1 regulates two routes including alkane and alcohol pathways for wax accumulation under salt stress.
Figure 4.
Overexpression of BnWIN1 enhanced epicuticular wax crystals accumulation under salt stress. Epicuticular wax crystals of leaf surface were observed under scanning electron microscopy. Forty-day-old plants grown in soil in pots were treated with 200 mM NaCl for 30 days and leaf samples were taken for observation. Arrows indicate wax crystals.
Figure 5.
Effect of BnWIN1 on wax content and compositions under salt stress. (a) Total wax content in leaves of BnWIN1-OE and WT plants under control and salt stress conditions. Values are means ± SD (n = 5). (b) Wax compositions in leaves of BnWIN1-OE and WT plants under control and salt stress conditions. Values are means ± SD (n = 5). Forty-day-old plants grown in soil in pots were treated without (control) or with 200 mM NaCl for 30 days. Epicuticular waxes were extracted from leaf surfaces and quantified by GC-MS analysis. * and ** denote significance at p < 0.05 and p < 0.01, respectively, compared with WT plants based on Student’s t test.
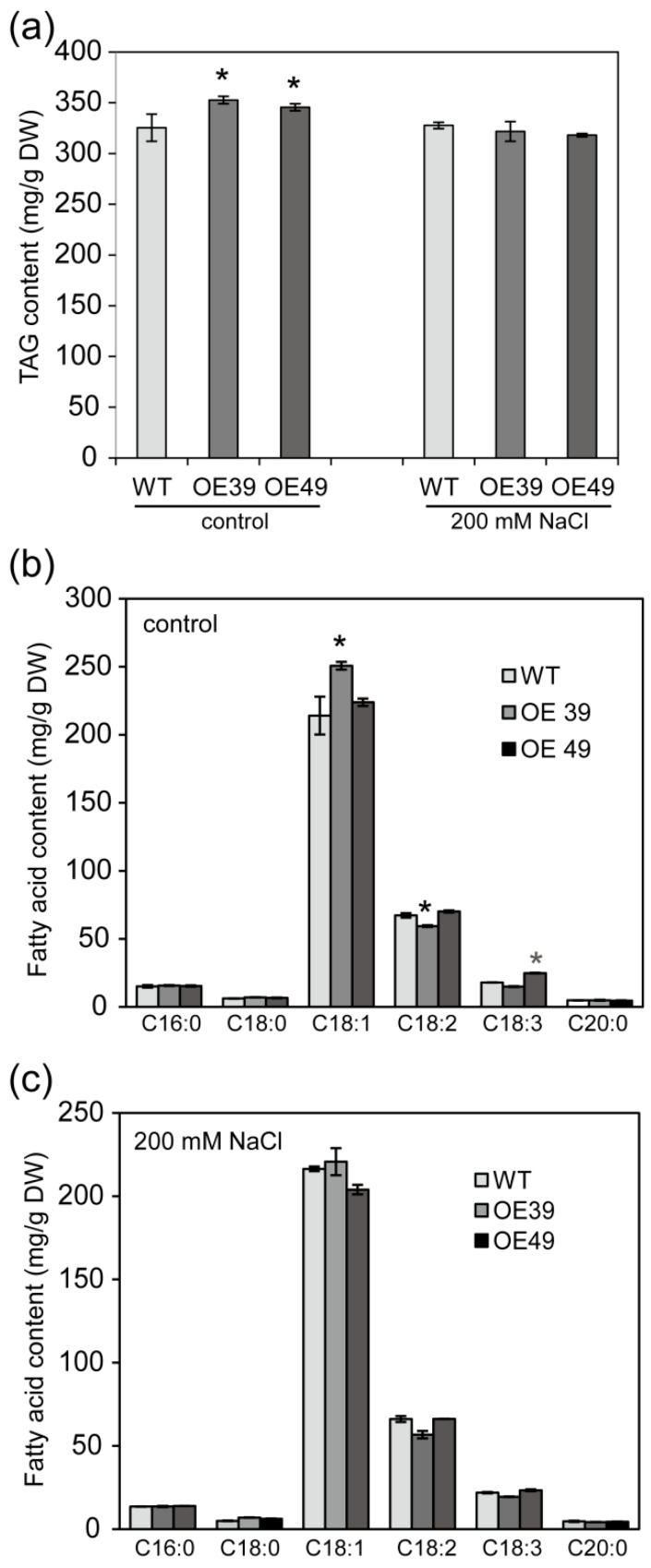
Intracellular and extracellular lipids share fatty acids as common precursors. To test whether enhanced wax accumulation by BnWIN1 led to reduced oil accumulation in seeds, the oil content was measured by GC analysis. Under salt stress, the total oil content of BnWIN1-OE seeds was not significantly different from WT seeds (Figure 6a,c). Under normal growth conditions, however, the total oil content of BnWIN1-OE seeds was increased by 8% as compared to WT seeds (Figure 6a). The increased oil content in the BnWIN1-OE seeds may have resulted from the increase in 18:1 or 18:3 fatty acids (Figure 6b). The results suggest that overexpressing BnWIN1 leads to enhanced overall lipids, thus, enhanced wax accumulation without a reduction of storage lipids under salt stress and enhanced oil accumulation under normal growth condition.
