Abstract
Genome packaging is an essential step to generate infectious HIV-1 virions and is mediated by interactions between the viral protein Gag and cis-acting elements in the full-length RNA. The sequence necessary and sufficient to allow RNA genome packaging into an HIV-1 particle has not been defined. Here, we used two distinct reporter systems to determine the HIV-1 sequence required for heterologous, non-viral RNAs to be packaged into viral particles. Although the 5’ untranslated region (UTR) of the HIV-1 RNA is known to be important for RNA packaging, we found that its ability to mediate packaging relies heavily on the context of the downstream sequences. Insertion of the 5’ UTR and the first 32-nt of gag into two different reporter RNAs is not sufficient to mediate the packaging of these RNA into HIV-1 particles. However, adding the 5’ half of the gag gene to the 5’ UTR strongly facilitates the packaging of two reporter RNAs; such RNAs can be packaged at >50% of the efficiencies of an HIV-1 near full-length vector. To further examine the role of the gag sequence in RNA packaging, we replaced the 5’ gag sequence in the HIV-1 genome with two codon-optimized gag sequences and found that such substitutions only resulted in a moderate decrease of RNA packaging efficiencies. Taken together, these results indicated that both HIV-1 5’ UTR and the 5’ gag sequence are required for efficient packaging of non-viral RNA into HIV-1 particles, although the gag sequence likely plays an indirect role in genome packaging.
Keywords: retrovirus, genome encapsidation, Gag:RNA interaction, luciferase, single virion analysis
Introduction
Most HIV-1 particles contain two copies of full-length viral RNA [1, 2]; this efficient genome packaging is mediated by the interactions between HIV-1 Gag polyprotein and cis-acting elements in full-length viral RNA [3–5]. All orthoretroviral Gag polyproteins contain matrix (MA), capsid (CA), and nucleocapsid (NC) domains. Additionally, the HIV-1 Gag polyprotein contains the p6 domain and two spacer peptides, SP1 and SP2, which are located between CA-NC and NC-p6 junctions, respectively. Of the Gag domains, NC plays an important role in retroviral RNA genome packaging [6–13]. Mutations in the HIV-1 NC domain, including those that alter the two CCHC zinc-chelating motifs, can reduce virion RNA packaging efficiency [10, 11, 13]. Additionally, complementation experiments have demonstrated that, of the thousands of Gag molecules assembled into each particle, a significant portion of the Gag (>17%) needs to contain functional NC before RNA can be packaged efficiently [14].
In the full-length retroviral RNAs, there are cis-acting sequences important for the encapsidation of the genome; such sequences are often referred to as the packaging signal. The packaging signal has been studied in multiple retroviruses including murine leukemia virus (MLV), spleen necrosis virus, Rous sarcoma virus (RSV), HIV-1, HIV-2, bovine leukemia virus (BLV), Mason Pfizer monkey virus (MPMV), and mouse mammary tumor virus [15–26]. In these retroviruses, sequences in the 5’ untranslated region (UTR) and often 5’ end of the gag gene are important for packaging. In MLV and RSV, the sequence necessary and sufficient for RNA packaging has been defined [15, 17]; when present in heterologous RNA, these sequences can mediate the encapsidation of heterologous RNA into MLV and RSV particles, respectively. Furthermore, the heterologous RNA containing the MLV packaging signal was encapsidated into viral particles as a dimer [27]. However, the sequence necessary and sufficient for efficient packaging of non-viral RNA into HIV-1 particles has not been defined.
Multiple studies have been performed to examine RNA elements important for HIV-1 RNA packaging ([18, 19, 28–32] and summarized in [33–38]). These studies showed that mutations in the 5’ UTR and gag, as well as other elements, affects HIV-1 RNA packaging. The 5’ UTR region of the viral RNA is highly structured and forms multiple stem-loop structures [39–44]; several studies suggest that many but not all of the 5’ UTR RNA structures are essential for genome packaging. For example, the trans-activation region (TAR), a stem-loop structure located at the very beginning of the HIV-1 RNA was suggested to be important for RNA packaging. However, later studies showed that another RNA stem-loop structure can partially replace the TAR element function, and it was suggested that TAR mainly provides stability to the RNA structure to facilitate genome packaging [32, 45–47]. Although it is generally agreed that sequences beyond the AUG of the gag gene can affect RNA packaging, the precise extent of the gag gene required for packaging has varied in different studies [32, 48]. Furthermore, translation of the gag gene is not required for its effect in enhancing RNA packaging [49]. The Gag-Pol ribosomal frameshift signal and the Rev response elements (RRE) were also suggested to be important for RNA packaging. Viral protein Rev binds to RRE and mediates the export of full-length and partially spliced HIV-1 RNAs [50–54]; Rev-RRE was suggested to play a role in HIV-1 RNA packaging as the lack of Rev or mutation of RRE affects genome packaging [55, 56]. However, other studies showed that the function of Rev-RRE can be replaced by the constitutive transport element (CTE) from MPMV [32, 57, 58]. The CTE from MPMV does not contain sequence or structural similarity with HIV-1 RRE; additionally, CTE mediates RNA export via the NXF1 pathway whereas the Rev-RRE complex mediates the RNA export via the CRM1 pathway [59–62]. Hence, the major effect of the Rev-RRE mutations is the lack of proper RNA export that leads to defects in genome packaging and the Rev-RRE complex does not directly participate in genome packaging [57, 58]. Similarly, the ribosomal frameshift signal that mediates the expression of Gag-Pol polyprotein was originally hypothesized to play an important role in RNA packaging but was later demonstrated to have no effect on RNA packaging [63, 64].
At this time, the minimal sequence necessary and sufficient to allow non-viral RNAs to be efficiently packaged into HIV-1 particles has not been defined. In this report, we sought to define the sequences required for non-viral reporter RNAs to be efficiently packaged into HIV-1 particles. We used a previously described single-virion analysis system [1] to visualize viral RNA in individual particles to determine the efficiency of HIV-1 RNA genome packaging. We performed systematic deletion/replacement of portions of the viral genome to define regions dispensable for RNA packaging. We then inserted HIV-1 sequences into reporter RNAs encoding either a firefly luciferase gene or a Renilla luciferase gene and examined the packaging efficiency of these RNAs. We found that including the 5’ UTR and the 5’ half of the gag gene sequence allows the non-viral reporter RNAs to be packaged efficiently into viral particles. However, the role of the gag gene sequences is likely to be to stabilize the RNA structure rather than contain specific Gag:RNA recognition sites. These results define the minimum HIV-1 RNA packaging signal and provide a better understanding of RNA genome encapsidation, a process essential for the generation of infectious HIV-1.
RESULTS
Experimental system used to examine HIV-1 RNA genome packaging efficiency.
