SUMMARY
3′ untranslated regions (3′ UTRs) of messenger RNAs (mRNAs) are best known to regulate mRNA-based processes, such as mRNA localization, mRNA stability, and translation. In addition, 3′ UTRs can establish 3′ UTR-mediated protein–protein interactions (PPIs), and thus can transmit genetic information encoded in 3′ UTRs to proteins. This function has been shown to regulate diverse protein features, including protein complex formation or posttranslational modifications, but is also expected to alter protein conformations. Therefore, 3′ UTR-mediated information transfer can regulate protein features that are not encoded in the amino acid sequence. This review summarizes both 3′ UTR functions—the regulation of mRNA and protein-based processes—and highlights how each 3′ UTR function was discovered with a focus on experimental approaches used and the concepts that were learned. This review also discusses novel approaches to study 3′ UTR functions in the future by taking advantage of recent advances in technology.
1. INTRODUCTION
Genetic information is stored in DNA and transmitted via messenger RNA (mRNA) to proteins (Crick 1958). For many years, it was thought that the information transfer from DNA to proteins happens exclusively through translation of the coding region of mRNAs into amino acids of proteins. Although it was well known that mRNAs also contain untranslated regions (UTRs) at their 5′ and 3′ ends, the prevailing view was that these regions largely regulate mRNA localization or protein abundance, either through the regulation of translation or stability of mRNAs.
With such a protein-centric view, it was surprising to find that the number of protein-coding genes was similar in the human genome and the genomes of more simple eukaryotic organisms (Lander et al. 2001; Mayr 2017). Also, protein size is largely similar across organisms (Milo and Phillips 2016). This points to a remarkable conservation of protein sequence and reveals the tremendous constraints that act on proteins. However, it also begs the question of what enables biological complexity of higher organisms. In this context, it is interesting to note that the sequence space occupied by 3′ UTRs has substantially expanded during the evolution of higher organisms and correlates with cellular complexity of organisms (Chen et al. 2012; Mayr 2016). Importantly, there are 3′ UTR regions ranging from a few to a few hundred nucleotides (nt) that are often highly conserved (Siepel et al. 2005; Xie et al. 2005). The expansion of 3′ UTR sequence during evolution, together with the recent finding that genetic information stored in 3′ UTRs can also be transmitted to proteins via the formation of 3′ UTR-mediated protein–protein interactions (PPIs) (Berkovits and Mayr 2015), suggests that 3′ UTRs may play important roles in the regulation of biological complexity. If this is so, why are 3′ UTRs not at the center of everyone’s attention?
One contributing factor is the scarcity of suitable and robust methods for the study of 3′ UTR functions as the availability of experimental approaches is often crucial for progress and discoveries. Compared with research on transcription or translation regulation, or on microRNA (miRNA)-mediated posttranscriptional regulation, research on miRNA-independent functions of 3′ UTRs lags far behind. Most functions of 3′ UTRs are mediated by RNA-binding proteins (RBPs). But, in contrast to miRNA target sites, the binding motifs for many RBPs are still not known (Baltz et al. 2012; Ray et al. 2013; Dominguez et al. 2018). And even if they are known, clear functional effects often require the motifs to be present several times. As the motifs are often spread out over large distances, functional motifs can often not be identified using deletion mutants (Besse et al. 2009; Kristjansdottir et al. 2015; Ma and Mayr 2018). Moreover, many RBPs bind to the same motif (Ray et al. 2013; Dominguez et al. 2018), and it is usually unknown whether they compete or cooperate (Hennig and Sattler 2015). Finally, more than half of human genes use alternative cleavage and polyadenylation to generate mRNA isoforms that differ only in their 3′ UTRs (Lianoglou et al. 2013). This makes research on 3′ UTR isoform–specific functions challenging as the amino acid sequences of the proteins generated from the alternative 3′ UTR isoforms are identical.
This review summarizes how research on 3′ UTR functions has been approached in the last 25 years, what has been learned, and how it could be addressed in the future. Several topics relevant for 3′ UTR biology are being reviewed elsewhere in this collection, including regulation by miRNAs, RNA editing, RNA structure, the identification of RBPs, and RNA–protein interactions. Also, several “noncanonical” functions of 3′ UTRs were reviewed recently and are not included here (Mayr 2017). Thus, this review is not comprehensive but tries to focus on experimental approaches and concepts to show how regulation by 3′ UTRs is accomplished.
2. 3′ UTRS THAT CONTAIN AU-RICH ELEMENTS REGULATE mRNA STABILITY
Nucleotide sequencing was developed in the 1970s and revealed the existence of untranslated sequence between the coding region and the polyadenylation signal. Having the DNA sequences from genes of different organisms in hand allowed the first comparative genomic analyses (Needleman and Wunsch 1970). As the rate of base substitution reflects the degree of functional constraint, the evaluation of sequence divergence across distant organisms then allowed the evaluation of functionally import specific elements. Although these early comparative genomic analyses found a higher degree of base substitutions in untranslated regions than in coding regions, they also revealed a high degree of sequence homology in 3′ UTRs (Miyata et al. 1980). Early on, one of the most intriguing findings was that 3′ UTR sequences of homologous genes coding for actin proteins are highly conserved across organisms, but the 3′ UTR sequences are highly divergent when similar actin isoforms with different functions or tissue distributions were compared (Yaffe et al. 1985). The high conservation indicated that 3′ UTRs have important regulatory roles, and the divergence of the 3′ UTR sequence of isoforms, on the other hand, suggested that 3′ UTRs contain additional genetic information to distinguish the functions of highly similar proteins.
The first indication that 3′ UTRs contain specific functional elements was found through investigation of the transforming ability of the c-fos gene (Miller et al. 1984). Whereas the viral fos (v-fos) gene was able to transform fibroblasts in culture, the cellular fos (c-fos) lacked this feature. Interestingly, the functional difference was not the result of a slightly different amino acid sequence of the two fos proteins. Instead, it was shown that c-fos could also transform fibroblasts when the 3′ UTR was omitted (Miller et al. 1984). This finding motivated the search for the responsible 3′ UTR cis-element and led to the discovery of AU-rich elements which were found to destabilize mRNAs (Meijlink et al. 1985). The proof for this function was obtained through transfer of the cis-element to an otherwise stable, heterologous mRNA, the β-actin mRNA (Shaw and Kamen 1986). At around the same time, it was noticed that AU-rich elements were predominantly found in 3′ UTRs of a certain class of genes that encodes short-lived factors, including cytokines, lymphokines, growth factors, and oncogenes (Caput et al. 1986). Intriguingly, the AU-rich elements were sometimes more conserved than the coding regions of these early response genes (Shaw and Kamen 1986). The identification of the trans-acting factors that bind to AU-rich elements, however, took many years and is still ongoing (Ray et al. 2013; Dominguez et al. 2018). Also, the mechanism of AU-rich element–induced mRNA decay was elucidated much later, as will be described below.
3. 3′ UTRs REGULATE mRNA LOCALIZATION
At around the time of discovery of AU-rich elements, 3′ UTRs were also identified as important regulators of subcellular mRNA localization. Early studies on the regulation of mRNA localization were mostly performed in fly or frog oocytes. But mRNA localization is not only essential in germ cells and early development as it also occurs in other polarized cells, including fibroblasts, myoblasts, and neurons (Lawrence and Singer 1986; Melton 1987). However, germ cells and the early stages of development are especially advantageous for RNA research as gene regulation in early animal development depends on maternally deposited mRNAs from oocytes before the onset of embryonic transcription. For example, the anterior body pattern of flies is specified predominantly by the protein product of the bicoid gene as mutants containing a disrupted gene lack the head and thorax (Macdonald and Struhl 1988). Bicoid protein localizes mainly to the anterior pole of the fly embryo and generates a protein gradient. As bicoid is a transcription factor, it drives expression of a large number of downstream genes. Importantly, the gradient in the embryo depends on prior localization of bicoid mRNA to the anterior pole of oocytes. Subcellular localization of bicoid mRNA to the anterior poles of oocytes and early embryos was revealed by RNA in situ hybridization. To identify the responsible cis-element, deletion constructs were tested using transgenic flies. RNase protection assays from the anterior and posterior poles of early fly embryos revealed that a 630 nt fragment of the bicoid 3′ UTR was necessary and sufficient for the subcellular mRNA localization (Macdonald and Struhl 1988).
