ABSTRACT
In humans, ribosome biogenesis mainly occurs in nucleoli following two alternative pre-rRNA processing pathways differing in the order in which cleavages take place but not by the sites of cleavage. To uncover the role of the nucleolar NAD+-dependent deacetylase sirtuin 7 in the synthesis of ribosomal subunits, pre-rRNA processing was analyzed after sirtinol-mediated inhibition of sirtuin 7 activity or depletion of sirtuin 7 protein. We thus reveal that sirtuin 7 activity is a critical regulator of processing of 45S, 32S and 30S pre-rRNAs. Sirtuin 7 protein is primarily essential to 45S pre-rRNA cleavage at site 2, which is the first step of processing pathway 2. Furthermore, we demonstrate that sirtuin 7 physically interacts with Nop56 and the GAR domain of fibrillarin, and propose that this could interfere with fibrillarin-dependent cleavage. Sirtuin 7 depletion results in the accumulation of 5′ extended forms of 32S pre-rRNA, and also influences the localization of fibrillarin. Thus, we establish a close relationship between sirtuin 7 and fibrillarin, which might determine the processing pathway used for ribosome biogenesis.
KEY WORDS: Pre-rRNA processing, Ribosome, Sirtinol, SIRT7, Nucleolus, Deacetylase
Summary: In humans, sirtuin 7 plays a key role in the synthesis of ribosomal subunits by controlling critical endonucleolytic cleavages of precursor ribosomal RNAs.
INTRODUCTION
Nucleoli are membrane-lacking nuclear bodies that constitute the ribosome factories of all eukaryotic cells (Brown and Gurdon, 1964). They are the sites where most steps of ribosome biogenesis take place, that is, in humans, RNA polymerase I-dependent transcription of the 47S precursor ribosomal RNA (47S pre-rRNA), processing of 47S pre-rRNA into mature 18S, 5.8S and 28S rRNAs, and assembly with ribosomal proteins and 5S rRNA (Hadjiolov, 1985). The organization and size of nucleoli are consequently directly related to ribosome production (Smetana and Busch, 1974). In mammalian cells, three nucleolar components can be discerned by electron microscopy: the fibrillar centers (FCs), the dense fibrillar component (DFC) and the granular component (GC). Ribosomal gene (rDNA) transcription takes place at the junction between the FCs and DFC, while processing of 47S pre-rRNA starts in the DFC and continues during its intra-nucleolar migration towards the GC (Cmarko et al., 2000). Accordingly, processing factors implicated in the early stages of pre-rRNA processing are localized in the DFC whereas those implicated in later stages are enriched in the GC (Hernandez-Verdun et al., 2010).
Several hundred pre-rRNA processing factors are involved in ribosome biogenesis (Tafforeau et al., 2013). Ribosomal proteins, non-ribosomal proteins and small nucleolar ribonucleoprotein complexes (snoRNPs) co- and post-transcriptionally associate with 47S pre-rRNAs to form the 90S pre-ribosomal particle (Grandi et al., 2002), also named the small subunit (SSU) processome (Dragon et al., 2002), which is rapidly processed into 40S and 60S pre-ribosomal particles. These pre-ribosomal particles are further matured to generate the 40S and 60S ribosomal subunits. The maturation of pre-ribosomal particles occurs concomitantly with pre-rRNA processing and modification. Pre-rRNAs undergo a huge number of modifications, which are mainly guided by small nucleolar (sno)RNAs. For instance, the most abundant pre-rRNA modifications, namely, 2′-O-methylation of the ribose and the isomerization of uridine to pseudouridine, are introduced by box C/D and box H/ACA snoRNPs, respectively (Sloan et al., 2017). Furthermore, the acetylation of two cytosine nucleotides, which is highly conserved in yeast, plant and human 18S rRNA, has recently been shown to be guided by two box C/D snoRNAs in yeast, namely snR4 and snR45 (Sharma et al., 2017). Besides modifications introduced by snoRNPs, eukaryotic ribosomes contain different base modifications introduced by base-modifying enzymes, such as base methylations introduced by RNA methyltransferases. Even if rRNA modifications occur throughout ribosome biogenesis, it is generally admitted that sno-RNA-guided modifications are mostly introduced in early pre-ribosomal complexes. Interestingly, partial rRNA modifications have recently been observed, which reveal subpopulations of ribosomes, possibly specialized in specific functions, suggesting that rRNA modifications can also be regulated (Sloan et al., 2017). Moreover, there is growing evidence that rRNA modifications play important roles in development, genetic diseases and cancer (Sloan et al., 2017). Ribosome diversification may also be based on incorporation of different ribosomal protein isoforms or post-translational modification of ribosomal proteins. The post-translational modifications may include acetylation, methylation, phosphorylation, ubiquitination and O-GlcNAcylation (Simsek and Barna, 2017; Shi and Barna, 2015).
The progressive processing of human 47S pre-rRNA is rather well documented (Henras et al., 2015; Mullineux and Lafontaine, 2012; Tafforeau et al., 2013). It comprises several steps (Fig. S1) that lead to the removal of 5′- and 3′-external transcribed spacers (ETS) and internal transcribed spacers (ITS) 1 and 2. Initial cleavages occur at sites 01 and 02 within 5′- and 3′-ETS and lead to 45S pre-rRNA whose maturation can follow either of two alternative pathways. The two pathways do not differ in the sites of cleavage but in the order in which cleavages take place. In pathway 1, the first cleavages take place in the 5′-ETS extremity at sites A0 and 1 whereas in pathway 2 the first cleavage occurs in ITS1 at site 2 (Henras et al., 2015; Mullineux and Lafontaine, 2012). Pathway 2 is the major processing pathway in HeLa cells. While our knowledge of ribosome biogenesis in humans was lagging behind and often based on that of budding yeast, Tafforeau et al. conducted a systematic screening of pre-rRNA processing factors (Tafforeau et al., 2013). Thereby, the authors first confirmed the involvement of well-known factors such as the four common core box C/D proteins, namely 15.5kD (also known as SNU13 or NHP2L1), Nop56, Nop58, the methyltransferase fibrillarin and the U3-55K (also known as RRP9). These proteins are known to be associated with U3 box C/D snoRNA to form the U3 box C/D snoRNP (Watkins et al., 2004) that is required for pre-rRNA processing (Langhendries et al., 2016). Beyond confirming the involvement of these proteins, the authors identified 286 nucleolar proteins required for pre-rRNA processing, depletion of which induces accumulation or reduction of pre-rRNAs. Among the new pre-rRNA processing factors, the NAD+-dependent deacetylase sirtuin 7 was already known to be involved in rDNA transcription in humans (Ford et al., 2006; Grob et al., 2009) and has since been reported to be essential for early pre-rRNA processing via deacetylation of U3-55K (Chen et al., 2016).
To further decipher the key role of sirtuin 7 in pre-rRNA processing, we here analyze the effects of sirtinol, a selective sirtuin inhibitor that effectively inhibits the sirtuin 7 catalytic activity, or of siRNA-mediated sirtuin 7 depletion. Sirtuin 7 involvement in early events of pre-rRNA processing was also clarified by comparing pre-rRNA profiles in cells depleted for sirtuin 7, fibrillarin or both sirtuin 7 and fibrillarin. First, we demonstrate that sirtuin 7 regulates 45S, 32S and 30S pre-rRNA processing, and is primarily involved in cleavage at site 2. Second, we establish that sirtuin 7 directly interacts with the GAR domain of fibrillarin and Nop56, and influences fibrillarin localization. We propose that sirtuin 7 interferes with fibrillarin-dependent cleavage and might be critical in determining which processing pathway ribosome biogenesis will follow.
RESULTS
In human cells, rDNA transcription results in the synthesis of the 47S pre-rRNA, which is cleaved at both ends to generate the 45S pre-rRNA and then processed by two alternative pathways to produce mature 18S, 5.8S and 28S rRNAs (Fig. S1; Mullineux and Lafontaine, 2012).
Sirtuin 7 activity is likely involved in the regulation of several pre-rRNA processing events
It was previously reported that the activity of the nucleolar sirtuin 7 is involved in rDNA transcription (Ford et al., 2006; Grob et al., 2009) and we speculated that sirtuin 7 activity may be implicated more generally in ribosome biogenesis. To address this question, we analyzed the fate of metabolically labeled pre-rRNAs in asynchronous HeLa cells treated or not treated with sirtinol (Fig. 1). Sirtinol is considered to selectively inhibit all human sirtuins; however, this was previously only verified by in vitro deacetylase assays for nuclear sirtuin 1 and cytoplasmic sirtuin 2 (Grozinger et al., 2001; Mai et al., 2005). Therefore, we first tested the effect of sirtinol on the capacity of sirtuin 7 to mediate deacetylation of the acetylated lysine 18 of histone H3 (H3K18-Ac) in vitro (Barber et al., 2012). To verify the specificity of our assay, we also tested the effect of EX-527, a selective inhibitor of sirtuin 1 (Fig. 1A). For this purpose, purified recombinant sirtuin 7 was preincubated with either 100 µM of sirtinol or EX-527 for 15 min at room temperature and in vitro deacetylase assays were performed on 100 µM fluorescent H3K18-Ac peptide in the presence of 6 mM NAD+ for 2 h at 37°C. Deacetylation of H3K18-Ac peptide was monitored and quantified as previously described (Duval et al., 2015) using reverse phase ultra-fast liquid chromatography (RP-UFLC). As observed in Fig. 1A, sirtinol significantly inhibited sirtuin 7 deacetylase activity in contrast to what was seen with EX-527, which induced, as expected, no significant effect on sirtuin 7. The inhibition of sirtuin 7 activity in sirtinol-treated cells was confirmed by performing the same in vitro deacetylase assay on sirtuin 7 immunoprecipitated from HEK-293T cells treated with sirtinol (Fig. S2). For this purpose, cells transfected with pCMV-Tag2-SIRT7 were treated with 100 µM sirtinol or vehicle (DMSO) for the last 6 h of culture. The in vitro deacetylase assay carried out after immunoprecipitation of Flag-SIRT7 showed that sirtuin 7 activity is clearly decreased by sirtinol treatment (Fig. S2). Thus, we confirm that sirtuin 7 activity is indeed inhibited in cells treated with sirtinol.
Fig. 1.
