Abstract
The cleavage and polyadenylation reaction is a crucial step in transcription termination and pre-mRNA maturation in human cells. Despite extensive research, the encoding of polyadenylation-mediated regulation of gene expression within the DNA sequence is not well understood. Here, we utilized a massively parallel reporter assay to inspect the effect of over 12,000 rationally designed polyadenylation sequences (PASs) on reporter gene expression and cleavage efficiency. We find that the PAS sequence can modulate gene expression by over five orders of magnitude. By using a uniquely designed scanning mutagenesis data set, we gain mechanistic insight into various modes of action by which the cleavage efficiency affects the sensitivity or robustness of the PAS to mutation. Furthermore, we employ motif discovery to identify both known and novel sequence motifs associated with PAS-mediated regulation. By leveraging the large scale of our data, we train a deep learning model for the highly accurate prediction of RNA levels from DNA sequence alone (R = 0.83). Moreover, we devise unique approaches for predicting exact cleavage sites for our reporter constructs and for endogenous transcripts. Taken together, our results expand our understanding of PAS-mediated regulation, and provide an unprecedented resource for analyzing and predicting PAS for regulatory genomics applications.
For the majority of human mRNAs the formation of the 3′ end is directed by interaction between trans-acting factors and cis-elements in the polyadenylation sequence (PAS) leading to cleavage and polyadenylation of the premature mRNA (Zhao et al. 1999; Matoulkova et al. 2012). Naturally, sequences upstream of the cleavage site, such as the canonical hexamer motif, fall within the 3′ UTR (Hu et al. 2005; Matoulkova et al. 2012). In addition, the premature mRNA includes sequences downstream from the cleavage site which may also contain cis-elements regulating the cleavage reaction (Zhao et al. 1999; Hu et al. 2005; Matoulkova et al. 2012). Thus, both sequences within the 3′ UTR and downstream from it are of interest when searching for sequence features affecting gene expression through modulating 3′ end processing.
The majority of previous research efforts to characterize cis-regulatory elements affecting cleavage and polyadenylation focused on bioinformatics analysis of mRNA 3′ end data (Legendre and Gautheret 2003; Zarudnaya et al. 2003; Hu et al. 2005) and on mutational analysis of individual transcripts (Hart et al. 1985; McDevitt et al. 1986; Zhang et al. 1986; Zhang and Cole 1987; Connelly and Manley 1988; Goodwin and Rottman 1992; Sittler et al. 1994; Moreira et al. 1995, 1998; Graveley and Gilmartin 1996; Antoniou et al. 1998; Natalizio 2002; Nunes et al. 2010; Yoon et al. 2012). These studies revealed a number of upstream elements (USEs) and downstream elements (DSEs) which were associated with polyadenylation site selection and efficiency. Moreover, it has been shown that different point mutants of the canonical hexamer have a varied effect on the cleavage and polyadenylation reaction (Sheets et al. 1990; Thomas and Saetrom 2012), even though in some cases the motif might not be required (Nunes et al. 2010). Despite the accumulating knowledge about the sequence features associated with polyadenylation, the prediction of functional polyadenylation sites is still limited to classification of input sequences according to their predicted propensity to serve as PASs, as opposed to prediction of exact cleavage sites (Legendre and Gautheret 2003; Cheng et al. 2006; Magana-Mora et al. 2017). Therefore, a comprehensive study of 3′ UTR and downstream sequences in the context of cleavage and polyadenylation efficiency may greatly advance our understanding of this crucial process.
Some research efforts employed machine learning and deep learning approaches to prediction of alternative polyadenylation events and classification of sequences as PASs (Cheng et al. 2006; Akhtar et al. 2010; Chang et al. 2011; Gao et al. 2018; Leung et al. 2018; Bogard et al. 2019). The deep learning approaches highlight the usefulness of convolutional neural networks (CNNs) for regulatory genomics and provide valuable predictions for PAS classification and isoform choice. However, the quality of input data is of the utmost importance for this kind of approach. A large data set of diverse reporter constructs, not limited to a small number of contexts, can thus contribute to model performance and generalizability. Moreover, a broadly applicable approach for accurate cleavage site prediction is yet to be established.
Recent progress in large-scale DNA synthesis promoted the establishment of massively parallel reporter assays (MPRAs) (Sharon et al. 2012; Goodman et al. 2013; Kheradpour et al. 2013; Mogno et al. 2013; Smith et al. 2013; Noderer et al. 2014; Muerdter et al. 2015; Rosenberg et al. 2015; Weingarten-Gabbay et al. 2016; Vainberg Slutskin et al. 2018), which were employed in the study of the regulatory outcomes of extensive variant libraries. Moreover, intelligent design followed by systematic analysis of the MPRA data has been previously shown to advance the understanding of multiple regulatory processes (Sharon et al. 2012, 2014; Smith et al. 2013; Lubliner et al. 2015; Shalem et al. 2015; Weingarten-Gabbay et al. 2016; Vainberg Slutskin et al. 2018). However, given the numerous regulatory processes in which 3′ UTRs are involved, a major gap remains in our ability to predict RNA levels from the relevant DNA sequences.
Here, we set out to advance our mechanistic understanding of PAS-mediated gene expression regulation by applying a MPRA approach developed in our lab. We leverage the intelligent design of our large-scale library to quantify the effect of numerous PAS features on RNA levels. We utilize a unique scanning mutagenesis approach to identify several mechanisms by which PASs regulate expression levels. Moreover, we quantify the effect of defined PAS features on expression and find associated known and novel regulatory motifs. Finally, we apply deep learning models for the accurate prediction of RNA levels and cleavage efficiency from DNA sequence alone within our library as well as prediction of exact endogenous cleavage sites. Taken together, our analysis and models boost our understanding of PAS-mediated regulation of gene expression and promote applications in the field of regulatory genomics.
Results
High-throughput measurement of expression levels and cleavage maps for over 12,000 PASs
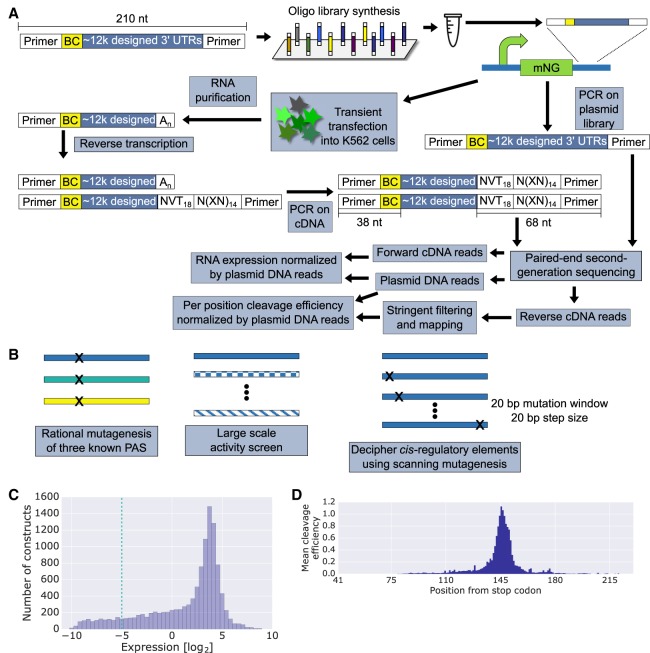
To get a quantitative measure for the effect of PASs on expression levels and cleavage efficiency, we adopted an MPRA approach previously used in our lab (Vainberg Slutskin et al. 2018). We designed 12,339, 210-nucleotide (nt)-long oligonucleotides, which are comprised of constant and variable regions (Methods). In our design, we included both systematically mutated sequences as well as native sequences from human and viral genomes. To measure the expression levels and cleavage efficiency, we transiently transfected our library of reporter constructs into K562 cells. The mRNA produced from the reporter was reverse-transcribed with a poly(T) primer and amplified with gene specific primers for paired-end second-generation sequencing. We used the shorter forward reads to map the construct barcode to our reference sequences and the longer reverse reads to map the cleavage sites for each construct. In addition, the plasmid library DNA was also amplified, and we used the plasmid DNA counts together with the cDNA forward and reverse reads to calculate the normalized RNA levels and per position cleavage efficiency, respectively, for each construct (Fig. 1A; Methods). We find that the expression measurements are highly reproducible between technical replicates (R = 0.99, P < 10−10) (Supplemental Fig. S1A). Furthermore, we estimated the technical noise of our system by examining groups of 10 constructs with identical sequences except for the DNA barcode and find that the median relative standard deviation (RSD) was 1.1% (Supplemental Fig. S1B), indicating that our system exhibits low technical noise. We note that the range of expression levels spanned by these constructs is over 5000-fold. Finally, we examined the cleavage efficiency in similar groups of 10 constructs and find high agreement across the different barcodes (Supplemental Fig. S1C–E).
Figure 1.