4. 3′ UTRs REGULATE TRANSLATION OF mRNAs
mRNA localization allows spatial as well as temporal regulation of protein production. In neurons, 3′ UTRs are well-known to regulate local protein synthesis in dendrites and synapses (Miller et al. 2002; An et al. 2008; Martin and Ephrussi 2009). But, in addition to the spatial organization of protein production, 3′ UTRs are also important regulators of temporal protein production. The initial observation in the discovery that 3′ UTRs regulate translation was a discrepancy between oskar mRNA and protein expression in fly oocytes. oskar mRNA determines the posterior fly body pattern (Ephrussi et al. 1991; Kim-Ha et al. 1993). Although oskar mRNA was present throughout oogenesis, oskar protein was only detectable after localization of oskar mRNA to the posterior pole (Kim-Ha et al. 1995). An ultraviolet (UV) cross-linking assay identified the RBP Bruno as the responsible factor for translation regulation. Binding of Bruno to oskar mRNA prevents premature translation of oskar protein and is necessary for oskar expression at the correct developmental stage as oskar mRNA is translated only after reaching the posterior pole of the oocytes (Kim-Ha et al. 1995). The exact mechanism of translation repression of oskar mRNA by Bruno was elucidated much later (Chekulaeva et al. 2006).
5. RBPs BIND TO 3′ UTR cis-ELEMENTS AND MEDIATE MULTIPLE AND OFTEN OPPOSING 3′ UTR FUNCTIONS
Initially, AU-rich elements were considered to be mRNA decay elements. Then, they were shown to also repress translation (Shaw and Kamen 1986; Kruys et al. 1989). After that, it was found that AU-rich elements can also increase protein production (Lindstein et al. 1989; Kontoyiannis et al. 1999). For example, in resting immune cells, AU-rich elements repress translation, but after lipopolysaccharide (LPS) stimulation of T cells, they were required for fast induction of protein expression (Lindstein et al. 1989; Kontoyiannis et al. 1999). This led to the notion that not the cis-element itself, but rather the binding of specific trans-acting factors, determines the function of a particular 3′ UTR cis-element.
This regulatory principle was nicely shown by deletion of the 62-nt-long, conserved AU-rich element of tumor necrosis factor (TNF)-α in the mouse (Kontoyiannis et al. 1999). Importantly, the remaining gene regulation of TNF-α expression was preserved because the endogenous promoter and polyadenylation signal were retained. Sole absence of the cis-regulatory AU-rich element resulted in inflammatory arthritis and inflammatory bowel disease with death of the mice at 5 to 12 weeks. In unstimulated conditions, AU-rich elements destabilize mRNAs. The AU-rich element of TNF-α represses translation permanently in nonhematopoietic cells and transiently in hematopoietic cells. Importantly, their function is not only repressive, as after activation by LPS they initially increase mRNA stability and translation in hematopoietic cells, whereas in later stages, they become repressive. In summary, AU-rich elements were found to be important regulatory elements that enable fast and highly dynamic regulation of protein production with a sharp on and off switch (Kontoyiannis et al. 1999).
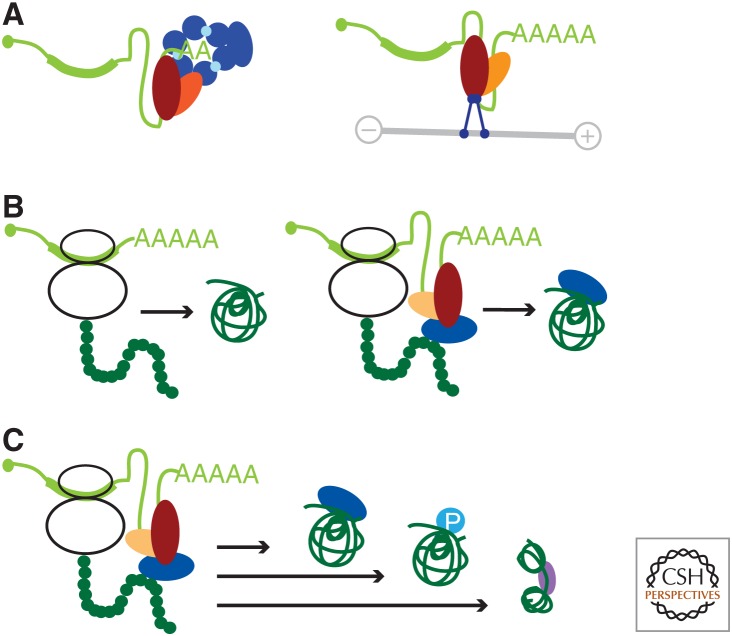
There are currently more than 10 known RBPs that bind to AU-rich elements (Brennan and Steitz 2001; Chen et al. 2001; Barreau et al. 2005; Lebedeva et al. 2011). The binding of tristetraprolin (TTP) or KHSRP leads to destabilization through recruitment of the exosome complex that degrades mRNAs (Chen et al. 2001; Lykke-Andersen and Wagner 2005). In contrast, binding of HuR stabilizes AU-rich containing mRNAs, likely through its inability to recruit the exosome (Chen et al. 2001). These findings revealed an important regulatory concept for 3′ UTR functions: RBPs bind to 3′ UTR cis-elements and serve as adapters for the recruitment of effector proteins. The recruited effector proteins are responsible for the observed effects (Fig. 1A). The adapter function of RBPs is critical as effector proteins are unable to directly bind to AU-rich-containing mRNAs (Chen et al. 2001).
Figure 1.
Functions of 3′ untranslated regions (UTRs). RNA-binding proteins (RBPs) bind to the 3′ UTR and recruit diverse effector proteins that determine 3′ UTR functions. (A) 3′ UTRs regulate processes at the messenger RNA (mRNA) level. RBPs (red, orange) bind to 3′ UTRs of mRNAs (light green) and recruit diverse effector proteins. The recruitment of the exosome (blue) results in mRNA destabilization (left panel), whereas the recruitment of a motor protein (blue) results in the regulation of mRNA localization using movement on a microtubule (gray line; right panel). (B) 3′ UTRs regulate protein features by mediating 3′ UTR-dependent protein–protein interactions (PPIs). Alternative 3′ UTRs can determine alternative protein functions despite encoding proteins with identical amino acid sequences. This results from 3′ UTR-dependent PPIs that are mediated only by the long 3′ UTR isoform (right panel) and not by the short 3′ UTR isoform (left panel). The RBPs that bind to the 3′ UTR, as well as the recruited effector protein, are color-coded as in A. (C) 3′ UTRs regulate diverse protein features by mediating 3′ UTR-dependent PPIs. This can result in 3′ UTR-dependent protein complex formation, 3′ UTR-dependent posttranslational modifications (P), and 3′ UTR-dependent protein folding.
3′ UTR elements can accomplish opposing effects in environments where different signaling pathways are active. This can be caused by altering the relative abundance or activity of RBPs that bind to the same cis-element. Thus, the function of a particular 3′ UTR cis-element is “context”-dependent, as competition and cooperativity between RBPs will ultimately determine the functional outcome of a specific 3′ UTR (Mayr 2017).
Similarly, a single RBP can accomplish diverse functions at different stages of development. In early development, translation is predominantly regulated through poly(A) tail length, which is determined by the RBP CPEB (cytoplasmic polyadenylation element [CPE]-binding protein) (Richter 2007). CPEB bound to the CPE can regulate mRNA polyadenylation, deadenylation, and translation. CPEB achieves these functions through binding to several different factors. It simultaneously binds to the deadenylase poly(A)-specific ribonuclease (PARN) and to the poly(A)polymerase Gld2 (PAPD4). In oocytes, mRNAs that contain the CPE have short poly(A) tails resulting in translation inhibition as in these cells the activity of PARN is higher than the activity of PAPD4 (Kim and Richter 2006). After oocyte fertilization, CPEB is phosphorylated, which leads to exclusion of PARN from the complex. This is followed by poly(A) tail lengthening, leading to translation initiation (Mendez et al. 2000). In summary, during early development, temporal regulation of protein synthesis occurs via the 3′ UTR and is mediated by the RBP CPEB and its associated effector proteins. Therefore, to elucidate different mechanisms of action, the associated effector proteins of RBPs need to be identified and studied.
6. MORE THAN HALF OF HUMAN GENES GENERATE ALTERNATIVE 3′ UTRs
Transcription of DNA by RNA polymerase II produces primary transcripts that need to be processed to generate mature mRNAs. The steps of mRNA processing include addition of a 5′ cap, splicing of introns and cleavage/polyadenylation at the 3′ end (reviewed in Tian and Manley 2017). Transcription does not stop at the 3′ ends of mRNAs as primary transcripts are extended beyond the polyadenylation signal by several thousand nucleotides (Core et al. 2008). Some factors of the cleavage and polyadenylation machinery travel with RNA polymerase II and the encounter of a functional polyadenylation signal results in endonucleolytic cleavage, followed by the addition of a poly(A) tail (Chan et al. 2014; Schonemann et al. 2014). Thus, 3′-end cleavage and polyadenylation determine 3′ UTR length as well as the cis-elements present within a 3′ UTR.