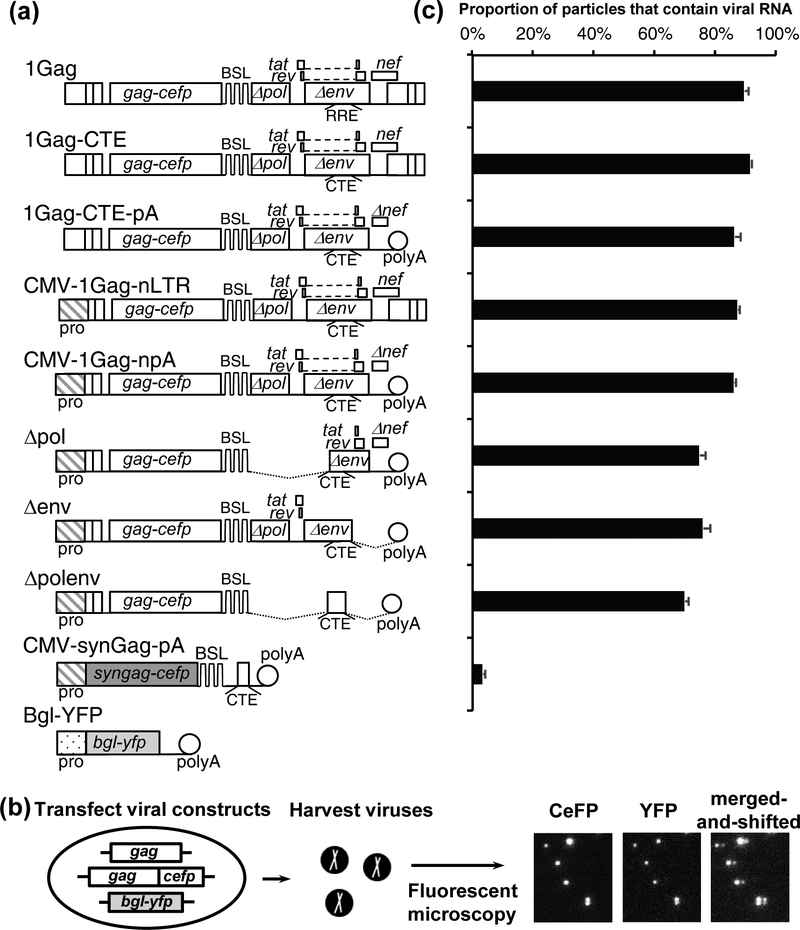
We used two previously described, NL4–3-based constructs to perform single-virion analysis. The general structure of one of the clones, 1Gag, is shown in Fig. 1a. This construct contains all of the cis-acting elements required for viral replication and expresses Gag fused to cerulean fluorescent protein (Gag-CeFP), Tat, Rev, and Nef, whereas portions of the pol, env, vif, vpr, and vpu gene were deleted. Additionally, a set of stem-loop sequences (BSL) recognized by bacterial protein BglG is located in the pol gene; thus, only the full-length, unspliced viral RNA contains the BSL. Construct 1GagΔCeFP, has the same structure as 1Gag but expresses an untagged Gag. For each of the constructs described below, a pair of plasmids were used, one that expresses Gag-CeFP and another that expresses untagged Gag; for brevity, only the Gag-CeFP version of the constructs are shown in all figures.
Fig.1.
System used to identify HIV-1 sequences important to RNA packaging. (a) General structures of constructs. Although only the constructs expressing Gag-CeFP fusion proteins are shown, each construct has another version that expresses untagged Gag; both plasmids were co-expressed in all experiments. (b) Outline of the experimental protocol and examples of the images. Gag and RNA signals were detected in CeFP and YFP channels, respectively. In the merged-and-shifted image, the image from the YFP channel was shifted 5-pixel to the right. (c) RNA genome packaging efficiencies of particles generated by HIV-1 constructs. RNA packaging efficiency was calculated by the proportion of viral particles identified by the CeFP signals that contain RNA (YFP) signals. Results from three independent experiments are summarized; error bars indicate standard deviations.
Single-virion analysis was performed by transfecting 293T cells with 1Gag, 1GagΔCeFP, and Bgl-YFP, which encodes a truncated BglG fused to yellow fluorescent protein (YFP). HIV-1 particles were harvested and visualized using fluorescent microscopy (Fig. 1b). The coexpression of Gag and Gag-CeFP allows the formation of morphologically normal HIV-1 particles [1, 2, 65, 66] and the detection of viral particles by their CeFP signals. The BSL in the viral RNA allows specific binding of Bgl-YFP to the full-length viral RNA genome; thus, the viral RNA genome is detected by the YFP signal. RNA packaging efficiency, the proportion of the viral particles containing viral RNA genomes, can be determined by comparing the CeFP signals and YFP signals from >1,000 particles in each sample. Using this method, we have previously shown that most HIV-1 particles contain full-length viral RNAs [1]. Consistent with previous studies, in this report, we found that most of the viral particles derived from 1Gag contain viral RNA (89% ± 1.4%; average and standard deviation from three experiments; Fig. 1c).
In this report, we sought to determine the minimal sequences required to efficiently package RNA into HIV-1 particles. For this purpose, we tested the effects of replacing three cis-acting elements, 5’ U3, RRE, and 3’ LTR, with non-HIV-1 sequences, and examined the efficiency of viral RNA packaging in particles generated from these constructs. First, we used a set of previously described vectors, 1Gag-CTE (Fig. 1) and 1GagΔCeFP-CTE [57], in which RREs were replaced with the CTE from MPMV [67]. Consistent with previous results, we found that RNAs from these CTE-containing HIV-1 constructs were packaged efficiently; ~91% of the particles contain viral RNA (Fig.1c). Similarly, replacing the 3’ LTR with the polyA signal from SV40, the 5’U3 with a CMV promoter, or both elements did not affect RNA packaging efficiencies (86% ± 1.9%, 87% ± 0.6%, 86% ± 0.5%, respectively; Fig. 1c).
To examine whether RNA sequences at the 3’ half of the viral genome contribute to genome packaging, we generated three mutants based on CMV-1Gag-npA (Fig. 1a). In construct Δpol, all of the pol sequences, the first exons of tat and rev, and a 5’ portion of the env were deleted; in construct Δenv, the 3’ portion of env, the second exons of tat and rev, 3’ UTR, and a portion of the nef gene were deleted; in construct Δpolenv all HIV-1 sequences 3’ to the gag gene were deleted. As shown in Fig. 1c, deletions of sequences 3’ to the gag gene had minor effects on RNA packaging efficiency; RNAs derived from Δpol, Δenv, and Δpolenv were packaged with the efficiencies of 75% ± 2%, 76% ± 2%, and 70% ± 1%, respectively. In contrast, RNAs derived from a codon-optimized Gag expression construct, CMV-synGag-pA (Fig. 1a), were rarely incorporated into viral particles (3% ± 0.3%; Fig. 1c) even though this construct encodes a functional gag gene. The codon-optimized gag sequence was derived from a previously described construct pSYNGP, which was modified based on the HXB2 molecular clone; compared with the NL4–3 sequence, the codon-optimized gag gene contains 78% nucleotide sequence identity and 96% amino acid sequence identity; a comparison of the nucleotide sequences is shown in Supplemental Figure 1. These results confirmed that the cis-acting elements in the viral RNA, and not the ability to serve as template for Gag translation, are important for efficient genome packaging. Furthermore, cis-acting elements important to HIV-1 RNA packaging reside primarily in 5’ UTR and/or gag gene sequences.
Sequences in the 5’ UTR and 5’ gag affect HIV-1 RNA packaging.
