Abstract
The single cell is considered the basic unit of biology, and the pursuit of understanding how heterogeneous populations of cells can functionally coexist in tissues, organisms, microbial ecosystems, and even cancer, makes them the subject of intense study. Next-generation sequencing (NGS) of RNA and DNA has opened a new frontier of (single)-cell biology. Hundreds to millions of cells now can be assayed in parallel, providing the molecular profile of each cell in its milieu inexpensively and in a manner that can be analyzed mathematically. The goal of this article is to provide a high-level overview of single-cell sequencing for the nonexpert and show how its applications are influencing both basic and applied clinical studies in embryology, developmental genetics, and cancer.
As recently as 2 years ago, a review article about single-cell sequencing technologies would have mainly focused on methods for efficient extraction, types of nucleic acid amplification, coverage, and even the price/unit (i.e., cell or nuclei). Today, although these aspects are regarded as basic contributors for successful single-cell experimentation, researchers are concentrating more on applying the right method and the right mathematical analysis to the problem at hand rather than in the “art” of getting a single-cell genomics laboratory set up. This is largely the result of the advent of high-throughput droplet encapsulation and emulsion-based amplification of nucleic acids and, more recently, the implementation of commercial droplet-based (Drop-Seq) instruments in nearly every sequencing core. Indeed, when large numbers of cells are available, as from whole tumors or embryos, commercial microfluidic bead emulsion devices such as the 10X Chromium (10X Genomics) and BD Rhapsody can amplify tens of thousands of single-cell transcriptomes at once and have them ready for sequencing in little more than 1 day. This technology is being exploited to profile the expression states and lineages of cells across species, from flatworms to vertebrates.
In this article, we will attempt to provide a snapshot of how next-generation sequencing (NGS) is bringing subjects as complex as cancer, neurobiology, microbial ecology, and embryology to a new level of resolution. For additional detail, the overall topic has been thoroughly reviewed in several recent publications for cancer (Baslan and Hicks 2017; Tsoucas and Yuan 2017), neurobiology (Poulin et al. 2016), and microbiology (Woyke et al. 2017), and a series of key reviews on the topic has been assembled in Nature Reviews (see www.nature.com/collections/sxnwgntqsk).
Each single cell is unique in both time (e.g., cell cycle) and space (e.g., microenvironment, nearest neighbors). Single-cell sequencing has been developed to gain insight into that “uniqueness” and to unmask the distinctive molecular properties of cells, not by themselves but as part of the functional coherent populations that colonize tissues, organs, tumors, and microbial ecosystems. Traditionally, scientists named cell types according to their appearance or location in a tissue or organism. Single-cell sequencing has not only redefined the molecular classification of cells but also uncovered new and unsuspected cell types and opened the window to further understanding the spatiotemporal organization of cells from embryonic development to tumor progression to aging itself. Furthermore, the very nature of genomic and transcriptomic data and the constant development of very clever computational tools makes it possible to evidence the most incredibly nuanced distinctions among cells, their types and states, as well as their fate (e.g., differentiation, death, etc.) within complex tissues such as the brain or developing embryo.
SINGLE-CELL SEQUENCING APPLICATIONS
Entire areas of investigation are being invigorated by single-cell sequencing. The first is cancer research, where the long known, but seldom acknowledged in practice, heterogeneity of tumor tissue can be tracked at its most basic level, the single cell. A second area is sequencing complex microbial populations, either from an organism, say human gut or mouth, or from our physical environment. Recent advances in microbial cell genomics and transcriptomics have enabled the assignment of functional roles to members of the human microbiome for which there are not successful methods available for culturing them. These approaches paved the way to deeper understanding of the phenotypic variation that exists among genetically related strains, opening new opportunities for studies of immunogenic microorganisms in disease. Another important but often overlooked area is in vitro fertilization (IVF), in which various levels of genomic sequence from a single cell isolated by hand from a human embryo can provide evidence of trisomy 21 or a suspected genetic disease, and at the same time enhance the chances for successful conception (Xu et al. 2016). Currently, however, the greatest scientific breakthroughs are likely to come in neurobiology and developmental biology. Indeed, the ability to perform transcription studies on thousands to millions of cells in a few hours or days using microfluidic emulsion amplifications has revolutionized the study of cell lineage and the detailed structure of complex cell populations. The explosion of new data in these areas was foreseen by Linnarsson and colleagues in a prescient review published in 2013 (Shapiro et al. 2013). At that time, they predicted that it would be possible to obtain molecular data from thousands of cells and, further, that methods to combine genomics, transcriptomics, epigenomics, and proteomics would be common in the ensuing decade. It has, in fact, taken less than 5 years for these predictions to be realized.
SINGLE-CELL SEQUENCING METHODOLOGY
Methods for the genome-wide amplification of RNA and DNA were in development from the early 1990s (Van Gelder et al. 1990; Telenius et al. 1992; Dean et al. 2002). However, it was not until both a relatively accurate reference human genome was drafted and made readily available (Lander et al. 2001; Kent et al. 2002) and massively parallel short-read sequencing was developed that the concept of genome-wide, single-cell sequencing began to be explored.
The first report of whole-transcriptome sequencing (Tang et al. 2009) used the ABI SOLID sequencing platform on complementary DNA (cDNA) prepared from a small number of mouse oocytes and blastomeres demonstrating clear regulatory effects of mutations in the microRNA (miRNA) silencing pathways. The first genome-wide copy number profiling of single tumor cells (Navin 2011) used flow sorting of 200 individual nuclei (then a lot) isolated from multiple sectors of two breast tumors and sequencing using the Illumina Genome Analyzer instrument. The results revealed the power of genomic copy number profiling to trace tumor lineage and to distinguish tumor cells from genomically normal (euploid) stroma. It further revealed the stability of clonal structure within a tumor as measured by copy number “breakpoints” while at the same time demonstrating stepwise evolution of the tumor genome through both increase in ploidy and the systematic increase in copy number alterations (CNAs) within the clonal structure of the lineage.
In those initial reports, the genomic DNA (gDNA) of single cells/nuclei were amplified, and the sequencing “libraries” (collection of DNA fragments prepared for sequencing) were constructed virtually one by one for each cell. In the wake of those initial reports, a wide variety of methods were described for both RNA and DNA sequencing, implementing important improvements in efficiency and reproducibility and cost. By 2013, single-cell sequencing was named “Method of the Year” by the journal Nature Methods.
SINGLE-CELL SEQUENCING PROTOCOLS
There are more than 300 publications describing single-cell protocols and ∼50 different data analysis methods listed in a review on the Illumina website (see www.illumina.com). In reality, these various “single-cell sequencing protocols” are not sequencing protocols at all. In the main, they are simply the means to isolate different subsets of RNA or DNA in a manner that they can be converted into standard short-read NGS libraries. An Illumina sequencing library is made up of 300–500 bp fragments of the target sequence flanked by hybrid specialized adaptor sequences that the instrument uses to prime the sequencing reactions. Libraries created from bulk tissue (DNA or RNA derived from thousands to millions of cells) usually contain very large numbers of “unique” fragments (a measurement of these number of fragments is “library complexity”). On the other hand, libraries made from single cells have a much lower number of unique fragments and therefore their library complexity is limited. A single mammalian cell may contain as few as 300,000 messenger RNA (mRNA) molecules and ∼10–30 pg of total mRNA (and ∼7 pg DNA). For single-cell sequencing, the massive output of a single unit, or “lane” of the sequence is much more than is needed to fully sequence all those unique fragments many times over. Thus, several single-cell libraries can be run together or “multiplexed” on a single lane as long as each library is constructed with adaptors that carry unique oligonucleotide sequences or “barcodes,” ultimately enabling each output sequence to be assigned to a specific cell during subsequent informatic analysis. In addition, in current practice, most sequencing adaptors also contain 8–10 nucleotides of completely random sequence, called unique molecular identifiers (UMIs) that can be used to informatically identify and (optionally) remove PCR duplicates, namely, replicates of the original unique molecules, eliminate errors introduced during the amplification of nuclei acids and sequencing stages, and to tag individual original transcripts allowing to “count” their abundance (i.e., expression level). The UMI concept was envisioned in several publications (Casbon et al. 2011; Fu et al. 2011; Kivioja et al. 2012; Shiroguchi et al. 2012) before it being used successfully by Islam et al. (2014) for removing duplicates when counting mRNA molecules in a mouse model and is now standard practice in most sequencing protocols, whether for validating point mutations in high-depth sequencing of bulk DNA, or in molecular counting applications such as transcriptome profiling.
SINGLE-CELL REVERSE TRANSCRIPTION (RT)
The central goal of single-cell transcriptome profiling is to accurately quantify the levels of specific messages in each cell and compare individual cells to a population to generate molecular phenotypes. NGS provides a mechanism for achieving that goal because each sequence read can be used as a tag for its template molecule. Huge numbers of short reads permit counting for millions of individual templates. The whole-transcriptome amplification method that is the basis for most transcription profiling was worked out in the Sandberg and Linnarsson Laboratories at the Karolinska Institute Stockholm and described in three important papers (Islam et al. 2011, 2014; Ramsköld et al. 2012). In the first paper used, the goal of the Ramsköld et al. paper was to generate full-length cDNA using an oligo(dT) primer carrying a PCR primer using Maloney murine leukemia virus (MMLV) reverse transcriptase, which adds a small stretch of cytosine residues to the 5′ end of the first strand cDNA. Strand switching is accomplished by the addition of a reverse primer sequence carrying a sample barcode along with a 3′dG sequence that hybridizes to the cytosine overhang and initiates second-strand synthesis going through to the end of the first strand. They named the method Smart-Seq. In the Islam et al. (2014) variation, the first- and second-strand primers carry a random UMI along with the sample barcode. After PCR amplification in the Islam et al. protocol, the ends of the PCR products were biotinylated before a fragmentation, and the fragments representing the 5′ and 3′ ends of the original transcript were captured on streptavidin beads and sequenced. Strand of origin was preserved and identifiable through the sequence and location of the barcode and the UMI was used to filter out duplicates, thereby providing a very accurate counting of the original mRNA molecules.
