Abstract
Induction and reversal of chromatin silencing is critical for successful development, tissue homeostasis, and the derivation of induced pluripotent stem cells (iPSCs). X-Chromosome inactivation (XCI) and reactivation (XCR) in female cells represent chromosome-wide transitions between active and inactive chromatin states. Although XCI has long been studied, providing important insights into gene regulation, the dynamics and mechanisms underlying the reversal of stable chromatin silencing of X-linked genes are much less understood. Here, we use allele-specific transcriptomics to study XCR during mouse iPSC reprogramming in order to elucidate the timing and mechanisms of chromosome-wide reversal of gene silencing. We show that XCR is hierarchical, with subsets of genes reactivating early, late, and very late during reprogramming. Early genes are activated before the onset of late pluripotency genes activation. Early genes are located genomically closer to genes that escape XCI, unlike genes reactivating late. Early genes also show increased pluripotency transcription factor (TF) binding. We also reveal that histone deacetylases (HDACs) restrict XCR in reprogramming intermediates and that the severe hypoacetylation state of the inactive X Chromosome (Xi) persists until late reprogramming stages. Altogether, these results reveal the timing of transcriptional activation of monoallelically repressed genes during iPSC reprogramming, and suggest that allelic activation involves the combined action of chromatin topology, pluripotency TFs, and chromatin regulators. These findings are important for our understanding of gene silencing, maintenance of cell identity, reprogramming, and disease.
Development, tissue homeostasis, and the derivation of induced pluripotent stem cells (iPSCs) depend on the accurate establishment, maintenance, and reversal of chromatin silencing. Although the formation of facultative heterochromatin has been extensively studied (Żylicz et al. 2019), it remains unclear how epigenetic memory of stable gene silencing is reversed by transcription factors (TFs) and accompanying chromatin mechanisms. Chromosome-wide transitions between active and inactive chromatin states are excellently modeled by X-Chromosome inactivation (XCI) and reactivation (XCR) in female mammals (Sado and Brockdorff 2013; Marks et al. 2015; Cantone and Fisher 2017; Postlmayr and Wutz 2017; Robert Finestra and Gribnau 2017; Galupa and Heard 2018; Janiszewski et al. 2018). XCI involves epigenetic regulators, including long noncoding RNAs (lncRNAs), chromatin modifications, and changes in chromosome topology, and leads to the establishment and maintenance of stable gene silencing (Giorgetti et al. 2016; Jégu et al. 2017; Żylicz et al. 2019). However, little is known about how cells erase epigenetic memory of stable silenced chromatin.
XCI is the rapid, chromosome-wide silencing of an entire X Chromosome during female mammalian development. It ensures dosage compensation between XX female and XY male cells (Lyon 1961). Moreover, XCI involves allelic gene regulation resulting in monoallelic expression, a phenomenon shared with several autosomal genes, such as imprinted genes (Gendrel et al. 2016). In early mouse embryos, imprinted XCI (iXCI), which always inactivates the paternal X Chromosome, takes place at the four-cell stage. It is followed by XCR, in the inner cell mass (ICM) of the blastocyst (Mak et al. 2004; Okamoto et al. 2004; Borensztein et al. 2017a, b). Then, random XCI (rXCI) of one of the two X Chromosomes is induced to establish dosage compensation in the epiblast (Lyon 1961).
The XCI process is initiated by the lncRNA Xist and leads to the removal of active histone marks such as histone acetylation and the accumulation of repressive histone marks such as histone H3 lysine 27 trimethylation (H3K27me3) on the inactive X Chromosome (Xi) (Kaslow and Migeon 1987; Plath et al. 2003; Silva et al. 2003; de la Cruz et al. 2005; Zhao et al. 2008; Marks et al. 2009). Moreover, recent studies on neural progenitor cells showed that, upon inactivation, the Xi undergoes conformational changes that include both attenuation of topologically associating domains (TADs) and the subsequent folding into two silenced mega-domains (Giorgetti et al. 2016). A subset of X-linked genes escapes XCI and maintains biallelic expression. These genes, termed “escapees,” bypass the suppressive effects of Xist RNA and repressive protein complexes and are located within residual TAD-like clusters on the Xi (Balaton and Brown 2016; Giorgetti et al. 2016). XCI, therefore, provides a paradigmatic example of chromosome-wide gene silencing stably maintained in somatic cells. The precise relationship between epigenetic modifications on the randomly inactivated X Chromosome and the stability and reversibility of gene silencing remains unclear.
The epigenetic memory of the Xi can be erased by the process of XCR. During development, XCR takes place in the epiblast in mouse and during the formation of female primordial germ cells in mouse and human (Mak et al. 2004; Okamoto et al. 2004; Chuva de Sousa Lopes et al. 2008). Despite its importance, much less is known about XCR compared with XCI. In contrast to XCI, XCR leads to the silencing of Xist, increased expression of antisense lncRNA Tsix, loss of repressive chromatin marks, recruitment of active chromatin modifications, and subsequent chromosome-wide gene reactivation (Pasque and Plath 2015; Talon et al. 2019). Previous studies during mouse development have shown that the reversal of iXCI is a gradual process over 24 h. XCR initiates before the loss of Xist and is partially restricted by H3K27me3, which is actively removed by KDM6A (also known as UTX) H3K27 histone demethylase in order to activate slowly reactivating genes (Borensztein et al. 2017a). However, the dynamics and mechanisms that mediate reversal of rXCI, as opposed to the reversal of iXCI, remain to be elucidated.
Reprogramming female somatic cells to iPSCs induces chromosome-wide reversal of gene silencing after rXCI. Previous studies on reprogramming to iPSCs have shown that XCR occurs late (Maherali et al. 2007), after the silencing of Xist and the activation of pluripotency markers such as NANOG and DPPA4 (Payer et al. 2013; Pasque et al. 2014; Schiebinger et al. 2019). Xist deletion did not affect these kinetics; however, its ectopic expression caused a delay in XCR, suggesting that silencing of Xist is required for XCR (Pasque et al. 2014). Additionally, inducing pluripotency by cell fusion of human female fibroblasts with mouse embryonic stem cells (ESCs) leads to partial XCR (Cantone et al. 2016). These studies suggest that there might be different levels of susceptibility of silenced X-linked genes to reactivation (Cantone et al. 2016, 2017). Nevertheless, it is still unknown whether the kinetics of reactivation during somatic reprogramming to iPSCs vary for different genes and which factors and chromatin features enable or restrict XCR.
Pluripotency has been strongly linked to XCR. The presence of two active X Chromosomes is considered a conserved hallmark of naïve pluripotency in mice and humans (Deuve and Avner 2011; Boroviak and Nichols 2017). Previous studies have established a clear link between the presence of a robust pluripotency network and stable suppression of Xist expression (Navarro et al. 2008; Donohoe et al. 2009; Silva et al. 2009; Sousa et al. 2018). However, the mechanisms linking pluripotency factors to Xist repression, and XCR, are unclear. One study proposed that pluripotency factors repress Xist by direct binding to Xist intron 1 (Navarro et al. 2008). However, deletion of this region had little effect on XCI or XCR (Minkovsky et al. 2013). As a result, it is still not known how pluripotency factors mediate XCR and biallelic X-linked gene expression. Given the importance of TFs in transcriptional regulation, one hypothesis is that pluripotency TFs may, in addition to repressing Xist via unknown regulatory regions, also directly bind X-linked genes for their transcriptional activation in the pluripotent state, but until now no evidence to lend support for such a model has been reported.
The reversibility of gene silencing might also depend on the architecture of the Xi. Escapee genes have been shown to be located outside of the two repressive mega-domains of the Xi, which allows their biallelic activity (Giorgetti et al. 2016). This raises the possibility that the location of repressed genes in 3D space might be linked to the stability of gene silencing. Yet, it remains unknown whether the location of genes on the X Chromosome, relative to suppressed compartments, is relevant for the reversal dynamics of rXCI.
In this study, we define the kinetics of chromosome-wide X-linked gene reactivation during reprogramming of mouse somatic cells to iPSCs. We ask which genomic features and epigenetic marks correlate with the timing of X-linked gene reactivation. In addition, we aim to identify the mechanisms that may enable or restrict reversal of gene silencing during cell fate conversion. We also test how functional interference with chromatin pathways influences XCR upon entry into pluripotency. This study identifies gene regulatory principles that may ensure the stability of repressed chromatin and may also facilitate stable maintenance of cellular identity. Finally, this study provides a framework for how TFs induce reversal of stable gene silencing by overcoming active chromatin barriers in order to activate transcription and erase epigenetic memory.