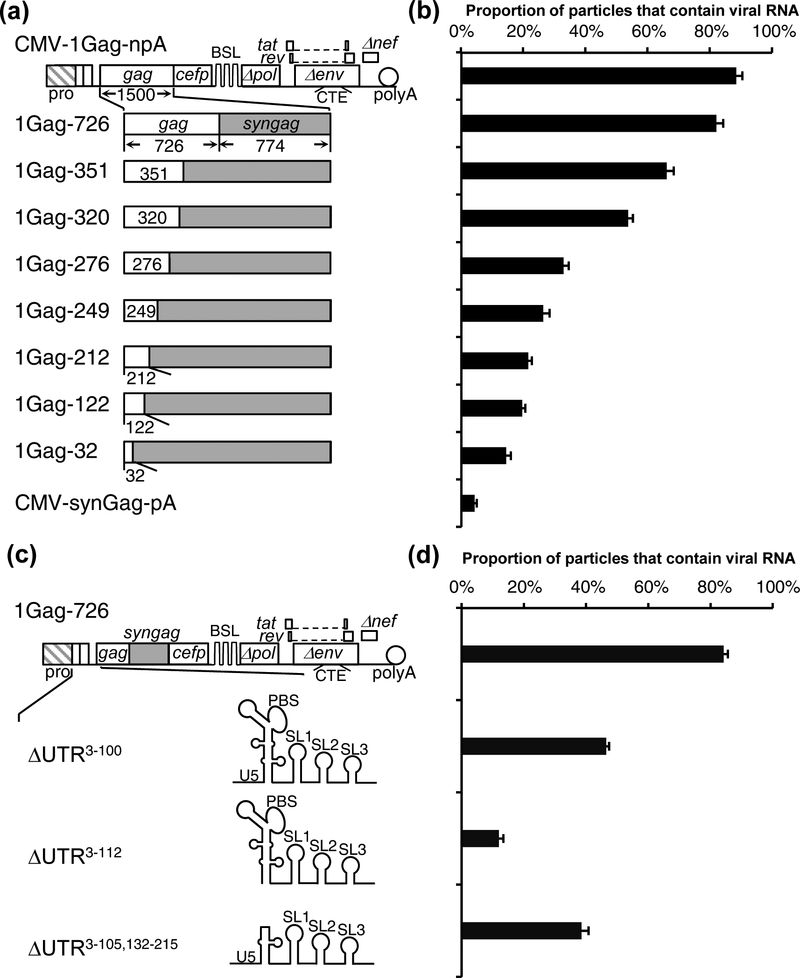
To determine whether sequences in the gag gene affect RNA packaging, we generated a series of mutants based on CMV-1Gag-npA (Fig. 2a). In these mutants, a portion of the gag was replaced with sequences from the codon optimized gag of CMV-synGag-pA (Fig. 1) so that the chimeric gag genes encode functional Gag proteins. The amino acid coding region in the NL4–3 gag gene is 1500-bp in length. The number of HIV-1 nucleotides retained on the 5’ end of the gag gene is reflected in the name of the construct; for example, 1Gag-726 retains the 5’ 726-bp of the HIV-1 (NL4–3) gag sequence (Fig. 2a). We generated 8 sets of constructs expressing chimeric gag genes and performed single-virion analyses to determine the genome packaging efficiencies. Despite the synonymous mutations, we found that all of the chimeric gag genes generated similar amounts of CeFP particles (data not shown); however, the RNA genome packaging efficiencies varied between these constructs (Fig. 2b). In particles generated from 1Gag-726, RNA genome packaging efficiency is 82% ± 2%, similar to that from CMV-1Gag-npA (88% ± 2%). In contrast, RNA genome packaging is inefficient in particles generated by 1Gag-32; only 14% ± 1% of the particles contained viral RNA genomes. Constructs containing 351-nt to 122-nt of HIV-1 gag sequences exhibited a gradient of packaging efficiencies; those with longer HIV-1 gag sequences were generally packaged more efficiently, indicating that the effects of the gag gene appeared to be dispersed over this region. These studies indicated that the nucleotide sequences of the 5’ half of the gag gene affects HIV-1 RNA genome packaging efficiency.
Fig. 2.
Determining the gag gene and the 5’ UTR sequence important for HIV-1 RNA packaging. (a) General structures of HIV-1 constructs containing a hybrid HIV-1 (NL4–3) gag/syngag gene with a portion of the HIV-1 gag replaced by codon-optimized sequences. Codon optimized sequences are shown as grey boxes. Numbers in the name of each construct indicate the length of NL4–3 gag sequence. (b) RNA genome packaging efficiencies of viral particles generated by HIV-1 constructs containing hybrid gag/syngag genes. Results from six independent experiments are summarized; error bars indicate standard deviations. (c) General structures of HIV-1 constructs containing 5’ UTR deletions. The numbers in the name of each construct indicate deleted sequences; the 1st base of the NL4–3 RNA transcript is defined as 1. (d) RNA genome packaging efficiencies of viral particles generated by HIV-1 constructs with mutations in 5’ UTR. Results from three independent experiments are summarized; error bars indicate the standard deviations.
The 5’ UTR of the HIV-1 RNA has been shown to be important for RNA packaging although some reports suggested that not all of the elements are required for efficient encapsidation. To determine the sequences in the 5’ UTR that are required for efficient encapsidation, we generated deletion mutants and determined the packaging efficiency of RNAs derived from these constructs (Fig. 2c); the deleted sequences are reflected in the names of the constructs. For simplicity, Figure 2 uses the RNA element structure and nomenclature that are commonly used in similar studies, although a recent study has suggested that the sequence often referred to as stem loop 2 (SL2) may not form a stem loop and may instead be used to stabilize a three-way junction structure [40].
We first tested deletion mutant that lack both TAR and polyA stem-loop structures (ΔUTR3−100) and found deleting these sequences resulted in a ~2-fold decrease in packaging efficiency. Particles derived from 1Gag-726 control and ΔUTR3−100 packaged RNA genomes at 83% ± 1% and 46% ± 1%, respectively. However, further deletions removing part of the U5 stem (ΔUTR3−112) results in drastic reduction of packaging efficiency; particles derived from this construct packaged RNA genome at 12% ± 1.4% (Fig. 2d). It has been suggested that the upper half of the PBS stem loop is not required for packaging; we generated a construct (ΔUTR3−105, 132−215) in which the TAR, polyA, and a portion of the PBS stem-loop was deleted and found that 38% ± 2% particles generated from this constructs packaged viral RNA.
These studies suggest that deletion of most of the elements in the 5’ UTR decreases RNA packaging. Therefore, we used the complete 5’ UTR and part of gag sequence to examine the sequence necessary and sufficient for heterologous RNA packaging into HIV-1 particles.
Sequence required to package a heterologous non-viral RNA into HIV-1 particle.
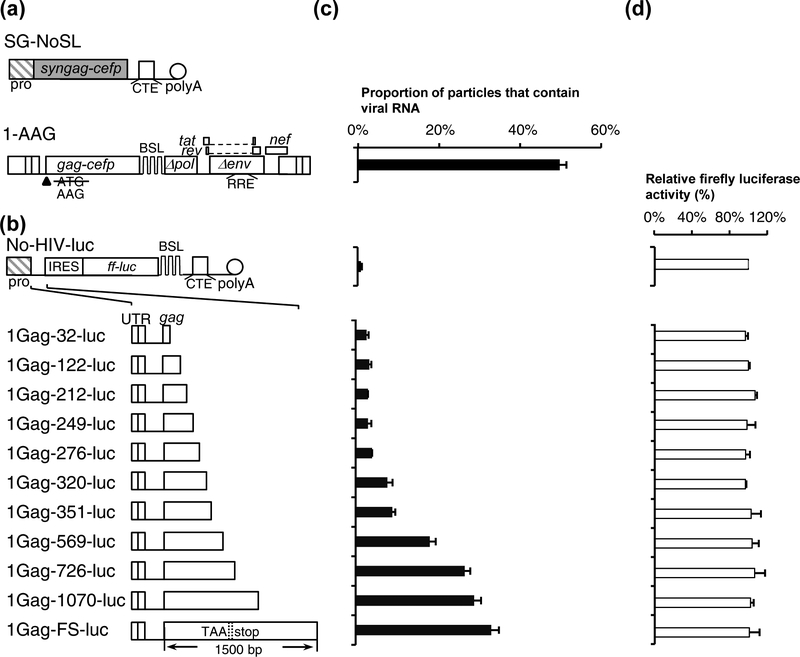
The studies above described the roles of various sequences in RNA encapsidation in the context of the viral genome. To determine the sequence necessary and sufficient for efficient genome encapsidation, we inserted various HIV-1 sequences into a non-HIV-1 reporter RNA and examined the efficiencies with which these RNAs were packaged. For this purpose, we cotransfected a reporter RNA construct, Bgl-YFP, and SG-NoSL (Fig. 3a) that expresses codon-optimized Gag and GagCeFP, harvested viral particles and performed imaging analyses. To assess this system, we first determined the packaging efficiency of RNA from HIV-1 construct 1-AAG (Fig. 3a), which is similar to the 1Gag construct described in Fig. 1 but contains an AAG instead of an AUG translational start codon in the beginning of gag. In a previous report, we have shown that the 1-AAG construct does not express functional Gag protein but expresses an RNA that can be efficiently packaged [49]. We found that when cotransfected with SG-NoSL and Bgl-YFP, our positive control 1-AAG RNA was packaged at 49% ± 2%, lower than those from systems described in Fig. 1 and Fig. 2. The Gag expression constructs are codon-optimized and lack the long HIV-1 5’ UTR and other viral sequences, hence the Gag proteins are generally expressed better from these constructs than those from the HIV-1 constructs. As a result, there may be more particles without viral RNAs.
