Abstract
Acquisition of effector functions in T cells is guided by transcription factors, including NF‐κB, that itself is tightly controlled by inhibitory proteins. The atypical NF‐κB inhibitor, IκBNS, is involved in the development of Th1, Th17, and regulatory T (Treg) cells. However, it remained unclear to which extend IκBNS contributed to the acquisition of effector function in T cells specifically responding to a pathogen during in vivo infection. Tracking of adoptively transferred T cells in Listeria monocytogenes infected mice antigen‐specific activation of CD4+ T cells following in vivo pathogen encounter to strongly rely on IκBNS. While IκBNS was largely dispensable for the acquisition of cytotoxic effector function in CD8+ T cells, IκBNS‐deficient Th1 effector cells exhibited significantly reduced proliferation, marked changes in the pattern of activation marker expression, and reduced production of the Th1‐cell cytokines IFN‐γ, IL‐2, and TNF‐α. Complementary in vitro analyses using cells from novel reporter and inducible knockout mice revealed that IκBNS predominantly affects the early phase of Th1‐cell differentiation while its function in terminally differentiated cells appears to be negligible. Our data suggest IκBNS as a potential target to modulate specifically CD4+ T‐cell responses.
Keywords: CD4+ T cells, IκBNS, In vivo infection, Listeria monocytogenes, Th1 cell differentiation
Introduction
The development and function of immune cells are regulated by a variety of transcription factors including NF‐κB. NF‐κB acts as a molecular switch that regulates many immunological processes including cell proliferation, activation of immune cells, and regulation of inflammation 1. The activation of NF‐κB is controlled by inhibitory proteins, such as the classical NF‐κB inhibitor IκBα, which retains NF‐κB in the cytoplasm by masking the nuclear‐localization sequence 1. Next to the cytoplasmic inhibitory proteins, a second class of atypical NF‐κB inhibitors comprising Bcl3, IκBζ, IκBη, and IκBNS exists in the nucleus, which either have activating or suppressing functions on NF‐κB‐mediated gene expression 2. The NF‐κB family of transcription factors contributes to Th1 differentiation. For instance, mice expressing a nondegradable IκBα mutant exhibit reduced IFN‐ɣ production and Th1 responses 3, 4. With respect to atypical NF‐κB inhibitors, Bcl‐3‐deficient mice exhibit impaired Th1 responses toward intracellular pathogens 5, 6. In contrast, T‐cell‐specific deletion of IκBζ results in increased IFN‐ɣ expression 7. IκBζ counteracts RelA/p65 activity at the Ifng locus and TGF‐β‐induced IκBζ represses Ifng promoter activity by reducing acetylation of histones associated with the Ifng locus 7. Similar to IκBζ, which interacts with chromatin‐modifying enzymes 8, 9, Bcl‐3 acts as a bridge to nuclear coregulators 10, 11.
IκBNS was first described in thymocytes in the context of negative selection 12. IκBNS is also expressed in different T‐cell subsets such as Th1 cells, regulatory T‐cell precursors, and Th17 cells 13, 14, 15. Of note, IκBNS −/− mice exhibit reduced numbers of Tregs, because IκBNS acts in concert with c‐Rel and p50 to regulate Foxp3 expression, thereby, mediating the transition of Treg precursors into mature Treg cells 14. In terms of T‐cell development and function, both IκBNS‐deficient CD4+ and CD8+ T cells, exhibit a proliferation defect upon in vitro TCR stimulation 13, 16. Moreover, IκBNS‐deficient T cells show decreased secretion of IL‐2 following in vitro stimulation and IκBNS −/− Th1 cells produce less IFN‐γ 13, 15. Furthermore, IκBNS is critical for the development of effector functions in Th17 cells both in vitro and in vivo 15. While together these data indicate a crucial role of IκBNS in the development and function of different T‐cell subsets, no data are available on how IκBNS‐deficiency affects the activation, proliferation, and effector function of T cells specifically responding to a pathogen‐derived antigen during in vivo infection.
Results and discussion
IκBNS fosters CD4+ T‐cell activation and Th1 cytokine induction during Listeria monocytogenes infection
To specify the role of IκBNS in CD4+ T‐cell activation following in vivo pathogen recognition, we combined systemic infection with ovalbumin‐expressing Listeria monocytogenes (LM‐OVA) and adoptive transfer of IκBNS sufficient or deficient TCR‐transgenic OT‐II CD4+ T cells (Fig. 1A). Analysis of reisolated cells revealed the first genotype‐specific differences in the spleen on day 3 post infection (Fig. 1B). Of note, by day 5, LM‐OVA‐specific IκBNS +/+ CD4+ T cells underwent extensive proliferation, which was impaired in CD4+ T cells lacking IκBNS (Fig. 1B). Analyzing the proliferated CD4+ T cells for the expression of activation markers and Th1‐related effector cytokines revealed striking differences between the genotypes (Fig. 2). Here, lack of IκBNS resulted in reduced frequency of LM‐OVA‐specific CD4+ T cells expressing the activation markers CD44 and PD‐1, as well as the Th1‐effector cytokines IFN‐γ, IL‐2, and TNF‐α. We conclude that IκBNS is required for CD4+ T‐cell activation and expansion in in vivo infectious settings and is critically involved in Th1‐cell differentiation.
