Abstract
Background
In early-stage pancreatic cancer, there are currently no biomarkers to guide selection of therapeutic options. This prospective biomarker trial evaluated the feasibility and potential clinical utility of circulating tumor DNA (ctDNA) analysis to inform adjuvant therapy decision making.
Materials and methods
Patients considered by the multidisciplinary team to have resectable pancreatic adenocarcinoma were enrolled. Pre- and post-operative samples for ctDNA analysis were collected. PCR-based-SafeSeqS assays were used to identify mutations at codon 12, 13 and 61 of KRAS in the primary pancreatic tumor and to detect ctDNA. Results of ctDNA analysis were correlated with CA19-9, recurrence-free and overall survival (OS). Patient management was per standard of care, blinded to ctDNA data.
Results
Of 112 patients consented pre-operatively, 81 (72%) underwent resection. KRAS mutations were identified in 91% (38/42) of available tumor samples. Of available plasma samples (N = 42), KRAS mutated ctDNA was detected in 62% (23/37) pre-operative and 37% (13/35) post-operative cases. At a median follow-up of 38.4 months, ctDNA detection in the pre-operative setting was associated with inferior recurrence-free survival (RFS) [hazard ratio (HR) 4.1; P = 0.002)] and OS (HR 4.1; P = 0.015). Detectable ctDNA following curative intent resection was associated with inferior RFS (HR 5.4; P < 0.0001) and OS (HR 4.0; P = 0.003). Recurrence occurred in 13/13 (100%) patients with detectable ctDNA post-operatively, including in seven that received gemcitabine-based adjuvant chemotherapy.
Conclusion
ctDNA studies in localized pancreatic cancer are challenging, with a substantial number of patients not able to undergo resection, not having sufficient tumor tissue for analysis or not completing per protocol sample collection. ctDNA analysis, pre- and/or post-surgery, is a promising prognostic marker. Studies of ctDNA guided therapy are justified, including of treatment intensification strategies for patients with detectable ctDNA post-operatively who appear at very high risk of recurrence despite gemcitabine-based adjuvant therapy.
Keywords: circulating tumor DNA, pancreatic ductal adenocarcinoma, pancreatic cancer, liquid biopsy, biomarkers, adjuvant therapy
Key Message
ctDNA detection pre-surgery is associated with inferior pancreatic cancer survival outcomes. Disease progression occurred, despite standard adjuvant therapy, in all patients with detectable ctDNA post-surgery. While no detectable ctDNA post-operatively was associated with reduced recurrence rates. Novel adjuvant strategies in patients with ctDNA detectable post-surgery should be explored.
Introduction
Only 20% of pancreatic ductal adenocarcinoma (PDAC) patients are suitable for curative intent resection at initial presentation. Relapse occurs in 80%–85% of cases despite ‘curative’ resection [1]. The use of adjuvant gemcitabine with capecitabine or FOLFIRINOX both improve overall survival (OS) compared with gemcitabine alone [2, 3]. Preliminary results from the APACT trial, demonstrate a nominal survival benefit from the combination of gemcitabine with Abraxane compared with gemcitabine alone, with no improvement in recurrence-free survival (RFS) [4].
Circulating tumor DNA (ctDNA), arises from somatic tumor DNA fragments released into the blood circulation during cell death [5]. ctDNA is highly tumor specific and can accurately detect the presence of metastatic [6, 7] and minimal residual disease across many solid tumors [7, 8] at a threshold ratio of 10 000 : 1 normal background DNA to mutant allele ctDNA [5, 9]. This level of sensitivity, ease of access from peripheral blood and the short half-life of ctDNA (≈2 h) [10] make ctDNA an ideal dynamic biomarker of tumor burden that can be monitored throughout treatment [7].
The KRAS gene is mutated in over 90% of PDAC tumors [11]. The use of the noninvasive blood-based biomarker ctDNA focusing on mutations in the KRAS gene [11], could be used to identify PDAC patients most likely to benefit from intensification of treatment pre- and/or post-operatively.
