This Outlook discusses the findings by Hustedt et al. in this issue of Genes & Development that report the identification of HROB, a novel factor required for loading MCM8–9 onto HR intermediates to facilitate DNA repair synthesis.
Keywords: CRISPR screens, cisplatin, DNA damage, DNA repair, DNA synthesis, germ cells, helicase, homologous recombination, infertility
Abstract
Homologous recombination (HR) is an important route for repairing DNA double-strand breaks (DSBs). The early stages of HR are well understood, but later stages remain mysterious. In this issue of Genes & Development, Hustedt and colleagues (pp. 1397–1415) reveal HROB as a new player in HR required for recruitment of the MCM8–9 complex, which is paralogous to the MCM2–7 replicative helicase. HROB functions closely with MCM8–9 to promote postsynaptic DNA repair synthesis. This study sheds valuable light on late events in HR and suggests that HROB may load MCM8–9 onto HR intermediates to facilitate the DNA unwinding required for DNA repair synthesis.
Homologous recombination (HR) is a major route for repairing DNA double-strand breaks (DSBs) (Prakash et al. 2015). HR starts with resection of broken DNA ends, involving nucleases and helicases. The resulting overhangs, coated by RAD51 to yield a nucleoprotein filament, invade the intact sister chromatid, searching for a homologous sequence. The pairing of RAD51-coated single-stranded DNA (ssDNA) with the donor strand (termed synapsis) results in the displacement of the strand complementary to the donor (forming a structure called a “D loop”) and the establishment of a RAD51-bound heteroduplex species that primes DNA repair synthesis (Fig. 1). The postsynaptic stages of HR are poorly understood, but a number of proteins are involved at this stage specifcially. For example, the MCM8 and MCM9 proteins, which are related to the subunits of the hexameric MCM2–7 replicative helicase, have been implicated in postsynaptic DNA synthesis. In contrast to MCM2–7, the MCM8–9 complex is dispensable for bulk DNA replication (Griffin and Trakselis 2019). The residual DNA synthesis that is maintained in MCM2-depleted cells requires MCM8–9, but this reflects DNA repair synthesis occurring during HR-mediated repair of the high level of DSBs that occur upon MCM2 depletion (Natsume et al. 2017). Importantly, MCM8–9 was shown to act downstream from RAD51 in the HR-mediated repair of DSBs induced by nuclease overexpression, in meiotic HR, and during the HR stage of DNA interstrand cross-link (ICL) repair (Lutzmann et al. 2012; Nishimura et al. 2012; Natsume et al. 2017). The dominant mode of ICL repair is initiated by collision of replisomes with ICLs, resulting in programmed formation of DSBs that are repaired by HR. Crucially, point mutations in MCM8–9 predicted to abolish helicase activity suppress ICL repair, suggesting that helicase activity is important for HR (Nishimura et al. 2012). One explanation is that MCM8–9 might facilitate postsynaptic DNA repair synthesis by unwinding D loops and enabling extension of the invading RAD51-coated DNA end (Fig. 1).
Figure 1.
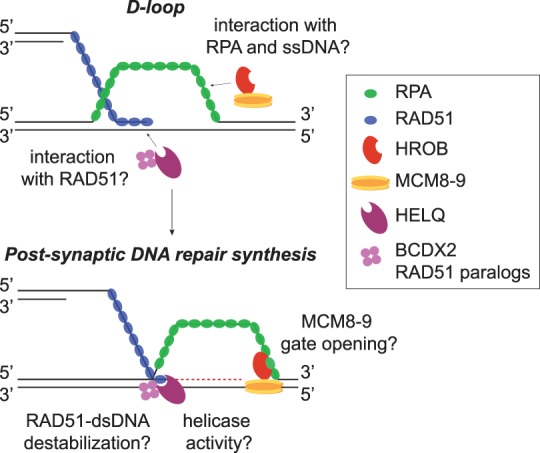
Model for the concerted actions of HROB–MCM8–MCM and HELQ in postsynpatic DNA synthesis during HR.
In this issue of Genes & Development, Hustedt et al. (2019) identify a new factor—HROB (HR OB-fold)—that functions closely with MCM8–9 in HR. Rationalizing that HR defects should cause sensitivity to inhibitors of ATR kinase and PARP, the investigators mined published CRISPR screens to identify new genes whose deletion sensitizes to both drug classes. This approach highlighted the uncharacterized C17ORF53 gene (renamed HROB) that, when disrupted in human cells or mice, causes pronounced HR defects. Disruption of mouse Hrob, for example, showed defects in meiosis and gametogenesis. Human HROB-deleted cells are defective in the HR step of ICL repair, and, whereas RAD51 loading is normal, RAD51 foci persist, hinting that HROB acts at a postsynaptic HR stage downstream from RAD51, similar to MCM8–9. In this light, HROB is epistatic with MCM8 and MCM9 in ICL repair and in HR-mediated repair of DSBs in a reporter cassette. Furthermore, residual DNA synthesis in MCM2-depleted cells, which depends on MCM8–9 (Natsume et al. 2017), requires HROB, with HROB and MCM8–9 acting epistatically in this context. A breakthrough came with the discovery that HROB recruits MCM8–9 to DNA lesions. HROB was found to physically interact with MCM8–9, and, excitingly, siRNA-mediated depletion of HROB compromised the accumulation of MCM8 at DNA repair foci but not vice versa. These data make a compelling case for HROB as an important HR factor acting with MCM8–9 to promote postsynaptic DNA repair synthesis. The investigators suggest that HROB acts by directly loading MCM8–9 onto HR intermediates, which draws interesting mechanistic parallels with MCM2–7, whose loading involves transient opening of a gate in the MCM2–7 hexamer (Deegan and Diffley 2016). It is not yet clear whether the MCM8–9 complex is hexameric in nature, nor is it known whether gate opening, perhaps mediated by HROB, is required for loading.