Figure 1.
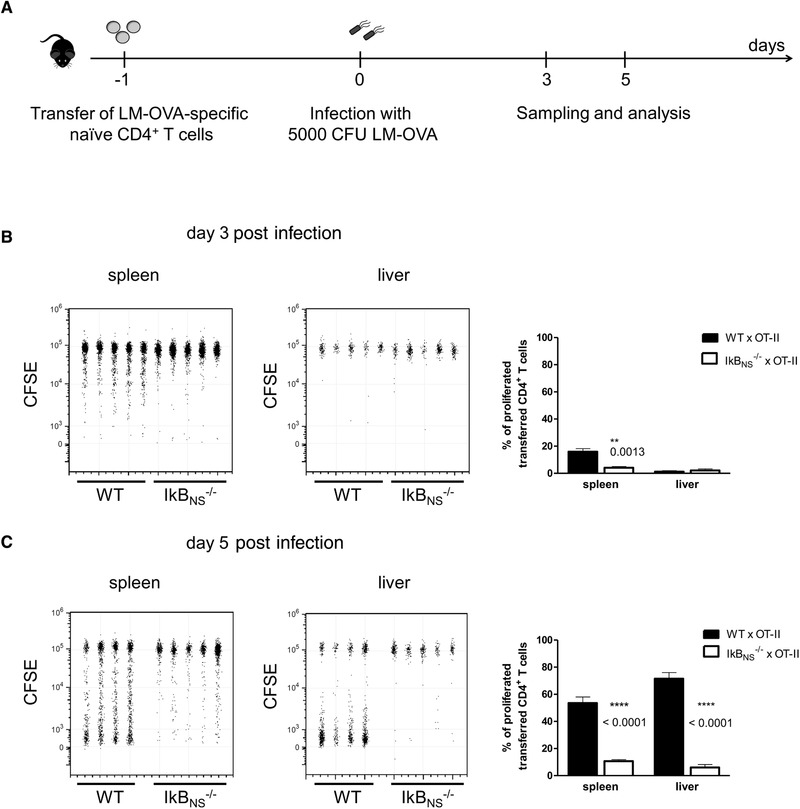
Proliferation of adoptively transferred LM‐OVA‐specific CD4+ T cells at different times post infection. (A) Experimental setup. 3 × 106 CD4+ T cells from Thy1.1+ OT‐II x WT and OT‐II x IκBNS −/− were transferred into C57BL/6 mice. One day post transfer recipients were infected with 5 × 103 LM‐OVA. CD4+ T cells from spleen and liver were analyzed for CFSE loss at the indicated time post infection by flow cytometry. (B, C) Flow cytometry data are representative for two (day 3) or three (day 5) independent experiments with similar outcome with n = 4–5 individually analyzed mice/group and data were constrained to alive singlet Thy1.1+ CD4+ T cells and are shown in columns side‐by‐side in a concatenated qualitative dot plot in which each column represents data of an individual mouse. The summary plots are depicted as mean ± SEM of 4–5 individually analyzed mice/group and indicate the percentages of proliferated (CFSElow) transferred CD4+ T cells in spleen and liver samples. Statistics were performed using two‐tailed unpaired student's t‐test. **p < 0.01, ****p < 0.0001.
Figure 2.
