Abstract
Genomic instability is a hallmark of cancer, and often is the result of altered DNA repair capacities in tumour cells. DNA damage repair defects are common in different cancer types; these alterations can also induce tumour-specific vulnerabilities that can be exploited therapeutically. In 2009, a first-in-man clinical trial of the poly(ADP-ribose) polymerase (PARP) inhibitor olaparib clinically validated the synthetic lethal interaction between inhibition of PARP1, a key sensor of DNA damage, and BRCA1/BRCA2 deficiency. In this review, we summarize a decade of PARP inhibitor clinical development, a work that has resulted in the registration of several PARP inhibitors in breast (olaparib and talazoparib) and ovarian cancer (olaparib, niraparib and rucaparib, either alone or following platinum chemotherapy as maintenance therapy). Over the past 10 years, our knowledge on the mechanism of action of PARP inhibitor as well as how tumours become resistant has been extended, and we summarise this work here. We also discuss opportunities for expanding the precision medicine approach with PARP inhibitors, identifying a wider population who could benefit from this drug class. This includes developing and validating better predictive biomarkers for patient stratification, mainly based on homologous recombination defects beyond BRCA1/BRCA2 mutations, identifying DNA repair deficient tumours in other cancer types such as prostate or pancreatic cancer, or by designing combination therapies with PARP inhibitors.
Keywords: PARP inhibitors, DNA repair, clinical trials
Key Message
Poly(ADP-ribose) polymerase (PARP) inhibitors are active against homologous recombination-defective tumours. Over the last decade, clinical trials led to the approval of several PARP inhibitors in breast and ovarian cancer. Here, we review precision medicine approaches with PARP inhibitors, and opportunities for a wider population benefiting from this drug class, based on new knowledge on tumour genomics and DNA repair.
A deeper understanding of the molecular make up of tumours has brought opportunities to develop more precise treatments, targeting tumour vulnerabilities. Inhibitors of poly(ADP-ribose) polymerase (PARP) are examples of the path towards precision medicine. In this manuscript, we review the clinical development of PARP inhibitors (PARPi) across tumour types, leading to approvals in breast and ovarian cancer, and analyse opportunities ahead to optimize the use of this drug class.
Fast-track for translational research: the launch of PARPi clinical development
In 2005, two seminal studies demonstrated that tumour cells lacking BRCA1 or BRCA2, key tumour suppressor proteins involved in double-strand DNA break (DSB) repair by homologous recombination (HR), are selectively sensitive to small molecule inhibitors of the PARP family of DNA repair enzymes [1, 2]. The model proposed was based on the concept of synthetic lethality: loss of either of two genes is not lethal per se, but concomitant inactivation leads to cell death. PARP1, the major target of PARPi, is primarily involved in the repair of single-strand DNA breaks (SSBs); PARP1 inhibition alone is not lethal as the DNA lesions caused by these drugs can be repaired by other DNA repair pathways, specifically HR. In contrast, in the absence of BRCA1/2 and therefore defective HR, the DNA lesions caused by PARPi are not repaired and cause cytotoxicity.
In these studies, decreasing PARP1 levels by RNA interference also resulted in a significant reduction of cell survival selectively in BRCA1- and BRCA2-deficient cells [1]. Cancer cell lines lacking BRCA1/2 were also sensitive to inhibitors of PARP1, whereas cells with only heterozygous loss of BRCA1/2 genes or those without BRCA1/2 defects were not. In a back-to-back publication, depletion of BRCA2 using short-interfering RNA (siRNA) sensitized cancer cell lines to PARP inhibition [2]. Later studies demonstrated how loss of other tumour suppressor DNA repair proteins, many of which are involved in HR, also caused sensitization to PARPi [3–5].
PARPi were originally developed for cancer treatment as radio- and chemo-sensitizing drugs, but the aforementioned preclinical observations supported the development of PARPi as single agents for the treatment of BRCA1/2-defective cancers. In 2005, most of the data on BRCA1/2 related to the role of these genes as risk susceptibility factors for familial breast and ovarian cancers. Given this, germline BRCA1/2 (gBRCA1/2) mutation carriers with cancer were the initial target population to test the PARPi-BRCA synthetic lethal hypothesis in the clinic. A first-in-human clinical trial of KU-0059436 (KuDOS Pharmaceuticals/AstraZeneca, later named AZD-2281/olaparib) was conducted to establish a recommended dose and to generate preliminary data in a biomarker-defined population [6, 7]. In this proof-of-concept trial, pharmacokinetics and pharmacodynamics [in peripheral mononuclear blood cells (PBMC), hair follicles, and tumour samples) studies were used to optimize the dose-escalation and expansion phases. Expansion cohorts only included patients with gBRCA1/2 mutations. Doses of 60 mg or more twice daily of olaparib resulted in >90% PARP1 inhibition in PBMCs, suggesting biological activity at low doses. Dose-limiting toxicities of fatigue, somnolence and thrombocytopenia led to establishing 400 mg of olaparib capsules twice daily as the maximum tolerated dose. A modified tablet formulation with enhanced bioavailability was later developed; the current olaparib approved dose is 300 mg tablet twice a day [8]. Importantly, gBRCA1/2 mutation carriers did not experience enhanced toxicities, supporting the hypothesis of a cancer-specific vulnerability. Overall, 21 gBRCA1/2 mutation carriers were enrolled and evaluated for response, with radiological responses in eight patients with ovarian cancer and one with breast cancer, and a prostate cancer patient with a sustained PSA response.
This rapid translation of preclinical studies into promising clinical data triggered the development of several PARPi in different tumour types.
Mechanisms of action of PARPi: beyond synthetic lethality
PARP1 is a DNA damage sensor and signal transducer that binds to DNA breaks and then synthesises poly(ADP-ribose) (PAR) chains on target proteins (PARylation) in the vicinity of the DNA break and itself (autoPARylation). These PAR chains lead to the recruitment of additional DNA repair effectors that complete the DNA repair process. In its non-DNA bound state, PARP1 has minimal catalytic activity due to an auto-inhibitory helical domain (HD) interaction with its catalytic domain [9]. When PARP1 binds DNA, via zinc finger domains, a conformational change in the PARP1 protein relieves the autoinhibitory interaction between the HD and the catalytic domain, allowing nicotinamide adenine dinucleotide (β-NAD+), the PARP1 co-factor, to bind the active site of the enzyme. PARP1 then uses the hydrolysis of β-NAD+ to catalyse the transfer of ADP-ribose moieties on to target proteins. This PARylation of proteins in the vicinity of the DNA breaks then likely mediates DNA repair by modifying chromatin structure (e.g. via histone-PARylation) and by localizing DNA repair effectors (e.g. XRCC1). PARP1 autoPARylation eventually leads to its own release from the site of DNA damage [9, 10].
