Abstract
Intravital microscopic imaging can uncover fundamental aspects of immune cell behavior in real time in both healthy and pathological states. Here we discuss approaches for single cell imaging of adaptive and innate immune cells to explore how they migrate, communicate and mediate regulatory or effector functions in various tissues throughout the body. We further review how intravital single cell imaging can be used to study drug effects on immune cells.
INTRODUCTION
We are at a point in immunology where we appreciate the complexity of the relationships between immune cell populations and disease states. Unravelling this complexity requires a palette of research tools to investigate immune cell subpopulations and their functions. Selecting the proper tools for a specific application and model requires knowledge of each technology’s strengths and limitations. The immunology field has historically focused on classifying immune cells by first isolating and then studying them using multiplexed cytometry. More recently, the introduction of single cell transcriptomics has given us the ability to collect unbiased information on all cell populations. In contrast to these ex vivo approaches, intravital imaging enables direct visualization of immune cells and their functions in various tissues in vivo, without the need for isolation and selection procedures that can introduce bias. Intravital imaging is particularly suited to dynamically visualize immune cells over time, often revealing previously unknown behaviors. Indeed, many immune functions are heavily dependent on cell migration and cell-cell contacts, both of which can be faithfully captured by single cell resolution intravital imaging. Spatial organization can dictate the effective functionality of immune cells, which is readily apparent from imaging studies but may not be detected using traditional cell profiling methods. Furthermore, the ability to quantitatively measure kinetics of behaviors provides key insights into immune processes and even potentiates mathematical modeling of emergent behaviors. Here, we focus on how immunologists can use intravital single cell imaging approaches to complement super-resolution imaging in isolated cells (1) and imaging at the whole body level (2).
THE TOOLBOX
A number of intravital imaging approaches exist and their basics and general implementations have been covered already (3–5). This section therefore focuses on specifically adapting these technologies to observe how immune cells travel, function and interact in live mice.
What are the different intravital imaging approaches available, and what are their advantages and limitations?
Today’s two main approaches for single cell imaging utilize either confocal laser scanning or multiphoton microscopy. The systems are generally implemented on upright microscopes, and several configurations are now commercially available. Most intravital imaging setups for mice now have an upright configuration, although in some cases, inverted systems are also used. The latter may be beneficial for dual purpose cell/tissue imaging systems or for exteriorized organs which assume a flat configuration on the glass. Confocal microscopy set-ups are usually less expensive and represent a good all-around technique for much of the imaging done today (6). These systems use an array of solid state lasers for excitation and matched laser/filter sets can demultiplex fluorophore signals, similar to flow cytometry. The downsides of confocal imaging are higher autofluorescence and scattering, which generally limit imaging depths to <100 μm, and typically in the range of 20–50 μm. Furthermore, shorter wavelength channels have higher phototoxicity, although this is often a lesser concern for in vivo imaging than it is for in vitro imaging.
Multiphoton laser scanning microscopy (7) bypasses the limitations of confocal microscopy using more expensive and tunable Ti:sapphire lasers that operate in the near-infrared range. Localized nonlinear excitation based on two-photon absorption allows for superior tissue penetration at higher wavelengths and less out-of-focus excitation. One of the major ways in which multiphoton imaging reduces phototoxicity and improves resolution is by inherent optical sectioning due to the more restricted photon excitation volume. Typical penetration depths are in the 200–300 μm range for most organs, except in the brain and cleared tissues where deeper imaging depths can be achieved. Multiphoton microscopy can induce photobleaching but again, this is often less of a concern for in vivo imaging. One minor disadvantage of the multiphoton system is that many of the fluorophores used in flow cytometry, epifluorescence and confocal microscopy experiments have not been characterized in the multiphoton setup.
What are the key requirements for a single cell imaging experiment?
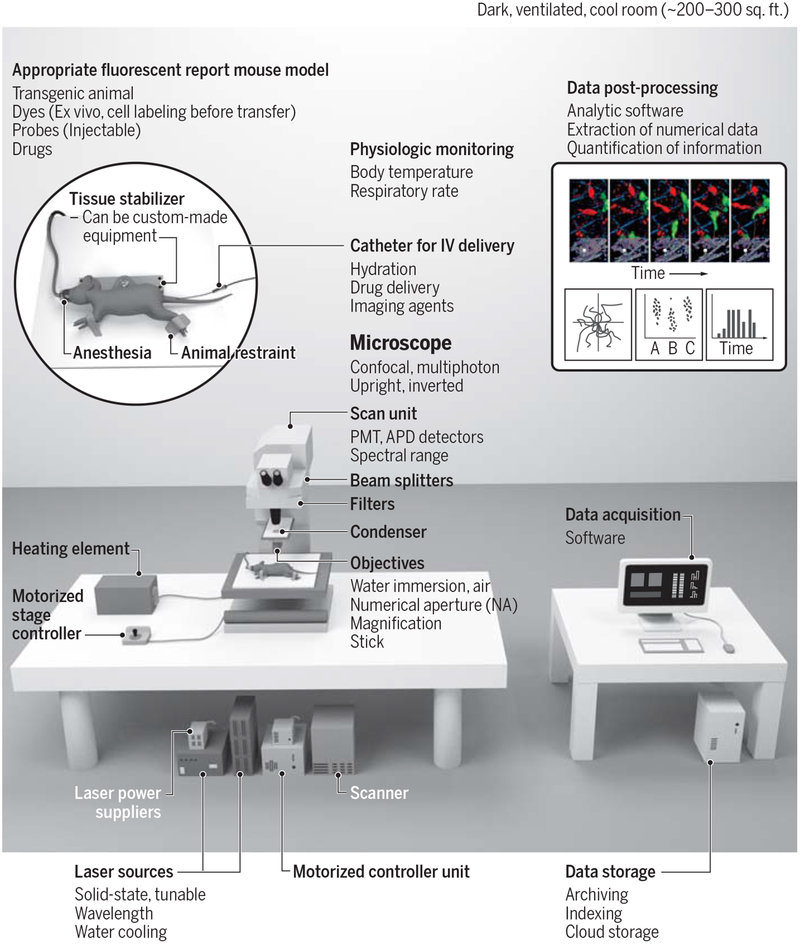
To perform state-of-the-art single cell 3D imaging one requires i) an integrated imaging system in a dark and appropriately cooled room (see supplemental information in (4)); ii) suitable fluorescent reporter mouse models with either exteriorized organs (8) or implanted window chambers (9); iii) motion suppression techniques (10); iv) physiologic support modules and v) data processing and analytical software (Fig. 1). Physiologic support, including feedback temperature controls, are critical to maintaining homeostasis and hydration in immobilized and ventilated animals. This support is most commonly achieved by using warming plates, immobilization chambers and continuous vital sign monitoring while the animal is anesthetized. Temperature control in exteriorized organs is especially critical to preserve cell motility.
Fig. 1 – Diagram of an intravital imaging setup.
Left: the equipment required for conducting an intravital imaging experiment includes an appropriate fluorescent mouse model, a microscope (shown here is an upright multiphoton microscope), laser sources, physiological monitoring, an appropriate anesthesia setup, and a tissue stabilizer to prevent motion artifacts during imaging. This image illustrates imaging in a dorsal skinfold window chamber; other tissues can be imaged (see Figure 2), each of them requiring their own methods of tissue preparation, stabilization and monitoring. Right: data acquisition, followed by data storage and data post-processing to extract quantitative information. Credit: A. Kitterman/AAAS
Quality control measurements include calibrating the imaging system with fluorescent bead phantoms, correcting artifacts, detecting channel bleed-through, determining the stability of reporter expression levels and properly managing data outliers. Data processing and analysis is a major part of a single cell imaging experiment. Various software packages automate the analysis of high-dimensional data, although manual curation is still often necessary. Common software packages that support image processing, segmentation, cell measurements and profiling features include CellProfiler, Fiji, Imaris and microscope vendor packages. Matlab and Python are programming languages that are also utilized to implement post-processing algorithms and quantitative analysis, whereas widely available packages such as GraphPad Prism or CellProfilerAnalyst are used for statistical analyses of extracted features across study groups, findings and datasets.