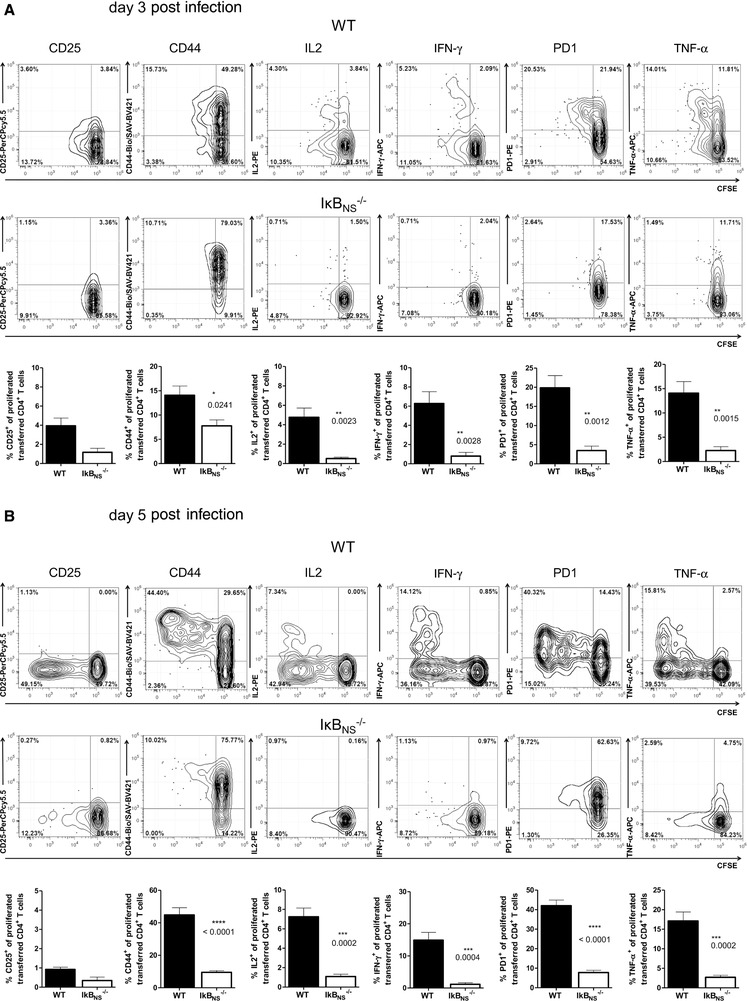
Phenotype of adoptively transferred LM‐OVA‐specific CD4+ T cells after LM‐OVA infection. Adoptive transfer of WT and IκBNS −/− OT‐II CD4+ T cells was performed as described in Figure 1. (A) 3 and (B) 5 days post LM‐OVA infection splenic lymphocytes were analyzed by flow cytometry. Flow cytometry analysis was constrained to alive singlet Thy1.1+CD4+ T cells. Data are depicted as mean +/− SEM (n = 4–5 individually analyzed mice/group) from two (day 3) or three (day 5) independent experiments with similar outcome. Upper rows: representative contour plots with 5% probability with outliers for CD25, CD44, IFN‐γ, IL‐2, PD‐1, and TNF‐α versus CFSE from OT‐II CD4+ T cells. Lower rows: summary plots indicate percentages of marker‐positive T cells within the CFSElow fraction. Statistics were performed using two‐tailed unpaired student‘s t‐test. *p < 0.05, **p < 0.01, ***p < 0.001.
IκBNS affects the early phase of Th1‐cell differentiation
To investigate in more detail IκBNS dependency of Th1‐cell differentiation, we used the newly generated reporter mouse NfkbidlacZ that contains a LacZ cassette within the IκBNS‐encoding Nfkbid gene and expresses ß‐galactosidase under the control of the Nfkbid promoter. After confirmation that the NfkbidlacZ mouse represents a faithful reporter to quantify Nfkbid gene expression (Supporting Information Fig. 1), we analyzed the kinetics of Nfkbid promoter activity under Th1‐polarizing conditions. Promoter activity increased until day 3 following T‐cell activation (Fig. 3A), suggesting that IκBNS is especially important for the early phase of Th1‐cell differentiation. To further confirm this, we utilized NfkbidFL/FL × RosaCreERT2 mice, which allow for targeted deletion of Nfkbid at time of interest following T‐cell stimulation. After confirming complete loss of IκBNS in CD4+ T cells within 48 h following stimulation (Fig. 3B), we evaluated at which phase of Th1‐cell differentiation IκBNS is required. Both, IFN‐γ and CD44 expression were reduced in IκBNS −/− CD4+ T cells when IκBNS‐deficiency was induced early (day 2) during Th1‐cell differentiation (Fig. 3C) but were not affected by adding tamoxifen on day 4 (Fig. 3C). Notably, the expression of T‐bet, which is crucial for Th1‐cell differentiation 17 was not affected in the absence of IκBNS (Fig. 3C), suggesting that other early factors of Th1‐cell differentiation are affected. Since IFN‐ɣ amplifies Th1‐cell differentiation, we suspect that the reduced IFN‐ɣ expression in the absence of IκBNS is responsible for reduced Th1 responses. As IκBζ and IκBNS have opposing activities on IL‐6 expression in macrophages 16, 18, it is tempting to speculate that a similar counteracting function operates in Th1 cells with respect to IFN‐ɣ expression.
Figure 3.