Approximately 10 years after the discovery that 3′ UTRs regulate several aspects of gene expression, a literature survey revealed that many genes produce alternative mRNA isoforms that differ in their 3′ ends. At the time, 95 genes were known to generate alternative 3′ UTRs, whereas 31 genes were known that used intronic polyadenylation sites to change the carboxyl terminus of the protein as well as the 3′ UTR (Edwalds-Gilbert et al. 1997). At around the same time, new technologies that allowed for shotgun sequencing of complementary DNA (cDNA) libraries enabled the creation of expressed sequence tag (EST) databases. The mining of these databases revealed that a sizable fraction of the human and mouse transcriptomes use alternative polyadenylation signals to generate mRNAs with alternative 3′ UTRs (Gautheret et al. 1998). The development of deep sequencing technology enabled the establishment of methods that were able to map all the 3′ ends of mRNAs transcriptome-wide (Derti et al. 2012; Hoque et al. 2013; Lianoglou et al. 2013; Gruber et al. 2016). These 3′-end sequencing methods revealed that more than half of human and mouse genes generate alternative mRNA isoforms that differ in their 3′ UTRs but encode proteins with identical amino acid sequences.
7. ALTERNATIVE 3′ UTR ISOFORM RATIOS ARE CELL TYPE–SPECIFIC AND CHANGE ON ACTIVATION OF SIGNALING PATHWAYS
3′-end sequencing methods provide information on 3′ UTR length and measure alternative 3′ UTR isoform levels quantitatively. 3′-end sequencing of samples from diverse tissues revealed that the location of functional polyadenylation sites does not vary across cell types, but the expression ratios of alternative 3′ UTR isoforms are highly tissue- and cell type–specific (Lianoglou et al. 2013). 3′-end sequencing data are available in databases and can be accessed and browsed (3′-seq, see cbio.mskcc.org/leslielab/ApA/atlas; polyA_DB, see exon.njms.rutgers.edu/polya_db/v3/; polyAsite, see polyasite.unibas.ch) (Lianoglou et al. 2013; Gruber et al. 2016; Wang et al. 2017; Singh et al. 2018).
Compared with RNA-seq data that exist for a large variety of samples, the availability of 3′-end sequencing data is still limited. Therefore, it would be very helpful if the expression of alternative 3′ UTRs could be extracted from RNA-seq data. Although this has been tried by several research groups (Masamha et al. 2014; Chang et al. 2015; Shenker et al. 2015; Grassi et al. 2016), the data obtained need to be used with caution as RNA-seq read coverage over 3′ UTRs is highly variable. If strict criteria are used to identify differences in 3′ UTR isoform expression, only a small number of changes (10%–15%) that are identified by 3′-end sequencing methods will be found by RNA-seq. On the other hand, less stringent analyses contain many artifacts, shown by the fact that there is little overlap in the locations of 3′-ends inferred from the different methods. Also, the direction of change (shortening or lengthening of 3′ UTRs) agreed only in 15% of the events considered to be significant by each method. Therefore, currently, 3′-end sequencing is the preferred method to determine alternative 3′ UTR expression.
In addition to being cell type–specific, alternative 3′ UTR isoform expression responds to receptor stimulation and subsequent activation of signaling pathways as was shown during immune cell stimulation or synaptic activation. A change in 3′ UTR isoform ratio can be achieved through exposure of the cells to extracellular stimuli and can be mimicked experimentally by the addition of growth factors, cytokines, or neurotransmitters (Flavell et al. 2008; Sandberg et al. 2008; Rhinn et al. 2012; Gruber et al. 2014; Jia et al. 2017). Expression ratios of alternative 3′ UTRs also change during normal development and differentiation (Lackford et al. 2014; Brumbaugh et al. 2018; Freimer et al. 2018) as well as in disease (Mayr and Bartel 2009; Rhinn et al. 2012; Batra et al. 2014; Masamha et al. 2014). A dramatic change in 3′ UTR length was recently found during mouse oocyte maturation from the germinal vesicle stage to metaphase II, which takes 9 hours and results in substantial lengthening of 3′ UTRs (Freimer et al. 2018). For ∼20% of genes, the functional consequence of changes in alternative 3′ UTR isoform expression is a difference in protein levels, as the availability of binding sites of miRNAs and RBPs is altered (Gupta et al. 2014; Nam et al. 2014; Brumbaugh et al. 2018).
8. 3′ UTRs REGULATE PROTEIN–PROTEIN INTERACTIONS
Alternative 3′ UTRs are widespread and the 3′ UTR sequences have expanded during the evolution of higher organisms (Mayr 2017). Therefore, we wondered whether 3′ UTRs might be able to regulate cellular processes beyond protein abundance. In this context, we discovered that some PPIs are established only when one of the interaction partners is recruited by the 3′ UTR (Fig. 1B) (Berkovits and Mayr 2015). This was first shown for the PPI between SET and CD47 and was then expanded to additional plasma membrane proteins in human cells and yeast (Berkovits and Mayr 2015; Chartron et al. 2016; Ma and Mayr 2018).
CD47 membrane protein is generated from alternative mRNA isoforms either containing a short or long 3′ UTR (SU or LU) (Lianoglou et al. 2013). The encoded proteins have identical amino acid sequences and are called CD47-SU and CD47-LU, respectively (Berkovits and Mayr 2015). It was found that only CD47-LU was able to interact with the protein SET in a 3′ UTR-dependent manner. As a result, CD47-SU and CD47-LU reside in different protein complexes, and thus, can have different functions. SET binding to CD47-LU protein resulted in more efficient plasma membrane localization, protected cells better from phagocytosis by macrophages, activated RAC1, enabled formation of lamellipodia, and improved cell survival after exposure to γ-irradiation (Berkovits and Mayr 2015). This list of functions is likely not exhaustive as RAC1 activation has multiple downstream consequences, including the activation of signaling pathways and increased cell migration.
The fact that 3′ UTRs are able to regulate protein complex formation and determine protein functions indicated that genetic information encoded in 3′ UTRs can be transmitted to proteins. This information transfer can regulate protein features not encoded in the amino acid sequence. As the sequence space contained within 3′ UTRs has expanded during the evolution of higher organisms, 3′ UTRs may contribute to the higher organismal complexity by providing additional information regarding proteins, including information about PPIs, posttranslational modifications, protein conformations, and protein multifunctionality (Fig. 1B,C). Thus, information about protein features can be genetically encoded in the untranslated mRNA sequence. It has not been shown yet that 3′ UTRs can regulate protein conformations but this is likely to occur as binding of the 3′ UTR-dependent interaction partner to the amino terminus of the nascent peptide chain could change the folding of the nascent protein.
Following the discovery of this new 3′ UTR function, it remained unclear how widespread this function is. It seems that 3′ UTR-mediated PPIs are common as they also occur on cytosolic and nuclear proteins (Halbach et al. 2009; Duncan and Mata 2011; Lee and Mayr 2017). This was shown in detail for the 3′ UTR-mediated interaction between IQGAP1 and the E3 ubiquitin ligase BIRC3, also called cIAP2 (Lee and Mayr 2017). BIRC3 protein is generated by alternative 3′ UTRs. Again, BIRC3 protein that was generated from the long 3′ UTR isoform was called BIRC3-LU, whereas BIRC3 protein encoded by the short 3′ UTR isoform was called BIRC3-SU. The long BIRC3 3′ UTR facilitated the assembly of a protein complex consisting of BIRC3-LU, the kinase scaffold IQGAP1, and the Ras-GTPase RALA. Importantly, this signaling complex could not be formed by BIRC3-SU. The 3′ UTR-dependent signaling complex associates with the G-protein-coupled receptor CXCR4 and influences plasma membrane expression of CXCR4 through the regulation of receptor recycling after ligand binding (Lee and Mayr 2017). The 3′ UTR-dependent recruitment of the signaling complex to CXCR4 also had biological consequences, as it was shown to be necessary for CXCR4-mediated B-cell migration. Importantly, despite similar overall BIRC3 mRNA levels observed in normal and malignant B cells, BIRC3-LU is up-regulated relative to BIRC3-SU in malignant B cells. This up-regulation is predicted to promote leukemia cell survival in vivo, as CXCR4-dependent B-cell migration was shown to allow the leukemia cells to home to bone marrow niches, which provide survival and proliferation signals (Lee and Mayr 2017).
Last, the 3′ UTR-mediated protein complex formation involving the E3 ubiquitin ligase BIRC3-LU has additional molecular consequences as it controls substrate specificity of the enzyme. Posttranslational modification of RALA was accomplished only by BIRC3-LU and not by BIRC3-SU, as substrate modification required prior protein interaction (Lee and Mayr 2017).
