Abstract
Mutations of KRAS, NRAS, BRAF and DNA mismatch repair (MMR) status have become an important part of the assessment of patients with colorectal cancer (CRC), while respective clinicopathologic features and prognostic significance in specific stages and related detection strategies remain unclear. We retrospectively analyzed clinicopathologic features and prognosis of 1,834 patients with Stage I–IV colorectal adenocarcinoma. Mutations in KRAS, NRAS and BRAF and DNA MMR status were determined. The mutation rates of KRAS, NRAS and BRAF were 46.4, 3.2 and 3.5%, respectively, and the mismatch repair gene deletion (dMMR) rate was 5.6%. In a multivariate analysis, female, advanced age, tumor type histology, mucinous carcinoma and positive tumor deposits were associated with a high KRAS mutation rate. A high BRAF mutation rate was associated with female, poor differentiation, lymphovascular invasion and positive tumor deposits. Factors associated with high dMMR rates included low age, large tumor size, poor differentiation, Stages I–III. Tumor site was independently associated with KRAS mutation, BRAF mutation and dMMR. KRAS and BRAF mutations were independent risk factors for shorter overall survival (OS) in Stage IV tumors but not in Stage I–III tumors. NRAS mutation was an independent risk factor for shorter OS in Stage I–II tumors. dMMR was independently associated with longer OS in Stage III tumors.
Keywords: KRAS, NRAS, RAF, MMR, colorectal cancer
Short abstract
What's new?
Mutations in KRAS, NRAS, BRAF and DNA mismatch repair (MMR) status are important biomarkers in the assessment of patients with colorectal cancer (CRC). However, the clinicopathologic features associated with these mutations—and their impact on prognosis—are unclear, especially at earlier stages of CRC. In this large Chinese study, the authors analyzed variables such as gender, age, tumor histology, lymphovascular invasion, etc., that were associated with particular oncogene mutations and overall survival. These results should provide guidance for improved clinical strategies and enhance the usefulness of these biomarkers.
Introduction
KRAS, NRAS and BRAF and DNA mismatch repair (MMR) status have become important biomarkers to evaluate colorectal cancer (CRC). KRAS mutations are widely observed in patients with resistance to antiepidermal growth factor receptor (EGFR) therapy and associated with poor prognosis in advanced or recurrent CRC.1, 2, 3 NRAS mutations are rare and the clinicopathologic features, prognosis and treatment approaches for patients with NRAS mutations are unclear.4, 5 BRAF mutations are known as an indicator of poor prognosis and negative predictive biomarkers of anti‐EGFR therapy in advanced CRC.6, 7, 8, 9, 10
Detection strategies and clinical significances of these genes for tumors at specific stages remain unclear since most studies and guidelines focus on patients with recurrence or metastasis and typically detect one or two genes instead of including all the biomarkers above. Accordingly, the prognostic value of mutations at relatively early stages and utility of gene detection as a supplement to the TNM staging system are unclear.
We conducted a large retrospective study of cases with KRAS, NRAS, BRAF and MMR data at Fudan University Shanghai Cancer Center over the past 5 years to explore clinicopathologic features and prognosis. The results of our study can provide guidance for development of clinical strategies for gene detection.
Materials and Methods
Patients
A database of patients underwent surgical treatment at the Department of Colorectal Surgery at the Shanghai Cancer Center from January 2013 to June 2018 was retrospectively reviewed. Gene information was found in 2,340 patients and 506 of them were confirmed with incomplete information of gene detection or clinicopathologic features. In total, 1,834 patients were included in the analysis. The treatment plans were designed based on the updated Chinese Ministry of Health guidelines for diagnosis and treatment of CRC and international guidelines.
Our study was conducted according to the principles of the Declaration of Helsinki and was approved by the Research Ethics Committee of the Fudan University Shanghai Cancer Center in China. All patients provided written informed consent for the use of their cancer tissue blocks for molecular analyses.
Mutation screening
The Department of Pathology of Fudan University Shanghai Cancer Center performed mutation detection in all cases using surgical cancer tissues. Sequencing was performed in 1,374 cases. KRAS exons 2–4, NRAS exons 2–4 and BRAF exon 15 were evaluated by bidirectional sequence using ABI 3730XL and a BigDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA). Three independent experiments were performed to confirm the positive samples. DNA from the other 460 patients was tested using the AmoyDx KRAS/NRAS/BRAF Mutations Detection Kit (Amoy Diagnostics, Xiamen, China) under the principle of the amplification refractory mutation system (ARMS), covering the detection of KRAS mutations (exons 2–4), NRAS mutations (exons 2–4) and BRAF V600 mutations (exon 15). All results were confirmed according to the criterion suggested by the manufacturer.
Immunohistochemistry
Mismatch repair gene deletion (dMMR) was determined by the absence of protein expression for any one of several genes, including hMLH1, hMSH2, hMSH6 or hPMS2. Immunohistochemistry was performed using the fully automated BenchMark ULTRA platform (Ventana Medical Systems, Inc., Tucson, AZ). Normal tissues adjacent to the tumor or lymphocytes in the stroma served as internal positive controls. Each result was confirmed by at least two experienced pathologists.