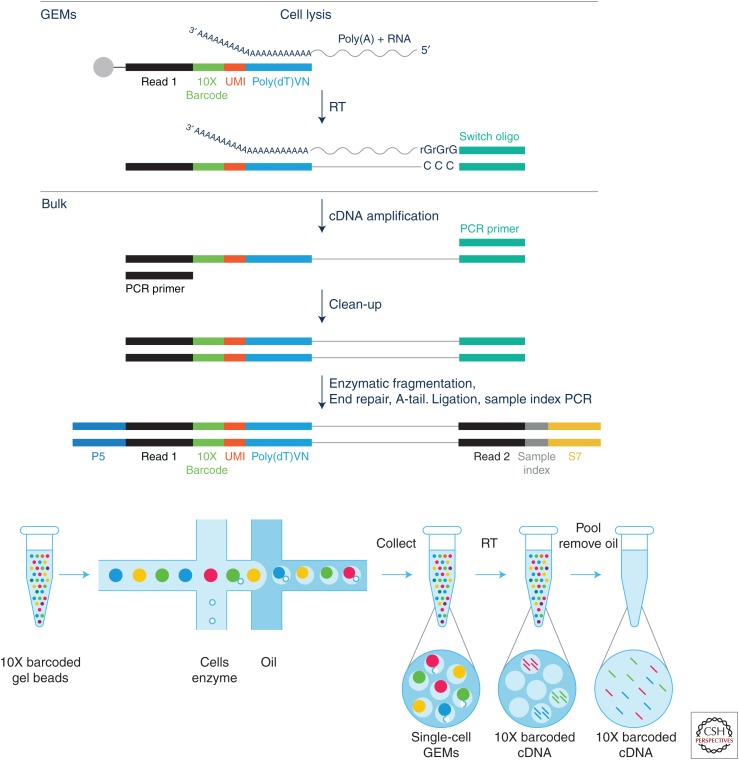
The strand-switching method using mouse mammary tumor virus (MMTV) RT requires no ligation, captures strand of origin, and is highly adaptable as the first- and second-strand primer sequences can be altered at will. Many variations on the strand-switching method are commercially available as SMART-Seq kits from Takara Bio USA (formerly ClonTech), including 3′ end capture and full-length cDNA as well as kits for capturing RNA from formalin-fixed, paraffin-embedded (FFPE) samples. Sensitivity for single cells seems to be at the level of 5–10 molecules/cell. The RNA amplification process is shown schematically in Figure 1.
Figure 1.
Schematic diagrams of single-cell RNA amplification using the strand-switching methodology, showing the molecular steps in generating double-stranded cDNA with sample barcodes and unique molecular identifiers (UMIs) (top); and the application of the basic method in the 10X high-throughput droplet/bead capture microfluidic mode (bottom). (Figures courtesy of 10X Genomics.)
It is important to note that it is not necessary to sequence the entire message to generate single-cell transcriptomic profiles. This is essentially a “counting” exercise more than a sequencing strategy in the traditional sense. For most molecular phenotyping studies, the sequencing reads are simply tags for counting up the number of each RNA molecule present in the cell. Filtering for duplicates using the UMI sequences ensure that each initial reverse transcript is only counted once, adding further stringency to the counting process. Although some Smart-Seq applications use random primers to initiate cDNA synthesis and thus can cover an entire message sequence, it would not yield complete coverage and most of the effort would be spent on only the highest frequency messages. For coverage at the base-pair level to identify mutations, a targeted protocol rather than a whole-transcriptome protocol would be favored.
DROPLET CAPTURE
Once it became clear that genomic and transcriptomic profiling of single cells was achievable in individual test tubes, the quest for high-throughput methods was underway. The C-1 amplification instrument, marketed by Fluidigm in 2011 was a step forward, but its maximum capacity of 100 cells did not present a significant improvement over what a bench scientist could do manually using individual tubes. However, the copublication of two breakthrough microfluidic applications by Evan Macosko and Allon Klein quickly changed the landscape for indexing mRNA from large populations of cells. Drop-Seq (Macosko et al. 2015) and InDrops (Klein et al. 2015) from the McCarroll and Kirschner laboratories at Harvard Medical School not only demonstrated reliable indexing of mRNA from thousands of single cells in a single run but also made the protocols and reagents accessible for all researchers. Both methods were based on earlier work from David Weitz's laboratory demonstrating encapsulation of reagents into droplets that were stable when mixed in emulsions. This method makes use of a microfluidic device to combine a single cell lysed in a nanoliter droplet with a hydrogel containing primers and reagents or a solid bead with primers synthesized on the surface. The InDrops (“indexing droplets”) method relies on lysing individual cells and capturing mRNA in nanoliter aqueous droplets that are mixed one by one in the microfluidic chamber with hydrogel beads containing barcoded DNA primers. After hybridization, the droplets are broken and the first strand cDNA reaction proceeds in bulk, with indexed sequences resolved after sequencing. In the Drop-Seq method, the DNA primers are synthesized on the bead surface using a “split and pool” method so that the sequence on each bead has a constant PCR handle, a unique 12-base barcode, and each primer contains a UMI plus the oligo(dT) to capture the mRNA through its poly(A) sequence and initiates cDNA synthesis during the RT reaction. After breaking the droplets, the captured mRNA is reverse-transcribed using template switching and the double-stranded RNA is amplified and sequenced, thus labeling the RNA from each cell with both a sample barcode and UMI.
In a similar fashion, the Klein group used their newly developed method, InDrop, to study the lineages of embryological development in two familiar vertebrate models, the zebrafish (Farrell et al. 2018; Wagner et al. 2018) and Xenopus, the western clawed toad (Briggs et al. 2018). In both of these efforts, the first steps were to create a pseudotemporal sequence of events by profiling single cells isolated from sequential stages of embryo development. Analyzing the transcriptome profiles of 60,000 to 139,000 single cells, they were able to visualize lineage trees leading from pluripotent stem cells to mature tissues and to track the temporal-spatial dynamics of the maturing populations.
These methods have recently become widely accessible through automation and commercialization by BD Bioscience as the BD Rhapsody instrument and by 10X Genomics as the Chromium instrument. A schematic of the 10X Chromium method for single-cell RNA (scRNA) sample barcoding and RNA amplification with UMI is shown in Figure 1 (bottom panel).
ANALYTICAL TOOLS FOR SINGLE-CELL TRANSCRIPTOMICS
Once single-cell transcriptome libraries have been created, much of the work to glean relevant information relies on the proper analysis and interpretation of the resulting data. Early methods for analysis relied on simple application of tools created for bulk datasets, with a few extra quality control (QC) steps implemented to remove poorly amplified or degraded libraries. Newer methods include more sophisticated statistical tools for QC and for the subsequent steps to measure transcript abundance, smooth or impute the expression profiles, and cluster the cells into similar groups of related and/or novel cell types.
The extra QC steps needed for single-cell RNA-Seq (scRNA-Seq) must include an awareness of how single-cell libraries differ from traditional bulk libraries. Populations of cells with very similar gross cellular phenotypes might show large differences in their transcriptomes for biological reasons such as stochastic transcription or unsynchronized cell-cycle stages. Unfortunately, these biologically driven differences can look very similar to technical artifacts caused by insufficient capture or shallow sequencing depth (Grün and van Oudenaarden 2015). Several calibration schemes have been implemented to try to mitigate these challenges, such as using spike-in controls to model technical noise and as a function of transcript abundance (Brennecke et al. 2013; Ding et al. 2015). The use of transcript UMIs, as discussed above, can help mitigate the effects of PCR amplification artifacts. Even with the use of UMIs and spike-ins, transcript dropout events are notoriously difficult to remove, such that many groups have resorted to imputing the expression profiles of genes with anomalously low counts by interpolating to match the expression profiles from similar cells (Li and Li 2018; van Dijk et al. 2018).
Following proper normalization and quality control, the hope is that novel clusters of cells will emerge as independent groups on a cluster map. Clustering methods work best when the set of input genes are preselected for those most likely to mark different cell types, for example, those that are well expressed and also show large differences between different cells. Selection methods include calculating the most variable genes (Klein et al. 2015; Macosko et al. 2015), determining the genes that are significantly different between known cell types (Shalek et al. 2013), or analyzing genes that contribute strongly to the first few principal components (Satija et al. 2015). Once a gene list has been selected, these are used as input for clustering algorithms that attempt to identify both how many clusters are present as well as who belongs to each cluster. Popular methods include matrix factorization approaches, k-means clustering approaches, as well as standard hierarchical clustering. Choosing the “best” clustering algorithm can depend on the underlying biology of the datasets. Although many scRNA-Seq analysis tools are optimized for mixed populations of distinct cells (Kharchenko et al. 2014; Grün and van Oudenaarden 2015; Haghverdi et al. 2015; Satija et al. 2015; Xu and Su 2015; Zeisel et al. 2015), their methods differ strongly from those developed to analyze time-series datasets that assume a smooth distribution between cell types, such as those involving development trajectories (Bendall et al. 2014; Marco et al. 2014; Trapnell et al. 2014; Setty et al. 2016). In practice, datasets often include a mixture of cells from distinct cell types as well as related subclusters with significant overlap. Therefore, trial analysis with both types of methods might be necessary before identifying what is best for a given experiment (Haghverdi et al. 2015).
