In this study, Lee et al. use both in vitro and in vivo approaches to elucidate the regulation of PRC2. They demonstrate a novel PRC2 self-regulatory mechanism through its EZH1/2-mediated automethylation activity.
Keywords: automethylation, H3K27M, PRC2, pediatric glioma, Polycomb
Abstract
The histone methyltransferase activity of PRC2 is central to the formation of H3K27me3-decorated facultative heterochromatin and gene silencing. In addition, PRC2 has been shown to automethylate its core subunits, EZH1/EZH2 and SUZ12. Here, we identify the lysine residues at which EZH1/EZH2 are automethylated with EZH2-K510 and EZH2-K514 being the major such sites in vivo. Automethylated EZH2/PRC2 exhibits a higher level of histone methyltransferase activity and is required for attaining proper cellular levels of H3K27me3. While occurring independently of PRC2 recruitment to chromatin, automethylation promotes PRC2 accessibility to the histone H3 tail. Intriguingly, EZH2 automethylation is significantly reduced in diffuse intrinsic pontine glioma (DIPG) cells that carry a lysine-to-methionine substitution in histone H3 (H3K27M), but not in cells that carry either EZH2 or EED mutants that abrogate PRC2 allosteric activation, indicating that H3K27M impairs the intrinsic activity of PRC2. Our study demonstrates a PRC2 self-regulatory mechanism through its EZH1/2-mediated automethylation activity.
In mammals, the catalysis of methylation on Lys27 of histone H3 (H3K27) is central to the formation of Polycomb-repressive chromatin domains associated with gene silencing. Polycomb-repressive complex 2 (PRC2) is the only known mammalian methyltransferase that catalyzes mono-, di-, and trimethylation of H3K27 (H3K27me1/me2/me3). The PRC2 core complex comprises EED, SUZ12, and two mutually exclusive and interchangeable catalytic subunits, EZH1 and EZH2. In isolation, the catalytic SET domain of EZH1 or EZH2 is in an autoinhibitory state (Wu et al. 2013), but EZH1/2 interaction with EED and SUZ12 relieves this autoinhibition, thereby fostering its histone methyltransferase (HMT) activity (Cao et al. 2002; Kuzmichev et al. 2002; Jiao and Liu 2015 ; Justin et al. 2016). Upon assembly of this minimally active PRC2 core complex, PRC2 engages in a positive feedback mechanism whereby its EED subunit binds to the product of PRC2 catalysis, H3K27me3, inducing an allosteric activation of the complex that significantly promotes EZH2 catalytic activity (Margueron et al. 2009). This “write and read” mechanism constitutes the molecular basis by which PRC2 forms extensive chromatin domains comprising H3K27me2/me3 that foster gene repression upon each cycle of cell division (Hansen et al. 2008; Oksuz et al. 2018; Reinberg and Vales 2018). Disruption of this feedback loop by point mutations in the key regions of EZH2 and EED results in a global loss of H3K27me2/me3 and notably, these mutations were found in human pathological conditions including developmental diseases and cancer (Imagawa et al. 2017; Lee et al. 2018b).
In addition to the allosteric activation deficient mutations, many other loss-of-function mutations in the PRC2 core subunits are frequently found to abrogate cellular levels of H3K27me3 in specific cancer subtypes, such as T-cell acute lymphoblastic leukemia (T-ALL) and malignant peripheral nerve sheath tumors (MPNST) (Ntziachristos et al. 2012; Lee et al. 2014). In contrast, gain-of-function mutations in the catalytic pocket of the SET domain of EZH2, which specifically promote the conversion of H3K27me2 to H3K27me3 (Morin et al. 2010; McCabe et al. 2012), were shown to be oncogenic drivers of diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (Morin et al. 2010; McCabe et al. 2012). More recently, a lysine-to-methionine mutation (H3K27M) of the PRC2 substrate, H3K27, was found to function as a dominant-negative mutant against PRC2 activity, leading to a global loss in H3K27me2/me3 in diffuse intrinsic pontine glioma (DIPG), a subtype of high-grade pediatric glioma (Chan et al. 2013; Lewis et al. 2013). Strikingly, biochemical studies demonstrate that after contact with and release from H3K27M, PRC2 manifests a decreased catalytic activity, indicating that H3K27M exerts a persistent, long-lasting inhibitory effect on PRC2 (Stafford et al. 2018). The mechanistic basis of this effect is yet to be determined and may prove pivotal to understanding the pathogenesis of DIPG.
In addition to genetic mutations that dysregulate PRC2 activity, several different proteins have been reported to associate with PRC2 and regulate its activity. Recent advances provide insights into the roles of some of the PRC2 accessory factors, such as AEBP2 (Lee et al. 2018a), JARID2 (Son et al. 2013; Sanulli et al. 2015), and Polycomb-like (PCL) proteins (Li et al. 2017; Oksuz et al. 2018), which stimulate PRC2 activity and/or facilitate its recruitment to chromatin. However, the function(s) of other accessory factors including EPOP/C17ORF96 (Beringer et al. 2016; Liefke et al. 2016), C10ORF12 (Alekseyenko et al. 2014), and PALI (LCOR and C10ORF12 fusion protein) (Conway et al. 2018) remains poorly understood. As these candidates have been isolated from transformed or ES cells, it is important to examine further their potential cell-type specific roles in postdevelopmental lineages (Yu et al. 2019).
Among these regulatory factors, JARID2 stands alone as being itself subject to PRC2-mediated methylation, specifically at its Lys116 residue (JARID2-K116me). Similar to H3K27me3, JARID2-K116me3 binds to the EED aromatic cage and elicits an allosteric activation of PRC2 (Sanulli et al. 2015). Intriguingly, PRC2 also exhibits an automethyltransferase activity directed towards its own core subunits, EZH2 and SUZ12 (Müller et al. 2002; Wang et al. 2017, 2019). Notably, this automethylation occurs intramolecularly (Wang et al. 2019). As many other enzymes harbor intrinsic regulatory mechanisms through self-modification (e.g,. autophosphorylation or automethylation) (Wang and Wu 2002; Chin et al. 2007; Dillon et al. 2013), we examined the functional impact of PRC2 automethylation in this study.
Results
Human PRC2 automethylates EZH1/2 and SUZ12 in vitro
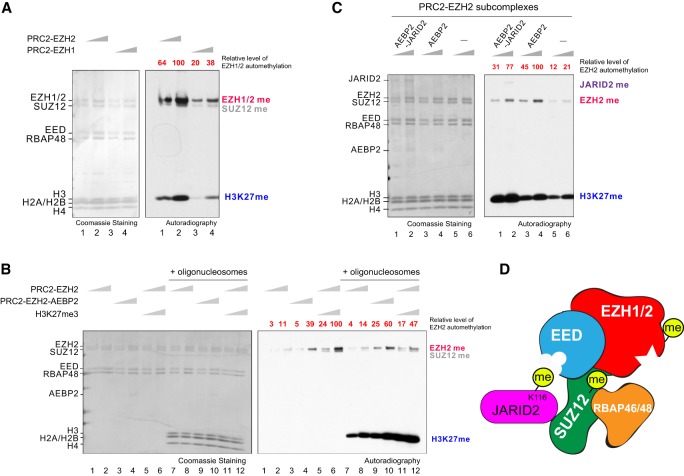
To begin investigations into the role of PRC2 automethylation, we first performed an in vitro methyltransferase (MT) assay using recombinant human PRC2 complex composed of EZH2, SUZ12, EED, and RBAP48 (Supplemental Fig. 1A) and monitored the efficiency of this reaction after the addition of the methyl donor [3H]-SAM. In agreement with previous findings (Müller et al. 2002; Wang et al. 2017), PRC2 methylates EZH2 and SUZ12 in the presense or absence of nucleosomal substrates (Fig. 1A; Supplemental Fig. 1B). Based on recent findings showing that this methylation occurs intramolecularly (Wang et al. 2019), we also indicate this catalysis as automethylation hereafter. While PRC2 can comprise either an EZH1 or EZH2 homolog, previous findings show that PRC2-EZH1 exhibits a considerably weaker HMT activity relative to that of PRC2–EZH2 (Margueron et al. 2008; Son et al. 2013; Lee et al. 2018a). Unsurprisingly, automethylation of EZH1 is considerably weaker than that of its EZH2 counterpart (Fig. 1A; Supplemental Fig. 1A,B). Nonetheless, the addition of the PRC2 accessory factor, AEBP2, or an allosteric activator, H3K27me3-peptide, increased EZH2 automethylation (Fig. 1B, lanes 3–6) in accordance with their respectively distinctive stimulatory effects on PRC2 HMT activity (Lee et al. 2018a). SUZ12 methylation was also enhanced by the H3K27me3-peptide, but surprisingly, was dampened by AEBP2 (Fig. 1B, lanes 3–6). Based on recent structural studies (Kasinath et al. 2018), the latter result could be due to the steric occlusion of potential SUZ12 methylation site(s) from the catalytic pocket when AEBP2 is complexed with PRC2 (Chen et al. 2018). In contrast to the stimulatory effect of AEBP2 on EZH2 automethylation, the presence of JARID2 led to a reduction (Fig. 1C, cf. lanes 1,2 and 3,4), likely due to a competition between these two nonhistone substrates, JARID2-K116 (Sanulli et al. 2015) and EZH2 automethylation sites, within the same complex.