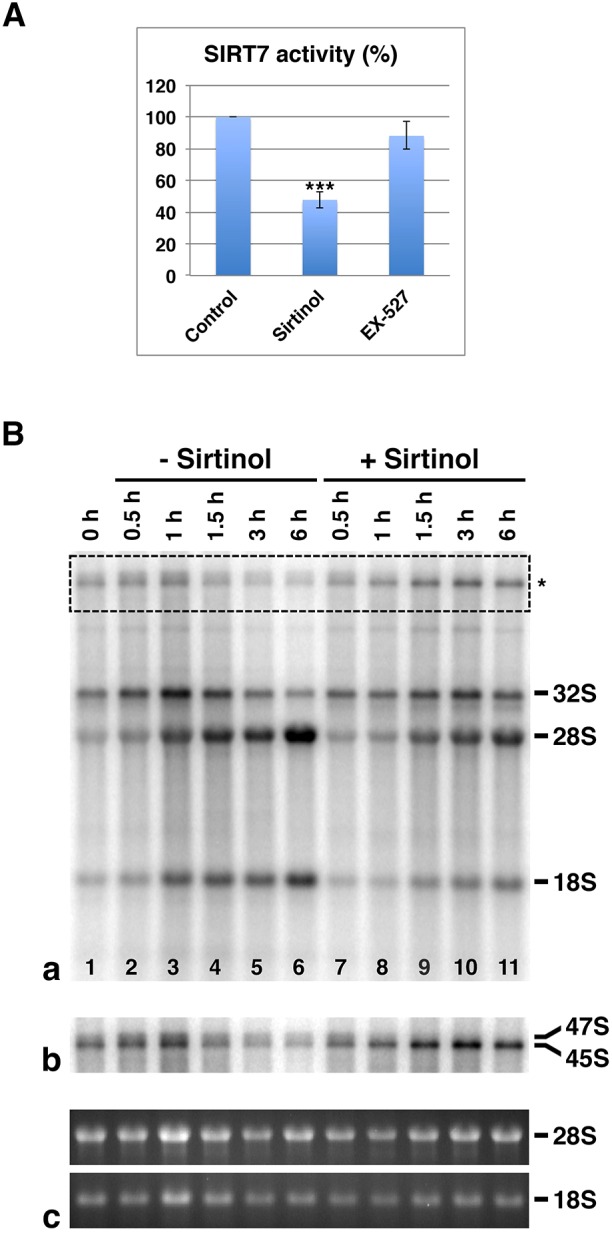
Sirtuin 7 activity is not only involved in the regulation of rDNA transcription. (A) In vitro sirtuin 7 deacetylase assays using fluorescent H3K18-Ac peptides performed with or without preincubation of full-length recombinant human sirtuin 7 with sirtinol or EX-527. Results represent mean±s.d. (n=3). ***P<0.001 compared with control (ANOVA and Dunnett's t-test). (B) Asynchronous HeLa cells were pulse labeled with [32P]orthophosphate for 2 h and chased in nonradioactive medium with or without sirtinol for various times up to 6 h. Total RNAs were isolated, resolved on the same 1% agarose formaldehyde gel and detected by ethidium bromide staining. The RNAs were then transferred to a positively charged membrane and UV cross-linked. (a) Autoradiography of newly synthesized RNAs before chase (lane 1) and after increasing times of chase in the absence (lanes 2–6) and in the presence of sirtinol (lanes 7–11). (b) The upper part of the autoradiography (asterisk) after applying a global intensity adjustment. (c) Detection of RNAs loaded on the gel by ethidium bromide staining.
Then, to analyze the fate of pre-rRNAs in sirtinol-treated cells, asynchronous HeLa cells were pulse labeled with [32P] orthophosphate in the absence of sirtinol and chased with or without sirtinol for various times up to 6 h. Total RNA was isolated from cells, separated by electrophoresis, transferred to positively charged membranes, and autoradiography was performed (Fig. 1Ba). Results were consistent with a decrease in rDNA transcription in sirtinol-treated cells as indicated by the fact that global [32P] incorporation (i.e. newly synthesized rRNAs), was higher in untreated (Fig. 1Ba, lanes 2–6) than in treated (Fig. 1Ba, lanes 7–11) cells. However, the decrease of [32P]-incorporation in sirtinol-treated cells contrasted with the increase of [32P]-labeled 45S pre-rRNA, that is the lower band of the doublet observed in the upper part of the autoradiographic image (Fig. 1Ba, asterisk). This increase was more evident after applying a global intensity adjustment (Fig. 1Bb) and could not be explained by a variation in the amounts of RNA loaded in the gel, as shown by the mature 28S and 18S rRNAs stained with ethidium bromide in the gel (Fig. 1Bc). This suggested that the processing of 45S pre-rRNA (Fig. S1) is impaired by sirtinol treatment.
To further investigate sirtinol interference with pre-rRNA processing, total RNAs were analyzed by quantitative (q)PCR following treatment of HeLa cells for 6 h with sirtinol or either of two different rDNA transcription inhibitors, namely, CX-5461 and AMD (Fig. 2A). Primers ETS1 and ETS2 (Fig. S1) allowed us to quantify unprocessed 47S pre-rRNA, while primers ETS3 and ETS4 were used to quantify 47S–30S pre-rRNAs (47S, 45S and 30S pre-rRNAs), and thus assess alterations in the first steps of pre-rRNA processing. Quantification of 47S pre-rRNA levels revealed that sirtinol induces a decrease of 47S pre-rRNA similar to CX-5461 but less pronounced than AMD (Fig. 2Aa). Given the involvement of sirtuin 7 activity in rDNA transcription (Ford et al., 2006; Grob et al., 2009), the decrease most probably reflects inhibition of rDNA transcription. However, since the amount of 47S pre-rRNA is directly dependent on 47S pre-rRNA synthesis and processing, the sirtinol-induced decrease could therefore be modulated by a sirtinol-induced effect on 47S pre-rRNA processing. Quantification of 47S–30S pre-rRNAs showed that, unlike CX-5461 and AMD, sirtinol does not induce a global decrease of 47S–30S pre-rRNAs despite the decrease of 47S pre-rRNA (Fig. 2Ab). These results collectively demonstrate the inhibition of pre-rRNA processing following sirtinol treatment.
Fig. 2.
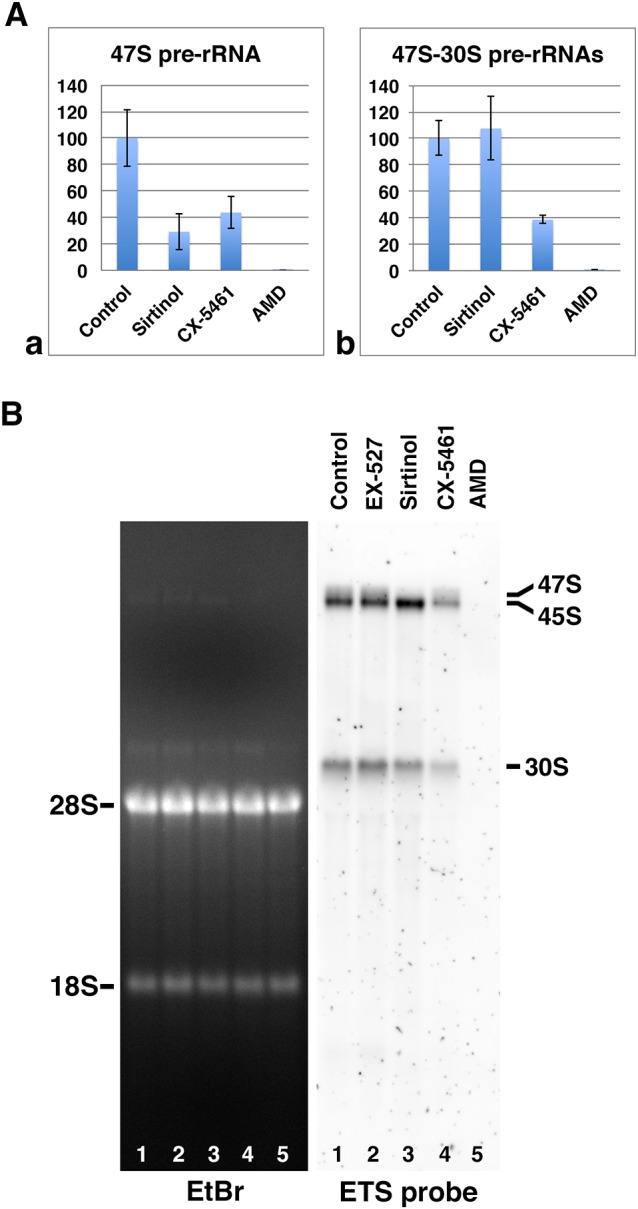
Sirtuin 7 activity is involved in the regulation of pre-rRNA processing. (A) RT-qPCR analyses of total RNAs prepared from HeLa cells treated or not treated with sirtinol, CX-5461 or AMD. Primers ETS1 and ETS2 were used to quantify 47S pre-rRNA (Aa) and primers ETS3 and ETS4 to quantify 47S–30S pre-rRNAs (Ab). Results represent mean±s.d. for three independent experiments. (B) Total RNAs were prepared from HeLa cells treated or not treated with EX-527, sirtinol, CX-5461 or AMD, analyzed in a gel (EtBr), blotted and hybridized with the ETS probe. The 45S pre-rRNA corresponds to the lower band of the doublet observed in the upper part of the northern blot.
To further clarify the inhibitory effect of sirtinol on pre-rRNA processing, total RNAs prepared from HeLa cells treated for 6 h with EX-527, sirtinol, CX-5461 or AMD were analyzed by northern blotting with ETS (Fig. 2B) and 5.8S+ (Fig. S3A) probes. Use of an ETS probe allowed detection of 47S, 45S and 30S pre-rRNAs, while a 5.8S+ probe allowed detection of 47S, 45S, 43S, 41S, 32S and 12S pre-rRNAs, as well as that of mature 5.8S rRNA (Fig. S1). Thereby, we saw that EX-527 has no obvious effect on pre-rRNA processing, as evidenced by comparison of the patterns obtained from extracts prepared from untreated and EX-527-treated cells (Fig. 2B; Fig. S3A, lanes 1 and 2). Conversely, sirtinol treatment clearly induced a decrease of 47S pre-rRNA and an accumulation of 45S pre-rRNA (Fig. 2B; Fig. S3A, lane 3). We did not observe a delay in 47S pre-rRNA processing in sirtinol-treated HeLa cells that would be consistent with defective cleavage at site 01/A′ (Henras et al., 2015), which we had anticipated based on a previously reported in vitro processing assay using L1210 cells treated with nicotinamide, another sirtuin inhibitor (Chen et al., 2016). This discrepancy invites further study and could denote a difference in the regulation of the cleavage at this site between mice and humans. In sirtinol-treated HeLa cells, the level of 30S pre-rRNA appeared only slightly lower than in untreated cells (Fig. 2B, lanes 1 and 3), and higher than in cells treated with rDNA transcription inhibitors (Fig. 2B, lanes 3–5). Considering the sirtinol-induced decrease in rDNA transcription and increase of 45S pre-rRNA half-life, the processing rate of 30S pre-rRNA was also most likely reduced. In addition, the abundance of 32S pre-rRNA was similar in sirtinol- and in CX-5461-treated HeLa cells, whereas 12S pre-rRNA was significantly reduced in sirtinol-treated cells (Fig. S3A, lanes 3 and 4). This reflects a sirtinol-induced reduction in the rate of 32S pre-rRNA processing. These combined results indicate that sirtinol treatment interferes with several pre-rRNA processing events.