Statistical analysis
All statistical analyses were performed by SPSS version 25.0 (IBM Corporation, Armonk, NY). Chi‐squared tests or Fisher's exact tests for categorical variables were used to compare the mutation status and clinical features. The Kolmogorov–Smirnov test was used to verify the normal distribution assumptions. The exploratory comparison of normally distributed and nonnormally distributed independent groups was performed using t‐tests and Mann–Whitney U tests (two groups). Overall survival (OS) was defined as the period of time between the first surgery and death from any cause. Analyses identifying prognostic predictors are performed using Cox proportional hazard models. Ten to fifteen predictors are necessary to proceed with multivariate survival analysis, whereby the selection for independent factors in the multivariate model was based on the univariate results. Log‐rank tests were employed to identify the associations between OS and predictors and all results are visualized by survival curves using the Kaplan–Meier method. A two‐sided p‐value <0.05 was considered statistically significant.
Results
Patients and mutations
Basic information for 1,834 patients is summarized in Table 1. One case of both KRAS and NRAS mutations, two cases of KRAS and BRAF mutations and three cases of NRAS and BRAF mutations were excluded from the prognostic analysis.
Table 1.
Clinical characteristics of 1,834 patients
| Variables | n (%) |
|---|---|
| Sex | |
| Male | 1,088 (59.3) |
| Female | 746 (40.7) |
| Age | 60.2 ± 11.9 |
| Tumor site | |
| Cecum | 43 (2.3) |
| Ascending colon | 277 (15.1) |
| Hepatic flexure | 70 (3.8) |
| Transverse colon | 76 (4.1) |
| Splenic flexure | 40 (2.2) |
| Descending flexure | 71 (3.9) |
| Sigmoid colon | 437 (23.8) |
| Rectum | 805 (43.9) |
| Multisite tumors | 15 (0.8) |
| Tumor size (cm) | 4.3 ± 1.9 |
| TNM stage | |
| I | 192 (10.5) |
| II | 502 (27.4) |
| III | 758 (41.3) |
| IV | 382 (20.8) |
| Histological | |
| Ulcer type | 1,219 (66.5) |
| Tumor type | 532 (29.0) |
| Invasive type | 83 (4.5) |
| Pathology | |
| Adenocarcinoma | 1,645 (89.7) |
| Mucinous carcinoma | 189 (10.3) |
| Differentiation | |
| G3–G4 | 557 (30.4) |
| G1–G2 | 1,277 (69.6) |
| Lymphovascular Invasion + | 698 (38.1) |
| Perineural Invasion + | 694 (37.8) |
| Extranodal tumor deposit + | 401 (21.9) |
| KRAS mutant | 851 (46.4) |
| NRAS mutant | 58 (3.2) |
| BRAF mutant | 65 (3.5) |
| dMMR | 102 (5.6) |
Clinicopathologic features
Univariate analyses of clinicopathologic features according to mutations in KRAS, NRAS and BRAF and DNA MMR status are listed in Table 2.
Table 2.
Univariate analysis of clinicopathologic features
| Variables | KRAS | NRAS | BRAF | MMR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild‐type | Mutant | Wild‐type | Mutant | Wild‐type | Mutant | pMMR | dMMR | p‐value | ||||
| n = 983 | n = 851 | p‐value | n = 1,776 | n = 58 | p‐value | n = 1,769 | n = 65 | p‐value | n = 1,732 | n = 102 | ||
| Sex | 0.008 | 0.70 | 0.001 | 0.47 | ||||||||
| Male | 611 (56.2) | 477 (43.8) | 1,055 (97.0) | 33 (3.0) | 1,062 (97.6) | 26 (2.4) | 1,024 (94.1) | 64 (5.9) | ||||
| Female | 372 (49.9) | 374 (50.1) | 721 (96.7) | 25 (3.3) | 707 (94.8) | 39 (5.2) | 708 (95.0) | 38 (5.0) | ||||
| Age | 59.5 ± 11.9 | 60.8 ± 11.9 | 0.023 | 60.1 ± 12.0 | 61.8 ± 9.4 | 0.17 | 60.1 ± 11.8 | 60.0 ± 13.9 | 0.96 | 60 ± 11.7 | 55.2 ± 14.1 | 0.001 |
| Tumor site1 | 0.061 | 0.170 | 0.001 | 0.001 | ||||||||
| Cecum | 15 (34.9) | 28 (65.1) | 41 (95.3) | 2 (4.7) | 41 (95.3) | 2 (4.7) | 39 (90.7) | 4 (9.3) | ||||
| Ascending colon | 115 (41.5) | 162 (58.5) | 269 (97.1) | 8 (2.9) | 256 (92.4) | 21 (7.6) | 242 (87.4) | 35 (12.6) | ||||
| Hepatic flexure | 34 (48.6) | 36 (51.4) | 69 (98.6) | 1 (1.4) | 65 (92.9) | 5 (7.1) | 56 (80.0) | 14 (20.0) | ||||
| Transverse colon | 43 (56.6) | 33 (43.4) | 75 (98.7) | 1 (1.3) | 73 (96.1) | 3 (3.9) | 63 (82.9) | 13 (17.1) | ||||
| Splenic flexure | 22 (55.0) | 18 (45.0) | 40 (100) | 0 (0) | 38 (95.0) | 2 (5.0) | 35 (87.5) | 5 (12.5) | ||||