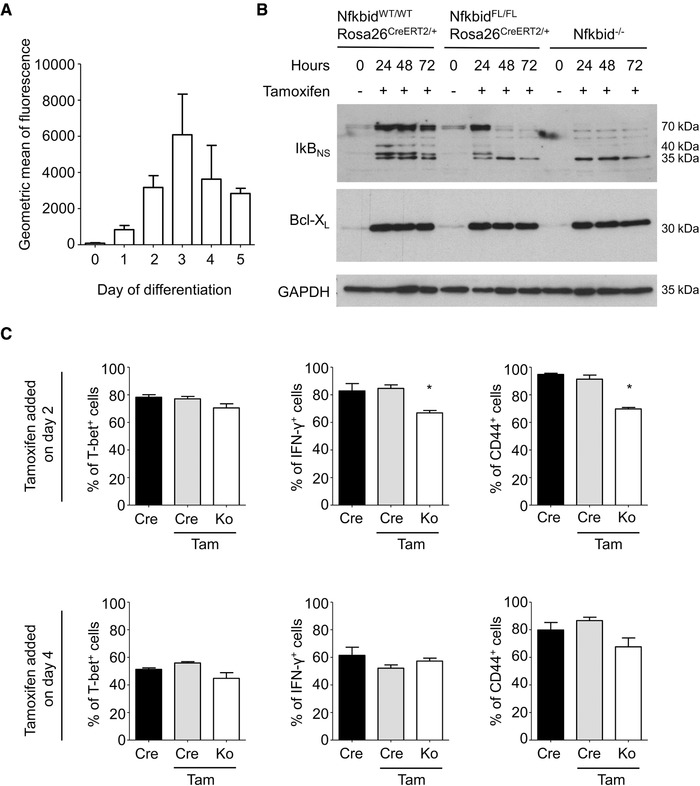
Conditional deletion of Nfkbid during Th1 differentiation. (A) Nfkbid promoter activity in CD4+ T cells from NfkbidLacZ mice during Th1 differentiation day 0 (unstimulated) and day 1–5 following α‐CD3/α‐CD28 stimulation in the presence of α‐IL‐4 and α‐IL‐12. Data are shown as mean +/− SEM (n = 3) and are representative of three independent experiments with n = 1 mouse per experiment. Chromophores: pacific blue for CD4 and FITC for lacZ. (B) Kinetic of IκBNS protein expression in CD4+ T cells from spleen and lymph nodes after tamoxifen administration. NfkbidWT/WTRosa26CreERT2/+: WT control; NfkbidFl/FLRosa26CreERT2/+: conditional deletion of Nfkbid; IκBNS −/−: negative control. Cells were stimulated with anti‐CD3 (2 µg/mL plate bound), anti‐CD28 (2 µg/mL in suspension), and IL‐2 (50 ng/mL). Data are representative of two independent experiments with n = 2 mice per experiment. (C) Conditional deletions of Nfkbid during Th1 differentiation. Cre: NfkbidWT/WTRosa26CreERT2/+ sample without addition of tamoxifen; Cre Tam: same sample with addition of tamoxifen. Ko: tamoxifen‐treated NfkbidFl/FLRosa26CreERT2/+ sample (conditional deletion). Data are shown as mean +/− SEM and are pooled from six (IFN‐γ) or three (T‐bet, CD44) independent experiments with n = 1 mouse per condition (Cre, Ko) per experiment. Statistics were performed using two‐tailed unpaired student‘s t‐test (Mann–Whitney test). The p values are 0.026 for IFN‐γ and 0.045 for CD44. Chromophores: APC for IFN‐γ, FITC for T‐bet, and PE‐Cy7 for CD44. Gating: see Supporting Information Figure 7.
Another hint regarding a potential mechanism underlying the impact of IκBNS on Th1‐cell differentiation comes from data published by Touma et al., who showed that the F‐box protein family member Fbxo17 is highly overexpressed in resting and activated IκBNS −/− T cells 13, a finding we confirmed in OT‐II CD4+ T cells (data not shown). F‐box proteins are the substrate‐binding subunits of SCF E3 ubiquitin ligases involved in regulation of cell cycle and proliferation 19. Fbxo17 regulates proteasomal degradation of glycogen synthase kinase‐3β (GSK3β) 20. Since GSK3 isoforms are crucial for Th1 differentiation 21, increased Fbxo17 expression in IκBNS −/− T cells might explain impaired Th1 differentiation. The exact mechanism of IκBNS and/or Fbxo17 in Th1 differentiation remains, however, unresolved and will be the focus of future studies.
Listeria monocytogenes induces a Th1‐dominated response with almost no Th17 cells and Th2‐cell responses even actively being suppressed by pathogen‐derived factors 22, 23. In line with data obtained in the in vitro Th1 differentiation, IκBNS‐deficient CD4+ T cells failed to upregulate CD44 and to produce the Th1‐effector cytokines IFN‐γ, IL‐2, and TNF‐α following in vivo pathogen encounter (Fig. 2). IκBNS‐dependency of IL‐2 and IFN‐γ production in CD4+ T cells has been described before 13 and we recently showed that IκBNS‐deficiency impairs CD4+ T‐cell proliferation during in vitro Th‐cell differentiation and is critically involved in the development of Th1 and Th17 cells 15. However, the impact of IκBNS on T‐cell differentiation appears to be context dependent. While EAE induction in IκBNS −/− mice did not affect the Th1‐cell pool 24, the DSS colitis model uncovered increased IFN‐γ expression in IκBNS −/− CD4+ T cells 15, 16. In contrast, intestinal infection of IκBNS‐deficient mice with Citrobacter rodentium resulted in reduced frequencies of IFN‐γ‐producing CD4+ T cells in spleen but not in colon and local lymph nodes 15. Of note, the present study is fundamentally different since we specifically track T cells responding to pathogen‐derived antigen in mice exhibiting an IκBNS‐sufficient immune system.