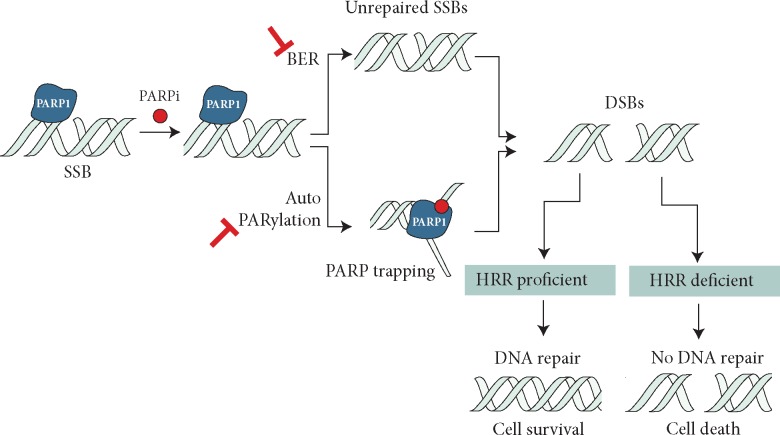
Pharmacological PARPi structurally mimic nicotinamide, and have two general effects: (i) catalytic inhibition of PARP1 (i.e. preventing PARylation) and (ii) locking or ‘trapping’ PARP1 on damaged DNA. Although the precise mechanisms that explain PARP1 trapping are still unclear, two have been proposed: (i) PARPi either prevents the release of PARP1 from DNA by inhibiting autoPARylation [11] or (ii) PARPi binding to the catalytic site causes allosteric changes in the PARP1 structure enhancing DNA avidity [3, 10, 12]. Either way, trapped PARP1 stalls the progress of replication forks (Figure 1). In normal, non-tumour cells, these stalled replication forks would be repaired by HR. In tumour cells that lack one of the key HR proteins, such as BRCA1, BRCA2, PALB2, or RAD51, cells use alternative DNA repair mechanisms in an attempt to repair DNA lesions caused by PARP inhibition, primarily through non-homologous or microhomology mediated end-joining (NHEJ or Alt-NHEJ). Rather than restoring the damaged DNA sequence back to its native form, the use of error-prone DNA repair pathways leads to a fragmentation of the genome that ultimately kills the cell.
Figure 1.
Proposed mechanisms for PARPi activity in HRR-deficient cells. PARP inhibition impairs repair of single strain breaks (SSBs) by disrupting the base excision repair (BER) pathway and also causing PARP1 trapping by inhibiting auto-PARylation and/or PARP release from DNA. These result in unresolved DNA double-strand breaks (DSBs) that in homologous recombination repair (HRR)-deficient cells lead to cell death.
Several PARPi in clinical development have different potencies as PARP1 catalytic inhibitors and as PARP-‘trappers’. It has been suggested that PARPi that are weak PARP1 trappers (e.g. veliparib), fail to elicit the same scale of synthetic lethality in pre-clinical models, compared with effective trappers (e.g. rucaparib, olaparib, talazoparib, niraparib) [13]. At present, no clinical trial has compared head-to-head different PARPi. Of note, iniparib (BiPar Sciences/Sanofi) was a compound developed as PARPi, but based on the alternative premise of altering PARP1 zinc-fingers (disturbing its activation by DNA breaks). After an unsuccessful phase III study in triple-negative breast cancer, preclinical work demonstrated that iniparib was not a bona fide PARPi [14].
In addition, PARP1 also binds directly to DNA to act as transcription factor and regulate chromatin structure remodelling. Recently, the role of PARP1 as transcriptional regulator for nuclear factors has raised some interest, due to the role of estrogen/progesterone and androgen receptors in breast and prostate cancer, respectively [15].
PARPi development across tumour types
Development of PARPi in ovarian cancer
Following the phase I trial, a phase II study assigned 33 patients with high-grade epithelial ovarian cancer (HGOC) and gBRCA mutations in a non-randomized manner to receiving olaparib at doses of either 100 (biologically active dose) or 400 mg (maximum tolerated dose) twice daily [16]. The primary end point was objective response rate (ORR), this being 33% at 400 mg b.i.d. and 13% at 100 mg b.i.d. PFS was also longer in the 400 mg cohort (median 5.8 versus 1.9 months). This dose–response relationship, was also suggested in another randomized trial (400 versus 200 mg), which also included a third arm of pegylated liposomal doxorubicin (PLD). Median PFS was 6.5 months (95% CI 5.5–10.1), 8.8 months (95% CI 5.4–9.2), and 7.1 months (95% CI 3.7–10.7) for the olaparib 200 mg, olaparib 400 mg, and PLD cohorts, respectively [17].
A multicentre phase II study enrolled 65 HGOC patients, regardless of gBRCA status. Confirmed RECIST responses with olaparib were seen in 41% and 24% of gBRCA mutation carriers and non-carriers, respectively [18], suggesting there could be a target population beyond gBRCA mutation carriers. Around 15% of HGOC are associated with gBRCA1/2 mutations (albeit regional differences). Some additional 6%–8% of patients can have tumour, not germline, BRCA1/2 mutations. Mutations in other HR genes such as RAD50, RAD51C/D, CDK12, or PALB2 are less common (<1%–3% each) [19].
All of these studies were conducted in populations previously exposed to platinum chemotherapy, the standard of care for ovarian cancer. There is a clear association between the platinum-sensitivity, defined based on the time gap from last platinum exposure to disease progression, and response to PARPi [20]. Platinum salts are DNA damaging agents and cause DNA cross-links that are in part repaired by HR. Hence, DNA repair deficient tumours are expected to be highly sensitive to platinum. At the same time, if resistant clones have restored DNA repair capacities, some cross-resistance with PARPi is expected [21]. Clinical trials in ovarian cancer shifted towards using PARPi as ‘maintenance’ therapy in patients who responded to platinum-based chemotherapy. The key initial study, olaparib's study 19, was carried out in high-grade serous ovarian cancer patients with platinum-sensitive relapse. This study met its primary end point, with improved PFS against placebo (8.4 versus 4.8 m, HR 0.35), even more pronounced in patients with germline/somatic BRCA1/2 mutations (11.2 versus 4.3 m, HR 0.18) [22, 23]. This led to the approval of olaparib as maintenance treatment of BRCA1/2-mutated patients in 2014; in 2018, the approval was expanded to all platinum-sensitive patients, regardless of BRCA1/2 status. The confirmatory phase III trial, SOLO-2 using the new tablet formulation in gBRCA1/2 mutation carriers confirmed these results (median PFS 19.1 versus 5.5 m for olaparib and placebo, respectively) [24]. In 2018, the phase III SOLO1 trial demonstrated an unprecedented benefit for olaparib versus placebo (HR PFS 0.30) in the maintenance setting after first-line platinum-based chemotherapy in FIGO stage III/IV high-grade ovarian cancer [25]. Based on these studies, olaparib is now also approved by the FDA for first-line maintenance of germline/somatic BRCA1/2-mutated HGOC (including fallopian tube or primary peritoneal cancer). Beyond the maintenance setting, olaparib is also FDA-approved for the treatment of gBRCA1/2-mutated ovarian cancer progressing to three or more prior lines of chemotherapy.