| Descending flexure | 46 (64.8) | 25 (35.2) | 70 (98.6) | 1 (1.4) | 67 (94.4) | 4 (5.6) | 63 (88.7) | 8 (11.3) | ||||
| Sigmoid colon | 279 (63.8) | 158 (36.2) | 423 (96.8) | 14 (3.2) | 427 (97.7) | 10 (2.3) | 432 (98.9) | 5 (1.1) | ||||
| Rectum | 423 (52.5) | 382 (47.5) | 774 (96.1) | 31 (3.9) | 787 (97.8) | 18 (2.2) | 790 (98.1) | 15 (1.9) | ||||
| Tumor size (cm) | 4.2 ± 1.9 | 4.4 ± 2.0 | 0.026 | 4.3 ± 2.0 | 3.8 ± 1.9 | 0.049 | 4.3 ± 1.9 | 4.6 ± 1.9 | 0.26 | 4.2 ± 1.9 | 5.9 ± 3.0 | 0.001 |
| T stage | 0.56 | 0.76 | 0.80 | 0.82 | ||||||||
| T1 | 32 (57.1) | 24 (42.9) | 53 (94.7) | 3 (5.3) | 54 (96.5) | 2 (3.5) | 54 (96.5) | 2 (3.5) | ||||
| T2 | 135 (57.2) | 101 (42.8) | 230 (97.5) | 6 (2.5) | 228 (96.6) | 8 (3.4) | 223 (94.5) | 13 (5.5) | ||||
| T3 | 550 (52.5) | 498 (47.5) | 1,015 (96.9) | 33 (3.1) | 1,014 (96.7) | 34 (3.3) | 986 (94.1) | 62 (5.9) | ||||
| T4 | 266 (53.8) | 228 (46.2) | 478 (96.7) | 16 (3.3) | 473 (95.7) | 21 (4.3) | 469 (94.9) | 25 (5.1) | ||||
| N stage | 0.15 | 0.18 | 0.005 | 0.001 | ||||||||
| N0 | 421 (55.6) | 336 (44.4) | 738 (97.5) | 19 (2.5) | 741 (97.9) | 16 (2.1) | 693 (91.6) | 64 (8.4) | ||||
| N+ | 562 (52.2) | 515 (47.8) | 1,038 (96.4) | 39 (3.6) | 1,028 (95.4) | 49 (4.6) | 1,039 (96.5) | 38 (3.5) | ||||
| M stage | 0.22 | 0.76 | 0.044 | 0.002 | ||||||||
| M0 | 789 (54.3) | 663 (45.7) | 1,407 (96.9) | 45 (3.1) | 1,407 (96.8) | 45 (3.2) | 1,359 (93.6) | 93 (6.4) | ||||
| M1 | 194 (50.8) | 188 (49.2) | 369 (96.7) | 13 (3.3) | 362 (94.9) | 20 (5.1) | 373 (97.5) | 9 (2.5) | ||||
| TNM stage | 0.27 | 0.35 | 0.014 | 0.001 | ||||||||
| I | 115 (59.9) | 77 (40.1) | 184 (95.9) | 8 (4.1) | 184 (95.9) | 8 (4.1) | 181 (94.3) | 11 (5.7) | ||||
| II | 270 (53.7) | 232 (46.3) | 492 (98.0) | 10 (2.0 | 495 (98.6) | 7 (1.4) | 452 (90.1) | 50 (9.9) | ||||
| III | 404 (53.3) | 354 (46.7) | 731 (96.4) | 27 (3.6) | 728 (96.0) | 30 (4.0) | 726 (95.8) | 32 (4.2) | ||||
| IV | 194 (50.8) | 188 (49.2) | 369 (96.7) | 13 (3.3) | 362 (94.9) | 20 (5.1) | 373 (97.5) | 9 (2.5) | ||||
| Histological | 983 | 851 | 0.001 | 0.21 | 0.52 | 0.15 | ||||||
| Ulcer type | 691 (56.7) | 528 (43.3) | 1,177 (96.5) | 42 (3.5) | 1,176 (96.5) | 43 (3.5) | 1,155 (94.7) | 64 (5.3) | ||||
| Tumor type | 244 (45.9) | 288 (54.1) | 518 (97.4) | 14 (2.6) | 514 (96.6) | 18 (3.4) | 496 (93.2) | 36 (6.8) | ||||
| Invasive type | 48 (57.8) | 35 (42.5) | 81 (97.6) | 2 (2.4) | 79 (95.2) | 4 (4.8) | 81 (97.6) | 2 (2.4) | ||||
| Pathology | 0.001 | 0.68 | 0.17 | 0.001 | ||||||||
| Adenocarcinoma | 898 (54.6) | 747 (45.4) | 1,592 (97.1) | 53 (2.9) | 1,590 (96.7) | 55 (3.3) | 1,569 (95.4) | 76 (4.6) | ||||
| Mucinous carcinoma | 85 (45.2) | 104 (54.8) | 184 (97.4) | 5 (2.6) | 179 (94.7) | 10 (5.3) | 163 (86.2) | 26 (13.8) | ||||
| Differentiation | 0.17 | 0.70 | 0.001 | 0.001 | ||||||||
| G3–G4 | 291 (52.2) | 266 (47.8) | 541 (97.1) | 16 (2.9) | 518 (93.0) | 39 (7.0) | 502 (90.1) | 55 (9.9) | ||||
| G1–G2 | 692 (54.2) | 585 (45.8) | 1,235 (96.7) | 42 (3.3) | 1,251 (98.0) | 26 (2.0) | 1,230 (96.3) | 47 (3.7) | ||||
| Lymphovascular Invasion | 0.77 | 0.58 | 0.001 | 0.17 | ||||||||
| Negative | 615 (54.1) | 521 (45.9) | 1,101 (97.0) | 35 (3.0) | 1,108 (97.5) | 28 (2.5) | 1,064 (93.7) | 72 (6.3) | ||||
| Positive | 368 (52.7) | 330 (47.3) | 675 (96.7) | 23 (3.3) | 661 (94.7) | 37 (5.3) | 668 (95.7) | 30 (4.3) | ||||
| Perineural Invasion | 0.41 | 0.40 | 0.07 | 0.002 | ||||||||
| Negative | 618 (54.2) | 522 (45.8) | 1,107 (97.1) | 33 (2.9) | 1,106 (97.0) | 34 (3.0) | 1,062 (93.2) | 78 (6.8) | ||||
| Positive | 365 (52.6) | 329 (47.4) | 669 (96.3) | 25 (3.7) | 663 (95.5) | 31 (4.5) | 670 (96.5) | 24 (3.5) | ||||
| Extranodal tumor deposit | 0.055 | 0.52 | 0.001 | 0.002 | ||||||||
| Negative | 785 (54.8) | 648 (45.2) | 1,390 (97.0) | 43 (3.0) | 1,394 (97.3) | 39 (2.7) | 1,341 (93.6) | 92 (6.4) | ||||
| Positive | 198 (49.3) | 203 (50.7) | 386 (96.3) | 15 (3.7) | 375 (93.5) | 26 (6.5) | 391 (97.5) | 10 (2.5) | ||||
Another 15 patients with multisite tumors were excluded.
