Abstract
Background
Phosphoglucomutase (PGM), a key enzyme in the metabolism of glucose-1-phosphate and glucose-6-phosphate, has been found to be associated with proliferation, invasion, and metastasis of cancer. However, the expression and function of PGM5 in colorectal cancer (CRC) remains unknown.
Methods
We tested PGM5 mRNA and protein expression levels in 79 CRC tissue and their matched adjacent tissue samples by qRT-PCR and immunohistochemistry, respectively. Overall survival (OS) was estimated with the Kaplan–Meier method and compared between groups with the log-rank test. We performed multivariable Cox regression analyses to identify factors associated with CRC risk. The cell proliferation, migration and invasion abilities of CRC cells were detected by using CCK-8, Transwell migration and invasion assays, respectively.
Results
The PGM5 protein levels expression in CRC tissues were significantly lower than those in the adjacent tissues (t = 5.035, P < 0.001), and Kaplan–Meier analysis indicated that low PGM5 expression were significantly associated with poor overall survival (P = 0.0069). Univariate and multivariate analyses demonstrated that PGM5 was an independent risk factor for overall survival (hazard ratio = 0.3951, P = 0.014). PGM5 overexpression significantly inhibited the proliferation, invasion and migration abilities of CRC cells. On the contrary, knockdown of PGM5 promotes the invasion and migration of CRC cells.
Conclusions
PMG5 regulates proliferation, invasion, and migration in the CRC and decreased PGM5 is associated with poor prognosis. Therefore, PGM5 is a promising biomarker in CRC and decreased PGM5 may predict poor overall survival in patients with CRC.
Keywords: Phosphoglucomutase 5, Colorectal cancer, Overall survival
Background
Colorectal cancer (CRC) is the leading cause of death in the world and is a major public health problem with increasing incidence and mortality [1]. According to temporal trend analyses from 2000 to 2011, CRC remains a rapidly increasing cancer in China despite recent advances in its early detection and treatment [2].
Current primary treatment options depend on the stages of CRC. If patients are diagnosed at an early stage with small tumours, they have a better 5-year survival rate than those who are diagnosed at later stages, as most metastatic CRCs eventually cause death. However, many patients are already at an advanced stage at diagnosis in clinical practice. Therefore, reliable biomarkers are needed to help make clinical decisions throughout the patient’s disease trajectory to select the optimal oncological and surgical strategies.
Metastasis of cancer is associated with the tumour’s microenvironment. When energy metabolism remodelling occurs in the microenvironment, cell glycolysis activity is enhanced. Oxidative phosphorylation activity is then weakened, and the main energy source of the cell changes from oxidative phosphorylation to glycolysis, which is a new basic feature of tumour cells and tumours. Cell biosynthesis provides ‘raw materials’ and other functions, giving an evolutionary advantage to tumour cells [3]. Phosphoglucomutase (PGM) is a key enzyme in the metabolism of glucose-1-phosphate and glucose-6-phosphate [4], and PGM has been associated with the proliferation, invasion and metastasis of cancer [5, 6]. PGM5, also known as phosphoglycosidase-related protein (PGM-rp) or Aciculin, is localized on human chromosome 9 (9q21.11) and is represented by two closely related 60 kDa and 63 kDa subtypes [7]. PGM5 shows high expression in smooth muscle, skeletal muscle and heart muscle.
The Cancer Genome Atlas (TCGA) database suggested that PGM5 expression was significantly down-regulated in CRC. However, the expression and function of PGM5 in CRC remains unknown. Therefore, this study further confirmed the expression of PGM5 in CRC tissues and identified the function of PGM5 in CRC cell lines.
Materials and methods
The Cancer Genome Atlas data sets
TCGA-COAD and TCGA-READ using Ballgown cancerous and cancerous-adjacent datasets were compared to obtain the significant P value and the fold difference between the q-value and the gene fold change for differential gene screening. These values were obtained with the following conditions: a P-value less than 0.01, a q-value less than 0.05 and a fold change greater than 2. The results were normalized and log2-scaled using GEPIA’s online analysis tool (http://gepia.cancer-pku.cn/). The study was approved by the institutional review board of Liuzhou Municipal Liutie Central Hospital.