A high-throughput system for studying PAS-mediated regulation of gene expression. (A) A schematic representation of a massively parallel reporter assay measuring expression and cleavage efficiency of PAS reporters (Methods). Briefly, sequences are designed in silico, synthesized, and cloned into a reporter plasmid containing mNeonGreen. The plasmid is transiently transfected into K562 cells, from which RNA is extracted and reverse-transcribed with a poly(T) primer. N = any base synthesized randomly, V = any base except T synthesized randomly, X = A, C, G or T preselected as described in the Methods. The cDNA and plasmid DNA are amplified for paired-end second-generation sequencing. The barcodes of each library member are quantified in the forward cDNA reads and the plasmid DNA reads. These are used to calculate normalized expression. The reverse cDNA reads are mapped to their respective library members identified by the barcode in the forward DNA reads. Following stringent filtering, the cleavage efficiency distribution, normalized by the plasmid DNA reads, is calculated. (B) Library design is based on the three illustrated schemes. First, we mutated three known PASs by varying annotated regulatory elements and surrounding sequences. Second, we constructed a compendium of 6197 native PASs from annotated transcripts of viruses whose host is human and from K562 3′ end sequencing data. Finally, we applied scanning mutagenesis by mutating every 20-bp sequence in selected 629 native PASs (Methods). (C) A histogram depicting the distribution of RNA expression levels acquired by the methods in A. The −5 cutoff is used later to define positive and negative sets for motif analysis. (D) Per position mean cleavage efficiency calculated over all library variants. Positions are indicated as the distance from the mNeonGreen stop codon.
Here, we applied multiple library design approaches to quantify the effect of regulatory elements within PASs (Fig. 1B). First, we used rational mutagenesis of three known PASs from human immunodeficiency 1 virus (HIV1) (Bohnlein et al. 1989; Valsamakis et al. 1991), Simian virus 40 late (SVL) (Sadofsky et al. 1985; Schek et al. 1992; Bagga et al. 1995), and the synthetic polyadenylation sequence (SPA1) (Levitt et al. 1989). Next, we constructed a large set (6197 constructs) of native PASs from K562 3′ end sequencing (Lin et al. 2012) data and from viral genomes (Hulo et al. 2011; Brister et al. 2015) whose host is human. Finally, to perform an unbiased search for regulatory elements within PASs, we applied a scanning mutagenesis approach on a subset of the native PASs (Methods).
We subjected the library to our experimental pipeline and obtained expression and cleavage efficiency measurements for 97.3% and 69.3% of the designed constructs, respectively. The difference in the percentage of the constructs can be attributed to the higher coverage requirements for detecting cleavage (Methods; Supplemental Note 2). We find that the assayed constructs span over five orders of magnitude in expression levels (Fig. 1C). The expression distribution is highly skewed, with the majority of sequences exhibiting high expression, consistent with the design of the library, which included a large proportion of sequences highly likely to function as PASs. To get a sense of the positions at which the cleavage is most frequent, we plotted the mean cleavage efficiency distribution (Fig. 1D). We observe a clear peak at position 145, which can be attributed to centering our variable regions on the previously annotated cleavage sites in the native sequences included in our library. We conclude that our MPRA approach can measure the effect of 3′ UTR sequences on RNA expression levels and per position cleavage efficiency over a wide range of values.
Scanning mutagenesis reveals complex relationships between mutation position, cleavage efficiency maps, and expression levels
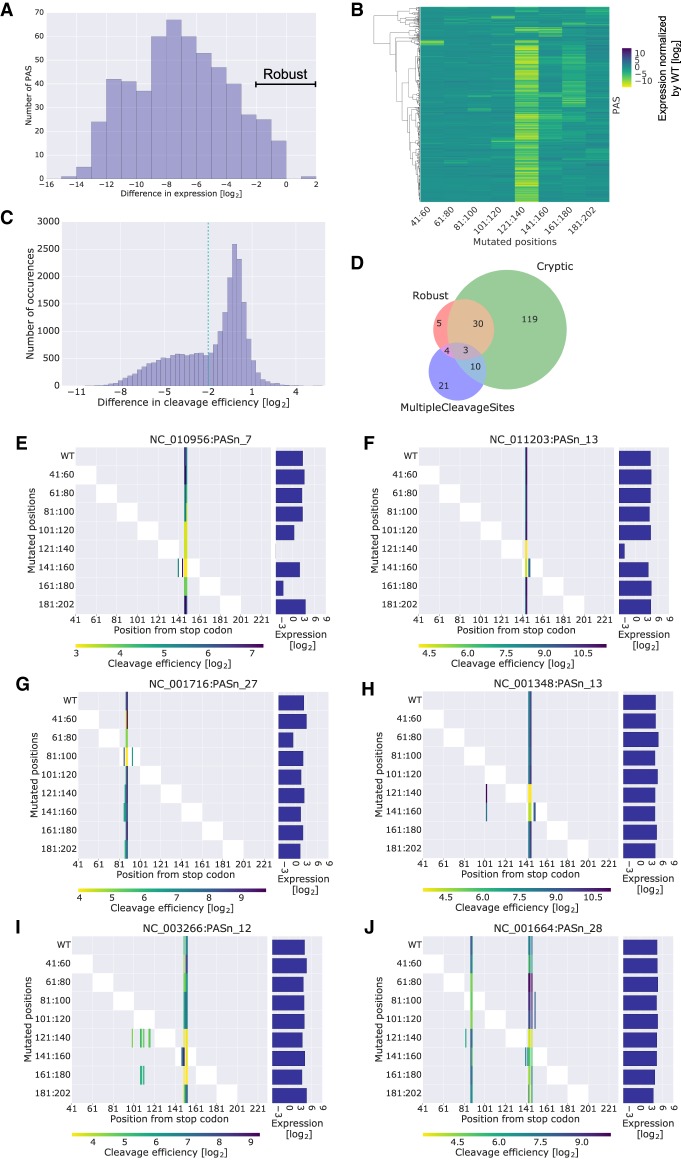
Scanning mutagenesis is an unbiased approach for discovery of regulatory sequences. Here, we mutated 20-bp blocks of 629 native PASs by replacing the native sequence with a random one (avoiding the introduction of certain sequences) (Methods) and measured the effect on expression and cleavage efficiency. We find that mutation blocks can decrease expression up to four orders of magnitude relative to wild type (WT), while in some cases there are no significant deviations in expression (Fig. 2A). Moreover, clustering of the changes in expression relative to WT upon mutation of the different blocks revealed clusters of different response patterns (Fig. 2B). The mutation block at positions 121:140 was frequently associated with the largest reduction in expression. Given that the most frequent cleavage was observed at position 145, this observation corresponds to the presence of an important upstream regulatory element. Moreover, positions 161:180 had a notable effect on expression, in line with a less frequent presence of a downstream regulatory element.
Figure 2.
Scanning mutagenesis reveals a mechanistic link between cleavage efficiency and expression levels. (A) Histogram of the difference in expression between the mutant with the lowest expression and the WT PAS for PASs with cleavage efficiency data. PASs with differences ≥ −2 are considered robust. (B) A clustered heat map where each row is a native PAS subjected to scanning mutagenesis and each column is a range of mutated 20 bp. The values are the expression of the mutant minus the expression of the WT for PASs with cleavage efficiency data. Rows were clustered with ward hierarchical clustering using a cosine distance. Two main clusters are observed, one that is robust to mutagenesis and one that is sensitive at certain mutation blocks. (C) Histogram of the differences in cleavage efficiency calculated per variant per cleaved WT position. The −2 cutoff is used later to define positive and negative sets for motif analysis. (D) PASs with cleavage efficiency data are classified based on their behavior in the scanning mutagenesis data. Robust PASs showed minimal changes in expression levels for all mutants, as annotated in A. PASs whose WT sequence has more than one cleavage site separated by at least 10 bases from one another are classified as MultipleCleavageSites. PASs for which at least one of their mutants has more cleavage sites than the WT sequence are classified as cryptic. (E–J) Visualization of cleavage efficiency and expression levels for example PASs. The right panel is a bar plot of expression levels. The left panel is a heat map of cleavage efficiencies, where each column is a position along the PAS sequence and each row refers to mutated positions within the PAS. The color bar corresponds to the measured cleavage efficiency for each variant at each position. The mutated positions are also visualized by the white blocks in the heat map. Missing cleavage efficiencies in the mutant variants at positions where the WT had a measured cleavage efficiency were imputed with the detection limit (see Methods). Titles correspond to RefSeq IDs of viral genomes followed by the number of the PAS as annotated in the GenBank record.
We found PASs that maintained robust expression levels at all mutation positions. We speculated that these results could be explained by changes in the cleavage efficiency maps. Thus, we examined these changes at each position of the WT sequence and find that mutations can result in reduction of up to three orders of magnitude in cleavage efficiency (Fig. 2C). When examining the combined measurements of expression and cleavage efficiency maps, we noticed that some of the robust cases can be associated with the presence of multiple cleavage sites, while others overlap with the presence of new cleavage sites in the mutant sequences (Fig. 2D). In cases where we do see reduction in expression in regions 121:140 and 161:180, it indeed corresponds to mutations upstream of and downstream from the cleavage site, respectively (Fig. 2E,F). Moreover, when the cleavage site occurred at other locations, we still observe a similar effect for the relative mutation blocks (Fig. 2G). In these cases, the reduction in expression corresponds to the local reduction in cleavage efficiency. In the more intriguing cases of robust expression of the given PAS upon scanning mutagenesis, we find occurrences of cryptic cleavage sites appearing in the mutant sequences that could explain the phenomenon (Fig. 2H,I). The cryptic cleavage sites could be focused (Fig. 2H) or dispersed (Fig. 2I) and occur upon mutagenesis of various regulatory regions. However, we note that the presence of cryptic sites is not sufficient for robustness as there are many PASs with cryptic sites that are not robust (Fig. 2D). We also find cases of alternative polyadenylation in the WT sequence (Fig. 2J), which could allow for compensation for the mutagenesis by increasing the cleavage efficiency of the PASs whose regulatory elements remained intact. We conclude that in some cases the mutation of regulatory regions upstream of and downstream from the cleavage site may result in decrease in expression, while in other cases expression levels may be robust to mutation owing to changes in the cleavage efficiency map.