Results of the multivariate analysis are summarized in Table 3. Only tumor size was associated with NRAS mutations in the univariate analysis. Therefore, NRAS mutations were excluded from the multivariate analysis. KRAS mutation rate was high for the following factors: female, advanced age, tumor type histology, mucinous carcinoma and positive tumor deposits. BRAF showed a high mutation rate in female, poor differentiation, lymphovascular invasion and positive tumor deposits. A high rate of dMMR was associated with low age, large tumor size, poor differentiation and Stages I–III. Tumor site was independently associated with KRAS mutation, BRAF mutation and dMMR.
Table 3.
Multivariate analysis of clinicopathologic features
| Variables | KRAS mutant | BRAF mutant | dMMR | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | p‐value | OR | 95%CI | p‐value | OR | 95%CI | p‐value | |
| Sex | |||||||||
| Female | 1 | Ref | 1 | Ref | |||||
| Male | 0.81 | 0.66–0.99 | 0.045 | 0.57 | 0.34–0.97 | 0.039 | |||
| Age | 1.01 | 1.01–1.02 | 0.005 | 0.97 | 0.95–0.98 | 0.001 | |||
| Tumor site | 0.92 | 0.88–0.96 | 0.001 | 0.81 | 0.73–0.90 | 0.001 | 0.71 | 0.64–0.78 | 0.001 |
| Tumor size | 1.29 | 1.17–1.42 | 0.001 | ||||||
| Histology | |||||||||
| Ulcer type | 1 | Ref | |||||||
| Tumor type | 1.63 | 1.31–2.04 | 0.001 | ||||||
| Invasive type | 0.91 | 0.53–1.56 | 0.726 | ||||||
| Pathology | |||||||||
| Adenocarcinoma | 0.66 | 0.47–0.94 | 0.021 | ||||||
| Mucinous carcinoma | 1 | Ref | |||||||
| Differentiation | |||||||||
| G3–G4 | 2.33 | 1.31–4.14 | 0.004 | 2.74 | 1.66–4.51 | 0.001 | |||
| G1–G2 | 1 | Ref | 1 | Ref | |||||
| Lymphovascular Invasion | 1.86 | 1.02–3.50 | 0.043 | ||||||
| Perineural Invasion | |||||||||
| Extranodal tumor deposit | 1.39 | 1.10–1.76 | 0.008 | 2.28 | 1.29–4.05 | 0.005 | |||
| TNM Stage | |||||||||
| I | 13.71 | 4.64–40.45 | 0.001 | ||||||
| II | 8.55 | 3.58–20.47 | 0.001 | ||||||
| III | 2.92 | 1.22–6.98 | 0.016 | ||||||
| IV | 1 | Ref | |||||||
Survival analysis
In a univariate analysis of Stage I–IV tumors, KRAS‐mutated tumors and BRAF‐mutated tumors were associated with a shorter OS compared to that of all‐wild‐type tumors. There was no significant difference in OS between NRAS‐mutated tumors and all‐wild‐type tumors (Fig. 1). Table 4 presents the results obtained from the Cox analysis of prognostic actors.
Figure 1.
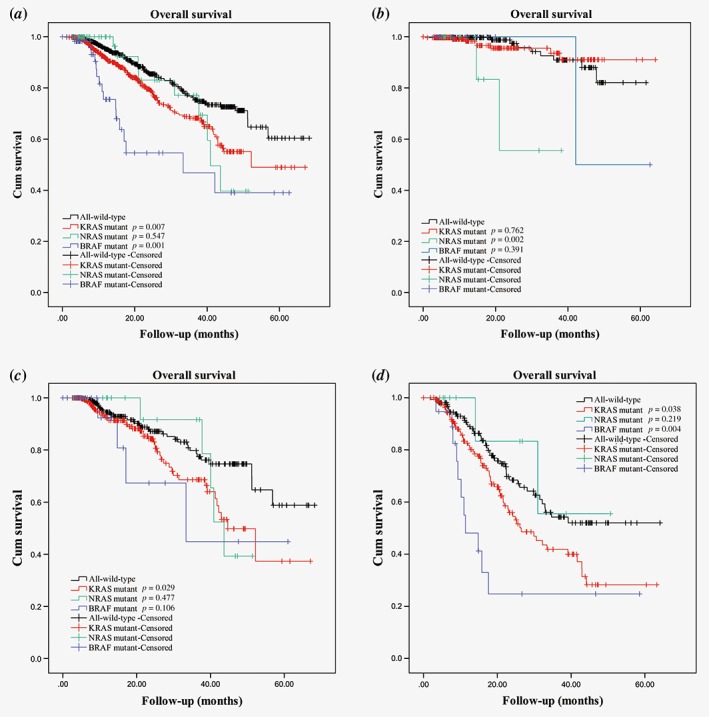
Kaplan–Meier analysis of OS at (a) Stages I–IV, (b) Stages I–II, (c) Stage III and (d) Stage IV. Each p‐value reflects its respective mutation compared with all‐wild‐type.