Immunohistochemistry
The tissues’ PGM5 expression levels were evaluated by immunohistochemistry (IHC) using CRC samples (Superchip Biotech, Shanghai, China). Anti-Aciculin antibody (1:50; Santa Cruz) was applied as the first antibody, and the avidin–biotin–peroxidase method was used to visualize immunoreactivity. Haematoxylin was used as the counterstain.
The semi-quantitative scanning approach used to evaluate PGM5 expression was based on the scores of the positive cell percentage and positive cell staining intensity. The image analysis software Aperio Image scope version 11.1 (Aperio Technologies, Leicabiosystems; AT) was used to determine the immunostaining intensities. Additionally, integrated optical density (IOD) values were obtained for the tumour areas in each section, and these values were counted and measured using Image-Pro Plus v6.0 software (Media Cybernetics, Inc., Bethesda, MD, USA).
IHC assessment was conducted independently by two investigators, and immunoreactivity in the cytoplasm was quantified as described in a previous study [8]. The staining index was evaluated as follows: staining index = staining intensity (SI) × percentage of positive cells (PP). PP was scored as follows: 0 (˂ 5% positively stained cells), 1 (5 to 24% positively stained cells), 2 (25 to 49% positively stained cells), 3 (50 to 74% positively stained cells) and 4 (75 to 100% positively stained cells). SI was scored as follows: 0 (negative staining), 1 (weak staining), 2 (moderate staining) and 3 (strong staining). The PGM5 expression cut-off value was identified as the median IHC score.
Cell lines and cell culture
The human CRC cell lines HT-29, DLD-1, HCT116, SW480, Caco-2 and LOVO, as well as the normal human colorectal cell FHC, were purchased from Hunan Fenghui Biotechnology Co., Ltd. (China). They were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA) containing 10% foetal bovine serum (FBS; HyClone, USA) at 37 °C in a humidified chamber supplemented with 5% CO2.
Real-time polymerase chain reaction test
The total RNA was extracted from the cell lines or tissues using Trizol reagent (Invitrogen, USA). Using a Reverse Transcription System Kit (Thermo #K1622, USA), cDNA was synthesized from the total RNA. The forward primers for PGM5 were as follows: 5′-TACAGCGTGGCGAAGACGGATAG-3′ and 5′-CTGCGTACAGTCTGAGGGTGGC-3′ for the reverse primers. PGM5 expression was analysed with the real-time polymerase chain reaction (RT-PCR) test using the SYBR Green PCR Kit (Thermo F-415XL, USA). β-Actin was used as a control, and the 2−ΔΔCt method for the relative quantitation of gene expression was used to determine miRNA expression levels.
Western blot analysis
Western blot analysis was performed for detection according to the standard protocol using peroxidase-conjugated secondary antibodies and the enhanced chemiluminescence system (Millipore, USA). The primary antibodies used were PGM5 (1:500; Novus, St. Louis, MO, USA) and beta-actin (1:2000; CST, Danvers, MA, USA, Catalogue #3700).
Cell transfection
Lipofectamin2000 (Invitrogen) was used to transfect siRNA NC, siRNA-1 (TCAATGAAGGTCCCTGTAT), siRNA-2 (GGACTCAGGACGTTGCAAT) and siRNA-3 (GGTCTGGCTCTCCATTATT) into the high-expression PGM5 cell line according to the instructions. The pcDNA3.1-EGFP-PGM5 cloning plasmid was constructed and transfected into the low-expression PGM5 cell line. About 48 h after transfection, RT-PCR was used to detect whether the cell transfection was successful. The overexpression and knockdown efficiency were shown in Additional file 1: Figure S1.
CCK-8 test
The cells were inoculated into a 96-well plate at 5 × 103 cells for each well, and five parallel holes were set in each group. The cells were cultured for 12, 24, 36, 48, 72 and 96 h. CCK-8 was added to each well at 37 °C in a 5% CO2 incubator, and the cells were incubated in the dark for 1 to 4 h. A 450-nm wavelength microplate reader was used to measure the OD (Optical Density) value at the same time point, and the effect of cell growth was analysed using the measured OD value.
