Abstract
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in children, but diagnosis is challenging due to limited availability of noninvasive biomarkers. Machine learning applied to high‐resolution metabolomics and clinical phenotype data offers a novel framework for developing a NAFLD screening panel in youth. Here, untargeted metabolomics by liquid chromatography–mass spectrometry was performed on plasma samples from a combined cross‐sectional sample of children and adolescents ages 2‐25 years old with NAFLD (n = 222) and without NAFLD (n = 337), confirmed by liver biopsy or magnetic resonance imaging. Anthropometrics, blood lipids, liver enzymes, and glucose and insulin metabolism were also assessed. A machine learning approach was applied to the metabolomics and clinical phenotype data sets, which were split into training and test sets, and included dimension reduction, feature selection, and classification model development. The selected metabolite features were the amino acids serine, leucine/isoleucine, and tryptophan; three putatively annotated compounds (dihydrothymine and two phospholipids); and two unknowns. The selected clinical phenotype variables were waist circumference, whole‐body insulin sensitivity index (WBISI) based on the oral glucose tolerance test, and blood triglycerides. The highest performing classification model was random forest, which had an area under the receiver operating characteristic curve (AUROC) of 0.94, sensitivity of 73%, and specificity of 97% for detecting NAFLD cases. A second classification model was developed using the homeostasis model assessment of insulin resistance substituted for the WBISI. Similarly, the highest performing classification model was random forest, which had an AUROC of 0.92, sensitivity of 73%, and specificity of 94%. Conclusion: The identified screening panel consisting of both metabolomics and clinical features has promising potential for screening for NAFLD in youth. Further development of this panel and independent validation testing in other cohorts are warranted.
Abbreviations
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- AUROC
area under the receiver operating curve
- BMI
body mass index
- CFS
correlation‐based feature search
- FPG
fasting plasma glucose
- HDL
high‐density lipoprotein
- HOMA‐IR
homeostasis model assessment of insulin resistance
- HRM
high‐resolution metabolomics
- IG
information gain
- LDL
low‐density lipoprotein
- LysoPC
lysophosphatidylcholine
- LysoPE
lysophosphatidylethanolamine
- m/z
mass‐to‐charge ratio
- MRI
magnetic resonance imaging
- NAFLD
nonalcoholic fatty liver disease
- OGTT
oral glucose tolerance test
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- RT
retention time
- SVM
support vector machine
- WBISI
whole‐body insulin sensitivity index
- WC
waist circumference
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in the United States, largely due to parallel rises in pediatric obesity.1, 2 The term NAFLD encompasses a range of disease severity from steatosis, defined as fat accumulation in at least 5% of hepatocytes, to nonalcoholic steatohepatitis, defined as steatosis with inflammation and hepatocyte ballooning. There are limited natural history data in children, but studies have shown that childhood NAFLD can result in cirrhosis, severe liver disease, and death.3, 4 NAFLD is also strongly associated with cardiometabolic disease risk factors, such as prediabetes and type 2 diabetes and atherogenic dyslipidemia.5, 6 Currently, NAFLD is a diagnosis of exclusion. Thus, there is a need for a sensitive and cost‐effective screening assay for pediatric NAFLD.
Machine learning can be used to examine patterns within a data set’s attributes to extract important relationships relative to a targeted class.7, 8 In combination with high‐resolution metabolomics (HRM), which is capable of measuring thousands of small molecule metabolites in biofluids, machine learning offers a potential solution for uncovering a novel combination of metabolites that differentiate NAFLD from non‐NAFLD cases. Here, we applied machine learning to high‐resolution liquid chromatography–mass spectrometry metabolomics data as well as clinical phenotype data. Our objective was to identify a panel of variables useful as a screening tool for pediatric NAFLD.
Patients and Methods
Study Population
The combined cohort for this study was a total of 559 patients ages 2‐25 years old who were diagnosed with or without NAFLD by one or both of the gold standard methods (magnetic resonance‐based imaging or liver biopsy) and who were enrolled in one of the three following studies: the Emory University Pediatric Liver Biopsy Data Repository (n = 172), the SweetBev Trial (n = 39), and the Yale Pediatric NAFLD Cohort (n = 348). The Emory Pediatric Liver Biopsy Cohort was a cross‐sectional study of patients enrolled from Children’s Healthcare of Atlanta before undergoing a clinically indicated liver biopsy for suspected liver disease or disease monitoring. Exclusion criteria were fever in the past 2 weeks, renal disease/insufficiency, or pregnancy. Participants provided a fasting blood sample before liver biopsy, which was processed and stored for future research, and shared their liver biopsy slides, which were blindly scored according to the Nonalcoholic Steatohepatitis Clinical Research Network criteria by two pathologists.9 NAFLD diagnosis was defined as having steatosis >5% on liver biopsy combined with a clinical diagnosis of NAFLD.
The Emory SweetBev Trial was conducted in Hispanic/Latino adolescents age 11‐18 years with overweight or obesity and tested the effect of a 4‐week reduction in dietary fructose intake on hepatic fat. Inclusion criteria were hepatic fat >10% and self‐reported habitual sugar‐sweetened beverage consumption. Exclusion criteria were currently attempting weight gain or loss, cirrhosis, renal disease/insufficiency, recent acute illness in the past 4 weeks, or pregnancy. More details of the sample are described elsewhere.10 For most participants (n = 29), plasma samples from baseline were used for metabolomics analysis in this study. For those without stored baseline samples, plasma samples collected at 2 weeks (n = 7) or 4 weeks (n = 3) were used. Hepatic fat was assessed by magnetic resonance spectroscopy (MRS) using described methods,11 and NAFLD diagnosis was defined as hepatic fat >5% based on MRS.