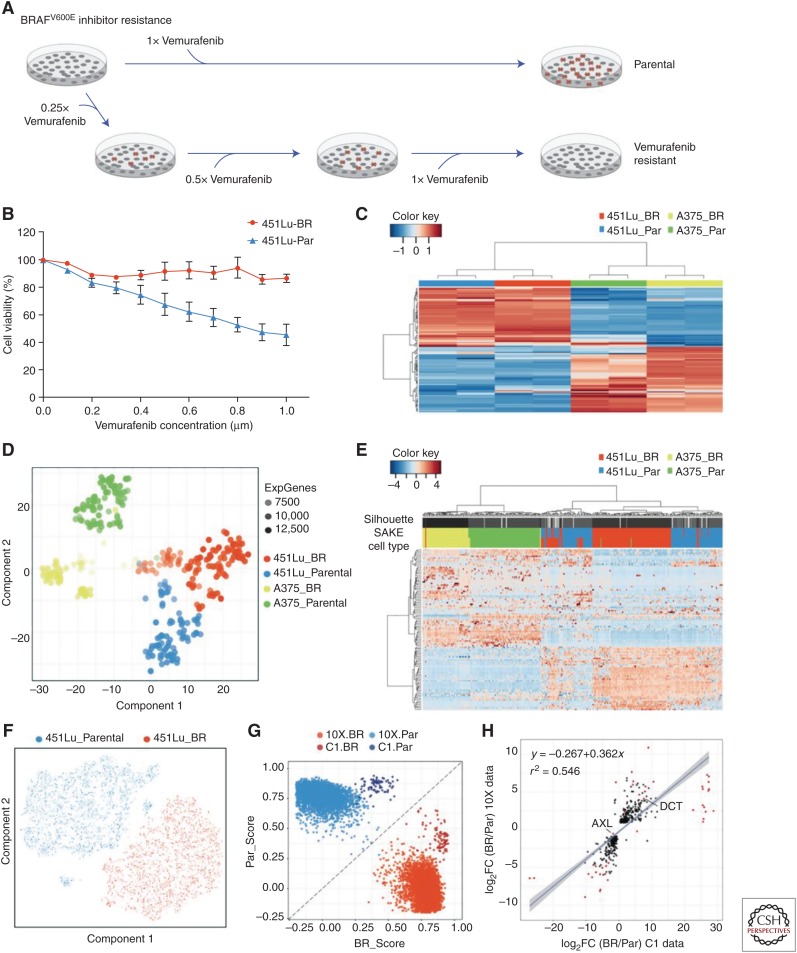
To address the challenges outlined above, Ho et al. (2018) designed the scRNA-Seq analysis and klustering evaluation (SAKE). SAKE provides several modules that include data preprocessing for quality control, sample clustering, t-distributed stochastic neighbor embedding (t-SNE) visualization of clusters, differential expression between clusters, and functional enrichment analysis. Comparing the performance of several published single-cell datasets, they showed that all of the scRNA-Seq analysis tools perform similarly for a wide range of sample types, despite each being algorithmically independent. However, SAKE performed best for very complex mixtures of cells with extensive substructure. Importantly, SAKE also includes quantitative statistics to evaluate the clustering results, a feature missing from most other methods (Deng et al. 2014; Ting et al. 2014; Zeisel et al. 2015), and to evaluate its performance by the ability to correctly identify clusters reported in these studies. Figure 2 presents an heuristic example of the manner in which these techniques can be used to rationalize scRNA-Seq data and to identify genes of interest by functional genomics (Box 1).
Figure 2.
Examples of a single-cell RNA (scRNA) sequence analysis to detect the molecular basis of drug resistance in melanoma. (A) Scheme for generating drug resistant derivatives. (B) Testing resistant cells. (C) Using signature melanoma signature genes to cluster bulk RNA-Seq data from cell lines. (D) t-SNE map was used to display the expression profiles of 400 single cells isolated using the Fluidigm C-1 instrument. (E) Heat map showing expression of highly expressed and highly variable genes. (F) t-SNE plot of scRNA-Seq of 6545 cells performed on a 10X Chromium instrument. (G) Direct comparison of Fluidigm cells and Chromium cells showing similar differences between resistant and parental but distinct patterns for the two methods. (H) Differential expression analysis identifies genes significantly altered in the resistant cell line derivative (for details, see Ho et al. 2018).
BOX 1.
ISOLATION AND AMPLIFICATION OF SINGLE CELLS
The range of methods that have been used to capture single cells for genomic analysis is as varied and nuanced as the range of experiments being pursued, but roughly they separate into two general classes according to whether the target cells are abundant, as from disaggregated tissue or cell lines, or rare, as circulating tumor cells (CTCs) in blood. Cells in suspension can be easily be separated by characteristic markers and dispensed into reaction tubes or multiwell plates by fluorescence-activated, cell-sorting FACS (Navin 2011; Baslan et al. 2015; Satija et al. 2015; Alexander et al. 2018) or distributed randomly by flowing across microwells (Yuan and Sims 2016; Gierahn et al. 2017). As of this writing, however, the high-throughput and reproducibility of microfluidic bead-capture methods for automated capture and amplification of single cells are rapidly becoming the standard for abundant cells in suspension.
By disaggregating cells from tissue, the spatial relationships are lost. In cases in which the exact source of cells is important, methods for coupling laser capture microscopy (LCM) with single-cell sequencing library preparation are available to capture cells from tissue preparations on slides (Nichterwitz et al. 2016). Another elegant approach for transcriptome mapping live neurons in situ was developed by Eberwine et al. (1992), in which a combined chemical and fluorescent probe injected into a cell can be photoactivated to “tag” mRNA molecules with biotin for later purification using streptavidin beads and sequenced (Lovatt et al. 2014).
Isolating very rare cells, as exemplified by CTCs from blood or bone marrow aspirates (typically one in a million nucleated cells) is much more difficult, and a wide array of commercial and noncommercial methods have been developed for this purpose. Among the commercially available methods, the CellSearch (Silicon Biosystems) system is a clinical application using magnetic capture of cells using ferrofluid nanoparticles conjugated to EpCAM antibodies to capture epithelial cells. Captured cell populations can be removed and individual cells isolated by micromanipulation or by using a coupled DEP Array system (Silicon Biosystems) to electrophoretically maneuver cells into reaction tubes for amplification. Other forms of automated fluorescence capture include the CellCelector (Automated Lab Systems, Gmbh) and the CyteFinder/Accucyte system (Rarecyte), which combines automated identification, cell picking, and amplification of blood samples. Alternatives to EpCAM as an enrichment method include separation by size using the ISET system (RareCells) and shape (Biofluidica).
Yet another nonselective method that has yielded an operational clinical assay for the androgen receptor variant AR-V7 (Scher et al. 2017) is the high-definition, single-cell analysis assay (HD-SCA) (Marrinucci et al. 2009) offered commercially by Epic Sciences (La Jolla, CA). This method arrays all nucleated cells on glass slides and uses both morphology and fluorescence markers to identify cells of interest. Cells can later be picked for amplification by micromanipulation and DNA copy number analysis (Dago et al. 2014; Greene et al. 2016).
t-SNE VISUALIZATION OF CLUSTERS
t-SNE (van der Maaten and Hinton 2008) has emerged as the most widely used visualization tool for single-cell transcriptomic studies, and is often used to represent data clustered and normalized by other methods, including Seurat, SAKE (Ho et al. 2018), and 10X Genomics Loupe Cell Ranger described (see support.10xgenomics.com/single-cell-vdj/software/visualization/latest/what-is-loupe-vdj-browser).
The goal of t-SNE plots is to represent the most relevant differences among points (e.g., single-cell genomic profiles) in high-dimensional space and represent them in a low-dimensional 2D or 3D scatterplot, such that, for example, clusters on a t-SNE plot might represent different cellular types as is shown in Figure 2. t-SNE implementations for gene expression datasets begin by reducing the very high dimensional expression matrix (m genes × n samples) into a lower dimensional set of principal components (PCs) that encode most of the variance in the dataset, typically 20–30 PCs. The t-SNE algorithm then calculates the distance between samples along each of these PCs and attempts to nonlinearly embed the PCs into a 2D or 3D representation that recapitulates relatedness of samples. This tends to work well when a filtered list of a few thousand genes that carry the most relevant information of cellular identity is used as the input to the t-SNE algorithms. A major distinction versus principal component analysis (PCA) is the addition of a user determined tunable parameter, “perplexity,” into the calculations. When perplexity is well chosen (typically 5–50, but well below the number of points) and the algorithm is allowed to iterate until saturation (typically ∼1000), t-SNE maps can be excellent tools for visualization of single-cell datasets. In addition, it has been demonstrated that the results from t-SNE may vary widely from different runs that do not saturate the perplexity of the embedded dimensions, such that cluster membership may not be reproducible from run to run (Wattenberg et al. 2016). A brief explanation of t-SNE for new users is presented elsewhere (see distill.pub/2016/misread-tsne).
HIGH-VOLUME SINGLE-CELL TRANSCRIPTOME STUDIES, THE “MEGACELL” EXPERIMENTS
Perhaps the most revolutionary application of scRNA-Seq has occurred only recently in the fields of developmental and organism biology, and of course, cancer. Using various forms of droplet capture and bead-based amplification, it is now possible to obtain transcriptome profiles from hundreds of thousands to millions of cells from embryos, whole organs, or tumors in a few experimental runs. The sheer amount and complexity of the data has spurred the development of new analytic methods that can create multidimensional arrays for mRNA profiles to reflect lineage relationships among cell types, from pluripotent stem cells to fully differentiated somatic cells. Of course, in dissociating the cells for high-volume sequencing, the original tissue structure is lost. The elegance of these “megacell” experiments is in using orthogonal methods to relate the transcriptomic profiles to cell type and cell location in the original specimen.
EMBRYOLOGY AND DEVELOPMENT: FLATWORMS, TOADS, FISH, AND REPTILES
In 2018, two groups synchronously published cell type atlases of the fresh water planarian (flatworm), Schmidtea mediterranea (Fincher et al. 2018; Plass et al. 2018). Planarians are multicell organisms with an organized body structure and distributed pluripotent stem cells. They are well known for their ability to regenerate complete animals from isolated parts and as such they are functionally immortal. The goal of these studies was to create an atlas of transcriptomic cell types keyed back to the known cell and tissue types in the animal and shed light on the lineage map of cells during development and regeneration. Using Drop-Seq to create libraries of tens of thousands of single cells followed by NGS, both groups obtained highly detailed and virtually complete transcriptome atlases of all the known cell types in the animal. Both groups used the Seurat R toolkit for single-cell genomics (Satija et al. 2015; Butler et al. 2018) to cluster the mRNA expression data and the clustered data was visualization with t-SNE (van der Maaten 2014). They then used a combination of known marker genes and in situ hybridization (ISH) on whole animals to classify the cell types cluster by cluster, identifying more than 20 known cell types and discovering several previously unknown cell types. This process also created a lineage map of planarian development.
Three simultaneous papers, also in 2018, took the atlas and lineage concept a step further into spatiotemporal analysis of vertebrate development by scRNA-Seq of tens of thousands of cells from staged embryos of zebrafish (Farrell et al. 2018; Wagner et al. 2018) and Xenopus (Wagner et al. 2018). Clustering the transcriptomic profiles by principle components over the embryonic time-points defines progressively branching tree-like maps of development in which the ultimate fates (e.g., organs, brain regions) can be identified by well-known markers. This work not only identifies previously unknown cell types and nodal branch points in the development process, but also provides a framework for comparative analysis of developmental steps across species and evolutionary time.
A final example reveals new insights into both neurobiology, evolutionary biology, and development. The origins and lineages of structures in the brain across the evolutionary spectrum has been an area of intense study for many decades. Tosches et al. (2018) built a transcriptomic atlas of cells comprising the basic brain component (pallium) where development varies widely from reptiles to birds to mammals. The transcriptomic profiles coupled with ISH enabled tracking the lineages with high resolution and present new opportunities for interpreting structure in terms of synaptic organization.