At present, two other PARPi have received approval for the treatment of ovarian cancer patients: niraparib and rucaparib.
Niraparib (previously MK4827, Merck/Tesaro) was first tested in a dose-escalation trial that established a recommended dose of 300 mg twice daily based on dose-limiting toxicities of fatigue, pneumonitis and thrombocytopenia [26]. Pharmacodynamic analyses confirmed PARP inhibition ≥50% at doses ≥80 mg/day, with responses observed ≥60 mg/day. Forty percent of 20 gBRCA1/2 mutation carriers with ovarian cancer achieved RECIST responses, with antitumour activity also documented in non-gBRCA associated platinum-sensitive cases. The development of niraparib in ovarian cancer has focussed on the post-platinum maintenance setting, although preliminary results of a phase II trial evaluating the drug in late-stage disease have been reported [27]. The phase III NOVA trial randomized 555 patients with platinum-sensitive recurrent HGOC to maintenance niraparib or placebo treatment after second or later line of platinum-based chemotherapy [28]. The study included two cohorts: gBRCA mutation carriers (median PFS 21 versus 5.5 months, P < 0.001) and non-gBRCA carriers (median PFS 9.3 versus 3.9 months, P < 0.001). In the latter, tumour samples were tested for genomic signatures associated with HR function defects [based on the presence of areas with loss-of-heterozygosity, large-scale state transitions (LST) and telomeric allele imbalance (TAI)]. Median PFS for the non-gBRCA carriers but signature-positive patients favoured niraparib (12.9 versus 3.8 months, P < 0.001). Patients without the HR-related signature still derived some benefit (median PFS 6.9 versus 3.8, P = 0.02). These data suggest that overall platinum-sensitivity status associates with PARPi sensitivity, although more benefit is seen in patients with canonical HR defects. This study led to niraparib being granted FDA and EMA approval in the maintenance setting, regardless of BRCA1/2 status.
Lastly, rucaparib (Clovis Oncology) was initially developed for IV administration but later evolved into an orally available drug, with a recommended dose of 600 mg twice daily [29, 30]. The ARIEL3 phase III trial evaluated rucaparib as maintenance treatment following platinum-based therapy in recurrent high-grade ovarian cancer, randomizing 564 women to rucaparib versus placebo (2 : 1) [31]. PFS was the primary end point, assessed in a step-down analysis for three predefined cohorts, namely, (i) germline/somatic BRCA1/2 mutation present, (ii) HR defect (HRD)-associated signature based on LOH, and (iii) the intent to treat population. The association between rucaparib sensitivity and an LOH-based signature had previously been identified in a phase II trial of rucaparib (ARIEL2) [32]. Rucaparib was superior to placebo in terms of PFS in all three cohorts (BRCA1/2-mutation positive: 16.6 versus 5.4 months P < 0.001; HRD signature: 13.6 versus 5.4 months, P=0.0001; and intention-to-treat population: 10.8 versus 5.4 months P=0.0001). Many of the BRCA1/2-WT, HRD-positive patients presented mutations in HR genes such as CHEK2, RAD51C, RAD51D, and RAD54L. These results led to rucaparib being FDA- and EMA-approved as maintenance treatment of HGOC regardless of BRCA1/2 status. In addition, rucaparib is also registered by the FDA and EMA for BRCA1/2-mutation associated ovarian cancer who have received at least 2 previous lines of chemotherapy [33].
Development of PARPi in breast cancer
In parallel to the initial ovarian studies, the biologically-active and the maximum-tolerated doses of olaparib were also tested in breast cancer in a non-randomized study [34]. This study demonstrated that olaparib (capsules, 400 mg b.i.d.) was effective and safe for patients with a gBRCA1/2 mutation, regardless of hormone receptor status. The ORR was 41% (11/27) and the main side-effects were fatigue and nausea (41% each), and anaemia (11% grades 3–4). Despite these initial encouraging results, several stumbling blocks delayed the clinical development in breast cancer [35]. In 2017, results from a randomized phase III trial (OlympiAD) in HER2-negative, gBRCA1/2 mutation-associated, metastatic breast cancer comparing olaparib with physician’s treatment choice (capecitabine, eribulin, vinorelbine, or gemcitabine) were reported [36]. Olaparib had superior median PFS [7.0 versus 4.2 months, HR 0.58 (95% CI 0.43–0.80)], with a delay on quality of life deterioration (HR 0.44; 95% CI 0.25–0.77), and a tolerable safety profile with only 4.7% rate of treatment discontinuation due to adverse events. This approach was validated in another phase III trial with talazoparib administered continuously compared with the same chemotherapy regimens [37], with a median PFS of 8.6 versus 5.6 months (HR 0.54, 95% CI 0.41–0.71). Of note, neither of the two trials were powered in their design to detect overall survival differences. Results from a phase III trial with niraparib and a similar design (BRAVO, NCT01905592) are awaited. Both olaparib and talazoparib are now approved by the FDA.
A phase II study with talazoparib (ABRAZO) [38] also demonstrated the association between antitumour activity of talazoparib and longer platinum-free interval.
Several trials are now investigating these therapies in earlier stages of the disease, including the adjuvant setting (NCT2032823), based on the hypothesis that fewer tumour evolution-related resistance mechanisms will be present in earlier stages of the disease. Talazoparib has also been tested as monotherapy in the neoadjuvant setting in patients with gBRCA1/2 mutation in a small phase II trial of 20 patients. Promisingly, the pCR was 53% and 63% when including residual cancer burden 0 and 1, respectively [39]. Conversely, veliparib, which has lower trapping potency, has not demonstrated significantly increased antitumour activity when combined with neoadjuvant platinum-based chemotherapy [40].
Expanding the indications for PARPi to other tumour types with HR defects
A basket trial of olaparib in patients with gBRCA1/2 mutations identified responding patients beyond the ovarian or breast cancer population, suggesting that other HR-defective tumours could be suitable for PARPi treatment [41]. Indeed, the phase I trials of olaparib, niraparib, and talozaparib have already documented responses beyond the two now approved indications. A recent study analysed the prevalence of BRCA1/2 alterations across tumour types beyond ovarian and breast cancer, in prostate, skin (non-melanoma), endometrial, pancreatic, and biliary duct cancers the prevalence was ≥5% [42]. Of those, clinical trials have been reported primarily for prostate and pancreatic cancer.