Figure 1.
PRC2 subunits can be automethylated. (A–C) Methyltransferase (MT) assays showing EZH1/2, SUZ12, and histone H3 are methylated; see the Materials and Methods for MT assay conditions. (Left) Coomassie blue staining of SDS-PAGE gels containing nucleosomes or PRC2 components was used to visualize the relative concentration of each component present in each reaction. (Right) The corresponding autoradiography image was shown for quantification. The relative levels of EZH2 automethylation were quantified and indicated at the top of the gel image (highlighted in red). (A) MT assays containing 30 or 60 nM PRC2-EZH2 or PRC2-EZH1, 2 µM 3H-SAM, and 300 nM oligonucleosomes. (B) MT assays with 30 or 60 nM PRC2-EZH2, PRC2-EZH2-AEBP2, or PRC2-EZH2 with 3 µM H3K27me3 peptide and incubated with 2 µM 3H-SAM in the absence (lanes 1–6) or presence (lanes 7–12) of 300 nM oligonucleosomes. (C) MT assays with 0 or 60 nM PRC2–EZH2–AEBP2 or PRC2–EZH2–AEBP2–JARID2 and incubated with 2 µM 3H-SAM in the presence of 300 nM oligonucleosomes.
Having established the conditions for PRC2 automethylation, we next examined the levels of EZH2 automethylation in the presence of oligonucleosomes (Fig. 1B, lanes 7–12). The addition of oligonucleosomes had little impact on EZH2 automethylation but surprisingly, dampened automethylation of EZH2 and SUZ12 in the presence of H3K27me3-peptide, suggesting that allosterically activated PRC2 shifts its substrate preference toward nucleosomes as a consequence of substrate competition (Fig. 1B, cf. lanes 5,6 and 11,12). Collectively, these results corroborated that PRC2 automethylates its core subunits, EZH2 and SUZ12. We found that EZH1 is also subject to automethylation and that while, in principle, PRC2 automethylation correlates with its levels of HMT activity, distinct PRC2 subcomplexes exhibit different levels of automethylation (Fig. 1D).
Automethylation of EZH1/2 resides in a key regulatory region between its SANT2L and CXC domains
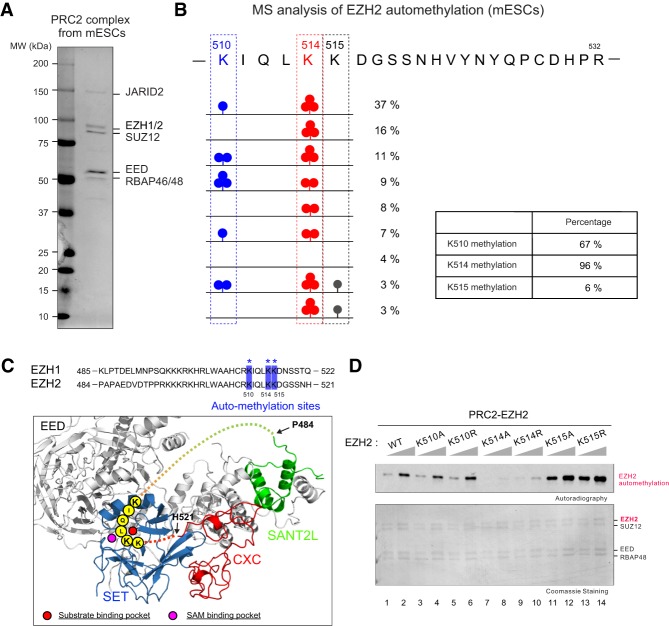
To better understand the functional role of PRC2-EZH1/2 automethylation, we purified PRC2 from mouse embryonic stem cells (mESCs) using Flag-based affinity purification (Fig. 2A; Supplemental Fig. 2A) from a previously constructed EED-deficient mESC line wherein N-terminally Flag-tagged EED was ectopically expressed (Lee et al. 2018b). Purified endogenous PRC2 from mESC was then subjected to Arg-C protease digestion and MS analysis to better preserve the lysine modifications. We identified three methylated lysine residues, all of which are conserved in EZH1 and EZH2, including EZH2-K510, EZH2-K514, and EZH2-K515, corresponding to EZH1-K511, EZH1-K515, EZH1-K516 (Fig. 2B; Supplemental Fig. 2B,C). These residues reside in an unstructured loop between the SANT2L and CXC domains of EZH1/2 (Fig. 2C). Notably, these lysine residues can be in close proximity to the substrate binding pocket of the SET domain (Fig. 2C), supporting the notion that EZH2 automethylation occurs in cis as shown in a recent study (Wang et al. 2019). Remarkably, 96% of EZH2-K514 and 67% of EZH2-K510 was either mono-, di-, or trimethylated (me1, me2, and me3, respectively) (Fig. 2B). However, only 6% of EZH2-K515 was methylated (Fig. 2B), indicating that K510 and K514 are the major sites of EZH2 automethylation. Furthermore, methylation of EZH2-K510 and EZH2-K515 was detected only in the presence of K514-methylation in cis (Fig. 2B; Supplemental Fig. 2B,D), suggesting that methylation of K514 is a prerequisite for that of K510 and K515.
Figure 2.
Identification of EZH2 methylation sites in mouse embryonic stem cells. (A,B) PRC2 complex was purified from Flag-tagged-EED E14 mouse embryonic stem cells (mESCs) to identify EZH2 automethylation site(s); see the Materials and Methods for purification details. (A) Purified PRC2 complex is shown in Coomassie blue-stained SDS-PAGE gel. (B) Mass spectrometry (MS) analysis of EZH2 methylation after Arg-C protease digestion (see the Materials and Methods). Illustration of EZH2 lysine residues within amino acids 510–532 and the percentages of their mono-, di-, and trimethylated states, as indicated. Boxed region shows overall percentages of methylation at the EZH2 lysine residues indicated. (C, top) Sequence alignment of human EZH1 and EZH2 between the SANT2L and CXC domains. Automethylation sites identified by MS are highlighted by blue squares and asterisks. (Bottom) Human PRC2 structure showing the position of the unstructured regulatory loop (dashed line) containing automethylation sites between EZH2-SANT2L and EZH2-CXC domains (modified from PDB: 5HYN). Note that the automethylation sites are in close proximity to the substrate binding pocket (red circle) and SAM binding pocket (pink circle). (D) MT assays containing 20 or 40 nM PRC2-EZH2, either WT or individually mutant in the automethylation sites. (Top image) The levels of methylation on EZH2 are shown by autoradiography. (Bottom image) Coomassie blue staining of SDS-PAGE gels containing PRC2 components was used to visualize the relative concentration of each component present in each reaction.
To corroborate that these residues are bonafide automethylation sites, we performed an MT assay on purified recombinant PRC2-EZH2 complexes that contain mutations of each EZH2 automethylation residue. We substituted each lysine (K) with either an alanine (A) or arginine (R), as the side chain of arginine preserves a positive charge similar to that of lysine but cannot be methylated by PRC2. While PRC2–EZH2K510A/R displayed little impact on overall automethylation signals, PRC2–EZH2K514A/R showed a dramatic reduction in automethylation (Fig. 2D). Surprisingly, PRC2–EZH2K515A/R exhibited increased automethylation (Fig. 2D), suggesting that K515 mutants enhance automethylation efficiency on K510 and K514. Thus, the primary sites of automethylation in EZH2 are K514 and K510, and K515 automethylation occurs concomitantly with K514 automethylation.