IMMUNE REPERTOIRE PROFILING
Application of single-cell sequencing for analyzing the transcriptomics of immune cells started with acquiring cells of known immune types through use of flow cytometry and antibodies against known markers. Innovations such as mass cytometry (CyTOF) increased the number of proteins that could be simultaneously used to study cells and made it possible to unravel more cell types quantitatively and qualitatively. However, the field has now progressed to unbiased analysis in which cells are probed without prior knowledge of their type. MARS-Seq (Jaitin et al. 2014) is one of the early papers that demonstrated that scRNA-Seq is a great tool for identification of different populations in heterogeneous tissues. Working with the spleen exposed to lipopolysaccharide (LPS), they were able to distinguish different types of immune cells, including B cells, macrophages, natural killer (NK) cells, dendritic cells, and were able to study the expression changes in untreated and treated samples. Soon, a plethora of papers leveraging scRNA-Seq and its ease to merge with other technologies highlighted how scRNA-Seq can be used to profile immune cells in cancer. Immune cell profiling has been extensively reviewed in Rosati et al. (2017), Seah et al. (2018), and Reece et al. (2016). T- and B-cell repertoire profiling, despite these advances, presents a challenge because of their inherent diversity. VDJ recombinations can result in millions of different T-cell receptor (TCR) chains that are generated in response to specific stimuli. Similarly, B cells also have variable regions, essential for building an adaptive immunity repertoire. This diversity can be studied using either gDNA or cDNA. There are several commercial options available from multiple companies such as BGI, Adaptive Biotechnologies, and iRepertoire, which probe different chains and regions from both starting materials. Gierahn et al. (2017) designed a Seq-Well device that is particularly useful for low numbers of cells. On the other hand, droplet-based sequencing methods have made it possible to analyze thousands of these cells at the same time. Azizi et al. (2018), using InDrop, have demonstrated the strength of paired gene expression and TCR sequencing and McDaniel et al. (2016) have reached the multimillion cell level in ultrahigh-throughput VDJ sequencing. Both BD Biosciences and 10X Genomics have several commercially available kits for either one of scRNA-Seq and TCR sequencing or both for use on the BD Rhapsody and the 10X Genomics Chromium instruments. These are great alternatives to setting up complex droplet microfluidic systems and also offer data analysis solutions. In these kits, the beads have the partial Read1, UMI, and 10X Genomics Barcode with the template switch oligo. This allows for full-length transcription of the mRNA and incorporates the barcode and UMI on the 5′ end, which is key for the dual application. The cDNA is amplified and divided for both applications. One half is fragmented and processed to prepare libraries for transcriptomics analysis and the other for T/B-cell repertoire investigation. Primers designed against the outer and inner constant region of variable chains are used for targeted PCR enrichment. These enriched fragments of different lengths are processed once again to make sequencing libraries. The libraries from both applications are also sequenced differently. The gene expression libraries are sequenced 26 cycles in read 1 to get the cell and UMI identities and 98 bases in read 2 to get mRNA insert sequence. The VDJ repertoire libraries are sequenced 150 bases in both reads to get the all the pertinent sequence data. The sequencing results can be easily analyzed in the software suite designed by 10X Genomics, which allows for clustering of cells, check enrichment of T/B cell clonotypes, and characterize the repertoire, making it an ingenious tool for immune profiling.
NOVEL APPLICATIONS OF MICROFLUIDIC DROPLET-BASED SEQUENCING
The power of genomics and transcriptomics at the single-cell level has spawned a startling number of variations on the single-cell sequencing paradigm with a dizzying array of cleverly acronymic sobriquets, including REAP-Seq, SLaAP-Seq, ATAC-Seq, G&T-Seq, Hi-C, and others. The available menu as of early 2017 was reviewed carefully by Tsoucas and Yuan (2017). As with the original scDNA and scRNA methods, some of these started out as “one cell per tube” preparations that were adapted to Drop-Seq, but most recent methods are developed directly for automated bead emulsion or microwell-based formats facilities (see Box 2).
BOX 2.
THE 10X CHROMIUM SYSTEM
The 10X Chromium system, built on their proprietary GemCode technology, has pushed the single-cell sequencing field into a new era. The high-throughput function, in which thousands of cells are barcoded in little time combined with the ability to be used for multiple applications, makes it a valuable asset in various areas of research.
Most of the droplet-based methods require the coencapsulation of a single cell and a barcoded bead/hydrogel microsphere. This is a double Poisson distribution loading resulting in low capture efficiency (7% InDrop, 12.8% Drop-Seq). However, 10X Chromium controller ensures that all droplets formed have a gel bead making the process a single Poisson distribution, greatly increasing the capture efficiency to 50%. This encapsulation is the core of different single-cell applications of 10X Chromium and is performed on a chip specific to each application. Samples, reagents, and partitioning oil are loaded onto the chip in designated wells. Gel beads in emulsion (GEMs), that is, the sample and barcoded gel bead in a single droplet, are collected from the recovery well and processed in PCR tubes to generate libraries for sequencing.
10X single-cell gene expression solution (scRNA-Seq) chip has the capacity to process up to eight samples at a time. The cell suspension volume and concentration are critical for obtaining the desired cell recovery and keeping the number of doublets low. The barcoded gel beads have a partial Illumina read 1, 16 nucleotide 10X barcode, 10 nucleotide unique molecular identifiers (UMIs), and 30 nucleotide poly(dT) primer sequence, in that order. This combination gives 10X technology a pool of ∼750,000 barcodes to separately index a large number of cells. A single such bead and one cell are encapsulated in a GEM with the reagents for reverse transcription (RT). These GEMs are transferred from chip into PCR strip in which the cells are lysed and mRNA is reverse transcribed into full-length complementary DNA (cDNA) (∼7000–9000 bp) with a cell barcode and UMI. Next, GEMs are dissolved to release cDNA, which is cleaned, size selected using SPRI beads, and quantified. Paired end sequencing libraries are constructed by adding Illumina compatible adapters to cDNA, which is fragmented to 300–700 bp. Also, these fragments are all from the 3′ end of cDNA because this is necessary to identify the origin of cDNA from the sample pool.
Single-cell copy number variation (CNV) uses the same principles but has cell bead, gel bead (CBGB) droplets instead of GEMs. The chip for this application is designed to handle up to four samples in one run. Cells are first encapsulated with cell matrix to generate cell beads (CBs). In CBs, the cell is lysed and nuclear proteins are degraded although genomic DNA remains trapped in the cell bead. These CBs undergo a further encapsulation step with barcoded gel beads and reagents to generate CBGBs. CBGBs are then harvested from the chip for amplification of DNA to generate barcoded sequencing libraries. All DNA from a single cell shares a common barcode. This enables separation of sequencing reads and assigning them to individual cells to generate CNVs and this processing of data can be performed easily using software tools from 10X. In addition to these single-cell applications, 10X Chromium can also be used for other applications such as single-cell immune profiling, genome, and exome sequencing. The barcodes on gel beads are different from one another and specific to each application.
REAP-Seq (RNA expression and protein sequencing) (Peterson et al. 2017) and CITE-Seq (Stoeckius et al. 2017), both introduced in 2017, are of some particular note among combination methods because of their novel use of the direct DNA polymerase activity of reverse transcriptase for identifying antibodies or other molecules that bind to specific cells and the broad applicability of the methods. The rationale for REAP-Seq and CITE-Seq is that mRNA expression does not necessarily predict protein expression, therefore measuring both simultaneously can provide a new level of information about cell states and subtypes. The mRNA transcriptomic component of REAP-Seq follows the general rules for droplet-based mRNA transcriptomics, but the protein component is assessed by conjugating specific oligonucleotide tags to antibodies against cell-surface proteins. The antibody tag is constructed with a poly(dA) sequence, followed by a sequence specific to each antibody and a PCR handle. After antibody binding to a cell mixture, the individual cells are encapsulated within a Drop-Seq bead using a microfluidic apparatus. After cell lysis, both mRNA and antibody tags hybridize to the bead primers (containing the cell barcode and UMI) and are extended by reverse transcriptase. After emulsion breakage, the tags and cDNA are amplified and sequenced.
In the initial REAP-Seq publication, 82 antibodies were used to characterize more than 7000 immunocytes by principal component analysis and t-SNE and identified a small subset of cells with the population of naïve CD8+ T cells. This outlier group was present in all donors indicating the presence a new population enriched in an intermediate step in megakaryocyte development. As of this writing, a variation on these methods using DNA aptamers (Apt-Seq) rather than antibodies (Delley et al. 2018) has also been published.
Yet, another variation exploiting the power of droplet-based single-cell methods in dissecting cellular circuitry is Perturb-Seq (Adamson et al. 2016; Dixit et al. 2016). In this method, the cells of interest are transduced with lentiviral vectors carrying multiple CRISPR-based perturbations and then subjected to an experimental stimulation. In the Broad example, CRISPR perturbations targeted transcription factors implicated in dendritic cell responses. After immune stimulation, the cells were processed by droplet-based transcriptomic analysis. The resulting sequences reveal both the specific perturbation(s) and the overall transcriptomic profile for each cell, yielding complex cause and effect networks that go far beyond the information gained by perturbing one target gene at a time.
SINGLE-CELL EPIGENETICS
Single-cell epigenetic analysis has been a long-standing goal that is now being approached, albeit with some difficulty. The basic issue is that epigenetic events cannot be directly read out by amplification with DNA polymerase, and the most common method for detecting CpG methylation, bisulfite treatment to convert unmethylated C residues to U, is very harsh and requires a large pool of starting DNA to be reliable. Several methods, including Hi-C (Nagano et al. 2013), pBAT (Miura et al. 2012), and scRRBS (single-cell reduced representation bisulfite sequencing) (Guo et al. 2013), have been published but their initial impact has been limited because of limited coverage of the methylated C residues in any single cell. In addition, because of the technical sensitivity of bisulfite treatment, these methods may be difficult to introduce into high-throughput droplet-based or microwell-based pipelines.