Migration and invasion assay
In the migration assay, the control and transfected cells at 5 × 105/well were seeded into the upper chambers containing DMEM with 1% FBS. About 600 µl of DMEM with 10% FBS was added to the lower chambers, and the cells were cultured at 37 °C in a CO2 incubator for 24 h. Afterwards, the upper chamber was removed, and the immigrated cells were carefully wiped from the membrane with a cotton swab. The cells were rinsed twice in a 37 °C pre-warmed PBS solution and were fixed with 4% paraformaldehyde for 30 min. Then, they were stained with haematoxylin for 5 min. Next, the cells were washed with distilled water and photographed under a microscope.
For the invasion assay, the upper chamber membranes were pre-coated with 50 µl of Matrigel (1.25 mg/ml). The control and transfected cells at 5 × 105/well were seeded into the upper chambers containing DMEM with 1% FBS. The bottom chambers were filled with DMEM with 10% FBS containing chemokine and were cultured at 37 °C in a CO2 incubator for 24 h. Afterwards, the cells on the upper side of the upper-chamber membranes were removed. The invasion cells on the lower side of the membranes were fixed with 4% paraformaldehyde for 30 min and stained with haematoxylin for 5 min. The chambers were carefully removed from the upper chamber substrate; they were then mounted and counted under a microscope. A 200-fold light microscope was used to select and average five field counts randomly.
Statistical analysis
All experiments were repeated three times independently, and the data were expressed as the mean ± SD. Statistical analysis was performed using SPSS 22.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 7 software. The relationship between PGM5 expression and clinicopathologic characteristics was measured using χ2 or Fisher’s exact tests. Overall survival (OS) was estimated using the Kaplan–Meier method. Univariate and multivariate survival analyses were performed using the Cox proportional hazard model with a forward stepwise procedure. A P-value less than 0.05 was considered statistically significant.
Results
Lower PGM5 expression in CRC
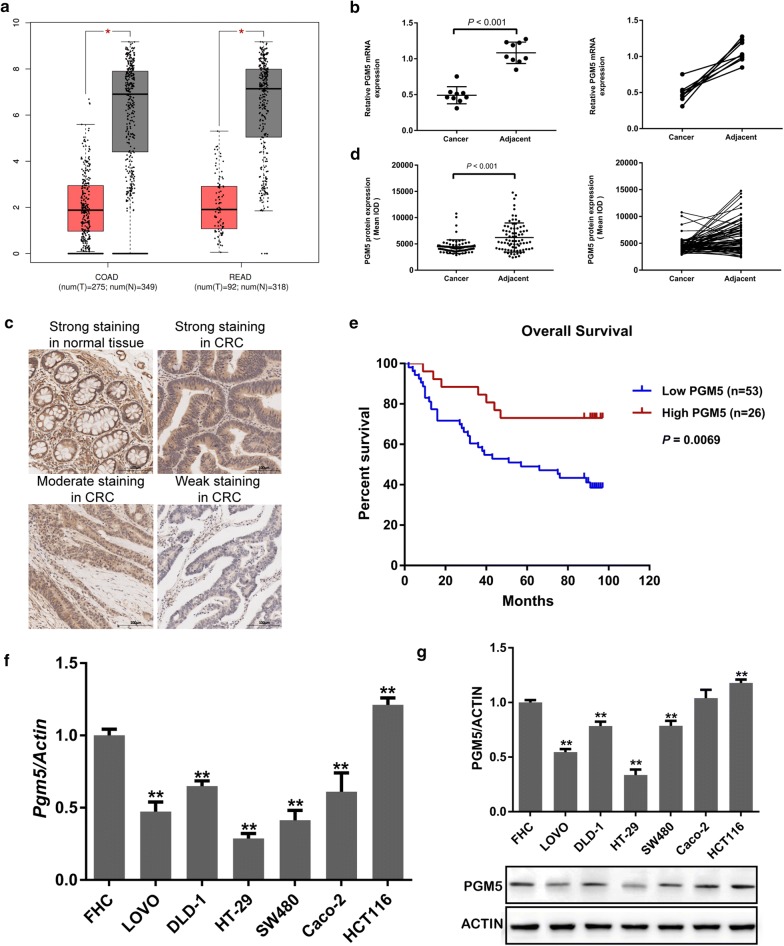
In TCGA database, PGM5 expression was found to be down-regulated significantly in gastrointestinal cancers, specifically COAD and READ (Fig. 1a). PGM5 expression in CRC was 0.17 times the expression in normal tissue (P < 0.0001). Nine pairs of clinical CRC tissues and matched adjacent tissues were selected to detect the expression levels of PGM5 miRNA and protein using RT-PCR. PGM5 miRNA expression in CRC tissues was significantly lower than in the adjacent tissues (t = 9.281, P < 0.001; Fig. 1b). To confirm these findings, PGM5 protein was examined using IHC in 79 CRC tissues (Fig. 1c), and IOD representing the expression level from 79 CRC tumours and the paired adjacent tissues was submitted to GraphPad Prism software. As shown in Fig. 1d, PGM5 expression was frequently lower in CRC tumour tissue than in the adjacent tissue (t = 5.035, P < 0.001). This suggested that the expression of PGM5 was down-regulated in CRC tissues.
