Abstract
Germ granules are hallmarks of all germ cells. Early ultrastructural studies in Drosophila first described these membraneless granules in the oocyte and early embryo as filled with amorphous to fibrillar material mixed with RNA. Genetic studies identified key protein components and specific mRNAs that regulate germ cell‐specific functions. More recently these ultrastructural studies have been complemented by biophysical analysis describing germ granules as phase‐transitioned condensates. In this review, we provide an overview that connects the composition of germ granules with their function in controlling germ cell specification, formation and migration, and illuminate these mysterious condensates as the gatekeepers of the next generation.
Keywords: germ granules; localized translation; mRNA clusters; Oskar; phase separation; RNA granules; RNA localization; vasa, nanos
1. GERM PLASM AND GERM GRANULES OF DROSOPHILA—FIRST DESCRIPTIONS
In his search for a “heritable substance” that is transmitted from generation to generation, German biologist August Weismann proposed in 1893 that the offspring owes its origin to a “peculiar substance of extremely complicated structure” called the germ plasm.1 It is this structure, he explained, that distinguishes cells that give rise to the next generation from those that produce the “perishable body.” Experimental support for such a germ cell determinant was later provided by Robert Hegner's pricking experiments where he removed the germ plasm from the beetle Calligrapha punctate and found that the resulting embryos lacked morphologically discernable germ cells.2 With microscopy techniques of the time, Hegner described the germ plasm as containing “special bodies,” which were dense and stained like “chromatin.”3 However, it was not until the early 1960s, when the detailed electron microscopic (EM) studies of Drosophila melanogaster and other Drosophilid embryos by Tony Mahowald revealed the existence of morphologically unique structures found within the germ plasm, called the germ granules (Figure 1A). Mahowald described these granules as round, membraneless bodies about 0.2 to 0.5 μm in diameter, which contained fibrous material and ribosomes and stained with nucleic acid markers.8 These studies also revealed that granules could have a defined structure, with the periphery more electron dense than the core, and could often closely associate with mitochondria9 (Figure 1A, B). While the morphology of granules changed during Drosophila development, and could even vary among Drosophila species, Mahowald noted that the electron‐dense, fibrous nature of germ granules was a hallmark of the germline lineage throughout the germline life cycle and shared among species.
Figure 1.
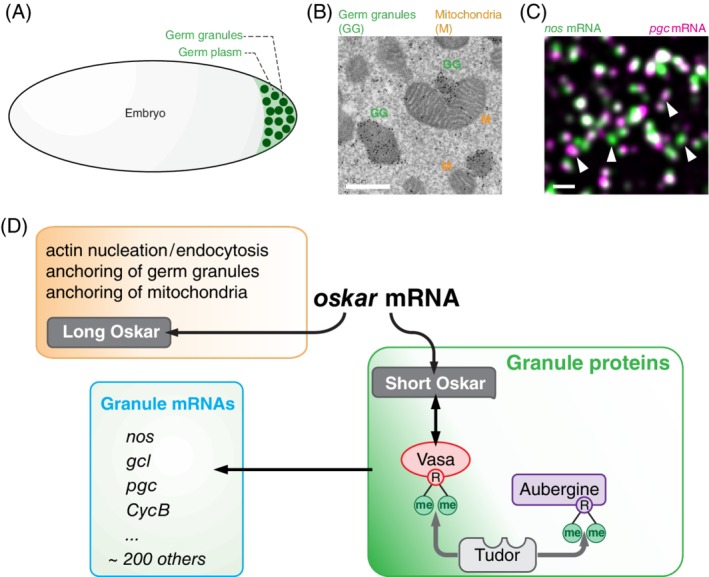
Formation of germ granules in Drosophila embryo (figure adapted from Reference 4. A, Germ granules form in the specialized cytoplasm called germ plasm at the embryo's posterior pole. B, An EM image showing that germ granules (labeled by immunogold particles staining Vasa protein; marked with green “GG”) are more electron dense than surrounding germ plasm and are closely associated with mitochondria (marked with orange “M”). C, Germ granules accumulate nos (green) and pgc (magenta) homotypic mRNA clusters,5, 6, 7 that are often colocalized within the same granule but that do not mix with each other.7 White arrows point at granules that are populated by only nos or pgc demonstrating that germ granules are heterogeneous in mRNA composition. D, oskar mRNA translates into Long and Short Osk isoforms that regulate distinct aspects of germ plasm and germ granules formation. R‐me indicates a methylated arginine. Scale bar in B is 500 nm and in C it is 1000 nm
Functionally, Mahowald and his postdoctoral fellow Karl Illmensee demonstrated the deterministic potential of the Drosophila germ plasm by transplanting it from the posterior pole, where germ cells form, to an ectopic anterior location in the embryo.10, 11 Nuclei located in the transplanted region formed cells with the morphology of “pole cells,” as the primordial germ cells (PGCs) in Drosophila are called. Moreover, these ectopic pole cells gave rise to functional germ cells after being transferred into a host embryo.10, 11 Some of the key proteins and mRNAs contained in germ plasm were later genetically identified as the products of so‐called maternal effect genes required maternally to regulate the assembly of germ plasm during oogenesis and the function of germ cells in the resulting embryo. Mutations in genes required for germ plasm assembly lead to a “grand‐childless” phenotype due to the loss of germ cells in the progeny of mutant mothers.12, 13, 14 Among these genes, oskar (osk) plays a central role in the organization of germ plasm and the formation of germ cells in the early embryo. An instructive role for Oskar protein akin to the germ plasm transplantation experiments of Mahowald and Illmensee was demonstrated genetically by expressing a transgene that encoded the open reading frame (ORF) of oskar with an RNA localization signal that anchored this transgene to the anterior pole of the embryo.14, 15 Thus, the anterior localization of osk's ORF and the resulting local production of Osk protein at the anterior pole was sufficient to attract other germ plasm components, leading to assembly of germ granules and specification of functional germ cells at the ectopic location.15 Interestingly, Oskar is not conserved beyond dipterans16 and is not a marker for all stages of germ line development (reviewed in Reference 4). However, orthologs of other germ plasm components are found throughout the animal kingdom and present in germ cells throughout their life cycle.4 Indeed, the core germ granule components Vasa, an ATP‐dependent RNA helicase, the translation factors Nanos, Pumilio and Dazl, Tudor (Tud), the founder of the Tudor domain family of proteins and Aubergine (Aub), a Piwi family Pi RNA‐binding protein,4 all have critical, evolutionary‐conserved roles in the germline across species. Therefore, deciphering the principles of germ granule formation and function in Drosophila allows us to understand the roles of these droplets in shaping the biology of germ cells in other organisms, including humans.
Early EM studies revealed that germ granules first appear as small and dense bodies at the posterior pole of the developing oocyte, are later inherited by the fertilized embryo and finally become engulfed by the newly formed pole cells. Modern molecular biology, microscopy and genetic tools revealed that these granules accumulate maternally provided messenger ribonucleic acids (mRNAs) and proteins critical for the establishment of PGCs and the germline; in their absence, germ cells do not form and the resultant embryo is sterile (reviewed in Reference 4). Thus, while Weismann was looking for the heritable substance, the DNA, he instead identified germ granules as the hallmark substance that provides continuity of the species, the subject discussed in this review.