Sequence determinants affecting polyadenylation-mediated regulation of reporter gene expression
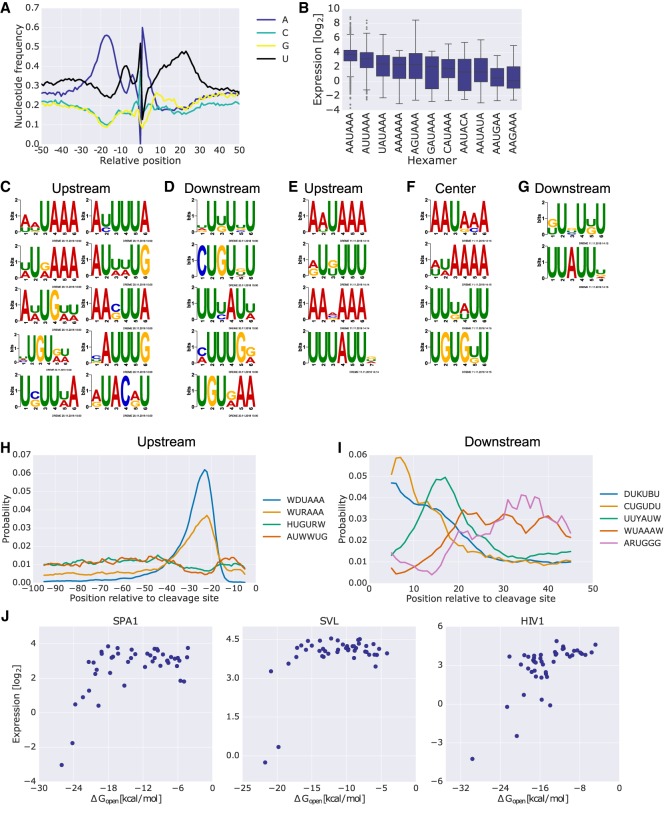
The cleavage and polyadenylation reaction is directed by interaction between trans-acting factors and cis-elements in the premature mRNA (Zhao et al. 1999; Hu et al. 2005; Matoulkova et al. 2012). Thus, we examined the sequences surrounding the cleavage sites measured by our assay. We find that sequences upstream of the cleavage site are mostly A-rich and U-rich, while sequences downstream were U-rich and slightly G-rich, as expected (Fig. 3A). Next, we quantified the effect of single nucleotide variants of the canonical hexamer motif AAUAAA in native constructs on expression (Fig. 3B). We find that the hexamer variants can span up to 12-fold in median expression levels. Moreover, the grouping by the hexamer variant alone is associated with different expression levels despite the varied sequences analyzed. We conclude that the hexamer variants have a considerable effect on expression although they do not fully explain the observed differences.
Figure 3.
Multiple PAS sequence features have a considerable effect on expression levels and cleavage site location. (A) The nucleotide frequencies surrounding the position of maximal cleavage efficiency of each variant. (B) Box plots comparing the expression levels of sequences grouped by the hexamer found upstream of the position of maximal cleavage efficiency of each native PAS. Only native PASs with a single variation of the hexamer were included. (C,D) Regulatory motifs found using DREME in 100 bp upstream of (C) and 50 bp downstream from (D) the position of maximal cleavage efficiency found in native sequences and also enriched in genomic sequences obtained from K562 3′ end sequencing data. The positive set consisted of library members with expression higher than 2−5, which had cleavage efficiency data. The negative set consisted of library members with expression lower than 2−5. The sequences were taken in corresponding length and orientation to the positive set but with respect to position 145. Only native library sequences were used for the analysis. Only motifs that were significantly enriched in a set of endogenous 3′ UTR sequences (using AME) are presented (enrichment P-value < 0.01) (Methods). (E–G) Regulatory motifs found using DREME in scanning mutagenesis data upstream of (E), overlapping (F), or downstream from (G) the position of maximal cleavage efficiency. Native and mutant 20-bp sequences were used as positive and negative sets, respectively. The regions for analysis were selected with respect to cleavage positions that showed a difference in cleavage efficiency smaller than 2−2. Only motifs that were significantly enriched in a set of endogenous 3′ UTR sequences (using AME) are presented (enrichment P-value < 0.01). All of the center motifs were enriched upstream, while only the bottom two were enriched downstream (Methods). (H,I) CentriMo analysis for the positional preference of each motif found upstream of (H) or downstream from (I) the analysis performed in C and D, respectively. The plot depicts positional distribution of the best match for each of the motifs for results with Fisher E-value < 0.01. Positions are indicated relative to the position of maximal cleavage efficiency. The motifs are indicated in the legend by their consensus sequence (Methods; Supplemental Fig. S2). (J) Expression as a function of ΔGopen, the change in ensemble free energy required to expose the canonical hexamer with an additional 15 bp upstream and downstream. The analysis was performed on rationally designed mutants of three PASs, SPA1 (left), SVL (center), and HIV1 (right).
To elucidate additional regulatory motifs, we turned to de novo motif discovery using DREME following two schemes (Bailey 2011). First, we analyzed sequences upstream of and downstream from the cleavage site for constructs with sufficiently high expression (Fig. 3C,D). Second, we leveraged our unique scanning mutagenesis data to identify sequence blocks that, when mutated, had a sufficiently large effect on cleavage efficiency (Fig. 3E–G). The analyzed blocks occurred upstream of, overlapping, or downstream from the measured cleavage site. To validate the discovered motifs, we tested for their enrichment using AME (McLeay and Bailey 2010) 100-bp upstream and 50-bp downstream endogenous cleavage sites identified using K562 3′ end sequencing data (Methods; Lin et al. 2012). The discovered motifs using the two schemes are in good agreement between them. Both highlight the AWUAAA hexamer motif and U-rich elements upstream of the cleavage site and GU-rich and U-rich elements downstream. Moreover, all of the motifs discovered in the scanning mutagenesis center subset are also enriched upstream of endogenous sites, while only the GU-rich and U-rich motifs are enriched downstream. These results indicate that these motifs may also occur in close proximity to the cleavage site. Taken together, our findings confirm the presence of polyadenylation cis-regulatory elements upstream of and downstream from the cleavage site.
We hypothesized that the discovered motifs may exhibit positional preference relative to the cleavage site. Therefore, we calculated the positional distribution of the best match for each of the motifs found upstream of or downstream from endogenous cleavage sites using CentriMo (Bailey and Machanick 2012). The plotted data revealed that certain motifs have a positional preference (Fig. 3H,I). The top upstream motif, which closely resembles the canonical hexamer, shows a clear preference for positions −10 to −40. However, we find an additional motif with a similar positional inclination, WURAAA, which shares some information with the canonical hexamer, yet is still different. Similar analysis for the downstream motifs also reveals positional preferences, as in the case of DUKUBU and CUGUDU enriched closer to the cleavage site and WUAAAW enriched further away from the cleavage site. We find similar results when examining the positional preference of the motifs from the scanning mutagenesis set (Supplemental Fig. S2). We conclude that the regulatory sequences governing the polyadenylation process are subject to preferences in their distribution surrounding the cleavage site.
Despite the importance of linear regulatory sequences, RNA secondary structure was shown to have an important role in 3′ UTR-mediated regulation of gene expression (Hans and Alwine 2000; Kertesz et al. 2007; Marín and Vaníček 2011; Wu and Bartel 2017; Vainberg Slutskin et al. 2018). To test for the regulatory effect of secondary structure surrounding the canonical hexamer, we mutated the sequences flanking the AAUAAA in three known PASs, generating 135 constructs. For each mutant, we calculated ΔGopen, the change in ensemble free energy required to expose the AAUAAA sequence. We find that very low ΔGopen values are required in order to have a negative effect, of over two orders of magnitude, on expression in all the three contexts tested (Fig. 3J). Moreover, in SPA1 we noted that below a certain threshold (ΔGopen = −18 kcal/mol) the reduction in expression is linear with respect to ΔGopen. We conclude that the effect of secondary structure surrounding the hexamer motif is context-specific and requires ΔGopen values below a particular threshold in order to be observed.