Fig. 1.
Downregulated PGM5 in CRC is associated with poor prognosis. a The PGM5 expression on Box Plotsacross all tumor samples and paired normal tissues in TCGA (Match TCGA normal and GTEx data, P < 0.001). Each dots represent expression of samples; b the expression level of PGM5 mRNA using RT-PCR; c representative images of PGM5 IHC staining in CRC and normal tissues (×200, scale bar 100 μm); d PGM5 protein expression in CRC tissues and adjacent tissues; e Kaplan–Meier analysis of the relationship between overall survival and PGM5 expression in patients with CRC; f The PGM5 expression in CRC cells. PGM phosphoglucomutase, CRC colorectal cancer, TCGA The Cancer Genome Atlas, IOD integrated optical density, qRT-PCR quantitative real-time PCR, IHC immunohistochemistry; **< 0.001
Decreased PGM5 in CRC associated with poor prognosis
To further evaluate PGM5 expression as a diagnostic marker for CRC, ROC curve analysis was performed, and the optimal cut-off value of PGM5 expression with the best discriminatory power was determined to be 6. Therefore, 79 immunostained CRC tissue samples were classified into two groups: high expression (index > 6, n = 26) and low expression (index ≤ 6, n = 53). As shown in Table 1, low PGM5 expression was significantly associated with lymph node metastasis (P = 0.041), clinical stage (P = 0.039) and poor survival (P = 0.005).
Table 1.
Associations of PGM5 expression with clinicopathological factors in CRC patients
| Variable | Number | PGM5 level | P value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 0.574 | |||
| < 65 | 27 | 17 (21.5%) | 10 (12.7%) | |
| ≥ 65 | 52 | 36 (45.6%) | 16 (20.3%) | |
| Gender | 0.314 | |||
| Male | 39 | 25 (31.6%) | 14 (17.7%) | |
| Female | 40 | 28 (35.4%) | 12 (15.2%) | |
| Tumor size (cm) | 0.083 | |||
| < 5 | 27 | 19 (24.1%) | 8 (10.1%) | |
| ≥ 5 | 52 | 34 (43.0%) | 18 (22.8%) | |
| Lymph node metastasis | 0.041 | |||
| N0 | 48 | 28 (35.4%) | 20 (25.3%) | |
| N1 | 22 | 16 (20.3%) | 6 (7.6%) | |
| N2 | 9 | 9 (11.4%) | 0 (24.1%) | |
| Clinical stage | 0.039 | |||
| I–II | 48 | 20 (25.3%) | 28 (35.4%) | |
| III–IV | 31 | 6 (7.6%) | 25 (31.6%) | |
| Survival | 0.005 | |||
| Live | 40 | 21 (26.6%) | 19 (24.1%) | |
| Dead | 39 | 32 (40.5%) | 7 (8.9%) | |
Italic values indicate significance of P value (P < 0.05)
PGM phosphoglucomutase, CRC colorectal cancer
However, a statistically significant association between PGM5 expression and age, gender or tumour size was not found. Thus, the prognostic significance of PGM5 expression for CRC patients was further investigated. The Kaplan–Meier analysis using the most efficient cut-off value of PGM5 expression indicated that low PGM5 expression was significantly associated with poor overall survival compared with high PGM5 expression (P = 0.0069; Fig. 1e). Furthermore, univariate analysis showed that gender (P = 0.026), lymph node metastasis (P < 0.001), clinical stage (P = 0.001) and PGM5 expression (P = 0.001) were significant predictors of overall survival in CRC. Multivariate analysis indicated that only PGM5 expression (P = 0.015) was an independent predictive factor for poor outcomes in CRC patients (Table 2). These findings suggested that low PGM5 expression in CRC is correlated with poor prognosis of patients.
