Summary
Background
Loop‐mediated isothermal amplification (LAMP) assays, which operate at a single temperature and require no postreaction processing, have been described for rapid species‐specific detection of numerous fungi. The technology has much less commonly been applied to identification of other key genetic traits such as fungicide resistance, and has not yet been applied to mating‐type determination in any fungus.
Objectives
To develop first LAMP assays for mating‐type identification in a fungus, in this instance with the saprophytic mould and human opportunistic pathogen Aspergillus fumigatus, a heterothallic ascomycete requiring isolates of opposite mating type (MAT1‐1, MAT1‐2) for sexual reproduction.
Methods
New LAMP primer sets, targeted to MAT gene sequences, were screened against 34 A fumigatus isolates (of known mating type) from diverse clinical, environmental and geographic sources to establish whether they could distinguish MAT1‐1 or MAT1‐2 genotypes.
Results and conclusions
The new assays, operating at a single temperature of 65°C, correctly identified the mating type of A fumigatus isolates in <20 minutes, and thus have numerous research and practical applications. Similar MAT LAMP assays could now be developed for other fungi of agricultural, environmental, industrial and/or medical importance.
Keywords: Aspergillus fumigatus, diagnostics, fungal pathogen, mating type, sexual reproduction
1. INTRODUCTION
The fungus Aspergillus fumigatus is a saprophytic mould commonly found on plant debris and in soil. It is also an opportunistic human pathogen causing allergic symptoms and life‐threatening invasive infections. The incidence of invasive aspergillosis (IA) has been increasing in recent years largely due to increased numbers of immunocompromised individuals in the population unable to fight off infection.1 For more than 145 years, A fumigatus was only known to reproduce asexually, although several signatures of cryptic sexuality were present, for example, presence and expression of mating (MAT) genes and evidence of gene recombination within natural populations.2 However, the breakthrough 2009 discovery of a functional sexual cycle3 had several implications including: (a) potentially explaining high genotypic diversity observed in populations; (b) production of sexually derived airborne ascospores possibly more resilient to unfavourable environmental conditions; and (c) generation of sexual progeny with potentially greater pathogenicity and/or reduced sensitivity to fungicides.4 Aspergillus fumigatus possesses a heterothallic (obligate outbreeding) mating system, with highly dissimilar stretches of DNA, termed “idiomorphs,” present in isolates of opposite mating type as is characteristic for heterothallic ascomycete species.5 Thus, MAT1‐1 isolates contain an alpha‐domain MAT1‐1‐1 gene whereas MAT1‐2 isolates contain a high‐mobility group MAT1‐2‐1 gene together with a recently described MAT1‐2‐4 gene.6 A multiplex PCR‐based assay for determination of mating type has previously been developed for A fumigatus.2
More recently, loop‐mediated isothermal amplification (LAMP) assays have become increasingly used for rapid species‐specific detection of numerous fungi, including A fumigatus.7 LAMP technology, first described by in 2000,8 typically involves 4‐6 primers in each reaction and has several purported advantages over PCR‐based diagnostics. These include faster reaction times, potentially improved sensitivity and specificity, increased tolerance of sample inhibitors, no requirement for additional postreaction processing (eg, resolving PCR products on agarose gels) and use of only a single constant reaction temperature thus raising the possibility of field‐based detection. Despite these advantages, LAMP assays have much less commonly been applied to detection of other key genetic traits such as fungicide resistance, one recent example being an assay targeted to a 34 bp tandem repeat in the cyp51A gene that has been associated with azole resistance in A fumigatus.9 To date, however, LAMP assays have not been used for rapid detection of different mating types in fungi. The objective of the present study was therefore to develop and evaluate for the first time whether LAMP assays could be used for the rapid identification of mating type in a fungus, with a focus here on the human opportunistic pathogen A fumigatus.
2. METHODS
2.1. Ethics statement
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required as the research in this article related to micro‐organisms.
2.2. Fungal isolates, DNA extraction and initial molecular characterisation
Details of A fumigatus isolates, including source material and geographic origin, are given in Table 2; all isolates are maintained as −80°C glycerol stocks at Rothamsted Research, UK. Genomic DNA was extracted from A fumigatus spores, harvested from one‐week old cultures grown on Sabouraud dextrose agar (Lab M Ltd) at 37°C, using a MasterPure yeast DNA purification kit (Epicentre) into a final volume of 100 μL TE buffer. DNA was quantified via nanodrop spectrophotometer and diluted to 10 ng/ μL using PCR‐grade water. The mating type of these isolates was first determined using the published multiplex PCR assay (Table 1).2 Amplicons were resolved on agarose gels, with 834 bp or 438 bp products amplified from MAT1‐1 or MAT1‐2 isolates, respectively.