Common approaches
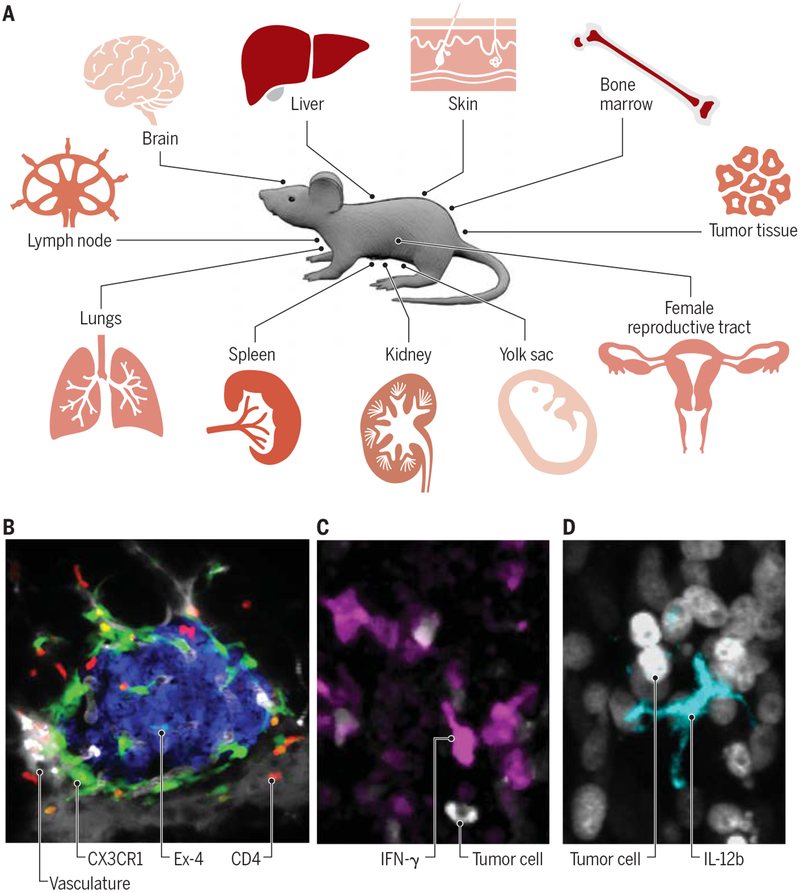
Prior to performing an imaging experiment, the questions to be answered should be clearly outlined and all experimental variables considered. What type of information can be retrieved, and how many parameters can be measured simultaneously? The answers to these questions will vary from organ to organ, but it is generally accepted that up to four (confocal) or six channels (multiphoton) can be acquired during a typical 3D acquisition. Most often, these channels are reserved for different immune cell populations (11), structural content (collagen fibers, vessels) or labeled drugs (12). Inhalation-anesthetized mice bearing window chambers generally tolerate repeat imaging well and can be analyzed on subsequent days unless non-survival surgery is performed on exteriorized organ preparations, in which case it is important to maintain the homeostatic environment with respect to blood flow and oxygen supply in addition to temperature. The typical imaging session varies in length but often lasts one to two hours unless slower phenomena are being studied. The most commonly imaged organ systems are tumors, lymph nodes and the skin but also include brain, bone marrow, kidney, liver, lungs, pancreas, reproductive tract, spleen and yolk sac (Fig. 2A–B).
Fig. 2 – Existing applications of immune cell imaging in various tissues.
(A) A wide range of mouse tissue sites is adapted for single cell imaging. (B) Intravital micrograph showcasing detection of two immune cell types, namely CD4+ T cells (red) and CX3CR1+ macrophages (green) in pancreatic islets. β-cells (blue) were visualized with exendin-4–like neopeptide conjugated to the fluorescent dye Se-Tau-647 and the vasculature was detected with 500 K MW dextran conjugated to a Pacific blue dye (grey). (C-D) Intravital micrographs showcasing detection of cytokines produced by immune cells in live mice, here in tumor tissues. Panel C shows IFN-γ-producing lymphocytes (magenta) and panel D shows IL-12b-producing myeloid cells (cyan). Tumor cells are also shown (grey). All scale bars represent 10 μm. Credit: A. Kitterman/AAAS
Window chamber models (9, 13) are particularly suited for intravital imaging because they can be easily accessed and immobilized. They also offer the possibility to investigate a wide range of pathological conditions, including autoimmunity (14), infection (15) and cancer (16). The installation of these immobilizing devices requires administration of a post-operative analgesic, often buprenorphine. Antibiotics or antimycotics can also be given to mitigate the risk of infection at the surgical site. Additional considerations may be taken concerning the housing and handling of the mice to avoid damaging the chamber if the animal is to be monitored over repeated imaging sessions. Typically, imaging is initiated several days after the chamber is implanted, i.e. when surgery-induced inflammation has resolved. However, it is always critical to asses whether the procedure used to prepare the mouse affects the readouts to be recorded during the imaging session; this should be done, at least in part, by imaging mice that underwent sham surgery. Detailed methods are available for intravital imaging of various organs such as lymph nodes (17), tumors (13, 18, 19), ear skin (20), abdominal tissues (21–23), lung (9) and heart (24).
INTRAVITAL IMAGING TO STUDY ADAPTIVE IMMUNITY
Cell migration and cell-cell interactions play pivotal roles in adaptive immunity. For instance, circulating naive T cells continuously home to secondary lymphoid organs, where they physically engage with antigen-presenting cells. Local encounters with cognate antigen can produce activated T cell progeny, including effector, central and tissue resident memory subsets, which gain distinct migratory and functional abilities. Cells that traffic to sites of inflammation can further interact with antigen-bearing and other cells, and mediate effector functions. Single cell imaging enables the direct study of all these dynamic processes in complex microenvironments and various tissues.
T cells
The least invasive methods to image T cells use genetic mouse models in which endogenous T cells selectively express a fluorescent protein reporter (Table 1). This approach can assess T cell infiltration in various tissues (11, 25) and bypasses ex vivo cell manipulation and transfer, which may affect cell behavior and fate. Indeed, endogenous T cells may behave differently from adoptively transferred ones. Ideally one would want to image endogenous T cells using mouse models that have T cell-specific reporters whereas adoptive transfer should be considered in special cases. Genetic mouse models can also be used for tracking T regulatory (Treg) cells, which has been performed in lymph nodes (26), bone marrow (27) and non-lymphoid tissues (28). In practice, however, intravital T cell tracking still often relies on adoptive cell transfer approaches. Here, T cells of interest are first purified from mice ubiquitously expressing a fluorescent protein reporter and then administered into recipient mice, so transferred cells and their progeny can be visualized within a tissue of interest (Table 1). This approach is convenient because it allows one to control the quantity of visible (exogenously added) cells in a given field of view. By contrast, the density of endogenous T cells in tissues such as lymph nodes is often quite high making it difficult to distinguish individual cells.
