Abstract
Dysregulated endogenous retroelements (EREs) are increasingly implicated in the initiation, progression, and immune surveillance of human cancer. However, incomplete knowledge of ERE activity limits mechanistic studies. By using pan-cancer de novo transcript assembly, we uncover the extent and complexity of ERE transcription. The current assembly doubled the number of previously annotated transcripts overlapping with long-terminal repeat (LTR) elements, several thousand of which were expressed specifically in one or a few related cancer types. Exemplified in melanoma, LTR-overlapping transcripts were highly predictable, disease prognostic, and closely linked with molecularly defined subtypes. They further showed the potential to affect disease-relevant genes, as well as produce novel cancer-specific antigenic peptides. This extended view of LTR elements provides the framework for functional validation of affected genes and targets for cancer immunotherapy.
The human genome hosts diverse families of endogenous retroelements (EREs), many of which have amplified their copies to staggering numbers (International Human Genome Sequencing Consortium 2001; de Koning et al. 2011). These comprise human endogenous retroviruses (HERVs) and mammalian apparent long-terminal repeat (LTR)-retrotransposons (MaLRs), distinguished by LTRs flanking the canonical proviral genomes and collectively referred to as LTR elements, and the larger group of non-LTR elements, which include long and short interspersed nuclear elements (LINEs and SINEs, respectively) and composite SINE-VNTR-Alu (SVA) elements (Burns and Boeke 2012; Feschotte and Gilbert 2012). The vast majority of human genomic ERE integrations are incomplete and mutated copies, incapable of replication. Nevertheless, many retain functional parts, including promoter or enhancer activities of the LTRs, splice donor and acceptor sites, polyadenylation sites, and even intact open reading frames (ORFs) for biologically active proteins, all of which have the potential to alter host physiology (Burns and Boeke 2012; Feschotte and Gilbert 2012; Kassiotis and Stoye 2016). Potential risks posed by functional components of ERE integrations are mitigated by epigenetic and splicing repression, largely preventing ERE expression or exonization (Feschotte and Gilbert 2012). However, this type of epigenetic control is reversible and often lost, particularly in the context of the altered chromatin landscape of cancer initiation and evolution (Baylin and Jones 2011; Flavahan et al. 2017).
Dysregulated EREs can affect cancer progression through distinct mechanisms (Romanish et al. 2010; Kassiotis and Stoye 2017). Examples include LTR-driven overexpression of proto-oncogenes, such as CSF1R overexpression in Hodgkin's lymphoma and anaplastic large-cell lymphoma (Lamprecht et al. 2010), or creation of truncated oncogenic forms of kinases through alternative splicing to an LTR element, such as anaplastic lymphoma kinase (ALK) in melanoma (Wiesner et al. 2015), and the erb-b2 receptor tyrosine kinase 4 (ERBB4) in ALK-negative anaplastic large-cell lymphoma (Scarfo et al. 2016). Genetic, pharmacologic, or cytokine-mediated transcriptional induction of LTR elements is incriminated in the activation of cancer cell–intrinsic innate immunity (Chiappinelli et al. 2015; Roulois et al. 2015; Goel et al. 2017; Cañadas et al. 2018; Sheng et al. 2018), and expression of LTR element clusters is linked with the strength of local antitumor immunity and the outcome of immunotherapy (Rooney et al. 2015; Desai et al. 2017; Smith et al. 2018). Moreover, canonical retroviral proteins or protein fragments encoded by a few distinct HERV-K, HERV-E, and HERV-H proviruses can be targeted by functionally relevant T cell and B cell responses (Schiavetti et al. 2002; Rakoff-Nahoum et al. 2006; Takahashi et al. 2008; Mullins and Linnebacher 2012; Cherkasova et al. 2016; Smith et al. 2018), but the potential of the multitude of noncanonical or chimeric ERE transcripts to generate cancer-specific antigenic epitopes has not yet been explored.
Despite their potential importance, complete understanding of the role of EREs in cancer is currently hampered by gaps in the annotation and quantitation of the transcriptional activity in cancer of the compendium of ERE-overlapping transcripts the human genome can produce. By using de novo assembly of cancer transcriptomes, we aimed to provide a comprehensive view of LTR element transcriptional behavior in human cancer.
Results
A comprehensive LTR retroelement transcriptome
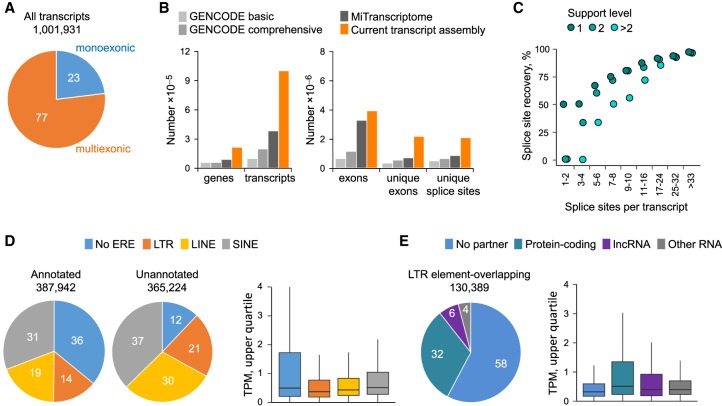
To identify and quantify unannotated or partially annotated LTR element–overlapping transcripts, we first de novo-assembled a comprehensive cancer transcriptome (Methods). To this end, RNA-seq reads from 768 patient samples, obtained from The Cancer Genome Atlas (TCGA) program and representing 31 cancer types (Supplemental Table S1), were used for genome-guided assembly. This process generated 1,001,931 contigs, the majority of which were multiexonic (Fig. 1A). Comparison with other comprehensive ab initio transcript assemblies, such as the Encyclopedia of DNA Elements subproject GENCODE (Frankish et al. 2019) and MiTranscriptome (Iyer et al. 2015), indicated considerably increased representation of genes, transcripts, unique exons, and unique splice sites in the current assembly (Fig. 1B). Moreover, comparison with splice sites in high-confidence GENCODE transcripts revealed recovery of 93% of all splice sites and an average of only one splice site missing per annotated transcript (Fig. 1C).
Figure 1.
Assembly, recovery, and expression of ERE-overlapping transcripts in tumors of diverse origins. (A) Total number and proportion of monoexonic or multiexonic de novo–assembled transcripts. (B) Comparison of the total number of genes, transcripts, exons, unique exons, and unique splice sites in the current transcript assembly with GENCODE (version 24) (Frankish et al. 2019) and MiTranscriptome (Iyer et al. 2015). Genes are defined here as nonoverlapping transcribed regions. (C) Completeness of the current transcript assembly, estimated by median recovery of splice sites annotated in GENCODE. The percentage of GENCODE recovered sites is plotted according to their support levels. Recovery of the 367,411 unique splice sites of high-confidence GENCODE transcripts was ∼93%. (D) Prior annotation status and ERE composition of the 753,166 transcripts out of the entire assembly that were expressed at one or more transcripts per million (TPM) in at least one sample (left) and expression levels of these transcripts according to their ERE composition (right). Transcripts were considered as previously annotated if all exons were present within GENCODE (v24 basic) and as ERE-overlapping if any exon overlapped with an ERE integration. For transcripts overlapping with multiple EREs, we assigned a hierarchical LTR, LINE, or SINE order. As overall expression level, we used the upper quartile TPM in the cancer type with highest expression for each transcript. (E) Breakdown of LTR element–overlapping transcripts (expressed at one or more TPM in at least one sample) according to overlap with protein-coding, lncRNA, or other RNA genes (left) and expression levels (upper quartile TPM in the cancer type with highest expression) or each type of LTR element–overlapping transcript (right).
To examine the representation of LTR or non-LTR elements in the cancer transcriptome, the assembled contigs were overlaid with a genomic repeat sequence annotation, generated through RepeatMasker (Smit et al. 2013–2015). We first concentrated on transcripts with expression level of one or more transcripts per million (TPM) in at least one sample (a total of 753,166 transcripts). These comprised both previously annotated (fully or partially) and unannotated transcripts in similar proportions (Fig. 1D). Transcripts overlapping EREs, particularly LTR elements or LINEs, were considerably enriched in the unannotated fraction (Fig. 1D). Expression of such transcripts was, on average, lower than of transcripts that did not include any EREs (Fig. 1D). Hence, the assembly captured transcripts using EREs, including those expressed at comparably lower levels.