Results
Allele-specific transcriptional analyses during somatic cell reprogramming to induced pluripotency
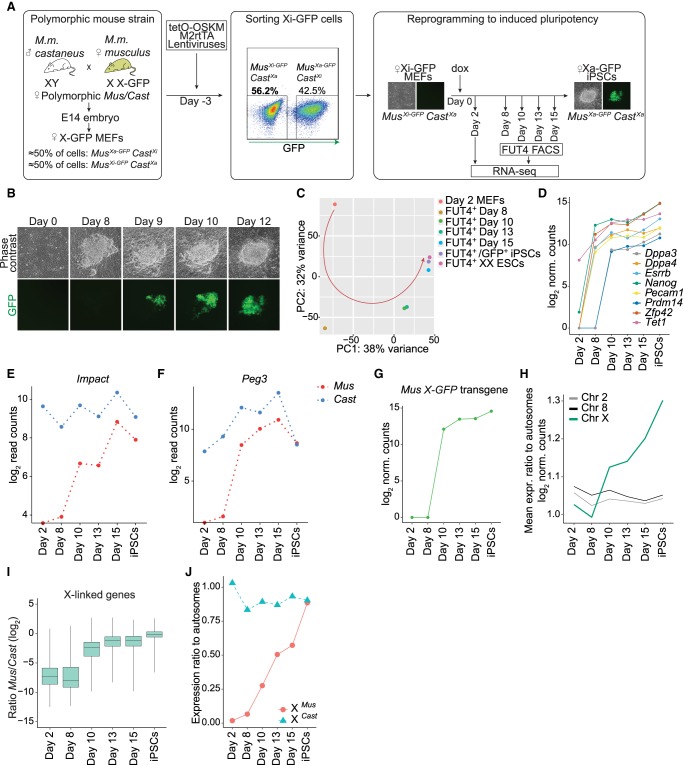
Overexpression of TFs Pou5f1 (also known as Oct4), Sox2, Klf4, and Myc (OSKM) in somatic cells leads to the induction of pluripotency and the reversal of transcriptional silencing of the Xi (Maherali et al. 2007). However, the precise timing and underlying mechanisms of chromosome-wide transcriptional activation of X-linked genes during the reversal of rXCI remain to be determined. To define the transcriptional dynamics of XCR, we established an inducible reprogramming mouse model suitable for allele-resolution transcriptome studies. We first isolated female mouse embryonic fibroblasts (MEFs) from highly polymorphic mouse strains originating from the cross between female Mus musculus musculus (Mus) with an X-GFP transgene on the X Chromosome (Pasque et al. 2011) and male Mus musculus castaneus (Cast) mice, carrying a high density of single-nucleotide polymorphisms (SNPs) spread throughout the genome (Fig. 1A). To ensure that the starting cells carry the same Xi, we used fluorescence-activated cell sorting (FACS) to specifically select only the female MEF cells with silenced X-GFP allele (GFP-negative cells) (Fig. 1A; Supplemental Fig. S1A,B). We then induced the reprogramming of Xi-GFP female MEFs into iPSCs by overexpression of OSKM. Reprogramming led to the appearance of iPSC colonies, which reactivated the Xi as judged by live imaging of GFP activation (Fig. 1B; Supplemental Fig. S1C).
Figure 1.
Allele-resolution system to study transcriptional reactivation of X-linked genes during reprogramming to pluripotency. (A) Schematic representation of the system used to trace XCR during reprogramming. (M2rtTA) Reverse tetracycline transactivator. (B) Phase contrast and fluorescent images of representative stages of reprogramming from day 0 to day 12, starting from female MusXi-GFP/CastXa MEFs to MusXa-GFP/CastXa iPSCs. GFP+ cells are increasing in number as a result of XCR. (C) Principal component analysis (PCA) of gene expression from different stages of reprogramming. Each colored dot represents a different time point: day 2 (orange), 8 (dark yellow), 10 (green), 13 (turquoise), 15 (blue), FUT4+/GFP+ iPSCs after four passages (purple), and female ESCs (pink). (D) Levels of pluripotency network gene expression (log2-transformed normalized read counts) during the time course of reprogramming. (E) Expression levels of Impact, an autosomal, paternally imprinted gene during factor-induced reprogramming. (F) As in E for Peg3. (G) X-GFP transgene expression during reprogramming (log2-transformed normalized read counts). (H) Mean expression ratio of Chromosome 2, 8, and X relative to autosomes (log2 normalized counts) during reprogramming. (I) X-linked genes expression ratio Mus/Cast (log2-transformed normalized read counts). See Methods. (J) Mean expression ratio of the MusXi allele (red) and CastXa allele to autosomes during reprogramming.
To define the kinetics of XCR with allele-resolution, we isolated, at different time points, reprogramming intermediates marked by reactivation of the cell surface marker FUT4 (also known as SSEA-1) (Supplemental Fig. S1C). FUT4 has been shown to mark cells poised for successful acquisition of the pluripotency program and XCR (Stadtfeld et al. 2008; Pasque et al. 2018; Schwarz et al. 2018). The first FUT4-positive (+) cells isolated did not show significant GFP fluorescence and then gradually reactivated GFP, whereas fully reprogrammed iPSCs were mostly GFP+ (Supplemental Fig. S1C). We applied full-transcript Smart-seq2 RNA-seq to populations of day 2 MEFs, FUT4+ reprogramming intermediates obtained at the first day that yielded sufficient number of FUT4+ cells (day 8), and subsequently day 10, day 13, and day 15 of reprogramming, as well as iPSCs and control ESCs (Supplemental Table S1). We observed gradual changes in the transcriptome of FUT4+ intermediates and found that, transcriptionally, day 15 intermediates closely resembled iPSCs and ESCs, indicating successful reprogramming (Fig. 1C).
Reprogramming to iPSCs and entrance into the pluripotency program are concomitant with the activation of key pluripotency genes (Buganim et al. 2012). Indeed, pluripotency markers such as Nanog, Esrrb, Zfp42, Pecam1, and Dppa4, as well as Tet1, were expressed in day 8 FUT4+ cells (Fig. 1D). However, Prdm14 and Dppa3 were activated only later, at day 10 (Fig. 1D). Next, we set out to verify whether this strategy allows the retention of allele-specific information by monitoring the allelic expression of imprinted genes (Impact and Peg3) during reprogramming. As expected, Impact and Peg3 were expressed exclusively from paternal alleles in MEFs (Kuroiwa et al. 1996; Hagiwara et al. 1997), validating monoallelic expression of these genes in somatic cells (Fig. 1E,F). We found that silenced Impact and Peg3 alleles became activated during reprogramming, indicating that imprints are erased during iPSC reprogramming. These results are in agreement with previous studies in iPSCs (Takikawa et al. 2013; Pasque et al. 2018). Thus, in this system, female somatic cells can be induced to reprogramming while enabling allele-resolution gene expression analyses.
XCR initiates early during entry into pluripotency
We next set out to precisely investigate when reactivation of the Xi takes place. First, we evaluated the expression of the maternally derived X-GFP allele. Transgenic GFP transcripts were first detected at day 10, together with GFP fluorescence, and following reactivation of several pluripotency TFs (Fig. 1G,D). Next, to achieve a better measurement of the erasure of Xi silencing, we calculated the mean expression ratio of genes on the X Chromosome relative to autosomes. We observed chromosome-wide XCR, reflected by a progressive increase in the X-to-autosome (X/A) gene expression ratio in FUT4+ intermediates starting at day 10 onward (Fig. 1H). In contrast, gene expression ratios for other chromosomes did not change over time (Chromosomes 2 or 8 shown as examples) (Fig. 1H). These results revealed up-regulation of gene expression from the X Chromosome during reprogramming to iPSCs. To determine if the increase in X/A gene expression ratio resulted from the reactivation of Xi rather than the up-regulation of a single active X Chromosome (Xa), we measured the average allelic expression ratios between Mus and Cast alleles on the X Chromosome throughout reprogramming (see Methods; Supplemental Tables S2, S3). Reactivation started after day 8 and was completed in late reprogramming stages, reaching, on average, equal expression levels between the two X Chromosomes in iPSCs (log2 ratio Mus/Cast median at day 13 = −1.148, day 15 = −1.143, iPSCs = −0.144) (Fig. 1I). In contrast, autosomal genes, on average, maintained similar allelic gene expression ratios throughout reprogramming, confirming increased X Chromosome dosage (Supplemental Fig. S1D). Allelic X/A expression ratios and comparing absolute gene expression levels for X Chromosomes and autosomes confirmed these findings (Fig. 1J; Supplemental Fig. S1E). These analyses are consistent with an increase in X Chromosome dosage in female iPSCs (Song et al. 2019).