Table 1:
Reporter systems to track different immune cells and their functions by single cell imaging*
| Cell Type/Other | Reporter | Notes | Examples |
|---|---|---|---|
| T Cells | GFP, CFP, DsRed | Requires adoptive cell transfer | (29) |
| PA-GFP | Photoactivatable fluorescent reporter useful for cell tracking; requires adoptive cell transfer | (52) | |
| DPE-GFP | A pan T cell marker, can also label pDCs | (25) | |
| NFAT-GFP, NFAT-YFP;H2B-mCherry | Readout of TCR signaling | (44) | |
| IFN-γ-YFP | Subset of activated T cells; can also be produced by NK cells | (25) | |
| IL-10-GFP | Subset of T cells; can also be expressed by macrophages | (44) | |
| FOXP3-GFP, FOXP3-mRFP | Labels regulatory T cells; FOXP3 is on the X chromosome, be aware of X chromosome inactivation in females | (27) | |
| Nur77-GFP | Labels T cells and also B cells upon antigen receptor engagement | (111) | |
| GranzymeB-TdTomato | A a marker of granule exocytosis in cytolytic T cells and NK cells | (112) | |
| Tbx21-ZsGreen | For assessing Th1 responses in T cells | (113) | |
| CD2-eGFP | Labels peripheral T cells and some NK subsets | (114) | |
| IL-17F-CreeYFP | Many variants exist for studying Th17 cells in vivo | (115) | |
| IL-4-eGFP | Used to assess Th2 immunity | (116) | |
| IL-21-GFP | Used to study the role of TFH cells | (117) | |
| B Cells | GFP, CFP, DsRed | Requires adoptive cell transfer | (118) |
| PA-GFP | Photoactivatable fluorescent reporter useful for cell tracking; requires adoptive cell transfer | (52) | |
| CD19-tdRFP | Expressed by numerous B cells, adoptive transfer may be necessary if tissues are densely infiltrated by B cells | (50) | |
| Bcl6-YFP | A B cell marker already validated for 2-photon microscopy; also labels TFH cells | (119) | |
| Blimp-1-YFP | Expressed by B cells and highly expressed in plasma cells | (120) | |
| AID-GFP | Subset of B cells | (121) | |
| Dendritic Cells | CD11c(ITGAX)-YFP, CD11c-mCherry | Cells other than DCs can express CD11c | (122) |
| XCR1-Venus/+ | Homozygous mice are null for XCR1 | (56) | |
| XCR1-KIKGR/+ | Xcr1 replaced with photoconvertible fluorescent protein Kikume Green-Red (KikGR) | (56) | |
| CXCL9-RFP, CXCL10-BFP | These chemokines can also be expressed by other cell types, including monocytes, macrophages, granulocytes and non-immune cells | (32) | |
| SIGLEC-H-GFP | Expressed by pDCs | (55) | |
| Flt3-BFP2 | This construct has also been incorporated into a genetic model used to visualize DCs, macrophages, CD4+ and CD8+ T cells simultaneously (Flt3-BFP2, Mertk-GFP-DTR, Cd4-tdTomato, Cd8a-tdTomato) | (11) | |
| Zbtb46-GFP | A cDC marker that may be expressed in erythroid and endothelial populations | (123) | |
| Monocytes/Macrophages | CX3CR1-GFP/+ | This chemokine receptor can also be expressed by T cells, NK, DC, and macrophages. Homozygous mice are null for CX3CR1 | (124) |
| CCR2-RFP/+ | Homozygous mice are null for CCR2 | (62) | |
| CX3CR1-GFP/+;CCR2-RFP/+ | Homozygous mice are null for CX3CR1 and/or CCR2 | (62) | |
| Mertk-GFP-DTR | Homozygous mice are null for MerTK | (11) | |
| c-fms-GFP | c-fms is also known as Csf-1 receptor | (61) | |
| MacBlue | A gal4-responsive UAS-ECFP cassette also under the Csf-1 receptor promoter | (125) | |
| Dye-conjugated Ferumoxytol | Administered to mice several hours before imaging | (25) | |
| Dye-conjugated dextrans and dextran nanoparticles | Administered to mice several hours before imaging; can be used to label blood vessels in short-term after administration | (72) | |
| Dye-conjugated anti-CD169 mAb | Used to label SCS macrophages | (126) | |
| Dye-conjugated anti-F4/80 mAb | A pan macrophage marker in mice | (62) | |
| Neutrophils | Dye-conjugated anti-Ly-6G mAb | Imaging is performed shortly after administration (~15 min) | (127) |
| Dye-conjugated anti-Gr-1 mAb | Imaging is performed shortly after administration (~15 min) | (127) | |
| LysM-GFP | Can also be expressed by macrophages | (78) | |
| Dye-conjugated anti-neutrophil elastase mAb | Imaging is performed shortly after administration (~ 15 min) | (86) | |
| CXCL12-mRFP | Can also be expressed by stromal cells | (81) | |
| SYTOX Green | Used to visualize NETs (extracellular DNA labeling); also demarcates cell death | (86) | |
| Innate Lymphocytes | GFP, CFP, DsRed | Requires adoptive cell transfer | (93) |
| CXCR6-GFP | Can be used to track ILCs and iNKT cells but is also expressed by some T cells | (90) | |
| Megakaryocytes and Platelets | PF4-cre tdTomato | Labels megakaryocytes as well as platelets, which are defined based on their smaller size | (89) |
| Dye-conjugated anti-CD49b mAb | Labels all platelets | (86) | |
| CD41-YFPki/+ | Labels a fraction of platelets | (86) | |
| HSC-Derived Cells | GFP, CFP, DsRed | Requires adoptive cell transfer and is used to track HSCs and their progeny | (85) |
| Non-Immune Components | Dye-conjugated anti-Lyve-1 mAb | Labels lymphatics | (46) |
| AngioSPARK | Labels blood vessels | (72) | |
| Second Harmonic Generation (SHG) | Applicable to multiphoton (not confocal) microscopy; enables visualization of collagen fibers | (92) | |
| IL-7-eCFP | Expressed by stromal cells in thymus and bone marrow | (45) | |
| GCaMP6s | Sensor of calcium flux; relevant in the context of e.g. virus-infected cells | (40) | |
| qDots | Bright, highly tunable fluorophores surface-engineered to maximize circulation | (128) | |
| Isolectin | Used to label endothelial wall of blood vessels | (129) | |
| Hoechst33342 and CellTracker dyes | Indiscriminately labels cell nuclei or cytosol; often used for adoptive cell transfer | (42) |
This list serves as a general survey of intravital imaging labeling strategies across cell types, however numerous Cre strains can additionally be incorporated into useable reporter systems.
Intravital imaging of adoptively transferred T cells has uncovered how naive (29, 30) and central memory (31, 32) T cells sense antigens and become activated in lymph nodes. For instance, imaging has revealed that naive lymph node CD8+ T cells are initially motile, which allows them to undergo multiple transient contacts with antigen-presenting DCs, but can then progressively decrease their motility to form stable contacts with DCs, at which time they also start producing effector cytokines. The T cells eventually resume their migration, begin to proliferate and can emigrate from the lymph node (29). The type of interaction that a CD8+ T cell undergoes with an antigen-presenting DC depends at least in part on the antigen dose. Whereas low antigen dose conditions can functionally prime CD8+ T cells, only high antigen dose may trigger stable T cell-DC contacts (33); these longer contacts may be necessary for the generation of full-fledged immune responses. Intravital lymph node imaging has further revealed that helper CD4+ and CD8+ T cells are largely compartmentalized during priming reactions; however, during later phases of lymphocyte maturation, these cells are brought together by some DCs. This process enables the delivery of CD4+ T cell help to CD8+ T cell responses, which is important for the generation of CD8+ T cell memory (30).
Intravital imaging in the context of vaccinia virus infection has further revealed the compartmentalization of naive and memory CD8+ T cells in lymph nodes. Whereas naive CD8+ T cells primarily locate in the cortex, memory CD8+ T cells distribute along the periphery of the lymph node (34). This distinction is likely relevant because it brings memory CD8+ T cells in close proximity to subcapsular sinus (SCS) macrophages, which are among the first lymph node cells to capture lymph-borne viruses. Therefore, it appears that memory CD8+ T cells have preferential access to antigen upon viral reinfection (31, 34). Other imaging studies have tracked T cells in nonlymphoid tissues. For example, skin and female reproductive tract imaging have defined antigen-specific T resident memory cell (TRM) activation and proliferation in response to viral infections (35, 36). TRM responses in the female reproductive tract can occur in the absence of DCs (36); however, it is unknown whether TRM responses in other tissues require these cells.
Additional imaging tools extend our ability to study T cell communication with neighboring cells at a deeper level, such that specific molecular outcomes can be identified (Table 1). For example, transgenic mice harboring a T cell receptor (TCR)-GFP fusion protein enable the visualization of TCR clustering and internalization following antigen encounter (37). Translocating nuclear factor of activated T cells (NFAT) from the cytoplasm to the nucleus is a sensitive readout of TCR signaling and can be assessed by intravital imaging using fluorescently labeled NFAT (38). The NFAT reporter is often used in combination with fluorescently labeled histone protein H2B, a nuclear marker. Other cell signaling mechanisms such as calcium flux can be detected using transgenic mice expressing the calcium sensor protein GCaMP6s to read out features such as in vivo T cell activation (39) or target cell killing (40).