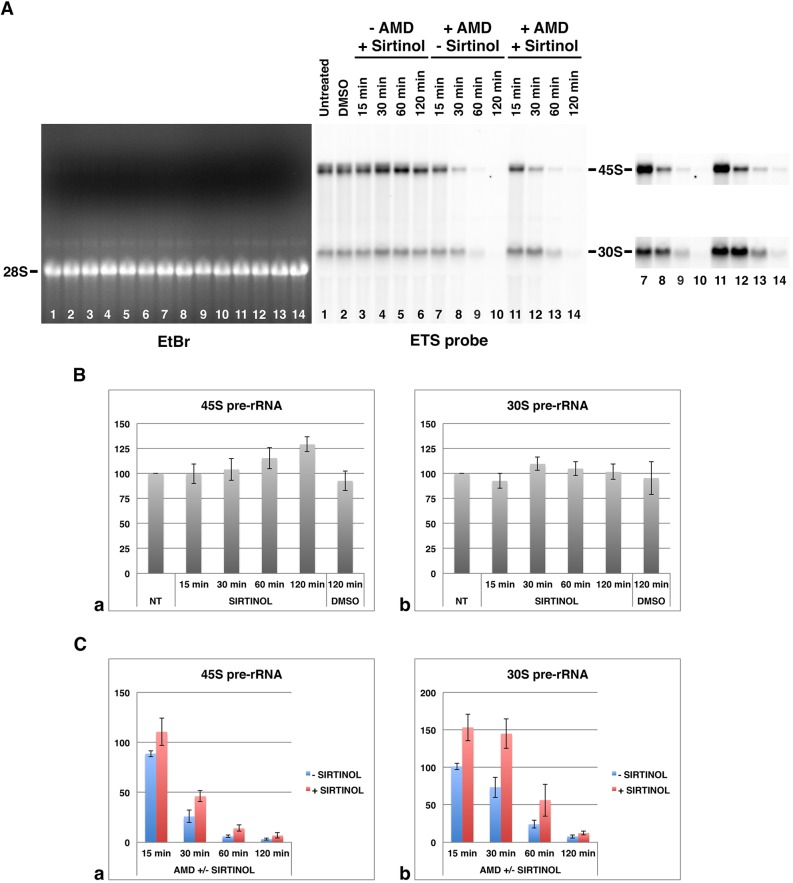
To confirm the effects of sirtinol on pre-rRNA processing events, we first analyzed pre-rRNAs present in HeLa cells treated with sirtinol for 15 to 120 min, by northern blotting using the ETS probe. As illustrated (Fig. 3A, lanes 1–6) and quantified for three independent experiments (Fig. 3Ba,b), neither 45S pre-rRNA nor 30S pre-rRNA significantly varied over the treatment. These results were consistent with the interpretation that the amounts of both pre-rRNAs resulted from opposite sirtinol-induced effects, namely, a sirtinol-induced inhibition of 47S pre-rRNA synthesis that would consequently induce a decrease of all pre-rRNAs, a sirtinol-induced defect of the 45S pre-rRNA processing that would consequently induce an increase of 45S pre-rRNA but a decrease of pre-rRNAs resulting from 45S pre-rRNA processing, and a sirtinol-induced defect of the 30S pre-rRNA processing that would induce an increase of 30S pre-rRNA. To more directly evaluate the effect of sirtinol on pre-rRNA maturation, rDNA transcription was suppressed by using AMD, which induces a total inhibition of rDNA transcription within 10 min (Popov et al., 2014). The levels of both 45S and 30S pre-rRNAs were then analyzed in the absence (Fig. 3A, lanes 7–10) or presence of sirtinol (Fig. 3A, lanes 11–14) for 15 to 120 min, and quantified for three independent experiments (Fig. 3Ca,b). Results confirmed a delay in the processing of 45S and 30S pre-rRNAs in the presence of sirtinol. Notably, the delay was more pronounced for 30S pre-rRNA than for 45S pre-rRNA (Fig. 3Cb) indicating that the 30S pre-rRNA processing is also directly modified in sirtinol-treated HeLa cells and not only influenced by the delay of 45S pre-rRNA processing. Thus, taken together, our results demonstrate that the sirtuin inhibitor sirtinol interferes with several pre-rRNA processing events, in particular processing of 45S, 32S and 30S pre-rRNAs.
Fig. 3.
Sirtuin 7 activity is involved in the regulation of both 45S and 30S pre-rRNA processing. (A) Total RNAs were prepared from untreated HeLa cells (lane 1), from DMSO treated HeLa cells (lane 2) and from HeLa cells treated with AMD (lanes 7–14) or not treated with AMD (lanes 3–6) in absence (lanes 7–10) or presence (lanes 3–6 and 11–14) of sirtinol for 15 to 120 min, analyzed in a gel (EtBr), and blotted and hybridized with the ETS probe. The intensity of signals corresponding to 45S and 30S pre-rRNAs (lanes 7–14) was linearly and globally adjusted, as shown on the right of the figure. The 45S pre-rRNA corresponds to the lower band of the doublet observed in the upper part of the northern blot. (B,C) Northern blot analyses and quantifications (mean±s.d., expressed in arbitrary units) of three independent experiments including the one presented in A. 45S and 30S pre-rRNAs were quantified during sirtinol treatment for 15 to 120 min (respectively, Ba and Bb) and during AMD treatment in the presence or absence of sirtinol for 15 to 120 min (respectively, Ca and Cb). The results of untreated cells were arbitrary normalized to 100. NT, not treated.
Sirtuin 7 protein is involved in the cleavage at site 2
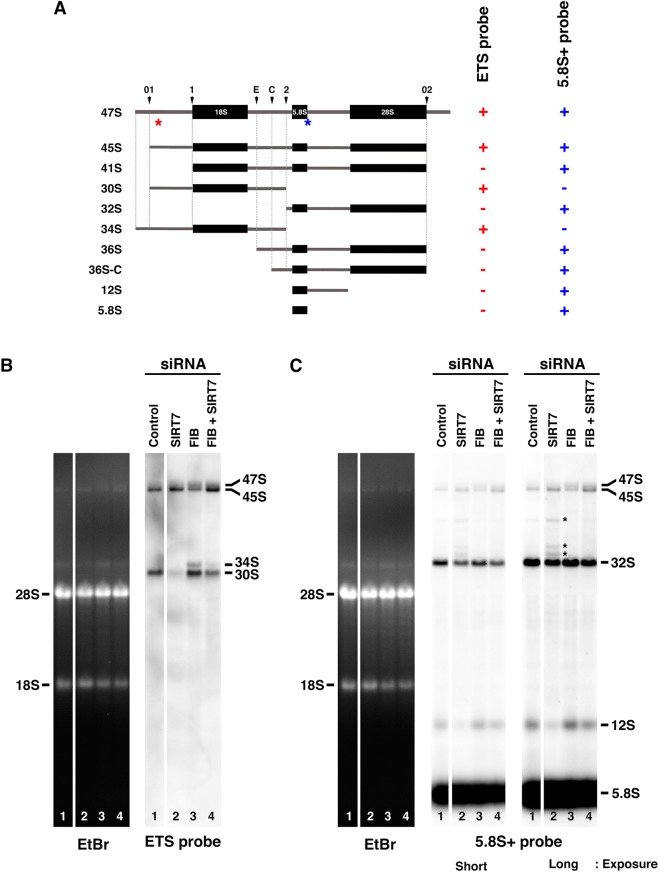
Because this is the only nucleolar sirtuin, sirtuin 7 is most likely implicated in the regulation of pre-rRNA processing. To further investigate the involvement of sirtuin 7 protein in the regulation, we analyzed the pre-rRNA processing by northern blotting with ETS and 5.8S+ probes in siRNA-transfected HeLa cells (Fig. 4A–C). Analyses were performed on total RNAs extracted from HeLa cells in which depletion had been performed for sirtuin 7 (Fig. 4B,C, lanes 1 and 2), fibrillarin (Fig. 4B,C, lane 3) or both sirtuin 7 and fibrillarin (Fig. 4B,C, lane 4). We also analyzed the pre-rRNA processing in siRNA-mediated sirtuin 1-depleted HeLa cells (Fig. S3C). The efficiencies of single or double depletions were verified by immunoblotting (Fig. S4A,B). Sirtuin 7 (SIRT7) and fibrillarin (FIB) siRNAs are specific as assessed by their absence of any effect on the nucleolar protein B23 (nucleophosmin, NPM) but interestingly, SIRT7 siRNAs induce an increase in fibrillarin, and FIB siRNAs a large decrease of sirtuin 7 (Fig. S4A). The obvious influences of sirtuin 7 on fibrillarin and conversely, of fibrillarin on sirtuin 7, might depict a functional interaction between the two proteins. The decrease of sirtuin 7 in fibrillarin-depleted HeLa cells was confirmed by immunofluorescence labeling (Fig. S4C). As recently reported (Iyer-Bierhoff et al., 2018), siRNA-mediated fibrillarin depletion induces an inhibition of rDNA transcription. Results obtained from northern blots showed that similar to what is seen upon sirtinol treatment, 45S pre-rRNA accumulates (Fig. 4B,C, lane 2) in HeLa cells depleted for sirtuin 7 when compared to control cells (Fig. 4B,C, lane 1). This increase in 45S pre-rRNA was accompanied by a significant decrease in 30S pre-rRNA (Fig. 4B, lane 2) suggesting that sirtuin 7 could be involved in the cleavage at site 2. This observation was strengthened by the use of the 5.8S+ probe showing the decreases of 32S and 12S pre-rRNAs in sirtuin 7-depleted HeLa cells (Fig. 4C, lane 2). Moreover, the use of 5.8S+ probe allowed us to also detect the appearance of three additional bands (Figs 4C, lane 2; Fig. S3B, lane 3, asterisks) corresponding to 41S, 36S and 36S-C pre-rRNAs. These pre-rRNAs correspond to 5′ extended forms of 32S pre-rRNA (Tafforeau et al., 2013; Fig. 4A) that accumulate upon a defective cleavage at site 2. As expected from previously reported results (Tafforeau et al., 2013), fibrillarin depletion induced accumulation of 47S and 30S pre-rRNAs and the appearance of 34S pre-rRNA (Fig. 4B, lane 3). This reflects the involvement of fibrillarin in the 5′ ETS processing of 47S and 30S pre-rRNAs. Notably, 45S pre-rRNA was observed to decrease, rather than to accumulate, in cells depleted for fibrillarin contrary to cells depleted for sirtuin 7 (Fig. 4B,C, compare lanes 2 and 3). Similar to what is seen in cells depleted for sirtuin 7, cells depleted for both sirtuin 7 and fibrillarin exhibited an increased amount of 45S pre-rRNA and decreased amounts of 30S and 32S pre-rRNAs (Fig. 4B,C, compare lanes 1 and 4) resulting from the defect in cleavage at site 2. The inference that cleavage at site 2 is dependent on sirtuin 7 was also supported by the significant decrease of 34S pre-rRNA in cells depleted for both sirtuin 7 and fibrillarin compared to cells depleted for fibrillarin only (Fig. 4B, lanes 3 and 4). Indeed, the accumulation of 34S pre-rRNA can only be observed when the processing of the 5′ extremity of 47S pre-rRNA fails but the cleavage at site 2 takes place (Fig. 4A).
Fig. 4.
Sirtuin 7 protein is involved in the cleavage at site 2. (A) Presentation of the pre-rRNAs mentioned in this manuscript. Arrowheads indicate the endonucleolytic sites. Asterisks indicate the position of the ETS (in red) and 5.8S+ (in blue) probes. The reactivity of both probes with each pre-rRNA is given on the right (+ or −). (B,C) Total RNAs were prepared from HeLa cells transfected with control siRNAs, or with siRNAs targeting sirtuin 7 or siRNAs targeting fibrillarin or siRNAs targeting both proteins. RNAs were separated by electrophoresis in a gel (EtBr) as two series, which were blotted and each series hybridized with the ETS probe (B) or with the 5.8S+ probe (C). To better appreciate the different bands revealed by the 5.8S+ probe, short and long exposures are presented and some bands are highlighted by asterisks. The 45S pre-rRNA corresponds to the lower band of the doublet observed in the upper part of the northern blot. The white spaces apparent in B and C correspond to the removal of the same lane from both series at the level of the gel and blot.