Highly accurate prediction of expression levels and cleavage sites from polyadenylation sequence
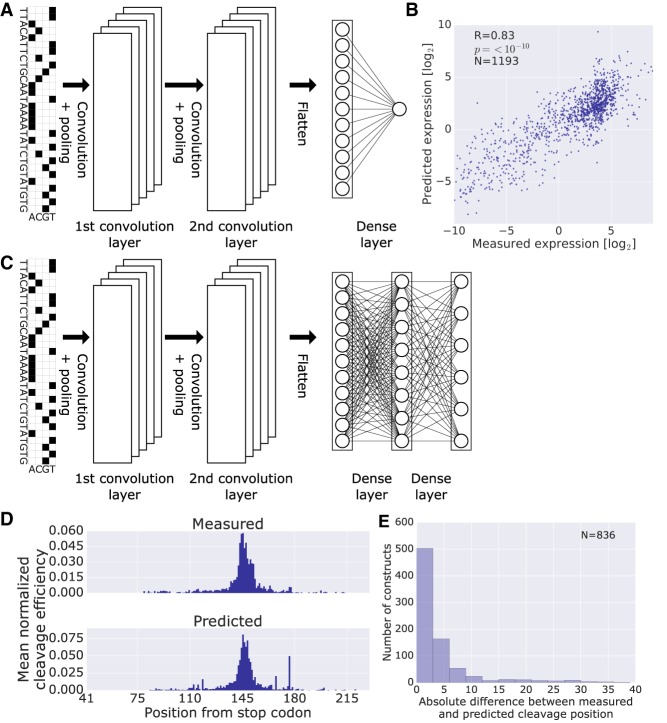
As our understanding of the polyadenylation regulatory code is not complete, we asked whether we could build a model that would learn directly from the input sequences, without carefully defined features. Thus, we chose to apply a CNN to predicting expression levels from DNA sequences directly (Fig. 4A). The input for the model is a 250-bp sequence encompassing the variable region, and the output is the predicted expression value. Our model predicts the observed expression with high accuracy both on training (R = 0.83) (Supplemental Fig. S3A) and held-out test data (R = 0.83) (Fig. 4B). Our CNN model showed superior performance over a gradient boosting tree-based model (R = 0.72 and R = 0.73 on training and test, respectively) (Methods; Supplemental Fig. S3B,C) and a k-mer elastic net model (R = 0.73 and R = 0.70 on training and test, respectively) (Methods; Supplemental Fig. S3D,E). These results indicate that the CNN can perform highly accurate predictions of expression from the DNA sequence alone, greatly outperforming simpler models.
Figure 4.
Prediction of expression levels and cleavage efficiencies from DNA sequence alone using CNNs. (A) Model architecture for prediction of expression levels. The input DNA sequence is one hot encoded and fed into a CNN composed of two convolutional layers and one dense layer with a final output of a single neuron with linear activation (Methods). (B) Scatter plot of predicted versus measured expression levels on held-out data. (C) Model architecture for prediction of cleavage efficiency maps. The input DNA sequence is one hot encoded and fed into a CNN composed of two convolutional layers and two dense layers with a final output of a vector of length 189, the number of positions considered. (D) Per position mean cleavage efficiency calculated over all the library members in the test set. For each member, the cleavage efficiencies were normalized by dividing by their sum, in order to facilitate comparison between the measured distribution and the one achieved by the model. (E) Histogram of the absolute differences between the measured and the most probable predicted cleavage site evaluated on library held-out test data. Only constructs with measured cleavage efficiency maps were used.
Next, we turned to the more challenging task of predicting the position at which cleavage happens. We adapted our CNN to output a vector corresponding to the predicted cleavage efficiency at each position (Fig. 4C). To assess model performance, we first compared the distributions of the mean normalized cleavage efficiencies (Fig. 4D; Supplemental Fig. S3F for training and test data, respectively). We observed that the distribution of the predicted values closely resembles that of the measured values. We then extracted the most probable position of cleavage from the cleavage efficiency maps and calculated the absolute difference between the measured and predicted most likely cleavage site for each construct. The distribution of absolute differences clearly shows good performance for the majority of constructs (Fig. 4E; Supplemental Fig. S3G for training and test data, respectively). These results demonstrate that cleavage efficiency maps can be obtained from the DNA sequence alone and that the most probable cleavage site can be reliably predicted.
Expression model interpretation and application to endogenous cleavage site prediction
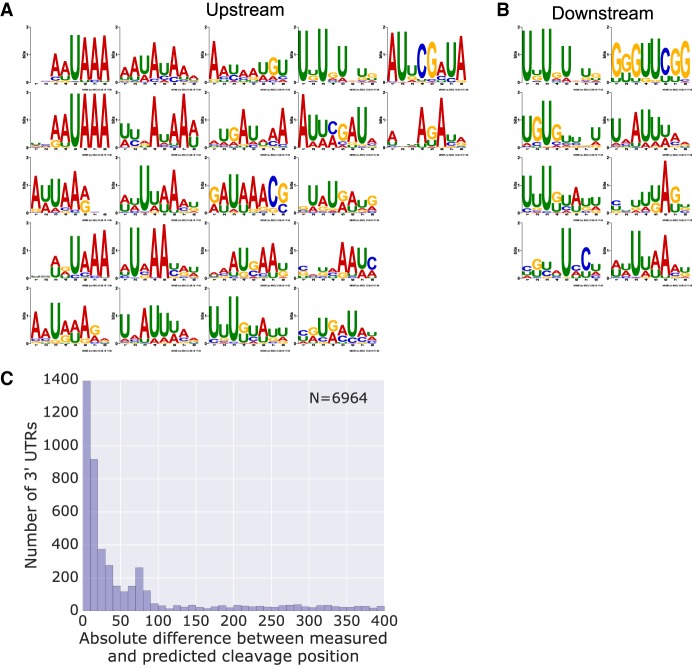
One of the challenges in applying CNNs to genomics in general and MPRAs in particular is model interpretation (Shrikumar et al. 2017). Here, we applied visualization and analysis of the learned convolutional filters to gain valuable insight into the sequence features underlying our expression predictions (Methods). To confirm that our model learns biologically relevant features, we checked for the enrichment of the motifs in the endogenous cleavage sites identified using K562 3′ end sequencing data. We found 22 and eight motifs enriched upstream and downstream, respectively (Figs. 5A,B). As in our de novo motif discovery analysis (Fig. 3C–G), among the upstream motifs we find motifs that closely resemble the AWUAAA hexamer as well as U-rich motifs, while among the downstream motifs we find GU-rich and U-rich motifs. We conclude that our model performance benefits from learning biologically relevant regulatory motifs.
Figure 5.
The expression model learns biologically relevant sequence motifs which contribute to highly accurate endogenous cleavage site prediction. (A,B) Motifs were constructed for each filter from first layer activations (Methods). The motifs were analyzed for enrichment upstream of (A) and downstream from (B) endogenous cleavage sites using AME. Only motifs with an enrichment P-value < 0.001 are presented. (C) Histogram of the absolute differences between the measured and the predicted cleavage site on a set of endogenous 3′ UTRs (Methods). Predictions were made by applying the expression model on windows of 250-bp sequences shifted by 1 bp at a time. The position at which the maximal expression was achieved was adjusted by 145, the most likely cleavage position within the reporter library. See also Supplemental Figure S4.
Another challenge in the study of polyadenylation is the accurate prediction of endogenous cleavage sites. Our expression model was trained using an MPRA designed to specifically quantify the effect of polyadenylation sequences on gene expression. Thus, we hypothesized that it could score endogenous sequences for their potential to serve as PASs. By walking along endogenous human 3′ UTRs at a single-base-pair resolution, we obtained a prediction score for each position, identified the position with the maximal predicted score, and calculated the most likely cleavage position (Methods). We computed the predicted position for the endogenous cleavage sites identified using K562 3′ end sequencing data (Methods) and examined the distribution of differences between measured and predicted cleavage sites (Fig. 5C). The predictions using our CNN model greatly outperform similar predictions generated using our gradient boosting tree or k-mer elastic net models as well as a previously published support vector machine-based model (Methods; Supplemental Fig. S4; Cheng et al. 2006). We conclude that our CNN expression model can be applied to prediction of endogenous cleavage sites with high accuracy.
Discussion
In this work, we systematically tested the quantitative effect of an unprecedented collection of PASs along with rationally designed mutations thereof on expression levels and cleavage efficiency maps. We used our measurements for our unique scanning mutagenesis data set to gain mechanistic insight into changes and robustness of expression levels upon mutation mediated by changes in cleavage efficiency. Moreover, we quantified the regulatory consequence of sequence features, such as canonical hexamer variability and accessibility. The large scale of our library and scanning mutagenesis data allowed us to identify regulatory motifs and their positional preferences within endogenous sequences associated with higher cleavage efficiency and gene expression levels. Furthermore, we developed a highly accurate machine learning approach, based on CNN, for predicting gene expression and cleavage efficiency maps directly from the input DNA sequence. Our CNN expression model performance can be explained, in part, by the learned biologically relevant regulatory motifs. Finally, we show that our expression model can be applied for highly accurate predictions of endogenous cleavage sites.