The Yale Pediatric NAFLD Cohort is a multi‐ethnic cohort study of children and adolescents with overweight or obesity recruited from the Yale Pediatric Obesity Clinic in New Haven, CT. Exclusion criteria were known liver diseases (except for NAFLD), alcohol consumption, and use of medications that alter glucose, lipid, or amino acid metabolism. Although some subjects were followed prospectively, for this study only data and samples from each participant’s first study visit were used, which included a stored fasting blood sample for metabolomics analysis among other phenotype assessments.12 A fast‐gradient magnetic resonance imaging (MRI) was performed to assess hepatic fat using methods based on the two‐point Dixon,13 and NAFLD diagnosis was defined as having hepatic fat >5%. The Institutional Review Board at Yale University approved all study procedures and the Institutional Review Board at Emory University approved this secondary analysis of the data and samples. All participants provided written informed consent and assent for their samples and data to be used for future research.
In the combined cohort, 222 participants were diagnosed with NAFLD and 337 were non‐NAFLD controls based on the biopsy or MRI criteria described above. Additional information for each parent study can be found in the Supporting Information.
HRM Analysis
HRM was performed on stored fasting plasma samples using described methods.14, 15 Briefly, plasma samples that had been frozen at −80°C since the time of collection were prepared and analyzed in batches of 40; each contained pooled human plasma (Q standard) at the start, middle, and end of each batch for quality control purposes. Samples were analyzed in triplicate using 10‐μL injections and hydrophilic interaction liquid chromatography with ultra‐high resolution mass spectrometry (Q‐Exactive HF Orbitrap, Thermo Scientific, San Jose, CA) and positive electrospray ionization. The mass spectrometer was operated at 120,000 resolution and in full‐scan mode in order to detect mass‐to‐charge ratios (m/z) ranging from 85 to 1,275. Raw data files were extracted and aligned by apLCMS with xMSanalyzer.16, 17 Spectral features were batch corrected by ComBat,18 filtered based on coefficient of variation, and averaged across triplicates based on Pearson’s correlation. The resulting data consisted of 13,013 metabolic features defined by an accurate mass m/z, retention time (RT), and ion abundance, which were then entered into further statistical analyses.
Metabolite Annotation and Identification
Metabolic features were computationally annotated using xMSannotator.19 This software performs accurate mass matching to common positive‐mode adducts in the Human Metabolome Database.20 For this study, an m/z tolerance of ±5 ppm and an RT threshold of 10 seconds were used to select matches. Additionally, xMSannotator uses a multilevel algorithm that assigns scores to matched metabolic feature, ranging from 0 (no confidence), to 3 (high confidence), based on adduct/isotope patterns, elemental or abundance ratio checks, and metabolic pathway‐level information. Features with no match or with a score of 0 or 1 in xMSannotator were level 4 “unknown” compounds according to criteria set by the Metabolomics Standards Initiative21; those with a score of 2 or greater in xMSannotator were at least level 2 “putatively annotated” compounds. Among these, annotations having m/z and RT of M+H adducts previously confirmed by comparing ion dissociation and elution time to reference standards22 were level 1 “identified” compounds.
Clinical Phenotype Assessments
Phenotype assessments collected for the combined cohort differed according to parent study and are summarized in the Supporting Information. For all participants, demographic information for age, sex, and race/ethnicity were collected by surveys or medical record review, and anthropometric measurements for height (cm) and weight (kg) were performed. Waist circumference (WC; cm) and hip circumference (cm) were also measured in the Emory Pediatric Liver Biopsy Cohort and the Yale Pediatric NAFLD Cohort, although only a subsample of the Emory Pediatric Liver Biopsy Cohort (n = 39) had these measurements because this was added to the protocol after enrollment started. For participants 2‐19 years, age‐ and sex‐adjusted body mass index (BMI) percentiles and z scores were calculated using the 2000 Centers for Disease Control and Prevention growth charts.23 For participants >19 years, we calculated BMI as weight (kg) divided by height in meters squared. Participants were grouped as follows: normal weight (BMI percentile, <85 or BMI, <25 kg/m2 for 2‐19 or 20‐25 years, respectively), overweight (BMI percentile, 85‐94 or BMI, 25‐29 kg/m2), and obese (BMI percentile, ≥95 or BMI, ≥30 kg/m2). Additionally, WC and hip circumference (cm) were measured for some participants (n = 377) in the Emory Pediatric Liver Biopsy Cohort and the Yale Pediatric NAFLD Cohort.
The following laboratory tests were performed in all three parent studies by routine methods: liver enzymes alanine aminotransferase (ALT) and aspartate aminotransferase (AST), fasting plasma glucose (FPG), and a blood lipid panel, including total cholesterol, blood triglycerides, high‐density lipoprotein (HDL) cholesterol, and low‐density lipoprotein (LDL) cholesterol. Fasting insulin was also measured in the Emory SweetBev Trial and the Yale Pediatric NAFLD Cohort and used with FPG to calculate the homeostasis model assessment of insulin resistance (HOMA‐IR).24 Hemoglobin A1C was measured in the Yale Pediatric NAFLD Cohort. A standard oral glucose tolerance test (OGTT) was also performed in the Yale Pediatric NAFLD Cohort and used to calculate 1) the whole‐body insulin sensitivity index (WBISI) based on the Matsuda index, which has been validated against the euglycemic‐hyperinsulinemic clamp in children and adolescents25; 2) the insulinogenic index (IGI) based on the formula Δinsulin (0‐30 minutes)/Δglucose (0‐30 minutes); 3) the disposition index based on the product of IGI and the WBISI. Participants with an FPG and/or 2‐hour postprandial glucose level from the OGTT were characterized as having type 2 diabetes mellitus, impaired fasting glucose, and/or impaired glucose tolerance based on criteria from the American Diabetes Association.26 Additional details of the laboratory methods used for each substudy are in the Supporting Information.