A different epigenetic methodology based on detecting open versus closed chromatin rather than CpG methylation relies on marking the chromatin regions accessible to a sequence conserving modifier. One such approach, ATAC-Seq (Buenrostro et al. 2015), makes use of the Tn5 transposase tagmentation process, often used to generate fragments for bulk NGS libraries, coupled with droplet-based amplification to identify genomic regions preferentially accessible to the transposase enzyme. The reported correspondence between the single-cell profiles of open chromatin near gene promoters and the data from bulk populations is remarkable. Very quickly, single-cell epigenetics has joined the realm of multi-omics, with a plethora of methods and acronymic names. Recently, 10X Genomics has come up with their own solution to probe the regulatory landscape of chromatin in hundreds to thousands of cells in a single sample. The recently launched Chromium Single Cell ATAC Solution allows use in bulk and in nuclei transposition reaction before using a microfluidic chip to partition nuclei into nanoliter-scale gel bead in emulsion (GEM) samples. A pool of ∼750,000 10X Genomics barcodes (GemCode technology) allows separately and uniquely indexing the transposed DNA of each individual cell. Libraries are generated and sequenced, and 10X Genomics barcodes are used to associate individual reads back to the individual partitions, and thereby, to each individual cell.
Single-cell epigenome data has limited utility without a functional readout, preferably in the form of transcription data. Combining single-cell transcriptomics with methylation data (scM&T-Seq) was demonstrated by Angermueller et al. (2016), using a modification of an earlier method for combined genomics and transcriptomics called scG&T-Seq (Macaulay et al. 2015). Hou et al. (2016) extended the multiplicity to combine genomic copy number profiling and bisulfite sequencing for detecting methylation with transcriptomics by gently lysing cells in individual tubes and separating the soluble RNA from the nucleus by centrifugation. Processing the nuclear DNA for bisulfite sequencing and amplification yields both genomic copy number variations (CNVs) data and methylation mapping data, although the RNA is prepared for a Smart-Seq-based transcriptomic analysis (Hou et al. 2016). Combining genomic analysis with methylation and a transcriptional readout will be extremely important for a new level of genomic exploration in cancer. Further advances in multi-omics undoubtedly await, likely involving some of the novel methods described above, and exemplified most recently by a report from Clark et al. (2018) on the combination of chromatin accessibility, DNA methylation, and transcription as scNMT-Seq.
APPLICATION IN NEUROBIOLOGY
Characterizing the cellular components of the complex ecosystem that is the brain is a prime application for single-cell genomic analysis, starting with the detection of specific gene expression in live rat neurons using antisense RNA ISH (Eberwine et al. 1992), to transcriptome in vivo analysis ([TIVA]; Lovatt et al. 2014) selection of RNA molecules for NGS, and now high-throughput RNA transcriptomics to both classify all the neuroanatomical cell types and trace the lineages back to stem cells (Dulken et al. 2017; also see Kester and van Oudenaarden 2018 for a detailed review on the general topic of transcriptomics and lineage).
The obvious applicability of scRNA-Seq to brain mapping and neuroanatomy has led to a literal “tsunami” of publications involving scRNA-Seq and there are far too many to address individually in this review. Furthermore, public interest in understanding the brain has led to the establishment and funding, by both the National Institutes of Health (NIH) and private foundations, of a large collection of brain transcriptome databases. Descriptions of the various databases have been collected in “The Brain Transcriptome Database: A User's Guide” by Kenneth Kwan and colleagues (Keil et al. 2018). This publication also provides details concerning access to the databases and an insightful review of the field.
As reviewed by Poulin et al. (2016), cellular identity is determined by the sum of all gene expression. Therefore, cellular classification must include expression profiling of all genes at the single-cell level. Further, to be maximally useful, the results must be rationalized within the framework of existing cell taxonomy and neural structure. Perhaps the most important privately funded effort to satisfy those needs and to combine scRNA-Seq data with neuroanatomy is focused on expanding the Allen Brain Atlas, an effort begun in 2001 and dedicated to characterize the cellular diversity of the brain at the molecular level (see brain-map.org). Efforts to complete the taxonomy of both human and mouse adult and developing brains are numerous, and more than 50 published and ongoing studies are listed on the Broad Institute Single Cell Portal along with instructions for accessing published data (see portals.broadinstitute.org/single_cell). As described in the Poulin et al. (2016) review, the resulting molecular atlas will not only identify cell types with high resolution, it will empower efforts aimed at mapping connectivity between specific neuronal cell types, determining the neuronal types contributing to behavior, and understanding selective degradation of neurons in age and disease.
The neuroscience literature also yields another example of the flexibility of the high-throughput scRNA-Seq methodology. High-throughput “megacell” experiments require disaggregation of tissue to obtain individual cells or nuclei and it is, of course, desirable that the cells be as close to the native state as possible. It has been a long-standing issue that artificial alterations in cell state would occur as a result of the process of disrupting living tissue and masking or contaminating the native transcription profile. In an attempt to reliably identify the expression of immediate-early genes (IEGs) after neuron activation in single cells without having them artificially activated by the disaggregation process at the single-cell level, Wu et al. (2017) adapted an old trick for analyzing transcription to high-throughput scRNA-Seq by freezing transcription with actinomycin-D before cell isolation (Keil et al. 2018). Dubbed Act-Seq, the method permitted identification of an array of IEGs and activation states in the amygdala and opens the way for linking stimulation with transcriptional state in a manner not previously possible.
MAPPING AND ANALYSIS FOR CNV PROFILING IN CANCER
Cancer is a genetic disease caused by mutations and rearrangements of the genome. It is also a disease of cell state. Alterations in the genome lead to alterations in the transcriptome and alterations in cell phenotype to a diseased state. Therefore, both transcriptomic and genomic information is required to fully characterize cancer at the single-cell level.
SINGLE-CELL DNA PROFILING OF TUMORS
Perhaps, the most extensive use of whole-genome single-cell DNA sequencing in cancer has been in studying tumor heterogeneity and using copy number profiling to identify subclonal complexity and tumor lineage. In this specialized application, the density of reads across the genome directly reflects CNVs resulting from gross genomic rearrangements and chromosome sorting mistakes during mitosis that lead to aberrant gains and losses that diverge from the diploid condition of the normal genome of somatic cells. Genomic rearrangements are a common feature of both hematologic and solid tumors, but the mitotic errors and sequential errors in recombination repair that lead to aberrant gains and losses of whole chromosome arms and the focal amplification of oncogenes are a hallmark of nearly all solid tumor tissue. We note that copy number variants are often called CNAs in cancerous tissue, but we will use the more common term CNV here.
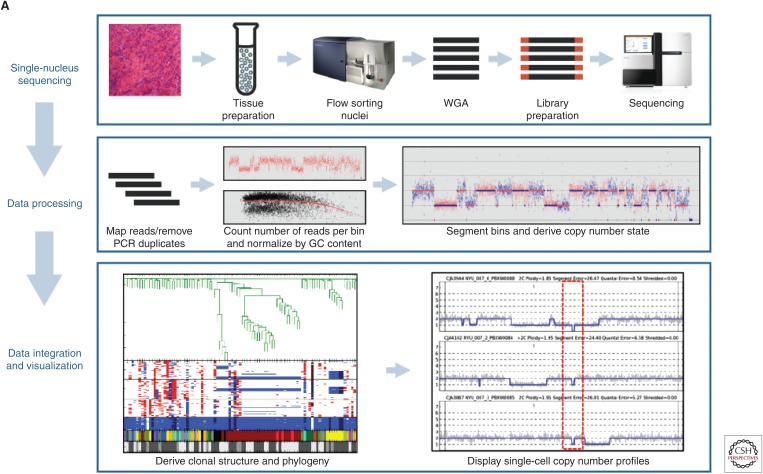
Although most sequencing methods are measured in read depth, or x-fold coverage of the genome, for the majority of single-cell CNV profiling the actual genome coverage required is actually minimal. Generally, there is no attempt to cover every base-pair as in deep sequencing of bulk tissue. There are limits to the coverage that can be obtained from just two sets of chromosomes using the amplification methods available, although studies with near whole-genome coverage have been reported using replicating premitotic cells (Wang et al. 2014), and improved high-coverage, single-cell amplification have been reported (Chen et al. 2017). A single cell contains up to thousands of copies of some mRNA molecules, but the nucleus contains only two copies (four strands) of each DNA molecule and the overall target sequence is 100 times larger, so collecting and amplifying a complete genome's worth of sequence information from single-cell DNA is much more difficult. However, as with the transcriptome important information can be gained from using NGS “reads” as “tags” for individual sequences. In the case of transcriptomics, the tags are used to count individual mRNA molecules to create a transcriptomic profile of the cell. For the genome, the most frequent approach is to use individual reads as tags representing segments of the genome. The density of tags across the genome is a very accurate measure of the relative copy number of each segment of the genome. This method was used by Navin (2011) to describe spatiotemporal evolution of a single tumor and by Alexander et al. (2018) to follow tumor evolution in prostate cancer biopsies. The segments are defined informatically by the user and the number of reads required to define a copy number profile is very low compared with base-pair level sequencing. Using sample barcodes, several hundred nuclei can be profiled on a single Illumina sequencing lane using ∼250,000 mapped reads per cell (Baslan et al. 2015). Details of the method and informatics analysis may be found in Baslan et al. (2012) and Kendall and Krasnitz (2014). Examples of typical results are shown in Figure 3.
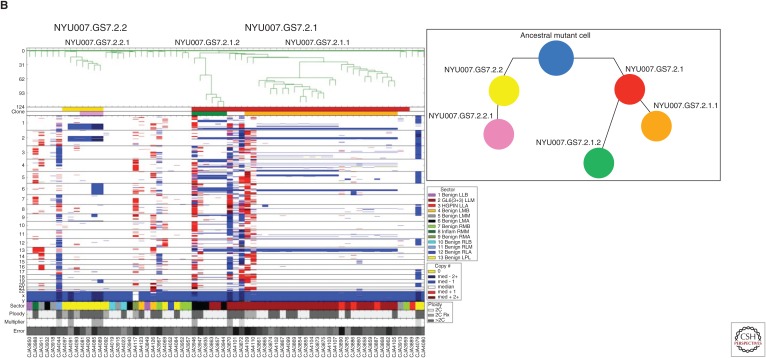
Figure 3.
The core components to the flow of sample and data processing by SNS. (A) Single-nucleus sequencing (top panel), through sequence read mapping and derivation of single-cell copy number profiles (middle panel), to clonal structure analysis and visualization (bottom panel). (B) Magnified view of clonal structure of a primary prostate cancer case contrasting the degree of genomic complexity between the two clones. There are two color tracks below dendrogram for clones and subclones and they are designated as follows: NYU007.GS7.2.1 (red) with its two subclones, NYU007.GS7.2.1.1 (green) and NYU007.GS7.2.1.2 (orange) and NYU007.GS7.2.2 (yellow) with its single subclone, NYU007.GS7.2.2.1 (pink). B shows inferred clonal progression for the clones using the same color scheme for clones and subclones as described in A. (From Alexander et al. 2018; reprinted courtesy of the American Association for Cancer Research.) (Note the ancestral mutant cell is designated by a blue circle.)
