Abstract
Despite the tremendous efforts for improving therapeutics of lung cancer patients, its prognosis remains disappointing. This can be largely attributed to the lack of comprehensive understanding of drug resistance leading to insufficient development of effective therapeutics in clinic. Based on the current progresses of lung cancer research, we classify drug resistance mechanisms into three different levels: molecular, cellular and pathological level. All these three levels have significantly contributed to the acquisition and evolution of drug resistance in clinic. Our understanding on drug resistance mechanisms has begun to change the way of clinical practice and improve patient prognosis. In this review, we focus on discussing the pathological changes linking to drug resistance as this has been largely overlooked in the past decades.
Keywords: Lung cancer, Drug resistance, Pathological transition
Background
Lung cancer can be classified into two main histological types: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). They account for approximately 15% and 85% of all lung cancers, respectively [1]. NSCLC can be further divided into three subtypes, namely lung adenocarcinoma (ADC), squamous cell carcinoma (SCC) and large cell carcinoma (LCC). These subtypes harbor different features, with distinct gene expression profiles [2] as well as lineage-specific biomarkers [3]. For example, lung ADC commonly express thyroid transcription factor-1 (TTF1, also known as NKX2-1) [4, 5], a p53-homologous nuclear protein mainly involved in basal cell commitment. LCC, as a pathologically heterogeneous entity which might represent solid ADC or non-keratinizing SCC, have no well-established biomarkers, yet [6]. In contrast to NSCLC, SCLC frequently express neuroendocrine markers, such as achaete-scute homologue 1 (ASCL1, also known as ASH1), neural cell adhesion molecule (NCAM) and synaptophysin (SYP) [7]. Lung ADC is frequently found at distal bronchioles [8], whereas SCC is often observed in more proximal airways [8]. Most lung ADC are considered as originating from alveolar type II (AT II) cells, club cells, or bronchial-alveolar stem cells (BASCs) [9], whereas SCC is observed at more proximal airway [6]. Most lung ADC are considered as originating from alveolar type II (AT II) cells, club cells, or bronchio-alveolar stem cells (BASCs) [7], whereas, lung SCC are mainly derived from basal cells located underneath trachea or bronchus epithelia [7]. SCLC arises from pulmonary neuro-endocrine (NE) cells and often spread along bronchi in a submucosal and circumferential fashion [8].
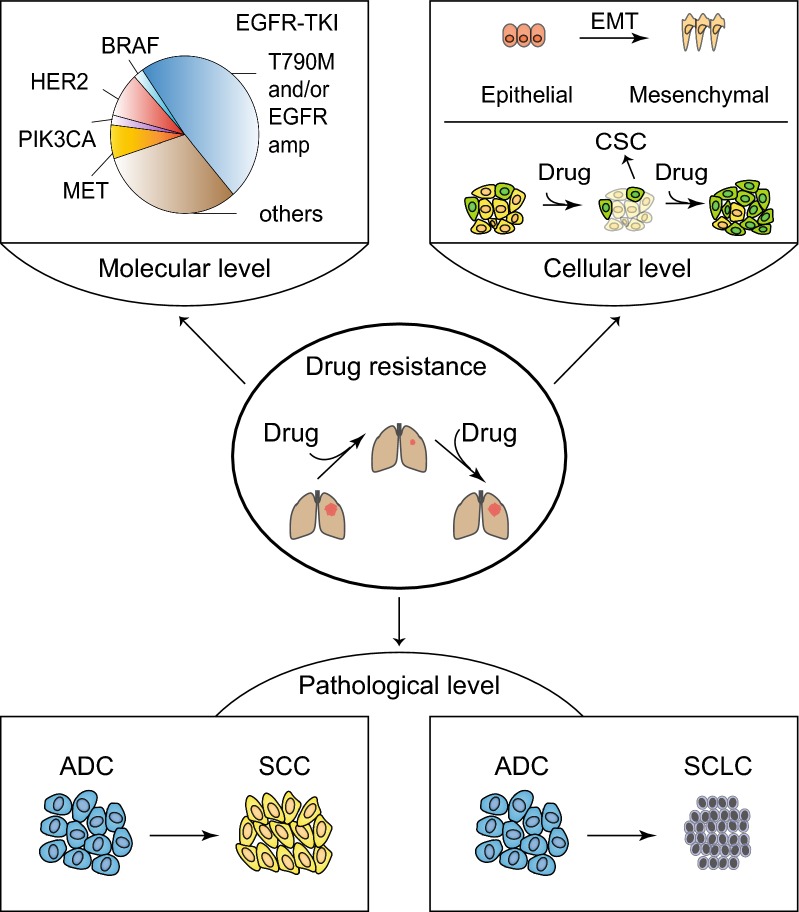
Despite of persistent medical efforts in last decades, lung cancer prognosis still remains disappointing, with a 5-year survival rate of approximately 15% [9]. This is in part attributed to the acquisition of early drug resistance. Understanding of drug resistance mechanisms hopefully improves therapeutic strategies and eventually changes clinical practice. We classify drug resistance mechanisms into three different levels: molecular, cellular and pathological level (Fig. 1). Although these three are closely linked with each other, changes in molecular level might occur in tumor initiation and development prior to those in the other two levels, which enable an early diagnosis with the usage of potential biomarkers. Previous studies have paid extensive attentions to the molecular and cellular level. In this review, we mainly focus on the pathological level which is largely unappreciated previously.
Fig. 1.
Three different levels of drug resistance mechanisms in lung cancer. Drug resistance develops at three different levels: molecular, cellular, and pathological level. Molecular level mechanism includes secondary EGFR T790M and MET amplification after the relapse from EGFR-TKI therapy. Cellular level mechanism mainly involves CSC and EMT. Pathological level mechanism includes the ADC-to-SCC transition and ADC or SCC-to-SCLC transition. EGFR epidermal growth factor receptor, EGFR-TKI epidermal growth factor receptor-tyrosine kinase inhibitor, BRAF serine/threonine-protein kinase B-raf, HER2 receptor tyrosine-protein kinase erbB-2, PIK3CA phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha, MET hepatocyte growth factor receptor, EMT epithelial-to-mesenchymal transition, CSC cell stem cell, ADC adenocarcinoma, SCC squamous cell carcinoma, SCLC small cell lung cancer
Drug resistance mechanisms at molecular and cellular level
Molecular changes are frequently detected in relapsed patients after clinical treatment including chemotherapy, targeted therapy and immunotherapy. There are multiple drug resistance mechanisms at molecular level limit the effectiveness of chemotherapy, e.g., the deregulation of genes involved in drug uptake, cell cycle, apoptosis, sphingolipid metabolism as well as intracellular drug sequestration [10]. Gardner et al. [11] recently show that the down-regulation of Schlafen 11 (SLFN 11) mediated by enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) and H3K27me3 modification induces DNA damage repair and thus enables SCLC chemo-resistance. Molecular alterations are also observed in relapsed patients after targeted therapy in lung cancers [12]. For example, during epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) treatment, secondary EGFR mutation T790M, MET proto-oncogene, receptor tyrosine kinase (MET, also known as hepatocyte growth factor receptor, HGFR) amplification, receptor tyrosine-protein kinase erbB-2 (ERBB2, also known as HER2) amplification as well as Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations are often detectable in relapsed lung cancer patients and known to contribute to drug resistance [13–15]. In the case of anaplastic lymphoma kinase (ALK)-rearranged NSCLC patients, ALK point mutations, KIT proto-oncogene receptor tyrosine kinase amplification, and other driver mutations are implicated for disease relapse [13]. During the immune checkpoint blockade treatment, neo-antigen landscape shows dynamic change which contributes to the resistance to immunotherapy [16]. These data together support an important role of molecular alterations in orchestrating drug resistance.
Drug resistance mechanisms at cellular level is mainly classified into two types: cancer stem cell (CSC)- and epithelial-to-mesenchymal transition (EMT)-mediated drug resistance. CSCs are considered to be highly plastic, resistant to chemotherapy and capable to seed new aggressive and chemo-resistant tumors in distant organs [17]. Great efforts have been paid to investigate the vulnerability of CSC with the purpose to overcome drug resistance. However, the findings of reversible transition between CSC and non-CSC make the specific targeting of CSC extremely difficult [17]. The non-CSC, frequently as the major component of malignant tumors, is also known to harbor strong stemness and plasticity [18]. Such stemness and plasticity allows non-CSC to de-differentiate into CSC under stressful environment [18, 19]. Such de-differentiation together with the reversible transition between CSC and non-CSC creates a huge hurdle for effective targeting either CSC or non-CSC alone [19].
The transition from epithelial cells to mesenchymal cells also reflects the strong plasticity of cancer cells, which is frequently implicated in drug resistance. In contrast to epithelial cells, mesenchymal cells tend to harbor strong transforming growth factor beta (TGFβ) and Wnt autocrine signaling [20]. The EMT often associates with down-regulation of multiple apoptotic signaling pathways, while it enhances drug efflux and slows cell proliferation [17]. The EMT activates several processes including of programmed death-ligand 1 (PD-L1) expression elevation and tumor suppressor region 1 (TSP-1) secretion elevation, which induces immune suppression and promotes immune escape [17, 21]. Besides, several transcription factors including snail family transcriptional repressor (SNAIL), twist family bHLH transcription factor (TWIST) and zinc finger E-box binding homeobox 1 (ZEB1) activate classical EMT-associated properties and induce anti-apoptotic and pro-survival phenotype supporting malignant progression [22]. All these EMT-associated features collectively promote cancer cell survival and help them escape from effective drug treatment. However, this doesn’t always turn out to be true. A recent study shows that EMT triggered by EGFR-TKI treatment associates with decreased PD-L1 expression, indicative of the complexity of the link between EMT and immune checkpoint regulation [23].