Transcriptional reactivation of X-linked genes during induction of pluripotency is gradual and takes several days
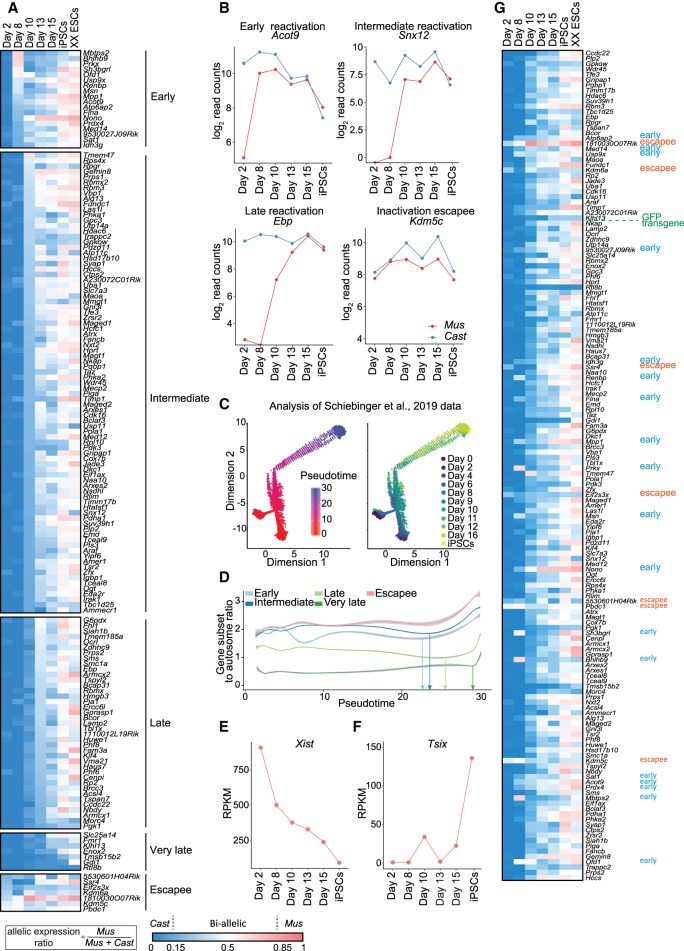
To reveal the dynamics of XCR, we generated heatmaps of X-linked allelic expression ratios over time. Specifically, we calculated the ratio of maternal-to-total reads for all informative and high-confidence X-linked genes that passed stringent SNP criteria (Supplemental Tables S2, S3). We extracted complete allelic information for 156 X-linked genes. This approach revealed that transcriptional activation of the Xi progresses with gene-specific kinetics (Fig. 2A). Several genes (11%, 18/156) were reactivated as early as day 8 of reprogramming, hereafter called “early” reactivated genes (Fig. 2A; Supplemental Fig. S2A). This contrasted with previous studies that reported reactivation of X-linked genes only much later in reprogramming, after activation of late pluripotency markers and complete Xist silencing (Pasque et al. 2014; Schiebinger et al. 2019). Other genes were delayed in transcriptional reactivation and could be segregated into different groups: with reactivation kinetics between day 8 and day 10 (“intermediate”), between day 10 and day 13 (“late”), and after day 13 (“very late”) (Fig. 2A). This is in contrast with the rapid reversal of the imprinted paternal Xi in the epiblast, which takes ∼24 h (Borensztein et al. 2017a).
Figure 2.
Different X-linked genes reactivate with different kinetics during factor-induced reprogramming to iPSCs. (A) X-linked genes ordered by reactivation timing at different time points of reprogramming within days 2 to 15 and in iPSCs and ESCs. Ratios were calculated by dividing maternal by total reads (Mus/Mus + Cast). The color gradient represents the parental origin of allelic expression, with Cast expression in blue (ratio < 0.15), Mus expression in red (ratio > 0.85), and a biallelic expression range of 0.15–0.85. Number of informative genes = 156. Genes were considered early if expressed biallelically at day 8, intermediate at day 10, late at day 13, very late at day 15, and escapees at day 2. (B) Gene expression levels (log2-transformed read counts) of representative X-linked genes for each reactivation class (early, intermediate, late, and escapee) during reprogramming. Parental origin of the allelic expression is indicated in blue for paternal origin (Cast) and in red for maternal origin (Mus). (C) Single-cell ordering along the inferred pseudotime trajectory constructed using Monocle. (Left) Single cells colored by the pseudotime; (right) single cells colored by the original collection timepoint. (D) Mean expression ratio of genes from different reactivation classes to autosomes as a function of pseudotime. The curve was derived using generalized additive model. (E) Xist expression levels (exon-to-intron ratio in reads per kilobase per million [RPKM]; see Methods) during reprogramming. (F) Tsix expression levels (exon to intron ratio RPKM, see methods) during reprogramming. (G) Gene reactivation kinetics ordered by genomic location of genes on the X Chromosome. Ratios were calculated as described in A.
By plotting the expression of selected paternal and maternal alleles, we observed that X-linked genes such as Acot9 very rapidly reactivate, illustrating early XCR events (Fig. 2B). On the other hand, Snx12 and Ebp displayed delayed activation of the previously inactive allele (Fig. 2B). We also recovered the biallelic expression of genes that escape XCI in the starting cells, including several known escapee genes (Fig. 2B; Supplemental Fig. S2A). In addition, we observed that the maternal X-GFP allele reactivated at day 10 of reprogramming, representing genes with delayed reactivation kinetics relative to early genes (Fig. 1G). An analysis of single-cell RNA-seq iPSC reprogramming data in a different reprogramming system and genetic background (Schiebinger et al. 2019) confirmed the finding that different groups of X-linked genes are activated at different times during pluripotency induction (Fig. 2C,D; Supplemental Fig. S2B,C). Our results reveal that different genes reactivate with different kinetics during XCR induced by iPSC reprogramming and that XCR is initiated earlier than previously thought and completed only after several days.
Gene reactivation in relation to Xist silencing
Because loss of the lncRNA Xist has been reported to be required for XCR (Pasque et al. 2014), we determined its expression kinetics. We observed gradual down-regulation of Xist starting from day 8, followed by the activation of its antagonist transcript Tsix in iPSCs (Fig. 2E,F). The down-regulation of Xist (Fig. 2E) suggests that the molecular machinery required to reverse the silenced state of the Xi is triggered as early as at day 8 during reprogramming but not yet completed. This is in agreement with complete Xist silencing taking place after Nanog reactivation (Pasque et al. 2014). Nevertheless, our time course analysis clearly indicated biallelic X-linked gene expression by day 8 of reprogramming (Fig. 2A,B; Supplemental Fig. S2A). Additionally, single-cell transcriptome analysis showed that reactivation of several early genes can take place in single cells still expressing high levels of Xist (Supplemental Fig. S2C–E). XCR before Xist loss has been reported in ICM (Borensztein et al. 2017a) but not yet during iPSC reprogramming. Our results suggest that XCR during reprogramming might be initiated before the complete loss of Xist transcripts. In addition, reactivation of early genes preceded the activation of the pluripotency-associated gene Prdm14, indicating that XCR initiation occurs before the activation of the entire pluripotency network (Figs. 1D, 2A). We did not detect strong biallelic expression of early genes in ESCs by RNA-FISH (Supplemental Fig. S3A,B). Altogether, these results indicate that reactivation of different X-linked genes might be mediated by different mechanisms and, for some genes, before the complete loss of Xist and/or full activation of the pluripotency network.
Gene reactivation kinetics relate to genomic and epigenomic features
We sought to identify features that help to explain the precise timing of X-linked gene reactivation. We first investigated whether there is a link between reactivation kinetics and location on the X Chromosome. To this end, we generated heat maps of allelic expression and ordered the genes according to their genomic location (Fig. 2G). Early genes were distributed throughout the chromosome. We detected two clusters of early reactivated genes. The first cluster contained Atp6ap2, Med14, and Usp9x. The second cluster contained Sat1, Acot9, and Prdx4.
The timing of transcriptional activation could not be explained by genomic distance to the Xist locus (Supplemental Fig. S4A). Moreover, there was no correlation between the timing of gene activation and the level of gene expression in ESCs (Supplemental Fig. S4A). However, the timing of reactivation was correlated with expression level from the Xa at the beginning of reprogramming (Fig. 2D; Supplemental Fig. S4A). The reactivation timing did not correlate with LINE-1 density, whereas very late genes might have a tendency to have reduced SINE1 repeats (Supplemental Fig. S4B). Next, we compared the timing of gene activation during reversal of rXCI with reactivation of iXCI (Borensztein et al. 2017a). Most of the genes that reactivate early or late were different in both systems (Supplemental Fig. S4C). We also compared the dynamics of XCR to the timing of rXCI (Marks et al. 2015). We did not observe any correlation between the timing of XCI and XCR during reprogramming (Supplemental Fig. S4C). Hence, the kinetics of XCR during reversal of rXCI are different from that of iXCI reprogramming and rXCI. Taken together, these results suggest that the reversal of rXCI differs from that of iXCI and is independent of the genomic distance of genes to the Xist locus but might be related to expression on the Xa before reprogramming.
Next, we determined whether the timing of reactivation could be associated with the presence or absence of chromatin marks on the Xi. We used allele-specific chromatin immunoprecipitation sequencing (ChIP-seq) data for H3K27me3, H3K36me3, and H3K4me3 in MEFs (Pinter et al. 2012) to determine the enrichment of chromatin marks around the transcription starting sites (TSSs) of early, intermediate, late, and very late reactivated genes. We found that on the Xi, H3K27me3 is clearly enriched on all classes of genes, but there were no clear differences between classes (Supplemental Fig. S4D). Thus, although H3K27me3 may restrict the activation of silenced genes on the Xi, it is not sufficient to explain the delay in reactivation of late genes. Additionally, on the Xi, H3K36me3 and H3K4me3 were depleted on all classes of genes. DNA methylation (Milagre et al. 2017) was also enriched on all classes of genes in MEFs (Supplemental Fig. S4E). Moreover, we did not observe significant differences in the density of CpG sites between different reactivation classes (Supplemental Fig. S4F). Altogether, this analysis revealed that different classes of genes do not appear to show significant differences in the chromatin marks examined on the Xi in MEFs. Therefore, the timing of XCR cannot be fully explained by different levels of these histone modifications on the Xi.