9. MECHANISM OF TRANSFER OF EFFECTOR PROTEINS FROM 3′ UTRs TO PROTEINS
How genetic information concerning protein features that go beyond the amino acid sequence is transmitted from mRNAs to proteins is still largely unclear. But the requirements for establishment of the 3′ UTR-mediated interaction between CD47-LU and SET proteins were recently identified (Ma and Mayr 2018). It was found that two RBPs that bind to AU-rich elements, HuR and TIS11B, are required for this process. HuR’s function is to recruit SET, whereas TIS11B creates a permissive environment that enables the 3′ UTR-mediated binding of SET to CD47 and other membrane proteins (Berkovits and Mayr 2015; Ma and Mayr 2018). TIS11B forms RNA granules that enrich membrane protein-encoding mRNAs containing AU-rich elements. TIS11B granules form a reticular meshwork that is intertwined with the endoplasmic reticulum. The functional interplay between TIS11B granules and the endoplasmic reticulum creates a subcellular compartment with a biophysically and biochemically environment distinct from that of the cytoplasm. Only within this special compartment are specific PPIs formed that cannot be established outside. This compartment enables 3′ UTR-mediated interaction of SET with CD47-LU, and also other membrane proteins (Ma and Mayr 2018). Moreover, these findings show that AU-rich elements have roles other than the control of protein abundance, as they regulate the formation of functionally relevant 3′ UTR-mediated PPIs.
10. 3′ UTR cis-ELEMENTS ARE USUALLY REPEATED AND OFTEN ACT SYNERGISTICALLY
There are several common features characteristic of 3′ UTR cis-elements. Generally, a motif needs only 3–8 nt to bind an RBP (Lambert et al. 2014). Such short motifs often occur multiple times in a given mRNA and can act in a synergistic or even cooperative manner (Kruys et al. 1989; Chao et al. 2010; Hennig et al. 2014; Hennig and Sattler 2015). The characteristics of individual 3′ UTR cis-elements are detailed in previous reviews (see Johnstone and Lasko 2001; Jambhekar and Derisi 2007).
However, mRNA localization signals tend to be much longer (50–500 nt), often contain stem-loops, and have no recognizable or recurrent motifs (Johnstone and Lasko 2001; Jambhekar and Derisi 2007). An exception to this rule is the zip-code element in the β-actin 3′ UTR, which initially was thought to be 54 nt long and is recognized by the RBP IGF2BP1, also called ZBP1 (Kislauskis et al. 1994). However, the motif was narrowed down to a bipartite element that required a spacer of defined length with each element recognized by the KH3 or KH4 domains of IGF2BP1 (Chao et al. 2010). Narrowing down the sequence to a motif was important as it enabled genome-wide screening for additional motif-containing 3′ UTRs (Patel et al. 2012).
The identification of cis-elements is important as it is required for the identification of RBPs that bind and mediate specific 3′ UTR functions. Traditionally, cis-elements were determined using deletion constructs (Macdonald and Struhl 1988; Kim-Ha et al. 1993; Kislauskis et al. 1994; Kim-Ha et al. 1995). However, often, the phenotypic effect of the entire 3′ UTR cannot be recapitulated using isolated parts of the 3′ UTR. This was shown for the translation regulation element of oskar mRNA and for the repressive element in the Hmga2 3′ UTR (Mayr et al. 2007; Besse et al. 2009; Kristjansdottir et al. 2015). To identify the element in the 2800-nt-long Hmga2 3′ UTR, which is able to fully recapitulate the repressive effect on protein production, a large set of 3′ UTR fragments was tested, but none conferred full activity. In the end, it was found that the cooperative action of three 3′ UTR subregions was necessary for the full repressive function (Kristjansdottir et al. 2015).
Another example is the 4200-nt long CD47 3′ UTR in which the cis-elements responsible for SET binding are distributed across the whole 3′ UTR and none of the tested fragments contained the full activity (Ma and Mayr 2018). To find the cis-element that fully recapitulates the desired phenotype, instead of using deletion mutants, 50–200-nt-long, diverse and unrelated 3′ UTR elements were screened. This new strategy identified the conserved AU-rich element from TNF-α as being fully able to recapitulate the effects of the full-length CD47 3′ UTR with respect to SET binding. Interestingly, comparable SET binding was accomplished whether 19 AUUUA motifs were spread out over the entire CD47 3′ UTR, or when six AU-rich elements were concatenated as occurs in the AU-rich element of TNF-α. This is similar to building arrays of transcription factor binding sites to study transcription factor functions. The identification of the short cis-element then enabled the discovery of the interacting RBPs using RNA-oligonucleotide pull-down followed by mass spectrometry (Ma and Mayr 2018).
In addition to AU-rich elements, many other cis-regulatory motifs are known (Ray et al. 2013; Dominguez et al. 2018), including those bound by IGF2BP1, Pumilio, PTB, and MBNL. However, AU-rich elements were the first cis-elements discovered and remain the most extensively studied.
11. PRIMARY CELLS SHOW MORE EXTENSIVE REGULATION OF 3′ UTR ISOFORM EXPRESSION THAN CELL LINES
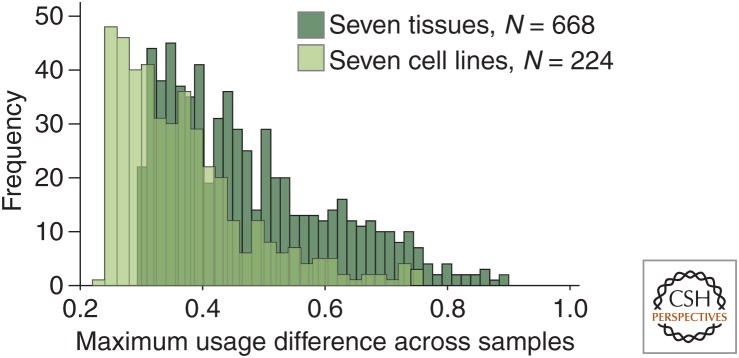
3′ UTR isoform ratios are substantially influenced by signaling pathways. Therefore, it was not surprising that primary cells show significantly greater changes in 3′ UTR isoform expression, when primary cells or tissues were compared with cell lines. In addition to the number of changes, the magnitude of change is higher in primary cells (Fig. 2) (Lianoglou et al. 2013). Alternative cleavage and polyadenylation can also occur in introns (Edwalds-Gilbert et al. 1997), and the recognition of intronic polyadenylation signals occurs substantially more frequently in primary cells than in cell lines (Lee et al., in press; Singh et al. 2018). Therefore, primary cells are beneficial for profiling alternative cleavage and polyadenylation.
Figure 2.
Primary cells show a higher number and a greater magnitude of cell type–specific changes in 3′ untranslated region (3′ UTR) isoform expression. 3′-seq data are from Lianoglou et al. (2013) and were used to calculate the maximum difference in usage of alternative polyadenylation sites across seven tissues and seven cell lines. Usage is the fraction of reads mapping to a single polyadenylation site out of all the reads mapping to the 3′ UTR. Shown are the 50% of genes with most variable 3′ UTR isoform expression. Only genes that generated two 3′ UTR isoforms were included in the analysis.
Because of the cell type–specific nature of 3′ UTR isoform expression, cell sorting into pure populations before profiling provides better quantification of cell type–specific 3′ UTR expression patterns than profiling complex tissues (Lianoglou et al. 2013). If sorting is not an option, cTag-PAPERCLIP was recently developed to measure 3′ UTR isoform expression of specific cell types in intact tissues. Based on co-IP of mRNA 3′ ends associated with green fluorescent protein (GFP)-tagged poly(A)-binding protein (PABP), the method requires mice that express tagged PABP in a tissue-specific manner (Hwang et al. 2017).
Metabolic differences between cell lines and primary cells not only cause differences in 3′ UTR isoform profiles, but also alter the regulatory effects of miRNAs and AU-rich elements (Kontoyiannis et al. 1999; La Rocca et al. 2015). For miRNAs to repress target mRNA expression, they need to be loaded into a megadalton-sized complex, called RISC (RNA-induced silencing complex). In cell lines and proliferating cells, most miRNAs reside in these large complexes. However, in primary cells, the majority of miRNAs are bound to Argonaute, a component of RISC, but are not part of the larger RISC complex (La Rocca et al. 2015). Assembly of the larger RISC complexes is mediated by the activation of signal transduction pathways, including the PI3K pathway. Thus, in primary cells, miRNAs are able to repress their targets only after pathway activation (La Rocca et al. 2015). This principle is likely to hold true for the function of RBPs, as signaling pathways may influence protein complex formation.
12. 3′ UTR-MEDIATED REGULATION IN VIVO
The powerful influence of 3′ UTRs on gene expression in whole organisms has been best shown using mouse models. One of the first examples was the c-fos transgenic mouse (Ruther et al. 1987). No mRNA expression was detectable when c-fos cDNA that contained its endogenous 3′ UTR and polyadenylation signal was expressed from a strong promoter. However, after replacement of the 3′ UTR with a retroviral long terminal repeat high mRNA expression was observed throughout tissues (Ruther et al. 1987). Thus, the c-fos 3′ UTR is necessary for repression of c-fos protein production in vivo.