Drug resistance mechanisms at pathological level
Pathological transition has recently been implicated in clinic. Observation of lung ADC transition to either SCC or SCLC has been reported in relapsed patients [24, 25]. Below we summarized the current progress and discuss their link to drug resistance (Fig. 2).
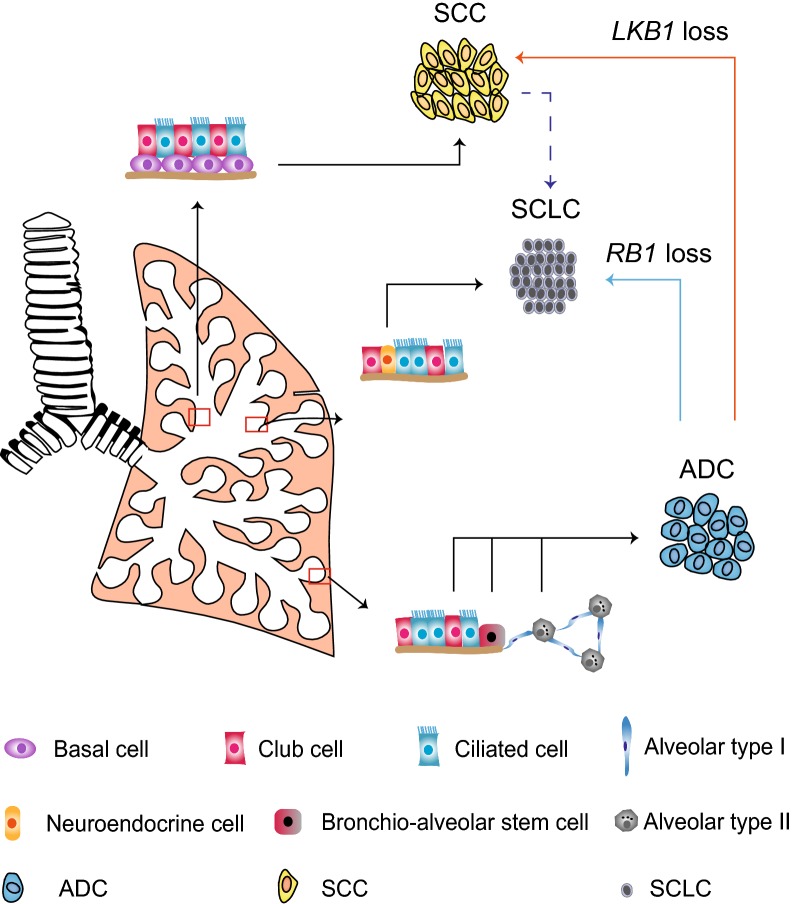
Fig. 2.
Pathological transition of different lung cancer subtypes. Lung cancer can be divided into two subtypes: NSCLC and SCLC. NSCLC can be further divided into three subtypes: ADC, SCC, and LCC. ADCs are considered to originate from alveolar type II cells, club cells or BASCs. SCC frequently found at more proximal airways is presumably derived from basal cells. SCLC is typically derived from neuroendocrine cells. Pathological transition is observed in clinic including lung ADC-to-SCC transdifferentiation and ADC or SCC-to-SCLC transition. Loss of LKB1 or RB1 potentially contributes to the squamous and SCLC transition, respectively. NSCLC non-small cell lung cancer, SCLC small cell lung cancer, ADC adenocarcinoma, SCC squamous cell carcinoma, LCC large cell carcinoma, BASC bronchio-alveolar stem cell, LKB1 liver kinase B1, RB1 retinoblastoma
ADC/SCC-to-SCLC transition promotes drug resistance
Two large cohort studies reveal that approximately 5% human lung cancer display the mixed pathology such as adenosquamous carcinoma (Ad-SCC), combined large cell neuroendocrine carcinoma (LCNEC) and combined SCLC [26, 27]. Combined SCLC accounts for about 2.2% of all lung cancer [28]. Previous studies show that the SCLC and non-SCLC components of combined SCLC often share exactly the same genetic mutations [29, 30], indicating that these two different pathological lesions might share the same cells of origin and/or exist potential pathological transition. Notably, most of these pathologically mixed cancer are observed at advanced stages [26, 27], indicating that the potential pathological transition might occur during late stage of malignancy progression.
The SCLC transition is also observed in clinic after patient relapse from molecular targeted therapy. This is initially found in a woman with lung adenocarcinoma [31]. First biopsy shows that her tumor harbors EGFR exon 19 deletion and erlotinib treatment shows partial response. After 18 months of treatment, the tumor mass progresses. The second biopsy is then performed and shows SCLC pathology with the original EGFR exon 19 deletion, indicative of the potential transition from ADC to SCLC and its link to drug resistance. In later studies, researchers provided solid evidence showing that the transition of EGFR-mutant ADC to SCLC serves as a drug resistance mechanism. Sequist et al. [32] find that about 14% (5/37 cases) drug-resistant ADC cases transit to SCLC and thereby the standard SCLC therapy overcomes such resistance. We summarized the SCLC transition cases with available clinical details in Table 1. Among the total 33 cases, there are 14 males and 19 females. Despite of those unknown smoking status, about 77% (20/26) of the patients are non-smokers. There seems no preference for gender and smoking status. Except for 8 cases with unknown mutation status, most patients (96%, 24/25) show exactly the same oncogenic EGFR mutations or ALK fusions in the first and second biopsies. Additional mutations such as phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) mutation, EGFR T790M are also detectable after relapse in 1 and 2 cases, respectively, indicative of the combined mechanisms at both molecular and pathological levels [13].
Table 1.
Characteristics of 33 relapsed lung ADC patients with SCLC transition
| Patient ID | Gender | Age | Smoking status | Therapy | 1st biopsya | 2nd biopsyb | References | ||
|---|---|---|---|---|---|---|---|---|---|
| Pathological status | Mutation status | Pathological status | Mutation status | ||||||
| 1 | M | 54 | NA | TKI | ADC | EGFR 19 del | SCLC | EGFR 19 del | [32] |
| 2 | F | 56 | NA | TKI | ADC | EGFR 19 del | SCLC | EGFR 19 del | [32] |
| 3 | F | 61 | NA | TKI | ADC | EGFR 19 del | SCLC | EGFR 19 del | [32] |
| 4 | F | 72 | N | Gef | ADC | EGFR 19 del | SCLC | EGFR 19 del | [91] |
| 5 | F | 46 | N | Erl | ADC | EGFR 19 del | SCLC | EGFR 19 del | [92] |
| 6 | F | 52 | N | Erl | ADC | EGFR 19 del | SCLC | EGFR 19 del | [93] |
| 7 | M | 80 | N | Ico | ADC | EGFR 19 del | SCLC | EGFR 19 del | [94] |
| 8 | F | 63 | N | Erl | ADC | EGFR 19 del | SCLC | EGFR 19 del | [95] |
| 9 | M | 46 | Y | Gef | ADC | EGFR 19 del | SCLC | EGFR 19 del | [96] |
| 10 | M | 49 | Y | Erl | ADC | EGFR 19 del and FGFR3 exon 17 deletion | SCLC | EGFR 19 del and FGFR3 exon 17 deletion | [97] |
| 11 | F | 60 | N | Gef | ADC | EGFR 19 del | SCLC | EGFR 19 del | [98] |
| 12 | M | 65 | N | Afa | ADC | EGFR 19 del | SCLC | EGFR 19 del | [99] |
| 13 | F | 37 | N | Gef | ADC | EGFR 19 del | SCLC | EGFR 19 del + T790M | [100] |
| 14 | F | 42 | N | Erl | ADC | EGFR 19 del | SCLC | EGFR 19 del + T790M | [101] |
| 15 | F | 49 | N | Gef | ADC | EGFR 19 del | SCLC | NA | [102] |
| 16 | M | 41 | Y | Gef | ADC | EGFR 19 del | SCLC + SCC | NA | [61] |
| 17 | M | 74 | Y | Gef | ADC | EGFR 19 del | SCLC | WT | [103] |
| 18 | F | 48 | NA | TKI | ADC | EGFR L858R | SCLC | EGFR L858R | [32] |
| 19 | F | 67 | NA | TKI | ADC | EGFR L858R | SCLC | EGFR L858R | [32] |
| 20 | F | 72 | N | Gef | ADC | EGFR L858R | SCLC | EGFR L858R | [104] |
| 21 | M | 46 | N | Gef | ADC | EGFR L858R | SCLC | EGFR L858R | [105] |
| 22 | M | 49 | Y | Gef | ADC | EGFR L858R | SCLC | EGFR L858R | [106] |
| 23 | F | 65 | N | Gef | ADC | EGFR L858R | SCLC | EGFR L858R | [107] |
| 24 | M | 73 | NA | Gef | ADC | EGFR L858R | SCLC | EGFR L858R | [108] |
| 25 | F | 40 | NA | TKI | ADC | EGFR L858R | SCLC | EGFR L858R and PIK3CA | [32] |
| 26 | M | 38 | N | Erl | ADC | EGFR L858R | SCLC | NA | [109] |
| 27 | M | 72 | Y | Crizo | ADC | ALK | SCLC | ALK | [33] |
| 28 | M | 67 | N | Alec | ADC | ALK | SCLC | ALK | [34] |
| 29 | F | 72 | N | Gef | ADC | WT | SCLC | NA | [35] |
| 30 | M | 61 | N | TKI | ADC | NA | SCLC | EGFR 19 del | [110] |
| 31 | F | 46 | N | Gef | ADC | NA | SCLC | EGFR 19 del | [111] |
| 32 | F | 45 | N | Erl- Gef | ADC | NA | SCLC | EGFR 19 del | [31] |
| 33 | F | 73 | N | Gef | ADC | NA | SCLC | EGFR L858R | [104] |
Y yes, N no, NA not available, M male, F female, ADC adenocarcinoma, SCC squamous cell carcinoma, SCLC small cell lung cancer, TKI tyrosine kinase inhibitor, Gef gefitinib, Erl erlotinib, Ico icotinib, Afa afatinib, Crizo crizotinib, Alec alectinib, EGFR epidermal growth factor receptor, EGFR 19 del EGFR exon 19 deletion, ALK anaplastic lymphoma kinase, WT wild-type
a1st biopsy: the first biopsy
b2nd biopsy: the second biopsy
Transition from ADC to SCLC is also observed in patients with ALK rearrangement. Two relapsed patients with ALK fusion after receiving alectinib or crizotinib treatment showed ADC-to-SCLC transition [33, 34]. Another patient with wild-type EGFR also showed the transition to SCLC after developing TKI resistance [35]. These data indicate that the transition to SCLC might be independent of various oncogenic drivers.