There are multiple challenges to undertaking biomarker studies in early PDAC, including the relatively small number of suitable participants undergoing surgery compared with more prevalent malignancies, a higher proportion of patients with inoperable disease identified at the time of surgery, and difficulties obtaining adequate PDAC tissue for analyses particularly if prior neoadjuvant therapy was undertaken. In this explorative biomarker study, we examined the feasibility of a blood-based biomarker study in localized pancreas cancer, including pre- and post-operative samples. We also examined the potential clinical utility of ctDNA as a prognostic marker to guide adjuvant therapy decisions in early-stage PDAC.
Patients and methods
This prospective multicenter biomarker trial recruited patients with early-stage operable pancreatic adenocarcinoma at centers in Australia, New Zealand and Singapore (Australian New Zealand Clinical Trials registry number ACTRN12612000763842) from January 2015 till June 2017. Matched tumor and blood samples were prospectively collected at diagnosis and at 4–8 weeks after surgery (before adjuvant chemotherapy), alongside clinico-pathologic, treatment and outcome data. All tumor tissue and blood analyses were conducted at Johns Hopkins University. All treatments were delivered as per standard of care and blinded to ctDNA results. Follow-up included 3-monthly clinical review for 2 years. The study was approved by the institutional review boards for human research at each institution. All participants provided informed written consent for the collection of specimens and data analysis. This report was written according to the Reporting recommendations for tumor MARKer prognostic studies (REMARK) [12].
Identification of somatic mutations in tumor tissue
Formalin fixed paraffin embedded tumor tissue were macro-dissected to ensure a neoplastic cellularity of >30%. The ctDNA was purified from 3.5 ml plasma using a QIASymphony DP DNA Midi Kit (cat. no. 937255) Hilden, Germany.
ctDNA analysis
A specifically designed PCR-based assay was used to identify somatic mutations at codon 12, 13 and 61 of the KRAS gene and in surrounding codons in the primary tumor [6]. The identified mutation was quantified in matching plasma using the Safe-Sequencing System (SafeSeqS) [9], which utilizes uniquely labeled molecular barcodes, for each template molecule. This minimizes replication errors that frequently occur in massively parallel sequencing. This technique can detect 1 mutant template amongst 10 000 normal templates, allowing detection and accurate quantification even at mutant-DNA levels as low as <0.1% of the total cell free DNA. Analysis for ctDNA was blinded to clinical data.
Algorithm for classifying ctDNA status
ctDNA was categorized as detectable (ctDNA-positive) or undetectable (ctDNA-negative) based on a permutation test (R package perm software version 3.2.3) comparing the mutational frequency in the sample of interest with the mutational frequency in healthy disease control individuals [7]. A P-value of 0.1 was chosen as the threshold to classify a sample of interest as ctDNA-positive (P < 0.1) or ctDNA-negative (P > 0.1). A specificity of at least 0.90 was considered desirable, and a P value of equal to 0.1 yielded 0.90 specificity when performing cross validation on the controls.
Statistical analysis
The primary outcome measure was RFS evaluated by standard RECIST criteria [13]. RFS was defined from date of surgery to first radiologic recurrence or death due to PDAC. OS was the secondary end point and defined from the date of diagnosis till death. Univariate and multivariate analyses were carried out with backward stepwise Cox regression modeling. Chi-squared or Fisher’s exact test and the Mann–Whitney (rank-sum) test were carried out. The Kaplan–Meier method was used to estimate recurrence and survival over time. Statistical analysis was carried out using GraphPad Prism version 7.0 (GraphPad Software Inc.) and R 3.4.2, where P values <0.05 were considered significant.