DNA repair gene alterations are common in metastatic prostate cancer (mPrC). Several genomic landscape studies have estimated that 8%–12% of all mPrC have BRCA2 mutations and homozygous deletions, with around half of these cases being linked to a germline mutation [43, 44]. When including other HR-related genes (such as PALB2, ATM, BRCA1, FANCA, etc.), up to 20%–25% of all mPrC harbour gene defects in different DDR pathways, which represents a significant enrichment when compared with localized, often indolent, prostate tumours [45]. The original phase I trial of niraparib included a small prostate cancer expansion cohort, with promising signs of clinical benefit [26]. Next, an investigator-initiated phase II study of olaparib in heavily pretreated mPrC patients documented responses in several patients. Using a combined definition of response including radiological responses, PSA falls, and CTC count conversions, 16/49 patients presented some degree of response. Retrospective genomic assessment of trial biopsies identified a strong association between antitumour activity and alterations in different DNA repair genes [46]. All eight patients with BRCA1/2 alterations responded to olaparib, many with durable responses for over 1 year, but mutations in genes such as ATM, PALB2, or FANCA were also observed in some responding patients. Similar data have been reported in a preliminary analysis of a phase II trial of rucaparib [47]. Based on these studies, registration trials of different PARPi in prostate cancer are now ongoing, although with different strategies when it comes to defining the putative predictive biomarker suite. Beyond their use as single agents in HR-defective tumours, PARP1 is an interesting target in prostate cancer based on the cross-talk between the androgen receptor (main oncogenic driver of prostate cancer) and DDR pathways; results from phase II combination trials of PARPi- and AR-targeting agents have now been reported [48] and have led to ongoing registration studies.
In the initial phase I study of talozaparib, two patients with pancreatic cancer and mutations in BRCA2 and PALB2, respectively, achieved radiological partial responses [49]. Platinum-based chemotherapy represents one of the main systemic treatments for advanced pancreatic cancer. Two recent prospective trials treated patients with stage III/IV pancreatic adenocarcinoma and germline BRCA1, BRCA2, or PALB2 mutations with veliparib or rucaparib. Response rates were 1/16 for veliparib [50] and 3/19 for rucaparib [51]. Of note, almost all patients in these trials had previously progressed on platinum-based chemotherapy. In pancreatic cancer, positive results of a phase III trial of olaparib for maintenance therapy after response to platinum chemotherapy, following the ovarian cancer model, have recently been announced. Other strategies currently being investigated include testing PARPi in combination with chemotherapy or as radiosensitizing agents.
Developing predictive biomarkers for PARPi
The analytical validation and clinical qualification of biomarkers able to stratify patients is of critical importance to deliver precision patient care with this drug class. During early phases of PARPi development, most efforts centred on detecting gBRCA1/2 mutations, initially using Sanger sequencing [52]. As interest has evolved beyond gBRCA1/2 mutations, multiplexed NGS assays of both germline and somatic DNA have become the preferred tool for patient identification [53, 54].
Multiplexed NGS panels assess a number of genes of interest (mostly exonic regions), either through amplification or capture-based technologies. Clinical implementation of multiplexed panels is, compared with wider whole-exome (WES) or whole-genome (WGS) sequencing, easier due to their lower cost and, critically, lower burden of bioinformatics requirements for data analysis. Accurate copy number assessment from targeted sequencing remains challenging, which is critical to demonstrating biallelic loss of DNA repair genes. This is, in part, due to intrinsic limitations in the technology but also relates to the need to integrate sample tumour purity, ploidy, and intratumour heterogeneity.
Wider WES or WGS assays add value by covering additional areas of the genome and by improving copy number and rearrangement calling. However, their applicability to clinical decision-making is still limited. Low-coverage WGS appears as a promising alternative for accurate and inexpensive copy number profiling, although it still requires significant bioinformatics analyses.
Incorporation of multiplexed biomarkers, such as DNA repair gene defects, has challenged some of the traditional concepts for biomarker validation in anticancer drug development. New genomic variants in the genes of interest are being identified, some of these being unique to individual patients. Hence, it is challenging to pursue clinical qualification of each of these separately in real time. Orthogonal methods to validate the impact on protein function of these findings is critical. Moreover, emergence of new variants also challenges traditional regulatory frameworks, demanding a continuous reassessment of the data in the post-approval setting [55]. Two main approaches could help circumvent the limitations of multiplexed biomarkers: (i) the identification of common genomic or transcriptional signatures in HR deficient tumours and (ii) assays capable of determining functional states of the HR pathway.
Genomic signatures or scars represent repetitive patterns of DNA alterations that translate an underlying biological condition [56]. Tumours deficient for HR prioritize NHEJ for repair of DSB, resulting in more errors ligating broken chromosomal ends. These tumours accumulate small insertions-deletions and loss-of-heterozygosity events [57]. Conversely, tumours with defective mismatch repair accumulate point mutations and tumours harbouring biallelic CDK12 loss develop a particular genomic profile characterized by repetitive tandem duplications, which increase neoantigen formation [58–60].
Several assays quantifying LOH events and/or TAI across the genome have been tested as predictive biomarkers of sensitivity to PARPi. Two of them have been approved by the FDA as companion diagnostics for PARPi in ovarian cancer: the ‘FoundationFocus CDx BRCA LOH’, evaluates the frequency of LOH events throughout the genome [57], and the ‘myChoice HRD’ (Myriad), a composite signature of LOH, TAI, and LST events. Most of the data on the predictive biomarker value of such signatures were generated in the randomized trials of niraparib and of rucaparib in ovarian cancer. The aim of such signatures is to identify patients likely to respond to PARPi by harbouring an intrinsic HR defect even when no obvious BRCA1/2 alterations are detected by NGS. However, these signatures present a critical limitation: the mutational/LOH patterns do not revert when a tumour has recovered HR function, so they may not be accurate to predict PARPi sensitivity in patients who have previously received and progressed on DNA damaging chemotherapy, such as platinum.
An alternative approach is to evaluate key DNA repair protein expression. ATM expression by IHC translates ATM genomic status and is associated with survival in patients with colorectal cancer treated with platinum [61]. Functional assays may directly inform on the capacity of the tumour cell to repair damage, translating upstream DNA/RNA alterations. Assays looking at accumulation of γH2AX (a marker of DNA damage) and RAD51 nuclear foci formation (indicating correct HR repair) are a gold standard for evaluating HR function in preclinical models. Their challenge raises from the need to be tested in samples with DNA damage and proliferating cells, since HR is cell cycle-related [62]. Functional assays evaluating γH2AX-RAD51 recently showed promising results in breast cancer biopsies, predicting PARPi sensitivity but also capturing the emergence of secondary resistances [63, 64].