A key aspect of cancer cell copy number profiling as shown in Figure 3 is the establishment of direct genetic lineage using the complexity of CNV gains and losses as “genomic barcoding.” Even as the complexity of the CNV landscape increases, the specific breakpoints of the CNVs remain stable and can be used as if they were novel SNPs to follow the history of the tumor or cell population. The Navin laboratory has used this method extensively in resolving tumor evolution and response to chemotherapy in breast cancer (Wang et al. 2014; Gao et al. 2016; Casasent et al. 2018). It should be noted that applications listed above were performed on purified nuclei or cells disaggregated from fresh or fresh-frozen tissue. Most pathology specimens, especially valuable archived specimens from clinical trials are preserved as FFPE tissue. With that in mind, Martelotto et al. (2017) developed a method for isolating and sequencing single nuclei from FFPE sections.
Once again, 10X Genomics has developed the Chromium single-cell CNV solution to determine genomic heterogeneity and map clonal evolution by profiling hundreds to thousands of cells in a single sample. This method is significantly more laborious than others developed by 10X Genomics. The method requires encapsulation of individual nuclei (nuclei suspensions are prepared beforehand) in a hydrogel matrix to generate cell beads on a microfluidic chip. The cell bead is treated to lyse the encapsulated cell and denature the gDNA. Then, a second microfluidic chip is used to separately index the gDNA of each individual cell in a way akin to other 10X Genomics protocols. The caveat with this method is that the low recovery rate of ∼15%–17% (of ∼3000 inputted, only ∼500 are recovered). Data resolution is relatively good (∼1–2 Mb) and can be improved by increasing the amount of sequencing/cell and, consequently, costs.
ESTIMATING COPY NUMBER FROM TRANSCRIPTOME DATA
In single-cell transcriptomic cancer studies using disaggregated cells, it is of great importance to distinguish actual malignant cells from tumor-associated stromal cells and other surrounding normal tissue cells in the sample. In the absence of specific protein markers, it is becoming common practice to use expression data from a population of single cells as a basis for inferring clonal genomic CNVs that identify the malignant aneuploid cells. Several methods for parallel genomic and transcriptomic analysis on single cells have been published (Dey et al. 2015; Macaulay et al. 2015); however, they require manual separation of DNA and RNA and are not amenable to high-throughput methods. In a study to identify a developmental hierarchy in dendroglioma, Tirosh et al. (2016a) used this method to separate the malignant populations from three tumors and then further analyzed their transcriptomic profiles to show that different development states existed within each CNV defined subclone, indicating a developmental hierarchy in the cancer independent of the genome. They used a sliding window of 100 genes across the genome to compare genome-wide expression levels among the cells and were able to define two populations; in one, the expression patterns matched known CNVs from bulk DNA sequencing. These were designated the malignant cells for the rest of the study. Although this method is based on inference and does not provide an accurate copy number profile of each cell, it can work well when enough supporting evidence (e.g., bulk CNV analysis) is available. A similar, but distinct, approach was successfully used by Müller et al. (2016) in studying glioblastoma multiforme (GBM) (Tirosh et al. 2016a), and software programs are available online at the Broad Institute website (see github.com/broadinstitute/inferCNV).
SINGLE-CELL TRANSCRIPTION STUDIES IN CANCER
Numerous groups have applied various scRNA-Seq methods to assess heterogeneity, tumor evolution, and cell-of-origin studies in tumors with relatively low throughput methods (Patel et al. 2014; Tirosh et al. 2016b; Li et al. 2017). Using high-throughput Drop-Seq methods, this field is being revolutionized. Targeted sequencing of thousands of single cells has enabled the identification of pathogenic mutations in patients in remission revealing complex clonal evolution in acute myeloid leukemia (AML) (Pellegrino et al. 2018). Using whole-transcriptome methods, other groups have cataloged the array of stromal cells in the lung cancer microenvironment (Lambrechts et al. 2018) and identified the cell of origin of kidney cancer (Young et al. 2018).
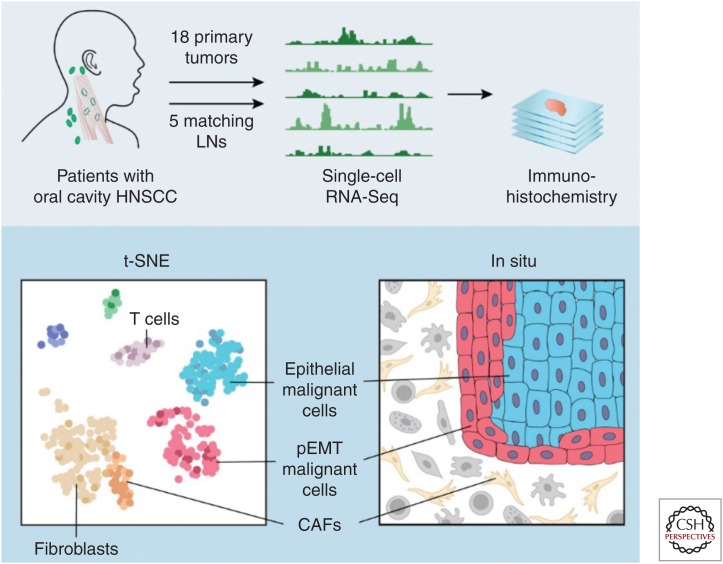
One study in particular demonstrates the power of multiplex single-cell analysis going beyond sequencing. A recent report from the laboratories of Derrick Lin, Aviv Regev, and Brad Bernstein, used single-cell transcription analysis, coupled with histology and cell biology to revise the subtypes of head and neck squamous cell cancer (HNSCC) and reveal the presence and location and function of novel tumor cell phenotypes related to metastasis in HNSCC (Puram et al. 2017).
Traditional molecular analyses of disaggregated tumor specimens are complicated by the variety of nonmalignant stromal and immunocytes that along with the tumor cells comprise the tumor tissue. However, rather than describing the average genomic signal from the bulk analysis of a homogenized tumor, the investigators performed scRNA sequencing on more than 6000 individual cells from 18 head and neck cancer patients to explore the diversity of cells within the all three populations, literally creating a molecular and cellular atlas of the disease. A schematic of the method taken from their paper is shown in Figure 4. Key to this single-cell study was the use of complementary genomic techniques, including inferred copy number profiling from scRNA-Seq data and expression phenotyping, with the aim of distinguishing individual malignant cells with aneuploid genomes from the copy number normal (euploid) but phenotypically distinct stromal populations, including cancer-associated fibroblasts and immunocytes. Once identified, analysis of the expression profiles by t-SNE revealed that the malignant (aneuploid) cells fell into separate clusters according to cancer subtype, whereas the stromal cells from all cases clustered together around identifiable cell phenotypes. Furthermore, the malignant cells defined three subtypes, rather than the four that were clinically recognized. The fourth category being identical to one of the others, but with an overabundance of fibroblasts that made it appear as a distinct subtype in bulk cell analysis. The project further became a tour de force when they used the RNA expression profiles to infer differing protein expression patterns among the tumor cells, and in fact defined a new epithelial mesenchymal transition (EMT)-like phenotype, which they call p(partial) EMT, which was inversely expressed relative to the epithelial profile of the tumor body. They then returned to the tumor itself by using antibodies made to those proteins to determine the positions of each cell type on mounted sections of primary tissue, showing that the pEMT cells created a layer on the growing front on the tumor facing the stroma and a concentration of cancer-associated fibroblasts (CAFs). This result lead to an examination of the ligands and receptors expressed at the tumor–stroma boundary, which revealed a corresponding overexpression of receptor–ligand pairs, indicating the activity of a pEMT induction pathway between the CAFs and boundary tumor cells. Thus, the study took disaggregated single-cell transcriptomics from molecular phenotyping, to histopathology to functional genomics and clinical subtyping. Although this study on a small number of HNSCC tumors is not definitive, it demonstrates the power that can be realized using single-cell omics coupled with functional genomics and classical clinical pathology.
Figure 4.
. Schematic of multiplex analysis of head and neck tumors. HNSCC, head and neck squamous cell cancer; LNs, lymph nodes; pEMT, partial epithelial mesenchymal transition; CAFs, cancer-associated fibroblasts. (From Puram et al. 2017; reprinted, with permission, from Elsevier © 2017.)
SINGLE-CELL SEQUENCING ON RARE AND ULTRA-RARE CELLS
As a polar opposite to the “megacell” experiments described above is the profiling of ultra-rare cells from liquid (blood) biopsies, in which information from a very few cells could provide near real-time information on the progress of cancer treatment. The killing force in cancer is metastasis, occurring often years after the primary tumor is discovered and excised. Yet, cancer treatment is most often based on biomarkers in the primary tumor alone. Biopsies of metastatic sites are infrequent, and necessarily incomplete because it is impossible to find, let alone biopsy, all of the metastatic sites in a progressing patient. One avenue to the real-time state of the tumor is the blood, either through circulating cell-free DNA or RNA, or through individual cells shed into the blood from the various metastatic sites. The key is identifying and then isolating cells that in most cases represent perhaps 10–100 nucleated cells in a standard blood draw containing up to 30 million nucleated white blood cells (WBCs).