Table 2.
Univariate and multivariate analysis
| Clinical features | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Gender | 1.749 (1.069–2.862) | 0.026 | 1.535 (0.897–2.627) | 0.118 |
| Age | 1.021 (0.996–1.047) | 0.098 | ||
| Tumor size | 1.416 (0.816–2.455) | 0.216 | ||
| Lymph node metastasis | 1.928 (1.419–2.621) | < 0.001 | 1.329 (0.704–2.507) | 0.380 |
| Clinical stage | 2.393 (1.403–4.080) | 0.001 | 1.387 (0.474–4.060) | 0.551 |
| PGM5 expression | 0.302 (0.150–0.611) | 0.001 | 0.395 (0.189–0.826) | 0.014 |
Italic values indicate significance of P value (P < 0.05)
PGM phosphoglucomutase, HR hazard ratio, CI confidence interval
PGM5 as a tumour suppressor in CRC
To evaluate the possible role of PGM5 in CRC and choose the CRC cell lines used for silencing or expressing PGM5, PGM5 expression was measured in six CRC cell lines as well as FHC cells. RT-PCR and Western blot analysis results indicated that PGM5 expression was highest in the HCT116 cell line (P = 0.005) and lowest in the HT-29 cell line (P < 0.001; Fig. 1f, g). The silencing effect of siRNA-3 was most significant among those of the control group and the negative control group (P = 0.001). Therefore, the HCT116 cell line was selected for silencing PGM5 using siRNA-3, and the HT-29 cell line was selected for overexpressing PGM5.
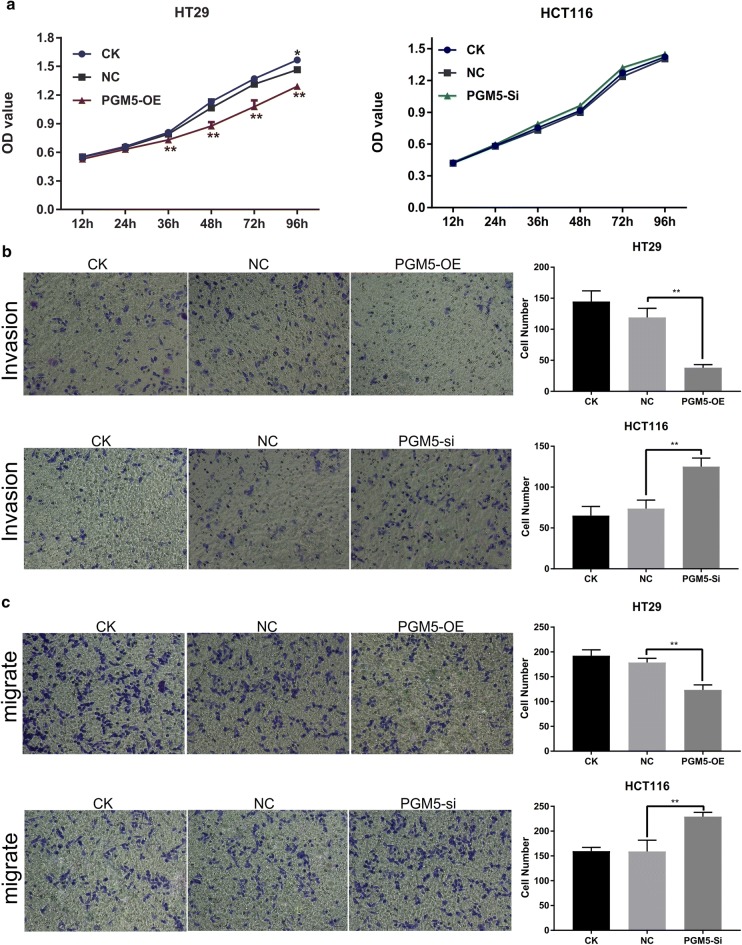
The up-regulation of PGM5 remarkably inhibited the cell’s proliferation (Fig. 2a), invasion (Fig. 2b) and migration abilities (Fig. 2c). PGM5 silencing did not significantly promote cell proliferation (Fig. 2a), but it remarkably promoted cell invasion and migration (Fig. 2b, c). These results suggested that PGM5 was a tumour suppressor in CRC.