Closer inspection of the subset of transcripts that contained at least one complete or partial LTR element revealed that more than half did not intersect any annotated gene (Fig. 1E). Approximately one-third of LTR element–overlapping transcripts spanned a protein-coding gene, whereas a much smaller proportion spanned long noncoding RNA (lncRNA) or other RNA genes (Fig. 1E). Expression of LTR element–overlapping transcripts was significantly higher if they also spanned protein-coding genes or lncRNA genes than if they were stand-alone LTR elements (Fig. 1E). These findings suggest that the current assembly was enriched for splice isoforms of annotated genes, as well as previously unannotated ERE-overlapping transcripts, likely owing to lack of bias against repetitive elements, compared with previous assemblies (Iyer et al. 2015; Frankish et al. 2019).
Cancer-specific LTR retroelement–overlapping transcripts
As the transcript assembly was based on cancer sample RNA-seq reads, it was expected to include transcripts that were expressed in a cancer-specific manner. To identify such cancer-specific LTR element–overlapping transcripts (referred to here as CLTs), we compared cancer samples with a collection of 811 samples from a wide variety of healthy tissues, using RNA-seq data obtained from TCGA, as well as the Genotype Tissue Expression (GTEx) Consortium (Supplemental Tables S2, S3). Transcripts were considered cancer-specific if they fulfilled the following criteria: (1) Their 75th percentile expression in a given cancer type was more than one TPM, three or more times the highest median expression in any healthy tissue, and three or more times the 90th percentile expression in the respective healthy tissue, and (2) their 90th percentile expression in healthy tissue samples was less than 10 TPM. This combination pruned the number of potential transcripts down to a total of 5923 transcripts containing complete or partial LTR elements and expressed in a cancer-specific manner (Supplemental Table S4).
As further validation, we intersected the identified CLTs with previously described examples of LTR element exaptation in the expression of oncogenes and cancer-associated lncRNAs (Babaian and Mager 2016; Jang et al. 2019). Although 73% (92 of 116) of precompiled loci (Babaian and Mager 2016; Jang et al. 2019) were present in the assembly, only 15 of them fulfilled the cancer specificity criteria we set (Supplemental Table S5), with the remaining either expressed also in healthy tissue or not expressed sufficiently recurrently in cancer patients.
Although a substantial number, the identified CLTs represented only a small proportion of genomic or transcriptionally used LTR elements. For example, of a total of 630,356 annotated genomic LTR elements, 108,946 (17.3%) were present in assembled transcripts and expressed at one or more TPM in at least one healthy or tumor sample. Of those, only 20,164 (3.2% of annotated LTR elements) were expressed specifically in cancer, with the remaining expressed either additionally (12.8%) or only (1.3%) in healthy tissues. When considering recurrence of expression between individuals, the 5923 CLTs expressed specifically and recurrently in cancer overlap with only 0.9% of annotated LTR elements, contrasting with a total of 66,277 LTR-overlapping transcripts expressed recurrently in healthy tissues (using 8.5% of annotated LTR elements). Therefore, LTR elements used specifically and recurrently in cancer represent one in 10 LTR elements used in healthy tissues and one in 100 genomic LTR elements.
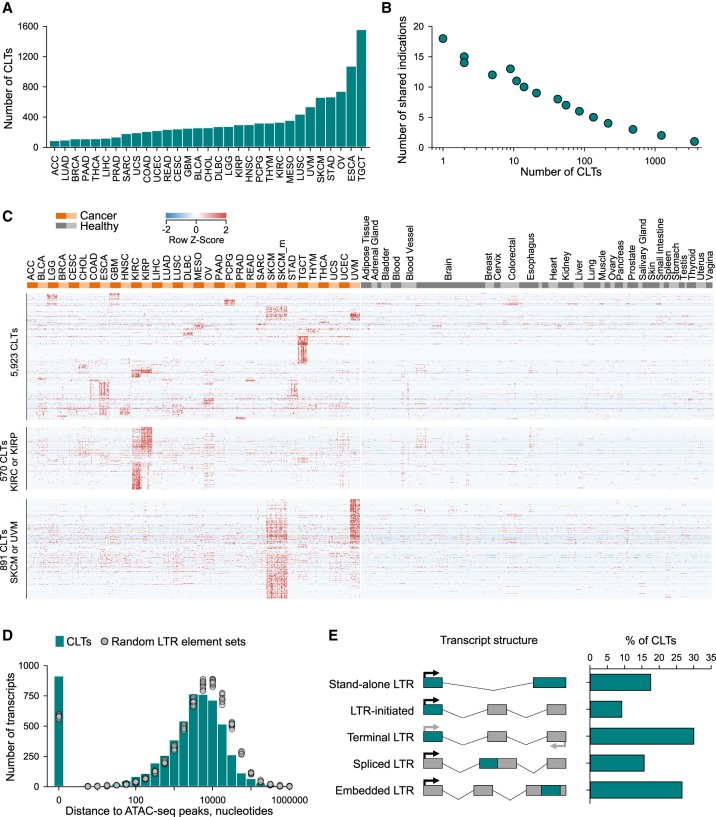
Whereas most cancer types showed significantly elevated expression of 100–300 of the identified 5923 transcripts, a few stood out, with testicular germ cell tumors (TGCTs) and esophageal carcinoma (ESCA) each expressing more than 1000 such CLTs (Fig. 2A). Conversely, the vast majority (>95%) of CLTs were expressed specifically in one or up to five different cancer types, and only 44 CLTs were shared by 10 or more cancer types (Fig. 2B), showing tissue-type specificity of LTR element utilization. Indeed, although cancer-specific, expression of CLTs clustered primarily according to tissue type, with most clusters restricted to one cancer type (Fig. 2C). Consequently, a considerable degree of overlap in CLT expression was observed in cancer types involving related tissues, such as kidney renal clear cell carcinoma (KIRC) and kidney renal papillary cell carcinoma (KIRP) or histotypes such as skin cutaneous melanoma (SKCM), both primary and metastatic, and uveal melanoma (UVM) (Fig. 2C). Tissue-restricted expression of CLTs was further supported by analysis of RNA-seq data from 933 cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE) (Supplemental Fig. S1), which are homogenous cell populations.
Figure 2.
Abundance of cancer-specific LTR element–overlapping transcripts. (A) Total number of CLTs identified per cancer type. (B) Overlap of CLT expression between cancer types, plotted as the number of CLTs against the number of cancer types sharing a given CLT. (C) Heatmap of expression values in cancer patient and healthy control samples of all 5923 identified CLTs (top), KIRC-specific and KIRP-specific CLTs (570) (middle), and SKCM-specific and UVM-specific CLTs (891) (bottom). (D) Proximity, in nucleotides, of the identified CLT TSS to ATAC-seq peaks. Also shown is the proximity of ATAC-seq peaks to the center of 10 random sets of similar numbers of LTR elements. (E) Composition of identified CLTs according to the indicated position of the LTR element in the transcript structure.
The distinct pattern of CLT expression among cancer types was suggestive of locus-specific epigenetic changes controlling LTR element activity. Indeed, transcription start sites (TSSs) of CLTs were in close proximity (P < 0.01) to assay for transposase-accessible chromatin using sequencing (ATAC-seq) peaks, identified in a recent mapping of enhancers and promoters in primary tumor samples (Fig. 2D; Corces et al. 2018), consistent with cancer specificity of CLT expression owing to activation of a local regulatory element. Consequently, transcriptional inclusion of LTR elements covered diverse LTR element families in similar proportions in each cancer type, with a few exceptions (Supplemental Fig. S2). These included prominent HERV-H element cluster in TCGTs and a few additional indications, such as colon adenocarcinoma (COAD) (Supplemental Fig. S2), consistent with prior reports (Pérot et al. 2015; Desai et al. 2017). They also included a HERV-K element cluster in prostate adenocarcinoma (PRAD) (Supplemental Fig. S2), consisting of both HERV-K (HML-2) and evolutionary older HERV-K elements in equal proportions (12 and 11 transcripts, respectively). Of note, the majority of the HERV-K (HML-2)–overlapping transcripts in PRAD derived from a single provirus on Chromosome 22q11.23 (HERVK[Chr22q11.23]), consistent with reported expression of this provirus in PRAD (Goering et al. 2015), and partially overlapped with recently identified lncRNA prostate cancer associated transcript 14 (PCAT14) (Shukla et al. 2016), indicating that PCAT14 is, in fact, a HERVK[Chr22q11.23] transcript (Supplemental Fig. S2).