2. ONTOGENY AND ORGANIZATION OF GERMPLASM
2.1. Localization of Oskar and other core germ plasm components
Germ granule formation in Drosophila is intricately linked to oocyte polarity and relies on the coordinated transport of a single mRNA, oskar, toward the posterior pole of the oocyte, where this mRNA remains localized throughout late oogenesis and early embryogenesis. To ensure the continuity of the species, Drosophila evolved an intricate mechanism by which osk reaches the posterior pole (described in detail4). Briefly, osk mRNA is synthesized by the nurse cells, which are sister, germline cells of the oocyte connected with the oocyte by large inter‐cellular bridges called ring canals. The nurse cells have large, polyploid nuclei that synthesize transcripts and proteins for the transcriptionally silent oocyte. Dynein motors transport osk and many other mRNAs into the growing oocyte along the minus‐end microtubule‐directed transport from nurse cells into the oocyte.17 Afterward and concurrent with repolarization of the oocyte microtubule network, osk transport particles shift preference toward the kinesin‐mediated, plus end‐directed transport, which leads to accumulation of osk mRNA at the oocyte's posterior pole. Concerted action of F‐actin and cortical microtubules establishes a sustained anchoring of osk transcripts at the posterior cortex.18, 19 However, this anchoring is not static. Instead, the granules display corralled movement or move directionally on the cytoskeletal network spanning several micrometers across the posterior cortex.18
Enroute from nurse cells into the oocyte, several osk molecules become packaged into large ribonucleoprotein particles (RNPs) containing the double‐stranded RNA‐binding protein Staufen and the exon junction complex components Mago nashi, Y14, eIF4AIII and Barentsz.20, 21, 22, 23, 24 Transport and mRNA localization to the posterior pole rely on the ability of osk mRNA to dimerize and are coordinated by cis‐acting sequences positioned within the osk 3′UTR and the first exon junction in osk. 25 Additional sequences in the osk 3′UTR are specifically recognized by translational regulatory proteins such as the RNA‐binding protein Bruno, which represses oskar translation during transport.26, 27, 28 The switch from minus‐ to plus‐end–directed microtubule transport depends on the germline‐specific isoform of tropomyosin.29, 30, 31 In addition to this early, motor‐dependent enrichment, osk accumulation also relies on a motor‐independent enrichment process during later stages of oogenesis and is mediated by the RNA‐binding proteins Rump and Lost.32 This accumulation of osk together with the continuous production of Oskar protein amplifies the amount of germ plasm available to form germ granules.5, 18, 32, 33
Upon localization, the repressive activity of Bruno is inhibited, allowing osk translation. Together these regulatory mechanisms ensure that oskar mRNA reaches the posterior pole with high efficiency and that Oskar protein is specifically synthesized there. Perhaps given the complex and possibly energy‐consuming nature of osk transport to the posterior pole, it is not surprising that only few mRNAs aside from osk use directed microtubule‐mediated transport to localize within the oocyte or are as tightly regulated as osk in the oocyte. For example, approximately 200 maternally provided mRNAs that enrich in germ plasm nucleated by Oskar34, 35 localize passively, through a diffusion and entrapment mechanisms (see below36). While this entrapment mechanism seems similar to the late stage localization of osk mRNA, osk accumulates in RNP particles that are distinct from germ granules, which contain the majority of localized mRNAs at the posterior pole.6, 7, 19
2.2. Organization of proteins in germ granules
Germ plasm is a specialized cytoplasm that forms at the embryo's posterior pole and is populated by core granule proteins Oskar, Vasa, Tud and Aub, a variety of RNA‐binding proteins involved in various aspects of RNA biology, maternally deposited mRNAs, piRNAs and mitochondria (Figure 1A‐D) (reviewed in Reference 37). While most of the RNA‐binding proteins enriched in germ plasm are also found elsewhere in the embryo, the core granule proteins are almost exclusively found only at the posterior pole.38, 39 The exact composition of germ plasm is not known, however, recent quantitative imaging data demonstrated that the vast majority of core germ plasm proteins (>94%) are condensed into granules, with very little of these proteins diffusing in the intergranular germ plasm space.38 Indeed, their concentration in the intergranular space is similar to their concentration in somatic regions.38 For germ plasm mRNAs, the majority of transcripts that enrich at the posterior remain dispersed in somatic regions of the embryo with only up to 4% of a particular mRNA enriched in germ plasm.7, 40 Despite this small fraction, however, these mRNAs become 8‐ to 10‐fold more concentrated upon localization.7 Some mRNAs that enrich in germ plasm also tend to preferentially accumulate in granules. For instance, whole‐mount in situ labeling of 59 mRNAs revealed that these transcripts interact with germ granules; they arrange as crescents surrounding the dividing PGC nuclei in older embryos, a spatial organization driven entirely by germ granule proteins (Figure 2A, see granule organization around nuclei of pole buds).41, 42 Interestingly, transcriptome analysis with microarrays of purified fluorescently labeled PGCs revealed that over 1700 different transcripts enrich in PGCs compared to the surrounding soma,46 suggesting that the diversity of germ plasm transcripts could be much higher than originally believed. It is unknown whether all these various transcripts associate with granules or instead with other unknown germ plasm components. Regardless, PGCs that remain transcriptionally silent longer than the surrounding soma (see below) could benefit from this enrichment as the diversity of localized mRNAs could provide quiescent PGCs with the necessary maternally provided material for PGCs to reach activation of their zygotic genome upon maternal to zygotic transition (MZT).
Figure 2.
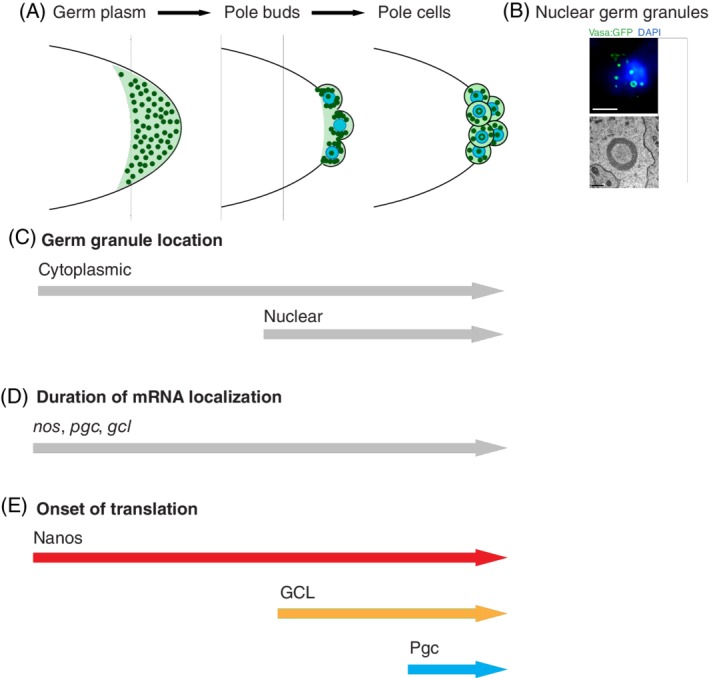
Spatial organization of germ granules and of granule‐associated posttranscriptional regulation through early embryo development. (A‐C) Initially, germ granules are uniformly distributed within germ plasm (0‐1.5 hours old fertilized eggs). Once the pole buds form at the posterior pole, germ granules then transport via dynein motors coupled to astral microtubules toward the centrosomes associated with pole bud nuclei.41 As such they become organized into crescents surrounding dividing nuclei in pole buds and pole cells (1.5‐3 hours old fertilized eggs) (A).41, 42 During this time, nuclear germ granules form that are also often hollow (B, C).38 Image in B shows a nucleus of a pole cell stained with DAPI (blue) that accumulates hollow nuclear germ granules stained with Vasa:GFP protein (green). EM image in B shows nuclear germ granules formed by Short Osk in Drosophila‐cultured cells lines. (D, E) nos, pgc and CycB mRNAs persistently localize in germ granules throughout early embryogenesis (D) but they nevertheless display distinct onsets of translation (E) to allow the body patterning of the early embryo (Nanos), cellularization of pole buds into primordial germ cells (Gcl) and transcriptional silencing of newly formed primordial germ cells (Pgc)42, 43, 44, 45 (figure adapted from Reference 42). Scale bar in B is 5 μm (fluorescent image) and 1 μm (EM image)