Acquisition of PARPi resistance
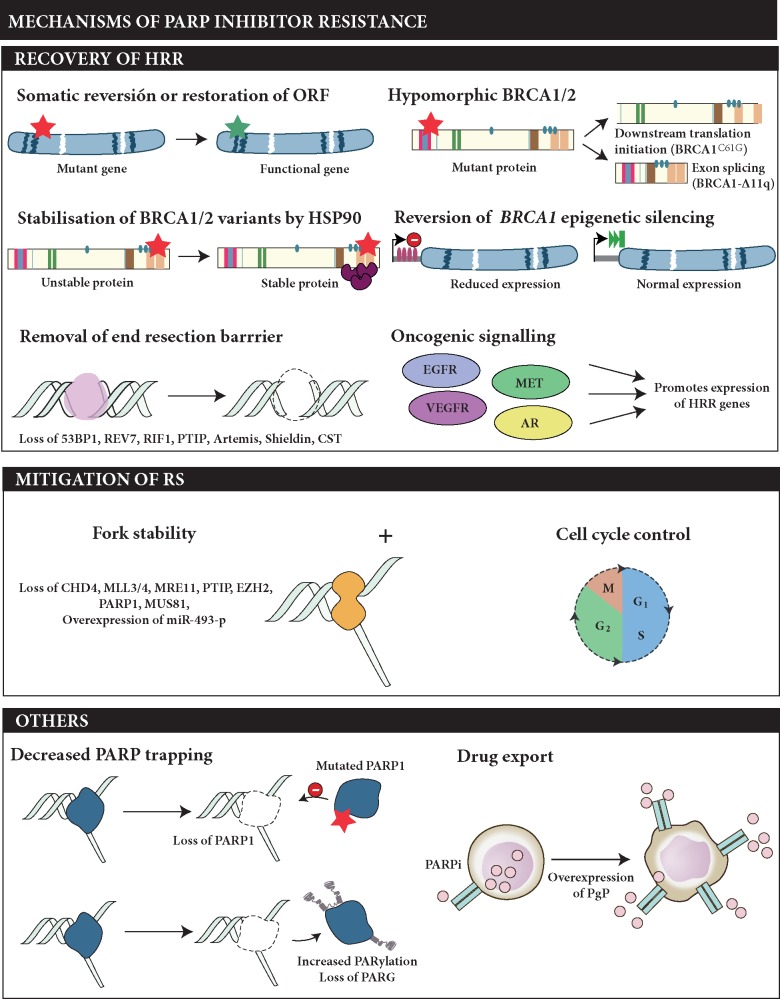
Numerous mechanisms of acquired resistance to PARPi have been described in pre-clinical and clinical studies [65, 66]; these can be grouped into three main classes (Figure 2).
Figure 2.
Described mechanisms of secondary resistance to PARP inhibitors. The potential mechanisms of PARPi resistance can be classified in three main groups: (i) those that result in restoration of homologous recombination repair (HRR), (ii) those leading to mitigation of replication stress (RS) commonly together with slower cell cycle progression, and (iii) other mechanisms not directly related with an HRR or RS pathway but that still alter the response to PARPi, such as mutations in PARP1 or drug effluxion pumps.
First, a number of different mechanisms result in restoration of HR function. The most common examples are secondary mutations restoring the open reading frame of HR repair genes (BRCA1/2, PALB2, RAD51C/D) in tumours with frameshift or nonsense mutations [67–70]. Expression of functional hypomorphic variants of BRCA1 has been associated with resistance to PARPi in patient-derived models [63, 71, 72]. Similarly, while epigenetic silencing of BRCA1 and RAD51C by hypermethylation of promoter regions results in PARPi sensitivity, demethylation is associated with mRNA re-expression and development of resistance [64, 73, 74]. There is also evidence in preclinical models of the co-existence of other mechanisms of resistance involving, for example, BRCA1 restoration with the removal of barriers to DNA end resection via loss of 53BP1 or proteins from the Shieldin complex, among others [75, 76].
An alternative mechanism for PARPi resistance is the protection of the replication fork, often combined with slowing cell cycle progression, as described in several preclinical studies. BRCA1, BRCA2 and PARP1, among others, play an important role in the protection of stalled replication forks, a critical step to enable replication fork repair after DNA damage [77–79]. In the absence of the aforementioned proteins, protected forks are extensively degraded, which leads to cell death. For example, BRCA2-mutant cells with loss of the MLL3/4 complex protein, were reported to become PARPi-resistant by fork protection through reduction of MRE11 recruitment to stalled forks [80]. Yazinski and colleagues further demonstrated that PARP-inhibitor resistant, BRCA1-deficient cells become dependent on ATR for survival [81].
Mutations in the DNA-binding domains of PARP1 represent other likely relevant mechanisms of resistance, although clinical data are sparse [82]. Similarly, mechanisms that increase PARylation of PARP1, such as loss of PARG, could lead to PARPi-resistance by decreasing PARP trapping [83].
Beyond mechanisms rewiring the DNA damage response, increased expression of ATP-binding cassette transporters, such as the P-glycoprotein efflux pump have been shown to reduce the efficacy of PARPi [84].
Closing remarks and future directions
The successful development of PARPi over the last 10 years has resulted in an effective therapeutic option being available for a subset of ovarian and breast cancers, with expansion to other biomarker-driven indications expected in the near future. These treatments exploit a tumour vulnerability, that in normal conditions makes these tumours more aggressive, and constitutes one of the prime examples of success in precision medicine to date.
We envision that in the years to come the utility of PARPi will expand further. First, by better understanding what alterations beyond BRCA1/2 sensitize different tumour types to PARP inhibition; this will require combining preclinical studies and clinical trial data. Secondly, by developing more precise assays to stratify sensitive patients, capturing different predictive biomarkers and, potentially, combinations of genomic events that when co-occurring together may be relevant. Facilitating the implementation of genomics in routine clinical practice will also lead to a wider population of patients being tested. To accomplish that aim, we need to provide better resources to physicians to access these technologies and to support genomic data analyses and interpretation.
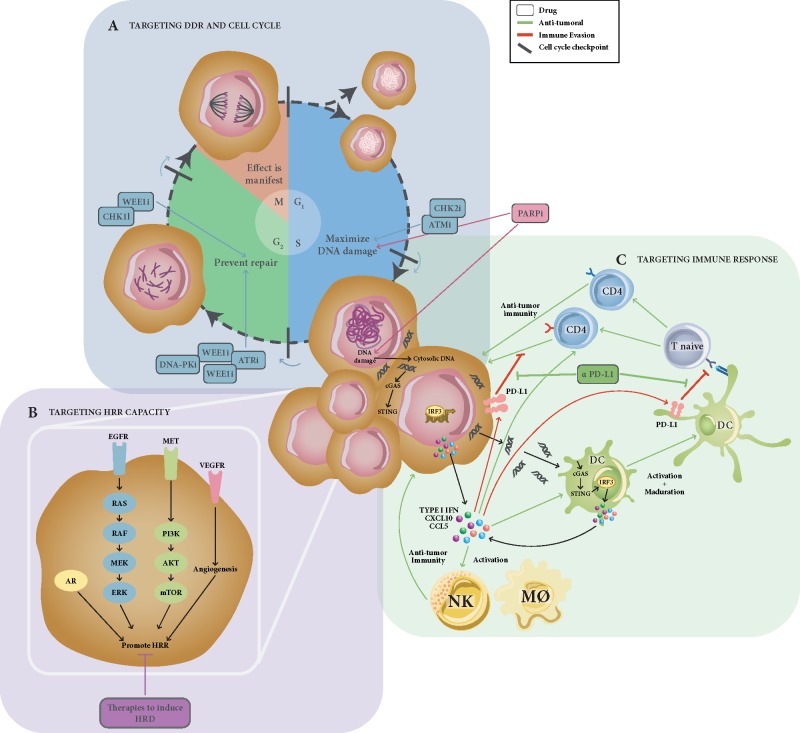
Lastly, rational drug combinations including PARPi may extend the patient population who may benefit from this drug class (Figure 3). Combinations with DNA damaging chemotherapy aim to maximize the effect of DNA damage, but they have been proved challenging due to overlapping toxicities [85]. Combination with DNA damaging radiation therapy should nevertheless be explored. Promising preliminary data have been reported combining PARPi with other targeted agents. This strategy could either aim to create synthetic lethal interactions by targeting several levels of the DNA repair machinery (i.e. combinations with ATR or HDAC inhibitors) [86, 87], take advantage of the relation between angiogenesis, hypoxia and DNA damage [88], or exploit the cross-talk between DDR and hormone receptor-driven pathways, such as ER and AR in breast and prostate cancer, respectively [89–91].