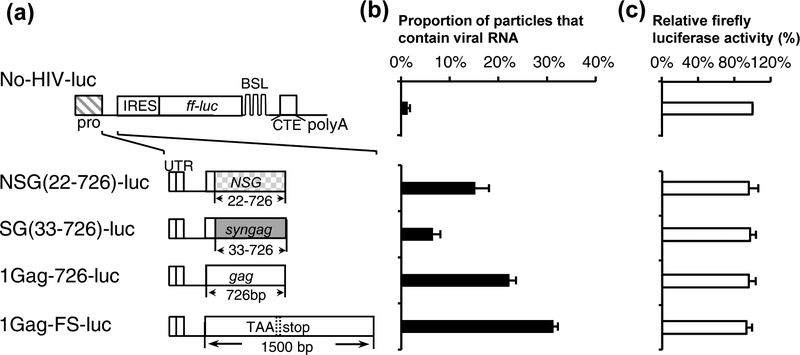
Fig. 3.
Delineating the HIV-1 sequence required to package a heterologous RNA into particles. (a) General structures of an HIV-1 helper and vector. Plasmid SG-NoSL expresses codon-optimized Gag-CeFP fusion protein whereas HIV-1 vector 1-AAG contains a mutation that abolished the gag translation start codon from AUG to AAG. (b) General structures of a firefly luciferase reporter constructs and the inserted HIV-1 sequences. The length of the gag sequence is indicated in the names of the constructs. (c) Packaging of heterologous RNAs. (d) Cellular expression of the reporter RNAs monitored by firefly luciferase activities. Firefly luciferase activities were normalized to that from No-HIV-luc. Results from three independent experiments are summarized; error bars indicate the standard deviations.
We then determined the HIV-1 sequence required to package a non-HIV-1 reporter RNA based on construct No-HIV-luc (Fig. 3b), which contains a CMV promoter that drives RNA transcription, an internal ribosomal entry site (IRES) from encephalomyocarditis virus followed by a firefly luciferase gene, a MPMV CTE element, and an SV40 poly A sequence. The firefly luciferase gene allows us to use the luciferase assay as a surrogate for RNA expression whereas the CTE element is included in the constructs to allow efficient RNA export as certain sequences in HIV-1 gag have been reported to have negative effects on nuclear export [68, 69]. We then generated a series of luciferase constructs containing various HIV-1 sequences (Fig. 3b); these plasmids were transfected into cells along with the codon-optimized Gag expression construct SG-NoSL and Bgl-YFP. Supernatants were harvested from transfected cells and used to determine RNA packaging efficiency (Fig. 3c); additionally, transfected cell lysates were generated and used to determine luciferase activity to monitor the expression of the luciferase containing plasmids.
We found that all of the tested reporter constructs resulted in similar levels of luciferase activity (Fig. 3d), indicating that their RNAs were similarly expressed; however, RNAs derived from these reporter constructs were packaged at different levels. As expected, in the absence of HIV sequence, reporter RNA (No-HIV-luc) was rarely present in the viral particles (~1%; Fig. 3c). When the entire 5’ UTR along with the first 32-nt of the gag gene was included (1Gag-32-luc), this RNA was not packaged efficiently and only 3% of the particles contained reporter RNA (Fig. 3c). We then gradually increased the amount gag sequence in the constructs; RNAs containing 5’ UTR plus 122-, 212-, 249-, and 276-nt of gag sequences were not packaged efficiently as only 3–4% of the particles contain reporter RNA. However, inserting additional gag sequences gradually improved the RNA packaging efficiencies; RNAs containing HIV-1 5’ UTR plus 320-, 351-, and 569-nt of gag were packaged at 8% ± 1%, 9% ± 1%, and 18% ± 1%, respectively. Furthermore, RNAs containing 5’ UTR plus 726-, 1070-, and the full-length gag were packaged at 26% ± 1%, 29% ± 2%, and 33% ± 2%, respectively; when compared with the HIV-1 construct 1-AAG, these RNAs are packaged at 54%, 59%, and 67% of the 1-AAG RNA, respectively. The full-length gag sequence in 1Gag-FS-luc contains a premature stop codon to abolish the expression of functional Gag. These studies indicate that the 5’ UTR and the very beginning of the gag sequences is not sufficient to mediate the efficient packaging of a heterologous RNA. Instead, heterologous RNA including the 5’ UTR and at least the first half of the gag gene sequence RNA can be packaged into the particles efficiently, within two-fold of a HIV-1 vector RNA (1-AAG). Thus, in addition to the 5’ UTR, a significant portion of the gag gene is required to mediate efficient heterologous RNA packaging.
Examining the role of the gag sequence in HIV-1 RNA packaging.
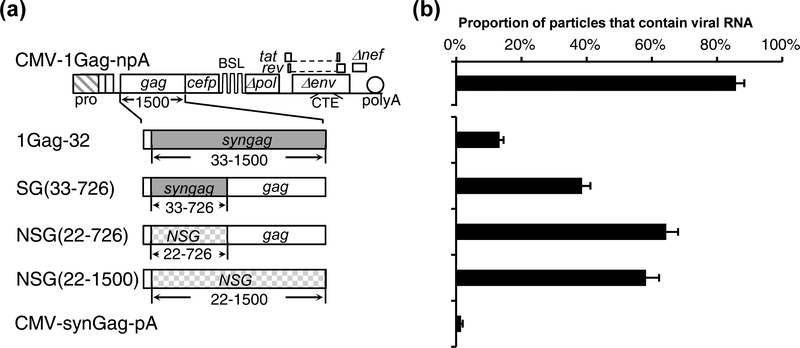
Our results described above showed that the 5’ half of the gag sequence is important to RNA packaging whereas the 3’ half of the gag sequence has little effect. In the aforementioned constructs, the modification/deletion starts from the 3’ end of the gag gene while retaining the 5’ end of the gag gene. To better define the role of the gag sequence in RNA packaging and to confirm our results, we generated and tested a construct SG(33–726) that contains codon-optimized 5’ gag sequence and HIV-1 3’ gag sequence; in this 5’ gag region, SG and HIV-1 gag share 71% nucleotide sequence homology (Supplemental Fig. 1). SG(33–726) was generated by replacing the 3’ half of the codon-optimized gag gene in 1Gag-32 (Fig. 2 and Fig. 4) with HIV-1 gag sequence (Fig. 4). To our surprise, replacing the 3’ half of the codon optimized gag with corresponding NL4–3 gag sequence recovered the RNA packaging efficiency from 13% ± 1% (1Gag-32) to 39% ± 3% [SG(33–726)] (Fig. 4b). However, our results, summarized in Fig. 2 and Fig. 3, showed that removing the 3’ gag sequence did not impact RNA packaging. We envisioned two possible explanations for these results: there may be redundant RNA elements in 5’ and 3’ gag that direct RNA packaging; alternatively, gag sequence may play an indirect role in RNA packaging, such as stabilizing RNA structures elsewhere that are important for viral RNA encapsidation. In the experiments described above and shown in Fig. 2, the HIV-1 gag sequence was replaced with that from a previously described codon-optimized expression construct pSYNGP [70]. We hypothesize that if the role of HIV-1 gag sequence is direct, such as elements in the viral RNA specifically interacting with the Gag protein to mediate packaging, then the function of gag RNA will not be easily replaced by other sequences. However, if the role of gag sequence is indirect, other sequences may be able to replace its function even though the sequence from the SYNGP plasmid cannot do so. To distinguish between these two possibilities, we tested whether another sequence can replace the function of the gag sequences. For this purpose, we generated a modified gag with synonymous mutations, referred to as new synonymous gag (NSG); the comparison of gag sequences from HIV-1, pSYNGP, and NSG is shown in Supplemental Fig. 1. NSG contains 71% nucleotide sequence identity with NL4–3 gag gene, and 73% nucleotide sequence identity with the codon-optimized gag gene from pSYNGP (Supplemental Fig. 1); the gag gene from NL4–3 and NSG encode the same amino acid sequence. We replaced the NL4–3 5’ gag sequences or most of the gag sequence in CMV-1Gag-npA with NSG to generate NSG(22–726) and NSG(22–1500), respectively (Fig. 4a) and performed single-virion analyses. We found that RNAs generated from these constructs were packaged efficiently at 66% ± 4% [NSG(22–726)] and 60% ± 4% [NSG(22–1500)]. The results from NSG(22–1500) are in sharp contrast with those from 1Gag-32; in both constructs, most of the gag genes contained synonymous mutations and yet the RNAs from these two constructs were packaged at very different efficiencies: 13% for 1Gag-32 and 60% for NSG(22–1500). These results suggest that the role of gag sequences in RNA packaging is indirect and the effects varied depending upon the sequence replacing the HIV-1 gag sequence.