The transition from SCC to SCLC is also associated with drug resistance in the clinic [28]. After receiving surgery or radiation and chemotherapy, a total of 16 SCC patients were found to have SCLC transition. The majority of transited SCLC (12/16, 75%) is found to locate at the same sites as the primary tumors.
Recent studies have begun to uncover the underlying mechanisms involved in the ADC-to-SCLC transition [25, 36]. Niederst et al. [25] found that retinoblastoma (RB) was universally lost in all transited SCLC. This is consistent with previous finding about the concurrent loss of RB and p53 alleles in most SCLC [37]. Consistently, Owen et al. [38] find that both RB and P53 deficiencies are required to reprogram lung epithelial cells to SCLC. Despite of the genomic evidence of EGFR mutations in transited SCLC, the expression of EGFR mutants are found to be remarkably decreased or even shut off [25]. Whether RB loss contributes to such down-regulation of EGFR level remains unknown. However, the decreased EGFR expression provides a reasonable explanation for the TKI resistance in transited SCLC [36].
Lung ADC-to-SCC transition links to drug resistance
Lung Ad-SCC is the major subtype of pathologically mixed lung cancer. Ad-SCC contains both adenomatous and squamous pathology [39] and accounts for approximately 60%–75% of all mixed lung cancer [26, 27]. Similar to combined SCLC, the adenomatous and squamous components in Ad-SCC frequently share the same genetic alterations [40–43], indicative of potential pathological transition. Up to date, about 22 reported cases support the link between the ADC-to-SCC transition (AST) and drug resistance (Table 2). Among these patients, the majority (81.8%) is female and 12 (66.7%) of them are non-smoker. Almost all of the transited SCC displays the same EGFR mutations as detected in ADC. EGFR T790M and PIK3CA mutations were also detected in 4 (18.2%) patients, indicative of complicate resistance mechanisms. Moreover, 2 ALK-fusion patients showed AST after the relapse from molecular targeted therapy. AST were also detected in two patients with wild-type EGFR. Except for molecular targeted therapy, AST was also found in patients treated with chemotherapy or immunotherapy. Two patients received chemotherapy and one received chemotherapy and immunotherapy were found to have SCC transition at relapse. These data convincingly support the important link between AST and drug resistance.
Table 2.
Characteristics of 22 relapsed lung ADC patients with potential squamous transition
| Patient ID | Gender | Age | Smoking status | Therapy | 1st biopsya | 2nd biopsyb | References | ||
|---|---|---|---|---|---|---|---|---|---|
| Pathological status | Mutation status | Pathological status | Mutation status | ||||||
| 1 | F | 79 | N | Chemotherapy | ADC | EGFR 19 del | SCC | EGFR 19 del | [112] |
| 2 | M | 43 | Y | Chemotherapy | ADC | EGFR 19 del | SCC | EGFR 19 del | [113] |
| 3 | F | 48 | N | Gef | ADC | EGFR 19 del | SCC | EGFR 19 del | [114] |
| 4 | F | 51 | NA | Gef | ADC | EGFR 19 del | SCC | EGFR 19 del | [115] |
| 5 | F | 58 | Y | Erl | ADC | EGFR 19 del | SCC | EGFR 19 del | [116] |
| 6 | F | 66 | N | Erl | ADC | EGFR 19 del | SCC | EGFR 19 del | [117] |
| 7 | F | 67 | NA | Afa | ADC | EGFR 19 del | SCC | EGFR 19 del and PIK3CA mutation | [118] |
| 8 | F | 40 | Y | Afa | ADC | EGFR 19 del | SCC | EGFR 19 del + T790M | [119] |
| 9 | F | 79 | N | Gef | ADC | EGFR 19 del | SCC | EGFR L858R + T790M | [120] |
| 10 | M | 41 | Y | Gef | ADC | EGFR 19 del | SCC + SCLC | NA | [61] |
| 11 | F | 52 | Y | Erl + Beva | ADC | EGFR 19 del | SCC | EGFR 19 del | [121] |
| 12 | F | 61 | N | Gef | ADC | EGFR L858R | SCC | EGFR L858R | [115] |
| 13 | M | 62 | N | Gef | ADC | EGFR L858R | SCC | EGFR L858R | [122] |
| 14 | F | 63 | N | Erl | ADC | EGFR L858R | SCC | EGFR L858R and PIK3CA | [123] |
| 15 | F | 74 | Y | Gef | ADC | EGFR L858R | SCC | EGFR L858R + T790M | [120] |
| 16 | M | 68 | Y | Erl | ADC | EGFR L858R | SCC | EGFR L858R + T790M | [124] |
| 17 | F | 43 | Y | Gef | ADC | EGFR L858R | SCC | EGFR L858R +S768I | [125] |
| 18 | F | 64 | N | Gef | ADC | EGFR L858R + T790M | SCC | EGFR L858R + T790M | [114] |
| 19 | F | 60 | Y | ALK TKI | ADC | ALK | SCC | ALK | [126] |
| 20 | F | 52 | N | Crizo/Alec | ADC | ALK | SCC | ALK | [127] |
| 21 | F | 63 | N | Erl | ADC | WT | SCC | EGFR L858R + T790M | [128] |
| 22 | M | 69 | N | Chemotherapy–immunotherapy | ADC | WT | SCC | NA | [129] |
Y yes, N no, NA not available, M male, F female, ADC adenocarcinoma, SCC squamous cell carcinoma, SCLC small cell lung cancer, EGFR epidermal growth factor receptor, TKI tyrosine kinase inhibitor, Gef gefitinib, Erl erlotinib, Afa afatinib, Crizo crizotinib, Alec alectinib, Ceri ceritinib, Beva bevacizumab, ALK anaplastic lymphoma kinase, WT wild type, EGFR 19 del EGFR exon 19 deletion
a1st biopsy: the first biopsy
b2nd biopsy: the second biopsy
Evidence from animal models supporting the ADC-to-SCC transition
Studies of the Genetically Engineered Mouse Models (GEMMs) have provided strong in vivo evidence in supporting the ADC-to-SCC transition [44, 45]. We and others have previously found that liver kinase B1 (LKB1, also named as STK11) is frequently mutated in human lung ADC, SCC as well as Ad-SCC [46, 47]. Inactivating mutations of LKB1 seem to be significantly concurrent with Kras mutations and confers lung ADC with strong malignant potential and promotes metastasis [46, 48]. Strikingly, Lkb1 deletion in KrasG12D GEMMs could make ADC progressively transition into SCC via metabolic reprogramming and excessive accumulation of reactive oxygen species (ROS) [49]. YAP, the major transcriptional co-factor of the Hippo pathway, functions as the barrier for AST. When Lkb1 is lost in lung ADC, YAP is activated and up-regulates ZEB2 expression, which in turn represses DNp63 transcription. During the malignant progression and when ADC grows big, the deficiency of extracellular matrix (ECM), e.g., decreased collagen deposition, fails to promote YAP activation and thus relieves ZEB2-mediated repression of DNp63 expression and eventually triggers the AST program [50].
The lysyl oxidase (LOX) family are responsible for cross-linking collagen and elastin, and thus importantly maintain the rigidity and structural stability of ECM [51]. The LOX family has five members including LOX, LOXL1, LOXL2, LOXL3 and LOXL4 with similar catalytic activities [51]. Previous study shows that LOX importantly regulates AST through ECM remodeling [44, 49]. During the AST process, LOX decreases with concurrent reduction of collagen disposition [44]. Pharmacological inhibition of LOX significantly accelerates the AST process in KrasG12D/Lkb1L/L (KL) model [44]. More importantly, long-term LOX inhibition could trigger AST even in KrasG12D/Trp53L/L (KP) mouse model, which is known to produce lung ADC only [52]. This highlights an essential role of LOX and ECM remodeling in AST, which is independent of LKB1 deficiency [52]. The transited SCC show strong resistance to LOX inhibition in contrast to lung ADC, consistent with the association of AST and drug resistance [52].