Figure 3.
Rational combinations of PARPi with other targeted agents. Hypothesis-driven combinations with PARP inhibitors are summarized; (A) combinations of PARPi with other compounds targeting alternatives DDR nodes aim to maximize accumulation DNA damage during G1 and S phases of the cell cycle, together with preventing its repair during G2 by minimizing the time to repair. This would lead to accumulation of DNA damage during mitosis and cell death. (B) Combinations with drugs targeting other biological pathways which have been shown to be modulated and/or to modulate HRR function, such as the PI3K/AKT pathway, RAS, VEGFR, and AR signalling pathways. (C) Rationale for developing PARPi-immunotherapy combinations; defects in DDR might increase genomic instability, leading to accumulation of mutations and, putatively, increased neoantigen production and T-cell activation. An alternative hypothesis supporting PARPi-immunotherapy combinations is the accumulation of cytosolic DNA induced by DDR defects, which would activate the innate immune system through the cGAS-STING pathway, inducing interferon-mediated response. This pro-inflammatory cascade would result in activation of NK cells and macrophages and the infiltration, proliferation and antitumour response of CD4+ and CD8+ T cells into the tumour. Paradoxically, the STING pathway also activates the expression of PD-L1 in tumour cells, therefore limiting the cytotoxic immune response, but potentially rendering the tumour sensitive to PD-L1 blockade (DC, dendritic cell; M∅, macrophage; NK, natural killer cell; Treg, regulatory T cell).
The advent of immune checkpoint inhibitors has transformed the management of several tumour types. There is a preclinical rational suggesting that PARP inhibition may trigger neoantigen and non-neoantigen-based mechanisms of tumour cell recognition by the immune system, making PARPi a potential partner for combination with immune checkpoint inhibitors [92–94]. A range of clinical trials exploring these combinations are now underway and may provide evidence of this clinical effect [95]. Optimizing dosage and scheduling of these combinations requires considering the time necessary for PARP inhibition to force tumour adaptation.
Funding
JM is supported by a Prostate Cancer Foundation Young Investigator Award and has received funding from Instituto de Salud Carlos III and FERO Foundation.
CJL and ANJT thank Breast Cancer Now and Cancer Research UK for funding the work in his laboratory. VS, CC, and MC-B have received funding support from Instituto de Salud Carlos III, Asociación Española Contra el Cáncer and FERO foundation. MC-B was supported by a PhD fellowship from AECC-JP Barcelona.
This work was supported by Prostate Cancer UK and the Movember Foundation through the London Movember Centre of Excellence (CEO13_2-002), the Prostate Cancer Foundation and through an Experimental Cancer Medical Centre (ECMC) grant from Cancer Research UK and the Department of Health (Ref: C12540/A25128). The authors acknowledge NHS funding to the NIHR Biomedical Research Centre at the Royal Marsden and The Institute of Cancer Research.
JSdB is a National Institute for Health Research (NIHR) Senior Investigator. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health.
Disclosure
JM has honoraria for consultancy for AstraZeneca, Roche and Janssen. CJL has received research funding from AstraZeneca, Merck KGaA, Artios. Received consultancy, SAB membership or honoraria payments from Sun Pharma, GLG, Merck KGaA, Vertex, AstraZeneca, Tango, 3rd Rock, Ono Pharma, Artios; and has stock in Tango. CJL is also a named inventor on patents describing the use of DNA repair inhibitors and stands to gain from the development as part of the ICR ‘Rewards to Inventors’ scheme. ANJT makes the following disclosures: received research funding from: AstraZeneca, Merck KGaA. Received consultancy, SAB membership or honoraria payments from: Merck KGaA, Vertex, AstraZeneca, Artios and is also a named inventor on patents describing the use of DNA repair inhibitors and stands to gain from the development as part of the ICR ‘Rewards to Inventors’ scheme. VS received research funding from AstraZeneca and Tesaro. AO has served as Advisor and received honorarium from Roche, AstraZeneca, Pharmamar, Clovis Oncology, Tesaro, Immunogen and Genmab. In addition, has received support for travel and/or accommodation from Roche, AstraZeneca, Clovis Oncology and Pharmamar. JB has served as advisor for AZ, Pfizer and Bristol Myers, and has received support for travel and/or accommodation from AstraZeneca and Pfizer. JdB has served as a consultant for Astellas, AstraZeneca, Bayer, Daiichi, Genentech, GSK, Janssen, Merck Serono, MSD, Orion, Pfizer Oncology, Sanofi-Aventis, Silicon Biosystems and Taiho. CJL, ANJT, SBK, and JdB are employees of The Institute of Cancer Research, which is a joint applicant for the patent entitled ‘DNA damage repair inhibitors for treatment of cancer.