In many additional mouse models, this principle was adopted as genes were expressed from their endogenous promoters, but their 3′ UTRs and polyadenylation signals were replaced with strong polyadenylation signals. Although the influence of 3′ UTRs on phenotypes can be assessed in these mice, researchers need to be aware that the resulting phenotype results from deletion of 3′ UTR cis-elements and overexpression of the protein through more efficient mRNA processing. The gene encoding the Camk2a protein kinase generates a short and long 3′ UTR isoform (Lianoglou et al. 2013). Replacement of the two isoforms by a single strong polyadenylation signal prevented Camk2a mRNA localization to the dendrites of hippocampal neurons. This impairs long-term memory formation as dendritic mRNA localization and local protein production are necessary for memory consolidation (Miller et al. 2002). The neurotrophic factor Bdnf is also encoded by an mRNA with a short or long 3′ UTR isoform. Bdnf is expressed in hippocampal neurons and exclusive disruption of the long 3′ UTR isoform results in impaired memory formation (An et al. 2008). Bdnf is also expressed in hypothalamic neurons and disruption of the long 3′ UTR isoform in these cells results in severe obesity and phenocopies a complete Bdnf knockout in neurons (Liao et al. 2012).
The 3′ UTR functions of additional growth factors, neurotrophic factors, and enzymes were studied in mice and included TGFβ1, Gdnf, and aldosterone synthase (Kakoki et al. 2004; Andressoo et al. 2006; Kakoki et al. 2013). The phenotypes observed range from organ abnormalities, increased blood pressure, to embryonic lethality. These experiments revealed that usage of different 3′ UTRs can change specific protein levels by 100-fold, which enables the generation of mice with graded mRNA expression levels ranging from 10% to 900% of normal. This is particularly useful if the complete gene knockout is embryonic-lethal (Kakoki et al. 2013).
13. NEW METHODS TO STUDY 3′ UTR FUNCTIONS
13.1. CRISPR Technology Facilitates New Approaches for Studying 3′ UTR Isoform–Specific Functions in Cell Lines and Organisms
From the 3′ UTR studies in mice, we learned that preserving regulation by the endogenous promoter and polyadenylation signal should be an experimental priority. Furthermore, whereas 3′ UTR isoform–specific functions have so far been studied mostly using tagged, 3′ UTR isoform–specific overexpression constructs (Sandberg et al. 2008; Mayr and Bartel 2009; Rhinn et al. 2012; Berkovits and Mayr 2015), recent advances in CRISPR (clustered regularly interspaced short palindromic repeats) technology allow more sophisticated manipulations of 3′ UTR isoforms in cell lines and animal models at endogenous loci (Jinek et al. 2012; Diao et al. 2017).
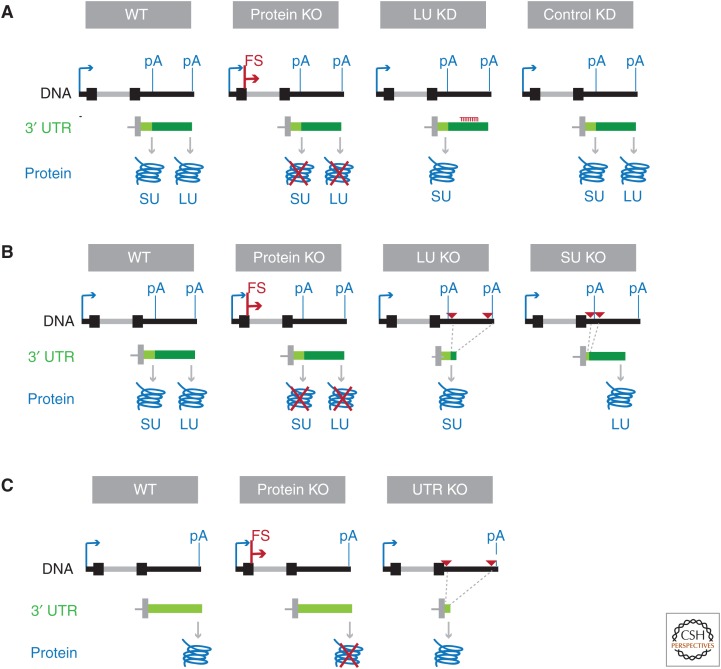
In the following paragraphs and in Figure 3, experimental approaches to study 3′ UTR functions using CRISPR technology are shown. If a gene generates alternative 3′ UTRs and the function of only the long 3′ UTR isoform is of interest, the strategy shown in Figure 3A is useful. In the experimental set-up, the following cell lines or mice are compared: (1) wild-type, (2) CRISPR-mediated disruption of the protein by adding a frame-shift in the coding region, (3) stable expression of a short hairpin RNA (shRNA) that targets the long 3′ UTR isoform, and (4) stable expression of a control shRNA. This approach was used to study the functions of CD47-LU and BIRC3-LU and is most suitable if the long 3′ UTR isoform is expressed at low levels, as its knockdown will not change overall protein levels much (Berkovits and Mayr 2015; Lee and Mayr 2017).
Figure 3.
Experimental approaches using CRISPR (clustered regularly interspaced short palindromic repeats) technology at the endogenous locus to study 3′ untranslated region (3′ UTR) functions in vivo. (A) Approach that allows phenotypes obtained for exclusive loss of the LU-generated protein compared with the total protein knockout (KO). Loss of the LU isoform is accomplished through short hairpin RNA (shRNA)-mediated knockdown (KD). At the DNA level, the arrow depicts the transcription start site. pA, Polyadenylation site; FS, frame-shift mutation. In the 3′ UTR panel, the last exon of the messenger RNA (mRNA) isoform is shown with the short 3′ UTR depicted in light green and the long 3′ UTR shown in dark green. The generated proteins are shown in blue. SU, Protein generated from the short 3′ UTR isoform; LU, protein generated from the long 3′ UTR isoform. (B) Approach that allows phenotypes obtained from the total protein KO to be compared to with exclusive expression of the protein generated from either the short (SU) or long 3′ UTR (LU) isoform. Shown as in A. The red triangles represent paired guide RNAs to delete 3′ UTR fragments. (C) Approach that allows the comparison of phenotypes obtained from the total protein KO to those generated by exclusive loss of regulatory elements in the 3′ UTR of genes that generate mRNAs with constitutive 3′ UTRs. Shown as in B. The regulation of mRNA abundance through the endogenous polyadenylation signal is preserved.
However, if the long 3′ UTR isoform is expressed at substantial levels and overall protein expression levels need to be preserved, a modified strategy, shown in Figure 3B, can be used. The phenotypes produced from the (1) wild-type and (2) protein knockout are compared with cell lines or animals that express only the (3) short or (4) long 3′ UTR isoforms. To obtain cells that express only the short 3′ UTR isoform, a pair of guide RNAs is used to delete the sequence between the proximal and distal polyadenylation signals. This moves the distal polyadenylation signal close to the proximal polyadenylation signal and should preserve overall protein expression as mRNA processing remains intact. To obtain cells that express only the long 3′ UTR isoform, the proximal polyadenylation site can either be deleted (using a pair of guide RNAs) or disrupted by point mutations (Li et al. 2017). As 3′ UTR deletions can also affect DNA-regulatory elements located at the 3′ end of genes (Sanjana et al. 2016), it is useful to include in the experiment tagged cDNA constructs that contain the coding region and either the short or long 3′ UTR isoform. Expression of these constructs will enable the rescue of the observed phenotypes and ensure that the phenotypes indeed are caused by the 3′ UTRs and not by deletion of DNA-regulatory elements (Lee and Mayr 2017).
To study the regulatory role of the 3′ UTR of a gene with a single 3′ UTR isoform, the wild-type and protein knockout can be compared with a cell line or animal with CRISPR-mediated deletion of the 3′ UTR (Fig. 3C). In this case, a pair of guide RNAs is used to delete the 3′ UTR sequence between the stop codon and the polyadenylation signal. However, ∼100 nucleotides of sequence located upstream of the polyadenylation signal should be preserved to enable proper mRNA processing from the endogenous polyadenylation signal (Zhao et al. 2017).
Instead of DNA manipulation, changes in mRNA processing through transfection of antisense morpholinos can also switch 3′ UTR isoform expression. This strategy has been used so far to alter inclusion or exclusion of alternative exons, including the splicing of terminal exons (Vorlova et al. 2011). However, similarly, alternative polyadenylation sites can be masked to shift alternative 3′ UTR isoform ratios (Marsollier et al. 2016).
13.2. 3′ UTR Isoform–Specific Translation
Several novel approaches were recently developed to study translation, but none of them allows assessment of translation in a 3′ UTR isoform–specific manner (reviewed in Chekulaeva and Landthaler 2016; King and Gerber 2016). Although it is now possible to monitor the translation of single endogenous mRNAs through simultaneous imaging of the mRNA and newly made protein, to do so requires disruption of the 3′ UTR by the addition of binding sites for either MS2 or PP7 coat protein (Chekulaeva and Landthaler 2016).