Chromatin analysis reveals the contribution of epigenetic regulation to AST process in KL model. The transited SCC are featured with the decrease of H3K27me3 level and the increase of H3K27ac and H3K4me3 levels, which might be involved in regulating several key squamous-associated genes such as Sox2, ∆Np63 and Ngfr [45]. EZH2, the methyltransferase responsible for catalyzing H3K27me3 [53], is highly expressed in transited SCC. Similar findings are observed in human lung SCC and the squamous component of human Ad-SCC [45].
Interestingly, Lkb1 loss together with ectopic SOX2 expression promotes the development of SCC, potentially through the progressive transition from ADC to SCC [54]. Simultaneous deletion of FoxA1/2 and Nkx2-1 in KRAS mouse model promotes the transition from ADC to SCC and these tumors are somehow different from those in KL model, and featured with keratinizing squamous cell carcinomas [55]. It remains interesting to see whether LKB1 is inactivated in this model and the squamous transition links to drug resistance.
Previous work has demonstrated that BASCs and club cells are the main cell types for squamous transition [45]. Up to date, most techniques used to isolate BASCs are based on FACS sorting. We recently take advantage of dual recombinant systems including Cre/LoxP and Dre/Rox systems to do the specific lineage-tracing of BASCs in vivo [56]. We found that BASCs are capable of differentiating into multiple cell lineages including club cells, ciliated cells, alveolar type I and type II cells in various lung injury models [56]. Future work will be interesting to illustrate whether BASC-derived tumors are prone to transdifferentiate into SCC when Lkb1 is deleted.
Conclusion
The drug resistance mechanisms can be classified into three different levels: molecular, cellular and pathological level. The ADC-to-SCLC transition and ADC-to-SCC transition are two major patterns for pathological transition in link to acquired drug resistance. A better understanding of drug resistance mechanisms will hopefully change the way of clinical practice and improve patient prognosis.
Perspectives
It’s well established that the pathology of lung cancer serves as an important factor for clinical management, e.g., lung ADC, SCC and SCLC are therapeutically treated differently. The link between pathological transition and drug resistance indicates that even in the era of targeted therapy, the importance of pathology should not be neglected. For example, in EGFR-mutant lung ADC, the median progression-free survival (PFS) after TKI treatment is about 10–13 months [57]. In contrast, the PFS for lung SCC with similar EGFR mutations is only 2.4 months [57]. Even more dramatic finding is that transited SCLC from EGFR-mutant ADC have almost no response to TKI, potentially due to the shut-down of EGFR transcription [36]. This could be explained by the ‘missing target’ theory, in which the therapeutic target disappears after long-term treatment and drug resistance development. It will be interesting to test how EGFR mutants are epigenetically regulated and how we could transcriptionally re-activate EGFR mutants, which might help develop novel therapeutic strategies to overcome drug resistance in these transited SCLC.
Recent studies have also indicated that therapeutic targets could “face off” between different pathologies, e.g., YAP works as proto-oncogene in lung ADC but tumor suppressor in SCC. In human lung ADC, YAP is highly expressed and associated with poor prognosis [58]. Consistently, ectopic YAP expression accelerates lung ADC progression in KrasG12D mouse model [59]. In contrast, YAP suppresses lung SCC progression potentially through down-regulation of the lineage-survival gene DNP63 [60]. We find that digitoxin is highly potent in suppressing SCC growth through YAP activation [60]. Understanding of the double faces of YAP in lung ADC and SCC will certainly help gain novel insights into the process of AST and pathological transition.
Interestingly, pathological transition is not limited to AST or ADC/SCC-to-SCLC transition. Although rare, one case report shows the transition of EGFR-mutated ADC to both SCC and SCLC [61]. Pathological transition is also observed when comparing the primary tumor with brain metastases [62]. For example, two patients with primary lung ADC show SCC or SCLC in brain metastasis whereas another patient with primary lung SCC shows ADC in brain metastasis. Transitions from ADC to LCNEC, or Ad-SCC to SCLC have also been reported [63, 64]. Obviously, our understanding of pathological transition is still very limited. Most case reports are from the EGFR-mutant lung cancer patients who are ethically treated with multiple biopsies. With the improvement of treatment strategies, multiple biopsies specimens will hopefully provide more insightful information about pathological transition.
Besides lung cancer, mixed pathologies are also observed in other types of cancer (Fig. 3), e.g., Ad-SCC are detectable in human colon cancer [65], prostate cancer [66–76] and pancreatic cancer [77]. The AST or AST-like process has been previously reported in pancreatic cancer [78], thyroid gland carcinoma [79–81] as well as gastric cancer [82]. Moreover, neuroendocrine differentiation in ADC has been reported in prostate cancer. After androgen receptor (AR) treatment in castration-resistant prostate cancer (CPRC), certain patients develop neuroendocrine small cell cancer (CRPC-NE) [83–85]. It is understandable that the malignant transformation of benign tumors induce drug resistance [86]. Interestingly, malignant tumors may sometimes become less aggressive or even benign after chemotherapy, such as neuroblastoma to ganglioneuroma transition [87] and malignant germ cell tumors to teratoma transition [88–90]. These transformed tumors are still growing but show the resistance to chemotherapy.
Fig. 3.
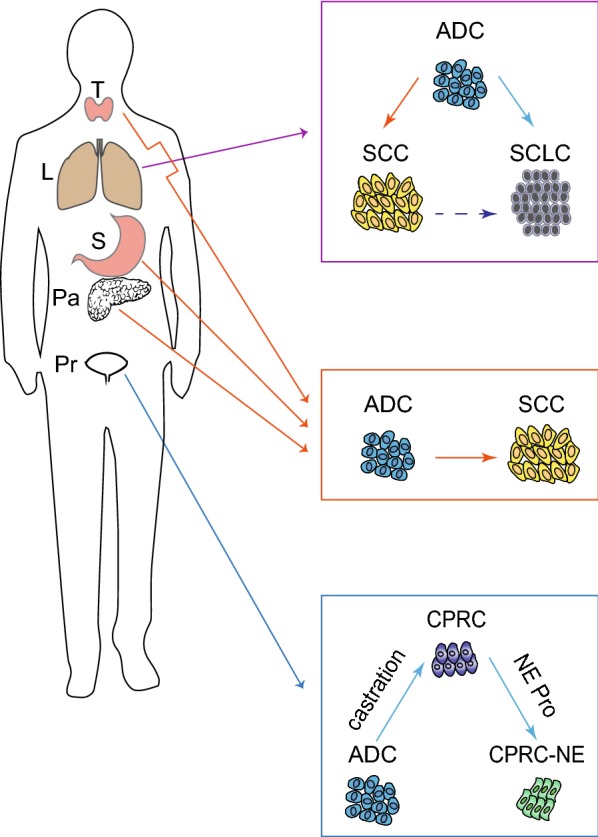
Pathological transition of different types of cancers. Pathological transition in lung cancer includes AST and ADC or SCC-to-SCLC transition. The AST or AST-like process is previously reported in thyroid gland carcinoma, pancreatic cancer as well as gastric cancer. Moreover, neuroendocrine differentiation in ADC has also been reported in prostate cancer. T thyroid gland, L lung, S stomach, Pa pancreas, Pr prostate, Pro proliferation, SCLC small cell lung cancer, ADC adenocarcinoma, SCC squamous cell carcinoma, AST ADC to SCC transition, CPRC castration-resistant prostate cancer, NE neuroendocrine
Together, these findings suggest that pathological transition might be more common than we previously thought. No doubt, the better understanding of pathological transition and the link with drug resistance will be beneficial for future clinic practice and eventually help cancer patients.
Abbreviations
- SCLC
small cell lung cancer
- NSCLC
non-small cell lung cancer
- ADC
adenocarcinoma
- SCC
squamous cell carcinoma
- LCC
large cell carcinoma
- TTF1
thyroid transcription factor-1
- DNP63
delta N tumor protein 63
- ASCL1
achaete-scute homologue 1
- NCAM
neural cell adhesion molecule
- SYP
synaptophysin
- AT II
alveolar type II
- BASC
bronchio-alveolar stem cell
- NE
neuro-endocrine
- SLFN 11
schlafen 11
- EZH2
enhancer of zeste 2 polycomb repressive complex 2 subunit
- EGFR
epidermal growth factor receptor
- MET
MET proto-oncogene, receptor tyrosine kinase (also known as HGFR)
- HGFR
hepatocyte growth factor receptor
- ERBB2
receptor tyrosine-protein kinase erbB-2 (also known as HER2)
- KRAS
kirsten rat sarcoma viral oncogene homolog
- ALK
anaplastic lymphoma kinase
- KIT
KIT proto-oncogene receptor tyrosine kinase
- TGFβ
transforming growth factor beta
- EMT
epithelial-to-mesenchymal transition
- PD-L1
programmed death-ligand 1
- TSP-1
tumor suppressor region 1
- SNAIL
snail family transcriptional repressor
- TWIST
wist family bHLH transcription factor
- ZEB1
zinc finger E-box binding homeobox 1
- Ad-SCC
adenosquamous carcinoma
- LCNEC
large cell neuroendocrine carcinoma
- RB
retinoblastoma
- AST
ADC-to-SCC transition
- GEMM
Genetically Engineered Mouse Model
- LKB1
liver kinase B1
- ROS
reactive oxygen species
- ECM
extracellular matrix
- LOX
lysyl oxidase
- KL
KrasG12D/Lkb1L/L
- KP
KrasG12D/Trp53L/L
- PFS
progression-free survival
- EGFR-TKI
EGFR-tyrosine kinase inhibitor
- CSC
cancer stem cell
- AR
androgen receptor
- CPRC
castration-resistant prostate cancer
- CRPC-NE
neuroendocrine small cell cancer
Authors’ contributions
YC and HJ conceived the idea and wrote the manuscript. WYT revised the background and clinical case report section. XT help revised the part about lung adenocarcinoma to squamous cell carcinoma transdifferentiation. All authors read and approved the final manuscript.