Machine Learning‐Based Analysis
Demographic and clinical characteristics of the sample were summarized using counts and frequencies for categorical variables and means and SDs or medians and interquartile ranges for continuous variables (Table 1). The metabolomics data set was randomly divided into a training set (two thirds of data) and testing set (one third of data) for developing and testing the NAFLD classification model, using the approach in Fig. 1. Partial least squares discriminant analysis (PLS‐DA) was used to reduce data set dimensionality. Metabolic features with a variable importance of projection (VIP) >1 were retained.27 A two‐step feature selection was performed on remaining features. The first step was a correlation‐based feature search (CFS) during which metabolic features were evaluated for their classification potential by Pearson’s correlation. The second step was information gain (IG) with a wrapper‐method support vector machine (SVM), during which metabolic features were evaluated by the amount of information gained and its importance to the SVM algorithm. Retained metabolic features were entered into classification analysis using logistic regression, Naive Bayes, and random forest. Performance of the metabolomics‐only model by each method was evaluated in the test and training sets by area under the receiver operating curve (AUROC), sensitivity, and specificity.
Table 1.
Sociodemographic and Health Characteristics of the Sample Stratified by NAFLD Status
| Non‐NAFLD (n = 337) | NAFLD (n = 222) | P value* | |||
|---|---|---|---|---|---|
| N Obs | Estimate | N (%) | Estimate | ||
| Age, mean (SD) | 337 | 13.7 (4.0) | 222 | 13.6 (2.9) | 0.687 |
| Sex, count (%) | |||||
| Male | 337 | 132 (39.1) | 222 | 135 (60.8) | <0.001 |
| Race/ethnicity, count (%) | |||||
| Non‐Hispanic black | 337 | 132 (38.9) | 222 | 25 (11.3) | <0.001 |
| Non‐Hispanic white | 128 (37.8) | 70 (31.5) | 0.16 | ||
| Hispanic | 66 (19.5) | 121 (54.5) | <0.001 | ||
| Asian/other | 10 (2.9) | 4 (1.8) | 0.75 | ||
| Weight status, count (%) | |||||
| Normal/underweight | 332 | 100 (29.4) | 222 | 4 (1.8) | <0.001 |
| Overweight | 45 (13.6) | 15 (6.8) | 0.027 | ||
| Obese | 187 (57.0) | 203 (91.4) | <0.001 | ||
| Waist circumference, mean (SD) | 237 | 96.0 (18.98) | 140 | 104.3 (17.91) | <0.001 |
| Hip circumference, mean (SD) | 236 | 104.9 (16.46) | 140 | 107.0 (16.93) | 0.24 |
| Fasting glucose (mg/dL), mean (SD) | 328 | 87.9 (12.7) | 220 | 88.3 (12.4) | 0.69 |
| 2‐Hour glucose (mg/dL), mean (SD) | 233 | 117.7 (29.7) | 115 | 127.2 (33.9) | 0.012 |
| Fasting insulin (mg//dL), mean (SD) | 238 | 27.8 (22.8) | 138 | 37.9 (21.5) | <0.001 |
| WBISI, mean (SD) | 226 | 2.4 (1.54) | 113 | 1.5 (0.79) | <0.001 |
| Insulinogenic index, mean (SD) | 218 | 4.7 (5.47) | 112 | 5.6 (4.58) | 0.15 |
| Disposition index, mean (SD) | 217 | 9.3 (16.64) | 112 | 7.1 (6.10) | 0.19 |
| HOMA‐IR, mean (SD) | 238 | 6.3 (5.38) | 138 | 8.7 (5.33) | <0.001 |
| HBA1C (%), mean (SD) | 204 | 5.5 (0.28) | 97 | 5.5 (0.34) | 0.07 |
| Diabetes diagnoses, count (%) | |||||
| Type 2 diabetes | 328 | 8 (2.7) | 220 | 4 (2.3) | 0.99 |
| Impaired glucose tolerance | 233 | 40 (17.2) | 115 | 26 (22.6) | 0.28 |
| Impaired fasting glucose | 328 | 26 (10.2) | 220 | 24 (14.0) | 0.69 |
| ALT (U/L), median (IQR) | 308 | 19 (28) | 204 | 24 (27) | <0.001 |
| AST (U/L), median (IQR) | 307 | 43 (65.3) | 203 | 32 (24.5) | <0.001 |
| Total‐C (mg/dL), mean (SD) | 304 | 158.4 (41.2) | 203 | 163.8 (37.9) | 0.13 |
| Triglycerides (mg/dL), mean (SD) | 304 | 89.3 (56.7) | 203 | 130.9 (71.6) | <0.001 |
| HDL‐C (mg/dL), mean (SD) | 304 | 46.7 (13.8) | 203 | 43.9 (10.9) | 0.010 |
| LDL‐C (mg/dL), mean (SD) | 304 | 93.8 (38.0) | 203 | 96.7 (32.3) | 0.34 |
P values calculated using Student t tests or chi‐square tests. For ALT and AST, nonparametric Mann‐Whitney U test was performed due to non‐normality of the variables. Bold values indicate clinically important differences between groups.
Abbreviations: C, cholesterol, HBA1C, hemoglobin A1C; IQR, interquartile range.
Figure 1.
Workflow of the machine learning‐based approach used for developing and testing the NAFLD screening panels. The proposed workflow consists of a dimension reduction technique, followed by a two‐step machine learning approach. After a potential subset of features is obtained, algorithm mapping with three classifiers is explored to identify an optimal model.