Table 2.
Validation of new Aspergillus fumigatus MAT LAMP assays by screening of isolates from diverse environmental sources and geographic localities
| Isolate | Source | Origin | MAT type (multiplex PCR)[Link] | LAMP detection time (min) | |
|---|---|---|---|---|---|
| MAT1‐1 assay | MAT1‐2 assay | ||||
| 47‐255 | Clinical | Europe | MAT1‐1 | 10‐11 | Negative |
| 47‐257 | Clinical | Europe | MAT1‐1 | 9‐10 | Negative |
| 47‐258 | Clinical | Europe | MAT1‐1 | 9‐10 | Negative |
| 47‐2 | Clinical | North America | MAT1‐1 | 9‐10 | Negative |
| C6‐UT1 | Food | Asia | MAT1‐1 | 15‐16 | Negative |
| C1‐2‐UT3 | Food | South America | MAT1‐1 | 9‐10 | Negative |
| C3‐UT1 | Food | South America | MAT1‐1 | 9‐10 | Negative |
| C3‐UT3 | Food | South America | MAT1‐1 | 9‐10 | Negative |
| O5‐5 | Plant | Africa | MAT1‐1 | 9‐10 | Negative |
| 18‐C6‐9 | Plant | Europe | MAT1‐1 | 9‐10 | Negative |
| 18‐C7‐8 | Plant | Europe | MAT1‐1 | 8‐9 | Negative |
| O9‐8 | Plant | Europe | MAT1‐1 | 12‐13 | Negative |
| T4‐1 | Plant | Europe | MAT1‐1 | 15‐16 | Negative |
| G4‐1 | Plant | South America | MAT1‐1 | 8‐9 | Negative |
| O10‐1 | Plant | South America | MAT1‐1 | 19‐20 | Negative |
| 1‐2.2‐B1 | Soil | Europe | MAT1‐1 | 9‐10 | Negative |
| 1‐2.2‐B2 | Soil | Europe | MAT1‐1 | 9‐10 | Negative |
| STNL1‐B1 | Soil | Europe | MAT1‐1 | 9‐10 | Negative |
| STNL1‐A8 | Soil | Europe | MAT1‐1 | 9‐10 | Negative |
| SWG1‐A9 | Soil | Europe | MAT1‐1 | 12‐13 | Negative |
| BKCb‐1 | Air | Europe | MAT1‐2 | Negative | 9‐10 |
| 47‐246 | Clinical | Europe | MAT1‐2 | Negative | 8‐9 |
| Af65 | Clinical | Europe | MAT1‐2 | Negative | 10‐11 |
| Af293 | Clinical | Europe | MAT1‐2 | Negative | 9‐10 |
| C5‐T8 | Food | Africa | MAT1‐2 | Negative | 9‐10 |
| 15‐37‐1 | Food | Asia | MAT1‐2 | Negative | 9‐10 |
| C1‐1‐T3 | Food | South America | MAT1‐2 | Negative | 9‐10 |
| C7‐T2 | Food | South America | MAT1‐2 | Negative | 9‐10 |
| C7‐UT1 | Food | South America | MAT1‐2 | Negative | 8‐9 |
| G2‐2 | Plant | Europe | MAT1‐2 | Negative | 7‐8 |
| SWF5‐C6 | Soil | Europe | MAT1‐2 | Negative | 7‐8 |
| PG1‐5 | Soil | Europe | MAT1‐2 | Negative | 9‐10 |
| WSN19‐3 | Soil | Europe | MAT1‐2 | Negative | 9‐10 |
| SWUK5‐A9 | Soil | Europe | MAT1‐2 | Negative | 8‐9 |
Determined by mating multiplex PCR assay.2
Table 1.