Funding
This work was supported by the National Basic Research Program of China (Grant No. 2017YFA0505501), Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB19020201), National Natural Science Foundation of China (Grant Nos. 81430066, 91731314, 31621003, 81872312, 81871875, 81802279).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Pubmed repository. For Table 1, we used one or a combination of the following terms in PubMed and selected articles on the basis of relevance to transition from ADC to SCLC: ‘non-small cell lung cancer’, ‘transition’, ‘transformation’, ‘transdifferentiation’, ‘adenocarcinoma’, ‘small cell carcinoma’ and ‘small cell lung cancer’. All dates and languages are included in the search. We only collect case reports with detailed clinical information. For Table 2, we used one or a combination of the following terms in PubMed and selected articles on the basis of relevance to transformation from ADC to SCC: ‘transition’, ‘transformation’, ‘transdifferentiation’, ‘adenocarcinoma’ and ‘squamous cell carcinoma’. All dates and languages are included in the search. We only collect case reports with detailed clinical information.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Yueqing Chen, Email: nkchenyueqing@163.com.
Waiying Yvonne Tang, Email: waiyingyvonnetang@hotmail.com.
Xinyuan Tong, Email: Tongxinyuan2013@sibcb.ac.cn.
Hongbin Ji, Email: hbji@sibcb.ac.cn.
References
- 1.Testa U, Castelli G, Pelosi E. Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers. 2018 doi: 10.3390/cancers10080248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujii T, Dracheva T, Player A, Chacko S, Clifford R, Strausberg RL, et al. A preliminary transcriptome map of non-small cell lung cancer. Can Res. 2002;62(12):3340. [PubMed] [Google Scholar]
- 3.Rekhtman N, Ang DC, Sima CS, Travis WD, Moreira AL. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod Pathol. 2011;24:1348. doi: 10.1038/modpathol.2011.92. [DOI] [PubMed] [Google Scholar]
- 4.Righi L, Graziano P, Fornari A, Rossi G, Barbareschi M, Cavazza A, et al. Immunohistochemical subtyping of nonsmall cell lung cancer not otherwise specified in fine-needle aspiration cytology: a retrospective study of 103 cases with surgical correlation. Cancer. 2011;117(15):3416–3423. doi: 10.1002/cncr.25830. [DOI] [PubMed] [Google Scholar]
- 5.Barlesi F, Pinot D, Legoffic A, Doddoli C, Chetaille B, Torre JP, et al. Positive thyroid transcription factor 1 staining strongly correlates with survival of patients with adenocarcinoma of the lung. Br J Cancer. 2005;93(4):450–452. doi: 10.1038/sj.bjc.6602717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanna JM, Onaitis MW. Cell of origin of lung cancer. J Carcinog. 2013;12:6. doi: 10.4103/1477-3163.109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer. 2017;17(12):725–737. doi: 10.1038/nrc.2017.87. [DOI] [PubMed] [Google Scholar]
- 9.Edwards BK, Noone A-M, Mariotto AB, Simard EP, Boscoe FP, Henley SJ, et al. Annual report to the nation on the status of cancer, 1975–2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer. 2014;120(9):1290–1314. doi: 10.1002/cncr.28509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kartal-Yandim M, Adan-Gokbulut A, Baran Y. Molecular mechanisms of drug resistance and its reversal in cancer. Crit Rev Biotechnol. 2016;36(4):716–726. doi: 10.3109/07388551.2015.1015957. [DOI] [PubMed] [Google Scholar]
- 11.Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell. 2017;31(2):286–299. doi: 10.1016/j.ccell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Tian C, Lv F, Wang J, Han W, Nie J, et al. Molecular analysis of cell-free DNA identifies distinct molecular features in patients with chemosensitive and chemorefractory small cell lung cancer. Cancer Commun (London, England) 2019;39(1):20. doi: 10.1186/s40880-019-0363-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11(8):473–481. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Zhu L, Xia B, Chen E, Zhao Q, Zhang X, et al. Epidermal growth factor receptor (EGFR) T790M mutation identified in plasma indicates failure sites and predicts clinical prognosis in non-small cell lung cancer progression during first-generation tyrosine kinase inhibitor therapy: a prospective observational study. Cancer Commun (London, England) 2018;38(1):28. doi: 10.1186/s40880-018-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Diao XY, Zhang X, Shao Q, Feng YF, An X, et al. Identification of genetic alterations associated with primary resistance to EGFR-TKIs in advanced non-small-cell lung cancer patients with EGFR sensitive mutations. Cancer Commun (London, England) 2019;39(1):7. doi: 10.1186/s40880-019-0354-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7(3):264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 19.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells—what challenges do they pose? Nat Rev Drug Discov. 2014;13(7):497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145(6):926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen TV, Hussmann D, Nielsen AL. PD-L1 and PD-L2 expression correlated genes in non-small-cell lung cancer. Cancer Commun (London, England) 2019;39(1):30. doi: 10.1186/s40880-019-0376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 23.Suda K, Rozeboom L, Rivard CJ, Yu H, Ellison K, Melnick MAC, et al. Therapy-induced E-cadherin downregulation alters expression of programmed death ligand-1 in lung cancer cells. Lung Cancer (Amsterdam, Netherlands) 2017;109:1–8. doi: 10.1016/j.lungcan.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou S, Han X, Ji H. Squamous transition of lung adenocarcinoma and drug resistance. Trends Cancer. 2016;2(9):463–466. doi: 10.1016/j.trecan.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. doi: 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng P, Hu C, Zhou L, Li Y, Huang L. Clinical characteristics and prognostic significance of 92 cases of patients with primary mixed-histology lung cancer. Mol Clin Oncol. 2013;1(5):863–868. doi: 10.3892/mco.2013.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruffini E, Rena O, Oliaro A, Filosso PL, Bongiovanni M, Arslanian A, et al. Lung tumors with mixed histologic pattern. Clinico-pathologic characteristics and prognostic significance. Eur J Cardiothorac Surg. 2002;22(5):701–707. doi: 10.1016/S1010-7940(02)00481-5. [DOI] [PubMed] [Google Scholar]
- 28.Ahmed T, Vial MR, Ost D, Stewart J, Hasan MA, Grosu HB. Non-small cell lung cancer transdifferentiation into small cell lung cancer: a case series. Lung Cancer (Amsterdam, Netherlands) 2018;122:220–223. doi: 10.1016/j.lungcan.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 29.Wagner PL, Kitabayashi N, Chen YT, Saqi A. Combined small cell lung carcinomas: genotypic and immunophenotypic analysis of the separate morphologic components. Am J Clin Pathol. 2009;131(3):376–382. doi: 10.1309/ajcpynpfl56pozqy. [DOI] [PubMed] [Google Scholar]
- 30.Siegele BJ, Shilo K, Chao BH, Carbone D, Zhao W, Ioffe O, et al. Epidermal growth factor receptor (EGFR) mutations in small cell lung cancers: two cases and a review of the literature. Lung Cancer (Amsterdam, Netherlands) 2016;95:65–72. doi: 10.1016/j.lungcan.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zakowski MF, Ladanyi M, Kris MG. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355(2):213–215. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 32.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cha YJ, Cho BC, Kim HR, Lee H-J, Shim HS. A case of ALK-rearranged adenocarcinoma with small cell carcinoma-like transformation and resistance to crizotinib. J Thorac Oncol. 2016;11(5):e55–e58. doi: 10.1016/j.jtho.2015.12.097. [DOI] [PubMed] [Google Scholar]
- 34.Fujita S, Masago K, Katakami N, Yatabe Y. Transformation to SCLC after treatment with the ALK inhibitor alectinib. J Thorac Oncol. 2016;11(6):e67–e72. doi: 10.1016/j.jtho.2015.12.105. [DOI] [PubMed] [Google Scholar]
- 35.Araki J, Okamoto I, Suto R, Ichikawa Y, Sasaki J. Efficacy of the tyrosine kinase inhibitor gefitinib in a patient with metastatic small cell lung cancer. Lung cancer (Amsterdam, Netherlands) 2005;48(1):141–144. doi: 10.1016/j.lungcan.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 36.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16(4):e165–e172. doi: 10.1016/s1470-2045(14)71180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JW, Lee JK, Sheu KM, Wang L, Balanis NG, Nguyen K, et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science. 2018;362(6410):91. doi: 10.1126/science.aat5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. WHO classification of tumours of the lung, pleura, thymus and heart. 4. Lyon: International Agency for Research on Cancer; 2015. p. 7. [DOI] [PubMed] [Google Scholar]