CD8+ T cells can also be “caught in the act” of effector activities. For instance, intravital imaging approaches have shown blood circulating CD8+ T effector cells to adhere onto liver sinusoids and engage in immune surveillance (41) and cytotoxic T cells to kill virus-infected cells in lymph nodes (40). The latter study used color-labeled viruses to visualize virus-infected cells and identified that T cell-mediated target cell killing in vivo may be much slower than anticipated from prior in vitro studies. Interestingly, the proximity of Treg can dramatically slow the kinetics of CD8+ T cell killing in vivo (42). Cytokines such as interferon-gamma (IFN-γ) and interleukin-10 (IL-10), both produced by T cells, can also be detected by intravital imaging. Because IFN-γ has a rapid on/off cycling (43), a common readout typically requires restimulation of T cells in vitro with an antigenic trigger. This method is useful to define whether given cells are equipped to produce IFN-γ but it cannot assess whether these cells actually produced the cytokine in vivo. By contrast, intravital imaging of IFN-γ-internal ribosome entry site-YFP (IFN-γ-IRES-YFP) reporter mice detects YFP, which is expressed by cells that have turned on IFN-γ production (25). YFP remains detectable even after IFN-γ production is turned off, which makes intravital imaging a particularly useful tool to detect IFN-γ pathway activation in vivo (Fig. 2C), although it cannot identify cells that are currently producing the cytokine. Imaging is also useful to detect cells producing IL-10, which was identified as a factor important for both viral containment and tissue protection following vaccinia virus infection (44). IL-7, which is required for T cell development, survival and memory development, can also be tracked in IL-7 reporter mice. Imaging of IL-7 reporter systems has provided useful information of the location and composition of IL-7-dependent immune cell niches (45).
B cells
Intravital B cell tracking, like T cell tracking, usually involves transgenic reporter mice or adoptively transferring fluorescent B cells into recipient mice (Table 1). Both approaches enable the study of B cells in different body compartments. In lymph nodes, intravital imaging has shed light into how B cells interact with various cellular and molecular components, including subcapsular sinus (SCS) macrophages (46), T follicular helper cells (TFH) (47) and type I interferon (39, 48), and how these dynamic interactions regulate the initiation of humoral immune responses. For example, in the context of infection, intravital imaging studies have revealed that lymph-borne viruses preferentially infect SCS macrophages, which then present viral particles to B cells to initiate humoral immune responses (46). Intravital imaging studies further identified B cells can express intercellular adhesion molecule 1 (ICAM1) and ICAM2, which are critical for forming stable contacts between B cells and TFH and subsequent development of humoral immunity (47). By contrast, imaging studies have shown that type I interferon inhibits antiviral B cell responses in lymph nodes, for example by recruiting inflammatory monocytes that produce nitric oxide (48) and by activating cytotoxic T lymphocytes that kill antiviral B cells (39). Indeed, not all T cell-B cell interactions in lymph nodes stimulate humoral immunity, as cytotoxic T lymphocytes can kill B cells under certain contexts (39, 42).
Intravital imaging has been further used to study B cells in other tissues. For example, intravital imaging revealed how splenic B cells shuttle antigens from the marginal zone and into B cell follicles (49). B cell imaging has also been extended to various bone compartments using novel imaging platforms that allow for long-term, longitudinal studies of bone marrow dynamics (50) and to non-lymphoid tissues such as the lungs and liver (51). For example, pulmonary intravital imaging has revealed that B cells can physically interact with aged neutrophils and contribute to their clearance locally (51).
By transferring B cells carrying the photoactivatable reporter PA-GFP, it is possible to tag these cells by multiphoton photoactivation within distinct micro-anatomical tissues such as the germinal center of a lymph node (52). This photoactivation system in situ was used to interrogate migratory behavior of B cells within germinal centers and identify that cell proliferation occurs in the dark zone of the germinal center after B cells are instructed by T cells in the light zone of the germinal center (52). The photoactivated cells can be monitored in vivo for up to several days or purified for additional analysis in vitro. In principle, this approach can be used to tag any adoptively transferred cell, regardless of its type, and be further extended to photoconvertible fluorescent reporters, such as Kaede and Dendra2, which shift from green to red fluorescence upon exposure to light of appropriate wavelength (4). These types of experiments enable investigators to have finer spatial resolution of migratory phenotypes as photoconversion of cells can identify distinct migratory properties that would otherwise be difficult to study. In addition, photoactivation models can be used in combination with single cell sequencing approaches to retain spatial information in high dimensional sequencing technologies (53).
INTRAVITAL IMAGING TO STUDY INNATE IMMUNITY
An increasing number of tools are available to image a variety of innate immune cell types, including dendritic cells (DCs), monocytes, macrophages, neutrophils and innate lymphocytes. Single cell imaging is most often used to understand innate immune cells’ interplay with their adaptive counterparts or to reveal their functions in development, steady state, health, and disease.
Dendritic Cells
Both genetic reporter and adoptively transferred DCs can be tracked by intravital imaging (Table 1). CD11c-YFP reporter mice have been used to yield insight into how endogenous DCs are spatially organized in lymph nodes, collect antigens, and promote T cell responses to these antigens (30, 54). However, other cell types including macrophages may express CD11c. Endogenous DCs can also be imaged using Flt3-BFP2 (11) or XCR1-Venus (55) reporter systems. Typically all DC subsets express Flt3, whereas macrophages and other cells usually don’t or do so at much lower levels. XCR1 expression is more selective for a subtype of DCs, whereas CD11c is expressed by DCs but also some macrophages and T cells. An XCR1KiKGR/+ genetic mouse model further enables photoconversion of XCR1+ DCs in vivo (56). These mice are convenient for tracking migratory DCs and distinguishing them from neighboring resident DCs. For instance, skin XCR1+ DCs photoconverted by violet-blue illumination can be tracked upon migration to skin-draining lymph nodes and discriminated from non-photoconverted DCs (56). This approach identified that skin XCR1+ DCs that migrate to draining lymph nodes gradually accumulate in deep medullary regions of the T cell zone, where they can prime naive CD8+ T cells (56). Painting the skin with a fluorescent dye, such as TRITC, may also distinguish recent skin DC migrants, which carry the dye, from other lymph node DCs (57), although it is formally possible that some dyes may percolate to lymph nodes independently of DCs or be transferred from one cell type to another. In the context of HSV-1 infection, TRITC-labeled DCs accumulate in the T cell zone of draining lymph nodes, where they interact predominantly with naive CD4+ but not CD8+ T cells. CD8+ T cell activation occurs approximately one day later and involves XCR1+ (TRITC–) DCs, suggesting that distinct DC subsets activate CD4+ and CD8+ T cells sequentially. Eventually, clusters of XCR1+ DCs and activated CD8+ T cells recruit the pre-activated helper CD4+ T cells, which provide licensing signals to enhance CD8+ T immunity (57). The formation of a “ménage-à-trois” between DCs, CD4+ and CD8+ T cells can also be promoted by clusters of DCs and CD4+ T cells that recruit naive CD8+ T cells (58).
Some DC functions can also be imaged. For example, a transgenic mouse reporting expression of the chemokines CXCL9 and CXCL10 (which bind the chemokine receptor CXCR3, expressed by T cells) has demonstrated that DC-derived CXCL10 facilitates interactions between DCs and naive CD4+ T cells in the T cell zone. Both CXCL10 and CXCL9 (produced by stromal cells) further guide activated CD4+ T cells away from the T cell zone and into interfollicular and medullary zones of the lymph node. This process is important for the development of the Th1 subtype of CD4+ T cells, presumably because it enables CD4+ T cells to receive stimuli from cell populations located in the periphery of the lymph node, including SCS macrophages (7, 32). Cytokines such as interleukin-12 (IL-12), which can be produced by DCs to activate T cells, is also detectable by intravital imaging (Fig. 2D).
Monocytes and Macrophages
Intravital imaging has been used to examine macrophages in various tissues, including lymph nodes (59), tumors (25, 60, 61), liver (62), lung (63), pancreas (11), kidney (64) and the brain (65). Macrophages can be viewed using genetic reporters such as MerTK-GFP (11) or after in vivo labeling with fluorescent nanomaterials (66) (Table 1). Furthermore, imaging has been employed to study several macrophage immune functions, including microenvironmental surveillance (67), bacteria capture (68), adaptive immune response stimulation (59), extracellular matrix remodeling (69) and tissue repair (62), as well as perhaps less expected functions such as shielding injured cells (70), exchanging cytoplasm with neighboring cells (71) and promoting vasculature bursting (72).