Results
Patient characteristics
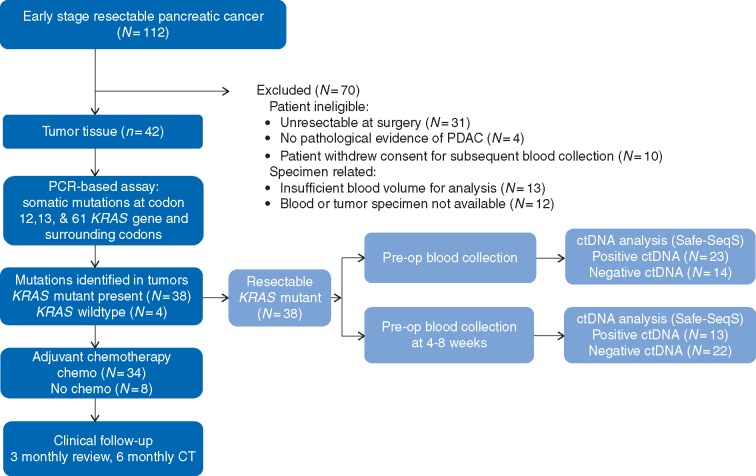
Of the 112 patients with PDAC consented pre-operatively, 81 (72%) proceeded to ‘curative’ resection, the remaining 31 (28%) cases having unresectable disease that was identified intra-operatively. Baseline patient characteristics are summarized in Table 1. Ultimately, 142 blood samples from 42 resected early-stage PDAC cases were analyzed in triplicate for ctDNA (Figure 1). The presence of a KRAS gene mutation was identified in 90% (38/42) of the tissue specimens. ctDNA analysis was carried out pre-operatively in 37 cases and post-operatively in 35, with insufficient tumor tissue available in 1 case, and insufficient plasma volume collected post-operatively in 3 cases. When detected, there was 100% concordance between the specific KRAS mutation codon identified in the tumor tissue and in the plasma KRAS mutant-ctDNA from the same patient.
Table 1.
Patient’s and tumor characteristics according to ctDNA status
| Clinico-pathologic features | Pre-op ctDNA-negative | Pre-op ctDNA-positive | P value | Post-op ctDNA-negative | Post-op ctDNA-positive | P value |
|---|---|---|---|---|---|---|
| N = 14 (%) | N = 23 (%) | N = 22 (%) | N = 13 (%) | |||
| Median age (years) | 66 | 65 | 66 | 65 | ||
| Age range | 48–79 | 48–81 | 47–79 | 48–77 | ||
| Sex | ||||||
| Male | 10 (71) | 13 (57) | 0.49 | 12 (55) | 9 (69) | 0.49 |
| Female | 4 (29) | 10 (43) | 10 (45) | 4 (31) | ||
| CA-19-9 < 36 U/ml | 6 (43) | 4 (17) | ||||
| CA-19-9 > 36 U/ml | 8 (57) | 19 (83) | 0.13 | |||
| Tumor location | ||||||
| Pancreas head | 10 (71) | 20 (87) | 0.64 | 16 (73) | 12 (92) | 0.37 |
| Pancreas body | 1 (7) | 3 (13) | 3 (14) | 1 (8) | ||
| Pancreas tail | 2 (15) | 0 (0) | 2 (9) | 0 (0) | ||
| Not reported | 1 (7) | 0 (0) | 1 (4) | 0 (0) | ||
| T stage | ||||||
| T1 | 0 (0) | 1 (4) | 0.43 | 0 (0) | 0 (0) | >0.99 |
| T2 | 2 (15) | 1 (4) | 2 (9) | 1 (8) | ||
| T3 | 12 (85) | 21 (91) | 20 (91) | 12 (92) | ||
| N status | ||||||
| N0 | 4 (29) | 2 (9) | 0.17 | 5 (23) | 1 (8) | 0.37 |
| N1 | 10 (71) | 21 (91) | 17 (77) | 12 (92) | ||
| R status | ||||||
| R0 | 5 (36) | 11 (48) | 0.12 | 12 (55) | 5 (38) | 0.15 |
| R1 | 9 (64) | 7 (30) | 10 (45) | 6 (46) | ||
| Rx | 0 (0) | 3 (13) | 0 (0) | 2 (16) | ||
| Lymphvascular invasion | ||||||
| Absent | 4 (29) | 6 (26) | 0.53 | 5 (23) | 3 (23) | 0.53 |
| Present | 10 (71) | 15 (65) | 16 (73) | 8 (61) | ||
| Not reported | 0 (0) | 2 (9) | 1 (4) | 2 (16) | ||
| Perineural invasion | ||||||
| Absent | 2 (15) | 6 (26) | 0.32 | 5 (23) | 2 (16) | 0.83 |
| Present | 12 (85) | 15 (65) | 16 (73) | 10 (76) | ||
| Not reported | 0 (0) | 2 (9) | 1 (4) | 1 (8) | ||
| Adjuvant chemotherapy | ||||||
| Yes | 13 (93) | 16 (70) | 21 (96) | 7 (54) | ||
| No | 1 (7) | 7 (30) | 1 (4) | 6 (46) |
Figure 1.