Automethylated EZH2 residues are critical for H3K27me3 catalysis, but not for PRC2 recruitment to chromain
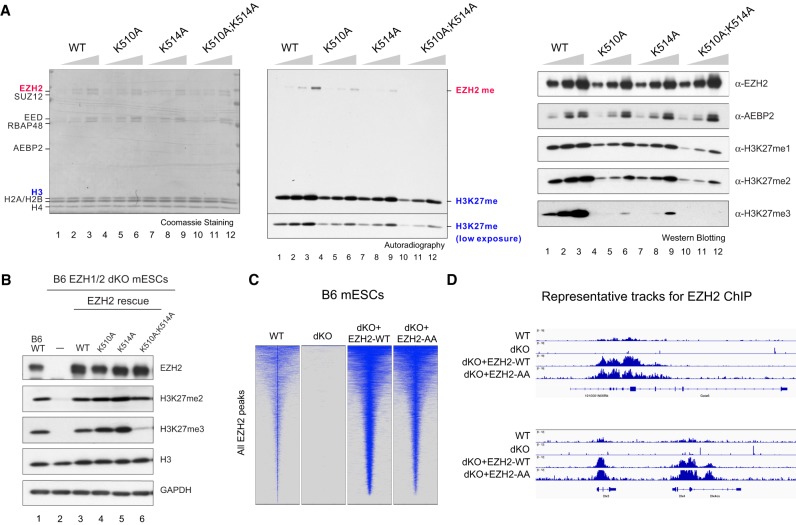
Next, we purified and examined the impact of EZH2 automethylation mutants on the HMT activity of PRC2 in complex with AEBP2 (Supplemental Fig. 3A), a common accessory factor found in many cell types (Kim et al. 2009). Intriguingly, while in the context of PRC2–AEBP2, both EZH2K510A and EZH2K514A had a partial effect on EZH2 automethylation, and the EZH2K510A;K514A double mutant exhibited an undetectable level of automethylation (Fig. 3A, left and middle), affirming that K510 and K514 are the predominant sites of automethylation in EZH2 (Fig. 2B), and suggesting that PRC2 association with accessory proteins regulates the substrate preference between these two sites. Importantly, the histone MT (HMT) activity of PRC2–AEBP2–EZH2K510A, PRC2–AEBP2–EZH2K514A, or PRC2–AEBP2–EZH2K510A;K514A was consistent and reflective of their intrinsic automethylation activity, with EZH2K510A and EZH2K514A manifesting a partial reduction and EZH2K510A;K514A displaying a synergistic and more profound defect in their respective HMT activity (Fig. 3A, middle). To further dissect the H3K27 methylation status under these assay conditions, we performed immunoblotting on aliquots of the HMT assays using antibodies specific to H3K27me1, H3K27me2, or H3K27me3. The PRC2–AEBP2 complex containing EZH2K510A, EZH2K514A, or EZH2K510A;K514A showed similarly modest reductions in H3K27me1 and H3K27me2 levels compared with wild-type PRC2–AEBP2 (Fig. 3A, right). However, a more prominent loss in H3K27me3 was observed in the HMT assay using PRC2–AEBP2–EZH2K510A;K514A relative to EZH2K510A or EZH2K514A (Fig. 3A, right).
Figure 3.
Residues automethylated in EZH2 are critical for PRC2 catalytic activity. (A) Methyltransferase assays using PRC2–EZH2–AEBP2 complexes containing either 15, 30, or 60 nM wild-type or mutant EZH2 as indicated and 300 nM oligonucleosomes as substrates. (Left) Coomassie blue staining of SDS–polyacrylamide gels containing nucleosomes or PRC2 components was used to visualize the relative concentration of each component present in each reaction. (Middle) Levels of EZH2 or histone H3 methylation are shown by autoradiography. (Right) Western blot analysis of the methyltransferase reaction components (EZH2 and AEBP2) and products (H3K27me1, H3K27me2, and H3K27me3). (B) Western blot analysis of EZH2, H3K27me2, H3K27me3, GAPDH, and total histone H3 levels in B6 mESCs, including WT, EZH1-KO/EZH2ΔSET (dKO), and EZH2 rescue conditions. (mESCs) Mouse embryonic stem cells. (C,D) EZH2 automethylation is not required for PRC2 deposition. (C) Heat maps representing EZH2 ChIP-seq peaks centered by maximum peak intensity at the genome-wide scale in ±10-kb windows within a 200-bp bin size. The genotypes of B6 mESCs for each experimental condition are indicated above. (dKO) EZH1-KO/EZH2ΔSET. (D) Representative track images for EZH2 (blue) ChIP-seq experiments performed in C for a select group of annotated genes. The UCSC annotations of exons and gene bodies are shown at the bottom. The scale of the peaks ranges from 0 to 18 reads per 10 million reads, with spike-in normalization using Drosophila chromatin and Drosophila H2A.X antibody in each ChIP reaction.
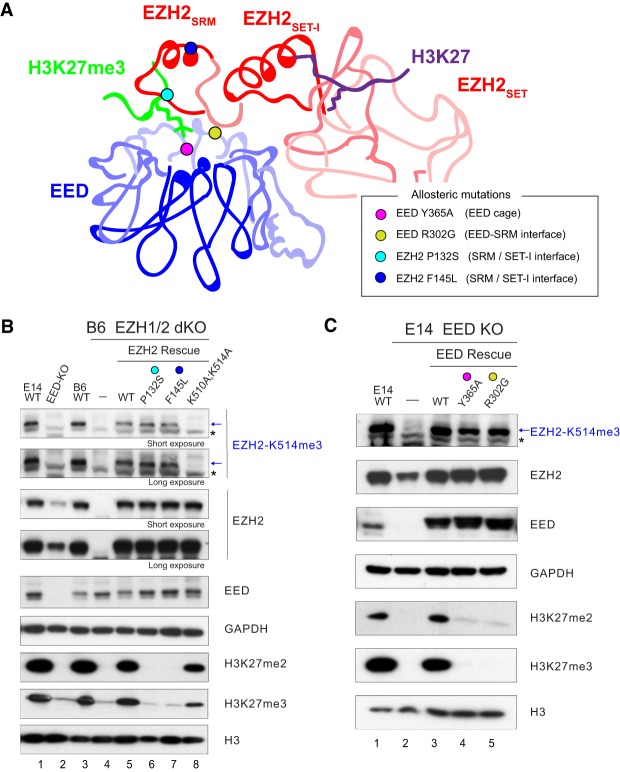
To ascertain whether these automethylation mutants exhibit a similar pattern of H3K27 methylation in vivo, we adopted a previously established system in which all H3K27 methylation is depleted in C57BL/6J (B6) mouse embryonic stem cells (mESCs) containing a knockout (KO) of both EZH1 and the SET domain of EZH2 (EZH1-KO/EZH2ΔSET, referred to here as EZH1/2 dKO) (Lee et al. 2018a). We rescued these EZH1/2 dKO mESCs with either EZH2WT or the EZH2 automethylation mutants by lentiviral transduction. As expected, EZH1/2 dKO mESCs showed a complete loss in H3K27me (Fig. 3B, lane 2), the levels of which were nearly fully restored upon rescue with EZH2WT (Fig. 3B, lane 3), consistent with our previous study (Lee et al. 2018a). Surprisingly, there was no reduction in H3K27me2/me3 upon rescue with either EZH2K510A or EZH2K514A (Fig. 3B, cf. lanes 3–5), in contrast to results from our in vitro HMT assays (Fig. 3A). Despite the restoration of H3K27me2 upon rescue with EZH2K510A;K514A, H3K27me3 remained very low (Fig. 3B, cf. lanes 3 and 6). Interestingly, a similar rescue experiment performed in 293T-Rex cells in which EZH2 was knocked down (KD), showed that H3K27me3 was partially restored by EZH2K510A and EZH2K514A, relative to EZH2WT (Supplemental Fig. 3B), whereas EZH2K510A;K514A was ineffectual. Thus, depending on the cellular context, the partial defect in HMT activity of EZH2K510A or EZH2514A might be compensated by other factors, such as the presence of accessory proteins or a high abundance of PRC2. Nonetheless, EZH2K510A;K514A exhibits a synergistic effect and is defective in H3K27me3 catalysis in vitro and in vivo. Of note, the reduction in H3K27me3 in cells comprising the EZH2 automethylation mutant was not due to an alteration in the subcellular distribution of EZH2 as the cellular and cytoplasmic content of EZH2 in these cells was similar to the WT case (Supplemental Fig. 3C).