- 40.Niho S, Yokose T, Kodama T, Nishiwaki Y, Mukai K. Clonal analysis of adenosquamous carcinoma of the lung. Jpn J Cancer Res. 1999;90(11):1244–1247. doi: 10.1111/j.1349-7006.1999.tb00703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang SM, Kang HJ, Shin JH, Kim H, Shin DH, Kim SK, et al. Identical epidermal growth factor receptor mutations in adenocarcinomatous and squamous cell carcinomatous components of adenosquamous carcinoma of the lung. Cancer. 2007;109(3):581–587. doi: 10.1002/cncr.22413. [DOI] [PubMed] [Google Scholar]
- 42.Jia X-L, Chen G. EGFR and KRAS mutations in Chinese patients with adenosquamous carcinoma of the lung. Lung Cancer (Amsterdam, Netherlands). 2011;74(3):396–400. doi: 10.1016/j.lungcan.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Tochigi N, Dacic S, Nikiforova M, Cieply KM, Yousem SA. Adenosquamous carcinoma of the lung: a microdissection study of KRAS and EGFR mutational and amplification status in a western patient population. Am J Clin Pathol. 2011;135(5):783–789. doi: 10.1309/AJCP08IQZAOGYLFL. [DOI] [PubMed] [Google Scholar]
- 44.Han X, Li F, Fang Z, Gao Y, Li F, Fang R, et al. Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nat Commun. 2014;5:3261. doi: 10.1038/ncomms4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, Fillmore Brainson C, Koyama S, Redig AJ, Chen T, Li S, et al. Lkb1 inactivation drives lung cancer lineage switching governed by polycomb repressive complex 2. Nat Commun. 2017;8:14922. doi: 10.1038/ncomms14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62(13):3659–3662. [PubMed] [Google Scholar]
- 48.Gao Y, Xiao Q, Ma H, Li L, Liu J, Feng Y, et al. LKB1 inhibits lung cancer progression through lysyl oxidase and extracellular matrix remodeling. Proc Natl Acad Sci USA. 2010;107(44):18892–18897. doi: 10.1073/pnas.1004952107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li F, Han X, Li F, Wang R, Wang H, Gao Y, et al. LKB1 inactivation elicits a redox imbalance to modulate non-small cell lung cancer plasticity and therapeutic response. Cancer Cell. 2015;27(5):698–711. doi: 10.1016/j.ccell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Y, Zhang W, Han X, Li F, Wang X, Wang R, et al. YAP inhibits squamous transdifferentiation of Lkb1-deficient lung adenocarcinoma through ZEB2-dependent DNp63 repression. Nat Commun. 2014;5:4629. doi: 10.1038/ncomms5629. [DOI] [PubMed] [Google Scholar]
- 51.Wang T-H, Hsia S-M, Shieh T-M. Lysyl oxidase and the tumor microenvironment. Int J Mol Sci. 2016;18(1):62. doi: 10.3390/ijms18010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao S, Han X, Tong X, Li F, Qin Z, Huang H-Y, et al. Lysyl oxidase inhibition drives the transdifferentiation from lung adenocarcinoma to squamous cell carcinoma in mice. bioRxiv. 2018 doi: 10.1101/314393. [DOI] [Google Scholar]
- 53.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukhopadhyay A, Berrett KC, Kc U, Clair PM, Pop SM, Carr SR, et al. Sox2 cooperates with Lkb1 loss in a mouse model of squamous cell lung cancer. Cell Rep. 2014;8(1):40–49. doi: 10.1016/j.celrep.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Camolotto SA, Pattabiraman S, Mosbruger TL, Jones A, Belova VK, Orstad G, et al. FoxA1 and FoxA2 drive gastric differentiation and suppress squamous identity in NKX2-1-negative lung cancer. eLife. 2018 doi: 10.7554/elife.38579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Q, Liu K, Cui G, Huang X, Yao S, Guo W, et al. Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat Genet. 2019;51(4):728–738. doi: 10.1038/s41588-019-0346-6. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, Shen Z, Li Z, Duan J, Fu S, Liu Z, et al. Activation of the BMP-BMPR pathway conferred resistance to EGFR-TKIs in lung squamous cell carcinoma patients with EGFR mutations. Proc Natl Acad Sci USA. 2015;112(32):9990–9995. doi: 10.1073/pnas.1510837112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci. 2010;101(5):1279–1285. doi: 10.1111/j.1349-7006.2010.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang W, Gao Y, Li F, Tong X, Ren Y, Han X, et al. YAP promotes malignant progression of Lkb1-deficient lung adenocarcinoma through downstream regulation of survivin. Cancer Res. 2015;75(21):4450–4457. doi: 10.1158/0008-5472.CAN-14-3396. [DOI] [PubMed] [Google Scholar]
- 60.Huang H, Zhang W, Pan Y, Gao Y, Deng L, Li F, et al. YAP suppresses lung squamous cell carcinoma progression via deregulation of the DNp63-GPX2 axis and ROS accumulation. Cancer Res. 2017;77(21):5769–5781. doi: 10.1158/0008-5472.CAN-17-0449. [DOI] [PubMed] [Google Scholar]
- 61.Yao Y, Zhu Z, Wu Y, Chai Y. Histologic transformation from adenocarcinoma to both small cell lung cancer and squamous cell carcinoma after treatment with gefitinib: a case report. Medicine. 2018;97(18):e0650. doi: 10.1097/md.0000000000010650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang M, Zhu X, Han X, Jing H, Han T, Li Q, et al. Histologic transformation of non-small-cell lung cancer in brain metastases. Int J Clin Oncol. 2019;24(4):375–384. doi: 10.1007/s10147-018-1369-1. [DOI] [PubMed] [Google Scholar]
- 63.Kogo M, Shimizu R, Uehara K, Takahashi Y, Kokubo M, Imai Y, et al. Transformation to large cell neuroendocrine carcinoma as acquired resistance mechanism of EGFR tyrosine kinase inhibitor. Lung Cancer (Amsterdam, Netherlands) 2015;90(2):364–368. doi: 10.1016/j.lungcan.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 64.Moriya R, Hokari S, Shibata S, Koizumi T, Tetsuka T, Ito K, et al. Histological transformation to large cell neuroendocrine carcinoma from lung adenocarcinoma harboring an EGFR mutation: an autopsy case report. Intern Med. 2017;56(15):2013–2017. doi: 10.2169/internalmedicine.56.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang DB, Oh JT, Jo HJ, Park WC. Primary adenosquamous carcinoma of the colon. J Korean Surg Soc. 2011;80(Suppl 1):S31–S35. doi: 10.4174/jkss.2011.80.Suppl1.S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bassler TJ, Jr, Orozco R, Bassler IC, Boyle LM, Bormes T. Adenosquamous carcinoma of the prostate: case report with DNA analysis, immunohistochemistry, and literature review. Urology. 1999;53(4):832–834. doi: 10.1016/S0090-4295(98)00418-X. [DOI] [PubMed] [Google Scholar]
- 67.Baydar DE, Kosemehmetoglu K, Akdogan B, Ozen H. Prostatic adenosquamous carcinoma metastasizing to testis. Sci World J. 2006;6:2491–2494. doi: 10.1100/tsw.2006.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gattuso P, Carson HJ, Candel A, Castelli MJ. Adenosquamous carcinoma of the prostate. Hum Pathol. 1995;26(1):123–126. doi: 10.1016/0046-8177(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 69.Accetta PA, Gardner WA., Jr Adenosquamous carcinoma of prostate. Urology. 1983;22(1):73–75. doi: 10.1016/0090-4295(83)90355-2. [DOI] [PubMed] [Google Scholar]
- 70.Devaney DM, Dorman A, Leader M. Adenosquamous carcinoma of the prostate: a case report. Hum Pathol. 1991;22(10):1046–1050. doi: 10.1016/0046-8177(91)90014-G. [DOI] [PubMed] [Google Scholar]
- 71.Egilmez T, Bal N, Guvel S, Kilinc F, Ozkardes H. Adenosquamous carcinoma of the prostate. Int J Urol. 2005;12(3):319–321. doi: 10.1111/j.1442-2042.2005.01036.x. [DOI] [PubMed] [Google Scholar]
- 72.Humphrey PA. Histological variants of prostatic carcinoma and their significance. Histopathology. 2012;60(1):59–74. doi: 10.1111/j.1365-2559.2011.04039.x. [DOI] [PubMed] [Google Scholar]
- 73.Ishigooka M, Yaguchi H, Tomaru M, Sasagawa I, Nakada T, Mitobe K. Mixed prostatic carcinoma containing malignant squamous element. Reports of two cases. Scand J Urol Nephrol. 1994;28(4):425–427. doi: 10.3109/00365599409180526. [DOI] [PubMed] [Google Scholar]
- 74.Mishra S, Goel H, Awasthi N, Puri A, Mahapatra R, Pal DK. Primary adenosquamous carcinoma of the prostate: a rare aggressive tumor. Clin Genitourin Cancer. 2014;12(1):e29–e31. doi: 10.1016/j.clgc.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 75.Moyana TN. Adenosquamous carcinoma of the prostate. Am J Surg Pathol. 1987;11(5):403–407. doi: 10.1097/00000478-198705000-00010. [DOI] [PubMed] [Google Scholar]