For the development of a combined metabolomics–clinical model, the clinical phenotype data were also analyzed by a two‐step feature selection. Before analysis, missing continuous data were imputed using the median population value of the attribute per the respected class. The two‐step feature selection step was first performed on all 26 demographic and clinical phenotype variables. The retained variables from this analysis were then combined with the retained metabolic features from above to assess their classification performance using logistic regression, Naive Bayes, and random forest. Due to the more intensive nature of performing an OGTT, we also repeated the two‐step feature selection and classification analysis using the clinical phenotype data without the OGTT measurements. Machine learning analysis was conducted in the Waikato Environment for Knowledge Analysis (WEKA, version 3.8), and all other statistical analyses were performed with R statistical package software (version 3.4.2).
Results
Characteristics of the sample are in Table 1. The NAFLD group compared to the non‐NAFLD group had a higher percentage of male subjects (61% vs. 39%, respectively), Hispanics (55% vs. 20%, respectively), and individuals with obesity (92% vs. 57%) (all P < 0.05). The NAFLD group also had significantly higher WC, HOMA‐IR, ALT, AST, and blood triglycerides and lower WBISI and HDL‐cholesterol compared to the non‐NAFLD group (all P < 0.05).
Machine Learning Analysis: Metabolomics‐Only Data
PLS‐DA was implemented for dimension reduction of the 13,008 metabolic features, resulting in 28 components being retained, which explained 95% cumulative variance, and 279 metabolic features being selected with a VIP score ≥1. The 279 metabolic features were then filtered by a two‐step feature selection. As a result of CFS, 222 metabolic features were removed and 57 were retained. The retained metabolic features are summarized in Supporting Table S1. As a result of IG‐SVM, 46 metabolic features were removed and 11 were retained, which are summarized in Table 2. This included the M+H adducts for serine, dihydrothymine, leucine/isoleucine, and tryptophan; the M+H adduct of lysophosphoethanolamine (LysoPE) 20:0; and the M+Na adduct of lysophosphotiylcholine (LysoPC) 18:1. The C13 isotopes of these adducts for leucine/isoleucine, tryptophan, and LysoPC (18:1) were also retained as well as two unknown metabolic features (m/z, 196.8651 and RT, 57; m/z, 434.6894 and RT, 55.3).
Table 2.
Annotated Identities for the 11 Metabolic Features From HRM Retained After Feature Selection
| m/z | Time (Seconds) | Compound Name | HMDB ID | Formula | MSI Level* | Adduct | Difference in Expression (NAFLD vs. Control) |
|---|---|---|---|---|---|---|---|
| 106.0499 | 81.1 | Serine | HMDB00187 | C3H7NO3 | 1 | M+H | ↓ |
| 129.0658 | 61.3 | Dihydrothymine | HMDB00079 | C5H8N2O2 | 2 | M+H | ↓ |
| 132.1019 | 44.9 | Leucine/isoleucine | HMDB00172 | C6H13NO2 | 1 | M+H | ↑ |
| 133.1052 | 45.9 | Leucine/isoleucine | HMDB00172 | C6H13NO2 | 1 | (M+1)+H | ↑ |
| 196.8651 | 57 | ‐ | ‐ | ‐ | 4 | ‐ | ↓ |
| 205.0972 | 45.8 | Tryptophan | HMDB00929 | C11N12N2O2 | 1 | M+H | ↑ |
| 206.1005 | 45.7 | Tryptophan | HMDB00929 | C11N12N2O2 | 1 | (M+1)+H | ↑ |
| 434.6894 | 55.3 | ‐ | ‐ | ‐ | 4 | ‐ | ↓ |
| 510.3552 | 37.6 | LysoPE(20:0) | HMDB11481 | C25H52NO7P | 2 | M+H | ↓ |
| 544.3377 | 39 | LysoPC(18:1) | HMDB02815 | C26H52NO7P | 2 | M+Na | ↓ |
| 545.3415 | 37.9 | LysoPC(18:1) | HMDB02815 | C26H52NO7P | 2 | (M+1)+Na | ↓ |
Confidence levels assigned according to MSI criteria, whereby level 1 is identified compounds, level 2 is putatively annotated compounds, level 3 is putatively characterized compound class, and level 4 is unknown compounds.
Abbreviations: HMDB, Human Metabolome Database; MSI, Metabolomics Standard Initiative.
The 11 selected metabolic features were then entered into a classification analysis using logistic regression, Naive Bayes, and random forest, which enabled us to examine different patterns within the data set between metabolic features and class. The training and test set evaluation metrics for the metabolomics‐only model are shown in Table 3. The three classifiers resulted in similar AUROCs, ranging from 0.85 to 0.86 in the training set and 0.82 to 0.84 in the test set (Fig. 2A). Naive Bayes had the highest sensitivity at 77% (training set) and 73% (test set), and random forest had the highest specificity at 84% (training set) and 87% (test set). As a sensitivity analysis, we tested classifier performance without the two unknown metabolic features. This resulted in minor changes in their evaluation metrics in the test set, with AUROCs of 0.81‐0.82, highest sensitivity of 75% using Naive Bayes, and highest specificity of 89% using random forest (Supporting Table S2).
Table 3.
Evaluation Metrics for the Metabolomics‐Only and the Combined Metabolomics–Clinical Models With and Without OGTT Variables Applied to the Training and Test Sets
| Classifier | Metabolomics‐Only Model | Combined Model #1 (With OGTT Variables) | Combined Model #2 (Without OGTT Variables) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| AUROC | Sensitivity | Specificity | AUROC | Sensitivity | Specificity | AUROC | Sensitivity | Specificity | |
| Training Set | |||||||||
| Naive Bayes | 0.847 | 77% | 78% | 0.880 | 82% | 77% | 0.866 | 81% | 81% |
| Random forest | 0.846 | 60% | 84% | 0.924 | 77% | 90% | 0.923 | 72% | 91% |
| Logistic | 0.860 | 68% | 83% | 0.875 | 75% | 87% | 0.876 | 73% | 88% |
| Test Set | |||||||||
| Naive Bayes | 0.825 | 73% | 78% | 0.848 | 73% | 75% | 0.842 | 73% | 76% |
| Random forest | 0.818 | 60% | 87% | 0.944 | 73% | 97% | 0.922 | 73% | 94% |
| Logistic | 0.839 | 69% | 81% | 0.863 | 72% | 83% | 0.858 | 70% | 81% |
Bold values indicate the classifier with the highest AUROC for each model in the training or test set.