Table 4.
Cox analysis of prognostic actors for OS in patients from Stage I to Stage IV
| Prognosis variables | Stages I–II | Stage III | Stage IV | |||
|---|---|---|---|---|---|---|
| p‐value | HR (95% CI) | p‐value | HR (95% CI) | p‐value | HR (95% CI) | |
| Sex (female) | 0.088 | 0.26 (0.06–1.22) | 0.20 | 0.74 (0.46–1.18) | 0.43 | 1.18 (0.79–1.76) |
| Age | 0.41 | 1.02 (0.97–1.07) | 0.86 | 1.00 (0.98–1.02) | 0.96 | 1.00 (0.98–1.02) |
| Tumor site | 0.79 | 0.97 (0.77–1.21) | 0.71 | 1.02 (0.92–1.13) | 0.013 | 0.91 (0.84–0.98) |
| Tumor size | 0.001 | 1.44 (1.21–1.73) | 0.001 | 1.40 (1.24–1.58) | 0.047 | 1.10 (1.01–1.21) |
| Histology | ||||||
| Tumor type | 0.98 | 1.02 (0.33–3.12) | 0.35 | 1.28 (0.76–2.15) | 0.94 | 0.94 (0.58–1.53) |
| Invasive | – | – | 0.90 | 0.94 (0.37–2.42) | 0.32 | 1.44 (0.70–2.99) |
| Pathology (Mucinous) | 0.83 | 0.75 (0.058–9.64) | 0.35 | 1.33 (0.74–2.42) | 0.48 | 0.79 (0.42–1.50) |
| Differentiation (G1–G2) | 0.98 | 1.02 (0.26–4.00) | 0.001 | 0.30 (0.19–0.49) | 0.007 | 0.59 (0.40–0.86) |
| Lymphovascular Invasion | 0.04 | 3.80 (1.06–13.54) | 0.18 | 1.43 (0.85–2.39) | 0.29 | 0.79 (0.51–1.22) |
| Perineural Invasion | 0.001 | 5.56 (2.01–15.41) | 0.008 | 1.88 (1.18–2.99) | 0.017 | 1.65 (1.09–2.49) |
| Extranodal tumor deposit | – | – | 0.006 | 1.89 (1.20–2.97) | 0.14 | 1.36 (0.91–2.04) |
| KRAS mutant | 0.76 | 1.20 (0.37–3.91) | 0.13 | 1.47 (0.89–2.42) | 0.022 | 1.60 (1.07–2.40) |
| NRAS mutant | 0.025 | 6.13 (1.25–30.01) | 0.071 | 2.29 (0.93–5.66) | 0.25 | 0.42 (0.10–1.81) |
| BRAF mutant | – | – | 0.29 | 1.78 (0.61–5.22) | 0.003 | 2.84 (1.43–5.67) |
| dMMR | 0.20 | 2.73 (0.60–12.47) | 0.008 | 0.12 (0.25–0.58) | 0.67 | 0.72 (0.16–3.26) |
Bold values indicate p‐values less than 0.05.
No differences in OS between KRAS‐mutated tumors and all‐wild‐type tumors of Stages I–II were detected in both the univariate analysis and multivariate analysis (Fig. 1 and Table 4). In Stage III, KRAS‐mutated tumors were associated with a shorter OS than that of all‐wild‐type tumors in a univariate analysis but not in a multivariate analysis. In Stage IV, we observed a significant difference in OS between KRAS‐mutated tumors and all‐wild‐type tumors in both a univariate analysis and multivariate analysis.
In Stages I–II, OS was shorter for NRAS‐mutated tumors than for all‐wild‐type tumors in both the univariate analysis and multivariate analysis. No similar difference was observed in Stage III tumors in the univariate analysis or multivariate analysis or in Stage IV tumors in the univariate analysis or multivariate analysis.
No statistically significant difference in OS between patients with BRAF‐mutated tumors and all‐wild‐type tumors was observed for Stages I–II and Stage III in the univariate analysis. For Stage IV, the OS of patients with BRAF‐mutated tumors was shorter than that of patients with all‐wild‐type tumors in both univariate analysis and multivariate analysis.
In the Cox analysis, dMMR was independently associated with longer OS in Stage III but not Stage I–II or Stage IV tumors.
Discussion
We retrospectively analyzed mutations in KRAS, NRAS and BRAF and DNA MMR status of 1,834 patients with colorectal adenocarcinoma during the past 5 years at our institution. Comprehensive information, including clinicopathologic features and prognosis, was gathered to explore the necessity and optimization of gene detection for tumors with different clinicopathologic features and stages.