At the posterior pole, osk mRNA translates into two protein isoforms: a longer 606 amino acid (aa) isoform called Long Oskar (Long Osk), which is translated from the first start codon, and a shorter, 467 aa isoform called Short Oskar (Short Osk) translated from the second start codon (Figure 1D).47 Interestingly, the two isoforms have different subcellular localizations and functions. Long Osk is found in close association with the egg cortex and is required for the sustained association of osk mRNA with the posterior pole and localization of mitochondria (see also below), while Short Osk is both necessary and sufficient for germ plasm assembly and germ cell formation (Figure 1D).19, 39, 47, 48, 49 Short Osk forms granules in the absence of other germ granule components in cultured Drosophila cells and human cells,38 indicating that condensation into a granule is an innate property of Oskar protein. Additionally, early Drosophila embryos form two types of germ granules: the cytoplasmic granules also called “polar granules” that associate with maternally deposited mRNAs and promote formation of PGCs, and nuclear germ granules that promote mitotic divisions of PGCs (Figure 2A‐C).38 Both granules are mostly round and nonmembrane bound, nucleated by Short Osk and recruit Vasa,8, 38, 49, 50, 51 while only cytoplasmic granules also recruit Aub, Tud and known granule‐enriched mRNAs.38 Nuclear germ granules are bigger and often appear hollow in EM and fluorescence images (Figure 2B),38, 50 and some cytoplasmic germ granules seem to have similar protein lucid cores. In both types of granules, this core appears less electron dense in EM images than the outside granule rim (Figure 2B).8, 38, 50, 51 A short nuclear localization sequence controls nuclear import of Osk and cotransport of Vasa into PGC nuclei where the two proteins condense into the same granules. As with cytoplasmic granules, the majority of Osk and Vasa is found in granules rather than freely diffusing in the nucleoplasm.38 How these nuclear granules regulate of PCG number is currently unknown.
Short Osk contains two, structurally discernable protein domains. The N‐terminus forms the LOTUS domain named after the Limkain (a human autoantigen whose function and binding partners are unknown), Oskar, and Tudor‐containing proteins 5 and 7 (TDRD5 and TDRD7 proteins, respectively), two mammalian members of the germline Tudor group.52 The C‐terminus is a novel RNA‐binding domain that shares similarity to SGNH hydrolases but lacks its enzymatic activity.53 Recent structural studies revealed that the LOTUS domain folds into a winged‐helix‐turn‐helix fold motif. The beta‐sheets of the LOTUS domain facilitate Osk dimerization while the extended helices interact with the C‐terminal RecA‐like helicase domain of Vasa (Figure 1D).54, 55, 56 Short Osk‐Vasa interaction stimulates Vasa's ATPase activity in the presence of single‐stranded and double‐stranded RNA and is required for posterior localization of Vasa.49, 55 It is unclear whether and how Osk and Vasa regulate RNA biology of granule mRNAs. For example, Osk could help recruit mRNAs to granules via its RNA‐binding domain. Indeed, in vitro and in vivo experiments demonstrated that Oskar interacts with germ granule‐enriched mRNAs nanos (nos), germ‐cell‐less (gcl), polar granule component (pgc) as well as its own mRNAs, albeit with low affinity and in a sequence unspecific manner.54, 57 Vasa on the other hand could unwind secondary RNA structures and RNA duplexes of localized mRNAs, possibly to facilitate their localization and translation in granules. The Osk‐Vasa complex may therefore play an instructive role in attracting mRNAs to the posterior pole.
In addition to mRNAs, germ granules recruit Tudor58 and Aubergine59 proteins (Figure 1D). Aub is a member of the Argonaute/Piwi family of proteins and binds to small piRNAs that regulate transposable elements but has also been implicated in mRNA binding (see below60). Tud consists of 11 Tudor domains, each able to bind symmetrically methylated arginines found in Aub. Tud tethers Aub via its Tudor domain to the germ plasm, as mutations that affect Tudor's ability to bind dimethylated arginines strongly reduce the localization of Aub to the posterior pole (Figure 1D).61, 62 Interestingly, not all Tudor domains bind Aubergine equally well and identical mutations in distinct Tudor domains have different effects on Aubergine localization.62 This finding suggests that the Tudor domains have additional, yet unknown specificities and can possibly organize multiple proteins containing dimethylated arginines. One such protein could be Vasa, also a methylated protein,63 suggesting that Vasa's persistent anchoring to granules could depend on both Osk and Tud. This possibility further suggests that Tudor's role in germ granule formation could be as a scaffolding protein. Indeed, EM studies of embryos expressing mutated versions of Tud protein revealed that in the absence of Tud protein, embryos form fewer granules that are also far less electron dense than their wild‐type counterparts.51, 58 Additionally, identical mutations in different Tudor domains also result in distinct appearance of germ granules,51 indicating that Tud plays a central role in maintaining the integrity and the morphology of germ granules in Drosophila (Figure 1D).
Oskar/Vasa and Tudor/Aubergine form specific complexes of high affinity interactions. However, these proteins also associate into large membraneless droplets that by other criteria such as high concentration, mobility of components and variable stochiometry resemble RNA droplets such as stress granules, processing bodies and germ granules called P‐granules of the round worm Caenorhabditis elegans (C. elegans). These RNA granules, including germ granules in Drosophila, are composed of diverse RNA‐binding proteins and mRNAs and form by phase separation, a process best described as oil‐and‐water demixing.37, 38, 64 Proteins that phase separate often contain intrinsically disordered regions (IDRs) (regions that do not adopt a particular protein fold) or low complexity sequences (LCs) (regions within a protein containing little diversity in amino acid composition).64, 65, 66, 67 Indeed, a 160 aa long IDR resides between the Osk's LOTUS and its RNA‐binding domain, while the first 47 aa of the LOTUS domain also harbors a LC sequence.38 When truncated versions of Oskar are expressed in Drosophila cultured cells, deletion of either the LOTUS domain, Osk domain, its IDR or LC fails to abolish condensation of Short Osk indicating that the four Short Osk regions act redundantly to form a granule. However, these protein truncations condense less efficiently indicating that these regions synergize to augment Short Osk condensation.38 Interestingly, Long Osk never forms granules be it in embryos or in cultured cells despite sharing all structural features with Short Osk.38, 39, 47 Therefore, the N‐terminal region of Long Osk interferes with the ability of the protein to condense. The biological relevance of this interference in embryos is not understood.
Biophysical studies further demonstrated that Drosophila germ granules display both liquid‐like and hydrogel‐like properties and are thus best described as phase‐transitioned condensates. Specifically, a fraction of core granule proteins readily exchanges with the granule environment while a fraction appears highly immobile and is retained within granules.38 Such properties are likely relevant as they can have profound functional consequences for the development of the germline. The liquid properties could enhance biochemical reactions occurring within granules while the more stable conformation could ensure that granule regulatory proteins persist throughout early embryonic development. Indeed, functional germ granules form during late oogenesis68 and persist through early embryogenesis, a process that lasts many hours when fertilization is delayed.69, 70 The persistence of granules and its components is fundamentally important for the formation of germ cells in Drosophila. For example, embryos of certain Osk and Tud alleles that fail to form granules or form only small granules are largely defective in RNA localization and do not form germ cells.14, 51, 58 Thus, robust mRNA localization within a liquid granule environment that affords biochemical reactions would support dynamic and prolonged posttranscriptional regulation locally at the posterior pole and enable synthesis of effector proteins necessary for germ cell fate and function (Figure 2A‐E).