Figure 6.
BnWIN1 enhanced seeds oil accumulation. (a) Oil (TAG) content of seeds. (b,c) Fatty acid compositions of seeds. Forty-day-old plants grown in soil in pots were treated without (control) or with 200 mM NaCl for 30 days, and seeds were collected from plants at the mature stage. Seed oil was analyzed by GC after methylation. Values are mean ± SD (n = 4 separate samples). * denote significance at p < 0.05 compared with WT plants based on Student’s t test.
2.4. BnWIN1 Bound to Promoter Regions of Genes Involved in FA, Wax, and Oil Biosynthesis
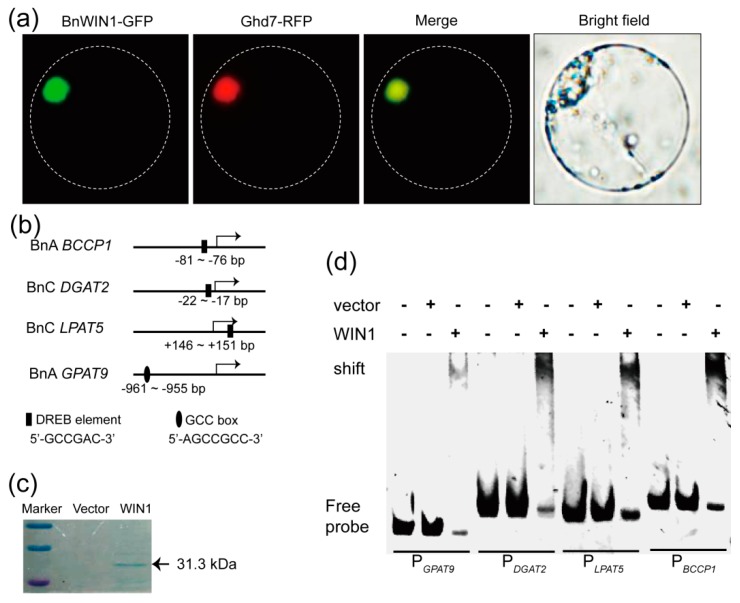
To test the subcellular localization of BnWIN1, the full length BnWIN1 coding sequence (CDS) was fused with GFP at C-terminus and was transiently expressed in Arabidopsis chloroplasts. The green fluorescent BnWIN1-GFP was visualized under a fluorescence microscope and was overlaid with nucleus marker Ghd7-RFP (Figure 7a), confirming BnWIN1 is localized to the nucleus.
Figure 7.
BnWIN1 bound to the promoter region of genes involved in lipid anabolism. (a) Nuclear localization of BnWIN1-GFP using BnWIN1-GFP transiently expressed in Arabidopsis protoplasts. Green fluorescence signal of BnWIN1-GFP was overlaid with nuclear red fluorescence (RFP) of the nuclear marker Ghd7-RFP. (b) Schematic representation of the cis-elements in promoter regions of target genes. (c) Purified recombinant BnWIN1 protein expressed in E. coli cells separated by SDS-PAGE gel and stained with Coomassie blue. (d) Electrophoretic mobility-shift assay of BnWIN1 binding to the promoter region of GPAT9, DGAT2, LPAT5, and BCCP1 containing GCC-box or DREB element.
The promoter regions of genes related to lipid anabolic process, such as BCCP1 in de novo FA synthesis [16], DGAT2 in wax esters [19], glycerol-3-phosphate acyltransferase 9 (GPAT9), and lysophosphatidic acid acyltransferase 5 (LPAT5) in oil synthesis [36,37,38], contain the DREB element (GCCGAC) or GCC box (AGCCGCC) (Figure 7b). To test whether BnWIN1 binds to the target DNA fragments, BnWIN1-His protein was expressed in E. coli and purified for binding assays (Figure 7c). The DNA fragments of PBCCP1, PDGAT2, PLPAT5, and PGPAT9containing the DREB element or GCC box in the promoter regions were amplified for electrophoretic mobility-shift assay (EMSA). When BnWIN1 protein was incubated with the DNA fragment PBCCP1(187 bp) amplified from the promoter region of BCCP1, the target DNA was bound to BnWIN1, as indicated by a shifted band at the top of gel visualized under UV light (Figure 7d). A similar gel shift was observed when BnWIN1 protein was incubated with the DNA fragments, PDGAT2(180 bp), PLPAT5(178 bp), and PGPAT9(156 bp), respectively (Figure 7d). By contrast, the empty vector without BnWIN1 had no such binding (Figure 7d). The results suggest that BnWIN1 binds to the promoter region of genes, including BCCP1, DGAT2, LPAT5, and GPAT9.