Relevant studies of relatively large populations are summarized in Table 5.5, 7, 8, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 The KRAS mutation rate in our study was similar to those in studies of Eastern and Western populations. As for NRAS, a low mutation rate was reported in most studies and only Stage IV cases are included. Interestingly, the BRAF mutation rate was lower in our study and other studies of Asian populations than in studies of Western populations. Turning now to DNA MMR status, immunohistochemical analyses and DNA sequencing are not 100% accurate for identifying dMMR/MSI. However, these two methods are highly consistent in results, that is, near 95%.23 It is difficult to definitively determine the difference in dMMR rate between Eastern and Western populations based on available studies.
Table 5.
Clinical studies reporting rates of KRAS, NRAS, and BRAF mutations and dMMR/MSI
| Year | Author | Journal | Number of centers | Area | Number of patients | Stage | KRAS mutant rate | NRAS mutant rate | BRAF mutant rate | dMMR/MSI rate |
|---|---|---|---|---|---|---|---|---|---|---|
| 2005 | Benatti et al. | Clin Cancer Res | 3 | Italy | 1,263 | I–IV | – | – | – | 20.3% |
| 2007 | Koopman et al. | Br J Cancer | Many (74) | The Netherlands | 515 | IV | – | – | – | 3.5% |
| 2009 | Des Guetz et al. | Eur J Cancer | Many | Many | 3,690 | II–III | – | – | – | 13.7% |
| 2010 | De Roock et al. | Lancet Oncol | 11 | Europe | 773 | IV | 40.0% | 2.6% | 4.7% | – |
| 2011 | Sinicrope et al. | J Natl Cancer Inst | Many | Europe and America | 2,141 | II–III | – | – | – | 16.1% |
| 2012 | Imamura et al. | Clin Cancer Res | Many | U.S | 1,261 | I–IV | 35.3% | – | 14.4% | – |
| 2013 | Douillard et al. | N Engl J Med | Many | Europe and America | 1,096 | IV | 45.6% | 7.5% | 8.3% | – |
| 2014 | Imamura et al. | Mol Cancer | Many | U.S | 1,267 | I–IV | 40.0% | 14.5% | – | |
| 2014 | Schirripa et al. | Int J Cancer | 1 | Italy | 786 | IV | 50.0% | 6.0% | 9.2% | – |
| 2016 | Summers et al. | Clin Cancer Res | Many | UK and Ireland | 2,157 | IV | 39.8% | 4.0% | 9.5% | 4.6% |
| 2011 | Yokota et al. | Br J Cancer | 1 | Japan | 229 | IV | 34.5% | ‐ | 6.5% | – |
| 2014 | Tong et al. | Cancer Biol Ther | 1 | HK, China | 1,506 | – | 44.5% | – | – | – |
| 2015 | Zhang et al. | Sci Rep | 3 | China | 1,110 | I‐IV | 45.4% | 3.9% | 3.1% | – |
| 2015 | Kawazoe et al. | BMC Cancer | 1 | Japan | 264 | IV | 37.9% | 4.2% | 5.4% | – |
| 2015 | Yan et al. | World J Gastroenterol | 1 | China | 538 | I–IV | 37.9% | – | 4.4% | 11.4% |
| Current study | Guo et al. | Int J Cancer | 1 | China | 1,854 | I–IV | 46.4% | 3.2% | 3.5% | 5.6% |
Clinicopathologic factors related to a high KRAS mutation rate in our study were female, advanced age, tumor type histology, mucinous carcinoma and positive tumor deposits. However, Imamura et al. reported different results in a study of 1,267 patients. In their study, KRAS mutations were associated with male, well‐moderate differentiation, absent‐minimal peritumoral lymphocytic reaction.16 Further studies are expected to explorer clinicopathologic features associated with KRAS mutations. No significant differences in clinicopathologic features were observed between NRAS‐mutated tumors and all‐wild‐type tumors in our study or other studies. A high BRAF mutation rate was associated with female, poor differentiation, lymphovascular invasion and positive tumor deposits in our study. Similar results were found in previous studies.6, 20 This is the first study to report that positive tumor deposits are independently related to BRAF mutations. Positive tumor deposits are associated with poor prognosis and have become a reference factor for TNM staging.24, 25 Our findings suggest that a high BRAF mutation rate is the main predictor of a poor prognosis in patients with positive tumor deposits. dMMR was relatively common in large, poor differentiated and Stage I–II tumors, as reported in previous studies.11, 22 However, we observed that the age of high incidence of dMMR was different in different population studies. Our results are consistent with other studies of Chinese populations, with a high dMMR rate in young individuals.26, 27 In studies of western populations, high dMMR rates appear to be associated with older age.28, 29, 30
In general, KRAS mutation rate decreased from right colon to left colon, but increased slightly in rectum. BRAF mutation rate was higher in right colon than left colon and lowest in rectum. The rate of dMMR increased gradually from cecum to hepatic flexure and then decreased from hepatic flexure to rectum. Yamauchi et al. reported that the rate of KRAS mutations was highest in cecal tumors and gradually decrease from cecal to transverse colon, but no obvious pattern of KRAS mutation rate was found from splenic flexure to rectum in their study.31 Rosty et al. and Imamura et al. confirmed the highest KRAS mutation rate in cecal tumors.16, 32 Different site distribution of KRAS mutations might be caused by the fact that only KRAS exon 2 (codons 12 and 13) was sequenced in these previous studies. Yamauchi et al. also reported that the rates of MSI‐high and BRAF mutations gradually increased from the rectum to ascending colon, followed by falls in the cecum, which was similar to the trends in our results.31
Considering that mutations in KRAS, NRAS and BRAF and DNA MMR statuses are not all routinely tested in many clinical institutions, we suggest a gene detection strategy to be developed in future studies based on the clinicopathologic differences described above.