2.3. Organization of mRNAs in granules
An estimated 200 mRNAs are specifically enriched in the germ plasm (Figure 1D).34 Most enriched mRNAs reach the posterior and anchor into germ granules during mid‐oogenesis by passive diffusion‐entrapment mechanisms (Figure 3).36 Here, transcriptionally active nurse cells dump their content into a quiescent and growing oocyte. The oocyte forms microtubule bundles, which cause cytoplasmic streaming in the oocyte that swirls the cytoplasm through the oocyte enabling mRNAs to entrap as they brush along the germ granules formed at the posterior pole (Figure 3).36 Deep‐sequencing studies have revealed that the initial entrapment of mRNAs into granules could be mediated by RNA:RNA interactions via short RNAs called piRNAs loaded into the Aubergine protein. In this model, partial complementarity is established between piRNAs and target mRNAs, which is sufficient to initially recruit and anchor a variety of transcripts in germ granules.60 For several mRNAs, the sequences proposed to mediate complementarity between piRNAs and target transcripts reside within the mRNAs' 3′ untranslated regions (UTRs).60 This result agrees well with previous findings using transgenically expressed reporter mRNAs, which demonstrated that sequences necessary and sufficient for posterior localization of nos, gcl and pgc reside within their 3′UTR.42, 43, 71 Additionally, recent high‐resolution microscopy studies have shed new light on the mechanism by which mRNAs such as nos, CycB, gcl and pgc may become highly enriched and organized within germ granules. While individual mRNAs localize as single transcripts, upon enrichment, however, between 2 to more than 40 mRNA molecules derived from the same gene organize into homotypic mRNA clusters within germ granules (Figure 1C).5, 6, 7 Importantly, these homotypic clusters occupy defined positions from the center to the periphery of granules, while the core granule proteins Osk, Vasa, Tud and Aub, that make the granules and recruit other granule components including mRNAs are homogeneously distributed within the same granule.7 These results suggest that the assembly of homotypic clusters is driven at least in part, by the granule mRNAs themselves, possibly through direct RNA‐RNA interactions. In support of a process mediated by RNA‐RNA interactions, mRNAs seem more likely to associate with each other than equally distributing across all granules, leaving some granules devoid of specific mRNA species (Figure 1C).5, 6, 7 Additionally, granule mRNAs display an ability to self‐recruit to granules,5 further supporting the possibility that the RNA‐RNA interactions could play a central role in mRNA enrichment and organization of mRNAs in Drosophila germ granules. Thus, a unified and fascinating theme is emerging from studies of RNA droplets in the fly: that RNA:RNA interactions could play a central role in the enrichment and organization of mRNAs in Drosophila germ granules.
Figure 3.
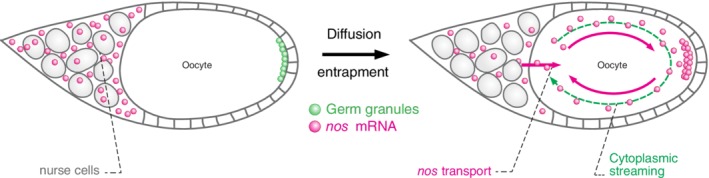
Mechanisms of mRNA enrichment to germ granules. mRNAs such as nos (pink) are transcribed in nurse cells during oogenesis and afterward dumped into a transcriptionally silent oocyte. Cytoplasmic streaming swirls these mRNA through the oocyte, which brush along the germ granules (green) formed at the posterior pole. mRNAs enrich in germ granules passively in a diffusion/entrapment‐dependent mechanism. Figure adapted from Reference 36
mRNA enrichment at the posterior pole within germ granules has important consequences for germ cell development and function. (a) Increased concentration. While only 3% to 4% of the total amount of a given mRNA is localized at the posterior pole, the final concentration in the granule is 8‐ to 10‐fold of that found elsewhere in the embryo.7, 40 (b) mRNA stability. Maternally synthesized mRNA and proteins deposited in the developing egg during oogenesis are degraded in the somatic regions of the embryo during the MZT, when the embryonic genome is activated. Germ plasm mRNAs are stabilized and protected from this degradation and MZT occurs at a later developmental stage in germ cells.46, 72, 73 (c) Translational regulation. A common feature of posteriorly localized mRNAs is that they are translated while the 96% of unlocalized counterparts distributed throughout the embryo that will give rise to future soma, are translationally repressed.42, 71 (d) mRNAs stored in the germ granules are translated temporally in the order of need (Figure 2E).42
For many mRNAs it has been shown that translational repression outside of germ granules depends on the RNA‐binding protein Smaug, which recruits the CCR4/NOT deadenylases to the respective mRNAs.74, 75 For nanos mRNA, translational activation has been linked to germ plasm localization via Oskar, which displaces Smaug from nos thereby preventing the deadenylation of nos CCR4/NOT complex and increasing the stability of the mRNA while simultaneously also relieving translational repression imposed by Smaug.76 It is unlikely that this is a general regulatory mechanism for germ plasm‐localized mRNAs as not all posteriorly localized mRNAs are Smaug targets.46 Additionally, the developmental time when posteriorly localized mRNAs are translated varies from immediately upon localization (nos), to later stages of development when nuclei enter the germ plasm (gcl), when germ cells form (pgc) or when germ cells reach the embryonic gonad (CycB) (Figure 2A,E).42 Curiously, the spatial organization of homotypic mRNA clusters within granules does not predict when an mRNA becomes translated nor how effectively it will be protected from somatic mRNA decay.7 Whether this mRNA self‐organization may influence other aspects of posttranscriptional control of localized transcripts is presently unclear.
2.4. Functions of germ plasm and germ granules
Components enriched in the germ plasm and germ granules control critical germ cell functions. Several of these functions can be directly linked to specific mRNAs, including those required for germ cell fate specification (nos), germ cell formation (gcl), transcriptional silencing (pgc), germ cell migration (tre1) and germ cell survival (wun2). Other germ cell functions, such as mitochondrial inheritance and germ cell genome integrity also rely on maternally synthesized factors. The latter, however, are not linked to a single gene but to the deposition of small RNAs, called piRNAs, which protect germ cells against transposable elements.
2.4.1. Nanos suppresses somatic fate in germ cells
Germ cell specification requires the function of the nanos gene. nanos translation is initiated specifically at the posterior pole as soon as the mRNA becomes localized (Figure 2A,D,E) and the resulting Nanos protein forms a posterior‐to‐anterior gradient. Germ granule localization of nos followed by local translation and formation of the gradient plays a critical role in establishing the anterior‐posterior segmental patterning of the embryo.43 Nanos together with its cofactor Pumilio inhibits the translation of maternal hunchback (hb) mRNA77 and the absence of Hb protein in the posterior region allows the expression of gap genes required for abdomen formation.78, 79 Thus, in the absence of maternally provided Nos, embryos fail to form an abdomen.13 Additionally, failure to localize nos and defects in posterior Nos protein translation account for the abdominal phenotype of the maternal effect genes required for germ plasm assembly (so called posterior group phenotype).13, 79 Nanos is not required for germ cell formation per se, but it is required to preserve a germ cell‐specific cell cycle program. As a consequence, in nanos mutant embryos, pole cells express somatic genes, undergo apoptosis, migrate aberrantly and lose their ability to give rise to functional germ cells.80, 81 How Nanos promotes germ cell specification remains unclear, however, recent experiments have provided some insight into this question. The tumor suppressor L(3)mbt was shown to secure somatic cellular identity in Drosophila ovaries and larval brains by repressing germline‐specific genes, including nanos. 82 Given its well‐documented role in translational regulation, it is likely that the primary role of Nanos is to repress factors that promote somatic development rather than to actively promote germ cell development, akin to its recently reported role in C. elegans. 83 Whether Nanos also has an independent role in actively promoting germ cell fate is less clear.