2.5. Overexpression of BnWIN1 Up-Regulated the Transcript Levels of Genes Related to FA, Wax, and Oil Biosynthesis
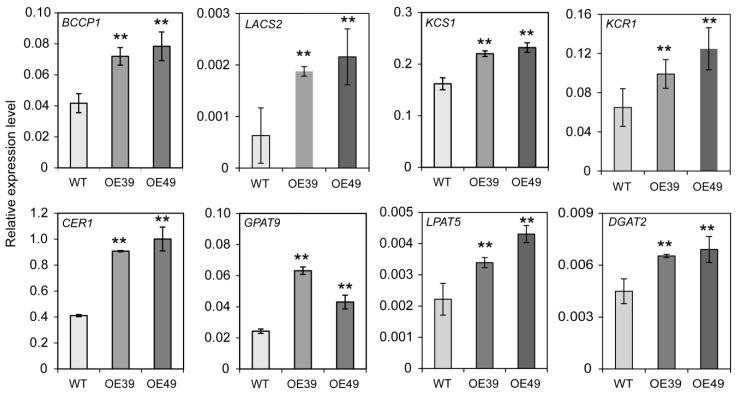
To investigate whether overexpression of BnWIN1 regulates the transcript level of genes involved in lipid anabolic process, the RNA was extracted from leaves and was analyzed by qRT-PCR. The BCCP1 transcript was up-regulated and the BCCP1 expression level in BnWIN1-OE plants was more than two-fold compared to WT (Figure 8). The transcript levels of LACS2, β-ketoacyl-CoA synthase 1 (KCS1), β-ketoacyl-CoA reductase 1 (KCR1), and CER1, responsible for fatty acid elongation and alkane production, were also substantially higher in BnWIN1-OE plants than in WT plants (Figure 8). Overexpressing BnWIN1 also led to increased expression level of DGAT2 involved in wax esters. Moreover, the mRNA level of GPAT9, encoding a key enzyme in oil synthesis, in BnWIN1-OE plants displayed a two-fold increase, as compared to WT (Figure 8). In addition, the expression level of LPAT5, a homolog to cotton At-GhLPPAT5 involved in oil synthesis [38], was also up-regulated in BnWIN1-OE plants (Figure 8). The results suggest that BnWIN1 synchronously regulates the induction of numerous genes in multiple lipid anabolic blocks including fatty de novo synthesis, fatty acid editing, and lipid assembly for both wax and oil accumulation in B. napus.
Figure 8.
BnWIN1 increased expression of genes related to lipid anabolism. Total RNA was extracted from 40-day-old plants treated with 200 mM NaCl for 12 h. The relative expression levels of genes were analyzed by real-time PCR normalized to β-Actin expression level. Values are means ± SD (n = 3 separated samples). ** denote significance at p < 0.01 compared with WT plants based on Student’s t test.
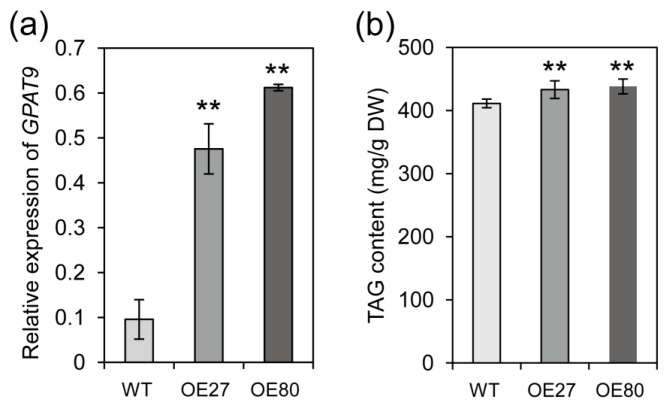
2.6. Overexpression of BnGPAT9 Enhanced Oil Accumulation in Seeds
One novel finding showed that BnWIN1 is also involved in oil accumulation through up-regulating BnGPAT9 expression. To verify the effect of BnGPAT9 on seed oil synthesis, BnGPAT9 was cloned and overexpressed in B. napus (Figure 9a). The oil content in BnGPAT9-OE seeds was significantly higher than in WT seeds (Figure 9b). The results suggest that BnWIN1 up-regulates BnGPAT9 expression to promote oil accumulation in B. napus.
Figure 9.
Overexpression of BnGPAT9 enhanced oil accumulation in seeds. (a) The BnGPAT9 expression level in transgenic plants confirmed by real-time PCR with β-Actin as an internal standard. Values are means ± SD (n = 3 separated samples). (b) Oil content of seeds of GPAT9-OE and WT plants. Seeds were collected from plants grown in field at the mature stage. The oil content of seeds was analyzed by GC after methylation. Values are means ± SD (n = 4). ** denote significance at p < 0.01 compared with WT based on Student’s t test.
3. Discussion
Given the nature of the complicated network of lipid metabolic processes and salt stress response, transcription factors play important roles by simultaneously regulating numerous genes. Here, we show that overexpression of a single gene encoding transcription factor BnWIN1 in B. napus could activate multiple genes to enhance wax production and improves plant tolerance to salt stress, whereas it promotes seed oil content under normal growth conditions. These results suggest that BnWIN1 has dual functions with wax and oil synthesis, depending on growth environments. Therefore, increased BnWIN1 expression in B. napus could have significant effects on salt stress tolerance and oil production, which open a potential avenue for the genetic improvement of oil crop plants.