Consort diagram of patient enrolment, specimen collection and clinical management.
Adjuvant therapy was administered following standard institutional protocols and blinded to ctDNA analysis. Eighty-one percent (34/42) of cases received adjuvant therapy. Nineteen patients were treated with single agent gemcitabine, nine received gemcitabine plus capecitabine and six received gemcitabine plus nab-paclitaxel. At a median follow-up of 38.4 months recurrence has occurred in 27 (65%) patients and 20 patients (48%) have died.
Prognostic value of pre-operative ctDNA
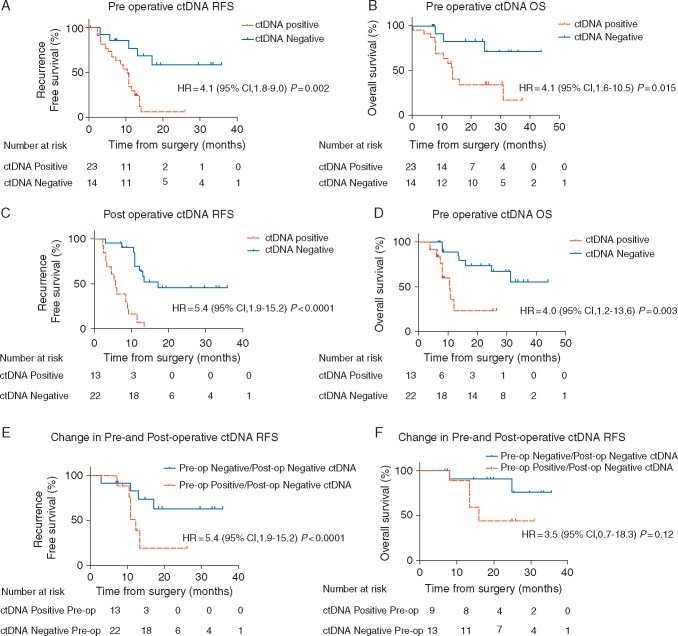
KRAS mutant ctDNA was detected in 62% (23/37) of patient samples pre-operatively, with 4 tumors being KRAS wildtype. Pre-operative detectable (positive) ctDNA when compared with the undetectable (negative) ctDNA cohort was associated with an increased risk of recurrence with a median RFS of 10.3 months versus RFS not reached [hazard ratio (HR) 4.1; 95% confidence interval (CI), 1.8–9.0; P = 0.002] (Figure 2A); and an inferior median OS of 13.6 months versus OS not reached (HR 4.1; 95% CI 1.6 to 10.5; P = 0.015) (Figure 2B). On multivariate and univariate analysis (Table 2), the presence of ctDNA in the pre-operative setting was the only variable that had a statistically significant association with recurrence (HR 4.1; 95% CI 1.4–12.1; P = 0.018) and with OS (HR 4.1; 95% CI 1.0–16.6; P = 0.049).
Figure 2.
(A) Kaplan–Meier estimates of recurrence-free survival (RFS) for all assessable patients undergoing curative intent surgery for early pancreatic cancer, stratified by pre-operative ctDNA status: detectable (positive) versus undetectable (negative). (B) Kaplan–Meier estimate for overall survival (OS) for matched patients, stratified by pre-operative ctDNA status. (C) Kaplan–Meier estimates for RFS, stratified by postoperative ctDNA status. (D) Kaplan–Meier estimates for OS, stratified by post-operative ctDNA status. (E, F) Kaplan–Meier estimates for RFS (E) and OS (F) for patients’ whose ctDNA status changes from detectable (positive) pre-operatively to undetectable (negative) post-operatively compared with patients whose ctDNA status remains undetectable (negative) both pre- and post-operatively.
Table 2.