In line with published data obtained with polyclonal T cells 13, we also found OT‐I CD8+ IκBNS −/− T‐cell proliferation to be impaired following in vitro antibody‐induced TCR stimulation (data not shown) while CD8+ T‐cell proliferation induced by antigen‐specific activation following in vivo pathogen encounter does not depend on IκBNS (Supporting Information Fig. 2). Moreover, with the exception of TNF‐α, IκBNS‐deficiency had no or only transient effect on the expression of all other markers analyzed (Supporting Information Fig. 3). Data from adoptive transfers suggest that IκBNS might affect cytotoxic function of CD8+ T cells during the early phase of infection, which is likely to be compensated by the WT adaptive immunity in the later infection phase (Supporting Information Fig. 4A). This was supported by LM‐OVA infections in WT and conventional IκBNS −/− mice that neither revealed differences in bacterial elimination (Supporting Information Fig. 4B) nor uncovered defects in the establishment of cytotoxic T‐cell responses from the polyclonal TCR pool (Supporting Information Fig. 4C). This at least in part excludes that the observed effects in IκBNS −/− CD8+ T cells are due to the transgenic OT‐I TCR specific for the model antigen expressed by LM‐OVA. We hypothesize that the effects of IκBNS‐deficiency in CD8+ T cells that are observed in vitro are compensated in vivo by the presence of host‐derived proinflammatory cytokines that are produced during infection. This is in line with our observation that genotype‐dependent differences in the activation pattern of CD8+ T cells are only transient. Interestingly, the percentage of TNFα‐producing LM‐OVA‐specific IκBNS −/− CD8+ T cells was significantly higher compared to IκBNS +/+ CD8+ T cells. There is evidence that IκBNS may function as suppressor of TNF‐α 25. To rule out the possibility that the fraction of TNF‐α+ CD8+ T cell is per se higher in IκBNS −/− mice, we analyzed its expression at 1 day post infection revealing even more TNF‐α+ CD8+ T cells in the IκBNS +/+ group (data not shown). Nevertheless, despite distinct changes in the activation pattern and kinetics, IκBNS −/− CD8+ T cells are fully capable of producing IFN‐γ and mice with IκBNS‐deficient T cells are competent in developing cytotoxic T‐cell responses against LM‐OVA (Supporting Information Fig. 4).
Concluding remarks
While being largely dispensable for the in vivo acquisition of effector function in CD8+ T cells, we unveiled IκBNS to play a pivotal role in the activation, expansion, and Th1‐cell differentiation of CD4+ T cells following in vivo pathogen encounter. In terms of timing, IκBNS affects the early phase of Th1‐cell differentiation but is dispensable in terminally differentiated Th1 cells.
Material and methods
Animals
IκBNS −/− mice (B6.129/SV‐NFKBID(tm1Clay)) 14, OT‐I x CD90.1 26, OT‐II x CD90.1 27, and IκBNS ‐/‐ mice crossed on OT‐I and OT‐II were bred and maintained under specific pathogen‐free conditions at the animal facilities of the Helmholtz Centre for Infection Research Braunschweig, Germany and the University Hospital Magdeburg, respectively. For genotyping of mice, see Supporting Information Figure 5. NfkbidlacZ (Nfkbidtm1a(EUCOMM)Wtsi) reporter mice were obtained from EUCOMM. NfkbidFl mice were generated by crossing NfkbidlacZ with FLP recombinase expressing mice and offsprings were crossed to B6.129‐Gt(ROSA)26Sortm1(cre/ERT2)Tyj / J mice 28 to create an inducible IκBNS −/− mouse. All experiments were approved by the Landesamt für Verbraucherschutz, Sachsen‐Anhalt (AZ 42502‐2‐1242).
Adoptive T‐cell transfers
OVA‐specific CD4+ and CD8+ T cells were isolated using CD4+ and CD8+ T‐cell isolation kits and an autoMACS (Miltenyi Biotec). Purity of cells was ≥90%. Cells were stained with CFSE (2.5 µM, Thermo Fisher Scientific) and 3 × 106 cells were injected in 200 µL sterile PBS (Gibco) into the tail vein of C57BL/6J recipient mice (Janvier Labs, Le Genest‐Saint‐Isle, France or Envigo, NM Horst, The Netherlands).
Bacterial infection
OVA‐expressing L. monocytogenes (LM‐OVA, strain 10403S) were grown overnight (37°C, 180 rpm) in BHI broth (BD Biosciences). A 1:5 dilution was prepared with fresh BHI and after 3 h bacteria were harvested and diluted in sterile PBS to establish an infection dose of 5 × 103 CFU/mouse. To determine CFU in spleen and livers organs were homogenized in 0.2% IGEPAL CA‐630 (Sigma‐Aldrich) lysis buffer and serial dilutions were plated on BHI agar plates to quantify colonies after incubation at 37°C for 24 h.