Abbreviations: AUROC, Area under the receiver operator curve; OGTT, Oral glucose tolerance test.
Figure 2.
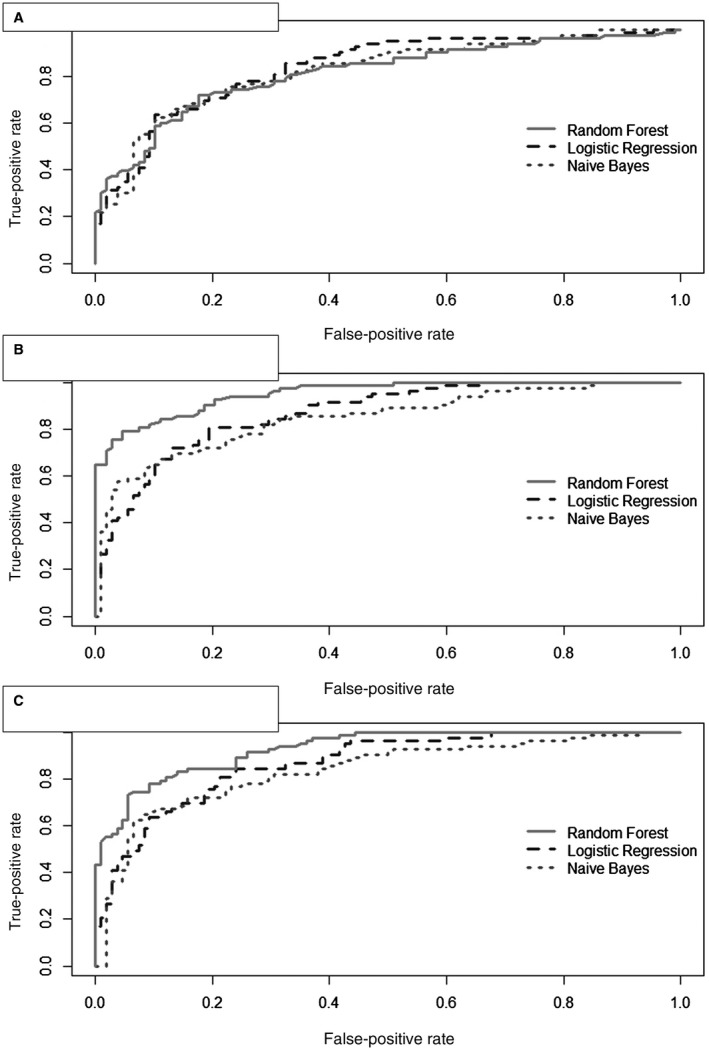
AUROC curves produced from random forest, logistic regression, and Naive Bayes classifiers. (A) Metabolomics‐only model, (B) combined metabolomics–clinical model with OGTT variables, (C) combined metabolomics–clinical model without OGTT variables.
Machine Learning Analysis: Combined Metabolomics–Clinical Data
The same two‐step feature selection analysis was applied to all 26 clinical phenotype variables. After CFS, 14 variables were retained, and after IG‐SVM, the following three variables were retained: WC, WBISI, and triglycerides. The training and test set evaluation metrics for the combined model, evaluated with the 11 metabolic features from above, are summarized in Table 3, and the AUROCs are shown in Fig. 2B. In the test set, random forest had the highest AUROC at 0.94 as well as the highest sensitivity at 73% and specificity at 97%.
A second analysis was conducted using 22 clinical phenotype variables, excluding the OGTT‐based measurements. After CFS, 12 variables were retained, and after IG‐SVM, the following three variables were retained: WC, HOMA‐IR, and triglycerides. The training and test set evaluation metrics for this second combined model are summarized in Table 3. In the test set, random forest had the highest AUROC at 0.92 and specificity at 94%. Naive Bayes and random forest had the same sensitivity of 73%. The AUROCs for each of the models are shown in Fig. 2C. Similar to above, we performed sensitivity analyses without the two unknown features for these models; this resulted in only slight reductions in their evaluation metrics in the test set (Supporting Table S2).
Discussion
Current approaches to screen for and diagnose NAFLD are inefficient and cumbersome. In this study, we applied a machine learning approach to a combination of HRM and clinical phenotype data to identify a set of variables with potential for use as a NAFLD screening panel in children and adolescents. The metabolomics‐only‐based model, which was developed using the training data set and consisted of 11 metabolic features, achieved a sensitivity of 73% and specificity of 84% in the test set. Further, with the addition of clinical phenotype variables to the machine learning workflow and model, we were able to achieve an even higher specificity of 97% in the test set.