We next examined whether CLT expression resulted from derepression of LTR element promoter activity or from alternative promoters, by mapping the position of the LTR element in the overall structure of the identified CLTs. Stand-alone LTR elements represented 17.2% of CLTs, whereas an LTR element was found at the TSS in 9% of the CLTs (LTR-initiated) (Fig. 2E), suggesting it acted as the promoter of those transcripts. A terminal LTR element was found in a further 30% of the transcripts, whose strand was not known (Fig. 2E), although it is likely that the LTR element is at the TSS of the majority of these transcripts. In a quarter of cases, an LTR element was embedded in one of the exons, typically the last exon with an extended untranslated region (UTR) (embedded LTR), and in 15% of CLTs, an LTR element provided at least one splice site (spliced LTR) (Fig. 2E). The latter transcripts were chimeric, encompassing an LTR element and either a protein-coding or lncRNA gene in equal proportions (48% and 52%, respectively).
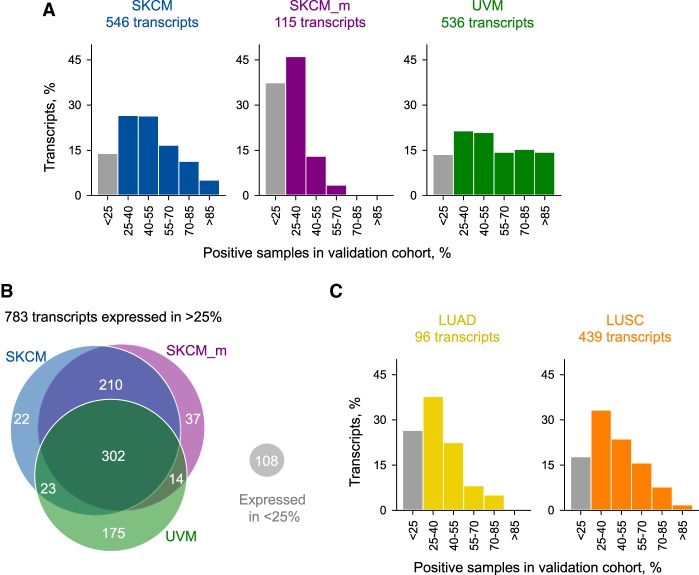
The structures of CLTs, as well as the specificity of their expression to individual cancer types, suggested a high degree of predictability of LTR element utilization through mechanisms, including LTR element–initiated transcription, as well as alternative splicing and inclusion of cryptic exons in transcripts initiated by alternative promoters. To confirm cancer specificity and prevalence of CLT expression, we extended our analysis to a larger cohort. For this purpose, we selected SKCM as an indication with well-characterized publicly available data and UVM as a related cancer histotype and analyzed a further 77 primary and 318 metastatic SKCM samples and 31 UVM samples. Of the 546 CLTs selected based on expression in more than a quarter of the primary SKCM discovery cohort, 470 (86%) were also expressed in more than a quarter of the primary SKCM validation cohort (Fig. 3A). In fact, primary SKCM-specific LTR element–overlapping transcripts were expressed on average in 60%, with some expressed in 98% of the analyzed samples (Fig. 3A). Highly comparable results were obtained also with the UVM validation cohort, in which 86% (463 of 536) of UVM-specific LTR element–overlapping transcripts were expressed above the threshold (Fig. 3A). Although still the majority (62%), only 72 of 115 CLTs expressed in the metastatic SKCM discovery cohort were expressed above the threshold also in the metastatic SKCM validation cohort (Fig. 3A). Moreover, in contrast to primary SKCM and UVM samples, only four CLTs were expressed in more than half and none were expressed in >70% of metastatic SKCM samples (Fig. 3A). Nevertheless, more than half (57%) of CLTs expressed in primary SKCM were also expressed in metastatic SKCM, and more than one-third (34%) were shared between SKCM and UVM (Fig. 3B). Similar validation rates were also obtained from analysis of further 395 lung adenocarcinoma (LUAD) and 338 lung squamous cell carcinoma (LUSC) samples, with 73% and 82% of CLTs identified based on expression in more than a quarter of each discovery cohort, also expressed in more than a quarter of the LUAD and LUSC validation cohorts, respectively (Fig. 3C). Validation rates in these cancer types were also comparable between LTR-initiated and other CLTs (Supplemental Fig. S3), indicating that LTR promoters can be used as recurrently between cancer patients as canonical promoters, in agreement with a recent independent study (Jang et al. 2019). Together, these results highlighted the abundance of LTR element–overlapping transcripts expressed specifically and predictably in cancer.
Figure 3.
Validation of CLT expression prevalence. (A) Percentage of CLTs expressed in each bin of percentage of positive samples in larger cohorts of primary SKCM (n = 77), metastatic SKCM (SKCM_m; n = 318), or UVM (n = 31). (B) Number and overlap between melanoma types of CLTs that were validated in the larger cohorts, that is, expressed in >25% of cancer patient samples in the validation cohort. (C) Percentage of CLTs expressed in each bin of percentage of positive samples in larger cohorts of LUAD (n = 395) or LUSC (n = 338). Samples were considered positive if transcript expression level was more than three times that of the highest median in any normal tissue.
Individual CLT expression patterns associated with melanoma subtypes and progression rate
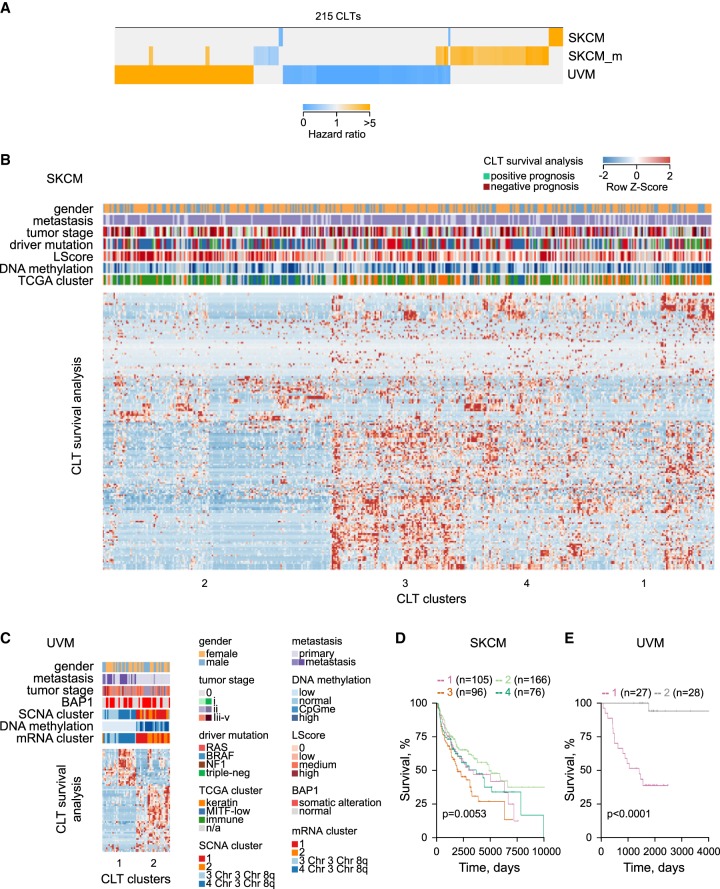
To further probe underlying reasons for cancer specificity of CLT expression, we investigated if expression of different CLTs associated with distinct cancer stages or subtypes. To this end, we focused on melanoma and looked for correlation between CLT expression with rates of melanoma progression or clinical and molecular subtypes. Of the 891 CLTs expressed in either SKCM or UVM, 215 (24%) were significantly associated with altered disease progression, revealed by hazard ratios for death between the 33rd and 66th expression percentiles (Fig. 4A). This association appeared to be disease specific, with expression levels of only few CLTs linked with the same outcome in primary or metastatic SKCM and UVM (Fig. 4A).