Cell preparation
Spleens and livers were squeezed through 100 µm cell strainers (Falcon), washed with PBS (300 × g, 10 min, 4°C) followed by erythrocyte lysis. Splenocytes were passed through a 30 µm cell strainer and resuspended in PBS. Liver cells were separated by density centrifugation in a 35% mixture of Easycoll (Biochrom) and PBS. After centrifugation (360 × g, 20 min, RT) cells were washed and resuspended in PBS.
Flow cytometric analyses
Flow cytometric analyses were performed in adherence to the “Guidelines for the use of flow cytometry and cell sorting in immunological studies” 29. Cells were stained with anti‐CD16/32 (Fc‐block) (BioLegend) and with Fixable Viability Dye‐eFluor 780 (eBioscience) to exclude dead cells. After washing, cells were incubated with the antibody mixture containing CD8a‐BV510 (53–6.7) or CD4‐BV510 (GK1.5), CD90.1‐Pe‐Cy7 (OX‐7), CD44‐Biotin (IM7), CD25‐PerCP‐Cy5.5 (PC61), and PD1‐PE (29F.1A12) (all BioLegend). In case of biotinylated antibodies, a second step with streptavidin‐BV421 (BioLegend) was performed. Transgenic T cells were identified by FACS as shown in Supporting Information Figure 6. For detection of intracellular cytokines, cells were stimulated with 1 µg/mL OVA323‐339 or OVA257‐264 peptide (synthesized at Helmholtz Centre for Infection Research Braunschweig, Germany). After 1 h brefeldin A (5 µg/mL, BioLegend) was added. After another 4 h, cells were washed and stained for extracellular markers and fixed with 2% PFA. Subsequently, cells were permeabilized (0.1% IGEPAL CA‐630 (Sigma‐Aldrich), 4 min, 4°C), washed and stained with anti‐IFN‐γ‐APC (XMG1.2), anti‐IL‐2‐PE (JES6‐5H4) or anti‐TNF‐α‐APC (MPC‐XT22) (BioLegend) for 30 min at 4°C. IκBNS‐dependency of cytotoxic function in CD8+ T cells was determined as described before 30. Cells were analyzed with a BD FACS Canto II, BD LSR II (BD Biosciences) or an Attune NxT Acoustic Focusing Cytometer (Thermo Fisher Scientific).
Th1‐cell differentiation
Naïve CD4+CD62L+CD25− T cells were isolated from spleens and lymph nodes by cell sorting (BD FACS Aria II (BD Biosciences) or Moflo (Beckman and Coulter)). Th1 differentiation was performed as described before 15. In case conditional deletion of Nfkbid was performed, 1 µM (Z)‐4‐Hydroxytamoxifen (Sigma–Aldrich) was added on day 2 and 4. On day 4 and 6, cells were restimulated with PMA (10 ng/mL) and ionomycin (1 µg/mL, both Sigma–Aldrich) for 4 h. After 2h stimulation, brefeldin A (10 µg/mL, Sigma–Aldrich) was added and subsequently cells were stained with LIVE/DEAD fixable blue (Life Technologies), for CD4‐Pacific Blue (RMA4‐5, BioLegend) and CD44‐PE‐Cy7 (IM7, eBioscience). Afterwards, the cells were fixed with Foxp3 staining buffer (Miltenyi Biotec) and stained for IFN‐γ‐APC (XMG1.2, BioLegend) and T‐bet‐FITC (4B10, BioLegend) according to manufacturer's recommendations.
Western blot analysis
Cells were lysed and immunoblot was performed as previously described 15. The following primary antibodies were used: IκBNS (rabbit, β‐actin (AC‐74, Sigma–Aldrich) 31, GAPDH (1E6D9, Proteintech), Bcl‐XL (clone 4, BD Biosciences).
Statistics
Results are expressed as mean ± SEM. Statistical analyses were performed with the GraphPad Prism 5.4 Software (La Jolla, CA, USA) and a p‐value below 0.05 was considered significant.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Abbreviation
- GSK3β
glycogen synthase kinase‐3β
Supporting information
Supporting Information
Acknowledgments
We thank Franziska Ewert and Sabrina Schumann for technical assistance. We thank Drs. Luka Cicin‐Sain and Thomas Schüler for helpful discussions. This work was supported by grants from the DFG (SCHM1586/6‐1) to Ingo Schmitz and project A23 of SFB854 to Dunja Bruder and Ingo Schmitz. Konstantinos Katsoulis‐Dimitriou was supported by President's Initiative and Networking Fund of the HGF (VH‐GS‐202). Sarah Frentzel and Konstantinos Katsoulis‐Dimitriou carried out the experiments and wrote the manuscript. Sarah Frentzel, Konstantinos Katsoulis‐Dimitriou, Andreas Jeron, Ingo Schmitz, and Dunja Bruder conceived the study, planned the experiments, and revised the manuscript.