Our previous findings showed that the activity of PRC2 is uncoupled from its recruitment to chromatin (Lee et al. 2018b). However, EZH2 automethylation mutants might harbor a dual defect in both catalytic activity and affinity with chromatin. To test this possibility, we performed a chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) to examine the genome-wide occupancy of EZH2 in WT and EZH1/2 dKO mESCs rescued with either EZH2WT or automethylation mutant EZH2. The vast majority of EZH2 peaks found in WT mESCs were depleted in dKO mESCs and recovered upon rescue with EZH2WT (Fig. 3C), albeit this ectopically expressed EZH2WT exhibited an increased occupancy, possibly due to its overexpression (Fig. 3B). Importantly, the EZH2K510A;K514A rescued cell line also exhibited a restoration of EZH2 peaks and to a similar extent as with the EZH2WT rescue (Fig. 3C,D). Taken together, the results demonstrated that EZH2 automethylation mutants appear to be specifically defective in H3K27me3 catalysis as they were not defective in chromatin recruitment.
EZH2 automethylation promotes PRC2 accessibility to histone H3K27
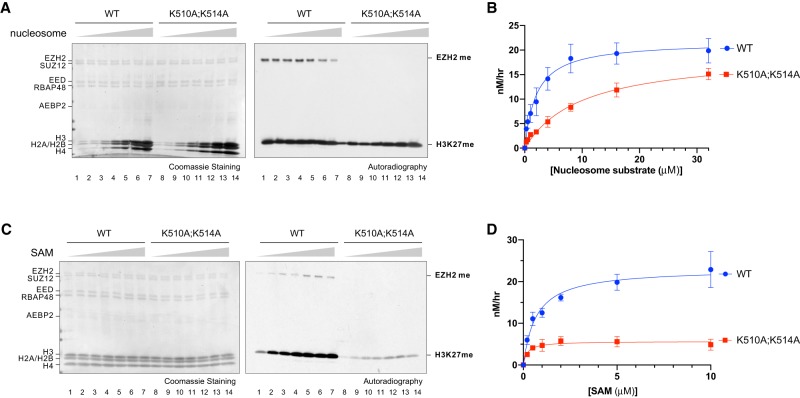
As the EZH2 automethylation loop is flexible and proximal to the SAM-binding and substrate-binding pockets (Fig. 2C), we reasoned that automethylation might promote PRC2 accessibility to SAM and/or nucleosomes, thereby regulating its HMT activity. To test these possibilities, we measured the levels of H3K27 methylation catalyzed on nucleosome substrates by PRC2–AEBP2–EZH2WT or PRC2–AEBP2–EZH2K510A;K514A. We first compared respective HMT activities in the presence of increasing amounts of nucleosomal substrates to an extent that saturates the reaction (Fig. 4A,B). We found that PRC2–AEBP2–EZH2K510A;K514A exhibited a significantly impaired HMT activity compared with PRC2–AEBP2–EZH2WT (Fig. 4B). Interestingly, the low HMT activity of PRC2–AEBP2–EZH2K510A;K514A was largely restored by increasing concentrations of nucleosomal substrates (Fig. 4A,B). A similar tendency was also observed in a time-course kinetic analysis using low, mid, or high concentrations of nucleosomal substrates (Supplemental Fig. 4A–C). Importantly, when the amounts of PRC2–AEBP2–EZH2WT and PRC2-AEBP2-EZH2K510A;K514A were fixed in the presence of increasing amounts of SAM, the HMT activity of PRC2–AEBP2–EZH2K510A;K514A remained low and could not be rescued with higher concentrations of SAM (Fig. 4C,D). Notably, while EZH2 automethylation gradually increased with increasing amounts of SAM (Fig. 4C), it declined at higher concentrations of nucleosomal substrates (Fig. 4A), indicating that upon reaching a threshold, nucleosomal substrates were able to compete with EZH2K510 and EZH2K514 for catalysis of methylation. These data point to the unstructured loop containing EZH2K510 and EZH2K514 being an intramolecular competitor for nucleosomal substrates such that automethylation relieves this autoinhibitory effect, enabling nucleosome access to the catalytic pocket of PRC2.
Figure 4.
Excessive nucleosomal substrates, but not SAM, rescued the catalytic defect of EZH2 automethylation mutants in vitro. (A–D) Enzyme kinetic analysis of PRC2–EZH2–AEBP2 complexes containing EZH2WT (WT) or EZH2K510A;K514A (K510A;K514A). (A) Methyltransferase assays using WT or K510A;K514A (60 nM) with increasing concentrations of oligonucleosomes (0.5, 1, 2, 4, 8, 16, 32 µM), and 2 µM 3H-SAM. (Left image) Coomassie blue staining of SDS-PAGE gels containing nucleosomes or PRC2 components was used to visualize the relative concentration of each component present in each reaction. (Right image) The levels of methylation on EZH2 or histone H3 are shown by autoradiography. (B) A plot of the relative amount of 3H-SAM incorporated into histone H3 after 60 min of incubation is shown. Error bars on the fitting curve represent the standard deviation of three replicates per titration point. The conversion of relative units is specified in the Materials and Methods (C) Methyltransferase assays using WT or K510A;K514A (60 nM) with increasing concentrations of SAM (0.2, 0.5, 1, 2, 5, 10, or 20 µM) and 15 µM H3K27me3 peptide, using oligonucleosomes (500 nM) as substrate. The left and right gel images are as described in A. (D) A plot of the quantification of the results obtained from MT assays shown in C, using increasing amounts of 3H-SAM and either WT or K510A;K514A. The coordinates of the plot are as described in B.
EZH2 automethylation precedes PRC2 allosteric activation
To further investigate the role of EZH2 automethylation in a cellular context, we generated an antibody that specifically recognizes methylated K514 of EZH2 (Supplemental Fig. 5A). For antibody production, we used a chemically trimethylated K514 (K514me3) version of an EZH2 peptide (504–521 amino acids) (Supplemental Fig. 5A), as EZH2-K514me3 was the most abundant automethylated state (70%) in PRC2 purified from mESCs (Fig. 2B). We demonstrated that the antibody is specific to EZH2-K514 methylation by Western blot, with the band corresponding to the molecular weight of EZH2 being absent in EZH1/2 dKO cells and restored by rescue with EZH2WT but not with EZH2K514A or the catalytically inactive mutant, EZH2Y731F (Y726F in human isoform c) (Supplemental Fig. 5A–D). In parallel, the antibody specificity was validated by immunoblotting recombinant PRC2 complexes that had been incubated with SAM; PRC2–AEBP2–EZH2K514A and PRC2–AEBP2–EZH2K510A;K514A exhibited a loss in signal compared with PRC2–AEBP2–EZH2WT and PRC2–AEBP2–EZH2K510A (Supplemental Fig. 5E).