The main feature of our machine learning approach was a two‐step feature selection consisting of CFS and IG‐SVM, which was applied to both the metabolomics and clinical phenotype data sets. CFS is a multivariate method of identifying subsets of features that are highly correlated with the class outcome and uncorrelated with each other.28 IG‐SVM was used to further analyze the remaining attributes by a univariate approach. IG is a filter method that eliminates irrelevant attributes in high‐dimensional data, whereas the SVM wrapper eliminates redundancy to decrease noise in the data. The IG‐SVM method has previously shown success for biomarker selection in high‐dimensional cancer gene data.29
Using these steps, 11 metabolic features were retained, of which five were identified as adducts of the amino acids serine, leucine/isoleucine, and tryptophan. Higher levels of branched chain and aromatic amino acids have been reported in children with NAFLD30, 31 and other cardiometabolic disease risk factors.32, 33, 34 The exact cause of these amino acid alterations remains an active area of research but may relate to higher protein consumption, impaired catabolism, and/or microbial amino acid metabolism. Additionally, adult studies have shown that serine levels are inversely associated with NAFLD severity,35, 36, 37, 38 possibly due to increased hepatic import/degradation for glutathione synthesis,39 although this relationship has not been extensively studied in youth. Another metabolic feature was putatively annotated as dihydrothymine, an intermediate in liver pyrimidine catabolism. Studies in animals have linked altered pyrimidine metabolism with increased hepatic lipid,40 providing plausibility for our finding in this study. The remaining features were annotated to two phospholipids, LysoPE (20:0) and LysoPC (18:1). Altered glycerophospholipid metabolism has been reported in both blood and liver tissue samples from patients with NAFLD41, 42, 43, 44, 45 and may relate to disruptions in the cytidine diphosphate‐choline pathway for PC synthesis or the PE to PC conversion pathway catalyzed by PE methyltransferase, both of which take place in the liver. Because there were multiple isomeric matches to these lysophospholipids, additional work is needed to confirm their identities as well as the identities of the two unknown metabolic features.
The machine learning‐based approach was also applied to the clinical phenotype data available for the cohort. Using all 26 variables, WC, WBISI, and blood triglycerides had the greatest potential to identify NAFLD. This aligns with research showing a strong association of NAFLD with metabolic syndrome, which refers to a cluster of metabolic abnormalities that increase risk for future cardiometabolic disease including central adiposity, impaired glucose tolerance, and dyslipidemia.46, 47, 48 Due to the additional time and cost required for an OGTT, the feature selection steps were reapplied to a subset of clinical phenotype variables not including OGTT measurements, and this revealed that WC, HOMA‐IR, and blood triglycerides had the greatest capability to identify NAFLD. This was not surprising because although HOMA‐IR and WBISI are calculated using different information, i.e., fasting versus OGTT‐based glucose and insulin measurements, respectively, both are generally reflective of an individual’s degree of insulin resistance, which is often a co‐occurring morbidity with NAFLD and has been shown in prior studies to be predictive of NAFLD among children with obesity.49
After feature selection, we tested several classification algorithms to identify the optimal algorithm for our screening model. For the metabolomics‐only model, the AUROCs of the classifiers were relatively similar, all greater than 0.8, suggesting that all 11 metabolic features were valuable in classifying NAFLD cases. When combined with WC, WBISI, and blood triglycerides, the evaluation metrics for all three classifiers improved, with the random forest algorithm showing the highest specificity of 97%, sensitivity of 73%, and an AUROC of 0.94 on the test set. The performance of this combined model is comparable to the OWLiver Care test, which is a serum‐based panel developed from 467 adults with and without liver biopsy‐proven NAFLD that achieved a specificity of 78%, sensitivity of 98%, and AUROC of 0.90.35 Compared to this test, all three classification algorithms in our study achieved higher specificity than sensitivity, possibly because the control group had patients with other non‐NAFLD liver diseases. When HOMA‐IR was substituted for WBISI, the random forest algorithm again had the greatest performance with specificity of 94%, sensitivity of 73%, and AUROC of 0.92. Thus, the evaluation metrics for this second combined model were improved from the metabolomics‐only model but inferior to the first combined model with OGTT‐based WBISI. However, the drop in AUROC was only from 0.94 to 0.92, suggesting that while WBISI may be a slightly stronger predictor of NALFD, HOMA‐IR could provide a more practical alternative to WBISI with minimal information loss.
This study was strengthened by its large and diverse sample of children, adolescents, and young adults recruited from two different geographic regions of the United States. All participants had a NAFLD diagnosis confirmed by one or both of the two most accurate methods, MRI/spectroscopy or liver biopsy, which increased our confidence in participant classifications. The use of an untargeted HRM assay allowed us to measure all possible metabolic features without an a priori hypothesis, offering greater potential to identify novel biomarkers.
Despite the promising results, there are limitations to this work. Because different methods (imaging or biopsy) were used for the classification of some participants, this may impact the internal validity of the model. The model was constructed and tested on the same population. Although hold‐out testing was performed, further studies are warranted to validate these models independently. Another limitation is the degree of missing data for some clinical variables. To address this, imputation was used and may have resulted in biased results. However, excluding subjects due to missing data would have resulted in loss of power due to a smaller sample size. Lastly, while we were able to annotate and/or identify most of the retained metabolic features with confidence, “unknown” features still remained and require further investigation. However, in sensitivity analyses excluding these features, the performance metrics of the panel was relatively unchanged, supporting its utility without their validation.
In conclusion, we were able to identify a panel of metabolites and clinical phenotype variables that offer potential as a screening tool for NAFLD by using HRM and clinical phenotype data combined with machine learning algorithms. In a clinic setting, this panel could be used as a first step in identifying children and adolescents who should be further evaluated for NAFLD diagnosis and/or staging using more invasive and expensive procedures. Further investigation to understand the biological significance of the chosen metabolic features as well as external validation of the proposed panel is warranted.
Supporting information
Supported in part by the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (grant numbers P30‐ES019776 and U2C‐ES026560 to D.P.J. and M.B.V.), the National Institute of Diabetes Digestive and Kidney Disease (grant numbers R01‐DK111038 to S.C. and R01‐DK114504 to N.S.), and the NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant numbers R01‐HD028016 to S.C. and R21‐HD089056 to M.B.V.).