2.4.2. The GCL protein regulates germ cell formation
Germ cells are the first cells formed in the Drosophila embryo. The early embryo begins its development without cell membranes. Instead, zygotic nuclei undergo rapid, synchronous divisions. Subsequently, nuclei line up along the egg cortex where they are transformed into “bona fide” cells. This occurs in two stages. Nuclei that reach the posterior pole cellularize first and give rise to pole cells, the PGCs, while several divisions later, the remaining nuclei are enveloped by cell membranes and generate all somatic tissues.44, 70, 84, 85 Pole cells form during the 10th nuclear division cycle during the time when GCL translates at the posterior pole (Figure 2A,D,E).42 At this stage, apical, actin‐filled membrane caps form around each nucleus where they basally constrict to separate the future germ cells from the rest of the embryo. GCL protein controls this basal constriction.44 Afterward, cellularized nuclei divide via the canonical anaphase constriction.44 Drosophila therefore exemplifies an extreme case of how germ line‐soma dichotomy is achieved. Not only are the two cell populations specified at different times during development, but also the cellular events leading to germ cell and somatic cell formation are strikingly different and are controlled by separate sets of genes. For example, germ cell formation and specification rely on maternally synthesized factors and occur even when zygotic mRNA transcription is blocked.44
A rate‐limiting component specific to pole cell formation is the Germ‐cell‐less protein. GCL is a BTB/BACK domain protein with homologs found from nematodes to humans and distinguished by the conserved GCL motif in the C‐terminal region.45 GCL is not directly involved in transcriptional silencing or centrosome segregation during germ cell formation as previously suggested.44, 86, 87 Instead, GCL functions as a substrate‐specific adaptor for the Cullin 3‐Ubiquitin‐ligase complex.45 GCL targets the receptor tyrosine kinase (RTK) Torso for degradation via the conserved GCL substrate‐binding motif. During pole cell formation, GCL translocates from the inner nuclear membrane to the cell membrane where it leads to the degradation of Torso. Amazingly, pole cell formation is fully restored in gcl, torso double mutants, indicating that Torso is the single critical target of GCL in the early embryo and involved in the formation of basal constriction promoted by GCL.44, 45 How Torso activation inhibits this constriction is not fully understood. Preliminary studies suggest that this process is independent of the conventional downstream components of the Torso RTK including MAP kinase‐mediated transcriptional activation of target genes.45 These studies have also suggested that additional, yet unknown germ plasm‐localized factors can promote basal furrow constrictions in the absence of gcl and torso.
2.4.3. Pgc represses transcription in germ cells
Major transcriptional activity in germ cells is not observed until they reach the gonad, although PGCs become transcriptionally competent shortly after they form.88 Polar granule component (Pgc) protein, which translates once PGCs cellularize (Figure 2A,D,E),42 is required for transcriptional silencing of primordial germ cells. Pgc mRNA was first suggested to encode a noncoding RNA, however, later studies demonstrated that instead it encodes a 71 amino‐acid peptide89, 90 This peptide blocks transcriptional elongation by inhibiting transcription elongation factor b (P‐TEFb).90 P‐TEFb promotes transcriptional elongation by phosphorylating the carboxy‐terminal repeat domain (CTD) of RNA polymerase II (RNAPII) at the Serine 2 position. In embryos derived from pgc mutant mothers, pole cells form but they now transcribe genes that are otherwise expressed only in somatic cells.91 As a result, maternally deposited, germ cell‐specific components are lost due to precocious activation of the MZT that at this developmental stage normally only takes place in the soma.92 Consequently, germ cells in pgc mutant embryos are unable to migrate during later developmental stages and instead undergo apoptosis.91 Thus, transcriptional silencing in germ cells maintains the germ line‐soma dichotomy by preventing the somatic program unfolding in primordial germ cells.
2.4.4. Tre1 and Wunen regulate germ cell survival and migration
Germ cells form at the posterior pole, and subsequently move toward the somatic tissues of the gonad, which are specified in the mesoderm of the abdomen. Successful survival and migration of germ cells is linked to reproductive success of the offspring. The number of germ cells formed depends on the overall amount of germ plasm. For example, reduction in the germ plasm organizer Osk reduces germ cell number while an increase in Oskar has the opposite effect.15, 93 The amount of germ plasm inherited is graded from the center of the posterior tip to the periphery: germ cells in the center inherit more germ plasm than those at the periphery. Not all germ cells reach the gonad and about 35% to 45% of germ cells are eliminated during migration. The ability to survive and reach the gonad is directly correlated with the amount of germ plasm inherited.94 Indeed, the levels of a single germ plasm‐localized mRNA, Wunen 2, which encodes a protein that is a homolog of mammalian lipid‐phosphate phosphatase 3, was shown to be a quantitative regulator of germ cell survival.94 Another regulator of germ cell migration, the G‐protein‐coupled receptor Tre1 is also synthesized maternally and deposited as a localized mRNA to the germ plasm and translated there. Tre1 receptor activation orients germ cells as they exit the gut and directs their migration toward the somatic gonad.95Thus, heterogeneity of germ plasm, inherited at the time primordial germ cells form, predetermines the success of these cells for future generations.
2.4.5. Mitochondria and germ plasm
The transmission of mitochondria through the germ line is an essential component of maternal inheritance. Mitochondria are passed from the mother to the progeny in most organisms and specific mechanisms exist to eliminate paternal mitochondria.96 Consequently, the mitochondria of PGCs will constitute the pool from which all mitochondria of the next generation originate. The original EM images suggested that germ granules and mitochondria could closely associate in the germ plasm.68 Subsequently, it was proposed that mitochondrial large and small ribosomal RNAs (mtrRNA), which are transcribed by the mitochondrial genome, were extra‐mitochondrial in the germ plasm and associated with germ granules.97 It was hypothesized that these extra‐mitochondrial mtrRNAs induced the assembly of “mitochondrial‐like” ribosomes within the germ plasm and directed translation of germ plasm mRNAs with mitochondrial codon usage.98, 99 While intriguing, recent high‐resolution imaging showed that mtrRNA localizes separate from germ granule markers such as nanos mRNA and is strictly confined to the mitochondria at the posterior pole as well as elsewhere in the embryo.48 This study shows that mitochondria are enriched at the posterior pole, consistent with the previous EM observations. Closer examination of the mechanisms by which mitochondria become enriched at the posterior pole showed that this localization requires the Long Osk (Figure 1D) and is independent of the germ plasm inducing function of Short Osk.48 Both Long Osk as well as just the 138 aa N‐terminal extension of Oskar, which distinguishes Long Osk from Short Osk, are sufficient to localize mitochondria to an ectopic location.48 Previous studies have suggested that Long Osk enhances the cortical recruitment and maintenance of osk mRNA at the posterior pole by mediating the formation of actin filaments and recruitment of endosomes.19, 100 The new studies further show that Long Osk also directly associates with actin cytoskeleton and that this function is critical for mitochondrial enrichment at the posterior pole.48 The functional significance of this mitochondrial enrichment in PGCs is unclear. Germ cells form in the absence of Long Osk on condition that Short Osk is provided separately, however, their number is reduced and they contain fewer mitochondria. This reduction in PGC number could be an indirect consequence of loss of Long Osk diminishing the ability of osk mRNA to anchor at the cortex, thereby translating less Short Osk and forming fewer germ granules. Alternatively, this finding may suggest a more direct requirement for increased mitochondrial activity during PGC formation. The later idea is supported by experiments that interfered with prokaryotic (and mitochondrial) translation specifically at the posterior pole and resulted in a reduction in germ cell number.99
Beyond a potential functional role during the formation of germ cells, enrichment of mitochondria could impact germ cell development in other ways. Germ granules are closely associated with mitochondria (Figure 1B) throughout their lifecycle and this association may be important for the success of germ cells. Recent experiments using heteroplasmic flies that simultaneously harbor wild‐type mitochondrial DNA and mutant DNA defective in mitochondrial oxidative phosphorylation showed that functional mitochondria are actively selected during early oogenesis.101 Thus, enrichment of mitochondria in germ plasm at the PGC stage may widen the bottle neck and increase the pool size from which good mitochondria can be selected for inheritance at a later developmental stage.