Figure 4.
Potential biological processes underlying melanoma association of melanoma-expressed CLTs. (A) Heatmap of hazard ratios, calculated by Cox regression model, of the 215 melanoma-expressed CLTs that were significantly associated with survival probability for each melanoma type of patients in the higher versus the lower expression tertiles. (B,C) Unsupervised clustering of 180 SKCM-prognostic CLTs (B) and 67 UVM-prognostic CLTs (C), according to their expression values (x-axis) and effect on survival probability (y-axis). Also annotated are TCGA-defined clinical and molecular subtypes: (LScore) lymphocyte infiltration score (The Cancer Genome Atlas Network 2015; Robertson et al. 2017). (D,E) Kaplan–Meier plots and P-values from Cox multi-regression model for patients stratified according to the four CLT clusters identified in SKCM (D) and the CLT two clusters identified in UVM (E).
Expression of prognostic CLTs in SKCM and UMV was independent from most clinical and genomic subtypes and known driver mutations but correlated well with molecular subclasses, previously identified by in-depth genomic and transcriptomic analyses (Fig. 4B–E; The Cancer Genome Atlas Network 2015; Robertson et al. 2017). Indeed, expression of CLTs associated with SKCM survival formed four distinct clusters, three of which matched clusters identified by TCGA (Fig. 4B,D). The cluster enriched in CLTs associated with the best prognosis (cluster 2) was enriched in the “immune” and “MITF-low” molecular signatures, whereas the cluster enriched in CLTs associated with worst prognosis (cluster 3) was enriched in the “keratin” signature (Fig. 4B). Similarly, expression of CLTs associated with UVM survival defined two clearly distinguishable and highly prognostic clusters (Fig. 4C,E). The cluster comprising all CLTs associated with better prognosis of UVM (cluster 2) was linked with low metastatic potential, Chr 3 and Chr 8q disomy, and preservations of normal DNA methylation patterns (Fig. 4C).
These results suggested that distinct and likely biologically relevant forces drive expression of CLT clusters in the different SKCM and UVM subtypes, a notion that was supported by detailed analysis of individual prognostic CLTs. One such example is the [TRPM1]MLT1A0 transcript, a truncated variant of TRPM1, which encodes melastatin (Supplemental Fig. S4). The [TRPM1]MLT1A0 transcript corresponds to TRPM1 variant 4 (TRPM1-210; NM_001252030.1), with the exception of an extended 3′ UTR, created by exonization of an intronic MLT1A0 LTR element, which serves as a terminal exon, replacing the last 24 exons of the canonical TRPM1 transcript (Supplemental Fig. S4). TRPM1 is a target of MITF, expressed in healthy, as well as transformed cutaneous and uveal melanocytes (Duncan et al. 1998; Miller et al. 2004), whereas [TRPM1]MLT1A0 was found principally expressed in SKCM and UVM (Supplemental Fig. S4). Levels of [TRPM1]MLT1A0 were directly proportional to those of TRPM1 in SKCM, and their down-regulation in a fraction of metastatic SKCM patients (Duncan et al. 1998) was linked with improved survival probability (Supplemental Fig. S4), as would expected for a MITF target (The Cancer Genome Atlas Network 2015). In UVM samples, however, higher expression of [TRPM1]MLT1A0 was at the expense of TRPM1 expression and linked with better prognosis (Supplemental Fig. S4).
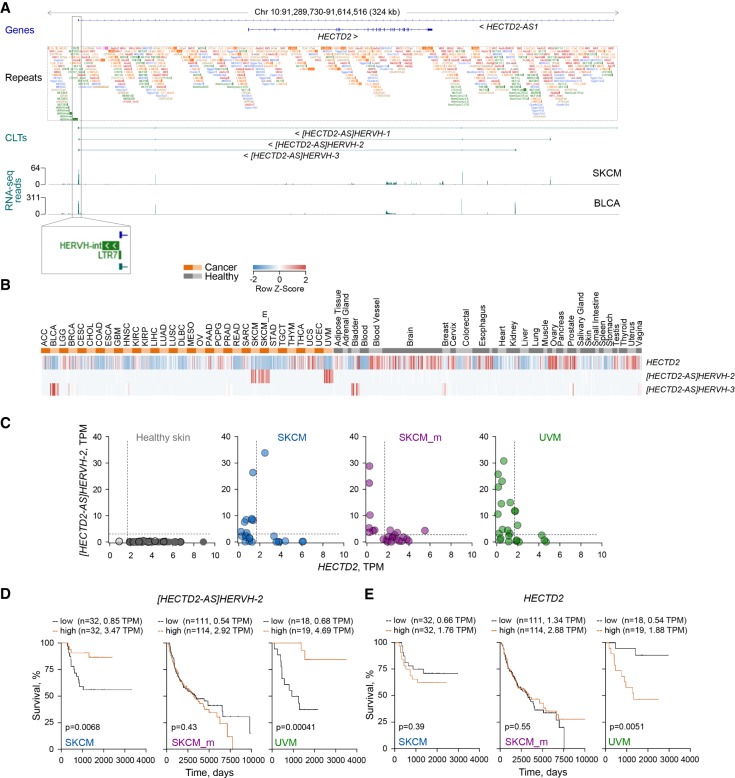
Context-dependent expression and association with survival probability was also observed for a novel transcript, [HECTD2-AS]HERVH-2, antisense to the HECTD2 locus (Fig. 5A–E). This locus was found to produce a long antisense transcript [HECTD2-AS]HERVH-1, partially matching the annotated HECTD2-AS1 lncRNA (NR_024467.1), but also two additional transcripts, [HECTD2-AS]HERVH-2 and [HECTD2-AS]HERVH-3, through use of alternative TSSs (Fig. 5A), further supported by promoter-based expression analyses (Supplemental Fig. S5; The FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014). All three transcripts skipped exon 2 of the annotated HECTD2-AS1 and terminated at a HERV-H LTR (Fig. 5A). [HECTD2-AS]HERVH-1 was not significantly expressed in any sample, but [HECTD2-AS]HERVH-2 was found highly expressed specifically in SKCM and UVM, and [HECTD2-AS]HERVH-3 was expressed in BLCA, as well as healthy bladder and reproductive tissues (Fig. 5B). Melanoma-specific expression of [HECTD2-AS]HERVH-2 was accompanied by loss of sense HECTD2 transcription (Fig. 5C), and a similar loss of the protein-coding sense transcript effect was mediated by antisense transcription of [HECTD2-AS]HERVH-3 in healthy bladder and BLCA (Supplemental Fig. S6), a pattern confirmed also in CCLE cell lines (Supplemental Fig. S7). The apparent switch from sense to antisense transcription was linked with better prognosis in primary SKCM and UVM, but not in metastatic SKCM, which showed higher expression of protein-coding HECTD2 across all patients (Fig. 5D,E).
Figure 5.
Down-regulation of HECTD2 expression by melanoma-specific antisense transcription of the [HECTD2-AS]HERVH-2 CLT. (A) GENCODE annotated transcripts at the indicated genomic location (genes), repeat content (repeats), CLTs and other selected transcripts at the same location in the current assembly (CLTs), and RNA-seq traces of representative SKCM and BCLA samples. (B) Heatmap of expression values in cancer patient and healthy control samples of HECTD2 and the two indicated antisense transcripts. (C) Anticorrelation of HECTD2 and [HECTD2-AS]HERVH-2 expression (TPM values). Each symbol is an individual patient or healthy control sample. (D,E) Kaplan-Meier plots and P-values from log-rank tests for melanoma patients stratified according to the higher versus the lower expression tertiles for [HECTD2-AS]HERVH-2 (D) and HECTD2 (E). The number of cases and the expression thresholds are also indicated in brackets.