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.201847961
References
- 1. Hayden, M. S. and Ghosh, S. , NF‐κB, the first quarter‐century: remarkable progress and outstanding questions. Genes Dev 2012. 26: 203–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schuster, M. , Annemann, M. , Plaza‐Sirvent, C. and Schmitz, I. , Atypical IκB proteins—nuclear modulators of NF‐κB signaling. Cell Commun. Signal. 2013. 11: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aronica, M. A. , Mora, A. L. , Mitchell, D. B. , Finn, P. W. , Johnson, J. E. , Sheller, J. R. and Boothby, M. R. , Preferential role for NF‐kappa B/Rel signaling in the type 1 but not type 2 T cell‐dependent immune response in vivo. J. Immunol. 1999. 163: 5116–5124. [PubMed] [Google Scholar]
- 4. Corn, R. A. , Aronica, M. A. , Zhang, F. , Tong, Y. , Stanley, S. A. , Kim, Se R. A. , Stephenson, L. et al., T cell‐intrinsic requirement for NF‐kappa B induction in postdifferentiation IFN‐gamma production and clonal expansion in a Th1 response. J. Immunol. 2003. 171: 1816–1824. [DOI] [PubMed] [Google Scholar]
- 5. Franzoso, G. , Carlson, L. , Scharton‐Kersten, T. , Shores, E. W. , Epstein, S. , Grinberg, A. , Tran, T. et al., Critical roles for the Bcl‐3 oncoprotein in T cell‐mediated immunity, splenic microarchitecture, and germinal center reactions. Immunity 1997. 6: 479–490. [DOI] [PubMed] [Google Scholar]
- 6. Schwarz, E. M. , Krimpenfort, P. , Berns, A. and Verma, I. M. , Immunological defects in mice with a targeted disruption in Bcl‐3. Genes Dev 1997. 11: 187–197. [DOI] [PubMed] [Google Scholar]
- 7. MaruYama, T. , Kobayashi, S. , Ogasawara, K. , Yoshimura, A. , Chen, W. and Muta, T. , Control of IFN‐γ production and regulatory function by the inducible nuclear protein IκB‐ζ in T cells. J. Leukoc. Biol. 2015. 98: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Totzke, G. , Essmann, F. , Pohlmann, S. , Lindenblatt, C. , Jänicke, R. U. and Schulze‐Osthoff, K. , A novel member of the IkappaB family, human IkappaB‐zeta, inhibits transactivation of p65 and its DNA binding. J. Biol. Chem. 2006. 281: 12645–12654. [DOI] [PubMed] [Google Scholar]
- 9. Zhang, Q. , Zhao, K. , Shen, Q. , Han, Y. , Gu, Y. , Li, X. , Zhao, D. et al., Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL‐6. Nature 2015. 525: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dechend, R. , Hirano, F. , Lehmann, K. , Heissmeyer, V. , Ansieau, S. , Wulczyn, F. G. , Scheidereit, C. et al., The Bcl‐3 oncoprotein acts as a bridging factor between NF‐kappaB/Rel and nuclear co‐regulators. Oncogene 1999. 18: 3316–3323. [DOI] [PubMed] [Google Scholar]
- 11. Wang, V. Y.‐F. , Huang, W. , Asagiri, M. , Spann, N. , Hoffmann, A. , Glass, C. and Ghosh, G. , The transcriptional specificity of NF‐κB dimers is coded within the κB DNA response elements. Cell Rep. 2012. 2: 824–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fiorini, E. , Schmitz, I. , Marissen, W. E. , Osborn, S. L. , Touma, M. , Sasada, T. , Reche, P. A. et al., Peptide‐induced negative selection of thymocytes activates transcription of an NF‐kappa B inhibitor. Mol Cell 2002. 9: 637–648. [DOI] [PubMed] [Google Scholar]
- 13. Touma, M. , Antonini, V. , Kumar, M. , Osborn, S. L. , Bobenchik, A. M. , Keskin, D. B. , Connolly, J. E. et al., Functional role for I kappa BNS in T cell cytokine regulation as revealed by targeted gene disruption. J. Immunol. 2007. 179: 1681–1692. [DOI] [PubMed] [Google Scholar]
- 14. Schuster, M. , Glauben, R. , Plaza‐Sirvent, C. , Schreiber, L. , Annemann, M. , Floess, S. , Kühl, A. A. et al., IκB(NS) protein mediates regulatory T cell development via induction of the Foxp3 transcription factor. Immunity 2012. 37: 998–1008. [DOI] [PubMed] [Google Scholar]
- 15. Annemann, M. , Wang, Z. , Plaza‐Sirvent, C. , Glauben, R. , Schuster, M. , Ewald Sander, F. , Mamareli, P. et al., IκBNS regulates murine Th17 differentiation during gut inflammation and infection. J. Immunol. 2015. 194: 2888–2898. [DOI] [PubMed] [Google Scholar]
- 16. Kuwata, H. , Matsumoto, M. , Atarashi, K. , Morishita, H. , Hirotani, T. , Koga, R. and Takeda, K. , IkappaBNS inhibits induction of a subset of Toll‐like receptor‐dependent genes and limits inflammation. Immunity 2006. 24: 41–51. [DOI] [PubMed] [Google Scholar]