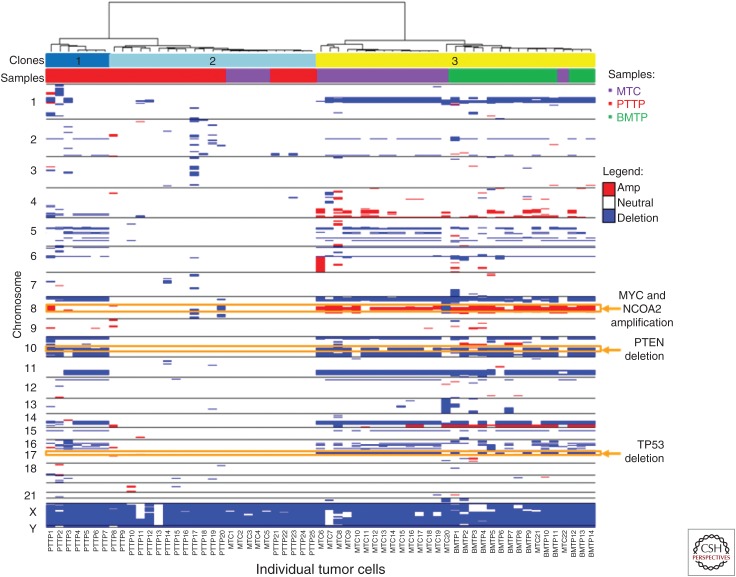
Capture and detailed analysis of circulating tumor cells (CTCs) has been a goal for two decades, and although at least 35 different technologies are in development, only one method, CellSearch, has gained Food and Drug Administration (FDA) approval for identifying cancer that is starting to progress. Single-cell DNA sequencing has presented an opportunity to advance the information gained from liquid biopsies and our understanding of metastasis itself. CellSearch is a well-developed method that captures putative tumor cells on the basis of the epithelial marker EpCam, but has been mainly used for simply enumerating EpCam-positive cells. Other parameters that have been used for capture include cell size or plasticity among others. Nonselective methods in which putative cells are identified and characterized in situ by immunofluorescent antibody tags have shown promise for both enumeration and molecular analysis at the single-cell level. In particular, the high-density, single-cell analysis or “no cell left behind assay” originally developed by Kuhn and colleagues (Marrinucci et al. 2009) and commercially developed by Epic Sciences (Werner et al. 2015; Greene et al. 2016) plates a full blood draw on slides and then identifies non-WBC with immunofluorohistochemistry. This method has been used to identify clonal relationships based on CNV profiles, follow changes in cell populations during therapy in castrate-resistant prostate cancer (Dago et al. 2014; Malihi et al. 2018), confirm vascular mimicry by tumor cells in small-cell lung cancer (Williamson et al. 2016), and confirm the identity of circulating cells in melanoma (Ruiz et al. 2015). An example of the utility of this method is shown in Figure 5, in which Malihi and colleagues used single-cell CNV profiling to track the lineage of circulating cells in a metastatic prostate cancer patient from the primary lesion to the bone marrow (Malihi et al. 2018). As with the prostate cancer biopsies shown in Figure 3, CNV profiling is key to the identification of actual tumor cells from genomically normal cells in the analysis of mixed populations, and is especially critical in identifying ultra-rare (one in a million) tumor cells from liquid biopsies. An automated version of this slide-based method based on the same principle has been developed by RareCyte (Seattle, WA).
Figure 5.
Tumor cell identification and clonal lineage by single-cell copy number variants (CNVs) profiling. Heat map and phylogenic tree of CNVs across the entire population of cells from metastatic circulation (MTC), bone marrow biopsy (BMTP), and primary resection (PTP). Sample type and clones are identified using color key. Three clones were identified: clone 1 consisting of prostate cells with hallmark alterations, clone 2 with few copy number alterations (CNAs), and clone 3 showing lineage relationship of bone marrow, primary, and circulation. Key genes such as MYC, NCOA2, PTEN, and TP53 have been highlighted by chromosome location across clone and sample type. (From Malihi et al. 2018; reprinted courtesy of the American Association for Cancer Research.)
CONCLUSIONS
As we experienced in the 2000s with NextGen Sequencing itself, automation has democratized single-cell sequencing. Single-cell sequencing in all of its variations has passed from the development stage in individual laboratories to a point where it can be applied by every laboratory and across every area of biology. Technical methods for single-cell genomic copy number and transcriptome profiling as well as targeted sequencing for mutation analysis are highly evolved and data can be presented in accepted formats. It has indeed progressed from the technology of the future to the technology of the present, and still more novel approaches are continually being realized and combined to create more insightful observations. The versatility in the cell-isolation methods has removed many previous barriers. We can analyze large populations, small populations, and even ultra-rare cells, without limits. Cell “atlases” of the type created by “megacell” projects described above will take advantage of all of these novel methods and will have an important role in understanding multicellular biology at increasingly detailed levels. A particularly important example is the effort sponsored by the Chan-Zuckerberg Initiative to create a complete human cell atlas based on multiplex molecular single-cell profiling of every cell type in the body (see www.chanzuckerberg.com/human-cell-atlas).
Such projects will undoubtedly lead to new insights into cell–cell and cell–organism dynamics in all life forms and may well be the keys to profound new understanding of the human condition and indeed life itself in both health and disease. We await these developments with great anticipation.
ACKNOWLEDGMENTS
The authors acknowledge the continuing support of single-cell sequencing from the Breast Cancer Research Foundation, Prostate Cancer Foundation, and Susan G. Komen for the Cure (J.H. and N.A.), support of the National Cancer Institute (NCI) and Leidos Biomedical Research (Contract No. HHSN261200800001E, contract agreements 12XS527 and 15X003), and of the NCI's USC Norris Comprehensive Cancer Center (CORE) Support (Grant No. 5P30CA014089- 40) to Prof. Peter Kuhn, USC. We also thank Jude Kendall, Alex Krasnitz, and Joan Alexander (Cold Spring Harbor Laboratory), Jens Durruthy (10X Genomics) for figures and analytical insights, and Peter Kuhn (USC) for support and enlightening discussions.
Footnotes
Editors: W. Richard McCombie, Elaine R. Mardis, James A. Knowles, and John D. McPherson
Additional Perspectives on Next-Generation Sequencing in Medicine available at www.perspectivesinmedicine.org
REFERENCES
- Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. 2016. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167: 1867–1882.e21. 10.1016/j.cell.2016.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Kendall J, McIndoo J, Rodgers L, Aboukhalil R, Levy D, Stepansky A, Sun G, Chobardjiev L, Riggs M, et al. 2018. Utility of single-cell genomics in diagnostic evaluation of prostate cancer. Cancer Res 78: 348–358. 10.1158/0008-5472.CAN-17-1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, Krueger F, Smallwood S, Ponting CP, Voet T, et al. 2016. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods 13: 229–232. 10.1038/nmeth.3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, et al. 2018. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174: 1293–1308.e36. 10.1016/j.cell.2018.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslan T, Hicks J. 2017. Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat Rev Cancer 17: 557–569. [DOI] [PubMed] [Google Scholar]
- Baslan T, Kendall J, Rodgers L, Cox H, Riggs M, Stepansky A, Troge J, Ravi K, Esposito D, Lakshmi B, et al. 2012. Genome-wide copy number analysis of single cells. Nat Protoc 7: 1024–1041. 10.1038/nprot.2012.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslan T, Kendall J, Ward B, Cox H, Leotta A, Rodgers L, Riggs M, D'Italia S, Sun G, Yong M, et al. 2015. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res 25: 714–724. 10.1101/gr.188060.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Davis KL, Amir el-AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nola GP, Pe'er D. 2014. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157: 714–725. 10.1016/j.cell.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke P, Anders S, Kim JK, Kolodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, et al. 2013. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods 10: 1093–1095. 10.1038/nmeth.2645 [DOI] [PubMed] [Google Scholar]
- Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, Klein AM. 2018. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360: eaar5780 10.1126/science.aar5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. 2015. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523: 486–490. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasent AK, Schalck A, Gao R, Sei E, Long A, Pangburn W, Casasent T, Meric-Bernstam F, Edgerton ME, Navin NE. 2018. Multiclonal invasion in breast tumors identified by topographic single cell sequencing. Cell 172: 205–217.e12. 10.1016/j.cell.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casbon JA, Osborne RJ, Brenner S, Lichtenstein CP. 2011. A method for counting PCR template molecules with application to next-generation sequencing. Nucleic Acids Res 39: e81 10.1093/nar/gkr217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Xing D, Tan L, Li H, Zhou G, Huang L, Xie XS. 2017. Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI). Science 356: 189–194. 10.1126/science.aak9787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Argelaguet R, Kapourani CA, Stubbs TM, Lee HJ, Alda-Catalinas C, Krueger F, Sanguinetti G, Kelsey G, Marioni JC, et al. 2018. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Commun 9: 781 10.1038/s41467-018-03149-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dago AE, Stepansky A, Carlsson A, Luttgen M, Kendall J, Baslan T, Kolatkar A, Wigler M, Bethel K, Gross ME, et al. 2014. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS ONE 9: e101777 10.1371/journal.pone.0101777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y et al. 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci 99: 5261–5266. 10.1073/pnas.082089499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delley CL, Liu L, Sarhan MF, Abate AR. 2018. Combined aptamer and transcriptome sequencing of single cells. Sci Rep 8: 2919 10.1038/s41598-018-21153-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Ramsköld D, Reinius B, Sandberg R. 2014. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343: 193–196. 10.1126/science.1245316 [DOI] [PubMed] [Google Scholar]
- Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. 2015. Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol 33: 285–289. 10.1038/nbt.3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Zheng L, Zhu Y, Li N, Jia H, Ai R, Wildberg A, Wang W. 2015. Normalization and noise reduction for single cell RNA-seq experiments. Bioinformatics 31: 2225–2227. 10.1093/bioinformatics/btv122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Pamas O, Li B, Chen J, Fulco CP, Jerby-Amon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. 2016. Perturb-Seq: Dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167: 1853–1866.e17. 10.1016/j.cell.2016.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken BW, Leeman DS, Boutet SC, Hebestreit K, Brunet A. 2017. Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep 18: 777–790. 10.1016/j.celrep.2016.12.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. 1992. Analysis of gene expression in single live neurons. Proc Natl Acad Sci 89: 3010–3014. 10.1073/pnas.89.7.3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF. 2018. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360: eaar3131 10.1126/science.aar3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW. 2018. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360: eaaq1736 10.1126/science.aaq1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu GK, Hu J, Wang PH, Fodor SP. 2011. Counting individual DNA molecules by the stochastic attachment of diverse labels. Proc Natl Acad Sci 108: 9026–9031. 10.1073/pnas.1017621108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, Tsai PC, Casasent A, Waters J, Zhang H, et al. 2016. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet 48: 1119–1130. 10.1038/ng.3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierahn TM, Wadsworth MH II, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK. 2017. Seq-Well: Portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods 14: 395–398. 10.1038/nmeth.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene SB, Dago AE, Leitz LJ, Wang Y, Lee J, Werner SL, Gendreau S, Patel P, Jia S, Zhang L, et al. 2016. Chromosomal instability estimation based on next generation sequencing and single cell genome wide copy number variation analysis. PLoS ONE 11: e0165089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, van Oudenaarden A. 2015. Design and analysis of single-cell sequencing experiments. Cell 163: 799–810. 10.1016/j.cell.2015.10.039 [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Wu X, Li X, Wen L, Tang F. 2013. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res 23: 2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Buettner F, Theis FJ. 2015. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31: 2989–2998. 10.1093/bioinformatics/btv325 [DOI] [PubMed] [Google Scholar]