Recurrence-free survival and overall survival analysis by clinico-pathologic variables and pre- and post-operative ctDNA status
| Pre-operative variable and RFS | ||||||
|---|---|---|---|---|---|---|
| Variable | Univariate analysis |
Multivariate analysis |
||||
| HR | 95% CI | P | HR | 95% CI | P | |
| CA19-9 at diagnosis, <36 U/ml versus >36 U/ml | 1.6 | 0.7–3.5 | 0.31 | |||
| Tumor stage, T3/4 versus T1/2 | 4.4 | 0.6–32.9 | 0.145 | 6.4 | 0.7–57.5 | 0.100 |
| Lymph node status, N1 versus N0 | 1.7 | 0.6–5.0 | 0.318 | 0.8 | 0.2–3.2 | 0.731 |
| Resection margin status, R1 versus R0 | 1.3 | 0.6–3.0 | 0.488 | 2.1 | 0.5–8.7 | 0.325 |
| Lymphvascular invasion status, present versus absent | 2.3 | 0.8–6.8 | 0.130 | 1.1 | 0.3–4.0 | 0.887 |
| Perineural invasion status, present versus absent | 0.8 | 0.3–2.1 | 0.726 | 0.3 | 0.1–1.2 | 0.083 |
| ctDNA status, positive versus negative | 3.9 | 1.4–10.8 | 0.008 | 4.1 | 1.4–12.1 | 0.018 |
| Pre-operative variable and OS | ||||||
| Tumor stage, T3/4 versus T1/2 | 2.9 | 0.4–22.0 | 0.300 | |||
| Lymph node status, N1 versus N0 | 1.0 | 0.3–3.0 | 0.960 | 0.7 | 0.1–3.6 | 0.653 |
| Resection margin status, R1 versus R0 | 1.4 | 0.5–4.0 | 0.533 | 2.0 | 0.6–7.1 | 0.290 |
| Lymphvascular invasion status, present versus absent | 1.3 | 0.4–4.2 | 0.616 | |||
| Perineural invasion status, present versus absent | 0.8 | 0.3–2.4 | 0.715 | |||
| ctDNA status, positive versus negative | 3.6 | 1.0–13.1 | 0.051 | 4.1 | 1.0–16.6 | 0.049 |
| Post-operative variable & RFS | ||||||
| Tumor stage, T3/4 versus T1/2 | 4.4 | 0.6–32.9 | 0.145 | |||
| Lymph node status, N1 versus N0 | 1.7 | 0.6–5.0 | 0.318 | 0.2 | 0.0–1.1 | 0.068 |
| Resection margin status, R1 versus R0 | 1.3 | 0.6–3.0 | 0.488 | 2.4 | 0.6–10.1 | 0.221 |
| Lymphvascular invasion status, present versus absent | 2.3 | 0.8–6.8 | 0.130 | 59.6 | 3.0–1178.8 | 0.007 |
| Perineural invasion status, present versus absent | 0.8 | 0.3–2.1 | 0.726 | 0.1 | 0.0–0.5 | 0.009 |
| ctDNA status, positive versus negative | 5.4 | 1.9–15.2 | <0.0001 | 6.3 | 2.4–16.2 | <0.0001 |
| Post-operative variable & OS | ||||||
| Tumor stage, T3/4 versus T1/2 | 2.9 | 0.4–22.0 | 0.300 | |||
| Lymph node status, N1 versus N0 | 1.0 | 0.3–3.0 | 0.960 | 1.3 | 0.3–7.3 | 0.733 |
| Resection margin status, R1 versus R0 | 1.4 | 0.5–4.0 | 0.533 | 0.6 | 0.2–2.3 | 0.494 |
| Lymphvascular invasion status, present versus absent | 1.3 | 0.4–4.2 | 0.616 | |||
| Perineural invasion status, present versus absent | 0.8 | 0.3–2.4 | 0.715 | |||
| ctDNA status, positive versus negative | 5.5 | 1–17.4 | 0.004 | 7.5 | 2.1–27.7 | 0.002 |
Prognostic value of post-operative ctDNA
Post-operative ctDNA plasma specimens were collected at a median of 6.3 (range 4–8) weeks. The presence of detectable ctDNA following curative resection predicted for recurrence, with a 100% positive predictive value at a median follow-up of 38.4 months, with a specificity and sensitivity of 100% and 57%, respectively.