Another notable finding is that HROB deletion in cells lacking the DNA helicase HELQ causes a stronger HR defect than in the single mutants. HELQ was known to be involved in HR during ICL repair (Takata et al. 2013) and in HR-mediated meiotic DSB repair (Adelman and Boulton 2010; Ward et al. 2010). In these contexts, HELQ deletion causes persistent RAD51 foci (Adelman and Boulton 2010). The new study shows that HELQ and HROB–MCM8–MCM9 function redundantly, perhaps representing parallel pathways for promoting DNA synthesis downstream from RAD51 (Fig. 1). How might these two pathways converge at the molecular level? Perhaps the simplest model is that unwinding of HR intermediates, such as D loops, requires the helicase activities of both HELQ and MCM8–9 to enable extension of the RAD51-coated DNA end. Such a model predicts that helicase-inactivating mutations in HELQ would phenocopy, at least partly, the gene deletion. This remains to be tested. It is interesting that HELQ can remove RAD51 from dsDNA in vitro independently of helicase activity, possibly by direct RAD51 binding (Adelman and Boulton 2010; Ward et al. 2010). Furthermore, HELQ interacts with the BCDX2 complex of RAD51 paralogs that may also have a RAD51-stripping activity (Adelman et al. 2013; Takata et al. 2013). It is conceivable, therefore, that the ability of HELQ to strip RAD51 off the terminus of the invading end in the D loop, perhaps together with BCDX2, facilitates postsynaptic DNA synthesis through freeing the terminus for primer extension. Such a model might be suggested if mammalian cells with helicase-inactivating HELQ mutations were HR-proficient. Whatever the case, the identification of HROB, working together with MCM8–9 and HELQ in facilitating postsynaptic DNA repair synthesis, sheds light on a poorly undertood aspect of HR.
Acknowledgments
G.S. is supported by an EMBO Long-Term Postdoctoral Fellowship (ALTF 951-2018). Work in the Rouse laboratory is funded by the Medical Research Council (grant no. MC_UU_12016/1).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.331850.119.
References
- Adelman CA, Boulton SJ. 2010. Metabolism of postsynaptic recombination intermediates. FEBS Lett 584: 3709–3716. 10.1016/j.febslet.2010.05.023 [DOI] [PubMed] [Google Scholar]
- Adelman CA, Lolo RL, Birkbak NJ, Murina O, Matsuzaki K, Horejsi Z, Parmar K, Borel V, Skehel JM, Stamp G, et al. 2013. HELQ promotes RAD51 paralogue-dependent repair to avert germ cell loss and tumorigenesis. Nature 502: 381–384. 10.1038/nature12565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deegan TD, Diffley JF. 2016. MCM: one ring to rule them all. Curr Opin Struct Biol 37: 145–151. 10.1016/j.sbi.2016.01.014 [DOI] [PubMed] [Google Scholar]
- Griffin WC, Trakselis MA. 2019. The MCM8/9 complex: A recent recruit to the roster of helicases involved in genome maintenance. DNA Repair (Amst) 76: 1–10. 10.1016/j.dnarep.2019.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt N, Saito Y, Zimmermann M, Álvarez-Quilón A, Setiaputra D, Adam S, McEwan A, Yuan JY, Olivieri M, Zhao Y, et al. 2019. Control of homologous recombination by the HROB–MCM8–MCM9 pathway. Genes Dev (this issue) 10.1101/gad.329508.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Grey C, Traver S, Ganier O, Maya-Mendoza A, Ranisavljevic N, Bernex F, Nishiyama A, Montel N, Gavois E, et al. 2012. MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol Cell 47: 523–534. 10.1016/j.molcel.2012.05.048 [DOI] [PubMed] [Google Scholar]
- Natsume T, Nishimura K, Minocherhomji S, Bhowmick R, Hickson ID, Kanemaki MT. 2017. Acute inactivation of the replicative helicase in human cells triggers MCM8–9-dependent DNA synthesis. Genes Dev 31: 816–829. 10.1101/gad.297663.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Ishiai M, Horikawa K, Fukagawa T, Takata M, Takisawa H, Kanemaki MT. 2012. Mcm8 and Mcm9 form a complex that functions in homologous recombination repair induced by DNA interstrand crosslinks. Mol Cell 47: 511–522. 10.1016/j.molcel.2012.05.047 [DOI] [PubMed] [Google Scholar]
- Prakash R, Zhang Y, Feng W, Jasin M. 2015. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol 7: a016600 10.1101/cshperspect.a016600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K, Reh S, Tomida J, Person MD, Wood RD. 2013. Human DNA helicase HELQ participates in DNA interstrand crosslink tolerance with ATR and RAD51 paralogs. Nat Commun 4: 2338 10.1038/ncomms3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JD, Muzzini DM, Petalcorin MI, Martinez-Perez E, Martin JS, Plevani P, Cassata G, Marini F, Boulton SJ. 2010. Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair. Mol Cell 37: 259–272. 10.1016/j.molcel.2009.12.026 [DOI] [PubMed] [Google Scholar]