- Ho YJ, Anaparthy N, Molik D, Mathew G, Aicher T, Patel A, Hicks J, Hammell MG. 2018. Single-cell RNA-seq analysis identifies markers of resistance to targeted BRAF inhibitors in melanoma cell populations. Genome Res 28: 1353–1363. 10.1101/gr.234062.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, Wu X, Wen L, Tang F, Huang Y, et al. 2016. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res 26: 304–319. 10.1038/cr.2016.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Kjällquist U, Moliner A, Zajac P, Fan JB, Lönnerberg P, Linnarsson S. 2011. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res 21: 1160–1167. 10.1101/gr.110882.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S. 2014. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 11: 163–166. 10.1038/nmeth.2772 [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. 2014. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343: 776–779. 10.1126/science.1247651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil JM, Qalieh A, Kwan KY. 2018. Brain transcriptome databases: A user's guide. J Neurosci 38: 2399–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall J, Krasnitz A. 2014. Computational methods for DNA copy-number analysis of tumors. Methods Mol Biol 1176: 243–259. 10.1007/978-1-4939-0992-6_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kester L, van Oudenaarden A. 2018. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 23: 166–179. 10.1016/j.stem.2018.04.014 [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, Scadden DT. 2014. Bayesian approach to single-cell differential expression analysis. Nat Methods 11: 740–742. 10.1038/nmeth.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivioja T, Vähärautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J. 2012. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods 9: 72–74. 10.1038/nmeth.1778 [DOI] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161: 1187–1201. 10.1016/j.cell.2015.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, Bassez A, Decaluwé H, Pircher A, Van den Eynde K, et al. 2018. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med 24: 1277–1289. 10.1038/s41591-018-0096-5 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewer K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Li WV, Li JJ. 2018. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat Commun 9: 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, Kong SL, Chua C, Hon LK, Tan WS, et al. 2017. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 49: 708–718. 10.1038/ng.3818 [DOI] [PubMed] [Google Scholar]
- Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, Fisher S, Francis C, Spaethling JM, Wolf JA, Grady MS, et al. 2014. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods 11: 190–196. 10.1038/nmeth.2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ, Goolam M, Saurat N, Coupland P, Shirley LM, et al. 2015. G&T-seq: Parallel sequencing of single-cell genomes and transcriptomes. Nat Methods 12: 519–522. 10.1038/nmeth.3370 [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malihi P, Morikado M, Welter L, Liu ST, Miller ET, Cadaneanu RM, Knudsen BS, Lewis MS, Carlsson A, Ruiz Velasco C, et al. 2018. Clonal diversity revealed by morphoproteomic and copy number profiles of single prostate cancer cells at diagnosis. Converg Sci Phys Oncol 4: 015003 10.1088/2057-1739/aaa00b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E, Karp RL, Guo G, Robson P, Hart AH, Trippa L, Guo-Cheng Y. 2014. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci 111: E5643–E5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrinucci D, Bethel K, Luttgen M, Bruce RH, Nieva J, Kuhn P. 2009. Circulating tumor cells from well-differentiated lung adenocarcinoma retain cytomorphologic features of primary tumor type. Arch Pathol Lab Med 133: 1468–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelotto LG, Baslan T, Kendall J, Geyer FC, Burke KA, Spraggon L, Piscuoglio S, Chadalavada K, Nanjangud G, Ng CK, et al. 2017. Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nat Med 23: 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel JR, DeKosky BJ, Tanno H, Ellington AD, Georgiou G. 2016. Ultra-high-throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat Protoc 11: 429–442. 10.1038/nprot.2016.024 [DOI] [PubMed] [Google Scholar]
- Miura F, Enomoto Y, Dairiki R, Ito T. 2012. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res 40: e136 10.1093/nar/gks454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Liu SJ, Di Lullo E, Malatesta M, Pollen AA, Nowakowski TJ, Kohanbash G, Aghi M, Kriegstein AR, Lim DA, et al. 2016. Single-cell sequencing maps gene expression to mutational phylogenies in PDGF- and EGF-driven gliomas. Mol Syst Biol 12: 889 10.15252/msb.20166969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. 2013. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin NN. 2011. Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94. 10.1038/nature09807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichterwitz S, Chen G, Aguila Benitez J, Yilmaz M, Storvall H, Cao M, Sandberg R, Deng Q, Hedlund E. 2016. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat Commun 7: 12139 10.1038/ncomms12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Nahed BV, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastomas. Science 344: 1396–1401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino M, Sciambi A, Treusch S, Durruthy-Durruthy R, Gokhale K, Jacob J, Chen TX, Geis JA, Oldham W, Matthews J, et al. 2018. High-throughput single-cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Res 28: 1345–1352. 10.1101/gr.232272.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S, Klappenbach JA. 2017. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol 35: 936–939. 10.1038/nbt.3973 [DOI] [PubMed] [Google Scholar]
- Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glažar P, Obermayer B, Theis FJ, Kocks C, Rajewsky N. 2018. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360: eaaq1723 10.1126/science.aaq1723 [DOI] [PubMed] [Google Scholar]
- Poulin JF, Tasic B, Hjerling-Leffler J, Trimarchi JM, Awatramani R. 2016. Disentangling neural cell diversity using single-cell transcriptomics. Nat Neurosci 19: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, et al. 2017. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171: 1611–1624.e24. 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. 2012. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 30: 777–782. 10.1038/nbt.2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece A, Xia B, Jiang Z, Noren B, McBride R, Oakey J. 2016. Microfluidic techniques for high throughput single cell analysis. Curr Opin Biotechnol 40: 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati E, Dowds CM, Liaskou E, Henriksen EKK, Karlsen TH, Franke A. 2017. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol 17: 61 10.1186/s12896-017-0379-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz C, Li J, Luttgen MS, Kolatkar A, Kendall JT, Flores E, Topp Z, Samlowski WE, McClay E, Bethel K, et al. 2015. Limited genomic heterogeneity of circulating melanoma cells in advanced stage patients. Phys Biol 9: 016008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33: 495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher HI, Graf RP, Schreiber NA, McLaughlin B, Lu D, Louw J, Danila DC, Dugan L, Johnson A, Heller G, et al. 2017. Nuclear-specific AR-V7 protein localization is necessary to guide treatment selection in metastatic castration-resistant prostate cancer. Eur Urol 71: 874–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah YFS, Hu H, Merten CA. 2018. Microfluidic single-cell technology in immunology and antibody screening. Mol Aspects Med 59: 47–61. 10.1016/j.mam.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Bendall S, Friedman N, Pe'er D. 2016. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol 34: 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. 2013. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498: 236–240. 10.1038/nature12172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. 2013. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet 14: 618–630. [DOI] [PubMed] [Google Scholar]
- Shiroguchi K, Jia TZ, Sims PA, Xie XS. 2012. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc Natl Acad Sci 109: 1347–1352. 10.1073/pnas.1118018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P. 2017. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14: 865–868. 10.1038/nmeth.4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. 2009. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6: 377–382. 10.1038/nmeth.1315 [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjöld M, Ponder BA, Tunnacliffe A. 1992. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerate primer. Genomics 13: 718–725. 10.1016/0888-7543(92)90147-K [DOI] [PubMed] [Google Scholar]
- Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, Xega K, et al. 2014. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep 8: 1905–1918. 10.1016/j.celrep.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. 2016a. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539: 309–313. 10.1038/nature20123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH II, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. 2016b. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352: 189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, Laurent G. 2018. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360: 881–888. 10.1126/science.aar4237 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32: 381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoucas D, Yuan GC. 2017. Recent progress in single-cell cancer genomics. Curr Opin Genet Dev 42: 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten LJP. 2014. Accelerating t-SNE using tree-based algorithms. J Mach Learn 15: 3221–3245. [Google Scholar]
- van der Maaten LJP, Hinton GE. 2008. Visualizing high-dimensional data using t-SNE. J Mach Learn 9: 2579–2605. [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. 2018. Recovering gene interactions from single-cell data using data diffusion. Cell 174: 716–729.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. 1990. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci 87: 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM. 2018. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360: 981–987. 10.1126/science.aar4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, et al. 2014. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512: 155–160. 10.1038/nature13600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattenberg M, Viegas F, Johnson I. 2016. How to use t-SNE effectively. Distill 10.23915/distill.00002 [DOI] [Google Scholar]
- Werner SL, Graf RP, Landers M, Valenta DT, Schroeder M, Greene SB, Bales N, Dittamore R, Marrinucci D. 2015. Analytical validation and capabilities of the epic CTC platform: Enrichment-free circulating tumour cell detection and characterization. J Circ Biomark 4: 3 10.5772/60725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SC, Metcalf RL, Trapani F, Mohan S, Antonello J, Abbott B, Leong HS, Chester CP, Simms N, Polanski R, et al. 2016. Vasculogenic mimicry in small cell lung cancer. Nat Commun 7: 13322 10.1038/ncomms13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Doud DFR, Schulz F. 2017. The trajectory of microbial single-cell sequencing. Nat Methods 14: 1045–1054. 10.1038/nmeth.4469 [DOI] [PubMed] [Google Scholar]
- Wu YE, Pan L, Zuo Y, Li X, Hong W. 2017. Detecting activated cell populations using single-cell RNA-Seq. Neuron 96: 313–329.e6. 10.1016/j.neuron.2017.09.026 [DOI] [PubMed] [Google Scholar]
- Xu C, Su Z. 2015. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics 31: 1974–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Fang R, Chen L, Chen D, Xiao JP, Yang W, Wang H, Song X, Ma T, Bo S, et al. 2016. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. Proc Natl Acad Sci 113: 11907–11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Mitchell TJ, Vieira Braga FA, Tran MGB, Stewart BJ, Ferdinand JR, Collord G, Botting RA, Popescu DM, Loudon KW, et al. 2018. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361: 594–599. 10.1126/science.aat1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Sims PA. 2016. An automated microwell platform for large-scale single cell RNA-Seq. Sci Rep 6: 33883 10.1038/srep33883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. 2015. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347: 1138–1142. 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]