- 76.Orhan D, Sak SD, Yaman O, Tulunay O, Gogus O. Adenosquamous carcinoma of the prostate. Br J Urol. 1996;78(4):646–647. doi: 10.1046/j.1464-410X.1996.20830.x. [DOI] [PubMed] [Google Scholar]
- 77.Regi P, Butturini G, Malleo G, Pedica F, D’Onofrio M, Bassi C. Clinicopathological features of adenosquamous pancreatic cancer. Langenbeck’s Arch Surg. 2011;396(2):217–222. doi: 10.1007/s00423-010-0677-3. [DOI] [PubMed] [Google Scholar]
- 78.Somerville TDD, Xu Y, Miyabayashi K, Tiriac H, Cleary CR, Maia-Silva D, et al. TP63-mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal adenocarcinoma. Cell Rep. 2018;25(7):1741–1755. doi: 10.1016/j.celrep.2018.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harada T, Shimaoka K, Yakumaru K, Ito K. Squamous cell carcinoma of the thyroid gland—transition from adenocarcinoma. J Surg Oncol. 1982;19(1):36–43. doi: 10.1002/jso.2930190111. [DOI] [PubMed] [Google Scholar]
- 80.Rausch T, Benhattar J, Sutter M, Andrejevic-Blant S. Thyroid carcinoma with papillary and squamous features: report of a case with histogenetic considerations. Pathol Res Pract. 2010;206(4):263–269. doi: 10.1016/j.prp.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 81.Gomi K, Kitagawa N, Usui Y, Tanaka M, Yoshida M, Hirata Y, et al. Papillary carcinoma with extensive squamous metaplasia arising from thyroglossal duct cyst in an 11-year-old girl: significance of differentiation from squamous cell carcinoma: a case report. J Pediatr Surg. 2011;46(4):e1–e4. doi: 10.1016/j.jpedsurg.2010.10.030. [DOI] [PubMed] [Google Scholar]
- 82.Ahn S, Bae GE, Kim K-M. Exuberant squamous metaplasia of the gastric mucosa in a patient with gastric adenocarcinoma. Diagn Pathol. 2015;10:46. doi: 10.1186/s13000-015-0281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zou M, Toivanen R, Mitrofanova A, Floch N, Hayati S, Sun Y, et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 2017;7(7):736–749. doi: 10.1158/2159-8290.cd-16-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alvarez MJ, Subramaniam PS, Tang LH, Grunn A, Aburi M, Rieckhof G, et al. A precision oncology approach to the pharmacological targeting of mechanistic dependencies in neuroendocrine tumors. Nat Genet. 2018;50(7):979–989. doi: 10.1038/s41588-018-0138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gong L, Yan Q, Zhang Y, Fang X, Liu B, Guan X. Cancer cell reprogramming: a promising therapy converting malignancy to benignity. Cancer Commun (London, England) 2019;39(1):48. doi: 10.1186/s40880-019-0393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miura K, Mineta H, Yokota N, Tsutsui Y. Olfactory neuroblastoma with epithelial and endocrine differentiation transformed into ganglioneuroma after chemoradiotherapy. Pathol Int. 2001;51(12):942–947. doi: 10.1046/j.1440-1827.2001.01300.x. [DOI] [PubMed] [Google Scholar]
- 88.Glass T, Cochrane DD, Rassekh SR, Goddard K, Hukin J. Growing teratoma syndrome in intracranial non-germinomatous germ cell tumors (iNGGCTs): a risk for secondary malignant transformation-a report of two cases. Child’s Nerv Syst. 2014;30(5):953–957. doi: 10.1007/s00381-013-2295-1. [DOI] [PubMed] [Google Scholar]
- 89.Kesler KA, Patel JB, Kruter LE, Birdas TJ, Rieger KM, Okereke IC, et al. The “growing teratoma syndrome” in primary mediastinal nonseminomatous germ cell tumors: criteria based on current practice. J Thorac Cardiovasc Surg. 2012;144(2):438–443. doi: 10.1016/j.jtcvs.2012.05.053. [DOI] [PubMed] [Google Scholar]
- 90.Ghaouti M, Roquet L, Fazzalari L, Sibert L, Sabourin JC. “Growing teratoma syndrome”: an unrecognized complication of treated germ cell tumors of the testis. Ann Pathol. 2013;33(6):402–405. doi: 10.1016/j.annpat.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 91.Okamoto I, Araki J, Suto R, Shimada M, Nakagawa K, Fukuoka M. EGFR mutation in gefitinib-responsive small-cell lung cancer. Ann Oncol. 2006;17(6):1028–1029. doi: 10.1093/annonc/mdj114. [DOI] [PubMed] [Google Scholar]
- 92.Popat S, Wotherspoon A, Nutting CM, Gonzalez D, Nicholson AG, O’Brien M. Transformation to “high grade” neuroendocrine carcinoma as an acquired drug resistance mechanism in EGFR-mutant lung adenocarcinoma. Lung Cancer (Amsterdam, Netherlands) 2013;80(1):1–4. doi: 10.1016/j.lungcan.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 93.Watanabe S, Sone T, Matsui T, Yamamura K, Tani M, Okazaki A, et al. Transformation to small-cell lung cancer following treatment with EGFR tyrosine kinase inhibitors in a patient with lung adenocarcinoma. Lung Cancer (Amsterdam, Netherlands) 2013;82(2):370–372. doi: 10.1016/j.lungcan.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y, Li XY, Tang Y, Xu Y, Guo WH, Li YC, et al. Rapid increase of serum neuron specific enolase level and tachyphylaxis of EGFR-tyrosine kinase inhibitor indicate small cell lung cancer transformation from EGFR positive lung adenocarcinoma? Lung Cancer (Amsterdam, Netherlands) 2013;81(2):302–305. doi: 10.1016/j.lungcan.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 95.Suda K, Murakami I, Sakai K, Mizuuchi H, Shimizu S, Sato K, et al. Small cell lung cancer transformation and T790M mutation: complimentary roles in acquired resistance to kinase inhibitors in lung cancer. Sci Rep. 2015;5:14447. doi: 10.1038/srep14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hui M, Uppin SG, Stalin BJ, Sadashivudu G. Histological transformation of adenocarcinoma to small cell carcinoma lung as a rare mechanism of resistance to epidermal growth factor receptor-tyrosine kinase inhibitors: report of a case with review of literature. Lung India. 2018;35(2):160–163. doi: 10.4103/lungindia.lungindia_347_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Santoni-Rugiu E, Grauslund M, Melchior LC, Costa JC, Sorensen JB, Urbanska EM. Heterogeneous resistance mechanisms in an EGFR exon 19-mutated non-small cell lung cancer patient treated with erlotinib: persistent FGFR3-mutation, localized transformation to EGFR-mutated SCLC, and acquired T790M EGFR-mutation. Lung Cancer (Amsterdam, Netherlands) 2017;113:14–17. doi: 10.1016/j.lungcan.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 98.Tokaca N, Wotherspoon A, Nicholson AG, Fotiadis N, Thompson L, Popat S. Lack of response to nivolumab in a patient with EGFR-mutant non-small cell lung cancer adenocarcinoma sub-type transformed to small cell lung cancer. Lung Cancer (Amsterdam, Netherlands) 2017;111:65–68. doi: 10.1016/j.lungcan.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 99.Wahab A, Kesari K, Chaudhary S, Khan M, Khan H, Smith S, et al. Sequential occurrence of small cell and non-small lung cancer in a male patient: is it a transformation? Cancer Biol Ther. 2017;18(12):940–943. doi: 10.1080/15384047.2017.1394546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu Y, Huang Z, Gong L, Fan Y. A case of resistance to tyrosine kinase inhibitor therapy: small cell carcinoma transformation concomitant with plasma-genotyped T790M positivity. Anticancer Drugs. 2017;28(9):1056–1061. doi: 10.1097/cad.0000000000000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Riel S, Thunnissen E, Heideman D, Smit EF, Biesma B. A patient with simultaneously appearing adenocarcinoma and small-cell lung carcinoma harbouring an identical EGFR exon 19 mutation. Ann Oncol. 2012;23(12):3188–3189. doi: 10.1093/annonc/mds525. [DOI] [PubMed] [Google Scholar]
- 102.Kim WJ, Kim S, Choi H, Chang J, Shin HJ, Park CK, et al. Histological transformation from non-small cell to small cell lung carcinoma after treatment with epidermal growth factor receptor-tyrosine kinase inhibitor. Thorac Cancer. 2015;6(6):800–804. doi: 10.1111/1759-7714.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alì G, Bruno R, Giordano M, Prediletto I, Marconi L, Zupo S, et al. Small cell lung cancer transformation and the T790M mutation: a case report of two acquired mechanisms of TKI resistance detected in a tumor rebiopsy and plasma sample of EGFR-mutant lung adenocarcinoma. Oncol Lett. 2016;12(5):4009–4012. doi: 10.3892/ol.2016.5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alam N, Gustafson KS, Ladanyi M, Zakowski MF, Kapoor A, Truskinovsky AM, et al. Small-cell carcinoma with an epidermal growth factor receptor mutation in a never-smoker with gefitinib-responsive adenocarcinoma of the lung. Clin Lung Cancer. 2010;11(5):E1–E4. doi: 10.3816/CLC.2010.n.046. [DOI] [PubMed] [Google Scholar]