Fig. 4.
Examining the impact of the 5’ gag sequence on viral RNA packaging. (a) General structures of the constructs. Vectors containing the previously described syngag sequences (grey boxes) and a new codon-optimized gag sequence (NSG; hatched boxes) contain SG and NSG in their names, respectively. Nucleotide and amino acid sequence comparisons of syngag and NSG are shown in Supplemental Fig.1. (b) RNA packaging efficiency. Results from three independent experiments are summarized; error bars indicate the standard deviations.
Determining the minimal HIV-1 sequence required for packaging of heterologous RNAs.
To examine whether the gag sequence in NSG can mediate the encapsidation of heterologous RNA, we inserted HIV-1 5’ UTR along with 5’ half of gag containing synonymous mutations into the luciferase reporter construct to generate NSG(22–726)-luc and SG(33–726)-luc (Fig. 5a). We then performed single virion analysis to determine RNA packaging efficiency (Fig. 5b) and luciferase assay to monitor the expression of these constructs (Fig. 5c). We found that RNAs from NSG(22–726)-luc and SG(33–726)-luc were packaged at 15% ± 3% and 7% ± 2%, respectively, which are lower than RNAs generated from 1Gag-726-luc (22% ± 1%) and 1Gag-FS-luc (31% ± 1%). Therefore, the relative packaging efficiencies of heterologous RNAs containing these sequences are similar to those in the context of the viral genome (shown in Fig. 4); RNAs containing NSG(22–726) sequences were packaged more efficiently than those containing SG(22–726), and less efficient than those containing HIV-1 5’ gag sequences.
Fig. 5.
Determining HIV-1 sequence required to package firefly luciferase reporter RNAs into particles. (a) General structures of plasmid expressing firefly luciferase reporter RNA and the inserted HIV-1 sequences. (b) Packaging of the reporter RNA. (c) Expression of reporter RNAs monitored by firefly luciferase activities standardized to that from No-HIV-luc. Results from three independent experiments are summarized; error bars indicate the standard deviations.
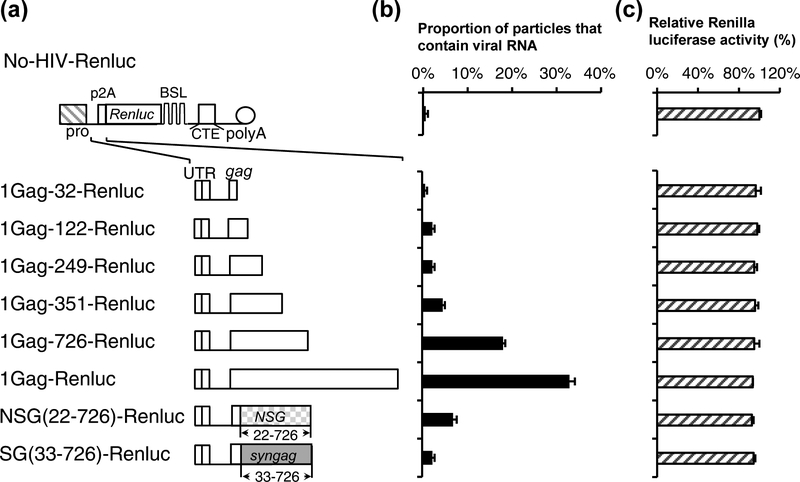
These studies indicate that the impact of HIV-1 5’ gag sequence is dependent upon the context of the RNA. Removing the HIV-1 5’ gag sequence severely affects packaging efficiency when the sequence is adjacent to the codon-optimized syngag sequence (Fig. 2) or the IRES-firefly luciferase gene (Fig. 3), but not when the sequence is next to the NSG sequence (Fig. 4 and Fig. 5). To better assess the role of HIV-1 5’ gag sequence in mediating heterologous RNA packaging, we used a second reporter RNA system to verify our results. This system is based on No-HIV-Renluc (Fig. 6a), which contains a ribosomal skipping p2A sequence derived from porcine teschovirus-1 and Renilla luciferase gene that is distinct from the firefly luciferase gene. There is no significant sequence homology or known conserved structures between IRES-luc and p2A-Renluc. We then tested various constructs expressing the Renilla luciferase gene by transfecting the constructs into cells along with Gag-expressing helper constructs and Bgl-YFP; viruses were harvested and used for image analyses to determine RNA packaging efficiency (Fig. 6b). Renilla luciferase activity was determined using the transfected cell lysates to monitor the expression of various constructs (Fig. 6c).
Fig. 6.
Determining HIV-1 sequence required to package Renilla luciferase reporter RNAs into particles. (a) General structures of plasmid expressing Renilla luciferase reporter RNA and the inserted HIV-1 sequences. (b) Packaging of the reporter RNA. (c) Expression of reporter RNAs monitored by Renilla luciferase activities standardized to that from No-HIV-Renluc. Results from three independent experiments are summarized; error bars indicate the standard deviations.
We found that in the Renilla luciferase system, RNA containing the 5’ UTR and the first 32 nt of HIV-1 gag was packaged at the background level (1%), whereas RNAs containing the 5’ UTR plus the first 726 and the full-length HIV-1 gag were packaged at 18% ± 0.5% and 33% ± 1%, respectively (Fig. 6b). These results indicate that 5’ UTR and the first 32 nt of HIV-1 gag is insufficient in mediating RNA packaging of the Renluc construct and that the HIV-1 5’ gag sequences need to be included for RNA packaging. Renilla luciferase activities were similar in lysates generated from cells transfected with different constructs (Fig. 6c). We have also tested the packaging of RNAs derived from NSG(22–726)-Renluc and SG(33–726)-Renluc; these RNAs were packaging at 7% ± 0.8% and 2% ± 0.5%, respectively.
Taken together, our results showed that HIV-1 5’ UTR and at least the 5’ half of the gag gene is required to mediate efficient packaging of heterologous reporter RNA into viral particles. However, the role of the gag gene sequences is likely to be indirect in nature.