As described above, siRNA-mediated sirtuin 7 depletion induced an accumulation of 41S, 36S and 36S-C pre-rRNAs not observed after sirtinol treatment (Fig. 4C; Fig. S3A,B, asterisks). These 5′ extended forms of 32S pre-rRNA are generated if 45S pre-rRNAs are cleaved at site 1, site E or site C without being cleaved at site 2 (Fig. 4A). In cells depleted for both sirtuin 7 and fibrillarin the accumulation was no longer observed and the level of 45S pre-rRNA was concomitantly increased (Fig. 4C, compare lanes 2 and 4, asterisks). In fibrillarin-depleted cells, neither the increase of 45S pre-rRNA nor the 5′ extended forms of 32S pre-rRNA were observed (Fig. 4C, compare lanes 2 and 3). These results collectively demonstrated that the presence of sirtuin 7 is not required for fibrillarin-dependent cleavage of 45S pre-rRNA. Thus, it is noteworthy to mention that in siRNA-mediated sirtuin 7-depleted cells, the amount of 45S pre-rRNA is the result of the active fibrillarin-dependent cleavages and of the defective cleavage at site 2. The decrease of 45S pre-rRNA caused by fibrillarin-dependent cleavages partly counteracts the increase generated by deficient pre-rRNA processing at site 2 as evidenced by comparison of the patterns obtained from extracts prepared from sirtuin 7-depleted cells, and from sirtuin 7 and fibrillarin co-depleted cells (Fig. 4B,C, compare lanes 2 and 4). Concerning the amount of 30S pre-rRNA in sirtuin 7-depleted cells, the decrease of 30S pre-rRNA that results from the deficiency of 45S pre-rRNA processing at site 2 is thus accentuated by the fibrillarin-dependent cleavages (Fig. 4B, compare lanes 2 and 4).
Strikingly, fibrillarin-dependent cleavage observed in the absence of sirtuin 7 protein was not observed after sirtinol-mediated inhibition of sirtuin 7 activity as assessed by the absence of 5′ extended forms of 32S pre-rRNA and by the clear stabilization of 45S and 30S pre-rRNAs (Figs 2 and 3; Fig. S3). It is therefore tempting to propose that beyond its regulatory catalytic activity, sirtuin 7 protein might physically prevent some cleavages, especially some fibrillarin-dependent cleavages. However, we cannot rule out the possibility that the absence of fibrillarin-dependent cleavages in sirtinol-treated cells is not linked to sirtinol-mediated inhibition of sirtuin 7. Nevertheless, the analysis of pre-rRNA processing in sirtuin 1-depleted HeLa cells did not show either the increase of 45S pre-rRNA or the decrease of 32S and 12S pre-rRNAs that are observed in sirtinol-treated cells (Fig. S3, compare A and C). In contrast, the 32S pre-rRNA level increased after sirtuin 1 was depleted, indicating that the sirtinol-induced effects may be difficult to correlate with sirtuin 1 inhibition. Whether or not the sirtuin 7 protein physically prevents certain fibrillarin-dependent cleavages remains an open question, but we establish here for the first time that the sirtuin 7 protein is involved in pre-rRNA cleavage at site 2.
Sirtuin 7 interacts with fibrillarin and Nop56
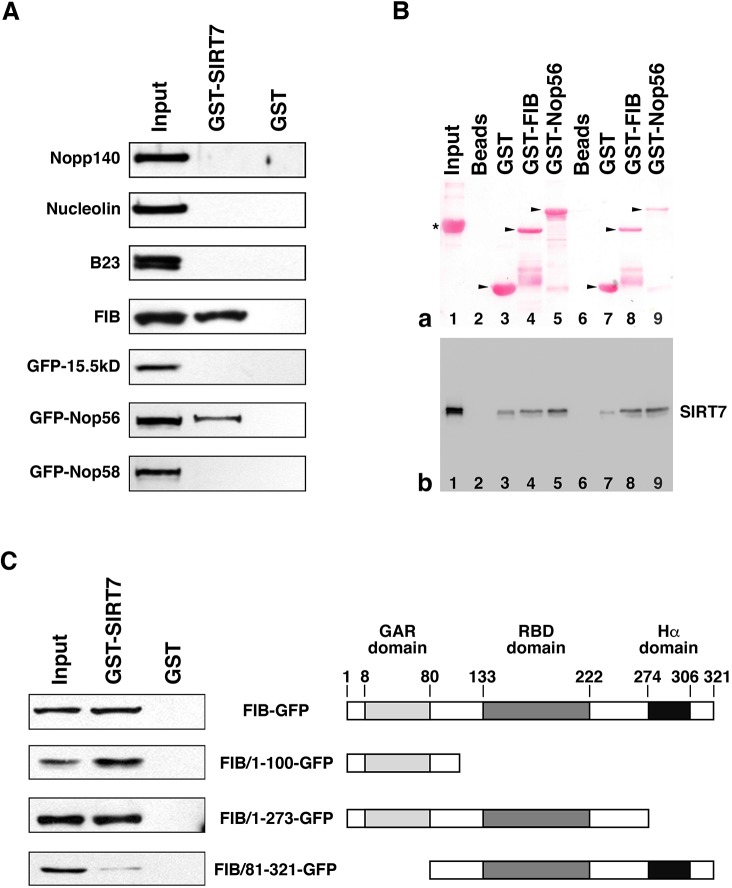
As mentioned above, beyond its regulatory activity, sirtuin 7 seemed to influence pre-rRNA processing by preventing some fibrillarin-dependent cleavages. To further understand the relationship between sirtuin 7 and fibrillarin, and between sirtuin 7 and other proteins known to be involved in pre-rRNA processing, we used glutathione S-tranferase (GST) pulldown experiments to examine their interactions (Fig. 5A). For this purpose, a GST–sirtuin 7 fusion was generated and pulldown assays were performed on whole-cell extracts prepared from HeLa cells or HeLa cells expressing 15.5kD, Nop56 or Nop58 tagged with green fluorescent protein (GFP). As shown in Fig. 5A, western blot analyses of the bound proteins revealed that sirtuin 7 interacts with fibrillarin and Nop56, but not with Nopp140 (also known as NOLC1), nucleolin, B23, 15.5kD and Nop58. Unfortunately, it was not possible to infer from these pulldown assays whether interactions are direct or indirect. Even so, it was possible to rule out that interactions only occur at the level of box C/D snoRNPs. Indeed, fibrillarin and Nop56 were pulled down, while other core box C/D proteins (15.5kD and Nop58) were not. However, given that fibrillarin and Nop56 interact independently of box C/D snoRNPs (Lechertier et al., 2009; McKeegan et al., 2007), the interaction of sirtuin 7 with one or other of these proteins could be indirect. To verify this point, pulldown assays were performed on purified recombinant sirtuin 7 (Fig. 5B) using GST–fibrillarin and GST–Nop56 fusions. The western blot analysis of bound sirtuin 7 (Fig. 5Bb) revealed that both GST fusions directly interact with sirtuin 7 more efficiently than GST, especially after overnight incubation (Fig. 5Bb, lanes 7–9). The same approach was used to determine the region within fibrillarin required for sirtuin 7–fibrillarin interaction. For this purpose, various fibrillarin truncation mutants were expressed in HeLa cells as GFP-fused proteins and GST–sirtuin 7 pulldown assays were performed to identify which fibrillarin mutants interacted with sirtuin 7 (Fig. 5C). The results showed that the N-terminal region (amino acids 1–100) including the glycine- and arginine-rich (GAR) domain (Aris and Blobel, 1991) governs interaction between fibrillarin and sirtuin 7, whereas the central and C–terminal regions comprising the RNA-binding (RBD) and α-helix (Hα) domains do not drive this interaction. The fact that the fibrillarin–sirtuin 7 interaction did not require the α-helix domain of fibrillarin (i.e. the domain needed for fibrillarin-Nop56 interaction; Lechertier et al., 2009) was in accordance with the fact that the fibrillarin–sirtuin 7 interaction takes place independently of the presence of Nop56.
Fig. 5.
Sirtuin 7 interacts with fibrillarin and Nop56. (A) GST and GST–SIRT7 pulldown assays were performed using whole-cell extracts prepared from HeLa cells or HeLa cells expressing either GFP–15.5kD, GFP–Nop56 or GFP–Nop58. 1% of extract used for each pulldown assay (Input) and 5% of the proteins eluted after GST–SIRT7 and after GST pulldown assays were submitted to 10% SDS-PAGE. Immunoblotting was performed using serum with specificity against Nopp140 and anti-nucleolin, anti-B23 and anti-fibrillarin antibodies for the pulldown assays performed using HeLa cells, and with anti-GFP antibody for the pulldown assays performed using HeLa cells expressing a GFP–protein fusion. (B) GST, GST–FIB and GST–Nop56 pulldown assays were performed using recombinant human sirtuin 7. (a) Ponceau Red staining of samples resolved by SDS-PAGE and transferred to nitrocellulose membrane. (b) Immunoblotting using anti-SIRT7 antibody. Lane 1: input control corresponding to 0.1 µg of recombinant sirtuin 7 and 10 µg of BSA. Lanes 2–5: buffer (lane 2, Beads) or the lysates containing GST (lane 3, GST), the GST–fibrillarin fusion (lane 4, GST–FIB) or the GST–Nop56 fusion (lane 5, GST–Nop56) were incubated with glutathione–agarose beads overnight and washed. The beads were then incubated for 2 h with 0.1 µg of recombinant sirtuin 7 and 10 µg of BSA, and washed. Lanes 6–9: buffer (lane 6, Beads) or lysates containing GST (lane 7, GST), the GST–fibrillarin fusion (lane 8, GST–FIB) or the GST–Nop56 fusion (lane 9, GST–Nop56) were incubated with glutathione–agarose beads for 1 h and washed. The beads were then incubated overnight with 0.1 µg of recombinant sirtuin 7 and 10 µg of BSA, and washed. (C) GST and GST–SIRT7 pulldown assays were performed using whole-cell extracts prepared from HeLa cells expressing either FIB–GFP, FIB/1-100–GFP, FIB/1-273–GFP or FIB/81-321–GFP. 1% of extract used for each pulldown assay (Input) and 5% of the proteins eluted after GST–SIRT7 and GST pulldown assays were resolved by 10% SDS-PAGE. Immunoblotting was performed using anti-GFP antibody. A schematic representation of domain structures of fibrillarin and fibrillarin truncation mutants is given on the right of the figure. The GAR, RBD and α-helix domains, and residue numbers are indicated above the diagram. Cropped immunoblots are shown in A and C. The asterisk corresponds to BSA and arrowheads point to the full-length GST fusion proteins in Ba.