Fig. 2.
PGM5 is a tumor suppressor in CRC. a Upregulation of PGM5 remarkably inhibits the proliferation in HT29; b upregulation of PGM5 remarkably inhibits the invasion in HT29 and silence of PGM5 remarkably promotes the invasion in HCT116; c upregulation of PGM5 remarkably inhibits the migration in HT29 and silence of PGM5 remarkably promotes the migration in HCT116. PGM phosphoglucomutase, CRC colorectal cancer, CK normal cell group, NC negative control, OE over expression, si silence. **< 0.001
Discussion
The PGM superfamily contains five proteins: PGM1, PGM2, PGM2L1, PGM3 and PGM5. Although these five proteins share a homology in sequence coding, their substrates or functions are different and have different biological effects. For example, PGM1 gene defects damage hepatocyte homeostasis via the central metabolic pathway and then affect the growth of liver cancer cells [9]. PGM3, on the other hand, is an N-acetylglucosamine triphosphatase involved in the biosynthesis of the amino alanine, and it plays an anti-cancer role. For example, sulforaphane can reduce PGM3 expression in prostate cancer cells by inducing apoptosis [6]. PGM5 has a concentrated expression where muscles are connected [10], and it participates in myofibril formation, maintenance and transformation [11]. PGM5 is a binding partner for dystrophin; it binds to the N- and C-termini of dystrophin and reduces their expression [12]. In addition, PGM5 expression is associated with bipolar disorder [13] and heart disease [11]. However, the expression and function of PGM5 in CRC is still unknown.
Only a few studies have been conducted on the expression and role of PGM5 in cancer. In the gene chip of prostate cancer, the PGM5 antisense RNA1 (PGM5-AS1) expression was found to be down-regulated [14]. PGM5 expression was similarly down-regulated in bladder cancer tissues, but validation in 34 pairs of tissues showed that PGM5 expression in cancer and adjacent tissues was not significantly different (up-regulation in 15 cases and down-regulation in 19 cases) [15]. Therefore, PGM5 is not consistently expressed in cancer with different tissue types, and it may be related to the muscle content.
Recent studies have shown that PGM5 is down-regulated in colorectal adenomas or adenocarcinomas [16]. In this study, PGM5 was found to be decreased significantly in human CRC and was correlated with poor overall survival. Moreover, decreased PGM5 was an independent predictive factor for poor outcomes in CRC patients. Therefore, the data suggested that PGM5 could play an important role in the pathogenesis and development of CRC. Functionally, the up-regulation of PGM5 was found to inhibit the proliferation, migration and invasion of CRC cells, while the down-regulation of PGM5 exerted a contrary role. This strongly suggested that PGM5 was a novel tumour suppressor in CRC.
The tumour metastasis process is complex and involves many factors, including not only the changes in the tumour cells themselves but also the changes in the surrounding microenvironment. The tumour microenvironment, a key factor in tumour metastasis, is a dynamic network composed of tumour cells, the extracellular matrix and interstitial tissues [17, 18]. Rapid proliferation is one of the characteristics of tumour cells and requires a large amount of biological matter and energy; this proliferation changes the metabolic pattern and performs metabolic remodelling [19]. Therefore, reconstructing the energy metabolism system is also one of the characteristics of tumours [20].