DISCUSSION
Although only comprising a small fraction of the cellular mRNA, HIV-1 full-length RNA is efficiently packaged during virus assembly. The interactions between viral protein Gag and the full-length RNA mediates the packaging of the HIV-1 genome. In this report, we defined the HIV-1 RNA sequence required for non-viral RNAs to be efficiently packaged into HIV-1 particles. Using two different non-viral reporter RNA systems, we found that the 5’ UTR and the first 32-nt of the gag gene sequence were insufficient to mediate the packaging of two heterologous RNAs. However, the presence of the HIV-1 5’ UTR along with the 5’ half of the gag gene can mediate efficient packaging of reporter RNAs.
Previous studies had performed mutational studies to define sequences important for HIV-1 RNA genome packaging [reviewed in [33, 34, 36, 37, 71, 72]]. One of the first such studies showed that deletion of a small stem-loop structure, often referred to as stem loop 3 (SL3), upstream of the gag AUG start codon results in the decrease of RNA genome packaging [18]. As a result, SL3 is sometimes referred to as the “packaging signal” for HIV-1, although SL3 is only one of the elements important for RNA packaging. More recently, another stretch of RNA from a portion of U5 to a little past the gag start codon was defined as the minimal RNA packaging signal [73]; we found that this element is necessary but not sufficient for mediating the encapsidation of two different heterologous RNAs (Fig. 3 and Fig. 6). Our results showed that sequences directly downstream of the 5’ UTR have a strong effect on RNA packaging; for example, replacing gag with different codon-optimized sequences results in different efficiencies of packaging (Fig. 2 and Fig. 4). Thus, although the previously described minimal element is functional in the context of the vector described in the previous study, it is insufficient to mediate the packaging of the two heterologous reporter RNAs examined in this report. It is noteworthy that HIV-1 assembly and RNA packaging have been examined using an in vitro micelle system [74]; in this system, a longer HIV-1 RNA, including part of the gag sequence, is packaged much more efficiently than a shorter HIV-1 RNA containing the 5’ UTR.
We found that adding the 5’ half (726 nt) of gag to the 5’ UTR improves RNA packaging. The sequence near the gag AUG start codon is known to be critical for HIV-1 RNA packaging: structural studies show that the AUG and sequences flanking the AUG form base-pairs with sequences in U5 and disrupting the base-pairing results in packaging defects [41, 43, 49]. However, the role of the rest of the 5’ gag sequence in RNA packaging is less clear. The gag gene sequence in other retroviruses have also been shown to affect RNA packaging. In BLV, it has been shown that a distinct stem-loop structure in gag, referred to as the secondary encapsidation signal, is important for RNA packaging [21, 75]. The mechanism by which MLV gag sequence enhances RNA packaging is less clear; one possible mechanism discussed is that the gag sequence might act as an insulator to protect the function of the 5’ UTR packaging signal [16].
Two sets of results in our report suggest a possible indirect role of 5’ gag in RNA packaging. First, we have placed four different sequences directly downstream of the 5’ UTR and the first 20–30 nt of gag: IRES-firefly luciferase, p2A-Renilla luciferase, a previously published codon-optimized HIV-1 gag sequence (syngag) and a second gag containing synonymous mutations (NSG). Of these, three of the RNAs packaged poorly (Fig. 2, 3, and 6), only the RNAs containing NSG sequences appeared to be packaged efficiently although not as well as those with gag sequences. For example, when placed in reporter RNA containing IRES-luc or p2A-Renluc, constructs with NSG(22–726) sequence were packaged at 68%, and 39% of those with 726-nt of NL4–3 5’ gag sequence, respectively (Fig. 5 and Fig. 6). Similarly, in the context of the viral genome, NSG(22–726) RNA was packaged at ~67% of those containing NL4–3 gag sequence (Fig. 4). The ability of a codon-optimized sequence to partially replace the 5’ gag function argues against its direct role in mediating packaging. Additionally, when increasing lengths of HIV-1 5’ gag sequences were replaced with those of codon optimized syngag sequences, we observed a gradual decrease of RNA packaging (Fig. 2), making it unlikely that a BLV-like “secondary encapsidation signal” exists in these sequences. Taken together, these data also do not support the presence of a distinct RNA structure or element in the 5’ gag from nt 32 to 726 that is involved in specific Gag: RNA interactions leading to packaging. Therefore, the presence of 5’ gag is likely to indirectly facilitate specific Gag:RNA interactions. We reasoned that genome encapsidation during the HIV-1 replication relies on the interactions between Gag and the full-length RNA. Therefore, the structure of the RNA involved in specific Gag interactions evolves as part of the full-length RNA and is likely to be affected by the neighboring sequence. We hypothesize that HIV-1 5’ gag sequence may ensure the folding of HIV-1 RNA to promote Gag:RNA interactions. Alternatively, the 5’ gag sequence may facilitate Gag:RNA interactions in a currently undefined manner. Additional studies are required to define the molecular mechanism behind the 5’ gag enhancement of RNA packaging.
MATERIALS AND METHODS
Molecular cloning of viral constructs.
All of the Gag expression constructs described in this report have two versions, one that expresses Gag-CeFP and one that expresses untagged Gag [1]. For simplicity, only the Gag-CeFP versions are illustrated in figures. HIV-1 constructs GagCeFP-BglSL and GagCeFP-BglSL-CTE have been described previously [1, 57, 76]; for clarity, they are referred to here as 1Gag and 1Gag-CTE, respectively. These constructs were derived from the NL4–3 molecular clone and contain inactivating deletions in pol, vif, vpr, vpu and env. The previously described Bgl-YFP plasmid encodes a truncated BglG protein fused to YFP [76]. The HIV-1 construct 1-AAG has been described previously and is similar to 1Gag but contains an inactivating ATG-to-AAG mutation at the Gag start codon [76].
The codon-optimized Gag-CeFP expression clone CMV-synGag-pA was modified from pSynGag-mCherry [77], a derivative of pSYNGP [70], by first replacing the mCherry fluorescent protein gene with a cefp gene; additionally, BSL, stem-loop sequences recognized by E.coli BglG protein, and the MPMV CTE were inserted downstream of the GagCeFP-coding sequence. Plasmid 1Gag-CTE-pA was generated by replacing the 3’ LTR of 1Gag-CTE with a fragment from pSYNGP that contains the SV40 polyA signal. Plasmid CMV-1Gag-nLTR was generated by replacing the 5’ U3 with CMV promoter with a PCR amplified DNA fragment; 5’ Race/primer extension reaction using a SMARTer® RACE 5’/3’ Kit (Clontech) was performed to ensure that transcription of HIV-1 RNA initiates at the correct position in this construct. CMV-1Gag-npA was derived from 1Gag-CTE-pA and CMV-1Gag-nLTR by combining the CMV promoter and SV40 polyA segments of these two plasmids.
Plasmid CMV-1Gag-npA was used to construct Δpol and Δenv by replacing a portion of the plasmid with a PCR fragment containing deletion of the sequences downstream of BSL to the beginning of CTE, or from the end of CTE to the beginning of nef, respectively. Plasmid Δpolenv was generated by combining the two deletions in Δpol and Δenv.
PCR was used to generate DNA fragments containing chimeric gag genes containing a portion of HIV-1 (NL4–3) sequence and a portion of codon-optimized gag sequence from pSYNGP. These DNA fragments generated by PCR were used to replace corresponding DNA fragments in CMV-1Gag-npA. This strategy was used to generate plasmids termed 1Gag-726 to 1Gag-32 and plasmid SG(33–726). Another DNA fragment was synthesized to contain synonymous mutations in the gag gene, referred to as NSG sequence; sequences from this DNA were used to generate all NSG plasmids. DNA fragments containing modified HIV-1 5’ UTR were generated by PCR and were used to replace corresponding sequences in 1Gag-726 to generate mutants with modified 5’ UTR sequences.