References
- 1. Farmer H, McCabe N, Lord CJ. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005; 434(7035): 917–921. [DOI] [PubMed] [Google Scholar]
- 2. Bryant HE, Schultz N, Thomas HD. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005; 434(7035): 913–917. [DOI] [PubMed] [Google Scholar]
- 3. Murai J, Huang SY, Das BB. et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res 2012; 72(21): 5588–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McCabe N, Turner NC, Lord CJ. et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 2006; 66(16): 8109–8115. [DOI] [PubMed] [Google Scholar]
- 5. Bajrami I, Frankum JR, Konde A. et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res 2014; 74(1): 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fong PC, Boss DS, Yap TA. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009; 361(2): 123–134. [DOI] [PubMed] [Google Scholar]
- 7. Yap TA, Sandhu SK, Workman P, de Bono JS.. Envisioning the future of early anticancer drug development. Nat Rev Cancer 2010; 10(7): 514–523. [DOI] [PubMed] [Google Scholar]
- 8. Mateo J, Moreno V, Gupta A. et al. An adaptive study to determine the optimal dose of the tablet formulation of the PARP inhibitor olaparib. Targ Oncol 2016; 11(3): 401–415. [DOI] [PubMed] [Google Scholar]
- 9. Langelier MF, Eisemann T, Riccio AA, Pascal JM.. PARP family enzymes: regulation and catalysis of the poly(ADP-ribose) posttranslational modification. Curr Opin Struct Biol 2018; 53: 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Murai J, Huang SY, Renaud A. et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther 2014; 13(2): 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hopkins TA, Shi Y, Rodriguez LE. et al. Mechanistic dissection of PARP1 trapping and the impact on in vivo tolerability and efficacy of PARP inhibitors. Mol Cancer Res 2015; 13(11): 1465–1477. [DOI] [PubMed] [Google Scholar]
- 12. Murai J, Zhang Y, Morris J. et al. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J Pharmacol Exp Ther 2014; 349(3): 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shen Y, Rehman FL, Feng Y. et al. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res 2013; 19(18): 5003–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patel AG, De Lorenzo SB, Flatten KS. et al. Failure of iniparib to inhibit poly(ADP-Ribose) polymerase in vitro. Clin Cancer Res 2012; 18(6): 1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schiewer MJ, Goodwin JF, Han S. et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov 2012; 2(12): 1134–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Audeh MW, Carmichael J, Penson RT. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 2010; 376(9737): 245–251. [DOI] [PubMed] [Google Scholar]
- 17. Kaye SB, Lubinski J, Matulonis U. et al. Phase II, open-label, randomized, multicenter study comparing the efficacy and safety of olaparib, a poly (ADP-ribose) polymerase inhibitor, and pegylated liposomal doxorubicin in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer. J Clin Oncol 2012; 30(4): 372–379. [DOI] [PubMed] [Google Scholar]
- 18. Gelmon KA, Tischkowitz M, Mackay H. et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol 2011; 12(9): 852–861. [DOI] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fong PC, Yap TA, Boss DS. et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol 2010; 28(15): 2512–2519. [DOI] [PubMed] [Google Scholar]
- 21. Ang JE, Gourley C, Powell CB. et al. Efficacy of chemotherapy in BRCA1/2 mutation carrier ovarian cancer in the setting of PARP inhibitor resistance: a multi-institutional study. Clin Cancer Res 2013; 19(19): 5485–5493. [DOI] [PubMed] [Google Scholar]
- 22. Ledermann J, Harter P, Gourley C. et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol 2014; 15(8): 852–861. [DOI] [PubMed] [Google Scholar]
- 23. Ledermann J, Harter P, Gourley C. et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med 2012; 366(15): 1382–1392. [DOI] [PubMed] [Google Scholar]
- 24. Pujade-Lauraine E, Ledermann JA, Selle F. et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 2017; 18(9): 1274–1284. [DOI] [PubMed] [Google Scholar]
- 25. Moore K, Colombo N, Scambia G. et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2018; 379(26): 2495–2505. [DOI] [PubMed] [Google Scholar]
- 26. Sandhu SK, Schelman WR, Wilding G. et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol 2013; 14(9): 882–892. [DOI] [PubMed] [Google Scholar]
- 27. Moore CM, Alvarez Secord A, Geller MA. et al. QUADRA: a phase 2, open-label, single-arm study to evaluate niraparib in patients (pts) with relapsed ovarian cancer (ROC) who have received ≥3 prior chemotherapy regimens. J Clin Oncol 2018; 36(Suppl 15): Abstr 5514. [Google Scholar]
- 28. Mirza MR, Monk BJ, Herrstedt J. et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med 2016; 375(22): 2154–2164. [DOI] [PubMed] [Google Scholar]
- 29. Wilson RH, Evans TJ, Middleton MR. et al. A phase I study of intravenous and oral rucaparib in combination with chemotherapy in patients with advanced solid tumours. Br J Cancer 2017; 116(7): 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kristeleit R, Shapiro GI, Burris HA. et al. A phase I-II study of the oral PARP inhibitor rucaparib in patients with germline BRCA1/2-mutated ovarian carcinoma or other solid tumors. Clin Cancer Res 2017; 23(15): 4095–4106. [DOI] [PubMed] [Google Scholar]
- 31. Coleman RL, Oza AM, Lorusso D. et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017; 390(10106): 1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Swisher EM, Lin KK, Oza AM. et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol 2017; 18(1): 75–87. [DOI] [PubMed] [Google Scholar]
- 33. Oza AM, Tinker AV, Oaknin A. et al. Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high-grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: integrated analysis of data from Study 10 and ARIEL2. Gynecol Oncol 2017; 147(2): 267–275. [DOI] [PubMed] [Google Scholar]
- 34. Tutt A, Robson M, Garber JE. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010; 376(9737): 235–244. [DOI] [PubMed] [Google Scholar]
- 35. Balmana J, Domchek SM, Tutt A, Garber JE.. Stumbling blocks on the path to personalized medicine in breast cancer: the case of PARP inhibitors for BRCA1/2-associated cancers. Cancer Discov 2011; 1: 29–34. [DOI] [PubMed] [Google Scholar]
- 36. Robson M, Im SA, Senkus E. et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 2017; 377(6): 523–533. [DOI] [PubMed] [Google Scholar]
- 37. Litton JK, Rugo HS, Ettl J. et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med 2018; 379(8): 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Turner NC, Telli ML, Rugo HS. et al. A phase II study of talazoparib after platinum or cytotoxic nonplatinum regimens in patients with advanced breast cancer and germline BRCA1/2 mutations (ABRAZO). Clin Cancer Res 2019; 25(9): 2717–2724. [DOI] [PubMed] [Google Scholar]
- 39. Litton JK, Scoggins M, Hess KR. et al. Neoadjuvant talazoparib (TALA) for operable breast cancer patients with a BRCA mutation (BRCA+). J Clin Oncol 2018; 36(Suppl 15): Abstr 508. [Google Scholar]
- 40. Loibl S, O'Shaughnessy J, Untch M. et al. Addition of the PARP inhibitor veliparib plus carboplatin or carboplatin alone to standard neoadjuvant chemotherapy in triple-negative breast cancer (BrighTNess): a randomised, phase 3 trial. Lancet Oncol 2018; 19(4): 497–509. [DOI] [PubMed] [Google Scholar]
- 41. Kaufman B, Shapira-Frommer R, Schmutzler RK. et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015; 33(3): 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Agarwal N, Sokol ES, Lara P. et al. Pan-cancer assessment of BRCA1/2 genomic alterations (GAs) by comprehensive genomic profiling (CGP) of tissue and circulating tumor DNA (ctDNA). Ann Oncol 2018; 29(Suppl 8): mdy269.049. [Google Scholar]
- 43. Robinson D, Van Allen EM, Wu YM. et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 162(2): 454. [DOI] [PubMed] [Google Scholar]
- 44. Pritchard CC, Mateo J, Walsh MF. et al. Inherited DNA-Repair gene mutations in men with metastatic prostate cancer. N Engl J Med 2016; 375(5): 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Armenia J, Wankowicz SAM, Liu D. et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018; 50(5): 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mateo J, Carreira S, Sandhu S. et al. DNA-Repair defects and olaparib in metastatic prostate cancer. N Engl J Med 2015; 373(18): 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abida W, Bryce AH, Vogelzang NJ. et al. Preliminary results from TRITON2: a phase 2 study of rucaparib in patients with metastatic castration-resistant prostate cancer (mCRPC) associated with homologous recombination repair (HRR) gene alterations. Ann Oncol 2018; 29: vii271–vii302. [Google Scholar]