2.4.6. Germ granules and the control of transposable elements through germline the life cycle
Transposable elements (TE) account for about one‐third of the Drosophila genome. Their activity is regulated in the germline by a gonad‐specific subclass of Argonaute proteins, called Piwi, Argonaute 3 and Aubergine, and the Piwi‐interacting (Pi) small RNAs (piRNAs). Immunity to TEs is transmitted maternally through the deposition of piRNAs in the germ plasm. Together with Piwi, maternally inherited piRNAs repress the transcription of TEs by recruiting heterochromatin to TE loci.102, 103, 104 With Aubergine, piRNAs slice TE transcripts in the cytoplasm and initiate an amplification loop, which generates new piRNAs in the sense and antisense orientation. According to the “ping‐pong” model, sense‐piRNAs and TE transcripts fuel the production of new TE‐complementary piRNAs, thereby providing continued and adaptable immunity against TE activity.105 Recent studies using immunoprecipitation followed by RNA sequencing suggested that in addition to targeting TEs, maternally deposited piRNAs associated with Aubergine and Tudor proteins also enriched for germ plasm localized mRNA.60 In this scenario, piRNA‐mRNA recognition would trap mRNAs in the germ plasm. In these experiments, piRNA‐mRNA pairs did not show any sequence specificity for germ plasm localized RNAs and it was therefore proposed that enrichment of localized mRNAs is achieved by special mRNA features such as longer 3′UTRs.60 While intriguing, other studies have suggested that Aubergine may bind directly and preferentially to germ plasm‐enriched mRNAs to stabilize them. In contrast, in the soma Aubergine/mRNA interactions lead to mRNA degradation.106, 107
3. CONCLUSION
Germ granules in Drosophila are the hubs for RNA biology: they enrich and posttranscriptionally regulate a subset of mRNAs crucial for the development of the germline to ensure fecundity of the offspring. A century of experimentation described Drosophila germ granules as electron dense, amorphous and nonmembrane bound organelles. Genetic analysis identified critical proteins and mRNAs enriched in germ granules, revealed their activity and regulatory mechanisms. Most recent biophysical studies have shown that germ granules form by phase separation. Yet, fundamental questions remain unaddressed. Is germ plasm functional or is the germline‐inducing activity restricted to germ granules? If so, what is its function? Is the composition of germ plasm different from the composition of germ granules? Aside from mRNA enrichment and storage, what additional functions, if any, do germ granules have, perhaps in regulating posttranscriptional reactions? How do homotypic mRNA clusters form in germ granules and how does their organization contribute to their biological function? What are the conserved and unique principles of composition, structure and function of germ granules compared to other droplets formed by phase separation? New quantitative, super‐resolution imaging studies combined with biochemistry and genetics are providing a framework to begin addressing some of these questions. These methods allow us to dissect the function of germ granules and explore how concentration of protein and mRNA components in granules establishes a highly specialized program that controls posttranscriptional regulation of mRNAs required for many aspects of early germ cell biology. As for the past century, with modern technology and approaches these mysterious condensates will continue to fascinate us.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
ACKNOWLEDGMENTS
We would like to acknowledge NYULH Microscopy Lab for providing electron microscopy services, whose resources are partially supported by Cancer Center Support Grant P30 CA016087‐38 (Laura and Isaac Perlmutter Cancer Center). This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development K99HD088675 grant awarded to T.T. RL is an HHMI investigator.
Trcek T, Lehmann R. Germ granules in Drosophila . Traffic. 2019;20:650–660. 10.1111/tra.12674
Present address Tatjana Trcek, Department of Biology, Johns Hopkins University.
Funding information Eunice Kennedy Shriver National Institute of Child Health and Human Development, Grant/Award Number: K99HD088675; Laura and Isaac Perlmutter Cancer Center
Peer Review The peer review history for this article is available at https://publons.com/publon/10.1111/tra.12674/
REFERENCES
- 1. Weismann A. Das Keimplasma: Eine Theorie der Vererbung, 1892. Translated by W. Newton Parker and Harriet Rönnfeldt as the Germ‐Plasm: a Theory of Heredity. New York, NY: Scribner; 1893. [Google Scholar]
- 2. Hegner RW. Effects of removing germ‐cell determinants from the eggs of some chrysomelid beetles. Preliminary report. Biological Bull. 1908;16:19‐26. [Google Scholar]
- 3. Hegner RW. Germ‐cell determinants and their significance. Am Nat. 1911;45(535):385‐397. [Google Scholar]
- 4. Lehmann R. Germ plasm biogenesis‐an Oskar‐centric perspective. Curr Top Dev Biol. 2016;116(pt A):679‐707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Niepielko MG, Eagle WVI, Gavis ER. Stochastic seeding coupled with mRNA self‐recruitment generates heterogeneous Drosophila germ granules. Curr Biol. 2018;28(12):1872‐1881.e1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Little SC, Sinsimer KS, Lee JJ, Wieschaus EF, Gavis ER. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat Cell Biol. 2015;17(5):558‐568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun. 2015;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mahowald AP. Fine structure of pole cells and polar granules in Drosophila melanogaster . J Exp Zool. 1962;151(3):201. [Google Scholar]
- 9. Mahowald AP. Polar granules of Drosophila. II. Ultrastructural changes during early embryogenesis. J Exp Zool. 1968;167(2):237‐261. [DOI] [PubMed] [Google Scholar]
- 10. Mahowald AP, Illmensee K, Turner FR. Interspecific transplantation of polar plasm between Drosophila embryos. J Cell Biol. 1976;70(2 pt 1):358‐373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci USA. 1974;71(4):1016‐1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boswell RE, Mahowald AP. Tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster . Cell. 1985;43(1):97‐104. [DOI] [PubMed] [Google Scholar]
- 13. Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 1989;121(1):101‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lehmann R, Nusslein‐Volhard C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell. 1986;47(1):141‐152. [DOI] [PubMed] [Google Scholar]
- 15. Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358(6385):387‐392. [DOI] [PubMed] [Google Scholar]
- 16. Ahuja A, Extavour CG. Patterns of molecular evolution of the germ line specification gene oskar suggest that a novel domain may contribute to functional divergence in Drosophila. Dev Genes Evol. 2014;224(2):65‐77. [DOI] [PubMed] [Google Scholar]
- 17. Bullock SL, Ish‐Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414(6864):611‐616. [DOI] [PubMed] [Google Scholar]
- 18. Sinsimer KS, Lee JJ, Thiberge SY, Gavis ER. Germ plasm anchoring is a dynamic state that requires persistent trafficking. Cell Rep. 2013;5(5):1169‐1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vanzo N, Oprins A, Xanthakis D, Ephrussi A, Rabouille C. Stimulation of endocytosis and actin dynamics by Oskar polarizes the Drosophila oocyte. Dev Cell. 2007;12(4):543‐555. [DOI] [PubMed] [Google Scholar]