Macrophages that accumulate in inflammatory sites typically originate from circulating monocytes, which can be detected using CX3CR1 reporter mice (73). The same mice can be used to track splenic reservoir monocytes and their deployment to the circulation in response to inflammatory cues (74). Additionally, CX3CR1-GFP;CCR2-RFP mice can be used to distinguish between monocyte/macrophage subsets that express these chemokine receptors at different levels (62). These mice were used to show that mature macrophages from the peritoneal cavity can migrate to the liver after acute injury (62), challenging the paradigm that macrophages are sessile immune cells. In contrast to macrophages in inflammatory sites, many macrophages found in resting adult tissue are established before birth. Imaging can also be used to track the development of the immune system through embryonic precursor populations by using CX3CR1 reporter models. These approaches have shown how yolk sac pre-macrophages migrate to the developing embryo and seed macrophage pools (75).
Neutrophils
Neutrophils have been imaged in various tissues such as liver (76), lung (77), skin (78), spleen (79), joints (80), trachea (81) and tumors (82). Methods to track neutrophils rely either on tagging them with fluorescent antibodies, such as anti-Ly-6G mAbs, or using genetically encoded reporters, such as LysM-GFP (Table 1). The antibodies used to tag neutrophils are given at lower doses than for cell depletion studies, and the neutrophils are imaged shortly (~15 min) after intravenous mAb administration. Such approaches require testing that the injected mAbs label the targeted cells without substantially altering their fate. Also, care must be taken in interpreting LysM-GFP studies because macrophages can also express LysM. Neutrophil extracellular traps (NETs), which are composed of DNA and are released by neutrophils to eliminate extracellular pathogens, can be visualized using nucleic acid-labeling agents (e.g. SYTOX Green) or antibodies against extracellular histones or neutrophil elastase (83). In sterile liver injury models, photoactivatable cell tracking approaches have started to reconstruct the journey of neutrophils. These studies identified that neutrophils, initially produced in the bone marrow, are recruited to the injury site where they mediate key effector functions, and then reenter the circulation and relocate to the lung and eventually the bone marrow where they are eliminated (76). The implications for neutrophil egress from injury sites, and homing back to the sites where they were produced, requires further study.
Other Hematopoietic Cells
Hematopoietic stem cells (HSCs), megakaryocytes, platelets, eosinophils and innate lymphocytes can also be tracked in living animals (Table 1). For example, intravital imaging of the mouse calvarium has helped characterize the location and composition of HSC niches in the bone marrow (27, 84). HSC progeny can also be tracked in extramedullary tissues (85). Additionally, CD41-YFP and Pf4-cre:mTmG transgenic mice are useful for detecting megakaryocytes and the platelets they produce. Intravital imaging studies have revealed platelets’ contributions to host defense. For example, they can promote the release of NETs to protect host cells from virus infection (86), collaborate with Kupffer cells to encase bacteria (87), help circulating T cells to probe for the presence of antigens (41), and migrate at sites of vascular injury to collect bacteria (88). Intravital imaging also identified the lung as an unexpected site of platelet production (89). Imaging has also been used to track eosinophils within mouse airways (85). Lastly, imaging studies have started to assess innate lymphocytes in various settings such as tissue repair (90), stroke (91), skin homeostasis (92), tumors (93), and patterning immune responses in lymph nodes (94); Kastenmüller et al., 2013, #93282}. Innate lymphocyte imaging largely relies on CXCR6 reporter mice; iNKT cells and ILC2 express this chemokine receptor, though other cell types may as well.
INTRAVITAL IMAGING TO STUDY DRUGS AND THEIR EFFECTS ON IMMUNE CELLS
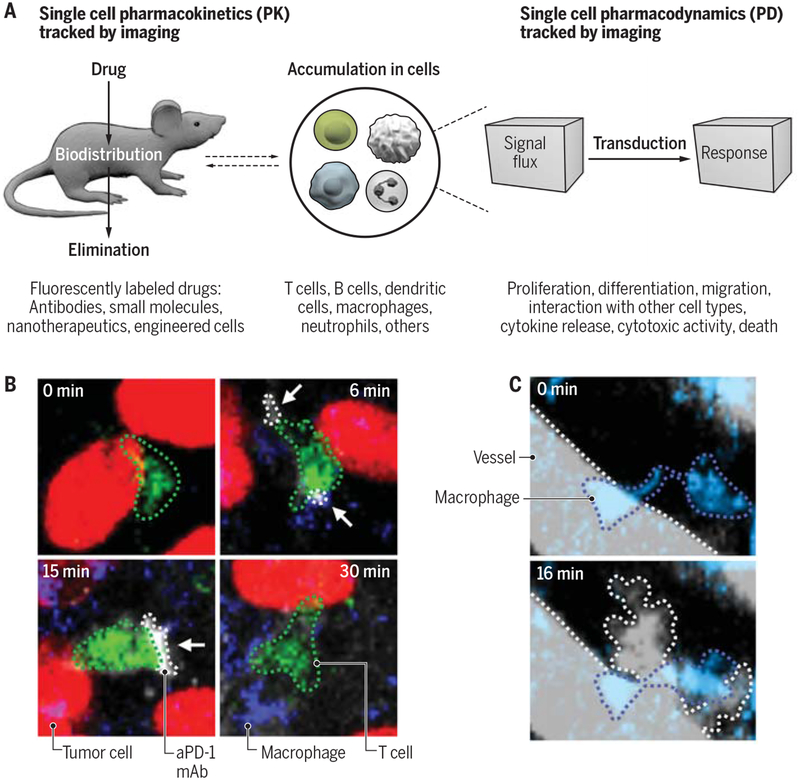
A major development in single cell imaging has been the ability to track drugs in complex tissue microenvironments (12). When combined with immune cell reporters, single cell imaging can uncover pharmacokinetics (PK) and pharmacodynamics (PD) at the single cell level with unparalleled insight into drug resistance (25) and mechanism of action (95) (Fig. 3A). For PK studies, fluorescently labeled drugs are imaged over time to follow their distribution and cell tropism. The approach requires validation that the fluorescent drug retains biological activity, which appears to be the case for antibody drugs and can also be achieved for small molecules. For example, single cell imaging has been used to uncover that the immune checkpoint blocker anti-PD-1 mAb initially binds its intended targets (i.e. PD-1 expressed on the surface of T cells) but can then be quickly captured by neighboring macrophages, which limits therapeutic efficacy (25) (Fig. 3B). For PD studies, all reporter systems presented in Table 1 could be deployed to assess a drug’s impact on immune components while additional reporters are emerging (12). For example, since IFN-γ is a key factor required for anti-tumor immunity, IFN-γ-eYFP reporter mice can be used to assess a central readout of successful immunotherapy response (25). PD studies have also shown that radiation therapy can prime tumor-associated macrophages to induce vascular bursts, which enhance drug uptake in the tumor microenvironment (Fig. 3C), and that chemotherapeutics can induce tumor infiltration by myeloid cells, which contribute to chemoresistance (96, 97). Currently, intravital imaging of drug responses focuses mostly on cancer, though existing approaches can be adapted for studies of host defense or autoimmunity.
Fig. 3 – Single cell imaging of drugs and their effects on immune cells.