Univariate analysis of six independent variables was carried out. Post-operative ctDNA status demonstrated a significant prognostic impact on RFS (HR 5.4; 95% CI 1.9–15.2; P < 0.0001) (Table 2). HR’s for other prognostic factors (Table 2) all trended in the expected direction but did not reach statistical significance, likely because of the sample size. On multivariate analysis, ctDNA detection remained a significant predictor of recurrence (HR 6.3; 95% CI 2.4–16.2; P ≤ 0.0001) and death (HR 7.5; 95% CI 2.1–27.7; P = 0.002).
Detectable (positive) ctDNA compared with undetectable ctDNA (negative) following curative resection was associated with a shorter time to recurrence of 5.4 versus 17.1 months (HR 5.4, CI 1.9–15.2; P < 0.001) (Figure 2C), and worse median OS of 10.6 months versus OS not reached (HR 4.0, CI 1.2–13.6; P = 0.003) (Figure 2D).
Of the 22 patients with undetectable ctDNA post-operatively, 10 patients have relapsed at a median follow-up of 38.4 months. Within the ctDNA-positive cohort, all 13 patients relapsed. In the post-operative ctDNA-positive cohort, the use of doublet chemotherapy showed a trend toward improved RFS with median RFS of 10.1 months compared with 5.1 months (HR 0.36, CI 0.1–1.6; P = 0.15) favoring doublet chemotherapy over gemcitabine alone (supplementary Figure S1, available at Annals of Oncology online).
Pre-operative versus post-operative results
Analyses of matched pre- and post-surgery plasma collected from the same patients, revealed that of the 23 of 42 (55%) patients that had detectable ctDNA before surgery there were 11 that still had detectable ctDNA post-surgery. Two patients with undetectable ctDNA levels before surgery, were found to have detectable ctDNA following surgery, bringing the total number of patients with detectable ctDNA following surgery to 13 (31%) of 42 patients. This corresponds to a 24% decrease in the proportion of patients with detectable ctDNA following surgery.
In patients with detectable ctDNA pre-surgery who then had undetectable ctDNA following surgery, the median RFS was 12.2 months. In the cohort with undetectable ctDNA both pre-and post-surgery median RFS was not reached (HR 3.2, CI 0.8–12.0; P = 0.05 versus detectable ctDNA pre-surgery) (Figure 2E). Where patient status went from detectable ctDNA pre-surgery to undetectable post-surgery median the OS was 15.9 months (HR 3.5, CI 0.7–18.3; P = 0.12) (Figure 2F).
Discussion
This is the first study in early-stage pancreatic cancer patients undergoing surgery with curative intent that was designed to collect and report on both pre- and post-operative ctDNA analyses. This study highlights the challenges faced in the conduct of such a study and the treatment of PDAC. Issues include managing resectable versus unresectable ‘early’ stage disease, access to sufficient tumor tissue for testing, and collecting blood samples according to strict protocols. Despite these challenges we demonstrated the validity of ctDNA as a liquid biopsy and tool to evaluate disease response and potentially guide therapy. We also demonstrated that ctDNA was detectable at diagnosis in 62% of early-stage KRAS mutated PDAC. This is consistent with other studies in the pre-operative setting that have reported detection rates of 31%–55% [6, 14–16].
Our data suggest a potential role for ctDNA as a prognostic marker in the pre-operative setting. Of the 23 patients with detectable ctDNA at diagnosis, 19 (83%) have recurred at a median follow-up of 38.4 months despite standard therapy being administered in 81%. These higher risk patients might be a population where neoadjuvant therapy strategies could be explored, rather than proceeding straight to surgery.
In the post-operative setting, our finding that ctDNA is detectable in 31% of patients following curative intent surgery is consistent with other small series. These reported a post-operative detection rate of 44% in 4/9 patients [17] and 45% post-operative detection rates in a sub-group analysis [15, 18] with a median time to recurrence of 9.9 months in 10 patients with detectable post-operative ctDNA (P = 0.019) [15]. In our series, the presence of detectable ctDNA was associated with an increased risk of relapse, with a median RFS of 10.3 months for pre-op ctDNA detection (P = 0.005) and 5.4 months for post-op ctDNA detection (P < 0.0001); and significantly worse OS compared with patients with undetectable ctDNA (P = 0.04 and P = 0.001, respectively).