- 20. Hachet O, Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr Biol. 2001;11(21):1666‐1674. [DOI] [PubMed] [Google Scholar]
- 21. Mohr SE, Dillon ST, Boswell RE. The RNA‐binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 2001;15(21):2886‐2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newmark PA, Boswell RE. The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development. 1994;120(5):1303‐1313. [DOI] [PubMed] [Google Scholar]
- 23. Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII‐containing complex required for mRNA localization and nonsense‐mediated mRNA decay. Nature. 2004;427(6976):753‐757. [DOI] [PubMed] [Google Scholar]
- 24. van Eeden FJ, Palacios IM, Petronczki M, Weston MJ, St Johnston D. Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J Cell Biol. 2001;154(3):511‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428(6986):959‐963. [DOI] [PubMed] [Google Scholar]
- 26. Castagnetti S, Hentze MW, Ephrussi A, Gebauer F. Control of oskar mRNA translation by Bruno in a novel cell‐free system from Drosophila ovaries. Development. 2000;127(5):1063‐1068. [DOI] [PubMed] [Google Scholar]
- 27. Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124(3):521‐533. [DOI] [PubMed] [Google Scholar]
- 28. Kim G, Pai CI, Sato K, Person MD, Nakamura A, Macdonald PM. Region‐specific activation of oskar mRNA translation by inhibition of Bruno‐mediated repression. PLoS Genet. 2015;11(2):e1004992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erdelyi M, Michon AM, Guichet A, Glotzer JB, Ephrussi A. Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature. 1995;377(6549):524‐527. [DOI] [PubMed] [Google Scholar]
- 30. Gaspar I, Sysoev V, Komissarov A, Ephrussi A. An RNA‐binding atypical tropomyosin recruits kinesin‐1 dynamically to oskar mRNPs. EMBO J. 2017;36(3):319‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Veeranan‐Karmegam R, Boggupalli DP, Liu G, Gonsalvez GB. A new isoform of Drosophila non‐muscle tropomyosin 1 interacts with Kinesin‐1 and functions in oskar mRNA localization. J Cell Sci. 2016;129(22):4252‐4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Glotzer JB, Saffrich R, Glotzer M, Ephrussi A. Cytoplasmic flows localize injected oskar RNA in Drosophila oocytes. Curr Biol. 1997;7(5):326‐337. [DOI] [PubMed] [Google Scholar]
- 33. Snee MJ, Harrison D, Yan N, Macdonald PM. A late phase of Oskar accumulation is crucial for posterior patterning of the Drosophila embryo, and is blocked by ectopic expression of Bruno. Differentiation. 2007;75(3):246‐255. [DOI] [PubMed] [Google Scholar]
- 34. Frise E, Hammonds AS, Celniker SE. Systematic image‐driven analysis of the spatial Drosophila embryonic expression landscape. Mol Syst Biol. 2010;6:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, Tomancak P. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13(14):1159‐1168. [DOI] [PubMed] [Google Scholar]
- 37. Voronina E, Seydoux G, Sassone‐Corsi P, Nagamori I. RNA granules in germ cells. Csh Perspect Biol. 2011;3(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kistler KE, Trcek T, Hurd TR, et al. Phase transitioned nuclear Oskar promotes cell division of Drosophila primordial germ cells. elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vanzo NF, Ephrussi A. Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development. 2002;129(15):3705‐3714. [DOI] [PubMed] [Google Scholar]
- 40. Bergsten SE, Gavis ER. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development. 1999;126(4):659‐669. [DOI] [PubMed] [Google Scholar]
- 41. Lerit DA, Gavis ER. Transport of germ plasm on astral microtubules directs germ cell development in Drosophila. Curr Biol. 2011;21(6):439‐448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rangan P, DeGennaro M, Jaime‐Bustamante K, Coux RX, Martinho RG, Lehmann R. Temporal and spatial control of germ‐plasm RNAs. Curr Biol. 2009;19(1):72‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71(2):301‐313. [DOI] [PubMed] [Google Scholar]
- 44. Cinalli RM, Lehmann R. A spindle‐independent cleavage pathway controls germ cell formation in Drosophila. Nat Cell Biol. 2013;15(7):839‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pae J, Cinalli RM, Marzio A, Pagano M, Lehmann R. GCL and CUL3 control the switch between cell lineages by mediating localized degradation of an RTK. Dev Cell. 2017;42(2):130‐142.e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Siddiqui NU, Li X, Luo H, et al. Genome‐wide analysis of the maternal‐to‐zygotic transition in Drosophila primordial germ cells. Genome Biol. 2012;13(2):R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Markussen FH, Michon AM, Breitwieser W, Ephrussi A. Translational control of Oskar generates short Osk, the isoform that induces pole plasm assembly. Development. 1995;121(11):3723‐3732. [DOI] [PubMed] [Google Scholar]
- 48. Hurd TR, Herrmann B, Sauerwald J, Sanny J, Grosch M, Lehmann R. Long Oskar controls mitochondrial inheritance in Drosophila melanogaster . Dev Cell. 2016;39(5):560‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Breitwieser W, Markussen FH, Horstmann H, Ephrussi A. Oskar protein interaction with vasa represents an essential step in polar granule assembly. Genes Dev. 1996;10(17):2179‐2188. [DOI] [PubMed] [Google Scholar]
- 50. Jones JR, Macdonald PM. Oskar controls morphology of polar granules and nuclear bodies in Drosophila. Development. 2007;134(2):233‐236. [DOI] [PubMed] [Google Scholar]
- 51. Arkov AL, Wang JY, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133(20):4053‐4062. [DOI] [PubMed] [Google Scholar]
- 52. Callebaut I, Mornon JP. LOTUS, a new domain associated with small RNA pathways in the germline. Bioinformatics. 2010;26(9):1140‐1144. [DOI] [PubMed] [Google Scholar]
- 53. Anantharaman V, Zhang D, Aravind L. OST‐HTH: a novel predicted RNA‐binding domain. Biol Direct. 2010;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jeske M, Bordi M, Glatt S, et al. The crystal structure of the Drosophila germline inducer Oskar identifies two domains with distinct vasa helicase‐ and RNA‐binding activities. Cell Rep. 2015;12(4):587‐598. [DOI] [PubMed] [Google Scholar]
- 55. Jeske M, Muller CW, Ephrussi A. The LOTUS domain is a conserved DEAD‐box RNA helicase regulator essential for the recruitment of vasa to the germ plasm and nuage. Genes Dev. 2017;31(9):939‐952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP‐dependent helicases. Cell. 1988;55(4):577‐587. [DOI] [PubMed] [Google Scholar]
- 57. Yang N, Yu Z, Hu M, Wang M, Lehmann R, Xu RM. Structure of Drosophila Oskar reveals a novel RNA binding protein. Proc Natl Acad Sci USA. 2015;112(37):11541‐11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40(3):164‐170. [DOI] [PubMed] [Google Scholar]
- 59. Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128(14):2823‐2832. [DOI] [PubMed] [Google Scholar]