The only currently available method that can inspect translation in a 3′ UTR isoform–specific manner uses ribo-tag or TRAP (translating ribosome affinity purification) (Doyle et al. 2008; Heiman et al. 2014; Kocabas et al. 2015). Ribo-tag/TRAP adds a tag to a ribosomal protein of the large ribosomal subunit in a cell type–specific manner by using either bac transgenics or the Cre-lox system in mice (Doyle et al. 2008; Heiman et al. 2014; Hupe et al. 2014), but can be applied to worms, flies, plants, and yeast (King and Gerber 2016). In addition to quantifying the transcriptome in a cell type–specific manner, the pull-down of ribosome-associated mRNAs also provides an approximation of the translatome without the necessity of purifying the cells. Ribo-tag/TRAP enables the identification of ribosome-associated transcripts in a 3′ UTR-specific manner if, in addition to RNA-seq, 3′-end sequencing is performed.
13.3. Temporal Regulation of Translation of Alternative 3′ UTR Isoforms
Application of the ribo-tag technique during meiosis of mouse oocytes revealed that translation of alternative 3′ UTR isoforms is regulated temporally (Yang et al. 2017). This was shown in detail for Cyclin B1, which expresses three 3′ UTR isoforms. Combining RNA-seq of ribosome-associated mRNAs with luciferase reporter assays revealed that the short 3′ UTR isoform is constitutively translated in all phases of meiosis. In contrast, the longer 3′ UTR isoforms—which contain binding sites for CPEB1—are translationally repressed in a CPEB1-dependent manner in dormant oocytes. However, during progression through meiosis and cell-cycle entry, these longer isoforms become polyadenylated, thus increasing loading into ribosomes, and leading to increased Cyclin B1 protein expression during oocyte maturation (Yang et al. 2017).
Another beautiful example of temporal regulation of translation of alternative 3′ UTR isoforms in nongerm cells was revealed by studying the differentiation of R cells into presynaptic terminals in Drosophila melanogaster (Zhang et al. 2016). This is a particularly good model system to study gene expression changes during differentiation as R cells differentiate in a synchronous fashion within 72 hours from growth cones to presynaptic terminals. Performing transcriptome and TRAP analysis during this time course revealed only modest changes in mRNA abundance, but a large number of alternative 3′ UTR isoforms changed their ribosome association in a temporal manner (Zhang et al. 2016). Moreover, mRNAs encoding presynaptic proteins changed their 3′ UTR isoform expression twice as often as randomly selected genes. Thus, changes in alternative 3′ UTR isoform expression as well as dynamic ribosome association of alternative transcripts are major contributors to the regulation of synaptic differentiation (Zhang et al. 2016).
A similar temporal regulation of translation of alternative 3′ UTR isoforms was observed in differentiated neurons during synaptic activation (Lau et al. 2010; Rhinn et al. 2012). In resting neurons, the short 3′ UTR isoforms of Bdnf and α-synuclein (Snca) are translated to maintain basal levels of protein expression, whereas the long 3′ UTR isoforms are translationally repressed. In contrast, upon activation, either through neuronal activity or exposure to dopamine, only the long 3′ UTR isoforms become associated with polyribosomes and are actively translated. Thus, the alternative 3′ UTRs of Bdnf or Snca are translated differentially in resting and activated neurons (Lau et al. 2010; Rhinn et al. 2012).
13.4. Identification of RNA-Binding Proteins That Cooperate with Each Other
In the end, the functions of 3′ UTRs are determined by the bound RBPs and their associated effector proteins. Whereas cross-linking immunoprecipitation (CLIP) identifies all mRNAs bound by a single RBP, complementary approaches detect all RBPs that bind single 3′ UTRs (Chu et al. 2015; Matia-Gonzalez et al. 2017).
However, the identification of functionally relevant RBP-binding sites is complicated as the motifs are often degenerate. Therefore, in vivo, strong and specific binding of an RBP is often achieved through oligomerization of the RBP, which enables binding to several sites simultaneously (Besse et al. 2009). Stronger binding and increased specificity can also be accomplished through cooperativity with another RBP (Hennig and Sattler 2015; Mayr 2017). However, the identification of cooperating pairs of RBPs has been accomplished for only a few select cases (Leeper et al. 2010; Campbell et al. 2012; Hennig et al. 2014; Hennig and Sattler 2015; Lee and Mayr 2017; Ma and Mayr 2018).
To approach this problem systematically, an experimental procedure based on CLIP was recently developed (Gregersen et al. 2014). First, the binding sites for the RBP MOV10 were identified by PAR-CLIP. Next, cross-linked cells were mildly treated with RNase to produce RNA fragments of ∼400 nt. To identify proteins on the MOV10-bound RNA fragments, the MOV10 IP was repeated and the enriched proteins were identified by mass spectrometry (Gregersen et al. 2014). This method found UPF1 to be the RBP mostly highly enriched on the MOV10-bound fragments and suggested synergistic action by the two RBPs.
13.5. Identification of Effector Proteins of RNA-Binding Proteins
It has been shown in many instances that RBPs can interact with diverse effector proteins. This suggests that RBPs have considerable multifunctionality that goes beyond the regulation of RNA metabolism. In some cases, the interaction partners of RBPs bind simultaneously, as shown for CPEB (Kim and Richter 2006). In other cases, the RBP was found to be part of different protein subcomplexes, and each subcomplex accomplished a different function. Different subcomplexes were identified using co-IP of 3′ UTR-mediated protein complexes or through affinity purification of native complexes (Hildebrandt et al. 2017; Lee and Mayr 2017).
One of the best-studied examples is HuR, which is known to regulate mRNA stability, alternative splicing, and alternative polyadenylation (Fan and Steitz 1998; Oktaba et al. 2015). In addition to regulating mRNA-based processes, HuR is necessary for 3′ UTR-mediated PPIs, and thus is involved in regulating surface expression of several membrane proteins (Berkovits and Mayr 2015; Lee and Mayr 2017; Ma and Mayr 2018). Last, UPF1, which is a well-known regulator of nonsense mediated decay, was discovered to also regulate protein decay of MYOD1, a master regulator of muscle differentiation (Feng et al. 2017).
These examples show that investigating the diverse functions of 3′ UTRs requires determination of the effector proteins of RBPs. One study identified RNA-dependent as well as RNA-independent protein-interaction partners of 12 RBPs, including hnRNPs, RBFOX2, and UPF1 (Brannan et al. 2016). Through intersection with known RBPs, the majority of interaction partners (72%) were found to be proteins other than RBPs. However, as this was not the focus of the study, it is currently unclear how many of the identified factors were true interaction partners.
RBPs have rich interactomes (Baltz et al. 2012). This may be owing to the enrichment of disordered regions, which are known to promote PPIs. To lower the rate of false-positive protein-interaction partners of RBPs, the following strategy was used to identify the protein-interaction partners of E3 ubiquitin ligases that contain domains for RNA binding (Hildebrandt et al. 2017). Stable isotope labeling with amino acids in cell culture (SILAC)-based mass spectrometry compared the interactomes of GFP and GFP-tagged RBPs after co-IP. Importantly, the two experimental conditions were already pooled before cell lysis and co-IP, rather than after cell lysis, which is usually the case. As a result, transient interactors reach an equilibrium consisting of light and heavy labeled forms, which should result in SILAC ratios close to background ratios (Hildebrandt et al. 2017). Moreover, performing replicate experiments allows identification of robust interaction partners (Brannan et al. 2016; Hildebrandt et al. 2017). For a specific RNA-binding ubiquitin ligase, between 9 and 42 stable protein-interaction partners were identified. The majority (69%) bound in an RNA-independent manner and belong to diverse functional categories (Hildebrandt et al. 2017), suggesting additional regulatory roles of RBPs beyond mRNA-based regulation.
14. FUTURE DIRECTIONS
As 3′ UTRs can regulate protein features, readouts for testing 3′ UTR functions need to go beyond the regulation of protein abundance.
As 3′ UTRs can accomplish a wide range of functions through the regulation of protein complex formation, the protein–interaction partners of RBPs need to be identified. Approximately half of identified PPIs do not appear to play a role in RNA biology (Hildebrandt et al. 2017). Therefore, the opportunity to study 3′ UTR functions goes beyond the regulation of protein abundance.
One of the greatest bottlenecks in the evaluation of 3′ UTR functions is the difficulty of reliably identifying responsible cis-elements, especially if the regulatory elements are distributed across the entire 3′ UTR. In cases in which 3′ UTR structure brings the scattered cis-elements together, a CRISPR deletion screen with high-resolution tiling across the entire 3′ UTR may help to identify such elements, as disruption of structure may suffice to detect a phenotype. Such a screen was initially developed to assess functional protein domains, but was recently also applied to discover enhancers or functional miRNA-binding sites (Shi et al. 2015; Sanjana et al. 2016; Wu et al. 2017).