Post-operative analysis of ctDNA appears most informative. Our preliminary data indicate that where ctDNA is detectable at diagnosis but becomes undetectable post-operatively that this is associated with a reduction in relapse risk compared with those where ctDNA remains detectable, but further adjuvant therapy measures should still be considered given the high recurrence risk. We would suggest that even with larger patient numbers and a prospective randomized trial it is unlikely that a ctDNA-negative group at such low risk of recurrence could be identified such that adjuvant therapy could be avoided.
Ours is the first series to examine the impact of adjuvant chemotherapy according to ctDNA status. Of the 13 patients with detectable ctDNA post-operatively, 5 received adjuvant gemcitabine alone and 2 received gemcitabine and capecitabine, the latter emerging as the standard of care following the ESPAC4 trial results that demonstrated improved median OS of 28 versus 25.5 months (P = 0.032) with gemcitabine with capecitabine compared with gemcitabine alone, and an improved median RFS of 13.9 versus 13.1 months (P = 0.082) [3]. Despite receiving adjuvant therapy all patients subsequently relapsed, providing preliminary evidence that current standard adjuvant therapy may not be of benefit in this very high-risk subset, and alternative adjuvant strategies need to be explored. Longitudinal monitoring using liquid biopsy samples through exoDNA and ctDNA provides both predictive and prognostic information that could be used for therapeutic stratification [19].
Results from the PRODIGE and APACT trials, evaluating the use of adjuvant FOLFIRINOX and adjuvant gemcitabine with nab-paclitaxel (Abraxane), respectively, against gemcitabine alone, (which did not stratify patients according to ctDNA status) reported that the use of adjuvant FOLFIRINOX compared with single agent gemcitabine significantly improved RFS (21.6 versus 12.8 months, P < 0.001) [2] whilst the use of gemcitabine with nab-paclitaxel compared with single agent gemcitabine did not [4]. However, a nominal improvement in OS was observed in the gemcitabine with nab-paclitaxel cohort [4]. Whether a subset of patients at higher risk of recurrence can be identified with a biomarker such as ctDNA post-operatively and then managed with a risk-adapted treatment strategy remains to be seen.
We have initiated a larger prospective clinical trial exploring a ctDNA informed approach to adjuvant treatment (CTRA U1111-1209-6200). Patients are randomized to standard of care treatment blinded to ctDNA analysis or to ctDNA informed treatment. This study will prospectively collect plasma and data to validate the findings of the current study regarding the impact of standard of care treatments in the ctDNA-positive and -negative patients. As suggested in the ASCO-CAP joint review on ctDNA analysis in patients with cancer, the most reliable method to demonstrate the clinical utility of ctDNA is through prospective clinical trials as a stand-alone diagnostic test, that include a patient cohort that resembles the intended-use population with defined entry criteria [20].
This present study had several important limitations: only a modest number of patients could be included in the major analysis, relatively small numbers in subsets of interest, the explorative study design and a missing validation cohort. The attrition rate partly reflects the nature of pancreatic cancer, where cases that initially appear to be resectable have inoperable disease at surgery. Our experience confirms that large biomarker studies in the setting of resectable pancreatic cancer can be challenging to complete, with cases being excluded due to a wide range of reasons. Future studies enrolling patients preoperatively should allow for a high proportion of patients not completing study related treatment and biomarker collection. Ensuring a minimum of 30 ml of whole blood is collected, can also prevent issues of insufficient sample for analysis.
In conclusion, we have shown the potential clinical utility of ctDNA analysis, before and after curative intent surgery, as a prognostic marker in patients with pancreatic cancer. These findings raise interesting possibilities around potential adjuvant and neo-adjuvant treatment strategies in ctDNA defined subsets, but these approaches need to be validated in prospective randomized clinical trials. It is crucial that studies involving ctDNA analysis should explore how to guide patient treatment rather than simply collecting plasma from patients in routine care, as demonstrating clinical utility and patient benefit from biomarker informed treatment is the end goal.