Using this antibody, we first tested whether EZH2 automethylation requires EED and SUZ12, as their assembly with EZH2 is required for HMT activity (Fig. 5A; Cao et al. 2002; Kuzmichev et al. 2002). To this end, we compared the level of EZH2-K514me3 in WT versus EED-KO mESCs (Fig. 5B, lanes 1,2). Despite the low abundance of EZH2 in EED-KO mESCs compared with WT, EZH2-K514me3 was not detectable even with higher exposures (Fig. 5B, lane 2), affirming that EED is required for the general catalytic activity of PRC2. Since the “write and read” mechanism that conveys allosteric activation of PRC2 is required for the maintenance of H3K27me2/me3 in vivo (Lee et al. 2018b), we tested whether allosteric activation is also critical for EZH2 automethylation. We monitored the automethylation of EZH2-K514 in EZH1/2 dKO mESCs rescued with either EZH2-WT or EZH2 mutants lacking key residues required for PRC2 allosteric activation (EZH2P132S or EZH2F145L) (Fig. 5A; Lee et al. 2018b). Surprisingly, the levels of EZH2-K514me3 were unaffected in EZH2P132S and EZH2F145L rescue cell lines compared with the EZH2WT rescue (Fig. 5B, cf. lanes 5 and 6,7). However, as expected, catalysis of H3K27me2/me3 was not detected in vivo, indicating that allosteric activation of PRC2 is dispensable for EZH2 automethylation. We then monitored the automethylation of EZH2-K514 in EED-KO mESCs rescued with EED-WT, or EED mutants lacking a key residue required for PRC2 allosteric activation (EEDR302G) (Fig. 5A; Lee et al. 2018b) or an EED aromatic cage mutant required for H3K27me3 recognition and PRC2 allosteric activation (EEDY365A) (Fig. 5A; Margueron et al. 2009; Oksuz et al. 2018). Similar to the other EZH2 allosteric mutants (Fig. 5B), the levels of EZH2-K514me3 were unaffected in EEDY365A and EEDR302G rescued cell lines compared with EEDWT (Fig. 5C, cf. lane 3 with lanes 4 and 5), reaffirming that allosteric activation of PRC2 is dispensable for EZH2 automethylation. Moreover, since trimethylated versions of H3K27 (H3K27me3) or JARID2-K116 (JARID2-K116me3) allosterically activate PRC2 through binding to the aromatic cage of its EED subunit (Margueron et al. 2009; Sanulli et al. 2015), we next tested whether methylated EZH2 functions similarly. To this end, we performed HMT assays using PRC2–AEBP2 with or without the EZH2 (504–521 amino acids) peptide containing K510me2/K514me3 or K514me3, and found it to be ineffectual (Supplemental Fig. 6). Together with the results from a similar study showing that EZH2 automethylation occurs intramolecularly (Wang et al. 2019), these data demonstrated that EZH2 automethylation reflects an intrinsic activity that precedes the allosteric activation of PRC2 and its subsequent catalysis on nucleosomal substrates.
Figure 5.
The level of EZH2 automethylation remains intact upon loss of allosteric activation. (A) A schematic illustration of EZH2 and EED structures, including an H3K27me3 peptide that interacts with the aromatic cage of EED as an allosteric stimulator and an H3 tail peptide present in the SET domain of EZH2 as substrate. Allosteric mutations in the EED cage (EED Y365A; pink dot), EED/EZH2-SRM interface (EED R302G; yellow dot), and SRM domain of EZH2 [P132S (turquoise dot) and F145L (blue dot)] are indicated. (B,C) EZH2 automethylation requires EED but occurs independently from allosteric activation of PRC2. Western blot analyses of EZH2-K514me3, EZH2, EED, GAPDH, H3K27me2, H3K27me3, and total histone H3 levels in E14 mESCs, either WT or EED-KO, and in B6 mESCs, either WT or EZH1/2 dKO with or without EZH2 rescue conditions. mESC, mouse embryonic stem cells. Asterisks indicate nonspecific bands.
EZH2 automethylation is diminished in cells expressing the H3K27M oncohistone
As the hallmark of H3K27M-DIPG is the global reduction in H3K27me2/me3, reflecting an H3K27M-mediated defect in PRC2 allosteric activation (Stafford et al. 2018; Harutyunyan et al. 2019), we wondered whether EZH2 automethylation remains intact in the presence of cellular H3K27M. We first utilized two previously constructed 293 T-Rex cell lines engineered to express H3.3-WT or H3.3-K27M in a doxycycline-inducible manner (Stafford et al. 2018) to assess the effect of H3K27M on EZH2 automethylation. Remarkably, the induction of H3.3-K27M, but not H3.3-WT, resulted not only in the expected reduction in H3K27me2/me3 levels, but also in reduced levels of EZH2-K514me3 (Fig. 6A). Next, we examined the EZH2-K514me3 levels in H3-WT and H3K27M glioma cell lines and observed a more profound loss in EZH2-K514me3 as well as H3K27me2/me3 in H3K27M-DIPG cells (Fig. 6B). These results suggested that the intrinsic activity of PRC2 is impaired by H3K27M and that this impairment is more profound than the disruption to allosteric activation alone.
Figure 6.
The level of EZH2 automethylation is significantly reduced in H3K27M containing cells. (A) EZH2 automethylation is significantly reduced in 293T-Rex cells containing doxycycline inducible H3.3K27M or H3.3K27WT. Western blot analyses of EZH2-K514me3, EZH2, GAPDH, H3K27me2, H3K27me3, and total histone H3 levels. Asterisks indicate nonspecific bands. (B) EZH2 automethylation is significantly reduced in diffuse intrinsic pontine gliomas (DIPGs) containing the histone H3K27M mutation. Western blot analysis of EZH2-K514me3, EZH2, GAPDH, H3K27me2, H3K27me3, and total histone H3 levels in the following glioma cell lines: H3K27M glioma (DIPG IV and DIPG XIII) and H3WT glioma (pcGBM2 and DIPG X). Asterisks indicate nonspecific bands.
Discussion
Automethylation has emerged as a newly appreciated regulatory mechanism for chromatin-modifying enzymes, such as in the case of human and Schizosaccharomyces pombe H3K9 methyltransferases, SUV39H1/2 and CLR4, respectively (Iglesias et al. 2018). However, in the case of CLR4, the lysine-to-alanine (K-to-A) mutants at the automethylated sites appear to be hyperactive compared with WT CLR4, while the lysine-to-arginine (K-to-R) mutants exhibit hypoactive activities (Iglesias et al. 2018). A proposed explanation for such seemingly contradictory observations is that arginine serves as a lysine mimic with a positively charged state that cannot be neutralized by methylation, thereby sustaining the occlusion of the active site of the SET domain of CLR4, while the K-to-A mutants fail to do so (Iglesias et al. 2018). In contrast to CLR4, the EZH2 K-to-A mutants at K510 and K514 automethylation sites are hypoactive in the context of PRC2, indicating that the charged state of K510 and K514 might not be critical to autoinhibition by the unstructured loop. Thus, although automethylation might be a widely conserved, self-regulatory feature of methyltransferases, the inherent operating mechanisms likely differ.
Interestingly, in contrast to the case of human PRC2, this unstructured loop containing EZH2 automethylation residues is not present in yeast Chaetomium thermophilum PRC2 (ctPRC2) (Jiao and Liu 2015). Unlike the case of H3K9 methyltransferases whose automethylated resides are conserved from yeast to human (Iglesias et al. 2018), automethylation of PRC2 might have been a newly evolved function. We speculate that the gain in PRC2 automethylation allows for a more efficient catalysis towards the highest methylation status of H3K27 as well as other potential nonhistone PRC2 substrates, to promote the stability of cellular identity in multicellular organisms.
Structural evidence for PRC2 automethylation is currently lacking as the automethylated loop is not observed by X-ray crystallography (Brooun et al. 2016; Justin et al. 2016). Yet, the biochemical data reported here strongly suggest that this unstructured loop functions as an intrinsic inhibitor that competes for nucleosomal substrate binding to the active pocket, and that automethylation at K510 and K514 of EZH2 displaces this loop, promoting PRC2 accessibility to nucleosomes. In addition, this loop contains a positively charged basic patch (495–502 amino acids, RKKKRKHR) (Fig. 2C), potentially interacting with nucleosomal DNA and, therefore, displacement of this loop would further increase nucleosome binding and enhance HMT activity. On the other hand, this basic patch may also interact with RNA, thereby inhibiting the activity of PRC2 (Long et al. 2017). Further investigation is warranted to elucidate the possible reciprocal relationship between the automethylation sites and the basic patch.
Since EZH2 automethylation occurs in cis (Wang et al. 2019), the intramolecular automethylation event should be significantly more efficient than the intermolecular histone H3K27 methylation event. Indeed, despite manifesting extremely low HMT activity in vitro and in vivo, the allosteric activation-deficient mutants of EZH2 retained an intact automethylation activity (Fig. 5). In light of this finding and our previous finding that allosterically activated PRC2 is more sensitive to the detrimental effects of interaction with the H3K27M oncohistone (Stafford et al. 2018), it is striking that H3K27M also impairs PRC2 automethylation, as shown here. Although relative to H3K27, PRC2 binds with higher affinity to H3K27M in biochemical assays, such H3K27M-mediated retention of PRC2 is not reflected in the context of steady-state chromatin as H3K27M does not colocalize with PRC2 in vivo (Mohammad et al. 2017; Piunti et al. 2017; Stafford et al. 2018). PRC2 interacts with H3K27M transiently on chromatin (Stafford et al. 2018) or possibly before being incorporated into chromatin. Such exposure to H3K27M might trigger a conformational change in PRC2 to a hypoautomethylated and hypoactive state (Stafford et al. 2018). Indeed, while the structure of PRC2 was initially solved by X-ray crystallography, a stable complex could only be derived in the presence of a SAM-competitive inhibitor (Brooun et al. 2016) or an H3K27M peptide (Jiao and Liu 2015; Justin et al. 2016), suggesting that the inhibited SET domain is in a more stable and thermodynamically favored state, which can be similarly triggered by the H3K27M oncohistone. Further structural and functional studies are certainly warranted to clarify the underlying mechanisms.