- 105.Jiang SY, Zhao J, Wang MZ, Huo Z, Zhang J, Zhong W, et al. Small-cell lung cancer transformation in patients with pulmonary adenocarcinoma: a case report and review of literature. Medicine. 2016;95(6):e2752. doi: 10.1097/md.0000000000002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin Q, Cai GP, Yang KY, Yang L, Chen CS, Li YP. Case report: small cell transformation and metastasis to the breast in a patient with lung adenocarcinoma following maintenance treatment with epidermal growth factor receptor tyrosine kinase inhibitors. BMC Cancer. 2016;16:593. doi: 10.1186/s12885-016-2623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma AT, Chan WK, Ma ES, Cheng T, Cheng PN. Small cell lung cancer with an epidermal growth factor receptor mutation in primary gefitinib-resistant adenocarcinoma of the lung. Acta Oncol (Stockholm, Sweden) 2012;51(4):557–559. doi: 10.3109/0284186x.2011.636757. [DOI] [PubMed] [Google Scholar]
- 108.Kudo K, Ohyanagi F, Horiike A, Miyauchi E, Yanagitani N, Hoshi R, et al. Clinicopathological findings of non-small-cell lung cancer with high serum progastrin-releasing peptide concentrations. Lung Cancer (Amsterdam, Netherlands) 2011;74(3):401–404. doi: 10.1016/j.lungcan.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 109.Liu Y. Small cell lung cancer transformation from EGFR-mutated lung adenocarcinoma: a case report and literatures review. Cancer Biol Ther. 2018 doi: 10.1080/15384047.2018.1435222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hwang KE, Jung JW, Oh SJ, Park MJ, Shon YJ, Choi KH, et al. Transformation to small cell lung cancer as an acquired resistance mechanism in EGFR-mutant lung adenocarcinoma: a case report of complete response to etoposide and cisplatin. Tumori. 2015;101(3):e96–e98. doi: 10.5301/tj.5000276. [DOI] [PubMed] [Google Scholar]
- 111.Morinaga R, Okamoto I, Furuta K, Kawano Y, Sekijima M, Dote K, et al. Sequential occurrence of non-small cell and small cell lung cancer with the same EGFR mutation. Lung Cancer (Amsterdam, Netherlands) 2007;58(3):411–413. doi: 10.1016/j.lungcan.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 112.Le T, Sailors J, Oliver DH, Mayer M, Hoskin S, Gerber DE. Histologic transformation of EGFR mutant lung adenocarcinoma without exposure to EGFR inhibition. Lung Cancer (Amsterdam, Netherlands) 2017;105:14–16. doi: 10.1016/j.lungcan.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Burkart J, Shilo K, Zhao W, Ozkan E, Ajam A, Otterson GA. Metastatic squamous cell carcinoma component from an adenosquamous carcinoma of the lung with identical epidermal growth factor receptor mutations. Case Rep Pulmonol. 2015;2015:4. doi: 10.1155/2015/283875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Haratani K, Hayashi H, Watanabe S, Kaneda H, Yoshida T, Takeda M, et al. Two cases of EGFR mutation-positive lung adenocarcinoma that transformed into squamous cell carcinoma: successful treatment of one case with rociletinib. Ann Oncol. 2016;27(1):200–202. doi: 10.1093/annonc/mdv495. [DOI] [PubMed] [Google Scholar]
- 115.Hsieh M-S, Jhuang J-Y, Hua S-F, Chou Y-H. Histologic evolution from adenocarcinoma to squamous cell carcinoma after gefitinib treatment. Ann Thorac Surg. 2015;99(1):316–319. doi: 10.1016/j.athoracsur.2014.02.075. [DOI] [PubMed] [Google Scholar]
- 116.Scher KS, Saldivar J-S, Fishbein M, Marchevsky A, Reckamp KL. EGFR-mutated lung cancer with T790M-acquired resistance in the brain and histologic transformation in the lung. J Natl Compr Cancer Netw. 2013;11(9):1040–1044. doi: 10.6004/jnccn.2013.0126. [DOI] [PubMed] [Google Scholar]
- 117.Levin PA, Mayer M, Hoskin S, Sailors J, Oliver DH, Gerber DE. Histologic transformation from adenocarcinoma to squamous cell carcinoma as a mechanism of resistance to EGFR inhibition. J Thorac Oncol. 2015;10(9):e86–e88. doi: 10.1097/JTO.0000000000000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Clery E, Pisapia P, Feliciano S, Vigliar E, Marano A, De Luca C, et al. There is still a role for cytology in the ‘liquid biopsy’ era. A lesson from a TKI-treated patient showing adenocarcinoma to squamous cell carcinoma transition during disease progression. J Clin Pathol. 2017;70(9):798. doi: 10.1136/jclinpath-2017-204370. [DOI] [PubMed] [Google Scholar]
- 119.Park HK, Seo Y, Choi YL, Ahn MJ, Han J. Metastatic squamous cell carcinoma from lung adenocarcinoma after epidermal growth factor receptor tyrosine kinase inhibitor therapy. J Pathol Transl Med. 2017;51(4):441–443. doi: 10.4132/jptm.2016.10.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jukna A, Montanari G, Mengoli MC, Cavazza A, Covi M, Barbieri F, et al. Squamous cell carcinoma “transformation” concurrent with secondary T790M mutation in resistant EGFR-mutated adenocarcinomas. J Thorac Oncol. 2016;11(4):e49–e51. doi: 10.1016/j.jtho.2015.12.096. [DOI] [PubMed] [Google Scholar]
- 121.Sato M, Matsui A, Shimoyama Y, Omote N, Morise M, Hase T, et al. An EGFR-mutated lung adenocarcinoma undergoing squamous cell carcinoma transformation exhibited a durable response to afatinib. Intern Med. 2018;57(23):3429–3432. doi: 10.2169/internalmedicine.0999-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shinohara S, Ichiki Y, Fukuichi Y, Honda Y, Kanayama M, Taira A, et al. Squamous cell carcinoma transformation from adenocarcinoma as an acquired resistance after the EGFR TKI therapy in (EGFR-mutated) non-small cell lung cancer. J Thorac Dis. 2018;10(7):E526–E531. doi: 10.21037/jtd.2018.06.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kuiper JL, Ronden MI, Becker A, Heideman DAM, van Hengel P, Ylstra B, et al. Transformation to a squamous cell carcinoma phenotype of an EGFR-mutated NSCLC patient after treatment with an EGFR-tyrosine kinase inhibitor. J Clin Pathol. 2015;68(4):320–321. doi: 10.1136/jclinpath-2015-202866. [DOI] [PubMed] [Google Scholar]
- 124.Izumi H, Yamasaki A, Ueda Y, Sumikawa T, Maeta H, Nakamoto S, et al. Squamous cell carcinoma transformation from EGFR-mutated lung adenocarcinoma: a case report and literature review. Clin Lung Cancer. 2018;19(1):e63–e66. doi: 10.1016/j.cllc.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 125.Longo L, Mengoli MC, Bertolini F, Bettelli S, Manfredini S, Rossi G. Synchronous occurrence of squamous-cell carcinoma “transformation” and EGFR exon 20 S768I mutation as a novel mechanism of resistance in EGFR-mutated lung adenocarcinoma. Lung Cancer (Amsterdam, Netherlands) 2017;103:24–26. doi: 10.1016/j.lungcan.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 126.Jay G, Jeffrey PG, Weijie M, Ken Y, Elizabeth HM, Megan ED, et al. Squamous cell transformation of primary lung adenocarcinoma in a patient with EML4-ALK fusion variant 5 refractory to ALK inhibitors. J Natl Compr Cancer Netw. 2019;17(4):297–301. doi: 10.6004/jnccn.2019.7291. [DOI] [PubMed] [Google Scholar]
- 127.Park S, Han J, Sun J-M. Histologic transformation of ALK-rearranged adenocarcinoma to squamous cell carcinoma after treatment with ALK inhibitor. Lung Cancer (Amsterdam, Netherlands) 2019;127:66–68. doi: 10.1016/j.lungcan.2018.11.027. [DOI] [PubMed] [Google Scholar]
- 128.Bugano DDG, Kalhor N, Zhang J, Neskey M, William WN. Squamous-cell transformation in a patient with lung adenocarcinoma receiving erlotinib: co-occurrence with T790M mutation. Cancer Treat Commun. 2015;4(Supplement C):34–36. doi: 10.1016/j.ctrc.2015.03.007. [DOI] [Google Scholar]
- 129.Hsu C-L, Chen K-Y, Kuo S-W, Chang Y-L. Histologic transformation in a patient with lung cancer treated with chemotherapy and pembrolizumab. J Thorac Oncol. 2017;12(6):e75–e76. doi: 10.1016/j.jtho.2017.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Pubmed repository. For Table 1, we used one or a combination of the following terms in PubMed and selected articles on the basis of relevance to transition from ADC to SCLC: ‘non-small cell lung cancer’, ‘transition’, ‘transformation’, ‘transdifferentiation’, ‘adenocarcinoma’, ‘small cell carcinoma’ and ‘small cell lung cancer’. All dates and languages are included in the search. We only collect case reports with detailed clinical information. For Table 2, we used one or a combination of the following terms in PubMed and selected articles on the basis of relevance to transformation from ADC to SCC: ‘transition’, ‘transformation’, ‘transdifferentiation’, ‘adenocarcinoma’ and ‘squamous cell carcinoma’. All dates and languages are included in the search. We only collect case reports with detailed clinical information.