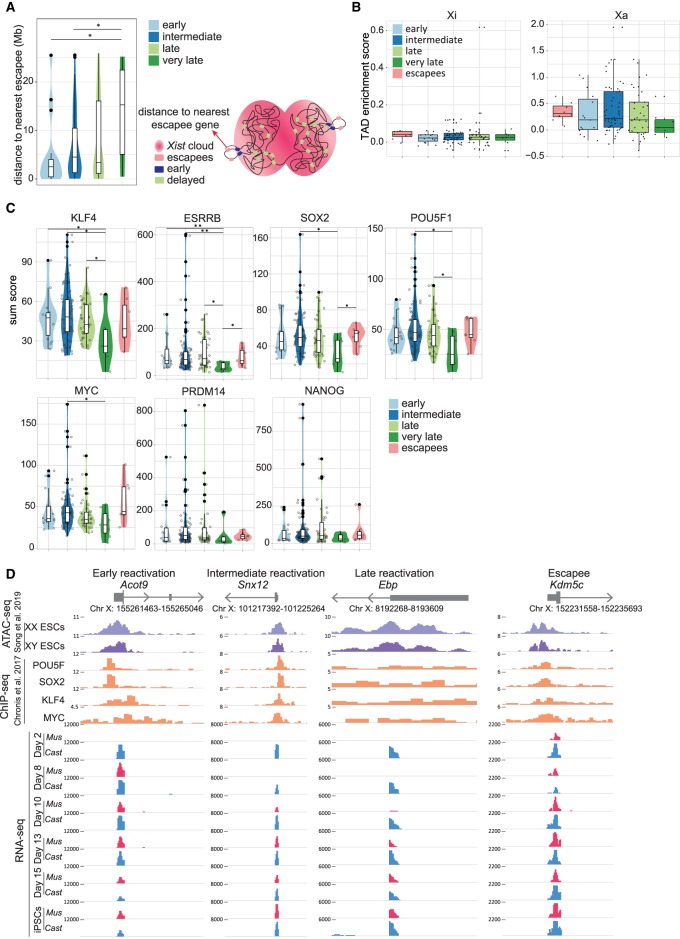
During XCI, silenced genes relocalize to the interior of the repressive Xi compartment, whereas escapee genes remain at the periphery of the Xist domain, outside of the silenced compartment occupied by the Xi. This allows them to avoid the silencing machinery (Bischoff et al. 1993; Eils et al. 1996; Chaumeil et al. 2006; Clemson et al. 2006; Chow et al. 2010; Splinter et al. 2011; Deng et al. 2015). Hence, we asked whether early reactivated genes are located closer to escapee genes, where they might reside in topologically favorable positions that facilitate their reactivation during reprogramming. To address this, we measured the average genomic distance between the different classes of genes and the nearest escapee gene. We found a clear and significant decrease in the genomic distance to nearest escapees for early and for intermediate genes compared with very late reactivated genes (respectively, P = 0.021 and P = 0.038 by Wilcoxon rank-sum test) (Fig. 3A). This trend was even more pronounced after lowering the stringency of SNP filtering criteria and thus increasing the number of detected genes (Supplemental Fig. S4G). Moreover, the two clusters of early genes each had a nearby escapee gene in the genomic sequence map (Fig. 2G). Examination of high-throughput chromosome conformation capture (Hi-C) data from MEFs (Gdula et al. 2019) did not reveal a difference in TAD strength between the different categories of genes on the Xi (Fig. 3B). Altogether, these results reveal that genes that reactivate early during iPSC reprogramming have significantly reduced genomic distance to escapee genes compared with very late genes. Overall, this suggests that the localization of a gene in 3D space relative to repressive chromatin domains may be related to the stability of gene silencing.
Figure 3.
Link between genomic and epigenomic features and XCR kinetics. (A) Violin diagram with the distance to nearest escapee (Mb) for each reactivation class (left). Schematic figure of distance to nearest escapee gene (right). The significant P-values of a Wilcoxon rank-sum test comparing the different gene reactivation classes are indicated with asterisks above the violin plots: (**) P-value = 0.001–0.01; (*) P-value = 0.01–0.05 = significant; (not significant) P-value ≥ 0.05. (B) TAD enrichment scores (with Z-score of contacts; see Methods) for genes from each reactivation class on the inactive X Chromosome (left) and active X Chromosome (right). (C) Violin plots indicating the sum score of enrichment levels of POU5F1, SOX2, KLF4, and MYC occupancy for each reactivation class (early, intermediate, late, very late, and escapees). The P-values are calculated as described above. (D) ATAC-seq signal for open chromatin in female and male ESCs; ChIP-seq signal for POU5F1, SOX2, KLF4, and MYC binding in male ESCs; and RNA-seq signal of allele resolution gene expression during reprogramming (blue for RNA-seq signal from Cast origin and red for Mus origin) for a representative gene corresponding to each reactivation class (early, intermediate, late, and escapee).
The timing of gene reactivation is linked to pluripotency TF binding
TFs are primary mediators of gene regulatory programs during development (Flavahan et al. 2017). It remains unclear how TFs mediate XCR during reprogramming and, in particular, if TFs may contribute to the differential reactivation kinetics of distinct X-linked genes. To address these questions, we set out to investigate whether differently reactivating genes have enrichment of distinct TF binding. TF binding prediction using iRegulon revealed an enrichment of the putative TF ZBTB33 (also known as KAISO) (Supplemental Fig. S4H). ZBTB33 has been implicated in the recruitment of repressive proteins; however more recently, ZBTB33 has also been associated with the recruitment of activating TFs (Blattler et al. 2013). Delayed genes were enriched for the YY1 motif (Supplemental Fig. S4H). YY1 has been implicated in recruitment of Xist RNA to the X Chromosome (Jeon and Lee 2011). We next sought to examine the enrichment levels of pluripotency TFs binding and used a previously published ChIP-seq data set with binding profiles of seven reprogramming TFs (POU5F1, SOX2, KLF4, MYC, ESRRB, PRDM14, NANOG) on the Xa in male ESCs (Chronis et al. 2017). We found that early genes showed a significantly higher enrichment of KLF4 and ESRRB binding at the TSS compared with very late reactivating genes (P = 0.046 and P = 0.009, respectively, by Wilcoxon rank-sum test) (Fig. 3C). Intermediate reactivating genes also showed a significant increase in the enrichment of KLF4, ESRRB, SOX2, POU5F1, and MYC compared with very late genes. Thus, Xi-linked genes might be targeted by pluripotency TFs for their transcriptional activation in iPSCs.
We next visualized TF enrichment at the TSS of Acot9 (early), Snx12 (intermediate), Ebp (late), and Kdm5c (escapee). In iPSCs, these genes are biallelically expressed and have accessible chromatin at their TSSs in ESCs (Fig. 3D). We found a clear enrichment of several pluripotency TFs binding at the TSS of Acot9, Snx12, and Kdm5c but not the late reactivated gene Ebp (Fig. 3D). These observations strengthen the possibility that pluripotency TFs might directly activate X-linked gene expression in iPSCs. Next, we asked whether pluripotency TFs possess the capacity to bind X-linked genes during the reprogramming process. We analyzed ChIP-seq data for SOX2 and POU5F1 in male FUT4+ reprogramming intermediates (Knaupp et al. 2017). Nevertheless, we found that several X-linked genes showed enrichment of SOX2 and POU5F1 binding at their promoters in reprogramming intermediates (Supplemental Fig. S4I). In addition, we confirmed the enrichment of POU5F1 and ESSRB binding to promoter regions of several X-linked genes at day 8 and day 15 of reprogramming by ChIP-PCR in our system (Supplemental Fig. S5A; Supplemental Table S4). We conclude that pluripotency TFs such as SOX2 and POU5F1 possess the ability to bind X-linked genes, at least on the Xa, during the reprogramming process. Altogether, pluripotency TFs may target early and intermediate genes more efficiently than late genes during the acquisition of the iPSC state.
XCR is restricted by the removal of active histone marks during reprogramming to iPSCs
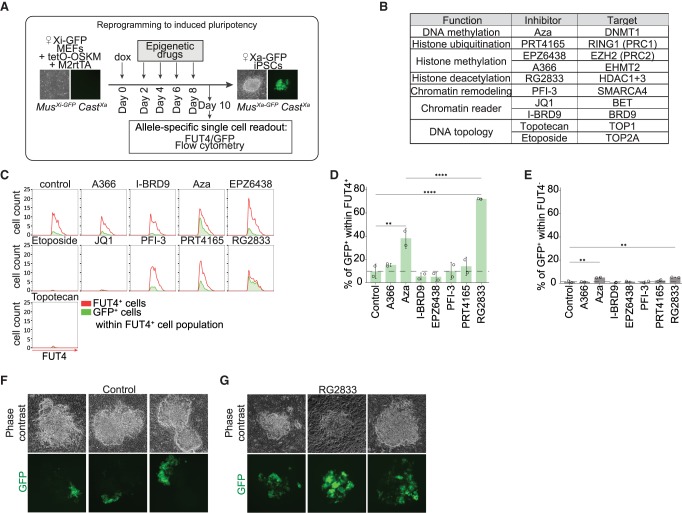
TF binding to gene regulatory elements mediates changes in chromatin structure. Hence, we aimed to identify chromatin pathways that functionally restrict or facilitate reversal of XCI during entry into the iPSC state. We used the X-GFP reporter to test whether interference with different chromatin pathways has an effect on transcriptional activation of nonearly genes (Fig. 1G). We induced reprogramming of Xi-GFP female MEFs into iPSCs and performed an epigenetic drug screen in order to identify chromatin regulators that might act as barriers or mediators of XCR (Fig. 4A). We included drugs that target several factors involved in chromatin regulation, such as DNA methylation, repressive histone modifications, as well as drugs involved in the regulation of DNA topology and chromatin remodeling (Fig. 4B; Bhatnagar et al. 2014; Minkovsky et al. 2015; Carrette et al. 2018). We recorded, after 10 d of reprogramming and continuous treatment with epigenetic drugs, the proportion of cells that activated Fut4 and GFP expression. This enabled us to define the effects of drug treatment on XCR in cells undergoing successful reprogramming (FUT4+ cells) versus nonproductive reprogramming intermediates (FUT4−).