Wax constituents differ in various plant species. VLC n-alkanes are predominant components accounting for more than 50% of the wax compounds in Arabidopsis leaves [12], tree tobacco leaves [39], and tomato fruits [40]. Cuticular wax in rice leaves is predominantly comprised of fatty acids, primary alcohols, and aldehydes [9,41]. Wax constituents in Medicago leaves mainly consist of alcohols, and overexpression of Medicago WXP1, an AP2 transcription factor, led to an increased C30 primary alcohol [33], indicating that WXP1 regulates the acyl-reduction pathway of wax biosynthesis. Overexpression of Arabidopsis WIN1-conferred plants enhanced cuticular wax accumulation with a pronounced increase in alkane components in leaves [7,8], suggesting that Arabidopsis WIN1 is predominantly involved in the decarbonylation pathway [8]. Our results from B. napus showed that overexpression of BnWIN1 led to increased C29-alkane, 31-alkane, C28- alcohol, and C29-alcohol species, as compared to WT. The results suggest that BnWIN1 plays important roles through regulating two routes, the alkane (decarbonylation) and alcohol (the acyl reduction) wax synthesis pathways. In addition, lipid profiling showed that levels of alkanes are much higher than those of fatty acids in wax mixture in B. napus leaves. It was suggested that cuticular alkanes play a greater role in water retention than VLCFAs under osmotic stress [42,43].
Fatty acids synthesized in plastids are activated by LACSs to produce acyl-CoAs for further elongation catalyzed by KCS and KCR, generating VLC acyl-CoAs [44,45]. The VLC acyl-CoAs are further reduced to aldehydes and decarbonylated to n-alkanes by the CER1/CER3 complex [20,25,26]. Loss of WIN1 led to down-regulation of genes involved in wax accumulation such as KCS1 and CER1 in Arabidopsis [7,8,46]. In agreement with the altered wax components, our results show that overexpression of BnWIN1 up-regulates synergistically the expression levels of numerous genes such as LACS2, KCS1, KCR1, and CER1, involved in fatty acid activation, elongation, and decarbonylation for wax accumulation under salt stress. The up-regulation of these wax biosynthesis-related genes can explain the wax accumulation, in particular the increase of VLC n-alkanes and alcohols in BnWIN1-OE plants under salt stress. Increased wax accumulation restricts surface permeability and water loss, thus enhances plants’ tolerance to osmotic stress.
Extracellular and intracellular lipids share fatty acid precursor pools. We also investigated whether wax accumulation could, directly or indirectly, lead to competition on substrate availability and, ultimately, oil accumulation in seeds. Our results showed that overexpressing BnWIN1 did not lead to reduced oil content under salt stress. Moreover, unlike what was found in Arabidopsis [7,8], overexpression of BnWIN1 in B. napus plants did not cause growth retardation. The different effects of BnWIN1 and AtWIN1 on allocations to intracellular and epidermal lipids suggest that overexpression BnWIN1 in B. napus may have less competition on fatty acids. WIN1 target genes, such as CER1, KCS1, and CER2, are exclusively related to extracellular lipid synthesis [7,8]. Our study indicates that BnWIN1 regulates directly the induction of genes involved in the synthesis of wax, fatty acids, and seed oils. BnWIN1 contains AP2 domains involved in DNA binding. Our results showed that BnWIN1 was localized to the nucleus, and EMSA data showed that BnWIN1 bound directly to the promoter region of BnBCCP1. BCCP is a subunit of ACCase complex that catalyzes rate-limiting reaction to produce malonyl-CoA, which provides a two-carbon donor for de novo FA synthesis [14,16] and also for acyl chain elongation to produce VLCFA [14]. Up-regulated BCCP1 expression in BnWIN1-OE plants promotes de novo fatty acid synthesis by push process and enhances substrate availability for overall lipids accumulation.
It was shown that Arabidopsis WIN1 is responsible for extracellular lipids [7,8]. This study unraveled the novel role of BnWIN1 in the oil accumulation in B. napus. Under normal growth condition, overexpresion of BnWIN1 leads to increased oil content in seeds, which is not found in Arabidopsis WIN1. To separate primary from secondary effects of BnWIN1 on oil accumulation, our results showed that the BnGPAT9 promoter region possesses a GCC box and BnGPAT9 expression was up-regulated in BnWIN1-OE plants. EMSA data showed that BnWIN1 bound directly to the promoter region of BnGPAT9, suggesting that BnGPAT9 is a direct target of BnWIN1. GPAT9 catalyzes the acylation at the sn-1 position of glycerol-3-phosphate (G3P) to produce LPA and was recently identified to be important for oil accumulation in Arabidopsis [36,37]. Our results from B. napus plants overexpressing BnGPAT9 demonstrate that BnGPAT9 plays a positive role in seed oil accumulation. In addition, BnWIN1 also binds to DGAT2 promoter region. DGAT2 was found to have dual functions for TAG assembly and wax accumulation [19,47,48]. These results suggest that the transcript regulation by BnWIN1 on lipid anabolic process is more comprehensive in B. napus as compared to its counterpart in Arabidopsis.