Plasmids containing firefly luciferase genes were generated as follows. First, a DNA fragment containing internal ribosomal entry site (IRES) from encephalomyocarditis virus and firefly luciferase (luc) gene was inserted into CMV-1Gag-dcefp-Δpolenv, which is similar to CMV-1Gag-Δpolenv except with the deletion of the cefp gene, to generate 1Gag-luc. Next, a stop codon was introduced into the gag gene to generate 1Gag-FS-luc. Plasmid 1Gag-FS-luc was used to generate plasmids containing shorter HIV-1 fragments by deletion, including No-HIV-luc, in which the HIV-1 5’ UTR and gag sequences were deleted. Plasmid 1Gag-luc was used to generate 1Gag-Renluc by replacing the IRES-firefly luciferase gene with p2A-Renilla luciferase gene. Deletions and replacements were made in 1Gag-Renluc to generate plasmids containing varying length of HIV-1 sequences, including No-HIV-Renluc, in which the HIV-1 5’ UTR and gag sequence were removed.
All cloning procedures were performed using standard methods; the general structures of the plasmids were characterized by restriction enzyme mapping; DNA sequencing was performed in all cloned DNA fragments that underwent PCR amplification.
Cell culture, DNA transfection and luciferase assay.
Human 293T cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (HyClone), penicillin (50 U/ml; Gibco) and streptomycin (50 μg/ml; Gibco). Cells were maintained in humidified 37°C incubators with 5% CO2. DNA transfection was performed using FuGENE® HD Transfection Reagent (Promega) according to manufacturer’s recommendations. Cell culture supernatants were harvested 36-h posttransfection, clarified through a 0.45-um-pore-size filter, and either stored at −80°C or used immediately for image acquisition. Firefly and Renilla luciferase activities were determined at 36-h posttransfection using Britelite Plus Reporter Gene Assay System (PerkinElmer) and Renilla Luciferase Assay System (Promega), respectively.
Single-virion analyses.
Clarified supernatants were mixed with polybrene (50 μg/ml, final concentration), placed on an 8-well μ-Slide (Ibidi), and centrifuged at low speed (1200 × g, S2096 Rotor, Allegra 21R Centrifuge, Beckman) for 15 min before imaging. Images were acquired using an inverted Nikon Eclipse Ti microscope with a 100× 1.40 numerical aperture oil objective, an X-Cite 120 system (EXFO Photonic Solution Inc.), an ANDOR technology iXon camera, and NIS element AR software (Nikon). The excitation and emission filter sets were 427/10 nm and 480/40 nm for CeFP, 504/12 nm and 542/27 nm for YFP. Gag particles were identified by CeFP signals whereas HIV-1 RNA genomes were identified by the YFP signals. Identification and localization of fluorescent protein signals were performed using custom MatLab programs. RNA packaging efficiency was determined by the proportion of CeFP signals that colocalized with YFP signals.
Supplementary Material
Supplemental Fig. 1. Alignment of nucleotide and amino acid sequences of HIV-1 gag, syngag and NSG. Nucleotide sequence alignment of the HIV-1 NL4–3 gag gene and a previously described codon-optimized syngag (a), two codon optimized sequences syngag and NSG (b), and NL4–3 gag and codon-optimized NSG (c). (d) Amino acid sequence alignment of the Gag proteins encoded by NL4–3 gag, NSG and syngag. Asterisks indicate amino acid residues that are identical in these three proteins.
Highlights.
The RNA sequence necessary and sufficient to mediate HIV-1 genome packaging has not been defined
The sequence required to package RNA into HIV-1 particles was determined Using two reporter RNAs
The HIV-1 5’ UTR and the first 32-nt of gag sequence are not sufficient to mediate packaging.
Reporter RNAs containing HIV-1 5’ UTR and the 5’ half of gag gene can be packaged into viral particles.
The role of the gag gene sequence is likely to be indirect but improves the Gag: 5’ UTR interaction.
Acknowledgement
We thank Drs. Jonathan Rawson, Steven Santos, Eric Freed for helpful discussions and/or critical reading of the manuscript. This work is supported by Intramural Research, National Institutes of Health, and by IATAP funding to W-S. H. and to V.K.P.
Abbreviations
- HIV-1
human immunodeficiency virus type 1
- HIV-2
human immunodeficiency virus type 2
- RSV
Rous sarcoma virus
- BLV
bovine leukemia virus
- MPMV
Mason Pfizer monkey virus
- UTR
untranslated region
- MA
matrix
- CA
capsid
- NC
nucleocapsid
- TAR
trans-activation region
- PBS
primer binding site
- RRE
Rev response elements
- CTE
constitutive transport element
- NXF1
nuclear RNA export factor 1
- CRM1
chromosomal maintenance 1
- CeFP
cerulean fluorescent protein
- BSL
stem-loop sequences recognized by antitermination protein BglG
- YFP
yellow fluorescent protein
- SV40
Simian virus 40
- CMV
cytomegalovirus
- IRES
internal ribosomal entry site
- luc
luciferase
- PCR
Polymerase chain reaction
References
- [1].Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, et al. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13535–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nikolaitchik OA, Dilley KA, Fu W, Gorelick RJ, Tai SH, Soheilian F, et al. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS pathogens. 2013;9:e1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Freed EO. HIV-1 assembly, release and maturation. Nature reviews. 2015;13:484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Abd El-Wahab EW, Smyth RP, Mailler E, Bernacchi S, Vivet-Boudou V, Hijnen M, et al. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat Commun. 2014;5:4304. [DOI] [PubMed] [Google Scholar]
- [5].Luban J, Goff SP. Binding of human immunodeficiency virus type 1 (HIV-1) RNA to recombinant HIV-1 gag polyprotein. Journal of virology. 1991;65:3203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Meric C, Gouilloud E, Spahr PF. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. Journal of virology. 1988;62:3328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Meric C, Goff SP. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. Journal of virology. 1989;63:1558–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Berkowitz RD, Ohagen A, Hoglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. Journal of virology. 1995;69:6445–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gorelick RJ, Henderson LE, Hanser JP, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:8420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gorelick RJ, Nigida SM Jr., Bess JW Jr., Arthur LO, Henderson LE, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. Journal of virology. 1990;64:3207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gorelick RJ, Chabot DJ, Rein A, Henderson LE, Arthur LO. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. Journal of virology. 1993;67:4027–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. Journal of virology. 1995;69:5716–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Y, Barklis E. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. Journal of virology. 1997;71:6765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nikolaitchik OA, Gorelick RJ, Leavitt MG, Pathak VK, Hu WS. Functional complementation of nucleocapsid and late domain PTAP mutants of human immunodeficiency virus type 1 during replication. Virology. 2008;375:539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Adam MA, Miller AD. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. Journal of virology. 1988;62:3802–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bender MA, Palmer TD, Gelinas RE, Miller AD. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. Journal of virology. 1987;61:1639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aronoff R, Linial M. Specificity of retroviral RNA packaging. Journal of virology. 1991;65:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. Journal of virology. 1989;63:4085–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5’ packaging signal of human immunodeficiency virus type 1. Journal of virology. 1995;69:2101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McCann EM, Lever AM. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. Journal of virology. 1997;71:4133–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mansky LM, Krueger AE, Temin HM. The bovine leukemia virus encapsidation signal is discontinuous and extends into the 5’ end of the gag gene. Journal of virology. 1995;69:3282–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Guesdon FM, Greatorex J, Rhee SR, Fisher R, Hunter E, Lever AM. Sequences in the 5’ leader of Mason-Pfizer monkey virus which affect viral particle production and genomic RNA packaging: development of MPMV packaging cell lines. Virology. 2001;288:81–8. [DOI] [PubMed] [Google Scholar]
- [23].Jaballah SA, Aktar SJ, Ali J, Phillip PS, Al Dhaheri NS, Jabeen A, et al. A G-C-rich palindromic structural motif and a stretch of single-stranded purines are required for optimal packaging of Mason-Pfizer monkey virus (MPMV) genomic RNA. Journal of molecular biology. 2010;401:996–1014. [DOI] [PubMed] [Google Scholar]
- [24].Kalloush RM, Vivet-Boudou V, Ali LM, Mustafa F, Marquet R, Rizvi TA. Packaging of Mason-Pfizer monkey virus (MPMV) genomic RNA depends upon conserved long-range interactions (LRIs) between U5 and gag sequences. RNA (New York, NY. 2016;22:905–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vile RG, Ali M, Hunter E, McClure MO. Identification of a generalised packaging sequence for D-type retroviruses and generation of a D-type retroviral vector. Virology. 1992;189:786–91. [DOI] [PubMed] [Google Scholar]