Potential conflict of interest: Dr. Vos consults for BMS, Intercept, Target Pharmasolutions, Boehringer Ingelheim, and ARMA and received grants from BMS and Immuron. The other authors have nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. Anderson EL, Howe LD, Jones HE, Higgins JP, Lawlor DA, Fraser A. The prevalence of non‐alcoholic fatty liver disease in children and adolescents: a systematic review and meta‐analysis. PLoS ONE 2015;10:e0140908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ogden CL, Carroll MD, Lawman HG, Fryar CD, Kruszon‐Moran D, Kit BK, et al. Trends in obesity prevalence among children and adolescents in the United States, 1988‐1994 through 2013‐2014. JAMA 2016;315:2292‐2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feldstein AE, Charatcharoenwitthaya P, Treeprasertsuk S, Benson JT, Enders FB, Angulo P. The natural history of non‐alcoholic fatty liver disease in children: a follow‐up study for up to 20 years. Gut 2009;58:1538‐1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cioffi CE, Welsh JA, Cleeton RL, Caltharp SA, Romero R, Wulkan ML, et al. Natural history of NAFLD diagnosed in childhood: a single‐center study. Children (Basel) 2017;4:pii:E34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jin R, Le NA, Cleeton R, Sun X, Cruz Munos J, Otvos J, et al. Amount of hepatic fat predicts cardiovascular risk independent of insulin resistance among Hispanic‐American adolescents. Lipids Health Dis 2015;14:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Newton KP, Hou J, Crimmins NA, Lavine JE, Barlow SE, Xanthakos SA, et al. Nonalcoholic Steatohepatitis Clinical Research Network. Prevalence of prediabetes and type 2 diabetes in children with nonalcoholic fatty liver disease. JAMA Pediatr 2016;170:e161971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kelleher JD, Namee BM, D'Arcy A. Fundamentals of Machine Learning for Predictive Data Analytics: Algorithms, Worked Examples, and Case Studies. Cambridge, MA: The MIT Press; 2015:624. [Google Scholar]
- 8. Swan AL, Mobasheri A, Allaway D, Liddell S, Bacardit J. Application of machine learning to proteomics data: classification and biomarker identification in postgenomics biology. OMICS 2013;17:595‐610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Nonalcoholic Steatohepatitis Clinical Research Network. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005;41:1313‐1321. [DOI] [PubMed] [Google Scholar]
- 10. Holzberg JR, Jin R, Le N‐A, Ziegler TR, Brunt EM, McClain CJ, et al. Plasminogen activator inhibitor‐1 predicts quantity of hepatic steatosis independent of insulin resistance and body weight. J Pediatr Gastroenterol Nutr 2016;62:819‐823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sharma P, Martin DR, Pineda N, Xu Q, Vos M, Anania F, et al. Quantitative analysis of T2‐correction in single‐voxel magnetic resonance spectroscopy of hepatic lipid fraction. J Magn Reson Imaging 2009;29:629‐635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tricò D, Caprio S, Rosaria Umano G, Pierpont B, Nouws J, Galderisi A, et al. Metabolic features of nonalcoholic fatty liver (NAFL) in obese adolescents: findings from a multiethnic cohort. Hepatology 2018;68:1376‐1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fishbein MH, Gardner KG, Potter CJ, Schmalbrock P, Smith MA. Introduction of fast MR imaging in the assessment of hepatic steatosis. Magn Reson Imaging 1997;15:287‐293. [DOI] [PubMed] [Google Scholar]
- 14. Walker DI, Lane KJ, Liu K, Uppal K, Patton AP, Durant JL, et al. Metabolomic assessment of exposure to near‐highway ultrafine particles. J Expo Sci Environ Epidemiol 2019;29:469‐483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Walker DI, Perry‐Walker K, Finnell RH, Pennell KD, Tran V, May RC, et al. Metabolome‐wide association study of anti‐epileptic drug treatment during pregnancy. Toxicol Appl Pharmacol 2019;363:122‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu T, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large‐scale, non‐targeted metabolomics data. BMC Bioinformatics 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu T, Park Y, Johnson JM, Jones DP. apLCMS–adaptive processing of high‐resolution LC/MS data. Bioinformatics 2009;25:1930‐1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118‐127. [DOI] [PubMed] [Google Scholar]
- 19. Uppal K, Walker DI, Jones DP. xMSannotator: An R package for network‐based annotation of high‐resolution metabolomics data. Anal Chem 2017;89:1063‐1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez‐Fresno R, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 2018;46:D608‐D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007;3:211‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Go YM, Walker DI, Liang Y, Uppal K, Soltow QA, Tran V, et al. Reference standardization for mass spectrometry and high‐resolution metabolomics applications to exposome research. Toxicol Sci 2015;148:531‐543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuczmarski RJ, Ogden CL, Guo SS, Grummer‐Strawn LM, Flegal KM, Mei Z, et al. 2000 CDC growth charts for the United States: methods and development. Vital Health Stat 2002;11:1‐190. [PubMed] [Google Scholar]
- 24. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta‐cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412‐419. [DOI] [PubMed] [Google Scholar]
- 25. Yeckel CW, Weiss R, Dziura J, Taksali SE, Dufour S, Burgert TS, et al. Validation of insulin sensitivity indices from oral glucose tolerance test parameters in obese children and adolescents. J Clin Endocrinol Metab 2004;89:1096‐1101. [DOI] [PubMed] [Google Scholar]
- 26. American Diabetes Association . 2. Classification and diagnosis of diabetes. Diabetes Care 2017;40(Suppl. 1):S11‐S24. [DOI] [PubMed] [Google Scholar]