In summary, current results showed that BnWIN1 enhanced wax accumulation and salt stress tolerance without apparent side effects on storage lipids and growth. Our data indicate that BnWIN1 regulates the homeostasis between extracellular and intracellular lipids and enhances oil accumulation under normal growth conditions, whereas it enhances wax accumulation to retain water under osmotic stress without at the expense of storage lipids. Our results shed light on the roles of BnWIN1 in lipid regulatory networks and its application in oil crop breeding to improve salt stress tolerance and oil accumulation.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Seeds of canola cultivar (cv.) Westar (Brassica napus L.) were sown in plates with moist filter paper or pots with soil. Two-week-old seedlings were transferred to pots containing soil or field under natural conditions during winter/spring seasons in Wuhan, China. For salt treatments, four-week-old seedlings of overexpression and wild type (WT) were transferred to the same pot containing soil and were watered regularly. After recovery for two weeks, the plants were treated with various concentrations (0, 150, 200 mM) of NaCl solution for 30 days in a growth room under 16 h light (25 °C)/8 h dark (20 °C), photosynthetic photon flux density of 200–300 mmol m−2 s−1, and 60% relative humidity.
4.2. RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted from Westar plant tissues such as roots, leaves, stems, siliques, and developing seeds using Transzol reagent (TransGen Biotech, Beijing, China). RNA extracts were treated with RNase-free DNaseI to remove contaminated DNA. The extracted RNA quality was checked and used as a template for first strand cDNA synthesis by reverse transcriptase using oligo-d (T) 18 primer according the manufacturer’s protocol (TransGen Biotech, Beijing, China). Quantitative real-time PCR was performed as described previously (Hong et al., 2018). Relative expression level was calculated by the comparative Ct method using Bnβ-Actin as an internal standard. The primers used are listed in Supplementary Table S1.
4.3. Gene Cloning, Vector Construction and Plant Transformation
To produce BnWIN1-OE B. napus lines, a cDNA pool was synthesized from mRNA extracted from leaves of cv. Westar plants by reverse transcription using a TIANscript RT Kit according to the manufacturer′s instructions (TansGene Biotech, Beijing, China). The full length of BnWIN1 coding sequence (CDS) was amplified from the cDNA pool by PCR using the primers BnWIN1 5′-GGATCCATGGTACAGACGAAGAAGTTC-3′ (forward) and BnWIN1 5′-GAGCTCGTTTGTATTGAGAAGCTCCTC-3′ (reverse) and then ligated into the binary vector pBI121 in BamH I and Sac I cutting site. The resultant construct was transformed into B. napus hypocotyls via agrobacterium GV3101 mediation according the method described previously [49]. Transgenic plants were first selected by resistance to kanamycin (50 µg/mL) and were confirmed by PCR using pBI121 vector sequence specific primer 5′-GATGGTTAGAGAGGCTTACGCA-3′ and BnWIN1 sequence specific primer 5′-GAGCTCGTTTGTATTGAGAAGCTCCTC-3′. GPAT9-OE plants were generated via a similar approach. The primers for GPAT9 CDS cloning are listed in Supplementary Table S1.
4.4. Subcellular Localization
The full length of BnWIN1 CDS was amplified from the cDNA pool of cv. Westar by PCR using primers 5′-GGTACCATGGTACAGACGAAGAAGTTC-3′ (forward) and 5′-GGATCCGTTTGTATTGAGAAGCTCCTC-3′ (reverse) and ligated into pCAMBIA1301s vector at the Kpn I and BamH I cutting site. The construct containing BnWIN1-GFP expression driven by the 35S promoter was transformed into protoplasts that were isolated from Arabidopsis leaves by polyethylene glycol (PEG)-mediated transient expression system. The construct was also introduced into agrobacterium GV3101, which was filtrated into the leaves of tobacco for 40–50 h for transient protein expression driven by the 35S promoter. The subcellular localization was visualized under a fluorescence microscope (Olympus, BX53, Tokyo, Japan) in Arabidopsis protoplasts for 12 h or tobacco leaves for 40–50 h after infection.
4.5. Cuticular Wax Observation by SEM
Leaf piece (1.0 cm2) was sampled and fixed with 2.5% glutaraldehyde at 4 °C for 48 h. The samples were dried and coated with palladium–gold in a sputter coater (JEOL JFC-1600, Akishima, Japan). The cuticular wax load was observed under scanning electron microscope (SEM) (QUANTA, Rotterdam, the Netherlands).