Contrasting prognostic association with primary SKCM or UVM and metastatic SKCM was observed also for the MLT1B[LINC00518]-1 transcript (Supplemental Fig. S8), which matches a transcript previously described as the SKCM-specific lncRNA LINC00518 (NR_027793.1) (Iyer et al. 2015) and which we found to be initiated by an MLT1B element in both SKCM and UVM (Supplemental Fig. S8). Better prognosis uniquely in primary SKCM was associated with higher expression of a standalone HERV-K integration, HERVK3[Chr19q13.43], despite its noticeable expression in many healthy tissues and further transcriptional induction in most cancer types (Supplemental Fig. S9). In comparison, survival probability specifically in metastatic SKCM positively correlated with higher expression of the well-studied MER41G[AIM2] transcript (Chuong et al. 2016) (Supplemental Fig. S10) and a novel transcript, [DRAIC]LTR67B, that partial overlaps with GENCODE transcript ENST00000559212 (DRAIC-210) (Supplemental Fig. S11), both showing an immune-related, rather than melanoma-specific, expression pattern.
Together, these results indicate diverse biological processes linking CLT expression clusters with disease progression, as well as the specificity of their transcriptional regulation in distinct cancer and tissue types, further increased in distinct disease subtypes.
Unique cancer-specific antigens predictably generated by CLTs
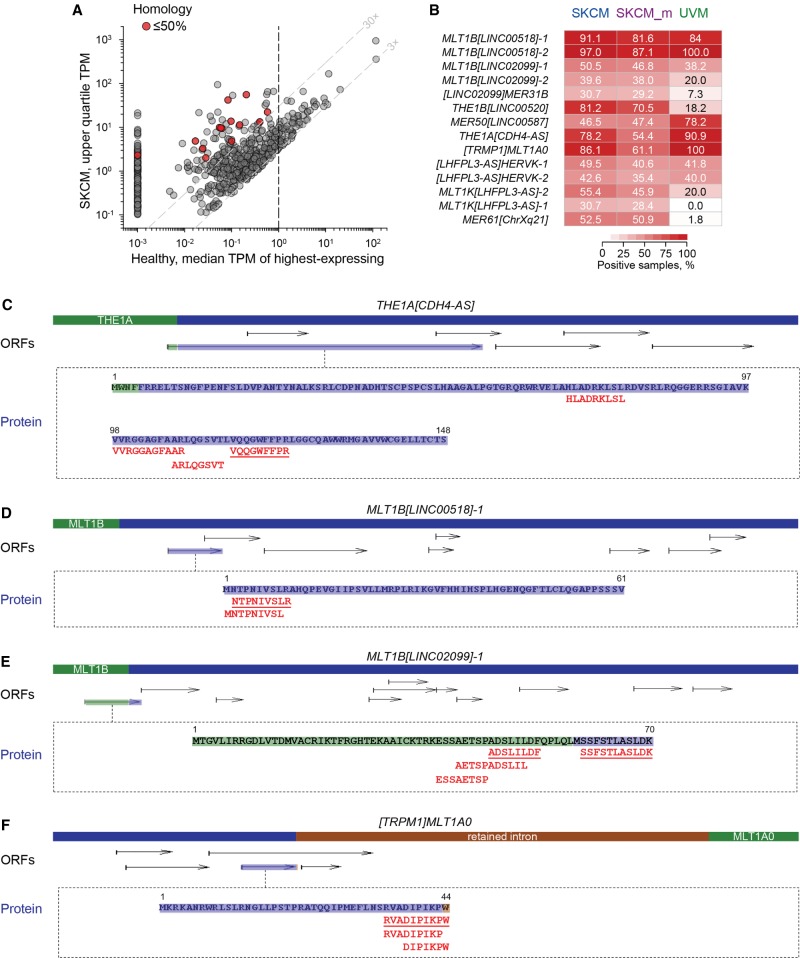
Regardless of the forces driving their expression in cancer, when expressed, CLTs may encode polypeptides that, irrespective of any intrinsic biological activity, are processed into unique antigenic peptides presented by MHC molecules. To identify CLTs with protein-coding potential, we used an ORF prediction algorithm, based on length and suitability of dicodon (hexamer) scores. We initiated our search in SKCM, ranking the transcripts according to expression and focusing on transcripts that contained one or more ORFs ≥300 nt. To ensure high cancer specificity, we eliminated CLTs with median levels of one or more TPM in the highest-expressing healthy tissue (Fig. 6A). Additionally, we removed transcripts whose largest ORF displayed >85% amino acid sequence identity with any other protein, annotated or predicted, that was potentially expressed in healthy tissues. This filter combination returned 14 CLTs, transcribed from eight different loci, each containing a unique ORF (0%–50% identity with other ORFs) (Fig. 6A,B). These SKCM-marking CLTs were expressed (one or more TPM) in between 46% and 99% of the extended SKCM cohort we have examined, and most of them were similarly expressed in UVM (Fig. 6B). The selected CLTs included the prognostic [TRPM1]MLT1A0, [HECTD2-AS]HERVH-2, and MLT1B[LINC00518]-1 transcripts and additional transcripts with similar structure. Several transcripts highly expressed specifically in melanoma overlapped LTR elements in the LHFPL3 locus, a gene whose expression was discordantly associated with survival of primary SKCM and UVM (Supplemental Fig. S12).
Figure 6.
Potential antigenicity of SKCM-specific CLTs. (A) Properties of selected CLTs (red circles) with unique protein-coding potential. Plotted is the expression level of SKCM CLTs with a predicted ORF ≥300 nt, in SKCM (upper quartile TPM) against the median TPM of the highest-expressing healthy tissue. Although the cut-off for CLT selection was ≤85% homology over the entire ORF length with any other ORF potentially expressed in healthy tissues, the final selected CLTs displayed 0%–50% homology. (B) Prevalence of expression of the indicted CLTs with unique protein-coding potential among primary SKCM (n = 101), metastatic SKCM (SKCM_m; n = 342), or UVM (n = 55) patients. Values are the percentages of patient that express each CLT at 0.5 or more TPM. (C–F) CLT structure, all predicted ORFs >75 nt (ORFs), where the ORF with evidence for translation is highlighted, and amino acid sequence of the latter ORF. Also shown is the sequence of MHC-eluted peptides uniquely mapping to each CLT product. Underlined peptides were confirmed by comparison with synthetic peptides.
To probe for protein production by the selected SKCM-specific CLTs, we searched through immunopeptidomic data previously generated from melanoma biopsies (Bassani-Sternberg et al. 2016). Although these CLTs were selected for the presence of at least one sequence-unique ORF, they could in principle contain additional or alternative ORFs. We therefore included the translation of any ORF of a minimum 75 nt from these selected CLTs in the search, which was performed using the Mascot search engine (Perkins et al. 1999). This analysis identified a number of MHC-eluted peptides, with Mascot scores higher or close to the respective homology scores (Supplemental Table S6), that mapped to ORFs from CLTs transcribed from four of the selected loci (Fig. 6C–F). To confirm correct assignment of the observed peptides, spectra from at least one peptide per ORF were compared, by spectral angle analyses (Tabb et al. 2003), with spectra generated with synthetic peptides (Supplemental Fig. S13). To obtain further support, the same immunopeptidomic data were searched with a different search engine, PEAKS, assisted by de novo peptide sequencing (Zhang et al. 2012). This analysis identified nine of the 13 Mascot-identified peptides as highly significant, with an average false-discovery rate (FDR) of 1.5% (Supplemental Table S6), providing independent validation of the observed spectra (Supplemental Fig. S14). Moreover, several of the identified peptides were predicted to bind at least one major HLA allele, and in most cases, they were also predicted to bind to the HLA allele (where known) of the patient in whom they were identified (Supplemental Fig. S15). Collectively, these results supported the antigenicity of the CLT translation products.
Peptides were identified from the largest predicted ORF of transcript THE1A[CDH4-AS] (Fig. 6C). This transcript, matching the annotated lncRNA RP11-429E11.2, was found to be initiated by a THE1A element in intron 3 of the CDH4 gene, driving transcription in the antisense orientation (Supplemental Fig. S16). The THE1A[CDH4-AS] transcript was very highly expressed specifically in SKCM and UVM but may also be expressed to a much lower degree in UV-exposed skin (Supplemental Fig. S16). Peptides were also identified from smaller ORFs of the prognostic transcript MLT1B[LINC00518]-1 and transcript MLT1B[LINC02099]-1 (Fig. 6D,E). The latter transcript was initiated by an MLT1B element and was one of three identified transcripts in the lncRNA LINC02099 locus, matching the annotated LINC02099-201 variant (Supplemental Fig. S17). Also identified were peptides derived from alternative translation of the prognostic [TRPM1]MLT1A0 transcript. Although translation of the largest ORF in transcript [TRPM1]MLT1A0 is supported by RefSeq evidence (annotated protein NP_001238959), eluted peptides were additionally derived from translation of a smaller embedded alternative ORF (Fig. 6F). Together, these results highlight the potential of CLT translation and presentation to contribute to the diversity of cancer-specific antigenic peptides.