Primer sets used in the present study
| Purpose/ Primer name | Primer sequence (5′ – 3′) | Source |
|---|---|---|
| New MAT1‐1‐specific LAMP assay: | ||
| AFMAT1F3 | CGGTTGGCGATATCGTGAA | Present study |
| AFMAT1B3 | GCCATCTGTCTCTTCAGGAG | |
| AFMAT1FIP | CAGCGAAGGCCATTGTGGAAGTTACTGGCTACGTGTCTGAGA | |
| AFMAT1BIP | ACGGCATTCAGATCACTGGCGCCACTTCAGGAGTTGCGAA | |
| AFMAT1LOOPF | TTGGTCCGTTCGTGTGGC | |
| AFMAT1LOOPB | ACGATGCCATTGTGACTGAC | |
| New MAT1‐2‐specific LAMP assay: | ||
| AFMAT2F3 | CCCGTCTTGGGTAAGTGTCT | Present study |
| AFMAT2B3 | GTGCGAAGGACTCAGTTACG | |
| AFMAT2FIP | CAACAGGTGCGCCAATGAGTGAGAGTTCCTCCTGAGCTTGA | |
| AFMAT2BIP | GCTCTCCGTGTTATGCGTACCCCAGCTTCACCGTGAGATGC | |
| AFMAT2LOOPF | CACTGTCATTCCGTGTTATCGG | |
| AFMAT2LOOPB | CAGCTTTTTCCGGAACAGCT | |
| Multiplex PCR mating‐type assay: | ||
| AFM1 | CCTTGACGCGATGGGGTGG | 2 |
| AFM2 | CGCTCCTCATCAGAACAACTCG | |
| AFM3 | CGGAAATCTGATGTCGCCACG | |
2.3. Design and validation of MAT LAMP assays
For the MAT1‐1 LAMP assay, MAT idiomorph sequence was downloaded from GenBank (Accession: AY898661 2), with LAMP primers targeted to the internal MAT1‐1‐1 gene. For the MAT1‐2 LAMP assay, MAT idiomorph sequence was sourced from the A fumigatus Ensembl genome (isolate AF293; gene ID: AFUA_3G06170), with LAMP primers targeted to the internal MAT1‐2‐1 gene. LAMP primer sets (Table 1) were designed using the free online software package PrimerExplorer (v. 5) with default settings.
For screening isolates against each of the MAT LAMP assays, 15 μL reactions contained 0.3 μL BIP primer (final concentration 2 μmol/L), 0.3 μL FIP primer (2 μmol/L), 0.15 μL LOOPB primer (1 μmol/L), 0.15 μL LOOPF primer (1 μmol/L), 0.3 μL B3 primer (0.2 μmol/L), 0.3 μL F3 primer (0.2 μmol/L) (Table 1), μL 7.5 μL isothermal mastermix (ISO‐001; Optigene) and 1 μL DNA template (10 ng total DNA). No‐template (PCR‐grade water) controls were included in each test run. LAMP assays were run at 65°C for 30 minutes (FAM fluorescence measured every 30 seconds), followed by a final dissociation step at 95°C for 1 minutes; 55°C for 30 seconds and 95°C for 30 seconds. Assays were run with a MX3000p qPCR system (Agilent), with data analysed using inbuilt 7500 SDS software (v.1.4; Applied Biosystems). Dissociation curves were checked manually after each run to confirm the presence of a single peak.
3. RESULTS
3.1. Development and validation of MAT LAMP assays
For all A fumigatus isolates tested, identical MAT genotype results were obtained using the previously described multiplex PCR assay2 (see Figure 1 for representative results) and the new MAT‐specific LAMP assays developed in the present study (Figure 2, Table 2). The new MAT1‐1 and MAT1‐2‐specific LAMP assays gave positive results within 10‐20 minutes (ie, clear amplification curves) only for isolates of corresponding MAT1‐1 or MAT1‐2 type, respectively (Figure 2A). Positive results obtained with each MAT‐specific LAMP assays gave single dissociation curves of c. 89.5°C (±0.3), indicating specific amplification of the targeted MAT gene regions (Figure 2B). No‐template (water) controls tested negative, that is, no amplification curves or dissociation plot peaks were observed (data not shown).
Figure 1.
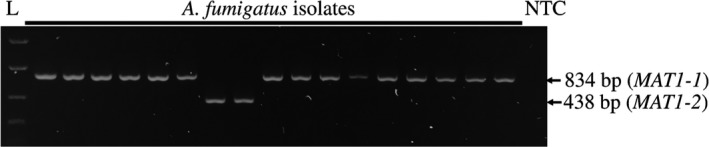
Representative results from screening of Aspergillus fumigatus isolates with the multiplex PCR mating‐type assay.2 MAT1‐1 and MAT1‐2 type isolates are distinguished by amplicons of 834 bp and 438 bp, respectively. “L” indicates Easyladder 1 (Bioline); NTC indicates no‐template control
Figure 2.