Supplementary Material
Acknowledgements
We thank all participating hospitals (Royal Melbourne Hospital, Western Health, Cabrini Health, Prince of Wales Hospital, Royal Brisbane Hospital, Eastern Health, Austin Health, Monash Medical Centre, Alfred Hospital, Auckland City Hospital, New Zealand, National Cancer Centre, Singapore) for patient enrolment and sample collection; M. Chapman, J. Nolan for administrative assistance; and the Victorian Cancer Biobank for sample processing.
Funding
This study was supported by National Institutes of Health (NIH) grants (CA57345, R37-CA43460, U01-CA152753 and P30-CA006973) and by the Virginia and DK Ludwig Fund for Cancer Research, the Conrad N. Hilton Foundation, the Sol Goldman Sequencing Facility at Johns Hopkins, Victorian Cancer Agency Clinical Research Fellowship (to JT), Walter & Eliza Hall Institute Hemstritch Centenary Fellowship (to BL). There are no applicable grant numbers for this funding.
Disclosure
The authors have declared no conflicts of interest.
References
- 1.Australian Institute of Health and Welfare (AIHW). Australian Cancer Incidence and Mortality (ACIM) books Canberra: AIHW. [Google Scholar]
- 2. Conroy T, Hammel P, Hebbar M. et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med 2018; 379(25): 2395–2406. [DOI] [PubMed] [Google Scholar]
- 3. Neopotelomos JPP, Palmer DH, Ghaneh P. et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised phase 3 trial. Lancet 2017; 389(10073): 1011–1024. [DOI] [PubMed] [Google Scholar]
- 4.Update-on-ABRAXANE-Combination-Therapy-in-the-Treatment-of-Metastatic-Triple-Negative-Breast-Cancer-and-Pancreatic-Cancer. 2019; https://ir.celgene.com/ (12 March 2019, date last accessed).
- 5. Diehl F, Li M, Dressman D. et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA 2005; 102(45): 16368–16373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bettegowda C, Sausen M, Leary RJ. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6(224): 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tie J, Kinde I, Wang Y. et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015; 26(8): 1715–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shaw JA, Brown J, Coombes RC. et al. Circulating tumor cells and plasma DNA analysis in patients with indeterminate early or metastatic breast cancer. Biomark Med 2011; 5(1): 87–91. [DOI] [PubMed] [Google Scholar]
- 9. Kinde I, Wu J, Papadopoulos N. et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA 2011; 108(23): 9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diehl F, Schmidt K, Choti MA. et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14(9): 985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Almoguera C, Shibata D, Forrester K. et al. Most human carcinomas of the exocrine pancreas contain mutant cK-ras genes. Cell 1988; 53(4): 549–554. [DOI] [PubMed] [Google Scholar]
- 12. Sauerbrei W, Taube SE, McShane LM. et al. Reporting recommendations for tumor marker prognostic studies (REMARK): an abridged explanation and elaboration. J Natl Cancer Inst 2018; 110(8): 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishino M, Jagannathan JP, Ramaiya NH, Van den Abbeele AD.. Revised RECIST guideline version 1.1: what oncologists want to know and what radiologists need to know. AJR Am J Roentgenol 2010; 195(2): 281–289. [DOI] [PubMed] [Google Scholar]
- 14. Hadano N, Murakami Y, Uemura K. et al. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br J Cancer 2016; 115(1): 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sausen M, Phallen J, Adleff V. et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 2015; 6(1): 7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee B, Cohen J, Lipton LR. et al. Potential role of circulating tumor DNA (ctDNA) in the early diagnosis and post-operative management of localised pancreatic cancer. Am Soc Clin Oncol 2017; 35(Suppl 15): 4101–4101. [Google Scholar]
- 17. Uemura T, Hibi K, Kaneko T. et al. Detection of K-ras mutations in the plasma DNA of pancreatic cancer patients. J Gastroenterol 2004; 39(1): 56–60. [DOI] [PubMed] [Google Scholar]
- 18. Pietrasz D, Pécuchet N, Garlan F. et al. Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin Cancer Res 2017; 23(1): 116–123. [DOI] [PubMed] [Google Scholar]
- 19. Bernard V, Kim DU, San Lucas FA. et al. Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology 2019; 156: 108–118.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Merker JD, Oxnard GR, Compton C. et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of American pathologists joint review. Arch Pathol Lab Med 2018; 142(10): 1242–1253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.