Figure 4.
Histone deacetylases restrict XCR during reprogramming to iPSCs. (A) A schematic representing experimental design of epigenetic drug inhibitor screen during reprogramming. (B) Inhibitors added individually at different time points during reprogramming with their function, name, and target molecule(s). (C) Histograms representing the flow cytometry analysis of the proportion of GFP+ cells within FUT4+ cells for each individual inhibitor at day 10 of reprogramming and vehicle control. The whole FUT4+ cell population is represented in red; population of GFP+ cells within FUT4+ cells, in green. (D) Proportion of GFP+ cells within the FUT4+ cell population for each individual inhibitor. The P-values of one-way ANOVA with Dunnett's multiple comparisons test comparing levels of GFP+ cells after treatment with each individual inhibitor and vehicle control are indicated with asterisks above the violin plots. The P-values of one-way ANOVA with Sidak's multiple comparisons test comparing levels of GFP+ cells treated with RG2833 and Aza are indicated with asterisks above the boxplot. (****) P-value < 0.0001; (***) P-value = 0.0001–0.001; (**) P-value = 0.001–0.01; (*) P-value = 0.01–0.05 = significant; (not significant) P-value ≥ 0.05. Error bars, SD. n = 2. (E) Proportion of GFP+ cells within FUT4− cell population for each individual inhibitor. The P-values of one-way ANOVA with Dunnett's multiple comparisons test comparing levels of GFP− cells after treatment with each individual drug inhibitor and vehicle control are indicated with asterisks above the boxplot. Error bars, SD. n = 2. (F) Phase contrast and fluorescent images of three representative colonies at day 10 of reprogramming after treatment with DMSO control during epigenetic drug screening. (G) Phase contrast and fluorescent images of three representative colonies at day 10 of reprogramming after treatment with RG2833, an inhibitor of HDAC1/3, during epigenetic drug screening.
We found that treatment with RG2833 or Aza both led to a significant increase in XCR rate within FUT4+ cells relative to the untreated control (Fig. 4C,D; Supplemental Fig. S6A). RG2833 is an HDAC3 and HDAC1 inhibitor, whereas Aza inhibits the maintenance DNA methyltransferase, DNMT1. The proportion of GFP+ cells after treatment with RG2833 and Aza was much higher in FUT4+ than in FUT4− cells, distinguishing effects of XCR in reprogramming versus loss of XCI maintenance in somatic cells (Fig. 4E). The effects of RG2833 were also clearly visible when following the reactivation of X-GFP transgene in iPSCs colonies after 10 d of reprogramming by microscopy (Fig. 4F,G). Western blot analysis confirmed the activity of the drug (Supplemental Fig. S6B). The increase in XCR upon Aza or RG2833 treatment was not the consequence of an increase in reprogramming efficiency, because we did not detect significant differences in the number of FUT4+ cells between conditions (Supplemental Fig. S6C). These results corroborated a previous study in which DNA methylation was reported to oppose XCR during the generation of iPSCs (Pasque et al. 2014). Thus, our screen recovered a known barrier to XCR but also identified HDACs as potential barriers to XCR during pluripotency induction. Recent work from Żylicz et al. (2019) revealed that HDAC3 activation on the X Chromosome initiates transcriptional silencing during initiation of XCI. Therefore, HDAC3 and/or HDAC1 may deacetylate silenced genes to oppose transcriptional activation on the Xi during pluripotency induction, acting as a barrier to XCR during iPSC reprogramming.
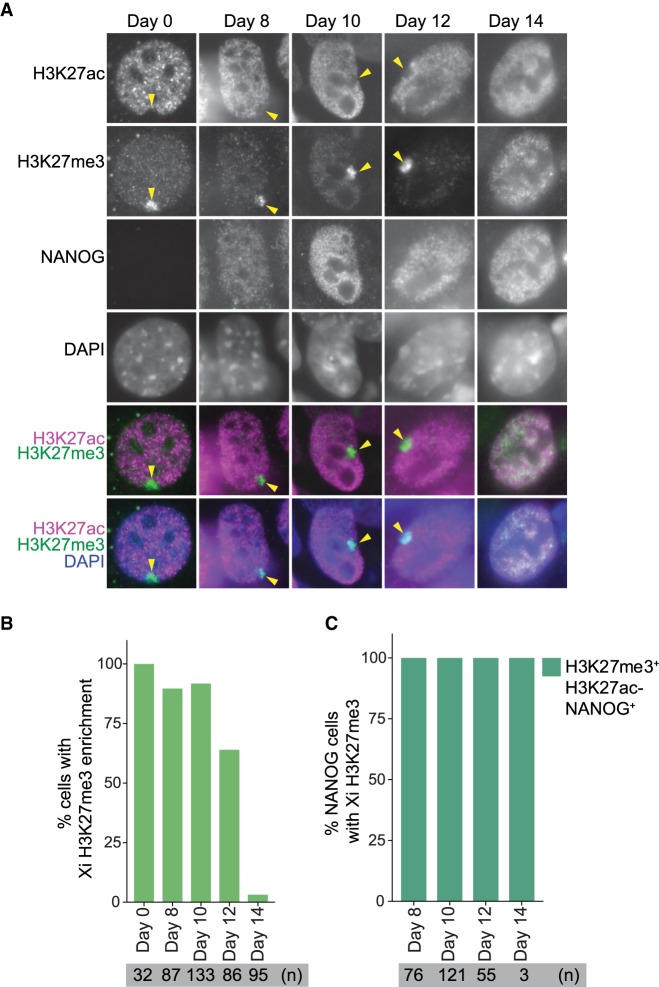
Chromatin acetylation on the Xi is prevented until late reprogramming stages
To explore the dynamics of histone acetylation of the Xi during reprogramming to induced pluripotency, we performed time course immunofluorescence analysis throughout reprogramming for H3K27ac, H3K27me3, and NANOG. In the starting MEFs, we found that the Xi was depleted of H3K27ac (Fig. 5A). During reprogramming, the hypoacetylated state of the Xi was maintained in NANOG+ cells when H3K27me3 enrichment on the Xi was still present (Fig. 5A–C; Supplemental Fig. S7A). Established iPSCs lost H3K27me3 enrichment and gained H3K27ac, in agreement with late global acetylation of the Xi during XCR. These results suggest that histone hypoacetylation on the Xi persists until the late stages of XCR and may be maintained by the constant action of histone deacetylases. We conclude that changes in chromatin states during XCR involve chromatin acetylation at a time when TFs may engage with chromatin of the Xi, leading to transcriptional activation. Altogether, our results provide a broader understanding of how TFs induce dynamic reversal of stable transcriptional silencing and overcome active barriers to transcriptional activation.
Figure 5.
Hypoacetylation of Xi persists until late stages of reprogramming. (A) Immunofluorescence analysis for H3K27ac (magenta in merge), H3K27me3 (green), and NANOG at different stages of reprogramming. DAPI staining (blue) marks nuclei. Xi H3K27me3 enrichment is marked with an arrowhead. (B) Proportion of cells with H3K27me3 enrichment during reprogramming. (C) Proportion of NANOG+ cells with H3K27me3 enrichment and H3K27 hypoacetylation during reprogramming.
Discussion
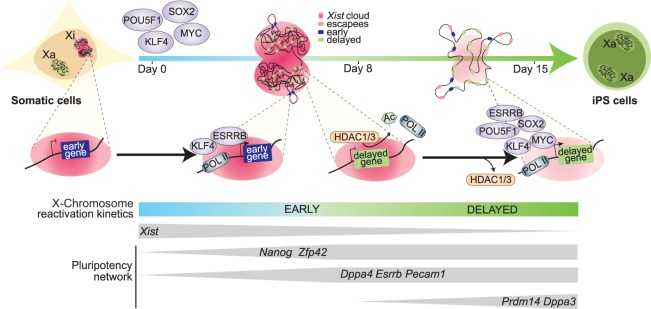
Reversal of epigenetic memory on the Xi during pluripotency induction is a paradigm for studying transcriptional activation of silenced chromatin. However, chromosome-wide allelic gene activation has not yet been completely understood. Furthermore, the relationship between pluripotency TFs and reversal of gene silencing has remained largely underexplored. We used transcriptomic and epigenomic approaches to define allele-resolution maps of chromosome-wide gene activation during reprogramming to iPSCs. This allowed us to focus on the exploration of the progressive nature of XCI reversal in reprogramming, in which different genes seem to show distinct levels of silencing stability and, as a result, reactivate with different kinetics (Fig. 6).