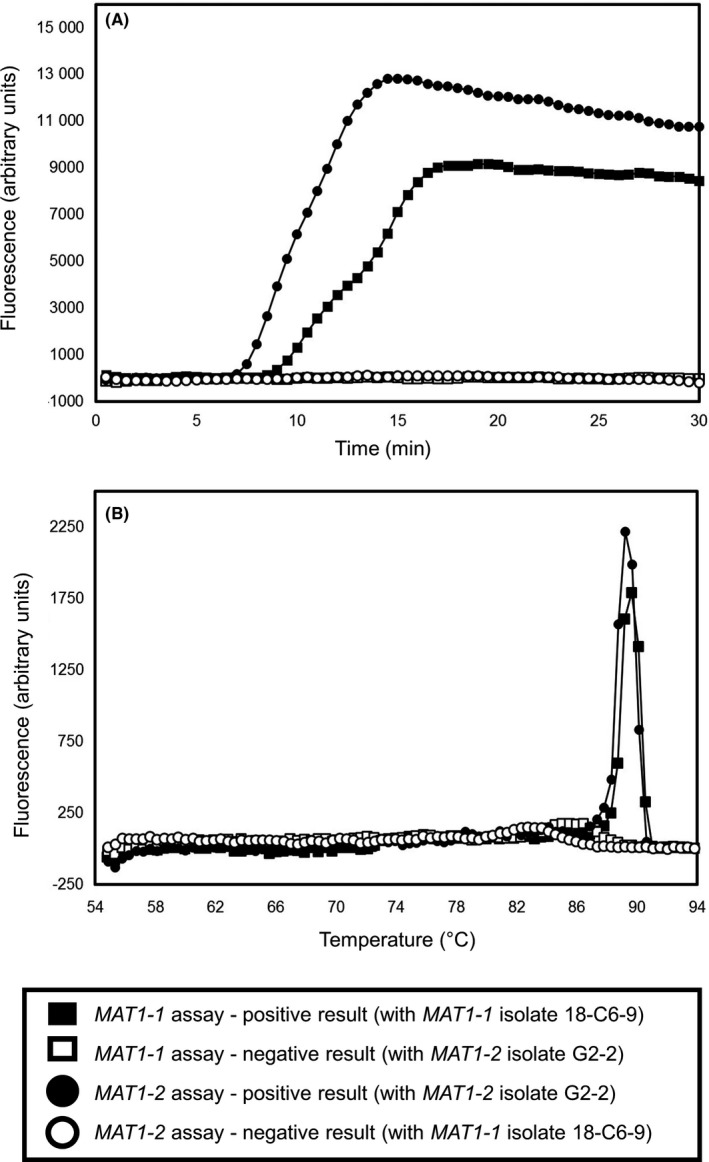
Representative results from screening of Aspergillus fumigatus isolates of MAT1‐1 (18‐C6‐9) or MAT1‐2 (G2‐2) type screened against the new MAT‐specific LAMP assays. Shown are (A) amplification plots and (B) dissociation plots. See base of figure for explanatory legend. No‐template (water) controls also gave negative results (data not shown)
4. DISCUSSION
This study reports the first use of LAMP technology to establish the mating‐type identity for a fungus, rapidly (within 20 minutes), as demonstrated here for isolates of A fumigatus. The MAT1‐1 and MAT1‐2‐specific LAMP assays appeared robust, being successfully applied to isolates of known opposite MAT type from a diverse range of clinical and environmental sources (air, food, plant and soil) and geographic locations (Africa, Asia, Europe and North and South America). These assays will be of use in research into the applied biology of this important human opportunistic pathogen. For example, they will allow the rapid set‐up of sexual crosses with isolates of known opposite MAT type, subsequent analysis of the MAT type inheritance of the progeny, and through progeny analysis the determination of the genetic basis of traits such as antifungal resistance and virulence.
It should now be possible to develop similar LAMP assays targeting MAT gene sequences to allow rapid mating‐type determination in other heterothallic fungi of medical [eg, Aspergillus lentulus—another causal agent of human aspergillosis10], agricultural [eg, Zymoseptoria tritici—cause of wheat Septoria leaf blotch11], environmental [eg, Hymenoscyphus fraxineus—cause of ash dieback12] and industrial [eg, Penicillium chrysogenum—used in penicillin production13] importance. Such assays could also provide a better understanding into the reproductive strategies of various fungal pathogens, providing insight into their evolutionary potential and possible risk of breakdown of disease management strategies.14 Furthermore, they could also be used to indirectly assess possible cryptic sexuality in fungi for which no sexual stage is yet known, given that frequency dependent selection operating on MAT genes generally, although not always, results in a 1:1 distribution of mating types.5, 14
CONFLICT OF INTEREST
No conflict of interest is declared.