We have performed a MPRA employing rational design and introduction of relatively long (162-nt) variable regions in order to study the effect of PASs on gene expression and cleavage efficiency in mammalian cells. We demonstrated that our method is highly reproducible in expression measurements across replicates (Supplemental Fig. S1A) and in cleavage efficiency measurements across internal controls (Supplemental Fig. S1C). However, we note a number of technical caveats which may still contribute to the noise in our data. First, we cannot exclude that the expression measurements of certain constructs are affected by regulatory elements other than PASs, such as miRNA seeds, AU-rich elements, Pumilio recognition sites, and others (Rabani et al. 2017; Vainberg Slutskin et al. 2018). However, as opposed to other approaches focused on analysis of endogenous data, our rational design approach biases our sequences to contain polyadenylation-related regulatory sequences. Second, a common issue with 3′ end sequencing protocols based on reverse transcription with a poly(T) primer is internal priming at poly(A) sequences other than the poly(A) tail, contributing to inaccurate quantification of cleavage efficiency. To reduce this risk, we performed reverse transcription at elevated temperatures (50°C). Despite these and other potential sources of noise in our data, we obtained highly quantitative and reliable measurements (Supplemental Fig. S1), indicating that the signal greatly surpasses the noise. We conclude that our assay measures both expression levels and cleavage efficiency maps simultaneously and reliably on a rationally designed library of 12,339 constructs.
Many of the previously studied data sets for PAS-mediated regulation of gene expressions were based on bioinformatics analysis (Legendre and Gautheret 2003; Zarudnaya et al. 2003; Hu et al. 2005), thus limited to examination of native sequences in their native contexts only. Our approach allowed us, for the first time, to isolate the effect of the PAS in a controlled sequence environment. This enabled us to quantify the effect of different PAS-associated regulatory elements on expression, thus highlighting their contribution to the polyadenylation regulatory code. Moreover, our exclusive scanning mutagenesis data provide an invaluable resource to study the complex relationship between sequence, cleavage efficiency, and expression. Thus, our technique allows us to gain mechanistic insight into PAS-mediated regulation of gene expression.
Using our scanning mutagenesis data set, we highlighted multiple potential modes of regulation of gene expression by PASs. Changes in the reporter expression levels, up to four orders of magnitude, can correspond to changes in cleavage efficiency and be explained by mutagenesis of regulatory elements at predetermined relative positions to the cleavage site (Fig. 2E–G). We highlight scenarios where, despite mutagenesis of regulatory regions, as evident by changes in cleavage efficiency at the WT positions, the construct expression levels remained robust. This robustness can be explained by the cryptic cleavage sites that rose upon mutations of sequences destructive to the native cleavage sites, thus providing evidence for the flexibility of polyadenylation-mediated regulation of gene expression. Moreover, our data introduced the possibility that a distal cleavage site can compensate for mutagenesis of a proximal one, and vice versa, when multiple cleavage sites are detected in the WT sequence. Such insight could not have been achieved via bioinformatics analysis or lower throughput methods and is highly dependent on our capability for rationally designing the library.
The scale of our library allowed us to assay thousands of designed and native sequences from viral and human genomes. Our results are in line with previous studies, as in the case of base frequencies surrounding the cleavage site (Wang et al. 2018) and the effect of the hexamer motif on expression (Deng et al. 2018). Minor differences in the local base frequencies and the ranking of hexamer mutants may be attributed to the differences in assayed PASs and specific methods used. Moreover, our analysis managed to derive both previously known (canonical hexamer, UGUA and U-rich elements upstream and GU-rich elements downstream) (Fig. 3) and novel (for example, ARUGGG) regulatory motifs in regions surrounding the cleavage site. Moreover, some of the motifs show considerable positional preference in endogenous PASs, suggesting that their location relative to the cleavage site may play a role in the regulatory process. Finally, we show that the RNA structure surrounding the canonical hexamer may have an effect of over two orders of magnitude on expression in a context- and threshold-specific manner. Thus, our results underline the complexity of polyadenylation-mediated regulation of gene expression.
A major aspect of our work is the development of accurate predictive models for polyadenylation-mediated regulation from DNA sequence alone. The models are built to receive as input 250 bp of DNA sequence of interest and predict the expression levels or the cleavage efficiency maps in our reporter system. The high accuracy of our expression model stems from the CNNs capability to learn biologically relevant sequence features on its own (Fig. 5A,B) as well as the nonlinear relationships between those features. The complexity of the deep learning approach for this task is justified by the higher performance of the CNN model when compared to the simpler gradient boosting tree and k-mer elastic net models. This superior performance in expression prediction has great potential, as shown in our endogenous cleavage site predictions (Fig. 5C). Taken together, our prediction and model interpretation results emphasize the biological applicability of our CNN models in particular and deep learning approaches for genomics in general.
Previous work applying deep learning approaches to polyadenylation was either applied to endogenous data (Gao et al. 2018; Leung et al. 2018) or to randomly mutated reporter constructs (Bogard et al. 2019). When learning on endogenous data, multiple factors other than the PASs, such as nucleosome composition and epigenetic modification (Lutz and Moreira 2011), can have an effect on polyadenylation. These factors may hinder model performance since they cannot be captured from the input 3′ UTR and downstream sequence. When learning from randomly mutated reporter constructs, the gained insight may be limited to the examined sequence contexts. In this work, we rationally select or mutate relatively long (162-nt) 3′ UTR inserts, allowing us to test a large variety of carefully designed sequences, thus contributing to model performance and generalizability. This approach complements previous efforts and provides additional insight, models, and applications.
In summary, we used a quantitative high-throughput assay to measure the regulatory effect of over 12,000 designed 3′ UTRs to decipher the rules of PAS-based regulation. We identified various modes of interaction between cleavage efficiency and expression levels in our unique scanning mutagenesis data. Furthermore, we analyzed sequence determinants of the cleavage site and identified features affecting expression levels. Moreover, we leveraged the scale of our library to construct predictive models for RNA levels and cleavage efficiency maps. Finally, we applied our expression model to predict exact endogenous cleavage sites using a unique approach. These results contribute to a systematic functional understanding of PAS-mediated gene regulation and pose a valuable resource for the regulatory genomics community.
Methods
Synthetic library design
General design notes
All the constructs were composed of an 18-nt forward primer, 12-nt barcode sequence, 162-nt variable region, and 18-nt reverse primer sequences. Unique primer sequences were used to facilitate targeted amplification of the 12,339 constructs pool from a larger library of 55,000 constructs. We made sure all sequences excluded restriction sites used for cloning.
Designing the rational mutagenesis set
We selected three PASs that were extensively studied and included PASs from human immunodeficiency 1 virus (HIV1, RefSeq ID K03455 bases 9512–9673) (Bohnlein et al. 1989; Valsamakis et al. 1991), Simian virus 40 late (SVL, RefSeq ID J02400 bases 2600–2761) (Sadofsky et al. 1985; Schek et al. 1992; Bagga et al. 1995), and the synthetic PAS (SPA1) based on the rabbit β-globin gene (Levitt et al. 1989). For each PAS, we mutated each annotated regulatory element as well as all of the annotated elements together by replacing the native sequence with a random one while avoiding the introduction of undesired sequences (Supplemental Note 1). In addition, we replaced the canonical hexamer with each of its point mutants in each of the native sequences. Moreover, the sequence upstream of and downstream from the hexamer was mutated to span five GC bins with three constructs in each bin (Supplemental Table S1).
Designing the compendium of native sequences
First, we selected annotated PASs in viral genomes whose host is human (NCBI viral genome resource [Brister et al. 2015] and ViralZone [Hulo et al. 2011]), resulting in 668 viral sequences from 48 viral genomes. Second, we used cleavage site data in K562 based on a 3′ end sequencing technique (Lin et al. 2012) to select 5529 sequences spanning different canonical hexamer sequences and gene expression levels. In addition, the HIV1, SVL, and SPA1 PASs were also included. The list of constructs containing the native sequences and the subset of constructs with a single hexamer upstream of the cleavage site (for Fig. 3B) are provided in Supplemental Tables S2 and S3, respectively.
Designing the scanning mutagenesis set
The mutagenesis was performed by mutating every nonoverlapping 20 bp in the candidate sequences. The sequence within the mutated block was replaced with a random sequence while avoiding the introduction of undesired sequences (Supplemental Note 1). The candidate sequences included 572 of the viral PASs, 17 sequences based on a literature search (Hart et al. 1985; McDevitt et al. 1986; Zhang et al. 1986; Zhang and Cole 1987; Connelly and Manley 1988; Goodwin and Rottman 1992; Sittler et al. 1994; Moreira et al. 1995, 1998; Graveley and Gilmartin 1996; Antoniou et al. 1998; Natalizio 2002; Zarudnaya et al. 2003; Nunes et al. 2010; Yoon et al. 2012), and 40 randomly selected sequences from the K562 data (Supplemental Table S4; Lin et al. 2012).
Designing a set of constructs with multiple barcodes
We selected 20 sequences expected to span a large range of expression levels. For each variant, we generated 10 different barcodes (Supplemental Table S5). Only one of the barcoded constructs was selected for all other downstream analysis.
Experimental procedures
Construction of the master plasmid
A previously assembled construct in our lab (Vainberg Slutskin et al. 2018) was modified to exclude the SV40 polyadenylation signal downstream from mNeonGreen by standard restriction and ligation cloning techniques. Correct clones were verified using Sanger sequencing.