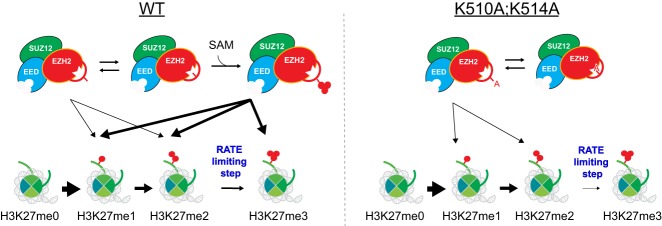
Notably, our data demonstrated that the major catalytic defect in EZH2 automethylation mutants appears to be in the conversion from H3K27me2 to H3K27me3 in vitro and in vivo. The competition between the automethylation loop and the histone H3 tail for entry into the EZH2 catalytic pocket is perhaps more pronounced in the case of H3K27me2-modified nucleosomes (Fig. 7). Given the unique role of H3K27me3 in gene repression and disease pathogenesis (Wigle et al. 2011; Yap et al. 2011; Lee et al. 2018b; Oksuz et al. 2018; Hübner et al. 2019; Jain et al. 2019), it is telling that automethylation of PRC2 mainly participates in the rate-limiting process of attaining the highest H3K27 methylation state (Fig. 7). Future investigations of PRC2 automethylation will likely elucidate how this regulatory step impacts H3K27me3 catalysis and its role in normal and disease states.
Figure 7.
A scheme showing a model by which EZH2 automethylation enhances the conversion of H3K27me2 to H3K27me3. (Left) In WT cells, PRC2 automethylates K510 and K514 of EZH2 which subsequently displaces the unstructured autoinhibitory loop and promotes PRC2 accessibility to nucleosomes. (Right) When EZH2 automethylation sites (EZH2-K510 and EZH2-K514) are mutated to alanine, automethylation fails and the loop may occlude the active site more frequently. This outcome significantly impairs the rate limiting step of PRC2 catalysis: conversion of H3K27me2 to H3K27me3.
Materials and methods
Cell culture
Mouse embryonic stem cells (mESCs) were grown in standard ESC medium containing Lif, 1 µM MEK1/2 inhibitor (PD0325901), and 3 µM GSK3 inhibitor (CHIR99021). 293T-Rex cells were grown in standard DMEM medium supplemented with 10% FBS, 1% nonessential amino acid, 1 mM Na pyruvate, and 1% penicillin/streptomycin.
Purification of protein using baculovirus expression system
To purify human PRC2 complexes, Flag-tagged-EED, EZH1, EZH2, SUZ12, RBAP48, Strep-tagged AEBP2 (short isoform), and HIS-tagged JARID2 were cloned independently into a baculovirus expression vector, pFASTBac1 (Invitrogen). EZH2 mutant constructs were generated by site-directed mutagenesis and mutations were confirmed by Sanger DNA sequencing. To purify the four-subunit PRC2 complex (Flag-tagged-EED, EZH1 or EZH2, SUZ12, and RBAP48), the five-subunit PRC2–EZH2–AEBP2 complex (four subunits plus Strep-tagged AEBP2), or the six-subunit PRC2-EZH2-AEBP2-JARID2 (four subunit plus Strep-tagged AEBP and His-tagged JARID2), the appropriate components were coexpressed in Sf9 cells by baculovirus infection. After 60 h of infection, Sf9 cells were resuspended in BC300 buffer (25 mM HEPES-NaOH at pH 7.5, 1 mM EDTA, 300 mM NaCl, 10% glycerol, 0.2 mM DTT, 0.1% NP-40) with protease inhibitors (1 mM phenylmethlysulfonyl fluoride [PMSF], 0.1 mM benzamidine, 1.25 mg/mL leupeptin, 0.625 mg/mL pepstatin A) and phosphatase inhibitors (20 mM NaF, 1 mM Na3VO4). Cells were lysed by sonication (Fisher Sonic Dismembrator model 100) and WT or mutant recombinant PRC2 was tandemly purified through Flag-M2 agarose beads (Sigma), Q sepharose beads (GE Healthcare), and glycerol gradient (15%–35%) sedimentation.
Nucleosome reconstitution
Recombinant histones were generated as previously described (Yun et al. 2012; Lee et al. 2013). Briefly, each core histone was expressed in Rosetta (DE3) cells (Novagen), extracted from inclusion bodies, and purified by sequential anion and cation chromatography. For refolding recombinant octamers, equal amounts of histones were mixed and dialyzed into refolding buffer (10 mM Tris-HCl at pH 7.5, 2 M NaCl, 1 mM EDTA, 5 mM β-mercaptoethanol). Octamers were further purified by size exclusion chromatography on a 24-mL Superdex 200 column (GE Healthcare) in refolding buffer. Recombinant oligonucleosomes were reconstituted by sequential salt dialysis of octamers and plasmid having 12 repeats of the 601-nucleosome positioning sequence.
HMT assay
Standard HMT assays were performed in a total volume of 15 µL containing HMT buffer (50 mM Tris-HCl at pH 8.5, 5 mM MgCl2, 4 mM DTT) with the indicated concentration of 3H-labeled S-adenosylmethionine (SAM) (Perkin Elmer), recombinant oligonucleosomes, and recombinant human PRC2 complexes. The reaction mixture was incubated for 60 min at 30°C and stopped by the addition of 4 µL of SDS buffer (0.2 M Tris-HCl at pH 6.8, 20% glycerol, 10% SDS, 10 mM β-mercaptoethanol, 0.05% Bromophenol blue). A titration of PRC2 (from 5 to 60 nM) was performed under these conditions to optimize the HMT reaction within a linear range, and the yield of each HMT reaction was measured using the following procedures. After HMT reactions, samples were incubated for 5 min at 95°C and separated on SDS-PAGE gels. The gels were then subjected to Coomassie blue staining for protein visualization or wet transfer of proteins to 0.45-µm PVDF membranes (Millipore). The radioactive signals were detected by exposure on autoradiography films (Denville Scientific).
Enzymatic kinetics analysis
Enzyme kinetics analysis was performed as described previously (Bratkowski et al. 2017). Briefly, bands corresponding to histone H3 were excised from the membrane and subjected to Scintillation counting. Disintegrations per minute (dpm) were converted to curies (Ci) and then converted to millimoles based on the specific activity of 3H-SAM, and enzyme activity data were then represented as velocity in nM/h. The assays included various ratios between unlabeled and 3H-labeled SAM, and relative activities were adjusted based on these ratios. Data points were fit to enzyme saturation curves and kinetics parameters and standard errors were calculated with the program GraphPad Prism 7 using Michaelis-Menton analysis.
Lentiviral production and delivery
WT or mutant EZH2 constructs were subcloned into the pLV-EF1-α-IRES-mCherry vector (Clontech) for lentiviral production and delivery. For the production of viral particles, lentiviral vectors containing 10 µg of WT or mutant EZH2 were cotransfected with 2.5 µg ofpcREV, 3 µg of BH-10, and 5 µg of pVSV-G packaging vectors into 293-FT cells. The virus-containing medium was collected 48 h after transfection and the target cells were spin infected. Polybrene was added to the viral medium at a concentration of 8 µg/mL. Infected cells were FACS-sorted for mCherry.
Purification of PRC2 complex from mESCs
To purify PRC2 complex from mESC, mESCs expressing Flag-tagged EED in an EED-deficient background were used (Lee et al. 2018b). Approximately 1 × 1010 cells were cultured and prepared for mESC nuclear extract. Cells were harvested and washed with PBS. Cells were lysed with intact nuclei in TMSD buffer (40 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 250 mM sucrose, 1 mM DTT, 0.02% NP-40) containing protease inhibitors and phosphatase inhibitors, as indicated above. The cell suspension was centrifuged at 800g for 10 min at 4°C. The pellets (nuclei) were resuspended in 20× cell pellet volume of BC400 buffer (20 mM Tris-HCl at pH 7.9, 400 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT, 0.02% NP-40) with protease inhibitors and phosphatase inhibitors and incubated for 1 h at 4°C. The cell suspension was centrifuged at 18,000g for 20 min at 4°C. The supernatant was collected as the nuclear extract. The extract was subjected to Flag affinity purification using Flag-M2 agarose beads and then further purified using Q Sepharose beads. The final purified protein complexes are shown in Figure 2A.