4.6. Wax Extraction and Analysis
Forty-day-old plants of OE and WT grown in the same pot containing soil were treated with or without 200 mM NaCl for 30 days. Leaf discs with same area (6.872 cm2) were sampled from leaves with comparable age and position of OE and WT plants and waxes were extracted from leaf surface with chloroform for 30 s with two repeats. The resultant extracts were dried under a stream of nitrogen and were dissolved in 100 µL of chloroform. Wax constituents containing 10 mg tetracosane internal standard were converted to their trimethylsilyl ethers and esters by adding 20 µL of N, N-bis-trimethylsilyltrifluoroacetamide and 20 µL of pyridine to the extracts and incubating the mixture for 40 min at 70 °C. The fatty acid derivatives were quantitatively analyzed by GC-FID (Agilent Technologies, Santa Clara, CA, USA) and GC-MS (Agilent gas chromatograph coupled to an Agilent 5973 N quadruple mass selective detector) at the Instrumental Analysis Center of Shanghai Jiao Tong University.
4.7. Electrophoretic Mobility-Shift Assay
The full length of BnWIN1 CDS was amplified from the cDNA pool of cv. Westar by PCR using forward primer 5′-GGATCCATGGTACAGACGAAGAAGTTC-3′ paired with reverse primer 5′- CTCGAGGTTTGTATTGAGAAGCTCCTC-3′ and then ligated to pET28a vector after digestion by BamH I and Xho I. The resultant construct was transformed into E. coli strain Rosetta (DE3) and the cells grown in Luria Bertani (LB) medium were induced by 0.6 mM isopropyl-β-D-thiogalactopyranoside (IPTG) to express BnWIN1 at 20 °C. Cells were lysed by sonication in a buffer (300 mM NaCl, 20 mM Tris-HCl, pH 8.0, 10 mM imidazole, 5% glycerol, 50 mM NaH2PO4) and the cell lysate was centrifuged at 12000 r/min for 20 min. BnWIN1 was purified using Ni-NTA resin according to the manufacturer′s instructions (Sangon, Shanghai, China). Protein from E. coli cells containing the pET28a vector only was used as a negative control. Meanwhile, the DNA fragments with approximately 200 bp in the promoter region of related genes such as BCCP1, DGAT2, GPAT9, and LPAT5 were amplified from cv. Westar DNA using the primers (Table S1). The amplified target DNA was incubated with purified BnWIN1 in binding buffer (20 mM Tris, pH 8.0, 250 mM NaCl, 2 mM MgCl2, 1% glycerol, 1 mg/mL BSA, 1 mM DTT) for 1 h at 4°C. The reaction mixture was separated on native PAGE (6%) and was visualized under UV light.
4.8. Seed Oil Measurement
Seeds were dried.in an oven at 60 °C for 3 h to measure dry weight. Seed oil was extracted and methylated with methanol and toluene containing 5% H2SO4 at 86 °C for 3 h for GC analysis using heptadecanoic acid as an internal standard. The GC running conditions were as follows: the injection port temperature was 180 °C, and the oven temperature was set at 180 °C for 2 min and was increased by 10°C/min up to 220 °C for 5 min. The temperature of the flame ionization detector was 280 °C with flow rates of 30,300, and 25 mL/min for hydrogen, air, and helium, respectively.
Abbreviations
| B. napus | Brasscia napus |
| BCCP | Biotin carboxyl carrier protein |
| CER | ECERIFERUM |
| DGAT | Diacylglycerol acyltransferase |
| EMSA | Electrophoretic mobility shift assay |
| FAE | Fatty acid elongase |
| FAS | Fatty acid synthase |
| EMSA | Electrophoretic mobility shift assay |
| FAE | Fatty acid elongase |
| FAS | Fatty acid synthase |
| GPAT | Glycerol-3-phosphate acyltransferase |
| KCR | β-Ketoacyl-CoA reductase |
| KCS | β-Ketoacyl-CoA synthase |
| LACS | Long chain acyl-CoA synthetase |
| LPAT | Lysophosphatidic acid acyltransferase |
| OE | Overexpression |
| TAG | Triacylglycerol |
| VLCFA | Very long chain fatty acid |
| WIN1 | WAX INDUCER1/SHINE1 |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/18/4435/s1.
Author Contributions
N.L. and J.C. performed most of the experiments and data analysis; T.W. and Q.L. assisted with vector construction, plant transformation and phenotype observations; P.C. assisted with genotyping, and salt stress treatments; C.J. assisted with gene expression and phylogenetic analysis; Y.H. and N.L. wrote the manuscript; Y.H. supervised this study.