Synthetic library cloning
The library was cloned using a technique previously established in our lab (Vainberg Slutskin et al. 2018). Briefly, a pool of 55,000 fully designed single-stranded 210-oligomers (Agilent Technologies), containing the 12,339 pool used in this study was amplified using specific primers with restriction site–containing tails, SpeI (Fw primer) and AscI (Rv primer). The underline represents the 18-nt complementary sequence to the ssOligos. The primers were: AATCTTCACTAGTAGCAATGGGGTTCGGTATGCGC (Fw primer), GCCTCGGCGCGCCAACTATCGTCTCGGGGAGCCTT (Rv primer). The amplified library was cloned into the master plasmid using high-efficiency restriction ligation using SpeI and AscI restriction sites followed by electroporation into Escherichia coli 10G electrocompetent cells (Lucigen). The cloned library was analyzed by colony PCR to ensure single insert ligation and purified (MACHEREY-NAGEL NucleoBond Xtra Maxi kit).
Transfection into K562 cells
Transient transfection of K562 cells was performed in two replicates using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer's protocol. The day of the transfection, 5 × 106 cells were plated in 10 mL of growth media without antibiotics and transfected using 20 µg of donor plasmid and 50 µL of Lipofectamine 2000. Once 4 h have passed, the cells were centrifuged and resuspended in 20 mL of complete growth media. Cells were harvested for RNA purification 24 h after transfection.
RNA purification and preparation for sequencing
Each of the replicates was harvested for RNA purification using a NucleoSpin RNA II kit (MACHEREY-NAGEL) according to the manufacturer's protocol. DNase-treated purified RNA was reverse-transcribed using the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific) with the designed poly(T) primer: GCTCAAGCCACGACGCTCTTCCGATCTNANCNGNTNANCNGNTNANCNGNANCNANTTTTTTTTTTTTTTTTTTVN, where N is any nucleotide and V is any nucleotide except T. The cDNA library was amplified with a forward gene-specific primer and a reverse primer complementary to the reverse transcription primer tail with KAPA HiFi ready mix X2 (KAPA Biosystems). In addition, the library was amplified with KAPA HiFi ready mix X2 (KAPA Biosystems) from the plasmid DNA used for the transient transfection. The amplified DNA was used for library preparation for second-generation sequencing (Supplemental Methods).
Computational analyses
Mapping second-generation sequencing reads
To determine the identity of the oligo after sequencing, a unique 12-mer barcode sequence was placed at the 5′ end of each variable region. Barcodes were designed to differ by 3 nt or more and to avoid the introduction of undesired sequences (Supplemental Note 1). For the cDNA, we obtained ∼50 and ∼43 million reads for replicate 1 and 2, respectively. For the plasmid DNA, we obtained ∼11 million reads.
For the cDNA replicates, we mapped the reads to an “artificial genome” in which each chromosome corresponds to a sample barcode. Each chromosome was composed of repeats of the 8-nt sample barcode, 18-nt constant region, 12-nt variant barcode, 4 nt from the variable region (42 nt total), and 60 “N's. We obtained paired-end NextSeq 500 reads in the length of 42 nt for R1 and 110 nt for R2. R1 reads shorter than 40 nt were discarded, while the rest were trimmed to a maximum of 42 nt and mapped to the artificial genome using NovoAlign aligner (http://www.novocraft.com/products/novoalign/), filtered for minimal mapping quality of 60 and for perfectly aligned reads for the length of 40–42 nt, and the number of reads for each designed oligo in each sample was counted. For the plasmid sample, we mapped the reads to a single “artificial chromosome” excluding the 8-nt sample barcode. We obtained the same paired-end NextSeq 500 reads and mapped them similarly to the cDNA replicates with the following differences. The R1 and R2 reads were combined and trimmed to a maximum of 34 nt, mapped to the artificial chromosome, and filtered for perfectly aligned reads for the length of 32–34 nt.
Computing RNA expression levels
To calculate RNA expression levels for a given variant, we required that it would have at least 10 DNA reads. For each variant, we calculated the log2 (cDNA reads + 1/plasmid DNA reads) as an estimate for normalized RNA levels. The one pseudocount in the numerator is added in order to account for the detection limit of the assay. Since the agreement between the replicates was high (R = 0.99, P < 10−10), we summed the reads between replicates for each variant and repeated the calculation for the normalized RNA levels. Only ∼4% of the variants that had at least 10 DNA reads had zero cDNA reads. For the rest of the constructs, the normalized RNA level was set to None (Supplemental Table S6).
Computing cleavage efficiency
For reads whose R1 was properly mapped, the R2 reads were filtered to perfectly match the pattern of the reverse transcription primer, CACGACGCTCTTCCGATCTNANCNGNTNANCNGNTNANCNGNANCNAN. Leading T nucleotides were stripped from the reads. Reads whose remaining length was <10 nt were discarded. For each variant, a FASTQ file of all of its remaining R2 reads was generated and mapped to a reference sequence of the variant followed by a 307-nt constant sequence from the plasmid backbone using NovoAlign aligner (http://www.novocraft.com/products/novoalign/) without soft clipping. For each variant, at each position, the number of reads with a perfect match for at least the first two nucleotides was counted and the data was arranged in a matrix where each row is a variant and each column is a position. To reduce noise in our measurements, we applied a number of filtering steps. First, positions with less than three reads were set to zero. Second, positions that got <10% of the reads that the variant received were set to zero. Finally, for constructs that remained with <50% of the reads they had before filtering, we set the entire row to zero. To calculate the cleavage efficiency, we calculated log2 (position cDNA reads/plasmid DNA reads) + 11 for each position for each variant that had a minimum of 10 DNA reads. The shift by 11 was applied in order to shift all values to the positive scale. Positions that resulted in a negative infinity following the log2 calculation were set back to zero (Supplemental Table S7).
Scanning mutagenesis imputation of mutant cleavage efficiencies
In our scanning mutagenesis data, mutant constructs with missing cleavage efficiency at a position corresponding to a WT cleavage site were imputed with the expected detection efficiency. The formula log2 (2/plasmid DNA reads) + 11 was used since, when calculating the cleavage efficiencies, we required a minimum of three cDNA reads per position per variant.
A set of endogenous 3′ UTRs for motif enrichment and cleavage site prediction
GENCODE V28lift37 comprehensive gene annotations for GRCh37 were downloaded from the UCSC Table Browser (Harrow et al. 2012). The published K562 3′ end sequencing data (Lin et al. 2012) were assigned with the GENCODE genomic annotations. Cleavage sites originating from 3′ UTR exons of coding genes and supported by more than 10 3′ end sequencing reads were selected. For each gene, a representative 3′ UTR sequence was selected starting at the 5′-most position. The extracted sequences began 250 bp upstream of the 3′ UTR start and extended to 1000 bp downstream from the cleavage site as indicated in the K562 3′ end sequencing data. The set of genes with unique sequences contained 6964 genes (Supplemental Table S8). GRCh37 was complete and adequate for the performed analysis; thus, GRCh38 would not significantly affect our conclusions.
Motif analysis
Motif discovery was performed using DREME 5.0.2 (Bailey 2011). Positive and negative sets were used as described in the text. DREME was run with the following parameters: -mink 6 -maxk 8 -g 20000 -norc -rna. The discovered motifs were subjected to enrichment analysis with AME 5.0.2 (McLeay and Bailey 2010) in the set of endogenous 3′ UTRs described above. Enrichment of upstream motifs was performed on 100-bp sequences upstream of the cleavage site with the next 100 bp serving as the negative set. Enrichment of downstream motifs was performed on 50-bp sequences downstream from the cleavage site with the sequence at 100–150 bp downstream serving as the negative set.
Analysis of positional preference for the discovered motifs was performed using CentriMo 5.0.2 (Bailey and Machanick 2012). CentriMo was run with the positive and negative data sets which were used for AME (McLeay and Bailey 2010) and with the following parameters: --norc –local. The CentriMo output was filtered for motifs with a Fisher E-value < 0.01, the per position counts were normalized by their sum to get probabilities and smoothed with a moving average filter with a window size of five.
Calculation of ΔGopen
To calculate ΔGopen, we used the RNAfold function from Vienna RNA 2.4.9 4 (Lorenz et al. 2011). We calculated the ensemble free energy for the examined sequence and subtracted from it the ensemble free energy with the constraint that the canonical hexamer along with 15-nt upstream and downstream regions are kept unpaired. The calculated difference is ΔGopen.
Prediction of expression levels
The data used for our model consisted of 11,822 sequences for which we quantified mRNA expression levels. The data excluded the subset of constructs which varied only in the barcode sequence. We used ∼90% of the data for training and ∼10% for held-out test data. When splitting the data, we stratified the variants by the mutagenesis scheme and made sure that scanning mutagenesis variants that share the same WT PAS are kept in the same data set. In a similar manner, we split the training data into 10-folds for 10-fold cross-validation (Supplemental Table S9).