- 17. Szabo, S. J. , Kim, S. T. , Costa, G. L. , Zhang, X. , Fathman, C. G. and Glimcher, L. H. , A novel transcription factor, T‐bet, directs Th1 lineage commitment. Cell 2000. 100: 655–669. [DOI] [PubMed] [Google Scholar]
- 18. Yamamoto, M. , Yamazaki, S. , Uematsu, S. , Sato, S. , Hemmi, H. , Hoshino, K. , Kaisho, T. et al., Regulation of Toll/IL‐1‐receptor‐mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature 2004. 430: 218–222. [DOI] [PubMed] [Google Scholar]
- 19. Skaar, J. R. , Pagan, J. K. and Pagano, M. , SCF ubiquitin ligase‐targeted therapies. Nat. Rev. Drug Discov. 2014. 13: 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suber, T. , Wei, J. , Jacko, A. M. , Nikolli, I. , Zhao, Y. , Zhao, J. and Mallampalli, R. K. , SCFFBXO17 E3 ligase modulates inflammation by regulating proteasomal degradation of glycogen synthase kinase‐3β in lung epithelia. J. Biol. Chem. 2017. 292: 7452–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beurel, E. , Kaidanovich‐Beilin, O. , Yeh, W.‐I. , Song, L. , Palomo, V. , Michalek, S. M. , Woodgett, J. R. et al., Regulation of Th1 cells and experimental autoimmune encephalomyelitis by glycogen synthase kinase‐3. J. Immunol. 2013. 190: 5000–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maruyama, S. , Kanoh, M. , Matsumoto, A. , Kuwahara, M. , Yamashita, M. and Asano, Y. , A novel function of interferon regulatory factor‐1: inhibition of Th2 cells by down‐regulating the Il4 gene during Listeria infection. Int. Immunol. 2015. 27: 143–152. [DOI] [PubMed] [Google Scholar]
- 23. Orgun, N. N. , Mathis, M. A. , Wilson, C. B. and Way, S. S. , Deviation from a strong Th1‐dominated to a modest Th17‐dominated CD4 T cell response in the absence of IL‐12p40 and type I IFNs sustains protective CD8 T cells. J. Immunol. 2008. 180: 4109–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kobayashi, S. , Hara, A. , Isagawa, T. , Manabe, I. , Takeda, K. and MaruYama, T. , The nuclear IκB family protein IκBNS influences the susceptibility to experimental autoimmune encephalomyelitis in a murine model. PLoS ONE 2014. 9: e110838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sierra‐Mondragón, E. , Gómez‐Chávez, F. , Murrieta‐Coxca, M. , Vázquez‐Sánchez, E. A. , Martínez‐Torres, I. , Cancino‐Díaz, M. E. , Rojas‐Espinosa, O. et al., Low expression of IL‐6 and TNF‐α correlates with the presence of the nuclear regulators of NF‐κB, IκBNS and BCL‐3, in the uterus of mice. Mol. Immunol. 2015. 68: 333–340. [DOI] [PubMed] [Google Scholar]
- 26. Clarke, S. R. , Barnden, M. , Kurts, C. , Carbone, F. R. , Miller, J. F. and Heath, W. R. , Characterization of the ovalbumin‐specific TCR transgenic line OT‐I: MHC elements for positive and negative selection. Immunol. Cell Biol. 2000. 78: 110–117. [DOI] [PubMed] [Google Scholar]
- 27. Robertson, J. M. , Jensen, P. E. and Evavold, B. D. , DO11.10 and OT‐II T cells recognize a C‐terminal ovalbumin 323–339 epitope. J. Immunol. 2000. 164: 4706–4712. [DOI] [PubMed] [Google Scholar]
- 28. Ventura, A. , Kirsch, D. G. , McLaughlin, M. E. , Tuveson, D. A. , Grimm, J. , Lintault, L. , Newman, J. et al., Restoration of p53 function leads to tumour regression in vivo. Nature 2007. 445: 661–665. [DOI] [PubMed] [Google Scholar]
- 29. Cossarizza, A. , Chang, H.‐D. , Radbruch, A. , Akdis, M. , Andrä, I. , Annunziato, F. , Bacher, P. et al., Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur. J. Immunol. 2017. 47: 1584–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Volckmar, J. , Gereke, M. , Ebensen, T. , Riese, P. , Philipsen, L. , Lienenklaus, S. , Wohlleber, D. et al., Targeted antigen delivery to dendritic cells elicits robust antiviral T cell‐mediated immunity in the liver. Sci. Rep. 2017. 7: 43985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schuster, M. , Plaza‐Sirvent, C. , Matthies, A.‐M. , Heise, U. , Jeron, A. , Bruder, D. , Visekruna, A. et al., c‐REL and IκBNS govern common and independent steps of regulatory T‐cell development from novel cD122‐expressing pre‐precursors. J. Immunol. 2017. 199: 920–930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