Poor prognosis and resistance to anti‐EGFR targeted therapy of KRAS mutations are defined in Stage IV patients.33, 34 However, in Stage I–III patients, the prognostic value of KRAS is controversial. Ogino et al. reported that the KRAS mutational status is not associated with DFS or OS in a study of 508 patients with Stage III CRC.35 Similarly, Roth et al. reported that KRAS mutations do not have major prognostic value based on a study of 1,404 patients with Stage II–III CRC.36 However, Hutchins et al. found that the risk of recurrence is significantly higher for KRAS mutants than wild‐type KRAS in the QUASAR study, which included 1,708 Stage II cases and 163 Stage III cases.37 Taieb et al. evaluated 4,411 patients with Stage III colorectal and found that BRAF or KRAS mutations were independently associated with a shorter time to recurrence, survival after recurrence and OS in patients with MSS, but not in MSI tumors.38 In the univariate analysis, prognosis was worse in KRAS mutants than all‐wild‐type cases and this could be explained by the increase in the rate of KRAS mutations as the tumor stage increased in our study (Table 2).
Poor prognosis for BRAF mutation has been widely reported in Stage IV cases.10 Similar results can be found in a few studies of Stage II or III patients.38, 39 Our statistical analyses showed that BRAF mutation is an independent risk factor for shorter OS only in Stage IV tumors but not Stage I–III tumors. The contradiction might be explained by the low incidence of BRAF mutation in the Asian population. Therefore, far fewer patients with Stage II–III BRAF‐mutated cancer were included in our study than in previous studies, resulting in statistically insignificant results.
Unlike KRAS and BRAF mutations, NRAS mutation is an independent risk factor for shorter OS in Stages I–II but not in Stage III or IV. A few studies have reported a poor prognosis associated with NRAS mutations.8, 40, 41 However, very few studies reported the prognostic value of NRAS mutations at specific stages. In a retrospective study of patients with Stage IV CRC, Schirripa et al. reported that NRAS mutations are associated with a shorter OS than all‐wild‐type cases and more patients with NRAS mutations at Stage IV were included in their study than in ours (47 vs. 13).5 Further studies of NRAS mutations are needed.
dMMR was an independent prognostic factor for a favorable prognosis for patients with Stage III cancer in our study. Similar conclusions are reached in a number of studies under certain conditions. Sinicrope et al. reported that dMMR is significantly associated with better survival after recurrence in patients with Stage III proximal colon cancers.42 In a study of 1,254 patients with Stage II–IV cancer, Klingbiel et al.43 reported that MSI‐H is associated with both longer relapse‐free survival and OS in Stage II patients and relapse‐free survival in Stage III patients. Differences in conclusions could be explained by different sample size and multivariate analysis methods.
In conclusion, mutations in KRAS, NRAS and BRAF and dMMR were associated with different clinicopathologic features. KRAS and BRAF mutations were independent risk factors for shorter OS in Stage IV tumors. NRAS mutations were an independent risk factor for shorter OS in Stage I–II tumors. dMMR was an independent protective factor for longer OS in Stage III tumors. The clinicopathologic features and prognostic values of these markers require further validation, especially in early‐stage patients.
Conflicts of interest: The authors have declared no conflicts of interest.
Data Availability Statement: The data that support the findings of our study are available from the corresponding author upon reasonable request.
Contributor Information
Fang‐Qi Liu, Email: liufq021@163.com.
Ye Xu, Email: xuye021@163.com.
Data availability statement
The data that support the findings of our study are available from the corresponding author upon reasonable request.
References
- 1. Lièvre A, Bachet J, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 2008;26:374–9. [DOI] [PubMed] [Google Scholar]
- 2. Amado RG, Wolf M, Peeters M, et al. Wild‐type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626–34. [DOI] [PubMed] [Google Scholar]
- 3. Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 2009;27:5931–7. [DOI] [PubMed] [Google Scholar]
- 4. Normanno N, Rachiglio AM, Lambiase M, et al. Heterogeneity of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer and potential effects on therapy in the CAPRI GOIM trial. Ann Oncol 2015;26:1710–4. [DOI] [PubMed] [Google Scholar]
- 5. Schirripa M, Cremolini C, Loupakis F, et al. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int J Cancer 2015;136:83–90. [DOI] [PubMed] [Google Scholar]
- 6. Fariña‐Sarasqueta A, van Lijnschoten G, Moerland E, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol 2010;21:2396–402. [DOI] [PubMed] [Google Scholar]
- 7. Yokota T, Ura T, Shibata N, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer 2011;104:856–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Douillard J, Oliner KS, Siena S, et al. Panitumumab‐FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023–34. [DOI] [PubMed] [Google Scholar]
- 9. Zlobec I, Bihl MP, Schwarb H, et al. Clinicopathological and protein characterization of BRAF‐ and K‐RAS‐mutated colorectal cancer and implications for prognosis. Int J Cancer 2010;127:367–80. [DOI] [PubMed] [Google Scholar]
- 10. Pietrantonio F, Petrelli F, Coinu A, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta‐analysis. Eur J Cancer 2015;51:587–94. [DOI] [PubMed] [Google Scholar]
- 11. Benatti P, Gafà R, Barana D, et al. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res 2005;11:8332–40. [DOI] [PubMed] [Google Scholar]
- 12. Des Guetz G, Schischmanoff O, Nicolas P, et al. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta‐analysis. Eur J Cancer 2009;45:1890–6. [DOI] [PubMed] [Google Scholar]