Sirtuin 7 influences the localization of fibrillarin
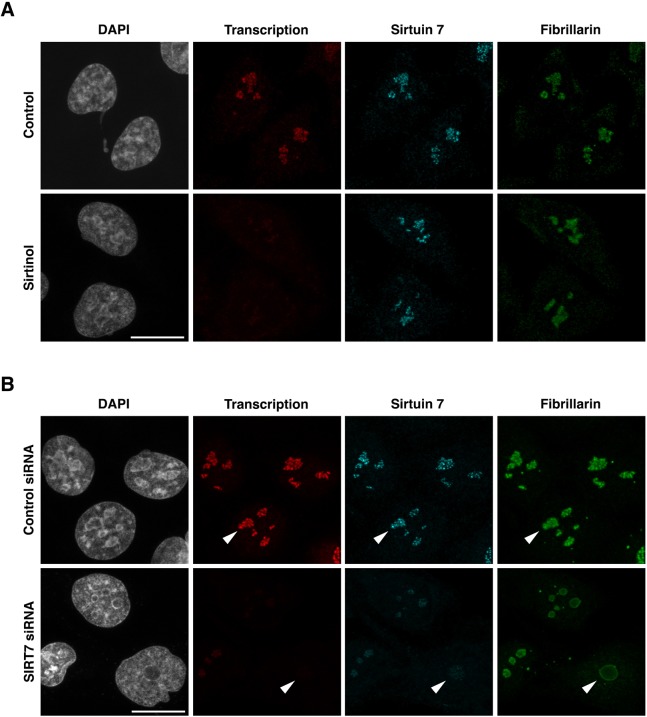
Pulldown assays revealed that sirtuin 7 interacts with fibrillarin, a DFC marker. This prompted us to analyze the influence of either sirtinol treatment or presence of sirtuin 7 in the localization of fibrillarin through immunofluorescence microscopy (Fig. 6A,B). To verify the inhibition of sirtuin 7 activity after sirtinol treatment, the resulting decrease in rDNA transcription was assessed by 5-fluorouridine (FU) metabolic labeling of RNAs synthesized for the last 20 min of culture (Fig. 6A, transcription). The effect of sirtuin 7 depletion on rDNA transcription was also detected by revealing FU metabolically labeled RNAs (Fig. 6B, transcription). As already reported (Grob et al., 2009), sirtuin 7 localized in control HeLa cells at the level of rDNA transcription sites as a punctate signal. The labeling was mainly colocalized with fibrillarin, as expected for a protein also involved in early cleavage of pre-rRNAs. More interestingly, the inhibition of sirtuin 7, despite causing a pronounced decrease in rDNA transcription, did not modify the localization of either sirtuin 7 or fibrillarin (Fig. 6A). Sirtuin 7 depletion using siRNAs induced a decrease in rDNA transcription and an expected decrease of the amount of sirtuin 7, as well as an unusual localization of fibrillarin associated with a decrease of fibrillarin signal (Fig. 6B, arrowheads). The more extensive the depletion of sirtuin 7, the more fibrillarin appeared as a diffuse signal reinforced at the nucleolar periphery rather than a punctate DFC signal. Therefore, as noted for the accumulation of 41S, 36S and 36S-C pre-rRNAs occurring in case of sirtuin 7 depletion (Fig. 4C, lane 2, asterisks) but not after sirtinol treatment (Fig. S3), the modified localization of fibrillarin was observed in HeLa cells transfected with siRNAs targeting sirtuin 7 (Fig. 6B) but not in HeLa cells treated with sirtinol (Fig. 6A). Consequently, the decrease and mislocalization of fibrillarin could not be solely the result of the decrease in rDNA transcription, and most likely highlighted the importance of sirtuin 7–fibrillarin interactions. However, considering the close relationship between nucleolar activity and nucleolar organization, the modified localization of fibrillarin in HeLa cells depleted for sirtuin 7 could not be restricted to fibrillarin, and in fact reveals a global change in the organization of the nucleolus, and more specifically of the DFC. To verify this point and to further assess the role of sirtuin 7, we analyzed the localization of another DFC marker, namely Nopp140 (Thiry et al., 2009). Unlike fibrillarin, Nopp140 appeared not to physically interact with sirtuin 7 (Fig. 5A) but it interacts with both box H/ACA and box C/D snoRNPs without being an integral part of either (Meier, 2005). As shown in Fig. S5, siRNA-mediated sirtuin 7 depletion induced not only a decrease of fibrillarin signal in nucleoli but also of the Nopp140 signal. This probably reflected the global involvement of sirtuin 7 in ribosome biogenesis. Nevertheless, in contrast to what was seen for fibrillarin, which appeared as a more diffuse signal reinforced at the nucleolar periphery, Nopp140 still appeared as a punctate signal (Fig. S5). It was noteworthy that the localizations of fibrillarin and Nopp140 in Cajal bodies (CBs) were maintained after siRNA-mediated sirtuin 7 depletion (Fig. S5A, arrowheads). The CBs were identified by the localization of the CB marker coilin (Fig. S5B, arrowheads). The difference between the fates of fibrillarin and Nopp140 in cells depleted for sirtuin 7 was evident from the loss of colocalization, as seen in the merged images. This can be logically linked to a specific defect in pre-rRNA processing such as the accumulation of 5′ extended forms of 32S pre-rRNA rather than a global change in the organization of the nucleolus resulting from a global defect in pre-rRNA processing. Although the mechanism behind the mislocalization of fibrillarin in cells depleted for sirtuin 7 is still unknown, the fact that no change of fibrillarin localization is observed after sirtinol treatment demonstrates that the inhibition of sirtuin 7 activity cannot be the main cause, and the effect most probably involves physical interaction between the sirtuin 7 and fibrillarin proteins.
Fig. 6.
Sirtuin 7 influences the localization of fibrillarin. (A) HeLa cells were treated (Sirtinol) or not treated (Control) and cultured in medium containing FU for the last 20 min of culture before being processed to reveal FU incorporation (Transcription) and to observe sirtuin 7 and fibrillarin. (B) HeLa cells were transfected with siRNA targeting sirtuin 7 (SIRT7 siRNA) or not (Control siRNA) and cultured in medium containing FU for the last 20 min of culture before being processed to reveal FU incorporation (Transcription) and to observe sirtuin 7 and fibrillarin. The arrowheads point to some nucleoli. Optical sections are shown. Scale bars: 10 μm.
DISCUSSION
The NAD+-dependent deacetylase sirtuin 7, which was previously known to be involved in rDNA transcription in humans (Ford et al., 2006; Grob et al., 2009), was recently reported to also be essential to the early cleavage steps of pre-rRNA processing (Chen et al., 2016). The authors proposed that sirtuin 7-dependent deacetylation of U3-55K enhances its binding to U3 snoRNA, which is a prerequisite for pre-rRNA processing. Here, we establish for the first time that sirtuin 7 activity is more generally involved in the processing of pre-rRNAs in HeLa cells, especially of 45S, 32S and 30S pre-rRNAs. Moreover, as assessed by the increase of 45S pre-rRNA and the concomitant decreases of 30S and 32S pre-rRNAs after sirtuin 7 depletion, sirtuin 7 protein is primarily involved in the cleavage at site 2. This was clearly demonstrated by the increase or appearance of 41S, 36S and 36S-C pre-rRNAs in HeLa cells depleted for sirtuin 7 and by the marked decrease of 34S pre-rRNA in HeLa cells depleted for both sirtuin 7 and fibrillarin compared to cells only depleted for fibrillarin. Indeed, the 5′ extended forms of 32S pre-rRNA accumulate when cleavage at site 2 is interrupted and 34S pre-rRNA is observed when the processing of the 5′ extremity of 47S pre-rRNA fails, but the cleavage at site 2 takes place (Tafforeau et al., 2013; Henras et al., 2015).
In humans, ribosome biogenesis involves the synthesis of 47S pre-rRNA, its processing and assembly with ribosomal proteins and 5S rRNA (Hadjiolov, 1985). During the progressive processing of 47S pre-rRNA (Henras et al., 2015; Mullineux and Lafontaine, 2012), ETS and ITSs 1 and 2 are removed according to two alternative pathways to give mature 18S, 5.8S and 28S rRNAs. Strikingly these two pathways seem not to differ in the sites of cleavage but only in the kinetics and order of cleavages. In pathway 1, the two first concomitant cleavages of 45S pre-rRNA take place in the 5′-ETS extremity at sites A0 and 1 leading to 41S pre-rRNA, whereas in pathway 2 the first cleavage occurs in ITS1 at site 2 and leads to 30S and 32S pre-rRNAs (Henras et al., 2015; Mullineux and Lafontaine, 2012). As shown by measuring the ratio between 41S and 30S pre-rRNAs, pathway 2 is the major pathway in HeLa cells (Henras et al., 2015), but the reason why this pathway is more frequently used is yet elusive. The fact that cleavage at site 2, that is the first step of pathway 2, is dependent on sirtuin 7 raised the possibility that cleavage pathway selection does not happen stochastically but in a regulated manner. Sirtuin 7 could promote the function of proteins involved in cleavage at site 2 and thus trigger the pathway 2. In agreement with this observation, sirtuin 7 protein might be not only necessary for an efficient cleavage of 45S pre-rRNA at site 2 but also to physically impede the fibrillarin-dependent cleavages at sites 1, E and C. This is inferred from the observation that siRNA-mediated sirtuin 7 depletion induced the increase or appearance of 41S, 36S and 36S–C pre-rRNAs in a fibrillarin-dependent manner, in contrast to what was seen upon sirtinol-mediated inhibition of sirtuin 7 activity. In the context of ribosome biogenesis, it is tempting to propose that the presence of sirtuin 7 protein allows the cleavage at site 2, which leads to the separation of the initial 90S pre-ribosomal particle into pre-40S and pre-60S particles. The 40S particle is then free of sirtuin 7 and consequently, fibrillarin-dependent cleavages of 30S pre-rRNA are no longer impeded. Although further studies are needed to prove whether the sirtuin 7 protein physically influences fibrillarin-dependent cleavages, we show here that sirtuin 7 protein is not required for these cleavages to occur. This may be deduced from comparison of the amounts of 45S and 30S pre-rRNAs in HeLa cells depleted for sirtuin 7 and in cells depleted for both sirtuin 7 and fibrillarin. Fibrillarin-dependent cleavages decrease half-lives of 45S and 30S pre-rRNAs in HeLa cells depleted for sirtuin 7 while 45S and 30S pre-rRNAs appeared clearly stabilized after sirtinol-mediated inhibition of sirtuin 7 activity. However, we cannot exclude the possibility that the clear stabilization of 45S and 30S pre-rRNAs in sirtinol-treated cells that is consistent with defective fibrillarin-dependent cleavages is not related to sirtuin 7 activity but to another sirtinol-induced effect. Nevertheless, the functional relationship between sirtuin 7 and fibrillarin seems to have been definitively established. First, sirtuin 7 and fibrillarin directly interact, and second, siRNA-mediated fibrillarin depletion induces also sirtuin 7 depletion. Moreover, fibrillarin was recently reported to be a substrate of sirtuin 7 deacetylase (Iyer-Bierhoff et al., 2018). Even though Iyer-Bierhoff et al. did not report the depletion of sirtuin 7 in siRNA-mediated fibrillarin-depleted cells, they reported, as here, that siRNA-mediated fibrillarin depletion compromises rDNA transcription to a similar extent to siRNA-mediated sirtuin 7 depletion (this study; Grob et al., 2009).