As the majority of RBPs bind with low affinity to mostly degenerate motifs located in 3′ UTRs, methods need to be developed that allow identification of binding sites for pairs of RBPs. Identification of cobound sites will be important for network analyses and may help to disentangle opposing functions known to be accomplished by RBPs.
ACKNOWLEDGMENTS
The author apologizes to all colleagues whose work could not be cited because of space constraints. She is grateful to all the members of the Mayr laboratory for helpful discussions and for critical reading of the manuscript, with special thanks to Sarah Tisdale. Funding for work on 3′ UTRs was obtained from the Innovator Award of the Damon Runyon-Rachleff Cancer Foundation and the Island Outreach Foundation, the Pershing Square Sohn Cancer Research Alliance, the MSC core grant (P30 CA008748), the National Cancer Institute (NCI) Grant U01-CA164190, and the National Institutes of Health (NIH) Director’s Pioneer Award (DP1-GM123454).
Footnotes
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
References
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. 2008. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134: 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andressoo JO, Mitchell JR, de Wit J, Hoogstraten D, Volker M, Toussaint W, Speksnijder E, Beems RB, van Steeg H, Jans J, et al. 2006. An Xpd mouse model for the combined xeroderma pigmentosum/Cockayne syndrome exhibiting both cancer predisposition and segmental progeria. Cancer Cell 10: 121–132. [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. 2005. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res 33: 7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R, Charizanis K, Manchanda M, Mohan A, Li M, Finn DJ, Goodwin M, Zhang C, Sobczak K, Thornton CA, et al. 2014. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol Cell 56: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, Mayr C. 2015. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse F, Lopez de Quinto S, Marchand V, Trucco A, Ephrussi A. 2009. Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev 23: 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan KW, Jin W, Huelga SC, Banks CA, Gilmore JM, Florens L, Washburn MP, Van Nostrand EL, Pratt GA, Schwinn MK, et al. 2016. SONAR discovers RNA-binding proteins from analysis of large-scale protein–protein interactomes. Mol Cell 64: 282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. 2001. HuR and mRNA stability. Cell Mol Life Sci 58: 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J, Di Stefano B, Wang X, Borkent M, Forouzmand E, Clowers KJ, Ji F, Schwarz BA, Kalocsay M, Elledge SJ, et al. 2018. Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell 172: 106–120.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell ZT, Bhimsaria D, Valley CT, Rodriguez-Martinez JA, Menichelli E, Williamson JR, Ansari AZ, Wickens M. 2012. Cooperativity in RNA–protein interactions: Global analysis of RNA binding specificity. Cell Rep 1: 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. 1986. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci 83: 1670–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR 3rd, Ule J, Manley JL, Shi Y. 2014. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 28: 2370–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JW, Zhang W, Yeh HS, de Jong EP, Jun S, Kim KH, Bae SS, Beckman K, Hwang TH, Kim KS, et al. 2015. mRNA 3′-UTR shortening is a molecular signature of mTORC1 activation. Nat Commun 6: 7218. [DOI] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. 2010. ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev 24: 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Hunt KC, Frydman J. 2016. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 536: 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Landthaler M. 2016. Eyes on translation. Mol Cell 63: 918–925. [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A. 2006. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124: 521–533. [DOI] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464. [DOI] [PubMed] [Google Scholar]
- Chen CY, Chen ST, Juan HF, Huang HC. 2012. Lengthening of 3′UTR increases with morphological complexity in animal evolution. Bioinformatics 28: 3178–3181. [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH. 1958. On protein synthesis. Symp Soc Exp Biol 12: 138–163. [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Fang R, Li B, Meng Z, Yu J, Qiu Y, Lin KC, Huang H, Liu T, Marina RJ, et al. 2017. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods 14: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D, Freese P, Alexis MS, Su A, Hochman M, Palden T, Bazile C, Lambert NJ, Van Nostrand EL, Pratt GA, et al. 2018. Sequence, structure, and context preferences of human RNA binding proteins. Mol Cell 70: 854–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. 2008. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135: 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CD, Mata J. 2011. Widespread cotranslational formation of protein complexes. PLoS Genet 7: e1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C. 1997. Alternative poly(A) site selection in complex transcription units: Means to an end? Nucleic Acids Res 25: 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. 1991. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37–50. [DOI] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 17: 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Jagannathan S, Bradley RK. 2017. The RNA surveillance factor UPF1 represses myogenesis via its E3 ubiquitin ligase activity. Mol Cell 67: 239–251.e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimer JW, Krishnakumar R, Cook MS, Blelloch R. 2018. Expression of alternative Ago2 isoform associated with loss of microRNA-driven translational repression in mouse oocytes. Curr Biol 28: 296–302.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret D, Poirot O, Lopez F, Audic S, Claverie JM. 1998. Alternate polyadenylation in human mRNAs: A large-scale analysis by EST clustering. Genome Res 8: 524–530. [DOI] [PubMed] [Google Scholar]
- Grassi E, Mariella E, Lembo A, Molineris I, Provero P. 2016. Roar: Detecting alternative polyadenylation with standard mRNA sequencing libraries. BMC Bioinformatics 17: 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. 2014. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell 54: 573–585. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Muller P, Schmidt A, Gruber AJ, Gumienny R, Mittal N, Jayachandran R, Pieters J, Keller W, et al. 2014. Global 3′ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat Commun 5: 5465. [DOI] [PubMed] [Google Scholar]
- Gruber AJ, Schmidt R, Gruber AR, Martin G, Ghosh S, Belmadani M, Keller W, Zavolan M. 2016. A comprehensive analysis of 3′ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res 26: 1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta I, Clauder-Munster S, Klaus B, Jarvelin AI, Aiyar RS, Benes V, Wilkening S, Huber W, Pelechano V, Steinmetz LM. 2014. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA–protein interactions. Mol Syst Biol 10: 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach A, Zhang H, Wengi A, Jablonska Z, Gruber IM, Halbeisen RE, Dehe PM, Kemmeren P, Holstege F, Geli V, et al. 2009. Cotranslational assembly of the yeast SET1C histone methyltransferase complex. EMBO J 28: 2959–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. 2014. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat Protoc 9: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig J, Sattler M. 2015. Deciphering the protein–RNA recognition code: Combining large-scale quantitative methods with structural biology. Bioessays 37: 899–908. [DOI] [PubMed] [Google Scholar]
- Hennig J, Gebauer F, Sattler M. 2014. Breaking the protein–RNA recognition code. Cell Cycle 13: 3619–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt A, Alanis-Lobato G, Voigt A, Zarnack K, Andrade-Navarro MA, Beli P, Konig J. 2017. Interaction profiling of RNA-binding ubiquitin ligases reveals a link between posttranscriptional regulation and the ubiquitin system. Sci Rep 7: 16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. 2013. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods 10: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupe M, Li MX, Gertow Gillner K, Adams RH, Stenman JM. 2014. Evaluation of TRAP-sequencing technology with a versatile conditional mouse model. Nucleic Acids Res 42: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Saito Y, Park CY, Blachere NE, Tajima Y, Fak JJ, Zucker-Scharff I, Darnell RB. 2017. cTag-PAPERCLIP reveals alternative polyadenylation promotes cell-type specific protein diversity and shifts Araf isoforms with microglia activation. Neuron 95: 1334–1349.e1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, Derisi JL. 2007. cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA 13: 625–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Yuan S, Wang Y, Fu Y, Ge Y, Ge Y, Lan X, Feng Y, Qiu F, Li P, et al. 2017. The role of alternative polyadenylation in the antiviral innate immune response. Nat Commun 8: 14605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. 2001. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet 35: 365–406. [DOI] [PubMed] [Google Scholar]
- Kakoki M, Tsai YS, Kim HS, Hatada S, Ciavatta DJ, Takahashi N, Arnold LW, Maeda N, Smithies O. 2004. Altering the expression in mice of genes by modifying their 3′ regions. Dev Cell 6: 597–606. [DOI] [PubMed] [Google Scholar]
- Kakoki M, Pochynyuk OM, Hathaway CM, Tomita H, Hagaman JR, Kim HS, Zaika OL, Mamenko M, Kayashima Y, Matsuki K, et al. 2013. Primary aldosteronism and impaired natriuresis in mice underexpressing TGFβ1. Proc Natl Acad Sci 110: 5600–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Richter JD. 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 24: 173–183. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Webster PJ, Smith JL, Macdonald PM. 1993. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development 119: 169–178. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. 1995. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81: 403–412. [DOI] [PubMed] [Google Scholar]