Funding
This work was funded by the National Key Basic Research Program of China (2015CB150204).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lu C., Napier J.A., Clemente T.E., Cahoon E.B. New frontiers in oilseed biotechnology: meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr. Opin. Biotechnol. 2011;22:252–259. doi: 10.1016/j.copbio.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Shahid M., Cai G., Zu F., Zhao Q., Qasim M.U., Hong Y., Fan C., Zhou Y. Comparative transcriptome analysis of developing seeds and silique wall reveals dynamic transcription networks for effective oil production in Brassica napus L. Int. J. Mol. Sci. 2019;20:E1982. doi: 10.3390/ijms20081982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 4.Vij S., Tyagi A.K. Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotechnol. J. 2007;5:361–380. doi: 10.1111/j.1467-7652.2007.00239.x. [DOI] [PubMed] [Google Scholar]
- 5.Kunst L., Samuels A.L. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 2003;42:51–80. doi: 10.1016/S0163-7827(02)00045-0. [DOI] [PubMed] [Google Scholar]
- 6.Ingram G., Nawrath C. The roles of the cuticle in plant development: organ adhesions and beyond. J. Exp. Bot. 2017;68:5307–5321. doi: 10.1093/jxb/erx313. [DOI] [PubMed] [Google Scholar]
- 7.Aharoni A., Dixit S., Jetter R., Thoenes E., van Arkel G., Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16:2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broun P., Poindexter P., Osborne E., Jiang C.Z., Riechmann J.L. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2004;101:4706–4711. doi: 10.1073/pnas.0305574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu X., Xiong L. Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc. Natl. Acad. Sci. USA. 2013;110:17790–17795. doi: 10.1073/pnas.1316412110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorb E.V., Gorb S.N. Anti-adhesive effects of plant wax coverage on insect attachment. J. Exp. Bot. 2017;68:5323–5337. doi: 10.1093/jxb/erx271. [DOI] [PubMed] [Google Scholar]
- 11.Ziv C., Zhao Z., Gao Y.G., Xia Y. Multifunctional roles of plant cticle during plant-pathogen interactions. Front. Plant Sci. 2018;9:1088. doi: 10.3389/fpls.2018.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernard A., Joubès J. Arabidopsis cuticular waxes: advances in regulation, synthesis, export and functions. Prog. Lipid Res. 2013;52:110–129. doi: 10.1016/j.plipres.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Samuels L., Kunst L., Jetter R. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008;59:683–707. doi: 10.1146/annurev.arplant.59.103006.093219. [DOI] [PubMed] [Google Scholar]
- 14.Li-Beisson Y., Shorrosh B., Beisson F. Acyl-lipid metabolism. In: Robt L., editor. Arabidopsis Book. Volume 11. The American Society of Plant Biologists; Rockville, MD, USA: 2013. pp. 1–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S.B., Suh M.C. Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol. Plant. 2013;6:246–249. doi: 10.1093/mp/sss159. [DOI] [PubMed] [Google Scholar]
- 16.Ohlrogge J., Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohlrogge J.B., Jaworski J.G. Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- 18.Haslam T.M., Haslam R., Thoraval D., Pascal S., Delude C., Domergue F., Fernández A.M., Beaudoin F., Napier J.A., Kunst L., et al. ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol. 2015;167:682–692. doi: 10.1104/pp.114.253195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F., Wu X., Lam P., Bird D., Zheng H., Samuels L., Jetter R., Kunst L. Identification of the wax ester synthase/acyl-coenzyme A: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 2008;148:97–107. doi: 10.1104/pp.108.123471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernard A., Domergue F., Pascal S., Jetter R., Renne C., Faure J.D., Haslam R.P., Napier J.A., Lessire R., Joubès J. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain-alkane synthesis complex. Plant Cell. 2012;24:3106–3118. doi: 10.1105/tpc.112.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunst L., Samuels L. Plant cuticles shine: advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 2009;12:721–727. doi: 10.1016/j.pbi.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Li Y., Beisson F., Ohlrogge J., Pollard M. Monoacylglycerols are components of root waxes and can be produced in the aerial cuticle by ectopic expression of a suberin-associated acyltransferase. Plant Physiol. 2007;144:1267–1277. doi: 10.1104/pp.107.099432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia Y., Nikolau B.J., Schnable P.S. Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell. 1996;8:1291–1304. doi: 10.1105/tpc.8.8.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millar A.A., Clemens S., Zachgo S., Giblin E.M., Taylor D.C., Kunst L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell. 1999;11:825–838. doi: 10.1105/tpc.11.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pascal S., Bernard A., Deslous P., Gronnier J., Fournier-Goss A., Domergue F., Rowland O., Joubès J. Arabidopsis CER1-LIKE1 functions in a cuticular very-long-chain alkane-forming complex. Plant Physiol. 2019;179:415–432. doi: 10.1104/pp.18.01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aarts M.G., Keijzer C.J., Stiekema W.J., Pereira A. Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell. 1995;7:2115–2127. doi: 10.1105/tpc.7.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haslam T.M., Mañas-Fernández A., Zhao L., Kunst L. Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol. 2012;160:1164–1174. doi: 10.1104/pp.112.201640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pascal S., Bernard A., Sorel M., Pervent M., Vile D., Haslam R.P., Napier J.A., Lessire R., Domergue F., Joubès J. The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J. 2013;73:733–746. doi: 10.1111/tpj.12060. [DOI] [PubMed] [Google Scholar]