Several mechanisms could explain how sirtuin 7 determines the maturation pathway. Sirtuin 7 might link transcription and processing of pre-rRNA, in a manner similar to some components of the SSU processome, which participate in pre-rRNA processing but are also required for efficient rDNA transcription (Gallagher et al., 2004; Prieto and McStay, 2007). Since modulation of RNA polymerase II and RNA polymerase I elongation rates respectively affect alternative splicing (de la Mata et al., 2003) and pre-rRNA processing (Schneider et al., 2007), the regulation of the RNA polymerase I-dependent transcription by sirtuin 7 could influence co-transcriptional interactions of pre-rRNA processing factors and thus influence pre-rRNA processing. The interactions observed for sirtuin 7, that is, the interactions with the transcription factor UBF (Grob et al., 2009) and with box C/D snoRNA-associated proteins, namely Nop56 and the methyltransferase fibrillarin (this study), are consistent with a dual regulatory role for sirtuin 7. Thus, sirtuin 7 interactions might facilitate, hamper or delay interactions of processing factors during the synthesis of 90S pre-ribosomal particles or within 90S pre-ribosomal particles. Sirtuin 7 might especially facilitate interactions of factors known to be necessary for efficient cleavage at site 2 such as NOP16 and MKI67IP proteins (Tafforeau et al., 2013), RPL26, PES1, BOP1, NOL12 and Nop52 (Henras et al., 2015; Preti et al., 2013; Sloan et al., 2013; Yoshikawa et al., 2015).
Nucleolar organization is directly related to ribosome production (Smetana and Busch, 1974) and pre-rRNA processing factors principally involved in the early stages of pre-rRNA processing are normally localized in the DFC (Hernandez-Verdun et al., 2010). Interestingly, the punctate signals corresponding to fibrillarin and Nopp140 are both decreased in HeLa cells depleted for sirtuin 7 but not in sirtinol-treated HeLa cells, even though rDNA transcription is clearly inhibited in both experimental conditions. Thus, the decrease of signals that denotes a modification of nucleolar organization cannot be related to inhibition of rDNA transcription but rather to defects in pre-rRNA processing. In addition to the global change in the organization of the nucleolus, depletion of sirtuin 7 induced a more specific effect on fibrillarin localization. Indeed, fibrillarin appeared as a more diffuse signal reinforced at the nucleolar periphery and no longer as a punctate signal. Remarkably, the modified localization of fibrillarin as well as the fibrillarin-dependent accumulation of the 5′ extended forms of 32S pre-rRNA occurred in HeLa cells depleted for sirtuin 7 but not after sirtinol-mediated inhibition of sirtuin 7 activity. Therefore, the mislocalization of fibrillarin revealed a specific change in the nucleolar organization and highlighted the importance of sirtuin 7–fibrillarin interaction in both fibrillarin localization and ribosome biogenesis.
The dynamics and localization of proteins depend on their interactions and, consequently, modifying the interactions of a given protein may induce a change not only in its dynamics and localization but also in the dynamics and localization of its partners (Lechertier et al., 2009). Given the central role of fibrillarin as one of the four core proteins present in all box C/D snoRNPs, it may be easily assumed that the sirtuin 7–fibrillarin interaction plays a major role in regulating different aspects of ribosome biogenesis. Even though further experiments are needed to precisely uncover the different roles of sirtuin 7, it is tempting to propose that sirtuin 7 is not only involved in the cleavages of pre-rRNAs but also in pre-rRNA modifications. Sirtuin 7–fibrillarin interaction may first interfere with synthesis of box C/D snoRNPs and thus with pre-rRNA modifications guided by box C/D snoRNAs. Sirtuin 7 might also influence the extent of rRNA modifications, especially 2′-O-methylations, and consequently influence ribosome biogenesis and function (Sloan et al., 2017).
Given that sirtuin 7 is associated with a growing number of diseases (Kiran et al., 2015), and that ribosomopathies are caused by mutations in genes coding for components of the ribosome, such as ribosomal proteins (RPs) or factors required for ribosome biogenesis, it may be hypothesized that mutated sirtuin 7 could cause or promote this kind of disease. Among ribosomopathies, Diamond–Blackfan anemia is associated with mutations of RP genes in more than 50% of patients. In agreement with this hypothesis, of these RP genes RPL26 appears to also be involved in cleavage of 45S pre-rRNA at site 2, as evidenced by the detection of extended forms of 32S pre-rRNA (36S and 36S-C pre-rRNAs) in patients (Gazda et al., 2012). Here, we establish that sirtuin 7 promotes 45S pre-rRNA cleavage at site 2 and consequently may determine the processing pathway to be followed. However, further studies are still needed to fully understand the precise role of the deacetylase sirtuin 7 in ribosome biogenesis and the influence of mutated sirtuin 7 on the development of diseases such as ribosomopathies. More generally, our knowledge of both the role of post-translational modifications in ribosome biogenesis and the upstream enzymes that control post-translational modifications is still very limited. Considering that the post-translational modifications that influence ribosome biogenesis control the expression of the entire cellular proteome, this topic is under-represented in contemporary research. Moreover, post-translational modifications could quantitatively, but also qualitatively, influence the biogenesis of ribosomes (Simsek and Barna, 2017).
MATERIALS AND METHODS
Cell culture, transfection and inhibitor treatments
HeLa and HEK-293T cells, recently authenticated and tested for contamination, were cultured in RPMI supplemented with 10% fetal calf serum (FCS) and 2 mM L-glutamine (GIBCO BRL) in a 5% CO2 incubator at 37°C. The cells were transfected with purified plasmids using effectene (Qiagen) or metafectene (Biontex) according to the manufacturer's instructions 24 h after seeding and were used 24–26 h after transfection. The drugs used were AMD (Sigma-Aldrich) at 0.1 µg/ml for 15 min to 6 h, sirtinol (Calbiochem) at 100 µM for 15 min to 6 h, EX-527 (Tocris Bioscience) at 1 µM for 6 h and CX-5461 (Selleckchem) at 5 µM for 6 h.
Antibodies
The human autoimmune sera with specificity against fibrillarin (O61; 1:3000) and Nopp140 (A10; 1:5000), and the rabbit polyclonal antibody recognizing sirtuin 7 (1:400) were previously described (Grob et al., 2009; Lechertier et al., 2007; Sirri et al., 2002). The rabbit polyclonal anti-B23 (1:15,000) antibody was kindly provided by Mark O. J. Olson, University of Mississippi Medical Center, Jackson, MS). The anti-fibrillarin [anti-FBL (126-140); 1:10,000], anti-α-tubulin (clone DM1A; 1:1000), anti-BrdU (clone BU-33; 1:500), anti-SIRT7 (S5947; 1:10,000) and anti-goat IgG-peroxidase (A5420; 1:3000) antibodies were from Sigma-Aldrich. The anti-B23 (C-19; 1:1000), anti-nucleolin (MS-3; 1:1000), anti-Nopp140 (E-7; 1:300), anti-fibrillarin (B-1; 1:5000), anti-SIRT7 (C-3; 1:10,000) and anti-SIRT1 (H-300; 1:1000) antibodies were from Santa Cruz Biotechnology. The anti-coilin antibody (612074; 1:500) was from BD Biosciences. The anti-histone H3 (17168-1-AP; 1:5000; and 96C10; 1:10,000) antibodies were from Proteintech and Cell Signaling, respectively. The anti-DDK (clone OTI4C5; 1:800) antibody was from OriGene. The alkaline phosphatase-conjugated anti-digoxigenin (11 093 274 910; 1:20,000) and the anti-GFP (11 814 460 001; 1:3000) antibodies were from Roche. The secondary antibodies coupled to Alexa Fluor 488 (109-545-088 and 111-545-144; 1:500), Alexa Fluor 594 (715-585-150; 1:500) and Alexa Fluor 647 (111-605-144 and 709-605-149; 1:500) were from Jackson ImmunoResearch Laboratories, Inc. and those coupled to horseradish peroxidase were from Jackson ImmunoResearch Laboratories (109-035-003, 111-035-003 and 115-035-003; 1:3000).
Primers
PCRs and/or real-time qPCRs were performed using oligonucleotides corresponding to human rDNA (Sirri et al., 2016). The forward oligonucleotides were: ETS1, 5′-GAGGTTGGGCCTCCGGATGC-3′; ETS3, 5′-CCTCTGACGCGGCAGACAGC-3′; ETS5, 5′-GTCGGTGTGGGGTTCGAGGC-3′; 5.8S1, 5′-CACTTCGAACGCACTTGCGG-3′; and 18S1, 5′-GTTCAAAGCAGGCCCGAGCC-3′. The reverse oligonucleotides were: ETS2, 5′-ACGCGCGAGAGAACAGCAGG-3′; ETS4: 5′-CTCCAGGAGCACCGCAAGGG-3′; ETS6, 5′-CCACCGCGATCGCTCACACG-3′; 5.8S2, 5′-CTGCGAGGGAACCCCCAGCC-3′; and 18S2, 5′-AGCGGCGCAATACGAATGCC-3′.
Probes
The ETS (nt +935 to +1082) and 5.8S+ (nt +6718 to +6845, numbers given relative to the transcription start site defined as the +1 position) probes corresponding to fragments of human rDNA were generated as digoxigenin-labeled using primers ETS5 and ETS6 and primers 5.8S1 and 5.8S2, respectively, and the PCR DIG probe synthesis kit (Roche, Sirri et al., 2016).
siRNA mediated mRNA knock down
The SIRT1 SMARTpool siRNA (SIRT1 siRNA) and the siControl RISC-free siRNA (Control siRNA) used as a control for siRNA transfection side effects were from Dharmacon. The siRNAs targeting sirtuin 7 (SIRT7 siRNA, GCCUGAAGGUUCUAAAGATT) and fibrillarin (FIB siRNA, GUCUUCAUUUGUCGAGGAATT) were from Eurogentec. The nucleolin targeting siRNA (nucleolin siRNA, SI02654925) was from Qiagen. The siRNAs were transfected at 10 nM using INTERFERin™ (Polyplus-transfection) according to the manufacturer's instructions. In the case of the double transfection of SIRT7 siRNA and FIB siRNA, both siRNAs were transfected at 10 nM. All siRNAs except for the nucleolin siRNA were transfected 24 h and 48 h after cell seeding. The nucleolin siRNA was transfected only once, at 24 h after cell seeding. At 48 h after the final transfection, the samples were subjected to immunofluorescence labeling or northern blot analysis. The efficiencies of single or double depletions were verified by immunoblotting using anti-SIRT7, anti-SIRT1, anti-fibrillarin, anti-B23 (Santa Cruz), anti-histone H3 (Proteintech and Cell Signaling) and anti-α-tubulin (Sigma-Aldrich) antibodies. The antibodies were visualized with suitable horseradish peroxidase-conjugated secondary antibodies and the immunoreactivity detected by chemiluminescence (GE Healthcare).