(A) Intravital imaging can reveal drugs’ PK (left) and PD (right) at single cell resolution. The examples listed below indicate which types of drugs, cells and responses can be detected and are relevant to immunology. (B) A PK study reveals that the immune checkpoint blocker anti-PD-1 mAb (aPD-1 mAb, gray) binds to T cells (green) only transiently in the tumor stroma (25). (C) A PD study reveals that radiation therapy primes tumor-associated macrophages (blue) to initiate vascular bursts (courtesy of M. Miller, Massachusetts General Hospital, Boston). Credit: A. Kitterman/AAAS
NEEDS AND EMERGING APPROACHES
Further improvement of intravital single cell imaging for an improved understanding of immunology will be possible with technological, biological and computational advances. Technological needs include active and passive methods of motion suppression (24) to image an increasing number of tissues; new chambers and surgical exteriorization methods (9, 19) for imaging in orthotopic organs; photoacoustic microscopy (98–100) for body-wide imaging; infrared imaging and three-photon imaging (98, 101) for deeper imaging (beyond 200 μm); and implantable microscope optics (102, 103) for imaging over days. Biological needs include new mouse models with fluorescent reporters in multiplex compatible formats for visualizing several immune cell (sub)types simultaneously, new signaling reporters––also in multiplex format––for more detailed understanding of immune cells’ molecular activities, as well as labeled antibodies and nanoprobes that are validated for intravital microscopy and could also be useful for clinical translation. Computational needs include the development of more efficient analytical software, which could be achieved by adapting deep learning and artificial intelligence solutions (104) and could also be used for whole organ mapping (105, 106). The combination for single cell imaging with single cell RNA sequencing or FISH should also offer deeper integration of physiological data at the cellular and molecular levels (53). Beyond these needs, a number of emerging technologies are on the horizon, including tissue clearing for whole organ imaging, photoactivatable optogenetic reporters and photoacoustic microscopy. These methods have considerable potential and applications for immune cell imaging. Tissue clearing methods (e.g. CLARITY, CUBIC, Ce3D) reduce light scattering and thus enable deep tissue imaging (107). These methods are still limited to excised and perfused whole organs but nevertheless represent a powerful method for 3D reconstruction of immune cell populations at the whole organ level (108, 109). Finally, photoacoustic microscopy is an innovative high-resolution, high-speed imaging methodology that can provide deeper, larger field-of-view optical contrast, especially when combined with cell-specific exogenous labeling agents. While much development has happened at the meso- and macroscopic scales (110), future developments will surely advance the field of immune cell imaging.
ACKNOWLEDGEMENTS
The work of M.J.P. is supported by US National Institutes of Health (NIH) grants R01-AI084880, R01-CA206890 and R01-CA2155565 and the Samana Cay MGH Research Scholar Fund. C.S.G. and S.P.A are supported by NIH grants F31-CA196035 and T32-CA79443, respectively. The work of R.W. is supported by NIH grants R33-CA202064, R01-CA206890 and U01CA206997. We apologize to the authors whose work we could not cite because of space constraints.
REFERENCES AND NOTES
- 1.Balzarotti F, Eilers Y, Gwosch KC, Gynnå AH, Westphal V, Stefani FD, Elf J, Hell SW, Science 355, 606 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Weissleder R, Schwaiger MC, Gambhir SS, Hricak H, Sci Transl Med 8, 355ps16 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Zipfel WR, Williams RM, Webb WW, Nat Biotechnol 21, 1369 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Pittet MJ, Weissleder R, Cell 147, 983 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellenbroek SI, van Rheenen J, Nat Rev Cancer 14, 406 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Jonkman J, Brown CM, J Biomol Tech 26, 54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hickman HD, Bennink JR, Yewdell JW, Cell Host Microbe 5, 13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrickson SE, Mempel TR, Mazo IB, Liu B, Artyomov MN, Zheng H, Peixoto A, Flynn M, Senman B, Junt T, Wong HC, Chakraborty AK, von Andrian UH, Sci Signal 1, pt2 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Entenberg D, Voiculescu S, Guo P, Borriello L, Wang Y, Karagiannis GS, Jones J, Baccay F, Oktay M, Condeelis J, Nat Methods 15, 73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinegoni C, Leon Swisher C, Fumene Feruglio P, Giedt RJ, Rousso DL, Stapleton S, Weissleder R, Nat Commun 7, 11077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohan JF, Kohler RH, Hill JA, Weissleder R, Mathis D, Benoist C, Proc Natl Acad Sci U S A 114, E7776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller MA, Weissleder R, Nat Rev Cancer 17, 399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alieva M, Ritsma L, Giedt RJ, Weissleder R, van Rheenen J, Intravital 3, e29917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ardigò M, Agozzino M, Longo C, Lallas A, Di Lernia V, Fabiano A, Conti A, Sperduti I, Argenziano G, Berardesca E, Pellacani G, J Eur Acad Dermatol Venereol 29, 2363 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Liese J, Rooijakkers SH, van Strijp JA, Novick RP, Dustin ML, Cell Microbiol 15, 891 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Fisher DT, Muhitch JB, Kim M, Doyen KC, Bogner PN, Evans SS, Skitzki JJ, Nat Commun 7, 10684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murooka TT, Mempel TR, Methods Mol Biol 757, 247 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Sckell A, Leunig M, Methods Mol Biol 1430, 251 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Kitamura T, Pollard JW, Vendrell M, Trends Biotechnol 35, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li JL, Goh CC, Keeble JL, Qin JS, Roediger B, Jain R, Wang Y, Chew WK, Weninger W, Ng LG, Nat Protoc 7, 221 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Ritsma L, Steller EJ, Ellenbroek SI, Kranenburg O, Borel Rinkes IH, van Rheenen J, Nat Protoc 8, 583 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Marques PE, Antunes MM, David BA, Pereira RV, Teixeira MM, Menezes GB, Nat Protoc 10, 258 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Ferrer M, Martin-Jaular L, Calvo M, del Portillo HA, J Vis Exp (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinegoni C, Aguirre AD, Lee S, Weissleder R, Nat Protoc 10, 1802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arlauckas SP, Garris CS, Kohler RH, Kitaoka M, Cuccarese MF, Yang KS, Miller MA, Carlson JC, Freeman GJ, Anthony RM, Weissleder R, Pittet MJ, Sci Transl Med 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Gerner MY, Van Panhuys N, Levine AG, Rudensky AY, Germain RN, Nature 528, 225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujisaki J, Wu J, Carlson AL, Silberstein L, Putheti P, Larocca R, Gao W, Saito TI, Lo Celso C, Tsuyuzaki H, Sato T, Côté D, Sykes M, Strom TB, Scadden DT, Lin CP, Nature 474, 216 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budhu S, Schaer DA, Li Y, Toledo-Crow R, Panageas K, Yang X, Zhong H, Houghton AN, Silverstein SC, Merghoub T, Wolchok JD, Sci Signal 10, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mempel TR, Henrickson SE, Von Andrian UH, Nature 427, 154 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Eickhoff S, Brewitz A, Gerner MY, Klauschen F, Komander K, Hemmi H, Garbi N, Kaisho T, Germain RN, Kastenmüller W, Cell 162, 1322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sung JH, Zhang H, Moseman EA, Alvarez D, Iannacone M, Henrickson SE, de la Torre JC, Groom JR, Luster AD, von Andrian UH, Cell 150, 1249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groom JR, Richmond J, Murooka TT, Sorensen EW, Sung JH, Bankert K, von Andrian UH, Moon JJ, Mempel TR, Luster AD, Immunity 37, 1091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henrickson SE, Mempel TR, Mazo IB, Liu