Figure 6.
Dynamics of XCR during somatic cell reprogramming to iPSCs. Timeline of XCR during reprogramming to induced pluripotency with factors that might function to contribute to the dynamic onset of genes reactivation from the inactive X Chromosome.
XCR is rapidly initiated during entry into pluripotency
We found that the machinery to induce silencing reversal starts to act early during the onset of reprogramming to iPSCs. The lncRNA Xist is down-regulated as cells enter the pluripotent state, when TFs such as Nanog become expressed. The early initiation of XCR is in contrast with previous reports that XCR takes place only very late during reprogramming to iPSCs (Pasque et al. 2014; Schiebinger et al. 2019). The differences might be because of the use of allele-specific RNA-seq compared with the previously used RNA-FISH and single-cell RNA-seq without allele specificity. Many studies indicated a role of pluripotency TFs in XCR induction (Minkovsky et al. 2013; Payer et al. 2013; Sousa et al. 2018). A robust naive pluripotency network in mouse ESCs has been shown to be required for complete suppression of Xist (Sousa et al. 2018). However, the mechanism through which these factors silence Xist is still unclear (Minkovsky et al. 2013). Our data suggest that pluripotency TFs may also be able to directly bind to regulatory elements of many X-linked genes during reprogramming and in ESCs. Although there might be different mechanisms at play, it is possible that pluripotency TFs bind in previously inaccessible chromatin, thereby opening up chromatin on the Xi, or suppress the silencing effect of the nuclear lamina, where the Xi is usually localized (Pollex and Heard 2019). Allele-resolution chromatin accessibility analyses could help further address these questions. The precise molecular mechanisms by which Xist down-regulation and XCR induction take place remain to be further explored. Additionally, the initiation of XCR during reprogramming is challenging to capture owing to the degree of heterogeneity associated with factor-induced iPSC reprogramming. To minimize the variability, our study provides information from FUT4 sorted cells that have been shown as a robust marker of cells poised to reprogram successfully (Schwarz et al. 2018). Moreover, single-cell analyses confirmed that different groups of X-linked genes are activated at different times. Furthermore, additional single-cell analyses might allow to pinpoint the most upstream events of XCR.
A subset of X-linked genes reactivates before pluripotency is fully established
In line with early XCR initiation, we found that a subset of genes can reactivate early, before the acquisition of a complete pluripotency network and perhaps even before complete loss of Xist transcripts, although this last conclusion may be owing to the sensitivity of the assays used. Nevertheless, our results are in agreement with studies of iXCI reactivation (Borensztein et al. 2017a). However, we found that reactivation of rXCI differs from that of iXCI in terms of the timing necessary to complete reactivation as well as the order of reactivating genes. This may be explained by different starting epigenetic states. Nevertheless, early reactivation in both cases suggests a specific mechanism that allows genes to reactivate rapidly. We hypothesize that the 3D architecture of the Xi might play an important role (Giorgetti et al. 2016). Analysis of Hi-C data did not support the idea that differences in TAD strength on the Xi help to explain XCR kinetics. However, our analysis may have been limited by sequencing depth. Moreover, it remains possible that genes closer to borders of the silent compartment might be more easily unfolded, becoming exposed to TFs during XCR. Indeed, we show that the linear genomic distance of early reactivating genes to escapees is shorter, and escapee genes are known to reside outside of megadomains (Jégu et al. 2017). This opens the possibility that the order of conformational changes during reprogramming might be linked with reactivation timing, which merits further exploration.
Many X-linked genes reactivate with a significant delay
Our data also show the protracted nature of XCR for most X-linked genes. After reactivation of early genes, at least 1 wk is required to complete activation of other loci. This suggests a possible mechanism in which suppressed chromatin has to be first remodeled in order to allow subsequent gene activation. We show, however, that the accumulation of histone marks such as H3K27me3 on the Xi of MEFs is insufficient to explain the delay in reactivation. Neither does the enrichment of active marks such as H3K4me3 and H3K36me3 on Xi explain early reactivation. This is in contrast with reactivation of iXCI, in which H3K27me3 enrichment on Xi had an impact on reactivation dynamics (Borensztein et al. 2017a). It is possible that during reactivation of rXCI, other repressive marks such as macroH2A or H2AK119ub are at play. Our study indicates that pluripotency TFs that accumulate during reprogramming to induced pluripotency might play a role in dictating gene activation kinetics. We found that on the Xa in ESCs, several pluripotency factors differentially bind to early and nonearly reactivating genes. This suggests that genes reactivating with the highest degree of delay might be less accessible to those TFs. The possible reasons for that, as discussed above, might be linked to 3D topology of the X Chromosome, chromatin accessibility, or other repressive features that obstruct TF binding sites. Allele-resolution chromatin accessibility and chromatin immunoprecipitation analyses during reprogramming will be needed for further clarification.
Histone deacetylases oppose rapid XCR
Here, using epigenetic drug screening with a GFP reporter for activation of delayed genes, we identified histone deacetylases as barriers to reactivation that contribute to the delayed nature of XCR. It has recently been shown that histone deacetylation is the most influencing suppressive chromatin mechanism during initiation of XCI (Żylicz et al. 2019), but the role of histone deacetylation in XCR during iPSC reprogramming was not known. We show that the inhibition of HDACs accelerates reactivation and thus highlights an important mechanism of silencing stability. The link between transcriptional activity and histone acetylation has been very well established and shown to be conserved throughout a wide range of species (Peserico and Simone 2011). In the context of XCI, synergistic effects of Xist coating, methylation of CpG islands, and hypoacetylation of histone H3 and H4 have been reported as features associated with the establishment and maintenance of the Xi in somatic cells (Csankovszki et al. 2001). Without the induction of pluripotency, however, inhibition of HDACs does not lead to complete reactivation, likely because DNA methylation and Xist still stabilize Xi (Carrette et al. 2018). It is possible that after XCI, HDACs remain bound to safeguard Xi silencing. Histone acetylation and deacetylation establish a regulatory balance, which changes depending on gene activity (Peserico and Simone 2011). We show here that the hypoacetylated state of the Xi persists until very late stages of reprogramming. Interesting remaining questions are whether HDACs continuously deacetylate the Xi, whether histone acetyltransferases are excluded from the Xi, and what are the precise dynamics of histone acetylation on the Xi during reprogramming.
In summary, our findings reveal relationships between genomic and epigenomic features and the reversal of gene silencing during cell fate reprogramming. Our results open up avenues for a better understanding of allelic gene regulation and epigenetic reprogramming.
Methods
Detailed information on cell culture conditions, immunofluorescence, flow cytometry, RNA-seq reads processing and filtering, Hi-C data processing, chromatin immunoprecipitation, qPCR, western blot, imaging and data processing, and visualization are provided in Supplemental Texts S1 through S12 and Supplemental Tables S2 and S3.
Cell lines
X-GFP reporter MEFs were derived from female E13.5 embryos hemizygous for the X-GFP transgenic allele (Hadjantonakis et al. 2001). These embryos resulted from the cross between female X-GFP Mus and male Cast lines (Pasque et al. 2011). Stem cell cassette (STEMCCA) MEFs refers to female MEFs carrying a single polycistronic reprogramming cassette with four reprogramming factors OSKM (referred to as tetO-OSKM) located in the Col1A locus together with a single copy of reverse tetracycline transactivator M2rtTA in the Rosa26 locus. iPSC controls were derived from reprogramming experiments in this study (described below). ESC-like colonies were picked at day 14 of reprogramming and cultured for four passages.
RNA-seq library preparation
RNA-seq library was prepared from low RNA input using an adapted Smart-seq2 protocol (Picelli et al. 2014). Libraries were pooled and sequenced at the VIB Nucleomics Core on a NextSeq 500 (Illumina) sequencer in high-output paired-end mode (2 × 75 bp), yielding on average 66 million reads per sample (Supplemental Table S1). Details are given in Supplemental Text S13.
Reactivation dynamics
For the calculation of allelic ratio in Figure 1I, Figure 2, A and G, and Supplemental Figure S2A, two additional filtering criteria were imposed. First, for each gene at a given time point, the sum of reads from both alleles had to be at least 40. Second, to unambiguously determine reactivation timing, only the genes that passed previous criterion across all time points were included. In Figure 1I, the ratio was calculated as the log2 ratio of Mus allele to Cast allele calculated for each gene. In Figure 2, A and G, the allelic ratio is represented as ratio of Mus to total: (Mus/(Mus + Cast). The heatmap was generated in the online software Morpheus (https://software.broadinstitute.org/morpheus).
Single-cell RNA-seq data analysis
Normalized expression counts for 65.781 cells, obtained by Schiebinger et al. (2019) were downloaded (GSE106340). Pseudotime (monocle, version 2.10.0 [Trapnell et al. 2014; Qiu et al. 2017a,b]) analysis was performed on the filtered data set. Further details are given in Supplemental Text S14.
Allele-specific ChIP-seq data analysis
For TF and histone mark enrichment analyses in regulatory regions of genes with different timing of reactivation, published ChIP-seq data were reanalyzed. The data sets used include the following: GSE90893 and GSE25409 (Ma et al. 2011; Chronis et al. 2017), pluripotency factors and chromatin marks in ESCs and MEFs; GSE GSE36905 (Pinter et al. 2012), histone marks in MEFs with allele-resolution. Raw ChIP-seq data were analyzed using the ChIP-seq pipeline from the Kundaje laboratory (version 0.3.0; https://github.com/kundajelab/atac_dnase_pipelines). Details are given in Supplemental Text S15.