Preparation of samples for mass spectrometry
PRC2 purified from mESCs (Fig. 2A) were reduced with 2 µL of 0.2 M dithiothreitol (pH 7.5) (Sigma) for 1 h at 57°C in 100 mM ammonium bicarbonate and subsequently alkylated with 2 µL of 0.5 M iodoacetamide (Sigma) for 45 min at room temperature in the dark. The sample was treated with 300 ng of Arg-C (Roche), and digestion proceeded overnight at room temperature. The digestion was halted by adding a slurry of R2 50 µm Poros beads (Applied Biosystems) in 5% formic acid and 0.2% trifluoroacetic acid (TFA) to the sample at a volume equal to that of the sample. The sample was allowed to shake for 2 h at 4°C. The beads were loaded onto C18 ziptips (Millipore) and equilibrated with 0.1% TFA using a microcentrifuge at 6000 rpm for 30 sec. The beads were washed with 0.5% acetic acid. Peptides were eluted with 40% acetonitrile in 0.5% acetic acid, followed by 80% acetonitrile in 0.5% acetic acid. The organic solvent was removed using a SpeedVac concentrator and the sample reconstituted in 0.5% acetic acid.
Mass spectrometry analysis
An aliquot of each sample was loaded onto an Acclaim PepMap trap column (2 cm × 75 µm) in line with an EASY-Spray analytical column (50 cm × 75 µm ID PepMap C18, 2 µm bead size) using the auto sampler of an EASY-nLC 1000 HPLC (Thermo Fisher Scientific) with solvent A consisting of 2% acetonitrile in 0.5% acetic acid and solvent B consisting of 80% acetonitrile in 0.5% acetic acid. The peptides were gradient eluted into a Thermo Fisher Scientific Orbitrap Fusion Lumos Mass Spectrometer using the following gradient: 5%–35% in 60 min, 35%–45% in 10 min, followed by 45%–100% in 10 min. High-resolution full MS spectra were recorded with a resolution of 120,000, an AGC target of 4e5, with a maximum ion time of 60 msec, and a scan range from 400 to 1500 m/z. The MS/MS spectra were collected using first a topN method for 3 sec with +2 and +3 ions subjected to HCD and higher charge states (4–10) to EThcD. The The MS/MS HCD spectra were collected with a resolution of 15,000, AGC target of 1e5, maximum ion time of 100 msec, one microscan, 2 m/z isolation window, and normalized collision energy (NCE) of 32. The MS/MS EThcD spectra were collected with a resolution of 15,000, AGC target of 1e5, maximum ion time of 100 msec, one microscan, 2 m/z isolation window, ETD reaction time of 30 msec, reagent target 2e5, and supplemental activation using an NCE of 32%. In addition, the m/z values of the +6 charge state for all possible methylated isoforms of peptide 510–532 were targeted for EThcD MS/MS using the same settings as described above but with a resolution of 30,000.
Data processing (MS)
The MS/MS spectra were searched against the Uniprot Homo sapiens reference proteome database (downloaded 10/2017) using Byonic (Bern et al. 2012). The Byonic search included variable modifications of oxidation on methionine, deamidation on asparagine and glutamine, acetylation and methylations on lysine, and a fixed modification of carbamidomethyl on cysteine. The fragment mass tolerance was set to 0.02 Da with a precursor tolerance of 10 ppm. A 1% FDR using a standard target-decoy database approach was used. Relative quantitation of methylated peptide residues 510–532 was performed using Byologic (Protein Metrics), which uses a semiautomatic label-free quantification approach. The area under the curve for all isoforms of a given peptide are summed and the percentage attributed to a specific isoform calculated.
ChIP-seq and data analysis
Standard ChIP experiments and data analysis were performed as previously described (Lee et al. 2018b). Briefly, 100 mg of sonicated chromatin was used in each ChIP reaction with 2.5 mg of anti-EZH2 antibody. One milligram of Drosophila chromatin and 0.1 mg of anti-Drosophila H2A.X antibody were added in each ChIP reaction as spike-in references. For ChIP-seq library preparation, up to 30 ng of immunoprecipitated DNA was end-repaired, A-tailed, and ligated to custom barcode adapters with T4 ligase. Libraries were sequenced on Illumina HiSeq. A custom barcoding system was used.
Cellular fractionation
Cells were harvested and washed with ice-cold PBS containing both protease and phosphatase inhibitors, as indicated above. Cells were then lysed in buffer A (10 mM Tris at pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.25% NP-40, 0.5 mM DTT) containing both protease and phosphatase inhibitors. The cell suspension was centrifuged at 1000g for 5 min at 4°C. The supernatant was collected as the “cytoplasmic” fraction. The pellets were resuspended in buffer C (20 mM Tris-HCl at pH 7.9, 1.5 MgCl2, 420 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT) with both protease and phosphatase inhibitors and incubated for 1 h at 4°C. The cell suspension was centrifuged at 18,000g for 20 min at 4°C. The supernatant was collected as the “nuclear extract” fraction. The pellet was then resuspended in unfolding buffer (6 M guanidine, 5 mM DTT) and incubated for 1 h at 25°C. The cell suspension was centrifuged at 18,000g for 20 min at 4°C. The supernatant was collected as the “nuclear pellet” fraction. This “nuclear pellet” fraction was dialyzed against buffer C.
Competing interest statement
D.R. is a cofounder of Constellation Biotechnology and Fulcrum Biotechnology.
Supplementary Material
Acknowledgments
We thank Dr. L. Vales for critical reading of the manuscript as well as past and current Reinberg laboratory members for critical comments and discussion, and L. Popoca, D. Hernandez, and H. Yang and for technical assistance. We also thank Dr. T. Cech for sharing and discussing unpublished data related to this study. We thank H. Zheng from the Biological Mass Spectrometry Facility at Robert Wood Johnson Medical School and Rutgers University for some mass spectrometry analyses. The New York University Flow Cytometry Core, Proteomics Laboratory, and Genome Technology Center were partially supported by the New York University School of Medicine and the Laura and Isaac Perlmutter Cancer Center support grant, National Cancer Institute (P30CA016087). The work in D.R.’s laboratory is supported by National Institutes of Health (NIH) grant R01CA199652, the Howard Hughes Medical Institute (HHMI), and the Making Headway Foundation St. Baldrick's Research Grant (189290). J-R.Y. is supported by the American Cancer Society (PF-17-035-01). J.G. is supported by the Vilcek Scholarship. G.L. is supported by the Making Headway Foundation St. Baldrick's Research Grant (189290). J.M.S. was supported as a Simons Foundation Junior Fellow and by NIH grant K99AA024837. The work in B.A.G.’s laboratory is supported by NIH grants R01GM110174 and P01CA196539.
Author contributions: C.-H.L., J.-R.Y., J.G., and D.R. conceptualized and designed the study. C.-H.L., J.-R.Y., J.G., G.L., and J.M.S. conducted the experiments. R.S.-M. and Y.J. performed the bioinformatics analysis. J.A., P.L., B.A.G., and B.U. performed mass spectrometry analysis. C.-H.L., J.-R.Y., J.G., and J.A. wrote the manuscript with assistance from B.U. and D.R.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.328773.119.