- 27. Akarachantachote N, Chadcham S, Saithanu K. Cutoff threshold of variable importance in projection for variable selection. Int J Pure Appl Math 2014;94:307‐322. [Google Scholar]
- 28. Hall MA. Correlation‐based feature selection for discrete and numeric class machine learning In: Proceedings of the 27th International Conference on Machine Learning; June 29‐July 2, 2000:359‐366; San Francisco, CA: Morgan Kaufmann Publishers, Inc; [Google Scholar]
- 29. Gao L, Ye M, Lu X, Huang D. Hybrid method based on information gain and support vector machine for gene selection in cancer classification. Genomics Proteomics Bioinformatics 2017;15:389‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin R, Banton S, Tran VT, Konomi JV, Li S, Jones DP, et al. Amino acid metabolism is altered in adolescents with nonalcoholic fatty liver disease‐an untargeted, high resolution metabolomics study. J Pediatr 2016;172:14‐19.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goffredo M, Santoro N, Tricò D, Giannini C, D’Adamo E, Zhao H, et al. A branched‐chain amino acid‐related metabolic signature characterizes obese adolescents with non‐alcoholic fatty liver disease. Nutrients 2017;9:pii: E642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wurtz P, Makinen VP, Soininen P, Kangas AJ, Tukiainen T, Kettunen J, et al. Metabolic signatures of insulin resistance in 7,098 young adults. Diabetes 2012;61:1372‐1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wurtz P, Soininen P, Kangas AJ, Ronnemaa T, Lehtimaki T, Kahonen M, et al. Branched‐chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care 2013;36:648‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao X, Gang X, Liu Y, Sun C, Han Q, Wang G. Using metabolomic profiles as biomarkers for insulin resistance in childhood obesity: a systematic review. J Diabetes Res 2016;2016:8160545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barr J, Caballeria J, Martinez‐Arranz I, Dominguez‐Diez A, Alonso C, Muntane J, et al. Obesity‐dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J Proteome Res 2012;11:2521‐2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bianchi G, Marchesini G, Brunetti N, Manicardi E, Montuschi F, Chianese R, et al. Impaired insulin‐mediated amino acid plasma disappearance in non‐alcoholic fatty liver disease: a feature of insulin resistance. Dig Liver Dis 2003;35:722‐727. [DOI] [PubMed] [Google Scholar]
- 37. Mardinoglu A, Agren R, Kampf C, Asplund A, Uhlen M, Nielsen J. Genome‐scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non‐alcoholic fatty liver disease. Nat Commun 2014;5:3083. [DOI] [PubMed] [Google Scholar]
- 38. Mardinoglu A, Bjornson E, Zhang C, Klevstig M, Soderlund S, Stahlman M, et al. Personal model‐assisted identification of NAD(+) and glutathione metabolism as intervention target in NAFLD. Mol Syst Biol 2017;13:916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gaggini M, Carli F, Rosso C, Buzzigoli E, Marietti M, Della Latta V, et al. Altered amino acid concentrations in NAFLD: impact of obesity and insulin resistance. Hepatology 2018;67:145‐158. [DOI] [PubMed] [Google Scholar]
- 40. Le TT, Ziemba A, Urasaki Y, Hayes E, Brotman S, Pizzorno G. Disruption of uridine homeostasis links liver pyrimidine metabolism to lipid accumulation. J Lipid Res 2013;54:1044‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Puri P, Baillie RA, Wiest MM, Mirshahi F, Choudhury J, Cheung O, et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007;46:1081‐1090. [DOI] [PubMed] [Google Scholar]
- 42. Han J, Dzierlenga AL, Lu Z, Billheimer DD, Torabzadeh E, Lake AD, et al. Metabolomic profiling distinction of human nonalcoholic fatty liver disease progression from a common rat model. Obesity (Silver Spring) 2017;25:1069‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lehmann R, Franken H, Dammeier S, Rosenbaum L, Kantartzis K, Peter A, et al. Circulating lysophosphatidylcholines are markers of a metabolically benign nonalcoholic fatty liver. Diabetes Care 2013;36:2331‐2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. García‐Cañaveras JC, Donato MT, Castell JV, Lahoz A. A comprehensive untargeted metabonomic analysis of human steatotic liver tissue by RP and HILIC chromatography coupled to mass spectrometry reveals important metabolic alterations. J Proteome Res 2011;10:4825‐4834. [DOI] [PubMed] [Google Scholar]
- 45. Barr J, Vázquez‐Chantada M, Alonso C, Pérez‐Cormenzana M, Mayo R, Galán A, et al. Liquid chromatography‐mass spectrometry‐based parallel metabolic profiling of human and mouse model serum reveals putative biomarkers associated with the progression of nonalcoholic fatty liver disease. J Proteome Res 2010;9:4501‐4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Patton HM, Yates K, Unalp‐Arida A, Behling CA, Huang TT, Rosenthal P, et al. Association between metabolic syndrome and liver histology among children with nonalcoholic fatty liver disease. Am J Gastroenterol 2010;105:2093‐2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pacifico L, Nobili V, Anania C, Verdecchia P, Chiesa C. Pediatric nonalcoholic fatty liver disease, metabolic syndrome and cardiovascular risk. World J Gastroenterol 2011;17:3082‐3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fu JF, Shi HB, Liu LR, Jiang P, Liang L, Wang CL, et al. Non‐alcoholic fatty liver disease: an early mediator predicting metabolic syndrome in obese children? World J Gastroenterol 2011;17:735‐742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prokopowicz Z, Malecka‐Tendera E, Matusik P. Predictive value of adiposity level, metabolic syndrome, and insulin resistance for the risk of nonalcoholic fatty liver disease diagnosis in obese children. Can J Gastroenterol Hepatol 2018;2018:9465784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