Correlation with LINE-1 and SINE
The table containing genomic coordinates of repeat elements (LINE-1 and SINE) was downloaded from the UCSC Table Browser; from the remasker table (“rmsk”) for the mm10 reference genome assembly. The density was calculated in windows of 200 kb centered around the TSSs of genes from the different reactivation timing categories.
DNA methylation analysis
For DNA methylation analysis with BS-seq, we used a published data set (Milagre et al. 2017), under the Gene Expression Omnibus (GEO) accession number GSE69823, containing Bismarck bedGraph coverage reports. SeqMonk (v1.45.1; https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) was used to pool three replicates to obtain threefold coverage. Next, probes were generated to span windows of 2000 bp upstream of and 500 bp downstream from the TSS of each gene. The average methylation levels per probe were then used to analyze methylation over genes from each reactivation kinetics category.
CpG site densities
The locations of CpG sites was extracted from the full-genome sequences for Mus musculus (mouse) as provided by UCSC (mm10, December 2011) and stored in Biostrings objects (https://www.bioconductor.org/packages/release/bioc/html/Biostrings.html). The density of CpG sites was measured within the window of ±5 kb from the TSSs of genes from different reactivation classes.
Motif discovery analysis
iRegulon was used for cis-regulatory sequence analysis (Janky et al. 2014). With a database with a motif collection of 9713 position weight matrices (PWM) and a track collection of 1120 ChIP-seq tracks, this tool was used to search for enriched TFs motifs in a putative regulatory region of 10 kb centered around the TSSs (−5/+5 kb relative to the TSS) of X-linked genes in early, intermediate, late, and very late reactivation kinetics. Delayed genes (intermediate, late, and very late) were pooled together. In addition to overrepresented motifs, iRegulon was also used for TF binding prediction (motif2TF algorithm). For TF prediction, the maximum false-discovery rate (FDR) on motif similarity used was FDR < 0.001.
Epidrug screen
Inhibitors were obtained from Selleckchem or Tocris and added to mouse ESC medium in concentrations as previously described (Supplemental Table S5; Carrette et al. 2018). Cells were treated with inhibitors starting from day 2 during reprogramming, and inhibitors were refreshed every other day by media change.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE126229.
Supplementary Material
Acknowledgments
We apologize to the authors we could not cite owing to space constraint. We thank Edith Heard, Maud Borensztein, Susana Chuva de Sousa Lopes, Hendrik Marks, Bernard Thienpont, Bernhard Payer, and Mitchell Guttman for discussions and support. We thank Jeannie T. Lee, Yevgeniya Li, and Ruslan Sadreyev for sharing ChIP-seq data. We thank the FACS Core KU Leuven/UZ Leuven, Genomics Core and KU Leuven Mouse facility, VIB Nucleomics Core, VIB/KU Leuven, and Leuven Stem Cell Institute. This work was supported by The Research Foundation–Flanders (FWO; Odysseus return grant G0F7716N to V.P.), the KU Leuven Research Fund (BOFZAP starting grant StG/15/021BF to V.P., C1 grant C14/16/077 to V.P., and project financing), a 12T1217N project by the Research Foundation–Flanders (FWO), and the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 665501, FWO PhD fellowship to A.J. (1158318N), and FWO-SB PhD fellowship to I.T. (1S72719N).
Author contributions: A.J., I.T., and V.P. designed experiments; A.J., I.T., J.S., N.D.G., C.P., L.V., F.R., O.M., and G.B. performed experiments; AJ., J.C., and S.C. analyzed sequencing data with input from I.T., S.K.T., and F.R. Writing was done by A.J., I.T., and V.P. with input from all authors. Funding was secured by V.P. and J.-C.M. Supervision was done by V.P.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.249706.119.
Freely available online through the Genome Research Open Access option.
References
- Balaton BP, Brown CJ. 2016. Escape artists of the X chromosome. Trends Genet 32: 348–359. 10.1016/j.tig.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Bhatnagar S, Zhu X, Ou J, Lin L, Chamberlain L, Zhu LJ, Wajapeyee N, Green MR. 2014. Genetic and pharmacological reactivation of the mammalian inactive X chromosome. Proc Natl Acad Sci 111: 12591–12598. 10.1073/pnas.1413620111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff A, Albers J, Kharboush I, Stelzer E, Cremer T, Cremer C. 1993. Differences of size and shape of active and inactive X-chromosome domains in human amniotic fluid cell nuclei. Microsc Res Tech 25: 68–77. 10.1002/jemt.1070250110 [DOI] [PubMed] [Google Scholar]
- Blattler A, Yao L, Wang Y, Ye Z, Jin VX, Farnham PJ. 2013. ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenetics Chromatin 6: 13 10.1186/1756-8935-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borensztein M, Okamoto I, Syx L, Guilbaud G, Picard C, Ancelin K, Galupa R, Diabangouaya P, Servant N, Barillot E, et al. 2017a. Contribution of epigenetic landscapes and transcription factors to X-chromosome reactivation in the inner cell mass. Nat Commun 8: 1297 10.1038/s41467-017-01415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borensztein M, Syx L, Ancelin K, Diabangouaya P, Picard C, Liu T, Liang JB, Vassilev I, Galupa R, Servant N, et al. 2017b. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol 24: 226–233. 10.1038/nsmb.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Nichols J. 2017. Primate embryogenesis predicts the hallmarks of human naïve pluripotency. Development 144: 175–186. 10.1242/dev.145177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. 2012. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150: 1209–1222. 10.1016/j.cell.2012.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantone I, Fisher AG. 2017. Human X chromosome inactivation and reactivation: implications for cell reprogramming and disease. Philos Trans R Soc Lond B Biol Sci 372: 20160358 10.1098/rstb.2016.0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantone I, Bagci H, Dormann D, Dharmalingam G, Nesterova T, Brockdorff N, Rougeulle C, Vallot C, Heard E, Chaligne R, et al. 2016. Ordered chromatin changes and human X chromosome reactivation by cell fusion-mediated pluripotent reprogramming. Nat Commun 7: 12354 10.1038/ncomms12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantone I, Dharmalingam G, Chan YW, Kohler AC, Lenhard B, Merkenschlager M, Fisher AG. 2017. Allele-specific analysis of cell fusion-mediated pluripotent reprograming reveals distinct and predictive susceptibilities of human X-linked genes to reactivation. Genome Biol 18: 2 10.1186/s13059-016-1136-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrette LLG, Wang CY, Wei C, Press W, Ma W, Kelleher RJ III, Lee JT. 2018. A mixed modality approach towards Xi reactivation for Rett syndrome and other X-linked disorders. Proc Natl Acad Sci 115: E668–E675. 10.1073/pnas.1715124115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev 20: 2223–2237. 10.1101/gad.380906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141: 956–969. 10.1016/j.cell.2010.04.042 [DOI] [PubMed] [Google Scholar]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K. 2017. Cooperative binding of transcription factors orchestrates reprogramming. Cell 168: 442–459.e20. 10.1016/j.cell.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes SM, Hayashi K, Shovlin TC, Mifsud W, Surani MA, McLaren A. 2008. X chromosome activity in mouse XX primordial germ cells. PLoS Genet 4: e30 10.1371/journal.pgen.0040030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. 2006. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci 103: 7688–7693. 10.1073/pnas.0601069103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. 2001. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol 153: 773–784. 10.1083/jcb.153.4.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz CC, Fang J, Plath K, Worringer KA, Nusinow DA, Zhang Y, Panning B. 2005. Developmental regulation of Suz12 localization. Chromosoma 114: 183–192. 10.1007/s00412-005-0008-6 [DOI] [PubMed] [Google Scholar]
- Deng X, Ma W, Ramani V, Hill A, Yang F, Ay F, Berletch JB, Blau CA, Shendure J, Duan Z, et al. 2015. Bipartite structure of the inactive mouse X chromosome. Genome Biol 16: 152 10.1186/s13059-015-0728-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuve JL, Avner P. 2011. The coupling of X-chromosome inactivation to pluripotency. Annu Rev Cell Dev Biol 27: 611–629. 10.1146/annurev-cellbio-092910-154020 [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. 2009. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature 460: 128–132. 10.1038/nature08098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eils R, Dietzel S, Bertin E, Schrock E, Speicher MR, Ried T, Robert-Nicoud M, Cremer C, Cremer T. 1996. Three-dimensional reconstruction of painted human interphase chromosomes: Active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J Cell Biol 135: 1427–1440. 10.1083/jcb.135.6.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, Bernstein BE. 2017. Epigenetic plasticity and the hallmarks of cancer. Science 357: eaal2380 10.1126/science.aal2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galupa R, Heard E. 2018. X-Chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu Rev Genet 52: 535–566. 10.1146/annurev-genet-120116-024611 [DOI] [PubMed] [Google Scholar]
- Gdula MR, Nesterova TB, Pintacuda G, Godwin J, Zhan Y, Ozadam H, McClellan M, Moralli D, Krueger F, Green CM, et al. 2019. The non-canonical SMC protein SmcHD1 antagonises TAD formation and compartmentalisation on the inactive X chromosome. Nat Commun 10: 30 10.1038/s41467-018-07907-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Marion-Poll L, Katoh K, Heard E. 2016. Random monoallelic expression of genes on autosomes: parallels with X-chromosome inactivation. Semin Cell Dev Biol 56: 100–110. 10.1016/j.semcdb.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535: 575–579. 10.1038/nature18589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Cox LL, Tam PP, Nagy A. 2001. An X-linked GFP transgene reveals unexpected paternal X-chromosome activity in trophoblastic giant cells of the mouse placenta. Genesis 29: 133–140. 10.1002/gene.1016 [DOI] [PubMed] [Google Scholar]