B, Artyomov MN, Zheng H, Peixoto A, Flynn M, Senman B, Junt T, Wong HC, Chakraborty AK, von Andrian UH, Sci Signal 1, pt2 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Kastenmüller W, Brandes M, Wang Z, Herz J, Egen JG, Germain RN, Immunity 38, 502 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, Alexandre YO, Gregory JL, Russell TA, Gebhardt T, Carbone FR, Tscharke DC, Heath WR, Mueller SN, Mackay LK, Nat Immunol 19, 183 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, Fonseca R, Burbach BJ, Hickman HD, Vezys V, Fife BT, Masopust D, Nat Immunol 19, 173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedman RS, Beemiller P, Sorensen CM, Jacobelli J, Krummel MF, J Exp Med 207, 2733 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marangoni F, Murooka TT, Manzo T, Kim EY, Carrizosa E, Elpek NM, Mempel TR, Immunity 38, 237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moseman EA, Wu T, de la Torre JC, Schwartzberg PL, McGavern DB, Sci Immunol 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halle S, Keyser KA, Stahl FR, Busche A, Marquardt A, Zheng X, Galla M, Heissmeyer V, Heller K, Boelter J, Wagner K, Bischoff Y, Martens R, Braun A, Werth K, Uvarovskii A, Kempf H, Meyer-Hermann M, Arens R, Kremer M, Sutter G, Messerle M, Förster R, Immunity 44, 233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guidotti LG, Inverso D, Sironi L, Di Lucia P, Fioravanti J, Ganzer L, Fiocchi A, Vacca M, Aiolfi R, Sammicheli S, Mainetti M, Cataudella T, Raimondi A, Gonzalez-Aseguinolaza G, Protzer U, Ruggeri ZM, Chisari FV, Isogawa M, Sitia G, Iannacone M, Cell 161, 486 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Mempel TR, Pittet MJ, Khazaie K, Weninger W, Weissleder R, von Boehmer H, von Andrian UH, Immunity 25, 129 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Slifka MK, Rodriguez F, Whitton JL, Nature 401, 76 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Cush SS, Reynoso GV, Kamenyeva O, Bennink JR, Yewdell JW, Hickman HD, PLoS Pathog 12, e1005493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazzucchelli RI, Warming S, Lawrence SM, Ishii M, Abshari M, Washington AV, Feigenbaum L, Warner AC, Sims DJ, Li WQ, Hixon JA, Gray DH, Rich BE, Morrow M, Anver MR, Cherry J, Naf D, Sternberg LR, McVicar DW, Farr AG, Germain RN, Rogers K, Jenkins NA, Copeland NG, Durum SK, PLoS One 4, e7637 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Junt T, Moseman EA, Iannacone M, Massberg S, Lang PA, Boes M, Fink K, Henrickson SE, Shayakhmetov DM, Di Paolo NC, van Rooijen N, Mempel TR, Whelan SP, von Andrian UH, Nature 450, 110 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Zaretsky I, Atrakchi O, Mazor RD, Stoler-Barak L, Biram A, Feigelson SW, Gitlin AD, Engelhardt B, Shulman Z, J Exp Med 214, 3435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sammicheli S, Kuka M, Di Lucia P, de Oya NJ, De Giovanni M, Fioravanti J, Cristofani C, Maganuco CG, Fallet B, Ganzer L, Sironi L, Mainetti M, Ostuni R, Larimore K, Greenberg PD, de la Torre JC, Guidotti LG, Iannacone M, Sci Immunol 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnon TI, Horton RM, Grigorova IL, Cyster JG, Nature 493, 684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reismann D, Stefanowski J, Günther R, Rakhymzhan A, Matthys R, Nützi R, Zehentmeier S, Schmidt-Bleek K, Petkau G, Chang HD, Naundorf S, Winter Y, Melchers F, Duda G, Hauser AE, Niesner RA, Nat Commun 8, 2153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JH, Podstawka J, Lou Y, Li L, Lee EKS, Divangahi M, Petri B, Jirik FR, Kelly MM, Yipp BG, Nat Immunol 19, 192 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC, Cell 143, 592 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Medaglia C, Giladi A, Stoler-Barak L, De Giovanni M, Salame TM, Biram A, David E, Li H, Iannacone M, Shulman Z, Amit I, Science 358, 1622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerner MY, Torabi-Parizi P, Germain RN, Immunity 42, 172 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Brewitz A, Eickhoff S, Dähling S, Quast T, Bedoui S, Kroczek RA, Kurts C, Garbi N, Barchet W, Iannacone M, Klauschen F, Kolanus W, Kaisho T, Colonna M, Germain RN, Kastenmüller W, Immunity 46, 205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitano M, Yamazaki C, Takumi A, Ikeno T, Hemmi H, Takahashi N, Shimizu K, Fraser SE, Hoshino K, Kaisho T, Okada T, Proc Natl Acad Sci U S A 113, 1044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hor JL, Whitney PG, Zaid A, Brooks AG, Heath WR, Mueller SN, Immunity 43, 554 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN, Nature 440, 890 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Sagoo P, Garcia Z, Breart B, Lemaître F, Michonneau D, Albert ML, Levy Y, Bousso P, Nat Med 22, 64 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Karagiannis GS, Pastoriza JM, Wang Y, Harney AS, Entenberg D, Pignatelli J, Sharma VP, Xue EA, Cheng E, D’Alfonso TM, Jones JG, Anampa J, Rohan TE, Sparano JA, Condeelis JS, Oktay MH, Sci Transl Med 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J, Cancer Res 67, 2649 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Wang J, Kubes P, Cell 165, 668 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Headley MB, Bins A, Nip A, Roberts EW, Looney MR, Gerard A, Krummel MF, Nature 531, 513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finsterbusch M, Hall P, Li A, Devi S, Westhorpe CL, Kitching AR, Hickey MJ, Proc Natl Acad Sci U S A 113, E5172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nayak D, Johnson KR, Heydari S, Roth TL, Zinselmeyer BH, McGavern DB, PLoS Pathog 9, e1003395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weissleder R, Nahrendorf M, Pittet MJ, Nat Mater 13, 125 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB, Nat Neurosci 8, 752 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Zeng Z, Surewaard BG, Wong CH, Geoghegan JA, Jenne CN, Kubes P, Cell Host Microbe 20, 99 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Madsen DH, Jürgensen HJ, Siersbæk MS, Kuczek DE, Grey Cloud L, Liu S, Behrendt N, Grøntved L, Weigert R, Bugge TH, Cell Rep 21, 3662 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nimmerjahn A, Kirchhoff F, Helmchen F, Science 308, 1314 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Roh-Johnson M, Shah AN, Stonick JA, Poudel KR, Kargl J, Yang GH, di Martino J, Hernandez RE, Gast CE, Zarour LR, Antoku S, Houghton AM, Bravo-Cordero JJ, Wong MH, Condeelis J, Moens CB, Dev Cell 43, 549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miller MA, Chandra R, Cuccarese MF, Pfirschke C, Engblom C, Stapleton S, Adhikary U, Kohler RH, Mohan JF, Pittet MJ, Weissleder R, Sci Transl Med 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, Cumano A, Geissmann F, Science 311, 83 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, Figueiredo JL, Kohler RH, Chudnovskiy A, Waterman P, Aikawa E, Mempel TR, Libby P, Weissleder R, Pittet MJ, Science 325, 612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stremmel C, Schuchert R, Wagner F, Thaler R, Weinberger T, Pick R, Mass E, Ishikawa-Ankerhold HC, Margraf A, Hutter S, Vagnozzi R, Klapproth S, Frampton J, Yona S, Scheiermann C, Molkentin JD, Jeschke U, Moser M, Sperandio M, Massberg S, Geissmann F, Schulz C, Nat Commun 9, 75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J, Hossain M, Thanabalasuriar A, Gunzer M, Meininger C, Kubes P, Science 358, 111 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Yipp BG, Kim JH, Lima R, Zbytnuik LD, Petri B, Swanlund N, Ho M, Szeto VG, Tak T, Koenderman L, Pickkers P, Tool ATJ, Kuijpers TW, van den Berg TK, Looney MR, Krummel MF, Kubes P, Sci Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lämmermann T, Afonso PV, Angermann BR, Wang JM, Kastenmüller W, Parent CA, Germain RN, Nature 498, 371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deniset JF, Surewaard BG, Lee WY, Kubes P, J Exp Med 214, 1333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miyabe Y, Miyabe C, Murooka TT, Kim EY, Newton GA, Kim ND, Haribabu B, Luscinskas FW, Mempel TR, Luster AD, Sci Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim K, Hyun YM, Lambert-Emo K, Capece T, Bae S, Miller