All deep learning predictions were made using a CNN designed with keras 2.0.6 (https://keras.io/) in Python 2.7.8. The basis for our architecture was inspired by previous deep learning work applied to genomics data (Zhou and Troyanskaya 2015; Angermueller et al. 2016; Kelley et al. 2016; Quang and Xie 2016; Gao et al. 2018; Leung et al. 2018; Bogard et al. 2019). The 250-nt input sequence, starting at the stop codon, was one hot encoded and fed into the model. The model consisted of a 1D convolution layer (num_filters = 64, kernel_size = 8) with relu activation, max pooling layer (pool_size = 2, strides = 1), dropout layer (rate = 0.5), 1D convolution layer (num_filters = 32, kernel_size = 6) with relu activation, max pooling layer (pool_size = 2, strides = 1), dropout layer (rate = 0.5), flatten layer, and a final dense layer with linear activation, and a single neuron output. L1 (0.0001) and L2 (0.0001) regularization was used in all the convolution and dense layers. The model was compiled with Adam optimizer (lr = 0.0015) and minimum squared error loss. The model was fitted with 75 epochs with a batch size of 256. The gradient boosting tree-based model was made using XGBoost (Chen and Guestrin 2016) 0.72.1 with default parameters except for “objective”: “reg:linear”, “n_estimators”: 300. The 250-nt input sequence, starting at the stop codon, was one hot encoded, flattened, and fed into the model. For the k-mer elastic net, we extracted nonoverlapping k-mer counts for k-mers of length one to six for each 250-nt sequence in the training and test data. The elastic net model (α = 2.15 × 10−2 and l1_ratio = 0.4) was implemented using scikit-learn 0.18.2 (Pedregosa et al. 2011). To assess the performance of each of the models on training data, we employed 10-fold cross-validation. To assess performance of our model on the held-out test data, we trained the model on all of our training data and predicted the values for the held-out test data. The model performance was evaluated by Pearson's correlation between the measured and predicted expression values. In addition to the described hyperparameters above, we tried numerous semirationally chosen hyperparameter combinations for each of the models and chose the set of parameters that produced optimal results in terms of loss curve behaviors in train and test data and the Pearson's correlation between the measured and predicted expression values.
Prediction of cleavage efficiencies
The data used for our model consisted of 11,822 sequences for which we quantified mRNA expression levels, excluding the duplicate barcodes for the multiple barcodes subset. The output label used was the 189 positions vector in which the cleavage sites were detected, starting from position 41 and ending at position 230 after the stop codon. Variants for which no cleavage sites were detected were still included with a vector consisting of only zeros.
All predictions were made using a CNN designed with keras 2.0.6 (https://keras.io/) in Python 2.7.8. The 250-nt input sequence was one hot encoded and fed into the model. The model consisted of a 1D convolution layer (num_filters = 128, kernel_ size = 12) with relu activation, max pooling layer (pool_size = 2, strides = 1), dropout layer (rate = 0.5), 1D convolution layer (num_ filters = 64, kernel_size = 8) with relu activation, max pooling layer (pool_ size = 2, strides = 1), dropout layer (rate = 0.5), flatten layer, dense layer (units = 4096) with relu activation, and a final dense layer with relu activation and 189 output neurons, each for a considered position of cleavage. L1 (0.0001) and L2 (0.0001) regularization was used in all the convolution and dense layers except for the last dense layer, where L1 (10−6) and L2(0) regularization was used. The model was compiled with Adam optimizer (lr = 0.0015) and Poisson loss. The model was fitted with 100 epochs with a batch size of 512. To assess the performance of our model on training data, we employed 10-fold cross-validation. To assess performance of our model on the held-out test data, we trained the model on all of our training data and predicted the values for the held-out test data. The model performance was evaluated by comparing the distribution of the per position mean cleavage efficiency of the predicted values and the measured ones. In addition, we preferred models that were able to predict cleavage efficiency for all the constructs that had measured values. Finally, for the constructs that had both measured and predicted cleavage efficiency maps, we examined the distribution of the absolute differences between predicted and measured cleavage positions. The cleavage positions were defined as the index of maximal measured or predicted values along the vector of 189 positions. In addition to the described hyperparameters above, we tried numerous semirationally chosen hyperparameter combinations and chose the set of parameters that produced optimal results in terms of loss curve behaviors in train and test data and the described model performance evaluation.
Expression model interpretation
To convert the first layer convolutional filters into sequence motifs, we fed the first layer with our test set data and acquired the layer activations. The layer activations were converted into motifs by searching for the position of maximal activation per sequence and extracting the DNA sequence corresponding to the kernel size (Alipanahi et al. 2015). Then, the sequences were converted into a motif object using Biopython 1.68, and the position weight matrices were converted into MEME format. The motifs were used for motif enrichment analysis using AME (McLeay and Bailey 2010) in the endogenous 3′ UTR data upstream of and downstream from the cleavage site as described for the motif analysis. Only motifs with enrichment P-value < 0.001 were reported.
Prediction of cleavage sites on endogenous 3′ UTRs
Each of the expression models was applied to the endogenous sequences described above in a base-by-base manner. The index of the highest score was added to 145, the most frequent cleavage site within the library, and designated as the predicted cleavage site. For polya_svm (Cheng et al. 2006) predictions, we supplied the endogenous sequences as input and set the min_score to zero in order to get all possible predictions. For each sequence, the position with the maximal score was designated as the predicted cleavage site.
Data access
The sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA548062. All other processed data can be found in the Supplemental Material. All the code required to train the predictors and to execute the predictions on the endogenous sequences has been submitted as Supplemental Code and is also available on GitHub (https://github.com/segallab/PolyApredictors).
Supplementary Material
Acknowledgments
We thank members of the Segal lab for useful discussions. E.S. is supported by the Crown Human Genome Center; D.L. Schwarz; J.N. Halpern; L. Steinberg; J. Benattar; Aliza Moussaieff; Adelis Foundation; and grants funded by the European Research Council and the Israel Science Foundation.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.247312.118.
References
- Akhtar MN, Bukhari SA, Fazal Z, Qamar R, Shahmuradov IA. 2010. POLYAR, a new computer program for prediction of poly(A) sites in human sequences. BMC Genomics 11: 646 10.1186/1471-2164-11-646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipanahi B, Delong A, Weirauch MT, Frey BJ. 2015. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol 33: 831–838. 10.1038/nbt.3300 [DOI] [PubMed] [Google Scholar]
- Angermueller C, Pärnamaa T, Parts L, Stegle O. 2016. Deep learning for computational biology. Mol Syst Biol 12: 878 10.15252/msb.20156651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou M, Geraghty F, Hurst J, Grosveld F. 1998. Efficient 3′-end formation of human β-globin mRNA in vivo requires sequences within the last intron but occurs independently of the splicing reaction. Nucleic Acids Res 26: 721–729. 10.1093/nar/26.3.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga PS, Ford LP, Chen F, Wilusz J. 1995. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res 23: 1625–1631. 10.1093/nar/23.9.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. 10.1093/bioinformatics/btr261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Machanick P. 2012. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40: e128 10.1093/nar/gks433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogard N, Linder J, Rosenberg AB, Seelig G. 2019. A deep neural network for predicting and engineering alternative polyadenylation. Cell 178: 91–106.e23. 10.1016/j.cell.2019.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnlein S, Hauber J, Cullen BR. 1989. Identification of a U5-specific sequence required for efficient polyadenylation within the human immunodeficiency virus long terminal repeat. J Virol 63: 421–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brister JR, Ako-Adjei D, Bao Y, Blinkova O. 2015. NCBI Viral Genomes Resource. Nucleic Acids Res 43: D571–D577. 10.1093/nar/gku1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T-H, Wu L-C, Chen Y-T, Huang H-D, Liu B-J, Cheng K-F, Horng J-T. 2011. Characterization and prediction of mRNA polyadenylation sites in human genes. Med Biol Eng Comput 49: 463–472. 10.1007/s11517-011-0732-4 [DOI] [PubMed] [Google Scholar]
- Chen T, Guestrin C. 2016. XGBoost: a scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 785–794. New York. [Google Scholar]
- Cheng Y, Miura RM, Tian B. 2006. Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics 22: 2320–2325. 10.1093/bioinformatics/btl394 [DOI] [PubMed] [Google Scholar]
- Connelly S, Manley JL. 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev 2: 440–452. 10.1101/gad.2.4.440 [DOI] [PubMed] [Google Scholar]
- Deng Z, Zhang S, Gu S, Ni X, Zeng W, Li X. 2018. Useful bicistronic reporter system for studying poly(A) site-defining cis elements and regulation of alternative polyadenylation. Int J Mol Sci 19: 279 10.3390/ijms19010279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang J, Wei Z, Hakonarson H. 2018. DeepPolyA: a convolutional neural network approach for polyadenylation site prediction. IEEE Access 6: 24340–24349. 10.1109/ACCESS.2018.2825996 [DOI] [Google Scholar]
- Goodman DB, Church GM, Kosuri S. 2013. Causes and effects of N-terminal codon bias in bacterial genes. Science 342: 475–479. 10.1126/science.1241934 [DOI] [PubMed] [Google Scholar]