AUTHOR CONTRIBUTIONS
KMK, NJH, PSD, JSW and BAF conceived the ideas; KMK and SA collected the data; KMK analysed the data; KMK led the writing; all authors critically reviewed the manuscript prior to submission.
ACKNOWLEDGEMENTS
These studies were partly supported by the Rothamsted Research Smart Crop Protection (SCP) strategic programme (BBS/OS/CP/000001) funded through the Biotechnology and Biological Sciences Research Council's Industrial Strategy Challenge Fund and by CropLife International through project 14209 ‘Investigating the sources and spread of azole resistant Aspergillus fumigatus strains in the environment'.
King KM, Hawkins NJ, Atkins S, Dyer PS, West JS, Fraaije BA. First application of loop‐mediated isothermal amplification (LAMP) assays for rapid identification of mating type in the heterothallic fungus Aspergillus fumigatus . Mycoses. 2019;62:812–817. 10.1111/myc.12959
REFERENCES
- 1. Dagenais T, Keller NP. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev. 2009;22:447‐465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paoletti M, Rydholm C, Schwier EU, et al. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus . Curr Biol. 2005;15:1242‐1248. [DOI] [PubMed] [Google Scholar]
- 3. O’Gorman CM, Fuller HT, Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus . Nature. 2009;457:471‐474. [DOI] [PubMed] [Google Scholar]
- 4. Zhang J, Snelders E, Zwaan BJ, et al. A novel environmental azole resistance mutation in Aspergillus fumigatus and a possible role of sexual reproduction in its emergence. mBio. 2017;8:e00791‐e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dyer PS, Inderbitzin P, Debuchy R. Mating‐type structure, function, regulation and evolution in the Pezizomycotina In: Wendland J, ed. Growth, Differentiation and Sexuality, The Mycota I, 3rd edn Switzerland: Springer International Publishing; 2016:351‐385. [Google Scholar]
- 6. Yu Y, Amich J, Will C, Eagle CE, Dyer PS, Krappmann S. The novel Aspergillus fumigatus MAT1‐2‐4 mating‐type gene is required for mating and cleistothecia formation. Fungal Biology and Genetics. 2017;108:1‐12. [DOI] [PubMed] [Google Scholar]
- 7. Tang Q, Tian S, Yu N, et al. Development and evaluation of a loop‐mediated isothermal amplification method for rapid detection of Aspergillus fumigatus . J Clin Microbiol. 2016;54:950‐955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Notomi T, Okayama H, Masubuchi H, et al. Loop‐mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu L‐S, Rodriguez‐Manzano J, Malpartida‐Cardenas K, et al. Rapid and sensitive detection of azole‐resistant Aspergillus fumigatus by tandem‐repeat loop‐mediated isothermal amplification. J Mol Diagn. 2018;. 10.1016/j.jmoldx.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Swilaiman SS, O'Gorman CM, Balajee SA, Dyer PS. Discovery of a Sexual Cycle in Aspergillus lentulus, a close relative of A fumigatus . Eukaryot Cell. 2013;12:962‐969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Waalwijk C, Mendes O, Verstappen E, de Waard MA, Kema G. Isolation and characterization of the mating‐type idiomorphs from the wheat septoria leaf blotch fungus Mycosphaerella graminicola . Fungal Genet Biol. 2002;35:277‐286. [DOI] [PubMed] [Google Scholar]
- 12. Gross A, Zaffarano PL, Duo A, Grünig CR. Reproductive mode and life cycle of the ash dieback pathogen Hymenoscyphus pseudoalbidus . Fungal Genet Biol. 2012;49:977‐986. [DOI] [PubMed] [Google Scholar]
- 13. Böhm J, Hoff B, O’Gorman CM, et al. Sexual reproduction and mating‐type–mediated strain development in the penicillin‐producing fungus Penicillium chrysogenum . Proc Natl Acad Sci. 2013;110:1476‐1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McDonald BA, Linde CC. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol. 2002;40:349‐379. [DOI] [PubMed] [Google Scholar]