Discussion
By assembling a comprehensive transcriptome, we extend our knowledge of LTR elements that are transcriptionally used and their potential involvement in human cancer. Although accumulating evidence incriminates ERVs and other EREs in multiple stages of cancer development and anticancer immunity (Romanish et al. 2010; Kassiotis and Stoye 2017), their impact is not yet fully appreciated owing to the lack of complete annotation and quantitation of LTR element–overlapping transcripts. Past efforts involving mapping reads to the reference genome have reduced representation of repeat-derived exons owing to exclusion of multimapping reads and loci. Our assembly mitigates this bias because the majority of the genomic repeats are not expressed in a particular indication, increasing the likelihood of unambiguously assigning a repeat-derived read to an individual transcript during transcriptome-based assembly. This resulted in substantially increased representation of unannotated or partially annotated transcripts derived from or overlapping with LTR and LINE elements, in the current assembly. Although cancer-specific ERE-overlapping transcripts might not be biologically more significant than those that do not overlap with EREs, they do represent the overwhelming majority of novel transcripts in this assembly and were therefore the focus of this study, starting with LTR-overlapping transcripts.
Global epigenetic changes may be necessary for the transcriptional utilization of LTR elements in cancer. However, our data suggest that alone these changes are not sufficient. Instead, utilization of distinct LTR elements is locus specific and does not necessitate activation of other LTR elements of the same family. A number of observations support this notion. Regardless of cancer specificity, the expression of chimeric transcripts was significantly higher than that of stand-alone LTR elements. Furthermore, chimeric transcripts were overrepresented in the CLT pool, as exemplified in melanoma CLTs, in which they were one order of magnitude more numerous than stand-alone LTR elements. Indeed, derepression of LTR promoter activity could account for transcriptional induction of only a minority of CLTs, whereas transcriptional inclusion of LTR elements by alternative splicing in transcripts driven by alternative promoters was more frequent. These findings suggest a combination of LTR and gene properties is responsible for cancer-specific transcriptional induction of chimeric CLTs and may also underlie their tissue-type specificity. This is further supported by the close association of CLT expression and molecular disease subtypes, defined based on protein-coding gene expression alone (The Cancer Genome Atlas Network 2015; Robertson et al. 2017). This close link substantially increases the predictability of CLT expression in their respective disease subtype, often exceeding 90% of samples analyzed here. It also underlies previously unappreciated shifts in CLT expression mirroring cancer progression or metastasis. For example, the lower extent of overlap in CLT expression between individual samples within and between metastatic SKCM cohorts indicated a considerably higher degree of heterogeneity in metastatic than in primary SKCM. Such shifts in CLT expression likely reflect changes in overall gene expression, such as down-regulation of TRPM1 and other MITF targets in metastatic SKCM (Duncan et al. 1998; The Cancer Genome Atlas Network 2015), or immune escape.
The combination of tissue and cancer specificity may also relate to cell lineage–specific transcription of CLTs. A confounding factor in determining cancer specificity of a given CLT by comparing its expression between tumor biopsies and healthy tissues is the relative proportion of tumor cell lineage in each sample. For example, a higher proportion of melanocytes in melanoma biopsies than in healthy skin might favor melanocyte-specific transcript selection. Indeed, expression of [HECTD2-AS]HERVH-1 was detected in a nontransformed melanocyte cell line, as well as in melanoma cell lines, and expression of THE1A[CDH4-AS] was also found in sun-exposed healthy skin. Nevertheless, melanocytes are a sizable constituent of healthy skin, with frequencies between 10% and 50% dependent on anatomical location (Cochran 1970), and melanoma biopsies also contain a substantial fraction of nontransformed stromal and immune cells, which can add up to 50%. Furthermore, although detectable in a nontransformed melanocyte cell line and in sun-exposed healthy skin, respectively, expression of [HECTD2-AS]HERVH-1 and THE1A[CDH4-AS] was still significantly elevated in melanoma cell lines and biopsies, supporting the notion that disregulated or exaggerated melanocyte-specific transcriptional patterns underlie melanoma specificity of at least some of the melanoma CLTs.
Although expression of most CLTs simply reflects the altered epigenetic, splicing, and transcriptional state of cancer, some will have functionally relevant implications. These include cis effects on gene transcriptional activity or splicing patterns, as represented by the HECTD2 locus. First identified as a prion disease susceptibility gene (Lloyd et al. 2009), HECTD2 encodes a ubiquitin E3 ligase that enhances the inflammatory response to innate stimulation (Coon et al. 2015). MicroRNA-mediated down-regulation of HECTD2 expression has been proposed to drive androgen independence in prostate cancer (Sun et al. 2014), and more recently, HECTD2 copy number alterations were suggested as drivers in neuroblastoma patients (Suo et al. 2018). However, the function of HECTD2 in cancer in general and in melanoma in particular remains to be elucidated. Its transcriptional regulation by antisense CLT transcription in melanoma revealed here clearly illustrates the potential of our assembly of novel CLTs to identify biologically relevant genes, as well as their regulation, which will provide numerous leads for further investigation.
Another functional implication of CLT expression is the potential to generate cancer-specific antigens for T cell recognition. Cancer cells present antigenic T cell epitopes from canonical and noncanonical polypeptides, showing different degrees of frequency and specificity (Schumacher and Schreiber 2015). These range from highly immunogenic neoantigens, generated by cancer-specific mutations restricted to one or few individuals, to partially immunologically tolerated antigens from nonmutated canonical proteins overexpressed in almost all cancer patients. It is becoming increasingly clear that the altered transcriptional state of cancer can generate novel noncanonical polypeptides without the need for somatic mutations (Kahles et al. 2018; Smart et al. 2018). These include alternative splicing events, such as intron retention (Smart et al. 2018) or mid-exon splicing (Kahles et al. 2018) of protein-coding genes, and, as we show here, transcriptional utilization of LTR elements. Our analysis uncovered several transcripts overlapping LTR elements that are highly prevalent in distinct cancer types, some expressed in nearly all patients examined. This high predictability of CLT expression contrasts with that of transcripts generated by other mutation-independent (Kahles et al. 2018; Smart et al. 2018) or mutation-driven processes (Schumacher and Schreiber 2015) that occur at relatively low frequency. Thus, our findings uncover a novel source of cancer antigens, with characteristics of expression specificity and intensity that predict a distinctive immunological profile. Although their antigenicity is shown here, further studies will be needed to establish their immunogenicity or therapeutic potential.
Despite their mutation-independent nature, CLTs have the potential to produce completely novel antigenic peptides, translated from LTR-initiated transcripts previously considered to lack coding potential (e.g., lncRNAs), chimeric ORFs resulting from LTR element exonization or alternative ORFs in LTR-overlapping transcripts, as well as ORFs in stand-alone LTR elements. Although protein products from these CLTs have not been previously observed, recent in-depth proteogenomic analysis of an Epstein-Barr virus–transformed B cell line (Laumont et al. 2016) or A431 cells and five healthy tissues (Zhu et al. 2018) provided evidence for the translation of similarly unconventional ORFs, including lncRNAs, alternative ORFs, and intronic sequences. Translation of pseudogenes and lncRNAs also showed tissue specificity (Zhu et al. 2018). Moreover, a recent analysis of the murine cancer immunopeptidome highlighted noncoding regions as the main source of cancer-specific antigens (Laumont et al. 2018). As our analysis of the melanoma immunopeptidome shows, such unconventional transcripts and ORFs therein hold great promise for cancer-specific antigens.
Although the annotated LTR transcriptome has been increasingly implicated in cancer (Romanish et al. 2010; Kassiotis and Stoye 2017), the broader view of LTR element transcriptional utilization offered by the current assembly reveals a much more extensive involvement of these elements than previously appreciated. We expect that the identification and quantitation of the cancer LTR transcriptome in this study will provide a framework for fully investigating and elucidating their role in cancer initiation, progression, and immune surveillance.