- Goodwin EC, Rottman FM. 1992. The 3′-flanking sequence of the bovine growth hormone gene contains novel elements required for efficient and accurate polyadenylation. J Biol Chem 267: 16330–16334. [PubMed] [Google Scholar]
- Graveley BR, Gilmartin GM. 1996. A common mechanism for the enhancement of mRNA 3′ processing by U3 sequences in two distantly related lentiviruses. J Virol 70: 1612–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans H, Alwine JC. 2000. Functionally significant secondary structure of the simian virus 40 late polyadenylation signal. Mol Cell Biol 20: 2926–2932. 10.1128/MCB.20.8.2926-2932.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart RP, McDevitt MA, Ali H, Nevins JR. 1985. Definition of essential sequences and functional equivalence of elements downstream of the adenovirus E2A and the early simian virus 40 polyadenylation sites. Mol Cell Biol 5: 2975–2983. 10.1128/MCB.5.11.2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J, Lutz, CS, Wilusz, J, Tian, B. 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11: 1485–1493. 10.1261/rna.2107305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo C, de Castro E, Masson P, Bougueleret L, Bairoch A, Xenarios I, Le Mercier P. 2011. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res 39: D576–D582. 10.1093/nar/gkq901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Snoek J, Rinn JL. 2016. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res 26: 990–999. 10.1101/gr.200535.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284. 10.1038/ng2135 [DOI] [PubMed] [Google Scholar]
- Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, Alston J, Mikkelsen TS, Kellis M. 2013. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res 23: 800–811. 10.1101/gr.144899.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Gautheret D. 2003. Sequence determinants in human polyadenylation site selection. BMC Genomics 4: 7 10.1186/1471-2164-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MKK, Delong A, Frey BJ. 2018. Inference of the human polyadenylation code. Bioinformatics 34: 2889–2898. 10.1093/bioinformatics/bty211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt N, Briggs D, Gil A, Proudfoot NJ. 1989. Definition of an efficient synthetic poly(A) site. Genes Dev 3: 1019–1025. 10.1101/gad.3.7.1019 [DOI] [PubMed] [Google Scholar]
- Lin Y, Li Z, Ozsolak F, Kim SW, Arango-Argoty G, Liu TT, Tenenbaum SA, Bailey T, Monaghan AP, Milos PM, et al. 2012. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res 40: 8460–8471. 10.1093/nar/gks637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubliner S, Regev I, Lotan-Pompan M, Edelheit S, Weinberger A, Segal E. 2015. Core promoter sequence in yeast is a major determinant of expression level. Genome Res 25: 1008–1117. 10.1101/gr.188193.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Moreira A. 2011. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip Rev RNA 2: 22–31. 10.1002/wrna.47 [DOI] [PubMed] [Google Scholar]
- Magana-Mora A, Kalkatawi M, Bajic VB. 2017. Omni-PolyA: a method and tool for accurate recognition of Poly(A) signals in human genomic DNA. BMC Genomics 18: 620 10.1186/s12864-017-4033-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín RM, Vaníček J. 2011. Efficient use of accessibility in microRNA target prediction. Nucleic Acids Res 39: 19–29. 10.1093/nar/gkq768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoulkova E, Michalova E, Vojtesek B, Hrstka R. 2012. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol 9: 563–576. 10.4161/rna.20231 [DOI] [PubMed] [Google Scholar]
- McDevitt MA, Hart RP, Wong WW, Nevins JR. 1986. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J 5: 2907–2913. 10.1002/j.1460-2075.1986.tb04586.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeay RC, Bailey TL. 2010. Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11: 165 10.1186/1471-2105-11-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogno I, Kwasnieski JC, Cohen BA. 2013. Massively parallel synthetic promoter assays reveal the in vivo effects of binding site variants. Genome Res 23: 1908–1915. 10.1101/gr.157891.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A, Wollerton M, Monks J, Proudfoot NJ. 1995. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J 14: 3809–3819. 10.1002/j.1460-2075.1995.tb00050.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ. 1998. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev 12: 2522–2534. 10.1101/gad.12.16.2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Boryń ŁM, Arnold CD. 2015. STARR-seq—Principles and applications. Genomics 106: 145–150. 10.1016/j.ygeno.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Natalizio BJ. 2002. Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J Biol Chem 277: 42733–42740. 10.1074/jbc.M208070200 [DOI] [PubMed] [Google Scholar]
- Noderer WL, Flockhart RJ, Bhaduri A, Diaz de Arce AJ, Zhang J, Khavari PA, Wang CL. 2014. Quantitative analysis of mammalian translation initiation sites by facs-seq. Mol Syst Biol 10: 748 10.15252/msb.20145136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes NM, Li W, Tian B, Furger A. 2010. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J 29: 1523–1536. 10.1038/emboj.2010.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12: 2825–2830. [Google Scholar]
- Quang D, Xie X. 2016. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res 44: e107 10.1093/nar/gkw226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Pieper L, Chew G-L, Schier AF. 2017. A massively parallel reporter assay of 3′ UTR sequences identifies in vivo rules for mRNA degradation. Mol Cell 68: 1083–1094.e5. 10.1016/j.molcel.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AB, Patwardhan RP, Shendure J, Seelig G. 2015. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell 163: 698–711. 10.1016/j.cell.2015.09.054 [DOI] [PubMed] [Google Scholar]
- Sadofsky M, Connelly S, Manley JL, Alwine JC. 1985. Identification of a sequence element on the 3′ side of AAUAAA which is necessary for simian virus 40 late mRNA 3′-end processing. Mol Cell Biol 5: 2713–2719. 10.1128/MCB.5.10.2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schek N, Cooke C, Alwine JC. 1992. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol Cell Biol 12: 5386–5393. 10.1128/MCB.12.12.5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sharon E, Lubliner S, Regev I, Lotan-Pompan M, Yakhini Z, Segal E. 2015. Systematic dissection of the sequence determinants of gene 3′ end mediated expression control. PLoS Genet 11: e1005147 10.1371/journal.pgen.1005147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E, Kalma Y, Sharp A, Raveh-Sadka T, Levo M, Zeevi D, Keren L, Yakhini Z, Weinberger A, Segal E. 2012. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol 30: 521–530. 10.1038/nbt.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E, van Dijk D, Kalma Y, Keren L, Manor O, Yakhini Z, Segal E. 2014. Probing the effect of promoters on noise in gene expression using thousands of designed sequences. Genome Res 24: 1698–1706. 10.1101/gr.168773.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP. 1990. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res 18: 5799–5805. 10.1093/nar/18.19.5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrikumar A, Greenside P, Kundaje A. 2017. Learning important features through propagating activation differences. arXiv:1704.02685 [cs.CV].
- Sittler A, Gallinaro H, Jacob M. 1994. Upstream and downstream cis-acting elements for cleavage at the L4 polyadenylation site of adenovirus-2. Nucleic Acids Res 22: 222–231. 10.1093/nar/22.2.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, Ovcharenko I, Ahituv N. 2013. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet 45: 1021–1028. 10.1038/ng.2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LF, Saetrom P. 2012. Single nucleotide polymorphisms can create alternative polyadenylation signals and affect gene expression through loss of microRNA-regulation. PLoS Comput Biol 8: e1002621 10.1371/journal.pcbi.1002621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg Slutskin I, Weingarten-Gabbay S, Nir R, Weinberger A, Segal E. 2018. Unraveling the determinants of microRNA mediated regulation using a massively parallel reporter assay. Nat Commun 9: 529 10.1038/s41467-018-02980-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A, Zeichner S, Carswell S, Alwine JC. 1991. The human immunodeficiency virus type 1 polyadenylylation signal: a 3′ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci 88: 2108–2112. 10.1073/pnas.88.6.2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zheng D, Yehia G, Tian B. 2018. A compendium of conserved cleavage and polyadenylation events in mammalian genes. Genome Res 28: 1427–1441. 10.1101/gr.237826.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. 2016. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351: aad4939 10.1126/science.aad4939 [DOI] [PubMed] [Google Scholar]
- Wu X, Bartel DP. 2017. Widespread influence of 3′-end structures on mammalian mRNA processing and stability. Cell 169: 905–917.e11. 10.1016/j.cell.2017.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon OK, Hsu TY, Im JH, Brem RB. 2012. Genetics and regulatory impact of alternative polyadenylation in human B-lymphoblastoid cells. PLoS Genet 8: e1002882 10.1371/journal.pgen.1002882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. 2003. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res 31: 1375–1386. 10.1093/nar/gkg241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cole CN. 1987. Identification of a complex associated with processing and polyadenylation in vitro of herpes simplex virus type 1 thymidine kinase precursor RNA. Mol Cell Biol 7: 3277–3286. 10.1128/MCB.7.9.3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Denome RM, Cole CN. 1986. Fine-structure analysis of the processing and polyadenylation region of the herpes simplex virus type 1 thymidine kinase gene by using linker scanning, internal deletion, and insertion mutations. Mol Cell Biol 6: 4611–4623. 10.1128/MCB.6.12.4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Troyanskaya OG. 2015. Predicting effects of noncoding variants with deep learning–based sequence model. Nat Methods 12: 931–934. 10.1038/nmeth.3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.