PGM is a key enzyme in glucose metabolism, and PGM deficiency causes abnormal metabolism of glycogen, which leads to the emergence of various diseases [21]. Specifically, PGM5 encodes a glucose phosphate mutase, which plays a key role in carbohydrate metabolism [7]. Thus, this study hypothesized that a decrease in PGM5 could lead to an imbalance in energy metabolism, a metabolic remodelling of tumour cells and an abnormal increase in the aerobic glycolysis pathway in the tumour cells.
The glycolytic pathway not only provides a substrate for the biosynthesis of tumour cell proliferation but also produces a large amount of lactic acid. This lactic acid is released outside the cell, and this acidic environment promotes the cell’s proliferation, invasion, migration and so on [22, 23]. Due to the changes in tumour cells’ energy metabolism pathways, more tumour cells are needed to absorb enough glucose to meet their energy, biosynthesis and redox requirements. This, in turn, promotes tumorigenesis, tumour proliferation and invasion [3].
This study did face some limitations. These included the relatively small size of the study and relatively small statistical power. However, the results from TCGA database supported this study’s results.
Conclusion
In conclusion, PGM5 was demonstrated to function as a tumour suppressor gene that is down-regulated in CRC. Moreover, lower expression of PGM5 was correlated with poorer prognosis in CRC patients. Functionally, the up-regulation of PGM5 was found to inhibit the proliferation, migration and invasion of CRC cells. Together, these findings suggested that PGM5 may serve as a potential novel predictive and therapeutic target for CRC.
Supplementary information
Additional file 1: Figure S1. (A) The silence efficiency in HT29 cell line; (B) The over expression efficiency in HCT116 cell line. OE: over expression; si: silence **< 0.001.
Acknowledgements
Not applicable.
Abbreviations
- PGM
phosphoglucomutase
- CRC
colorectal cancer
- TCGA
The Cancer Genome Atlas
- IOD
integrated optical density
- qRT-PCR
quantitative real-time PCR
Authors’ contributions
YFS and YQS designed the experiments; HHL, SL and LPP conducted the experiments; HHL, QQY and TWL provided research materials and methods; YFS and JLC analysed data; and YFS and YQS wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was financially supported by the National Natural Science Foundation of China (81760501); Guangxi Natural Science Foundation (2018GXNSFAA281034, 2018GXNSFAA294029, 2017GXNSFAA198041); Liuzhou Scientific Research and Technological Development Programs (2018BJ10506, 2018DH10504); Youth Science Foundation of Guangxi Medical University (GXMUYSF201730).
Availability of data and materials
The datasets used in this study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Liuzhou Municipal Liutie Central Hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yifan Sun, Haihua Long and Lin Sun contributed equally to this work, so they should be considered as co-first authors
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12935-019-0967-y.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 4.Muenks AG, Stiers KM, Beamer LJ. Sequence-structure relationships, expression profiles, and disease-associated mutations in the paralogs of phosphoglucomutase 1. PLoS ONE. 2017;12(8):e0183563. doi: 10.1371/journal.pone.0183563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis M, Kenny HA, Ashcroft B, Mukherjee A, Johnson A, Zhang Y, Helou Y, Batlle R, Liu X, Gutierrez N, et al. Fibroblasts mobilize tumor cell glycogen to promote proliferation and metastasis. Cell Metabol. 2018;29(1):141–155. doi: 10.1016/j.cmet.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee CH, Jeong SJ, Yun SM, Kim JH, Lee HJ, Ahn KS, Won SH, Kim HS, Lee HJ, Ahn KS, et al. Down-regulation of phosphoglucomutase 3 mediates sulforaphane-induced cell death in LNCaP prostate cancer cells. Proteome Sci. 2010;8:67. doi: 10.1186/1477-5956-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards YH, Putt W, Fox M, Ives JH. A novel human phosphoglucomutase (PGM5) maps to the centromeric region of chromosome 9. Genomics. 1995;30(2):350–353. doi: 10.1006/geno.1995.9866. [DOI] [PubMed] [Google Scholar]