Immunofluorescence labeling
To evaluate the level of the rDNA transcription, cells were cultured in medium containing 10 µM 5-fluorouridine (FU) for the last 20 min of culture. The cells were fixed with methanol for 20 min at −20°C, air-dried for 5 min and rehydrated with PBS for 5 min. FU incorporation was then detected using anti-BrdU antibody labeled with Alexa-Fluor-594-conjugated anti-mouse-IgG antibody. Sirtuin 7 was detected simultaneously using rabbit polyclonal antibody labeled with Alexa-Fluor-647- or with Alexa-Fluor-488-conjugated anti-rabbit antibody, and fibrillarin using the O61 serum labeled with Alexa-Fluor-488- or with Alexa-Fluor-647-conjugated anti-human-IgG antibody. In the absence of evaluation of rDNA transcription, cells were fixed in 4% (w/v) paraformaldehyde for 10 min at RT and permeabilized with 0.1% Triton X-100 for 10 min. Fibrillarin was detected using O61 serum labeled with Alexa-Fluor-488-conjugated anti-human-IgG antibody, and Nopp140 was detected using mouse monoclonal antibody labeled with Alexa-Fluor-594-conjugated anti-mouse-IgG antibody. Fibrillarin was also detected using O61 serum labeled with Alexa-Fluor-647-conjugated anti-human-IgG antibody together with coilin, detected using mouse monoclonal antibody labeled with Alexa-Fluor-594-conjugated anti-mouse-IgG antibody, and sirtuin 7, detected using rabbit polyclonal antibody labeled with Alexa-Fluor-488-conjugated anti-rabbit-IgG antibody. All preparations were mounted with the Fluoroshield antifading solution containing 4,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich). The cells were imaged by fluorescence microscopy performed using a Leica upright SP5 confocal microscope with a 63× objective and version 2.7.3 of the Leica Application Suite Advanced Fluorescence software. Multiple fluorophores were recorded sequentially at each Z-step. Images were merged using ImageJ software (NIH).
Pulldown experiments and immunoblotting
The GST, GST–SIRT7, GFP–15.5kD, GFP–Nop56, GFP-–Nop58, FIB–GFP and fibrillarin truncation mutant constructs (FIB/1-100–GFP, FIB/1-273–GFP and FIB/81-321–GFP) used for in vitro pulldown assays were previously described (Grob et al., 2009; Lechertier et al., 2007, 2009). GST–FIB was prepared by subcloning human cDNA into the EcoRI site of the pGEX4T1 (Amersham). GST–Nop56 was prepared by PCR-amplification from mouse cDNA (clone IMAGE 5006404) and subcloning into the BamHI/SalI sites of the pGEX4T1 (Amersham).
For the analysis of sirtuin 7 interactions, GST–SIRT7 proteins were overexpressed in Escherichia coli (BL21 DE3) and then induced in Staby™Switch auto-inducible medium (Eurogentec) according to manufacturer's instructions. The GST proteins were induced for 3 h with 0.1 mM isopropylthiogalactoside (IPTG) after overexpression in E. coli (BL21 DE3). Lysates were obtained by enzymatic reaction with lysosyme (100 µg/ml) and DNase I (20 ng/µl) followed by ultracentrifugation at 65,000 g for 30 min. The clarified lysates were incubated with glutathione–Sepharose beads (GE Healthcare) for 2 h at 4°C. GST and GST–SIRT7 proteins bound to gluthathione–Sepharose beads were washed four times with wash buffer 1 [50 mM Tris-HCl pH 8, 1 M NaCl, 10% glycerol and protease inhibitors (Complete, Roche)] and once with binding buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.2% NP40, 10% glycerol and Complete protease inhibitors). Proteins of HeLa cells and HeLa cells expressing either FIB–GFP, FIB/1-100–GFP, FIB/1-273–GFP or FIB/81-321–GFP) were extracted using lysis buffer (50 mM Tris-HCl pH 7.4, 500 mM NaCl, 1 mM EDTA, 1% NP40, 10% glycerol and Complete protease inhibitors). After centrifugation at 16,000 g for 15 min, the supernatants were adjusted to the binding buffer conditions and these whole-cell lysates were incubated with GST and GST–SIRT7 bound to glutathione–Sepharose beads. After gentle shaking overnight at 4°C, the beads were centrifuged at 500 g for 2 min and washed five times in wash buffer 2 (50 mM Tris-HCl pH 7.4, 300 mM NaCl, 1 mM EDTA, 0.2% NP40, 10% glycerol and Complete protease inhibitors). Proteins corresponding to cell lysates and proteins bound to beads were resuspended in SDS loading buffer (Laemmli, 1970), boiled for 5 min at 100°C, resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes (Protan, Schleicher & Schuell).
For the analysis of the interaction of fibrillarin or Nop56 with the recombinant human sirtuin 7 (Sigma-Aldrich), the GST, GST–FIB and GST–Nop56 proteins were overexpressed in E. coli (BL21 DE3) and induced overnight with 500 µM IPTG at 16°C. Cells were pelleted and resuspended in lysis buffer (50 mM Tris-HCl pH 7.4, 1% Triton X-100, 300 mM NaCl, 1 mg/ml lysozyme and Complete protease inhibitors) for 30 min at 4°C. Lysates were then sonicated on ice and centrifuged (15,000 g) for 30 min at 4°C. Clarified lysates (2 mg) were incubated with glutathione–agarose beads (G4510, Sigma-Aldrich) for 1 h or overnight at 4°C. The GST and GST fusion proteins bound to beads were washed four times with binding buffer (25 mM Tris-HCl pH 7.4, 0.1% Triton X-100, 150 mM NaCl, 5% glycerol and Complete protease inhibitors) and incubated with 600 µl of binding buffer containing 0.1 µg of recombinant human sirtuin 7 and 10 µg of bovine serum albumin (BSA; A9647, Sigma-Aldrich) for 2 h or overnight at 4°C. Beads were pelleted by centrifugation (400 g) for 3 min at 4°C and washed three times with wash buffer (50 mM Tris-HCl pH 7.4, 0.1% Triton X-100, 300 mM NaCl, 5% glycerol and Complete protease inhibitors) for 10 min at 4°C. Beads were then resuspended in SDS loading buffer (Laemmli, 1970), boiled for 5 min at 100°C, resolved by SDS-PAGE (12% gels) and transferred to nitrocellulose membrane (Protan, Schleicher & Schuell). A sample containing 0.1 µg of full-length human recombinant sirtuin 7 and 10 µg of BSA was included as an input control. For immunoblotting, the membranes were incubated to serum with specificity against Nopp140 (A10) or anti-nucleolin, anti-B23 (M. O. J. Olson), anti-fibrillarin (Sigma-Aldrich), anti-GFP (Roche), anti-SIRT7 (Santa Cruz Biotechnology) antibodies as described above. The membranes were then incubated with suitable horseradish peroxidase-conjugated secondary antibodies and the immunoreactivity detected by chemiluminescence (GE Healthcare).
In vitro sirtuin 7 deacetylase assay
To determine the effect of sirtuin inhibitors on sirtuin 7 deacetylase activity, in vitro assays were conducted based on the capacity of sirtuin 7 to deacetylate H3K18-Ac (Barber et al., 2012). The fluorescent H3K18-Ac peptide used (Proteogenix, France) was previously reported (Duval et al., 2015). Assays were performed in a total volume of 10 µl of deacetylase buffer (Tris-HCl 50 mM pH 8, 150 mM NaCl) with 0.1 µg full-length recombinant human sirtuin 7 (SRP5274, Sigma-Aldrich), 6 mM NAD+, 100 µM H3K18-Ac peptide, 0.6 µg yeast tRNA (Sigma-Aldrich) and 1 mM dithiothreitol (DTT). Sirtuin 7 was first pre-incubated with vehicle (DMSO), EX-527 (100 µM) or sirtinol (100 µM) for 15 min at room temperature. The reaction was then started with the addition of H3K18-Ac peptide, tRNA, NAD+ and DTT, incubated for 2 h at 37°C and stopped by adding 50 µl of HClO4 (15% in water). Samples were transferred in 96-wells ELISA plate (Thermo Fisher Scientific) and 5 µl of the mixture were injected into the RP-UFLC (Prominence Shimadzu UFLC system interfaced with LabSolutions software) for detection and quantification of fluorescent H3K18 and H3K18-Ac peptides as previously described (Duval et al., 2015).
SIRT7 immunoprecipitation and in vitro deacetylase assay
Flag–SIRT7 was prepared by subcloning human cDNA into the EcoRI/XhoI sites of the pCMV-Tag2A (Stratagene). At 24 h after being transfection with Flag-SIRT7 plasmid, HEK293T cells were treated with 100 µM sirtinol or vehicle (DMSO) for 6 h, and 20 mM sodium butyrate (NaBu) was added to the cell medium 30 min before the cells were harvested. Cells were then lysed with cell lysis buffer (PBS with 150 mM NaCl, pH 7.4, 1% Triton X-100, 20 mM NaBu and protease inhibitors) for 30 min at 4°C, briefly sonicated and clarified by centrifugation (15,000 g) for 15 min at 4°C. For Flag–SIRT7 immunoprecipitation, 800 µl of whole-cell extracts (1.63 mg/ml) were incubated at 4°C overnight with 1 µg of anti-DDK, 3 µM trichostatin A (TSA) and protease inhibitors. Samples were then mixed with 30 µl of protein G-Plus agarose beads (Santa Cruz Biotechnology) and incubated at 4°C for 2 h. Beads were pelleted by centrifugation (400 g) for 3 min at 4°C, washed twice with cell lysis buffer and twice with deacetylase buffer. A quarter of the beads were then resuspended in SDS loading buffer (Laemmli, 1970), boiled for 5 min at 100°C while the remaining beads were used for the in vitro deacetylase assay. They were incubated with 60 µl of deacetylase buffer containing 6 mM NAD+, 100 µM H3K18Ac peptide, 1 µg tRNA, 1 mM DTT, 1 µM TSA, 5 units of RNase inhibitor and protease inhibitors for 4 h at room temperature. At different time points (2 and 4 h), 20 µl of the solution was taken and mixed with 50 µl of HClO4 to stop the reaction, and 5 µl of the mix were injected into the RP-UFLC column for analysis. For the western blot analysis, the beads and 30 µg of each whole-cell extract were resuspended in SDS loading buffer, and resolved by SDS-PAGE on 4–15% gels (Bio-Rad) and transferred to nitrocellulose membrane. The membrane was then incubated with anti-SIRT7 antibody (Sigma-Aldrich) and suitable horseradish peroxidase-conjugated secondary antibody, and the immunoreactivity detected by chemiluminescence.
Metabolic labeling and RNA analysis
For 32P-labeling experiments, asynchronous HeLa cells were pulse labeled with [32P]orthophosphate (150 µCi/ml) in phosphate-free Minimum Essential Medium for 2 h and chased in nonradioactive medium in excesses of phosphate with or without sirtinol for various times up to 6 h. Total RNA was isolated from cells using NucleoSpin RNA (Macherey-Nagel) and 5 µg of the whole RNA was separated by electrophoresis in 1% agarose formaldehyde gels. The RNAs were then transferred to positively charged membranes (Roche) and UV cross-linked. Autoradiographic imaging was performed with a Typhoon 9400 (Amersham Biosciences).
Northern blot analysis
For northern blot analyses, total RNA was extracted from cells using NucleoSpin RNA, separated (4 μg) in 0.8% agarose formaldehyde gels and transferred to positively charged membranes (Roche). Hybridization was carried out using DIG Easy Hyb buffer (Roche) at 50°C. After washes, the probe was revealed using the DIGWash and Block Buffer Set (Roche) and the alkaline-phosphatase-conjugated anti-digoxigenin antibodies. Alkaline phosphatase activity was detected using the chemiluminescent substrate CDP-Star (Roche) and quantified with ImageJ software.