- King HA, Gerber AP. 2016. Translatome profiling: Methods for genome-scale analysis of mRNA translation. Brief Funct Genomics 15: 22–31. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. 1994. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J Cell Biol 127: 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabas A, Duarte T, Kumar S, Hynes MA. 2015. Widespread differential expression of coding region and 3′ UTR sequences in neurons and other tissues. Neuron 88: 1149–1156. [DOI] [PubMed] [Google Scholar]
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: Implications for joint and gut-associated immunopathologies. Immunity 10: 387–398. [DOI] [PubMed] [Google Scholar]
- Kristjansdottir K, Fogarty EA, Grimson A. 2015. Systematic analysis of the Hmga2 3′ UTR identifies many independent regulatory sequences and a novel interaction between distal sites. RNA 21: 1346–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruys V, Marinx O, Shaw G, Deschamps J, Huez G. 1989. Translational blockade imposed by cytokine-derived UA-rich sequences. Science 245: 852–855. [DOI] [PubMed] [Google Scholar]
- Lackford B, Yao C, Charles GM, Weng L, Zheng X, Choi EA, Xie X, Wan J, Xing Y, Freudenberg JM, et al. 2014. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J 33: 878–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. 2014. RNA Bind-n-Seq: Quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell 54: 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- La Rocca G, Olejniczak SH, Gonzalez AJ, Briskin D, Vidigal JA, Spraggon L, DeMatteo RG, Radler MR, Lindsten T, Ventura A, et al. 2015. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc Natl Acad Sci 112: 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AG, Irier HA, Gu J, Tian D, Ku L, Liu G, Xia M, Fritsch B, Zheng JQ, Dingledine R, et al. 2010. Distinct 3′UTRs differentially regulate activity-dependent translation of brain-derived neurotrophic factor (BDNF). Proc Natl Acad Sci 107: 15945–15950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. 1986. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45: 407–415. [DOI] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. 2011. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 43: 340–352. [DOI] [PubMed] [Google Scholar]
- Lee SH, Mayr C. 2017. 3′UTR-dependent signalosome assembly determines substrate specificity of BIRC3 E3 ligase during B cell migration. In EMBO conference, RNA localisation and local translation Barga, Italy. [Google Scholar]
- Lee SH, Singh I, Tisdale S, Abdel-Wahab O, Leslie CS, Mayr C. Widespread intronic polyadenylation inactivates tumor suppressor genes in leukemia. Nature 10.1038/s41586-018-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeper TC, Qu X, Lu C, Moore C, Varani G. 2010. Novel protein–protein contacts facilitate mRNA 3′-processing signal recognition by Rna15 and Hrp1. J Mol Biol 401: 334–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Harvey AR, Hodgetts SI, Fox AH. 2017. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA 23: 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. 2013. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 27: 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao GY, An JJ, Gharami K, Waterhouse EG, Vanevski F, Jones KR, Xu B. 2012. Dendritically targeted Bdnf mRNA is essential for energy balance and response to leptin. Nat Med 18: 564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstein T, June CH, Ledbetter JA, Stella G, Thompson CB. 1989. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science 244: 339–343. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Mayr C. 2018. A subcellular compartment defined by TIS granules and the ER enables 3′UTR-mediated protein–protein interactions. Society for Developmental Biology, 77th Annual Meeting, Portland, OR (July 20–24, 2018). [Google Scholar]
- Macdonald PM, Struhl G. 1988. cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature 336: 595–598. [DOI] [PubMed] [Google Scholar]
- Marsollier AC, Ciszewski L, Mariot V, Popplewell L, Voit T, Dickson G, Dumonceaux J. 2016. Antisense targeting of 3′ end elements involved in DUX4 mRNA processing is an efficient therapeutic strategy for facioscapulohumeral dystrophy: A new gene-silencing approach. Hum Mol Genet 25: 1468–1478. [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. 2009. mRNA localization: Gene expression in the spatial dimension. Cell 136: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ. 2014. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510: 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matia-Gonzalez AM, Iadevaia V, Gerber AP. 2017. A versatile tandem RNA isolation procedure to capture in vivo formed mRNA–protein complexes. Methods 118-119: 93–100. [DOI] [PubMed] [Google Scholar]
- Mayr C. 2016. Evolution and biological roles of alternative 3′UTRs. Trends Cell Biol 26: 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. 2017. Regulation by 3′-untranslated regions. Annu Rev Genet 51: 171–194. [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. 2007. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315: 1576–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijlink F, Curran T, Miller AD, Verma IM. 1985. Removal of a 67-base-pair sequence in the noncoding region of protooncogene fos converts it to a transforming gene. Proc Natl Acad Sci 82: 4987–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton DA. 1987. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature 328: 80–82. [DOI] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. 2000. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature 404: 302–307. [DOI] [PubMed] [Google Scholar]
- Miller AD, Curran T, Verma IM. 1984. c-fos protein can induce cellular transformation: A novel mechanism of activation of a cellular oncogene. Cell 36: 51–60. [DOI] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. 2002. Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron 36: 507–519. [DOI] [PubMed] [Google Scholar]
- Milo R, Phillips R. 2016. Cell biology by the numbers. Taylor & Francis, New York. [Google Scholar]
- Miyata T, Yasunaga T, Nishida T. 1980. Nucleotide sequence divergence and functional constraint in mRNA evolution. Proc Natl Acad Sci 77: 7328–7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JW, Rissland OS, Koppstein D, Abreu-Goodger C, Jan CH, Agarwal V, Yildirim MA, Rodriguez A, Bartel DP. 2014. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell 53: 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48: 443–453. [DOI] [PubMed] [Google Scholar]
- Oktaba K, Zhang W, Lotz TS, Jun DJ, Lemke SB, Ng SP, Esposito E, Levine M, Hilgers V. 2015. ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol Cell 57: 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, et al. 2012. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev 26: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn H, Qiang L, Yamashita T, Rhee D, Zolin A, Vanti W, Abeliovich A. 2012. Alternative α-synuclein transcript usage as a convergent mechanism in Parkinson’s disease pathology. Nat Commun 3: 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. 2007. CPEB: A life in translation. Trends Biochem Sci 32: 279–285. [DOI] [PubMed] [Google Scholar]
- Ruther U, Garber C, Komitowski D, Muller R, Wagner EF. 1987. Deregulated c-fos expression interferes with normal bone development in transgenic mice. Nature 325: 412–416. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Wright J, Zheng K, Shalem O, Fontanillas P, Joung J, Cheng C, Regev A, Zhang F. 2016. High-resolution interrogation of functional elements in the noncoding genome. Science 353: 1545–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonemann L, Kuhn U, Martin G, Schafer P, Gruber AR, Keller W, Zavolan M, Wahle E. 2014. Reconstitution of CPSF active in polyadenylation: Recognition of the polyadenylation signal by WDR33. Genes Dev 28: 2381–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G, Kamen R. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46: 659–667. [DOI] [PubMed] [Google Scholar]
- Shenker S, Miura P, Sanfilippo P, Lai EC. 2015. IsoSCM: Improved and alternative 3′ UTR annotation using multiple change-point inference. RNA 21: 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. 2015. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33: 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I, Lee SH, Sperling AS, Samur MK, Tai YT, Fulciniti M, Munshi NC, Mayr C, Leslie CL. 2018. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nat Commun 9: 1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlova S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L. 2011. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell 43: 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Nambiar R, Zheng D, Tian B. 2017. PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res 46: D315–D319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Ferry QRV, Baeumler TA, Michaels YS, Vitsios DM, Habib O, Arnold R, Jiang X, Maio S, Steinkraus BR, et al. 2017. In situ functional dissection of RNA cis-regulatory elements by multiplex CRISPR-Cas9 genome engineering. Nat Commun 8: 2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D, Nudel U, Mayer Y, Neuman S. 1985. Highly conserved sequences in the 3′ untranslated region of mRNAs coding for homologous proteins in distantly related species. Nucleic Acids Res 13: 3723–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yang CR, Han SJ, Daldello EM, Cho A, Martins JPS, Xia G, Conti M. 2017. Maternal mRNAs with distinct 3′ UTRs define the temporal pattern of Ccnb1 synthesis during mouse oocyte meiotic maturation. Genes Dev 31: 1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KX, Tan L, Pellegrini M, Zipursky SL, McEwen JM. 2016. Rapid changes in the translatome during the conversion of growth cones to synaptic terminals. Cell Rep 14: 1258–1271. [DOI] [PubMed] [Google Scholar]
- Zhao W, Siegel D, Biton A, Tonqueze OL, Zaitlen N, Ahituv N, Erle DJ. 2017. CRISPR-Cas9-mediated functional dissection of 3′-UTRs. Nucleic Acids Res 45: 10800–10810. [DOI] [PMC free article] [PubMed] [Google Scholar]