- 60. Vourekas A, Alexiou P, Vrettos N, Maragkakis M, Mourelatos Z. Sequence‐dependent but not sequence‐specific piRNA adhesion traps mRNAs to the germ plasm. Nature. 2016;531(7594):390‐394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kirino Y, Vourekas A, Sayed N, et al. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2010;16(1):70‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu H, Wang JY, Huang Y, et al. Structural basis for methylarginine‐dependent recognition of Aubergine by Tudor. Genes Dev. 2010;24(17):1876‐1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kirino Y, Vourekas A, Kim N, et al. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem. 2010;285(11):8148‐8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? EMBO J. 2016;35(15):1603‐1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Banani SF, Rice AM, Peeples WB, et al. Compositional control of phase‐separated cellular bodies. Cell. 2016;166(3):651‐663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lin Y, Protter DS, Rosen MK, Parker R. Formation and maturation of phase‐separated liquid droplets by RNA‐binding proteins. Mol Cell. 2015;60(2):208‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Protter DSW, Rao BS, Van Treeck B, et al. Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep. 2018;22(6):1401‐1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Illmensee K, Mahowald AP, Loomis MR. The ontogeny of germ plasm during oogenesis in Drosophila. Dev Biol. 1976;49(1):40‐65. [DOI] [PubMed] [Google Scholar]
- 69. Spradling A. Developmental Genetics of Oogenesis in the Development of Drosophila melanogaster. New York: Cold Spring Harbor Laboratory Press; 1993:1‐70. [Google Scholar]
- 70. Su TT, Campbell SD, O'Farrell PH. The cell cycle program in germ cells of the Drosophila embryo. Dev Biol. 1998;196(2):160‐170. [DOI] [PubMed] [Google Scholar]
- 71. Gavis ER, Lehmann R. Translational regulation of nanos by RNA localization. Nature. 1994;369(6478):315‐318. [DOI] [PubMed] [Google Scholar]
- 72. Thomsen S, Anders S, Janga SC, Huber W, Alonso CR. Genome‐wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 2010;11(9):R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bashirullah A, Halsell SR, Cooperstock RL, et al. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster . EMBO J. 1999;18(9):2610‐2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen L, Dumelie JG, Li X, et al. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA‐binding protein. Genome Biol. 2014;15(1):R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15(4):284‐294. [DOI] [PubMed] [Google Scholar]
- 76. Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133(22):4573‐4583. [DOI] [PubMed] [Google Scholar]
- 77. Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80(5):747‐756. [DOI] [PubMed] [Google Scholar]
- 78. Lehmann R, Nusslein‐Volhard C. Hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol. 1987;119(2):402‐417. [DOI] [PubMed] [Google Scholar]
- 79. Lehmann R, Nusslein‐Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112(3):679‐691. [DOI] [PubMed] [Google Scholar]
- 80. Asaoka‐Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol. 1999;1(7):431‐437. [DOI] [PubMed] [Google Scholar]
- 81. Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125(4):679‐690. [DOI] [PubMed] [Google Scholar]
- 82. Coux RX, Teixeira FK, Lehmann R. L(3)mbt and the LINT complex safeguard cellular identity in the Drosophila ovary. Development. 2018;145(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee CS, Lu T, Seydoux G. Nanos promotes epigenetic reprograming of the germline by down‐regulation of the THAP transcription factor LIN‐15B. elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31‐70. [DOI] [PubMed] [Google Scholar]
- 85. Campos‐Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Berlin, Germany: Springer‐Verlag; 1985. [Google Scholar]
- 86. Leatherman JL, Levin L, Boero J, Jongens TA. Germ cell‐less acts to repress transcription during the establishment of the Drosophila germ cell lineage. Curr Biol. 2002;12(19):1681‐1685. [DOI] [PubMed] [Google Scholar]
- 87. Lerit DA, Shebelut CW, Lawlor KJ, et al. Germ cell‐less promotes centrosome segregation to induce germ cell formation. Cell Rep. 2017;18(4):831‐839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8(4):243‐246. [DOI] [PubMed] [Google Scholar]
- 89. Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274(5295):2075‐2079. [DOI] [PubMed] [Google Scholar]
- 90. Hanyu‐Nakamura K, Sonobe‐Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P‐TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451(7179):730‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII‐dependent transcription in primordial germ cells. Curr Biol. 2004;14(2):159‐165. [DOI] [PubMed] [Google Scholar]
- 92. Hanyu‐Nakamura K, Matsuda K, Cohen SM, Nakamura A. Pgc suppresses the zygotically‐acting RNA decay pathway to protect germ plasm RNAs in the Drosophila embryo. Development. 2019;167(7). [DOI] [PubMed] [Google Scholar]
- 93. Smith JL, Wilson JE, Macdonald PM. Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell. 1992;70(5):849‐859. [DOI] [PubMed] [Google Scholar]
- 94. Slaidina M, Lehmann R. Quantitative differences in a single maternal factor determine survival probabilities among Drosophila germ cells. Curr Biol. 2017;27(2):291‐297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. LeBlanc MG, Lehmann R. Domain‐specific control of germ cell polarity and migration by multifunction Tre1 GPCR. J Cell Biol. 2017;216(9):2945‐2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yu Z, O'Farrell PH, Yakubovich N, DeLuca SZ. The mitochondrial DNA polymerase promotes elimination of paternal mitochondrial genomes. Curr Biol. 2017;27(7):1033‐1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kobayashi S, Amikura R, Okada M. Presence of mitochondrial large ribosomal RNA outside mitochondria in germ plasm of Drosophila melanogaster . Science. 1993;260(5113):1521‐1524. [DOI] [PubMed] [Google Scholar]
- 98. Amikura R, Kashikawa M, Nakamura A, Kobayashi S. Presence of mitochondria‐type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc Natl Acad Sci USA. 2001;98(16):9133‐9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Amikura R, Sato K, Kobayashi S. Role of mitochondrial ribosome‐dependent translation in germline formation in Drosophila embryos. Mech Dev. 2005;122(10):1087‐1093. [DOI] [PubMed] [Google Scholar]
- 100. Tanaka T, Nakamura A. The endocytic pathway acts downstream of Oskar in Drosophila germ plasm assembly. Development. 2008;135(6):1107‐1117. [DOI] [PubMed] [Google Scholar]
- 101. Lieber T, Jeedigunta SP, Palozzi JM, Lehmann R, Hurd TR. Mitochondrial fragmentation drives selective removal of deleterious mtDNA in the germline. Nature. 2019;570:380‐384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yu Y, Gu J, Jin Y, et al. Panoramix enforces piRNA‐dependent cotranscriptional silencing. Science. 2015;350(6258):339‐342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sienski G, Batki J, Senti KA, et al. Silencio/CG9754 connects the Piwi‐piRNA complex to the cellular heterochromatin machinery. Genes Dev. 2015;29(21):2258‐2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Teixeira FK, Okuniewska M, Malone CD, Coux RX, Rio DC, Lehmann R. piRNA‐mediated regulation of transposon alternative splicing in the soma and germ line. Nature. 2017;552(7684):268‐272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Aravin AA, Hannon GJ, Brennecke J. The Piwi‐piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761‐764. [DOI] [PubMed] [Google Scholar]
- 106. Barckmann B, Pierson S, Dufourt J, et al. Aubergine iCLIP reveals piRNA‐dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep. 2015;12(7):1205‐1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Dufourt J, Bontonou G, Chartier A, et al. piRNAs and Aubergine cooperate with Wispy poly(A) polymerase to stabilize mRNAs in the germ plasm. Nat Commun. 2017;8(1):1305. [DOI] [PMC free article] [PubMed] [Google Scholar]