RT-qPCR
Total RNA was quantified, electrophoresed to verify its quality and reverse transcribed (2 μg) with the Superscript First-Strand Synthesis System for RT-PCR (Invitrogen) using random hexamers as primers (125 ng). qPCR was performed using LightCycler 480 SYBR Green I Master mix and run on a LightCycler 480 II device (Roche). For each reaction, 1.5% synthesized cDNA and 1 μM of a pair of specific primers, either ETS1 and ETS2, or ETS3 and ETS4, were used. Normalization was performed using primers 18S1 and 18S2, which primarily amplify the cDNAs corresponding to mature 18S rRNA. Amplification efficiency for each assay was determined by running a standard dilution curve. Cycle threshold values and relative quantifications were calculated by the LightCycler 480 software.
Supplementary Material
Acknowledgements
The authors thank the imaging and real-time PCR platforms of the Institut de Biologie Paris Seine, Linh Chi Bui and the platform Bioprofiler of the facility ‘Métabolisme’ of the Unité de Biologie Fonctionnelle et Adaptative, and Oliver Brookes for critical reading of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: V.S., A.G., J.B., P.R.; Methodology: V.S., A.G., J.B., N.J., P.R.; Software: N.J.; Validation: V.S., A.G., J.B., P.R.; Formal analysis: V.S., J.B., P.R.; Investigation: V.S., A.G., J.B., N.J., P.R.; Data curation: P.R.; Writing - review & editing: P.R.; Supervision: V.S., P.R.
Funding
This study was supported by grants from the Centre National de la Recherche Scientifique and the Université Paris Diderot. A.G. is supported by a Wellcome Trust UK New Investigator Award No. WT102944. J.B. is supported by PhD fellowship from the Région Ile-de-France. Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.228601.supplemental
References
- Aris J. P. and Blobel G. (1991). cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl. Acad. Sci. USA 88, 931-935. 10.1073/pnas.88.3.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber M. F., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z., Tennen R. I., Paredes S., Young N. L., Chen K. et al. (2012). SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487, 114-118. 10.1038/nature11043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. D. and Gurdon J. B. (1964). Absence of ribosomal RNA Synthesis in the anucleolate mutant of Xenopus laevis. Proc. Natl. Acad. Sci. USA 51, 139-146. 10.1073/pnas.51.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Blank M. F., Iyer A., Huang B., Wang L., Grummt I. and Voit R. (2016). SIRT7-dependent deacetylation of the U3-55k protein controls pre-rRNA processing. Nat. Commun. 7, 10734 10.1038/ncomms10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko D., Verschure P. J., Rothblum L. I., Hernandez-Verdun D., Amalric F., van Driel R. and Fakan S. (2000). Ultrastructural analysis of nucleolar transcription in cells microinjected with 5-bromo-UTP. Histochem. Cell Biol. 113, 181-187. 10.1007/s004180050437 [DOI] [PubMed] [Google Scholar]
- de la Mata M., Alonso C. R., Kadener S., Fededa J. P., Blaustein M., Pelisch F., Cramer P., Bentley D. and Kornblihtt A. R. (2003). A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12, 525-532. 10.1016/j.molcel.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Dragon F., Gallagher J. E. G., Compagnone-Post P. A., Mitchell B. M., Porwancher K. A., Wehner K. A., Wormsley S., Settlage R. E., Shabanowitz J., Osheim Y. et al. (2002). A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417, 967 10.1038/nature00769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval R., Fritsch L., Bui L.-C., Berthelet J., Guidez F., Mathieu C., Dupret J.-M., Chomienne C., Ait-Si-Ali S. and Rodrigues-Lima F. (2015). An acetyltransferase assay for CREB-binding protein based on reverse phase–ultra-fast liquid chromatography of fluorescent histone H3 peptides. Anal. Biochem. 486, 35-37. 10.1016/j.ab.2015.06.024 [DOI] [PubMed] [Google Scholar]
- Ford E., Voit R., Liszt G., Magin C., Grummt I. and Guarente L. (2006). Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20, 1075-1080. 10.1101/gad.1399706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher J. E. G., Dunbar D. A., Granneman S., Mitchell B. M., Osheim Y., Beyer A. L. and Baserga S. J. (2004). RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18, 2506-2517. 10.1101/gad.1226604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda H. T., Preti M., Sheen M. R., O'Donohue M.-F., Vlachos A., Davies S. M., Kattamis A., Doherty L., Landowski M., Buros C. et al. (2012). Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in Diamond-Blackfan anemia. Hum. Mutat. 33, 1037-1044. 10.1002/humu.22081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Rybin V., Baßler J., Petfalski E., Strauß D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D. et al. (2002). 90S Pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10, 105-115. 10.1016/S1097-2765(02)00579-8 [DOI] [PubMed] [Google Scholar]
- Grob A., Roussel P., Wright J. E., McStay B., Hernandez-Verdun D. and Sirri V. (2009). Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell Sci. 122, 489-498. 10.1242/jcs.042382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger C. M., Chao E. D., Blackwell H. E., Moazed D. and Schreiber S. L. (2001). Identification of a class of small molecule inhibitors of the Sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 276, 38837-38843. 10.1074/jbc.M106779200 [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A. (1985). The Nucleolus and Ribosome Biogenesis. Wien: Springer-Verlag. [Google Scholar]
- Henras A. K., Plisson-Chastang C., O'Donohue M.-F., Chakraborty A. and Gleizes P.-E. (2015). An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6, 225-242. 10.1002/wrna.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D., Roussel P., Thiry M., Sirri V. and Lafontaine D. L. J. (2010). The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip. Rev. RNA 1, 415-431. 10.1002/wrna.39 [DOI] [PubMed] [Google Scholar]
- Iyer-Bierhoff A., Krogh N., Tessarz P., Ruppert T., Nielsen H. and Grummt I. (2018). SIRT7-dependent deacetylation of fibrillarin controls histone H2A methylation and rRNA synthesis during the cell cycle. Cell Rep. 25, 2946-2954.e5. 10.1016/j.celrep.2018.11.051 [DOI] [PubMed] [Google Scholar]
- Kiran S., Anwar T., Kiran M. and Ramakrishna G. (2015). Sirtuin 7 in cell proliferation, stress and disease: Rise of the Seventh Sirtuin! Cell . Signal. 27, 673-682. 10.1016/j.cellsig.2014.11.026 [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Langhendries J.-L., Nicolas E., Doumont G., Goldman S. and Lafontaine D. L. J. (2016). The human box C/D snoRNAs U3 and U8 are required for pre-rRNA processing and tumorigenesis. Oncotarget 7, 59519-59534. 10.18632/oncotarget.11148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechertier T., Sirri V., Hernandez-Verdun D. and Roussel P. (2007). A B23-interacting sequence as a tool to visualize protein interactions in a cellular context. J. Cell Sci. 120, 265-275. 10.1242/jcs.03345 [DOI] [PubMed] [Google Scholar]
- Lechertier T., Grob A., Hernandez-Verdun D. and Roussel P. (2009). Fibrillarin and Nop56 interact before being co-assembled in box C/D snoRNPs. Exp. Cell Res. 315, 928-942. 10.1016/j.yexcr.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Mai A., Massa S., Lavu S., Pezzi R., Simeoni S., Ragno R., Mariotti F. R., Chiani F., Camilloni G. and Sinclair D. A. (2005). Design, synthesis, and biological evaluation of sirtinol analogues as class III histone/protein deacetylase (Sirtuin) inhibitors. J. Med. Chem. 48, 7789-7795. 10.1021/jm050100l [DOI] [PubMed] [Google Scholar]
- McKeegan K. S., Debieux C. M., Boulon S., Bertrand E. and Watkins N. J. (2007). A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell. Biol. 27, 6782-6793. 10.1128/MCB.01097-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U. T. (2005). The many facets of H/ACA ribonucleoproteins. Chromosoma 114, 1-14. 10.1007/s00412-005-0333-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineux S.-T. and Lafontaine D. L. J. (2012). Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie 94, 1521-1532. 10.1016/j.biochi.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Popov A., Smirnov E., Kováčik L., Raška O., Hagen G., Stixová L. and Raška I. (2014). Duration of the first steps of the human rRNA processing. Nucleus 4, 134-141. 10.4161/nucl.23985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti M., O'Donohue M.-F., Montel-Lehry N., Bortolin-Cavaillé M.-L., Choesmel V. and Gleizes P.-E. (2013). Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res. 41, 4709-4723. 10.1093/nar/gkt160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J.-L. and McStay B. (2007). Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 21, 2041-2054. 10.1101/gad.436707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D. A., Michel A., Sikes M. L., Vu L., Dodd J. A., Salgia S., Osheim Y. N., Beyer A. L. and Nomura M. (2007). Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol. Cell 26, 217-229. 10.1016/j.molcel.2007.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Yang J., van Nues R., Watzinger P., Kötter P., Lafontaine D. L. J., Granneman S. and Entian K.-D. (2017). Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 13, e1006804 10.1371/journal.pgen.1006804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z. and Barna M. (2015). Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 31, 31-54. 10.1146/annurev-cellbio-100814-125346 [DOI] [PubMed] [Google Scholar]
- Simsek D. and Barna M. (2017). An emerging role for the ribosome as a nexus for post-translational modifications. Curr. Opin. Cell Biol. 45, 92-101. 10.1016/j.ceb.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V., Hernandez-Verdun D. and Roussel P. (2002). Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J. Cell Biol. 156, 969-981. 10.1083/jcb.200201024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V., Jourdan N., Hernandez-Verdun D. and Roussel P. (2016). Sharing of mitotic pre-ribosomal particles between daughter cells. J. Cell Sci. 129, 1592-1604. 10.1242/jcs.180521 [DOI] [PubMed] [Google Scholar]
- Sloan K. E., Mattijssen S., Lebaron S., Tollervey D., Pruijn G. J. M. and Watkins N. J. (2013). Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol. 200, 577-588. 10.1083/jcb.201207131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan K. E., Warda A. S., Sharma S., Entian K.-D., Lafontaine D. L. J. and Bohnsack M. T. (2017). Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138-1152. 10.1080/15476286.2016.1259781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetana K. and Busch H. (1974). The Nucleolus and nucleolar DNA. In The Cell Nucleus (ed. Busch H.), pp. 73-147. Academic Press. [Google Scholar]
- Tafforeau L., Zorbas C., Langhendries J.-L., Mullineux S.-T., Stamatopoulou V., Mullier R., Wacheul L. and Lafontaine D. L. J. (2013). The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol. Cell 51, 539-551. 10.1016/j.molcel.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Thiry M., Cheutin T., Lamaye F., Thelen N., Meier U. T., O'Donohue M.-F. and Ploton D. (2009). Localization of Nopp140 within mammalian cells during interphase and mitosis. Histochem. Cell Biol. 132, 129-140. 10.1007/s00418-009-0599-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N. J., Lemm I., Ingelfinger D., Schneider C., Hossbach M., Urlaub H. and Lührmann R. (2004). Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell 16, 789-798. 10.1016/j.molcel.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Ishikawa H., Izumikawa K., Miura Y., Hayano T., Isobe T., Simpson R. J. and Takahashi N. (2015). Human nucleolar protein Nop52 (RRP1/NNP-1) is involved in site 2 cleavage in internal transcribed spacer 1 of pre-rRNAs at early stages of ribosome biogenesis. Nucleic Acids Res. 43, 5524-5536. 10.1093/nar/gkv470 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.