- [26].Mustafa F, Al Amri D, Al Ali F, Al Sari N, Al Suwaidi S, Jayanth P, et al. Sequences within both the 5’ UTR and Gag are required for optimal in vivo packaging and propagation of mouse mammary tumor virus (MMTV) genomic RNA. PloS one. 2012;7:e47088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hibbert CS, Mirro J, Rein A. mRNA molecules containing murine leukemia virus packaging signals are encapsidated as dimers. Journal of virology. 2004;78:10927–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Aldovini A, Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. Journal of virology. 1990;64:1920–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Clever JL, Miranda D Jr., Parslow TG. RNA structure and packaging signals in the 5’ leader region of the human immunodeficiency virus type 1 genome. Journal of virology. 2002;76:12381–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Clever JL, Parslow TG. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. Journal of virology. 1997;71:3407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].McBride MS, Panganiban AT. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. Journal of virology. 1996;70:2963–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].McBride MS, Schwartz MD, Panganiban AT. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. Journal of virology. 1997;71:4544–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. [DOI] [PubMed] [Google Scholar]
- [34].Lever AM. HIV-1 RNA packaging. Advances in pharmacology (San Diego, Calif. 2007;55:1–32. [DOI] [PubMed] [Google Scholar]
- [35].Comas-Garcia M, Davis SR, Rein A. On the Selective Packaging of Genomic RNA by HIV-1. Viruses. 2016;8:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kuzembayeva M, Dilley K, Sardo L, Hu WS. Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology. 2014;454–455:362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nature reviews. 2004;2:461–72. [DOI] [PubMed] [Google Scholar]
- [38].Russell RS, Liang C, Wainberg MA. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology. 2004;1:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Structure Berkhout B. and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. [DOI] [PubMed] [Google Scholar]
- [40].Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, et al. Structure of the HIV-1 RNA packaging signal. Science (New York, NY. 2015;348:917–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, et al. NMR detection of structures in the HIV-1 5’-leader RNA that regulate genome packaging. Science (New York, NY. 2011;334:242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Berkhout B, van Wamel JL. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA (New York, NY. 2000;6:282–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr., Swanstrom R, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS biology. 2008;6:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Das AT, Harwig A, Vrolijk MM, Berkhout B. The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. Journal of virology. 2007;81:7742–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Das AT, Klaver B, Klasens BI, van Wamel JL, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. Journal of virology. 1997;71:2346–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Helga-Maria C, Hammarskjold ML, Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. Journal of virology. 1999;73:4127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Luban J, Goff SP. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. Journal of virology. 1994;68:3784–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nikolaitchik O, Rhodes TD, Ott D, Hu WS. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. Journal of virology. 2006;80:4691–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–7. [DOI] [PubMed] [Google Scholar]
- [51].Cochrane AW, Chen CH, Rosen CA. Specific interaction of the human immunodeficiency virus Rev protein with a structured region in the env mRNA. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Heaphy S, Dingwall C, Ernberg I, Gait MJ, Green SM, Karn J, et al. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990;60:685–93. [DOI] [PubMed] [Google Scholar]
- [53].Hope TJ, McDonald D, Huang XJ, Low J, Parslow TG. Mutational analysis of the human immunodeficiency virus type 1 Rev transactivator: essential residues near the amino terminus. Journal of virology. 1990;64:5360–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–83. [DOI] [PubMed] [Google Scholar]
- [55].Brandt S, Blissenbach M, Grewe B, Konietzny R, Grunwald T, Uberla K. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS pathogens. 2007;3:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cockrell AS, van Praag H, Santistevan N, Ma H, Kafri T. The HIV-1 Rev/RRE system is required for HIV-1 5’ UTR cis elements to augment encapsidation of heterologous RNA into HIV-1 viral particles. Retrovirology. 2011;8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Moore MD, Nikolaitchik OA, Chen J, Hammarskjold ML, Rekosh D, Hu WS. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS pathogens. 2009;5:e1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Blissenbach M, Grewe B, Hoffmann B, Brandt S, Uberla K. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. Journal of virology. 2010;84:6598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pasquinelli AE, Ernst RK, Lund E, Grimm C, Zapp ML, Rekosh D, et al. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. Embo J. 1997;16:7500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, et al. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Molecular cell. 1998;1:649–59. [DOI] [PubMed] [Google Scholar]
- [61].Fridell RA, Bogerd HP, Cullen BR. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:4421–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fritz CC, Green MR. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Current biology : CB. 1996;6:848–54. [DOI] [PubMed] [Google Scholar]
- [63].Chamanian M, Purzycka KJ, Wille PT, Ha JS, McDonald D, Gao Y, et al. A cis-Acting Element in Retroviral Genomic RNA Links Gag-Pol Ribosomal Frameshifting to Selective Viral RNA Encapsidation. Cell host & microbe. 2013;13:181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Nikolaitchik OA, Hu WS. Deciphering the role of the Gag-Pol ribosomal frameshift signal in HIV-1 RNA genome packaging. Journal of virology. 2014;88:4040–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Larson DR, Johnson MC, Webb WW, Vogt VM. Visualization of retrovirus budding with correlated light and electron microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, et al. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Schneider R, Campbell M, Nasioulas G, Felber BK, Pavlakis GN. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. Journal of virology. 1997;71:4892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber BK, Pavlakis GN. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. Journal of virology. 1992;66:7176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kotsopoulou E, Kim VN, Kingsman AJ, Kingsman SM, Mitrophanous KA. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. Journal of virology. 2000;74:4839–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lever AM. HIV RNA packaging and lentivirus-based vectors. Advances in pharmacology (San Diego, Calif. 2000;48:1–28. [DOI] [PubMed] [Google Scholar]
- [72].Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. Journal of molecular biology. 2011;410:609–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, et al. Identification of a minimal region of the HIV-1 5’-leader required for RNA dimerization, NC binding, and packaging. Journal of molecular biology. 2012;417:224–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Carlson LA, Bai Y, Keane SC, Doudna JA, Hurley JH. Reconstitution of selective HIV-1 RNA packaging in vitro by membrane-bound Gag assemblies. eLife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mansky LM, Wisniewski RM. The bovine leukemia virus encapsidation signal is composed of RNA secondary structures. Journal of virology. 1998;72:3196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chen J, Grunwald D, Sardo L, Galli A, Plisov S, Nikolaitchik OA, et al. Cytoplasmic HIV-1 RNA is mainly transported by diffusion in the presence or absence of Gag protein. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E5205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Burdick R, Smith JL, Chaipan C, Friew Y, Chen J, Venkatachari NJ, et al. P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. Journal of virology. 2010;84:10241–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Alignment of nucleotide and amino acid sequences of HIV-1 gag, syngag and NSG. Nucleotide sequence alignment of the HIV-1 NL4–3 gag gene and a previously described codon-optimized syngag (a), two codon optimized sequences syngag and NSG (b), and NL4–3 gag and codon-optimized NSG (c). (d) Amino acid sequence alignment of the Gag proteins encoded by NL4–3 gag, NSG and syngag. Asterisks indicate amino acid residues that are identical in these three proteins.