References
- Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI. 2014. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc Natl Acad Sci 111: 2488–2493. 10.1073/pnas.1400648111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer M, Pisano P, Di Carlo V, Blanco E, Chammas P, Vizán P, Gutiérrez A, Aranda S, Payer B, Wierer M, et al. 2016. EPOP functionally links elongin and polycomb in pluripotent stem cells. Mol Cell 64: 645–658. 10.1016/j.molcel.2016.10.018 [DOI] [PubMed] [Google Scholar]
- Bern M, Kil YJ, Becker C. 2012. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics 43: 13.20.1–13.20.14. 10.1002/0471250953.bi1320s40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratkowski M, Yang X, Liu X. 2017. Polycomb repressive complex 2 in an autoinhibited state. J Biol Chem 292: 13323–13332. 10.1074/jbc.M117.787572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooun A, Gajiwala KS, Deng Y-L, Liu W, Bolaños B, Bingham P, He Y-A, Diehl W, Grable N, Kung P-P, et al. 2016. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat Commun 7: 11384 10.1038/ncomms11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043. 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, David James C, Jenkins R, et al. 2013. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990. 10.1101/gad.217778.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Jiao L, Shubbar M, Yang X, Liu X. 2018. Unique Structural platforms of Suz12 dictate distinct classes of PRC2 for chromatin binding. Mol Cell 69: 840–852.e5. 10.1016/j.molcel.2018.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin HG, Estève PO, Pradhan M, Benner J, Patnaik D, Carey MF, Pradhan S. 2007. Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res 35: 7313–7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway E, Jerman E, Healy E, Ito S, Holoch D, Oliviero G, Deevy O, Glancy E, Fitzpatrick DJ, Mucha M, et al. 2018. A family of vertebrate-specific polycombs encoded by the LCOR/LCORL genes balance PRC2 subtype activities. Mol Cell 70: 408–421.e8. 10.1016/j.molcel.2018.03.005 [DOI] [PubMed] [Google Scholar]
- Dillon MBC, Rust HL, Thompson PR, Mowen K. 2013. Automethylation of protein arginine methyltransferase 8 regulates activity by impeding AdoMet sensitivity. J Biol Chem 288: 27872–27880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300. 10.1038/ncb1787 [DOI] [PubMed] [Google Scholar]
- Harutyunyan AS, Krug B, Chen H, Papillon-Cavanagh S, Zeinieh M, De Jay N, Deshmukh S, Chen CCL, Belle J, Mikael LG, et al. 2019. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat Commun 10: 1262 10.1038/s41467-019-09140-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner J-M, Müller T, Papageorgiou DN, Mauermann M, Krijgsveld J, Russell RB, Ellison DW, Pfister SM, Pajtler KW, Kool M. 2019. EZHIP/CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro Oncol 21: 878–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Currie MA, Jih G, Paulo JA, Siuti N, Kalocsay M, Gygi SP, Moazed D. 2018. Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability. Nature 560: 504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa E, Higashimoto K, Sakai Y, Numakura C, Okamoto N, Matsunaga S, Ryo A, Sato Y, Sanefuji M, Ihara K, et al. 2017. Mutations in genes encoding polycomb repressive complex 2 subunits cause Weaver syndrome. Hum Mutat 38: 637–648. 10.1002/humu.23200 [DOI] [PubMed] [Google Scholar]
- Jain SU, Do TJ, Lund PJ, Rashoff AQ, Diehl KL, Cieslik M, Bajic A, Juretic N, Deshmukh S, Venneti S, et al. 2019. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat Commun 10: 2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L, Liu X. 2015. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350: aac4383 10.1126/science.aac4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, De Marco V, Haire LF, Walker PA, Reinberg D, et al. 2016. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun 7: 11316 10.1038/ncomms11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinath V, Faini M, Poepsel S, Reif D, Feng XA, Stjepanovic G, Aebersold R, Nogales E. 2018. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359: 940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J. 2009. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res 37: 2940–2950. 10.1093/nar/gkp149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev 16: 2893–2905. 10.1101/gad.1035902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Wu J, Li B. 2013. Chromatin remodelers fine-tune h3k36me-directed deacetylation of neighbor nucleosomes by Rpd3S. Mol Cell 52: 255–263. 10.1016/j.molcel.2013.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Teckie S, Wiesner T, Ran L, Prieto Granada CN, Lin M, Zhu S, Cao Z, Liang Y, Sboner A, et al. 2014. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet 46: 1227–1232. 10.1038/ng.3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Holder M, Grau D, Saldaña-Meyer R, Yu JR, Ganai RA, Zhang J, Wang M, LeRoy G, Dobenecker MW, et al. 2018a. Distinct stimulatory mechanisms regulate the catalytic activity of polycomb repressive complex 2. Mol Cell 70: 435–448.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Yu JR, Kumar S, Jin Y, LeRoy G, Bhanu N, Kaneko S, Garcia BA, Hamilton AD, Reinberg D. 2018b. Allosteric activation dictates PRC2 activity independent of its recruitment to chromatin. Mol Cell 70: 422–434.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. 10.1126/science.1232245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liefke R, Jiang J, Kurland JV, Tian W, Deng P, Zhang W, He Q, Patel DJ, Bulyk ML, et al. 2017. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549: 287–291. 10.1038/nature23881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefke R, Karwacki-Neisius V, Shi Y, Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI, Aso T, Lane WS, Conaway JW, et al. 2016. EPOP interacts with elongin BC and USP7 to modulate the chromatin landscape. Mol Cell 64: 659–672. 10.1016/j.molcel.2016.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Bolanos B, Gong L, Liu W, Goodrich KJ, Yang X, Chen S, Gooding AR, Maegley KA, Gajiwala KS, et al. 2017. Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. Elife 6: e31558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. 2008. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32: 503–518. 10.1016/j.molcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. 10.1038/nature08398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A III, Diaz E, et al. 2012. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492: 108–112. 10.1038/nature11606 [DOI] [PubMed] [Google Scholar]
- Mohammad F, Weissmann S, Leblanc B, Pandey DP, Højfeldt JW, Comet I, Zheng C, Johansen JV, Rapin N, Porse BT, et al. 2017. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med 23: 483–492. [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185. 10.1038/ng.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208. 10.1016/S0092-8674(02)00976-5 [DOI] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. 2012. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 18: 298–301. 10.1038/nm.2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksuz O, Narendra V, Lee CH, Descostes N, LeRoy G, Raviram R, Blumenberg L, Karch K, Rocha PP, Garcia BA, et al. 2018. Capturing the onset of PRC2-mediated repressive domain formation. Mol Cell 70: 1149–1162.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, Rendleman EJ, Ma Q, Takahashi YH, Woodfin AR, et al. 2017. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med 23: 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D, Vales LD. 2018. Chromatin domains rich in inheritance. Science 361: 33–34. [DOI] [PubMed] [Google Scholar]
- Sanulli S, Justin N, Teissandier AA, Ancelin K, Portoso M, Caron M, Michaud A, Lombard BB, da Rocha ST, Offer J, et al. 2015. Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol Cell 57: 769–783. 10.1016/j.molcel.2014.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Shen SS, Margueron R, Reinberg D. 2013. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev 27: 2663–2677. 10.1101/gad.225888.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JM, Lee C-H, Voigt P, Descostes N, Saldaña-Meyer R, Yu J-R, Leroy G, Oksuz O, Chapman JR, Suarez F, et al. 2018. Multiple modes of PRC2 inhibition elicit global chromatin alterations in H3K27M pediatric glioma. Sci Adv 4: eaau5935 10.1126/sciadv.aau5935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-X, Wu J-W. 2002. Autophosphorylation kinetics of protein kinases. Biochem J 368: 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Paucek RD, Gooding AR, Brown ZZ, Ge EJ, Muir TW, Cech TR. 2017. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol 24: 1028–1038. 10.1038/nsmb.3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Long Y, Paucek RD, Gooding AR, Lee T, Burdorf R, Cech TR. 2019. Regulation of histone methylation by automethylation of PRC2. Genes Dev (this issue) 10.1101/gad.328849.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle TJ, Knutson SK, Jin L, Kuntz KW, Pollock RM, Richon VM, Copeland RA, Scott MP. 2011. The Y641C mutation of EZH2 alters substrate specificity for histone H3 lysine 27 methylation states. FEBS Lett 585: 3011–3014. 10.1016/j.febslet.2011.08.018 [DOI] [PubMed] [Google Scholar]
- Wu H, Zeng H, Dong A, Li F, He H, Senisterra G, Seitova A, Duan S, Brown PJ, Vedadi M, et al. 2013. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS One 8: e83737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB, Chu J, Berg T, Schapira M, Cheng SWG, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, et al. 2011. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117: 2451–2459. 10.1182/blood-2010-11-321208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J-R, Lee C-H, Oksuz O, Stafford JM, Reinberg D. 2019. PRC2 is high maintenance. Genes Dev 33: 903–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Ruan C, Huh JW, Li B. 2012. Reconstitution of modified chromatin templates for in vitro functional assays. Methods Mol Biol 833: 237–253. 10.1007/978-1-61779-477-3_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.