- 8.Hou Z, Guo K, Sun X, Hu F, Chen Q, Luo X, Wang G, Hu J, Sun L. TRIB2 functions as novel oncogene in colorectal cancer by blocking cellular senescence through AP4/p21 signaling. Mol Cancer. 2018;17(1):172. doi: 10.1186/s12943-018-0922-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroder PC, Segura V, Riezu JI, Sangro B, Mato JM, Prieto J, Santamaria E, Corrales FJ. A signature of six genes highlights defects on cell growth and specific metabolic pathways in murine and human hepatocellular carcinoma. Funct Integr Genomics. 2011;11(3):419–429. doi: 10.1007/s10142-011-0230-7. [DOI] [PubMed] [Google Scholar]
- 10.Belkin AM, Klimanskaya IV, Lukashev ME, Lilley K, Critchley DR, Koteliansky VE. A novel phosphoglucomutase-related protein is concentrated in adherens junctions of muscle and nonmuscle cells. J Cell Sci. 1994;107(Pt 1):159–173. doi: 10.1242/jcs.107.1.159. [DOI] [PubMed] [Google Scholar]
- 11.Molt S, Buhrdel JB, Yakovlev S, Schein P, Orfanos Z, Kirfel G, Winter L, Wiche G, van der Ven PF, Rottbauer W, et al. Aciculin interacts with filamin C and Xin and is essential for myofibril assembly, remodeling and maintenance. J Cell Sci. 2014;127(Pt 16):3578–3592. doi: 10.1242/jcs.152157. [DOI] [PubMed] [Google Scholar]
- 12.Wakayama Y, Inoue M, Kojima H, Murahashi M, Shibuya S, Yamashita S, Oniki H. Aciculin and its relation to dystrophin: immunocytochemical studies in human normal and Duchenne dystrophy quadriceps muscles. Acta Neuropathol. 2000;99(6):654–662. doi: 10.1007/s004010051176. [DOI] [PubMed] [Google Scholar]
- 13.Lee HJ, Woo HG, Greenwood TA, Kripke DF, Kelsoe JR. A genome-wide association study of seasonal pattern mania identifies NF1A as a possible susceptibility gene for bipolar disorder. J Affect Disord. 2013;145(2):200–207. doi: 10.1016/j.jad.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qianlin X, Menglin S, Tao D, Yanjun Z, Jun H, Jianghua Z. Screening for defferential genes of the prostate cancer and bioinformatics analysis of their interaction. China Oncol. 2017;27(03):169–176. [Google Scholar]
- 15.Dong H, Chenghua L, Yaping G, Xu Z, Guang Y, Tao S. Screening and verification for the relevant genes of bladder cancer. Acad J Chin PLA Med Sch. 2017;38(04):323–327. [Google Scholar]
- 16.Uzozie AC, Selevsek N, Wahlander A, Nanni P, Grossmann J, Weber A, Buffoli F, Marra G. Targeted proteomics for multiplexed verification of markers of colorectal tumorigenesis. Mol Cell Proteom. 2017;16(3):407–427. doi: 10.1074/mcp.M116.062273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spano D, Zollo M. Tumor microenvironment: a main actor in the metastasis process. Clin Exp Metas. 2012;29(4):381–395. doi: 10.1007/s10585-012-9457-5. [DOI] [PubMed] [Google Scholar]
- 18.McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16(8):717–727. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Tegtmeyer LC, Rust S, van Scherpenzeel M, Ng BG, Losfeld ME, Timal S, Raymond K, He P, Ichikawa M, Veltman J, et al. Multiple phenotypes in phosphoglucomutase 1 deficiency. N Engl J Med. 2014;370(6):533–542. doi: 10.1056/NEJMoa1206605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaneton B, Gottlieb E. PGAMgnam style: a glycolytic switch controls biosynthesis. Cancer Cell. 2012;22(5):565–566. doi: 10.1016/j.ccr.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Bohme I, Bosserhoff AK. Acidic tumor microenvironment in human melanoma. Pigment Cell Melanoma Res. 2016;29(5):508–523. doi: 10.1111/pcmr.12495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. (A) The silence efficiency in HT29 cell line; (B) The over expression efficiency in HCT116 cell line. OE: over expression; si: silence **< 0.001.
Data Availability Statement
The datasets used in this study are available from the corresponding author upon reasonable request.