Methods
Repeat region annotation
LTR and non-LTR elements were annotated as described previously (Attig et al. 2017) and in the Supplemental Methods.
Transcript assembly
RNA-seq reads from 24 patient samples from 31 primary and one metastatic (melanoma) cancer types (totaling 768 samples) were obtained from TCGA (Supplemental Table S1) and used to generate a pan-cancer transcriptome, as described in full in the Supplemental Methods (see also Supplemental File S1; Supplemental Code S3).
Cancer-specific transcript selection
TPMs were estimated for all transcripts with a custom Bash pipeline (Supplemental Code S1) using GNU parallel (Tange 2011) and Salmon, and expression within each cancer type was compared with expression across 811 healthy tissue samples (healthy tissue-matched controls for all cancer types, where available, from TCGA and the GTEx Consortium) (The GTEx Consortium 2015). RNA-seq data from both these consortia are based on poly(A) RNA, which, as opposed to total RNA, may affect the capture of certain classes of transcripts. However, poly(A) RNA selection was recently shown to limit detection only of satellite repeat transcripts, whereas transcripts from ERVs, LINEs, and SINEs were similarly represented in total and poly(A)-selected RNA (Solovyov et al. 2018). Moreover, the 3′ UTR and thus the poly(A) tail in the majority of CLTs are provided by the canonical fusion gene rather than the LTR element. Therefore, the impact of using poly(A)-selected RNA-seq samples on our ability to detect and quantify LTR-overlapping transcripts should be negligible.
Transcripts were considered expressed if detected at more than one TPM in any sample and as cancer-specific if the following criteria were fulfilled: (1) expressed in six or more of the 24 samples of each cancer type; (2) expressed at less than 10 TPM in ≥90% of all healthy tissue samples; (3) expressed in the cancer type of interest three or more times the median expression in any control tissue type; and (4) expressed in the cancer type of interest three or more times the 90th percentile of the respective healthy tissue, when available (Supplemental Code S2). In addition to these expression thresholds, transcript selection was based on manual inspection, excluding potentially misassembled contigs. When the DNA strand could not be unambiguously assigned (using a range of criteria, including overlap with annotated exons, directionality of LTRs, splice or polyadenylation sites), transcripts corresponding to both strands were considered.
Survival analysis and hazard ratio calculations
For survival analysis, we downloaded the complete cohorts of SKCM and UVM from TCGA, for which survival data were recorded (primary SKCM: 95; metastatic SKCM: 335; UVM: 54). To test if expression of a transcript of interest correlated with patient survival, we identified the patients in the bottom and top tercentile expression (“low” vs. “high” expression). Survival analysis was performed using the survfit function of the survival R package (v. 2.42) (R Core Team 2018), using overall survival time based on the days_to_death annotation or the last update of the annotation in March 2017 (i.e., 365 days × (2017 − year of birth) − the age-at-diagnosis in days). To compare curves between low and high expression tertiles, log-rank testing was used, and a Cox regression model was built to test the assumption of proportional hazards holds. Hazard odd ratios are given based on the Cox regression model. To compare survival between multiple expression clusters, the Cox regression model was used.
Unique protein-coding potential prediction
To identify CLTs with protein-coding potential, we ran an ORF prediction algorithm, based on length and suitability of dicodon (hexamer) scores. A HMM was trained on hexamers derived from Ensembl CDS annotations, and ORFs ≥300 nt were taken forward, where their sense hexamer score exceeded the antisense score. To identify unique protein sequences potentially encoded by CLTs, sequences translated from the largest ORF of selected CLTs were aligned with those translated from all ORFs ≥210 nt from the entire transcript assembly using TBLASTN (BLAST+ v2.3.0) without soft-masking and retaining alignments with an E-value >10−5. Uniqueness was defined as ≤85% amino acid sequence identity (over the entire length of the protein) with any other predicted protein, whether the latter was expressed or not. For CLT-encoded proteins that showed >85% sequence identity with one or more predicted proteins, we interrogated the expression pattern of transcripts encoding the similar proteins. Where these additional transcripts were also expressed in a cancer-specific manner (based on the criteria listed above), the respective CLT was retained. Where the additional transcripts were expressed in healthy tissue, the respective CLT was discarded.
Immunopeptidomic analyses
CLT-encoded HLA-presented peptides were identified by immunopeptidomic analyses of previously generated spectrometry data (accession number: PXD004894) (Bassani-Sternberg et al. 2016), as detailed in the Supplemental Methods (see also Supplemental File S2).
Competing interest statement
G.K. is a scientific cofounder of and consulting for ERVAXX and a member of its scientific advisory board. G.R.Y. consults for ERVAXX. N.T. is currently directing immunopeptidomics research at ERVAXX. Part of this work was included in a patent application (ERV-P2166EPp).
Supplementary Material
Acknowledgments
We thank the Scientific Computing and Peptide Chemistry facilities at the Francis Crick Institute for assistance. The results shown here are in whole or part based upon data generated by The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. This work benefited from data assembled by the CCLE consortium. This work was supported by the Francis Crick Institute (FC001099, FC001162), which receives its core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust, and by the Wellcome Trust (102898/B/13/Z).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.248922.119.
Freely available online through the Genome Research Open Access option.
References
- Attig J, Young GR, Stoye JP, Kassiotis G. 2017. Physiological and pathological transcriptional activation of endogenous retroelements assessed by RNA-sequencing of B lymphocytes. Front Microbiol 8: 2489 10.3389/fmicb.2017.02489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaian A, Mager DL. 2016. Endogenous retroviral promoter exaptation in human cancer. Mob DNA 7: 24 10.1186/s13100-016-0080-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassani-Sternberg M, Bräunlein E, Klar R, Engleitner T, Sinitcyn P, Audehm S, Straub M, Weber J, Slotta-Huspenina J, Specht K, et al. 2016. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 7: 13404 10.1038/ncomms13404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. 2011. A decade of exploring the cancer epigenome: biological and translational implications. Nat Rev Cancer 11: 726–734. 10.1038/nrc3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Boeke JD. 2012. Human transposon tectonics. Cell 149: 740–752. 10.1016/j.cell.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, Campisi M, Kuang Y, Zhang Y, Gjini E, et al. 2018. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med 24: 1143–1150. 10.1038/s41591-018-0116-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2015. Genomic classification of cutaneous melanoma. Cell 161: 1681–1696. 10.1016/j.cell.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova E, Scrivani C, Doh S, Weisman Q, Takahashi Y, Harashima N, Yokoyama H, Srinivasan R, Linehan WM, Lerman MI, et al. 2016. Detection of an immunogenic HERV-E envelope with selective expression in clear cell kidney cancer. Cancer Res 76: 2177–2185. 10.1158/0008-5472.CAN-15-3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162: 974–986. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351: 1083–1087. 10.1126/science.aad5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran AJ. 1970. The incidence of melanocytes in normal human skin. J Invest Dermatol 55: 65–70. 10.1111/1523-1747.ep12290550 [DOI] [PubMed] [Google Scholar]