- 29.Lu S., Song T., Kosma D.K., Parsons E.P., Rowland O., Jenks M.A. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009;59:553–564. doi: 10.1111/j.1365-313X.2009.03892.x. [DOI] [PubMed] [Google Scholar]
- 30.Bird D., Beisson F., Brigham A., Shin J., Greer S., Jetter R., Kunst L., Wu X., Yephremov A., Samuels L. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 2007;52:485–498. doi: 10.1111/j.1365-313X.2007.03252.x. [DOI] [PubMed] [Google Scholar]
- 31.Luo B., Xue X.Y., Hu W.L., Wang L.J., Chen X.Y. An ABC transporter gene of Arabidopsis thaliana, AtWBC11, is involved in cuticle development and prevention of organ fusion. Plant Cell Physiol. 2007;48:1790–1802. doi: 10.1093/pcp/pcm152. [DOI] [PubMed] [Google Scholar]
- 32.Suh M.C., Samuels A.L., Jetter R., Kunst L., Pollard M., Ohlrogge J., Beisson F. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 2005;139:1649–1665. doi: 10.1104/pp.105.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J.Y., Broeckling C.D., Blancaflor E.B., Sledge M.K., Sumner L.W., Wang Z.Y. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa) Plant J. 2005;42:689–707. doi: 10.1111/j.1365-313X.2005.02405.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Wan L., Zhang L., Zhang Z., Zhang H., Quan R., Zhou S., Huang R. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 2012;78:275–288. doi: 10.1007/s11103-011-9861-2. [DOI] [PubMed] [Google Scholar]
- 35.Park C.S., Go Y.S., Suh M.C. Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 2016;88:257–270. doi: 10.1111/tpj.13248. [DOI] [PubMed] [Google Scholar]
- 36.Shockey J., Regmi A., Cotton K., Adhikari N., Browse J., Bates P.D. Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. 2016;170:163–179. doi: 10.1104/pp.15.01563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singer S.D., Chen G., Mietkiewska E., Tomasi P., Jayawardhane K., Dyer J.M., Weselake R.J. Arabidopsis GPAT9 contributes to synthesis of intracellular glycerolipids glycerolipids but not surface lipids. J. Exp. Bot. 2016;67:4627–4638. doi: 10.1093/jxb/erw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang N., Ma J., Pei W., Wu M., Li H., Li X., Yu S., Zhang J., Yu J. A genome-wide analysis of the lysophosphatidate acyltransferase (LPAAT) gene family in cotton: organization, expression, sequence variation, and association with seed oil content and fiber quality. BMC Genomics. 2017;18:218. doi: 10.1186/s12864-017-3594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cameron K.D., Teece M.A., Smart L.B. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 2006;140:176–183. doi: 10.1104/pp.105.069724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leide J., Hildebrandt U., Reussing K., Riederer M., Vogg G. The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: effects of a deficiency in a b-ketoacyl-coenzyme a synthase (LeCER6) Plant Physiol. 2007;144:1667–1679. doi: 10.1104/pp.107.099481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao B., Cheng Z., Lei C., Xu F., Gao S., Ren Y., Wang J., Zhang X., Wang J., Wu F., et al. Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta. 2012;235:39–52. doi: 10.1007/s00425-011-1481-1. [DOI] [PubMed] [Google Scholar]
- 42.Kosma D.K., Bourdenx B., Bernard A., Parsons E.P., Lü S., Joubès J., Jenks M.A. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 2009;151:1918–1929. doi: 10.1104/pp.109.141911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo P.J., Lee S.B., Suh M.C., Park M.J., Go Y.S., Park C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell. 2011;23:1138–1152. doi: 10.1105/tpc.111.083485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Todd J., Post-Beittenmiller D., Jaworski J.G. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999;17:119–130. doi: 10.1046/j.1365-313X.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- 45.Beaudoin F., Wu X., Li F., Haslam R.P., Markham J.E., Zheng H., Napier J.A., Kunst L. Functional characterization of the Arabidopsis β-ketoacyl-Coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 2009;150:1174–1191. doi: 10.1104/pp.109.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oshima Y., Shikata M., Koyama T., Ohtsubo N., Mitsuda N., Ohme-Takagi M. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell. 2013;25:1609–1624. doi: 10.1105/tpc.113.110783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shockey J.M., Gidda S.K., Chapital D.C., Kuan J.C., Dhanoa P.K., Bland J.M., Rothstein S.J., Mullen R.T., Dyer J.M. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell. 2006;18:2294–2313. doi: 10.1105/tpc.106.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C., Iskandarov U., Klotz E.T., Stevens R.L., Cahoon R.E., Nazarenus T.J., Pereira S.L., Cahoon E.B. A thraustochytrid diacylglycerol acyltransferase 2 with broad substrate specificity strongly increases oleic acid content in engineered Arabidopsis thaliana seeds. J. Exp. Bot. 2013;64:3189–3200. doi: 10.1093/jxb/ert156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu S., Bahn S.C., Qu G., Qin H., Hong Y., Xu Q., Zhou Y., Hong Y., Wang X. Increased expression of phospholipase Dα1 in guard cells decreases water loss with improved seed production under drought in Brassica napus. Plant Biotechnol. J. 2013;11:380–389. doi: 10.1111/pbi.12028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.