- Hagiwara Y, Hirai M, Nishiyama K, Kanazawa I, Ueda T, Sakaki Y, Ito T. 1997. Screening for imprinted genes by allelic message display: identification of a paternally expressed gene Impact on mouse chromosome 18. Proc Natl Acad Sci 94: 9249–9254. 10.1073/pnas.94.17.9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiszewski A, Song J, Vanheer L, De Geest N, Pasque V. 2018. Dynamics of DNA methylation reprogramming influenced by X chromosome dosage in induced pluripotent stem cells. Epigenet Insights 11: 2516865718802931 10.1177/2516865718802931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janky R, Verfaillie A, Imrichová H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, et al. 2014. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 10: e1003731 10.1371/journal.pcbi.1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jégu T, Aeby E, Lee JT. 2017. The X chromosome in space. Nat Rev Genet 18: 377–389. 10.1038/nrg.2017.17 [DOI] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. 2011. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146: 119–133. 10.1016/j.cell.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslow DC, Migeon BR. 1987. DNA methylation stabilizes X chromosome inactivation in eutherians but not in marsupials: evidence for multistep maintenance of mammalian X dosage compensation. Proc Natl Acad Sci 84: 6210–6214. 10.1073/pnas.84.17.6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaupp AS, Buckberry S, Pflueger J, Lim SM, Ford E, Larcombe MR, Rossello FJ, de Mendoza A, Alaei S, Firas J, et al. 2017. Transient and permanent reconfiguration of chromatin and transcription factor occupancy drive reprogramming. Cell Stem Cell 21: 834–845.e6. 10.1016/j.stem.2017.11.007 [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li LL, Tada M, Suzuki R, Yokoyama M, Shiroishi T, Wakana S, et al. 1996. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nat Genet 12: 186–190. 10.1038/ng0296-186 [DOI] [PubMed] [Google Scholar]
- Lyon MF. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. 10.1038/190372a0 [DOI] [PubMed] [Google Scholar]
- Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. 2011. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat Struct Mol Biol 18: 120–127. 10.1038/nsmb.2000 [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. 2007. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1: 55–70. 10.1016/j.stem.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303: 666–669. 10.1126/science.1092674 [DOI] [PubMed] [Google Scholar]
- Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, Stunnenberg HG. 2009. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res 19: 1361–1373. 10.1101/gr.092643.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kerstens HH, Barakat TS, Splinter E, Dirks RA, van Mierlo G, Joshi O, Wang SY, Babak T, Albers CA, et al. 2015. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol 16: 149 10.1186/s13059-015-0698-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milagre I, Stubbs TM, King MR, Spindel J, Santos F, Krueger F, Bachman M, Segonds-Pichon A, Balasubramanian S, Andrews SR, et al. 2017. Gender differences in global but not targeted demethylation in iPSC reprogramming. Cell Rep 18: 1079–1089. 10.1016/j.celrep.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkovsky A, Barakat TS, Sellami N, Chin MH, Gunhanlar N, Gribnau J, Plath K. 2013. The pluripotency factor-bound intron 1 of Xist is dispensable for X chromosome inactivation and reactivation in vitro and in vivo. Cell Rep 3: 905–918. 10.1016/j.celrep.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkovsky A, Sahakyan A, Bonora G, Damoiseaux R, Dimitrova E, Rubbi L, Pellegrini M, Radu CG, Plath K. 2015. A high-throughput screen of inactive X chromosome reactivation identifies the enhancement of DNA demethylation by 5-aza-2′-dC upon inhibition of ribonucleotide reductase. Epigenetics Chromatin 8: 42 10.1186/s13072-015-0034-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. 2008. Molecular coupling of Xist regulation and pluripotency. Science 321: 1693–1695. 10.1126/science.1160952 [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303: 644–649. 10.1126/science.1092727 [DOI] [PubMed] [Google Scholar]
- Pasque V, Plath K. 2015. X chromosome reactivation in reprogramming and in development. Curr Opin Cell Biol 37: 75–83. 10.1016/j.ceb.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V, Gillich A, Garrett N, Gurdon JB. 2011. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J 30: 2373–2387. 10.1038/emboj.2011.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V, Tchieu J, Karnik R, Uyeda M, Sadhu Dimashkie A, Case D, Papp B, Bonora G, Patel S, Ho R, et al. 2014. X chromosome reactivation dynamics reveal stages of reprogramming to pluripotency. Cell 159: 1681–1697. 10.1016/j.cell.2014.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V, Karnik R, Chronis C, Petrella P, Langerman J, Bonora G, Song J, Vanheer L, Sadhu Dimashkie A, Meissner A, et al. 2018. X chromosome dosage influences DNA methylation dynamics during reprogramming to mouse iPSCs. Stem Cell Reports 10: 1537–1550. 10.1016/j.stemcr.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Rosenberg M, Yamaji M, Yabuta Y, Koyanagi-Aoi M, Hayashi K, Yamanaka S, Saitou M, Lee JT. 2013. Tsix RNA and the germline factor, PRDM14, link X reactivation and stem cell reprogramming. Mol Cell 52: 805–818. 10.1016/j.molcel.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peserico A, Simone C. 2011. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol 2011: 371832 10.1155/2011/371832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9: 171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Pinter SF, Sadreyev RI, Yildirim E, Jeon Y, Ohsumi TK, Borowsky M, Lee JT. 2012. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res 22: 1864–1876. 10.1101/gr.133751.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135. 10.1126/science.1084274 [DOI] [PubMed] [Google Scholar]
- Pollex T, Heard E. 2019. Nuclear positioning and pairing of X-chromosome inactivation centers are not primary determinants during initiation of random X-inactivation. Nat Genet 51: 285–295. 10.1038/s41588-018-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlmayr A, Wutz A. 2017. Insights into the establishment of chromatin states in pluripotent cells from studies of X inactivation. J Mol Biol 429: 1521–1531. 10.1016/j.jmb.2017.03.013 [DOI] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. 2017a. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14: 309–315. 10.1038/nmeth.4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. 2017b. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14: 979–982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert Finestra T, Gribnau J. 2017. X chromosome inactivation: silencing, topology and reactivation. Curr Opin Cell Biol 46: 54–61. 10.1016/j.ceb.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Sado T, Brockdorff N. 2013. Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos Trans R Soc Lond B Biol Sci 368: 20110325 10.1098/rstb.2011.0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, Gould J, Liu S, Lin S, Berube P, et al. 2019. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176: 928–943.e22. 10.1016/j.cell.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz BA, Cetinbas M, Clement K, Walsh RM, Cheloufi S, Gu H, Langkabel J, Kamiya A, Schorle H, Meissner A, et al. 2018. Prospective isolation of poised iPSC intermediates reveals principles of cellular reprogramming. Cell Stem Cell 23: 289–305.e5. 10.1016/j.stem.2018.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 Polycomb group complexes. Dev Cell 4: 481–495. 10.1016/S1534-5807(03)00068-6 [DOI] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. 2009. Nanog is the gateway to the pluripotent ground state. Cell 138: 722–737. 10.1016/j.cell.2009.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Janiszewski A, De Geest N, Vanheer L, Talon I, El Bakkali M, Oh T, Pasque V. 2019. X-chromosome dosage modulates multiple molecular and cellular properties of mouse pluripotent stem cells independently of global DNA methylation levels. Stem Cell Reports 12: 333–350. 10.1016/j.stemcr.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa EJ, Stuart HT, Bates LE, Ghorbani M, Nichols J, Dietmann S, Silva JCR. 2018. Exit from naive pluripotency induces a transient X chromosome inactivation-like state in males. Cell Stem Cell 22: 919–928.e6. 10.1016/j.stem.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, et al. 2011. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 25: 1371–1383. 10.1101/gad.633311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. 2008. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2: 230–240. 10.1016/j.stem.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takikawa S, Ray C, Wang X, Shamis Y, Wu TY, Li X. 2013. Genomic imprinting is variably lost during reprogramming of mouse iPS cells. Stem Cell Res 11: 861–873. 10.1016/j.scr.2013.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon I, Janiszewski A, Chappell J, Vanheer L, Pasque V. 2019. Recent advances in understanding the reversal of gene silencing during X chromosome reactivation. Front Cell Dev Biol 10.3389/fcell.2019.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32: 381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756. 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żylicz JJ, Bousard A, Žumer K, Dossin F, Mohammad E, da Rocha ST, Schwalb B, Syx L, Dingli F, Loew D, et al. 2019. The implication of early chromatin changes in X chromosome inactivation. Cell 176: 182–197.e23. 10.1016/j.cell.2018.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.