- Coon TA, McKelvey AC, Lear T, Rajbhandari S, Dunn SR, Connelly W, Zhao JY, Han S, Liu Y, Weathington NM, et al. 2015. The proinflammatory role of HECTD2 in innate immunity and experimental lung injury. Sci Transl Med 7: 295ra109 10.1126/scitranslmed.aab3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, Silva TC, Groeneveld C, Wong CK, Cho SW, et al. 2018. The chromatin accessibility landscape of primary human cancers. Science 362: eaav1898 10.1126/science.aav1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N, Sajed D, Arora KS, Solovyov A, Rajurkar M, Bledsoe JR, Sil S, Amri R, Tai E, MacKenzie OC, et al. 2017. Diverse repetitive element RNA expression defines epigenetic and immunologic features of colon cancer. JCI Insight 2: e91078 10.1172/jci.insight.91078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LM, Deeds J, Hunter J, Shao J, Holmgren LM, Woolf EA, Tepper RI, Shyjan AW. 1998. Down-regulation of the novel gene melastatin correlates with potential for melanoma metastasis. Cancer Res 58: 1515–1520. [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN PMI and CLST (DGT). 2014. A promoter-level mammalian expression atlas. Nature 507: 462–470. 10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13: 283–296. 10.1038/nrg3199 [DOI] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, Bernstein BE. 2017. Epigenetic plasticity and the hallmarks of cancer. Science 357: eaal2380 10.1126/science.aal2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47: D766–D773. 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, Khan N, Ubellacker JM, Xie S, Metzger-Filho O, et al. 2017. CDK4/6 inhibition triggers anti-tumour immunity. Nature 548: 471–475. 10.1038/nature23465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering W, Schmitt K, Dostert M, Schaal H, Deenen R, Mayer J, Schulz WA. 2015. Human endogenous retrovirus HERV-K(HML-2) activity in prostate cancer is dominated by a few loci. Prostate 75: 1958–1971. 10.1002/pros.23095 [DOI] [PubMed] [Google Scholar]
- The GTEx Consortium. 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. 2015. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47: 199–208. 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, Zhang D, Li D, Xing X, Kim S, et al. 2019. Transposable elements drive widespread expression of oncogenes in human cancers. Nat Genet 51: 611–617. 10.1038/s41588-019-0373-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahles A, Lehmann KV, Toussaint NC, Hüser M, Stark SG, Sachsenberg T, Stegle O, Kohlbacher O, Sander C; Cancer Genome Atlas Research Network, et al. 2018. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34: 211–224.e6. 10.1016/j.ccell.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiotis G, Stoye JP. 2016. Immune responses to endogenous retroelements: taking the bad with the good. Nat Rev Immunol 16: 207–219. 10.1038/nri.2016.27 [DOI] [PubMed] [Google Scholar]
- Kassiotis G, Stoye JP. 2017. Making a virtue of necessity: the pleiotropic role of human endogenous retroviruses in cancer. Philos Trans R Soc Lond B Biol Sci 372: 20160277 10.1098/rstb.2016.0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Köchert K, Bouhlel MA, Richter J, Soler E, et al. 2010. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med 16: 571–579, 571p 10.1038/nm.2129 [DOI] [PubMed] [Google Scholar]
- Laumont CM, Daouda T, Laverdure JP, Bonneil E, Caron-Lizotte O, Hardy MP, Granados DP, Durette C, Lemieux S, Thibault P, et al. 2016. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat Commun 7: 10238 10.1038/ncomms10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumont CM, Vincent K, Hesnard L, Audemard É, Bonneil É, Laverdure J-P, Gendron P, Courcelles M, Hardy M-P, Côté C, et al. 2018. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med 10: eaau5516 10.1126/scitranslmed.aau5516 [DOI] [PubMed] [Google Scholar]
- Lloyd SE, Maytham EG, Pota H, Grizenkova J, Molou E, Uphill J, Hummerich H, Whitfield J, Alpers MP, Mead S, et al. 2009. HECTD2 is associated with susceptibility to mouse and human prion disease. PLoS Genet 5: e1000383 10.1371/journal.pgen.1000383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE. 2004. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res 64: 509–516. 10.1158/0008-5472.CAN-03-2440 [DOI] [PubMed] [Google Scholar]
- Mullins CS, Linnebacher M. 2012. Endogenous retrovirus sequences as a novel class of tumor-specific antigens: an example of HERV-H env encoding strong CTL epitopes. Cancer Immunol Immunother 61: 1093–1100. 10.1007/s00262-011-1183-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567. [DOI] [PubMed] [Google Scholar]
- Pérot P, Mullins CS, Naville M, Bressan C, Hühns M, Gock M, Kühn F, Volff JN, Trillet-Lenoir V, Linnebacher M, et al. 2015. Expression of young HERV-H loci in the course of colorectal carcinoma and correlation with molecular subtypes. Oncotarget 6: 40095–40111. 10.18632/oncotarget.5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Rakoff-Nahoum S, Kuebler PJ, Heymann JJ, E Sheehy M, Ortiz GM, S Ogg G, Barbour JD, Lenz J, Steinfeld AD, Nixon DF. 2006. Detection of T lymphocytes specific for human endogenous retrovirus K (HERV-K) in patients with seminoma. AIDS Res Hum Retroviruses 22: 52–56. 10.1089/aid.2006.22.52 [DOI] [PubMed] [Google Scholar]
- Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, et al. 2017. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32: 204–220.e15. 10.1016/j.ccell.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanish MT, Cohen CJ, Mager DL. 2010. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin Cancer Biol 20: 246–253. 10.1016/j.semcancer.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. 2015. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160: 48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, et al. 2015. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162: 961–973. 10.1016/j.cell.2015.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarfo I, Pellegrino E, Mereu E, Kwee I, Agnelli L, Bergaggio E, Garaffo G, Vitale N, Caputo M, Machiorlatti R, et al. 2016. Identification of a new subclass of ALK-negative ALCL expressing aberrant levels of ERBB4 transcripts. Blood 127: 221–232. 10.1182/blood-2014-12-614503 [DOI] [PubMed] [Google Scholar]
- Schiavetti F, Thonnard J, Colau D, Boon T, Coulie PG. 2002. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res 62: 5510–5516. [PubMed] [Google Scholar]
- Schumacher TN, Schreiber RD. 2015. Neoantigens in cancer immunotherapy. Science 348: 69–74. 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, Li Y, Chen H, Yang H, Hsu PH, et al. 2018. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174: 549–563.e19. 10.1016/j.cell.2018.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Zhang X, Niknafs YS, Xiao L, Mehra R, Cieślik M, Ross A, Schaeffer E, Malik B, Guo S, et al. 2016. Identification and validation of PCAT14 as prognostic biomarker in prostate cancer. Neoplasia 18: 489–499. 10.1016/j.neo.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart AC, Margolis CA, Pimentel H, He MX, Miao D, Adeegbe D, Fugmann T, Wong KK, Van Allen EM. 2018. Intron retention is a source of neoepitopes in cancer. Nat Biotechnol 36: 1056–1058. 10.1038/nbt.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. www.repeatmasker.org.
- Smith CC, Beckermann KE, Bortone DS, de Cubas AA, Bixby LM, Lee SJ, Panda A, Ganesan S, Bhanot G, Wallen EM, et al. 2018. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest 128: 4804–4820. 10.1172/jci121476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyov A, Vabret N, Arora KS, Snyder A, Funt SA, Bajorin DF, Rosenberg JE, Bhardwaj N, Ting DT, Greenbaum BD. 2018. Global cancer transcriptome quantifies repeat element polarization between immunotherapy responsive and T cell suppressive classes. Cell Rep 23: 512–521. 10.1016/j.celrep.2018.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Wang X, He HH, Sweeney CJ, Liu SX, Brown M, Balk S, Lee GS, Kantoff PW. 2014. MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene 33: 2790–2800. 10.1038/onc.2013.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo C, Deng W, Vu TN, Li M, Shi L, Pawitan Y. 2018. Accumulation of potential driver genes with genomic alterations predicts survival of high-risk neuroblastoma patients. Biol Direct 13: 14 10.1186/s13062-018-0218-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, MacCoss MJ, Wu CC, Anderson SD, Yates JR, 3rd. 2003. Similarity among tandem mass spectra from proteomic experiments: detection, significance, and utility. Anal Chem 75: 2470–2477. 10.1021/ac026424o [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy JP, Hanada K, Mena O, Kurlander R, Tawab A, et al. 2008. Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J Clin Invest 118: 1099–1109. 10.1172/JCI34409C1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange O. 2011. GNU Parallel: the command-line power tool. USENIX Mag 36: 42–47. [Google Scholar]
- Wiesner T, Lee W, Obenauf AC, Ran L, Murali R, Zhang QF, Wong EW, Hu W, Scott SN, Shah RH, et al. 2015. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature 526: 453–457. 10.1038/nature15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, Zhang W, Zhang Z, Lajoie GA, Ma B. 2012. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics 11: M111.010587 10.1074/mcp.M111.010587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Orre LM, Johansson HJ, Huss M, Boekel J, Vesterlund M, Fernandez-Woodbridge A, Branca RMM, Lehtiö J. 2018. Discovery of coding regions in the human genome by integrated proteogenomics analysis workflow. Nat Commun 9: 903 10.1038/s41467-018-03311-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.