- 48. Clarke N, Wiechno P, Alekseev B. et al. Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 2018; 19(7): 975–986. [DOI] [PubMed] [Google Scholar]
- 49. de Bono J, Ramanathan RK, Mina L. et al. Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov 2017; 7(6): 620–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lowery MA, Kelsen DP, Capanu M. et al. Phase II trial of veliparib in patients with previously treated BRCA-mutated pancreas ductal adenocarcinoma. Eur J Cancer 2018; 89: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shroff RT, Hendifar A, McWilliams RR. et al. Rucaparib monotherapy in patients with pancreatic cancer and a known deleterious BRCA mutation. JCO Precis Oncol 2018; 2018. doi: 10.1200/PO.17.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sanger F, Coulson AR.. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 1975; 94(3): 441–448. [DOI] [PubMed] [Google Scholar]
- 53. Kurian AW, Hare EE, Mills MA. et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol 2014; 32(19): 2001–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zehir A, Benayed R, Shah RH. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017; 23(6): 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Evans BJ, Burke W, Jarvik GP.. The FDA and genomic tests–getting regulation right. N Engl J Med 2015; 372(23): 2258–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Alexandrov LB, Nik-Zainal S, Wedge DC. et al. Signatures of mutational processes in human cancer. Nature 2013; 500(7463): 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Davies H, Glodzik D, Morganella S. et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017; 23(4): 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu Y-M, Cieślik M, Lonigro RJ. et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 2018; 173(7): 1770–1782 e1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Quigley DA, Dang HX, Zhao SG. et al. Genomic Hallmarks and structural variation in metastatic prostate cancer. Cell 2018; 174(3): 758–769.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Joshi PM, Sutor SL, Huntoon CJ, Karnitz LM.. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J Biol Chem 2014; 289(13): 9247–9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sundar R, Miranda S, Rodrigues DN. et al. Ataxia telangiectasia mutated protein loss and benefit from oxaliplatin-based chemotherapy in colorectal cancer. Clin Colorectal Cancer 2018; 17(4): 280–284. [DOI] [PubMed] [Google Scholar]
- 62. Graeser M, McCarthy A, Lord CJ. et al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res 2010; 16(24): 6159–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cruz C, Castroviejo-Bermejo M, Gutierrez ES. et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol 2018; 29(5): 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Castroviejo-Bermejo M, Cruz C, Llop-Guevara A. et al. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med 2018; 10(12): e9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D'Andrea AD.. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov 2015; 5(11): 1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lord CJ, Ashworth A.. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med 2013; 19(11): 1381–1388. [DOI] [PubMed] [Google Scholar]
- 67. Barber LJ, Sandhu S, Chen L. et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J Pathol 2013; 229(3): 422–429. [DOI] [PubMed] [Google Scholar]
- 68. Norquist B, Wurz KA, Pennil CC. et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol 2011; 29(22): 3008–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Goodall J, Mateo J, Yuan W. et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov 2017; 7(9): 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kondrashova O, Nguyen M, Shield-Artin K. et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov 2017; 7(9): 984–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Drost R, Dhillon KK, van der Gulden H. et al. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J Clin Invest 2016; 126(8): 2903–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang Y, Bernhardy AJ, Cruz C. et al. The BRCA1-Delta11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res 2016; 76(9): 2778–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ter Brugge P, Kristel P, van der Burg E. et al. Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. J Natl Cancer Inst 2016; 108(11). doi: 10.1093/jnci/djw148. [DOI] [PubMed] [Google Scholar]
- 74. Kondrashova O, Topp M, Nesic K. et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun 2018; 9(1): 3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dev H, Chiang TW, Lescale C. et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat Cell Biol 2018; 20(8): 954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nacson J, Krais JJ, Bernhardy AJ. et al. BRCA1 mutation-specific responses to 53BP1 loss-induced homologous recombination and PARP inhibitor resistance. Cell Rep 2018; 25(5): 1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lomonosov M, Anand S, Sangrithi M. et al. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev 2003; 17(24): 3017–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schlacher K, Wu H, Jasin M.. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 2012; 22(1): 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Berti M, Ray Chaudhuri A, Thangavel S. et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol 2013; 20(3): 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ray Chaudhuri A, Callen E, Ding X. et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 2016; 535(7612): 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yazinski SA, Comaills V, Buisson R. et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev 2017; 31(3): 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pettitt SJ, Krastev DB, Brandsma I. et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat Commun 2018; 9: 1849.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gogola E, Duarte AA, de Ruiter JR. et al. Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell 2018; 33(6): 1078–1093.e1012. [DOI] [PubMed] [Google Scholar]
- 84. Rottenberg S, Jaspers JE, Kersbergen A. et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci USA 2008; 105(44): 17079–17084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Oza AM, Cibula D, Benzaquen AO. et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol 2015; 16(1): 87–97. [DOI] [PubMed] [Google Scholar]
- 86. Chao OS, Goodman OB Jr. Synergistic loss of prostate cancer cell viability by coinhibition of HDAC and PARP. Mol Cancer Res 2014; 12(12): 1755–1766. [DOI] [PubMed] [Google Scholar]
- 87. Kim H, George E, Ragland R. et al. Targeting the ATR/CHK1 axis with PARP inhibition results in tumor regression in BRCA-mutant ovarian cancer models. Clin Cancer Res 2017; 23(12): 3097–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liu JF, Barry WT, Birrer M. et al. Combination cediranib and olaparib versus olaparib alone for women with recurrent platinum-sensitive ovarian cancer: a randomised phase 2 study. Lancet Oncol 2014; 15(11): 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhang F, Wang Y, Wang L. et al. Poly(ADP-ribose) polymerase 1 is a key regulator of estrogen receptor alpha-dependent gene transcription. J Biol Chem 2013; 288(16): 11348–11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Goodwin JF, Kothari V, Drake JM. et al. DNA-PKcs-mediated transcriptional regulation drives prostate cancer progression and metastasis. Cancer Cell 2015; 28(1): 97–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Goodwin JF, Schiewer MJ, Dean JL. et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov 2013; 3(11): 1254–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jiao S, Xia W, Yamaguchi H. et al. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res 2017; 23(14): 3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Touat M, Sourisseau T, Dorvault N. et al. DNA repair deficiency sensitizes lung cancer cells to NAD+ biosynthesis blockade. J Clin Invest 2018; 128(4): 1671–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chabanon RM, Muirhead G, Krastev DB. et al. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J Clin Invest 2019; 129(3): 1211–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mouw KW, Goldberg MS, Konstantinopoulos PA, D'Andrea AD.. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov 2017; 7(7): 675–693. [DOI] [PMC free article] [PubMed] [Google Scholar]