R, Topham DJ, Kim M, Science 349, aaa4352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park J, Wysocki RW, Amoozgar Z, Maiorino L, Fein MR, Jorns J, Schott AF, Kinugasa-Katayama Y, Lee Y, Won NH, Nakasone ES, Hearn SA, Küttner V, Qiu J, Almeida AS, Perurena N, Kessenbrock K, Goldberg MS, Egeblad M, Sci Transl Med 8, 361ra138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kolaczkowska E, Jenne CN, Surewaard BG, Thanabalasuriar A, Lee WY, Sanz MJ, Mowen K, Opdenakker G, Kubes P, Nat Commun 6, 6673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mazo IB, Gutierrez-Ramos JC, Frenette PS, Hynes RO, Wagner DD, von Andrian UH, J Exp Med 188, 465 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cortez-Retamozo V, Etzrodt M, Newton A, Rauch PJ, Chudnovskiy A, Berger C, Ryan RJ, Iwamoto Y, Marinelli B, Gorbatov R, Forghani R, Novobrantseva TI, Koteliansky V, Figueiredo JL, Chen JW, Anderson DG, Nahrendorf M, Swirski FK, Weissleder R, Pittet MJ, Proc Natl Acad Sci U S A 109, 2491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jenne CN, Wong CH, Zemp FJ, McDonald B, Rahman MM, Forsyth PA, McFadden G, Kubes P, Cell Host Microbe 13, 169 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Wong CH, Jenne CN, Petri B, Chrobok NL, Kubes P, Nat Immunol 14, 785 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gaertner F, Ahmad Z, Rosenberger G, Fan S, Nicolai L, Busch B, Yavuz G, Luckner M, Ishikawa-Ankerhold H, Hennel R, Benechet A, Lorenz M, Chandraratne S, Schubert I, Helmer S, Striednig B, Stark K, Janko M, Böttcher RT, Verschoor A, Leon C, Gachet C, Gudermann T, Mederos Y Schnitzler M, Pincus Z, Iannacone M, Haas R, Wanner G, Lauber K, Sixt M, Massberg S, Cell 171, 1368 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Lefrançais E, Ortiz-Muñoz G, Caudrillier A, Mallavia B, Liu F, Sayah DM, Thornton EE, Headley MB, David T, Coughlin SR, Krummel MF, Leavitt AD, Passegué E, Looney MR, Nature 544, 105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liew PX, Lee WY, Kubes P, Immunity 47, 752 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Wong CH, Jenne CN, Lee WY, Léger C, Kubes P, Science 334, 101 (2011). [DOI] [PubMed] [Google Scholar]
- 92.Roediger B, Kyle R, Yip KH, Sumaria N, Guy TV, Kim BS, Mitchell AJ, Tay SS, Jain R, Forbes-Blom E, Chen X, Tong PL, Bolton HA, Artis D, Paul WE, de St Groth B. Fazekas , Grimbaldeston MA, Le Gros G, Weninger W, Nat Immunol 14, 564 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deguine J, Breart B, Lemaître F, Di Santo JP, Bousso P, Immunity 33, 632 (2010). [DOI] [PubMed] [Google Scholar]
- 94.Gaya M, Barral P, Burbage M, Aggarwal S, Montaner B, Warren Navia A, Aid M, Tsui C, Maldonado P, Nair U, Ghneim K, Fallon PG, Sekaly RP, Barouch DH, Shalek AK, Bruckbauer A, Strid J, Batista FD, Cell 172, 517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miller MA, Zheng YR, Gadde S, Pfirschke C, Zope H, Engblom C, Kohler RH, Iwamoto Y, Yang KS, Askevold B, Kolishetti N, Pittet M, Lippard SJ, Farokhzad OC, Weissleder R, Nat Commun 6, 8692 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nakasone ES, Askautrud HA, Kees T, Park JH, Plaks V, Ewald AJ, Fein M, Rasch MG, Tan YX, Qiu J, Park J, Sinha P, Bissell MJ, Frengen E, Werb Z, Egeblad M, Cancer Cell 21, 488 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Engblom C, Pfirschke C, Pittet MJ, Nat Rev Cancer 16, 447 (2016). [DOI] [PubMed] [Google Scholar]
- 98.Hong G, Diao S, Chang J, Antaris AL, Chen C, Zhang B, Zhao S, Atochin DN, Huang PL, Andreasson KI, Kuo CJ, Dai H, Nat Photonics 8, 723 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang LV, Hu S, Science 335, 1458 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yao J, Wang L, Yang JM, Maslov KI, Wong TT, Li L, Huang CH, Zou J, Wang LV, Nat Methods 12, 407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rowlands CJ, Park D, Bruns OT, Piatkevich KD, Fukumura D, Jain RK, Bawendi MG, Boyden ES, So PT, Light Sci Appl 6, e16255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hamel EJ, Grewe BF, Parker JG, Schnitzer MJ, Neuron 86, 140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim TH, Zhang Y, Lecoq J, Jung JC, Li J, Zeng H, Niell CM, Schnitzer MJ, Cell Rep 17, 3385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, Ferrero E, Agapow PM, Zietz M, Hoffman MM, Xie W, Rosen GL, Lengerich BJ, Israeli J, Lanchantin J, Woloszynek S, Carpenter AE, Shrikumar A, Xu J, Cofer EM, Lavender CA, Turaga SC, Alexandari AM, Lu Z, Harris DJ, DeCaprio D, Qi Y, Kundaje A, Peng Y, Wiley LK, Segler MHS, Boca SM, Swamidass SJ, Huang A, Gitter A, Greene CS, J R Soc Interface 15, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Economo MN, Clack NG, Lavis LD, Gerfen CR, Svoboda K, Myers EW, Chandrashekar J, Elife 5, e10566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tanaka N, Kanatani S, Tomer R, Sahlgren C, Kronqvist P, Kaczynska D, Louhivuori L, Kis L, Lindh C, Mitura P, Nature Biomedical Engineering 1, 796 (2017). [DOI] [PubMed] [Google Scholar]
- 107.Richardson DS, Lichtman JW, Cell 162, 246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cuccarese MF, Dubach JM, Pfirschke C, Engblom C, Garris C, Miller MA, Pittet MJ, Weissleder R, Nat Commun 8, 14293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li W, Germain RN, Gerner MY, Proc Natl Acad Sci U S A 114, E7321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ripoll J, Koberstein-Schwarz B, Ntziachristos V, Trends Biotechnol 33, 679 (2015). [DOI] [PubMed] [Google Scholar]
- 111.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA, J Exp Med 208, 1279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mouchacca P, Schmitt-Verhulst AM, Boyer C, PLoS One 8, e67239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, Guo L, Yagi R, Yamane H, Punkosdy G, Feigenbaum L, Zhao K, Paul WE, Immunity 37, 660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singbartl K, Thatte J, Smith ML, Wethmar K, Day K, Ley K, J Immunol 166, 7520 (2001). [DOI] [PubMed] [Google Scholar]
- 115.Croxford AL, Kurschus FC, Waisman A, J Immunol 182, 1237 (2009). [DOI] [PubMed] [Google Scholar]
- 116.Mohrs M, Shinkai K, Mohrs K, Locksley RM, Immunity 15, 303 (2001). [DOI] [PubMed] [Google Scholar]
- 117.Lüthje K, Kallies A, Shimohakamada Y, Belz GT, Light A, Tarlinton DM, Nutt SL, Nat Immunol 13, 491 (2012). [DOI] [PubMed] [Google Scholar]
- 118.Qi H, Egen JG, Huang AY, Germain RN, Science 312, 1672 (2006). [DOI] [PubMed] [Google Scholar]
- 119.Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, Okada T, Immunity 34, 961 (2011). [DOI] [PubMed] [Google Scholar]
- 120.Fooksman DR, Schwickert TA, Victora GD, Dustin ML, Nussenzweig MC, Skokos D, Immunity 33, 118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Crouch EE, Li Z, Takizawa M, Fichtner-Feigl S, Gourzi P, Montaño C, Feigenbaum L, Wilson P, Janz S, Papavasiliou FN, Casellas R, J Exp Med 204, 1145 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Engelhardt JJ, Boldajipour B, Beemiller P, Pandurangi P, Sorensen C, Werb Z, Egeblad M, Krummel MF, Cancer Cell 21, 402 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Satpathy AT, KC W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, Murphy TL, Murphy KM, J Exp Med 209, 1135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, Sarnacki S, Cumano A, Lauvau G, Geissmann F, Science 317, 666 (2007). [DOI] [PubMed] [Google Scholar]
- 125.Sauter KA, Pridans C, Sehgal A, Bain CC, Scott C, Moffat L, Rojo R, Stutchfield BM, Davies CL, Donaldson DS, Renault K, McColl BW, Mowat AM, Serrels A, Frame MC, Mabbott NA, Hume DA, PLoS One 9, e105429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gray EE, Friend S, Suzuki K, Phan TG, Cyster JG, PLoS One 7, e38258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cools-Lartigue J, Spicer J, McDonald B, Gowing S, Chow S, Giannias B, Bourdeau F, Kubes P, Ferri L, J Clin Invest (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jayagopal A, Russ PK, Haselton FR, Bioconjug Chem 18, 1424 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Darbousset R, Thomas GM, Mezouar S, Frère C, Bonier R, Mackman N, Renné T, Dignat-George F, Dubois C, Panicot-Dubois L, Blood 120, 2133 (2012). [DOI] [PubMed] [Google Scholar]