- 13. De Roock W, De Vriendt V, Normanno N, et al. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol 2011;12:594–603. [DOI] [PubMed] [Google Scholar]
- 14. Sinicrope FA, Foster NR, Thibodeau SN, et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5‐fluorouracil‐based adjuvant therapy. J Natl Cancer Inst 2011;103:863–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Imamura Y, Morikawa T, Liao X, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild‐type colorectal cancers. Clin Cancer Res 2012;18:4753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imamura Y, Lochhead P, Yamauchi M, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer 2014;13:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Summers MG, Smith CG, Maughan TS, et al. BRAF and NRAS locus‐specific variants have different outcomes on survival to colorectal cancer. Clin Cancer Res 2017;23:2742–9. [DOI] [PubMed] [Google Scholar]
- 18. Tong JH, Lung RW, Sin FM, et al. Characterization of rare transforming KRAS mutations in sporadic colorectal cancer. Cancer Biol Ther 2014;15:768–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang J, Zheng J, Yang Y, et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Sci Rep 2016;5:18678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kawazoe A, Shitara K, Fukuoka S, et al. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer 2015;15:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan W, Hu J, Xie L, et al. Prediction of biological behavior and prognosis of colorectal cancer patients by tumor MSI/MMR in the Chinese population. OncoTargets Ther 2016;9:7415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koopman M, Kortman GA, Mekenkamp L, et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer 2009;100:266–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takehara Y, Nagasaka T, Nyuya A, et al. Accuracy of four mononucleotide‐repeat markers for the identification of DNA mismatch‐repair deficiency in solid tumors. J Transl Med 2018;16:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tong L, Gao P, Wang Z, et al. Is the seventh edition of the UICC/AJCC TNM staging system reasonable for patients with tumor deposits in colorectal cancer? Ann Surg 2012;255:208–13. [DOI] [PubMed] [Google Scholar]
- 25. Nagtegaal ID, Tot T, Jayne DG, et al. Lymph nodes, tumor deposits, and TNM: are we getting better? J Clin Oncol 2011;29:2487–92. [DOI] [PubMed] [Google Scholar]
- 26. Zhang X, Ran W, Wu J, et al. Deficient mismatch repair and RAS mutation in colorectal carcinoma patients: a retrospective study in eastern China. PeerJ 2018;6:e4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li P, Xiao ZT, Braciak TA, et al. Impact of age and mismatch repair status on survival in colorectal cancer. Cancer Med 2017;6:975–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tanaka M, Nakajima T, Sugano K, et al. Mismatch repair deficiency in lynch syndrome‐associated colorectal adenomas is more prevalent in older patients. Histopathology 2016;69:322–8. [DOI] [PubMed] [Google Scholar]
- 29. Aparicio T, Schischmanoff O, Poupardin C, et al. High prevalence of deficient mismatch repair phenotype and the V600E BRAF mutation in elderly patients with colorectal cancer. J Geriatr Oncol 2014;5:384–8. [DOI] [PubMed] [Google Scholar]
- 30. Zaanan A, Shi Q, Taieb J, et al. Role of deficient DNA mismatch repair status in patients with stage III colon cancer treated with FOLFOX adjuvant chemotherapy: a pooled analysis from 2 randomized clinical trials. JAMA Oncol 2018;4:379–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 2012;61:847–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosty C, Young JP, Walsh MD, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol 2013;26:825–34. [DOI] [PubMed] [Google Scholar]
- 33. Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer 2013;119:4137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bokemeyer C, Cutsem EV, Rougier P, et al. Addition of cetuximab to chemotherapy as first‐line treatment for KRAS wild‐type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer 2012;48:1466–75. [DOI] [PubMed] [Google Scholar]
- 35. Ogino S, Meyerhardt JA, Irahara N, et al. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res 2009;15:7322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC‐3, EORTC 40993, SAKK 60‐00 trial. J Clin Oncol 2010;28:466–74. [DOI] [PubMed] [Google Scholar]
- 37. Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol 2011;29:1261–70. [DOI] [PubMed] [Google Scholar]
- 38. Taieb J, Le Malicot K, Shi Q, et al. Prognostic value of BRAF and KRAS mutations in MSI and MSS stage III colon cancer. J Natl Cancer Inst 2016;109:djw272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gavin PG, Colangelo LH, Fumagalli D, et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clin Cancer Res 2012;18:6531–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy‐refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753–62. [DOI] [PubMed] [Google Scholar]
- 41. Peeters M, Oliner KS, Parker A, et al. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clin Cancer Res 2013;19:1902–12. [DOI] [PubMed] [Google Scholar]
- 42. Sinicrope FA, Shi Q, Allegra CJ, et al. Association of DNA mismatch repair and mutations in BRAF and KRAS with survival after recurrence in stage III colon cancers from phase III adjuvant chemotherapy trials. JAMA Oncol 2017;3:472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klingbiel D, Saridaki Z, Roth AD, et al. Prognosis of stage II and III colon cancer treated with adjuvant 5‐fluorouracil or FOLFIRI in relation to microsatellite status: results of the PETACC‐3 trial. Ann Oncol 2015;26:126–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of our study are available from the corresponding author upon reasonable request.


