Abstract
Small‐cell lung cancer (SCLC) is an aggressive malignancy characterized by high cellular proliferation and early distant metastasis. Our study aimed to explore the effect of miR‐22‐3p (miR‐22, for short) on SCLC radiosensitivity and its molecular mechanisms. The expression level of miR‐22 was evaluated in a human normal lung epithelial cell line and a human SCLC cell line, and cell apoptosis and migration were detected. The expression of the miR‐22 direct target WRNIP1 mRNA and protein were explored. Five differentially expressed genes were detected. The miR‐22 expression in NCI‐H446 was significantly decreased, and miR‐22 overexpression significantly promoted cell apoptosis. miR‐22 overexpression could significantly inhibit the cell migration of SCLC cells, and miR‐22 had a negative regulatory effect on WRNIP1 mRNA and protein levels. KLK8 was downregulated, and the messenger RNA (mRNA) of four other genes (PC, SCUBE1, STC1, and GPM6A) was upregulated mRNA in cells overexpressing miR‐22, which was in accordance with the bioinformatics analysis. miR‐22 could enhance the radiosensitivity of SCLC by targeting WRNIP1.
Keywords: DEGs, miR‐22, radiosensitivity, SCLC, WRNIP1
1. INTRODUCTION
Lung cancer (LC) has the highest morbidity and mortality in the world, which seriously threatens human life and health.1 Small‐cell lung cancer (SCLC) is the main type of LC, which has the characteristics of short multiplication, rapid growth, and early‐onset metastasis.2 With the rapid development of stereotactic radiosurgery and radiotherapy technology, radiotherapy provides an effective strategy for SCLC treatment. A growing number of studies have revealed that SCLC is sensitive to radiotherapy. SCLC is prone to relapse, resulting in radioresistance in the late stage of radiotherapy; thus the effect of radiotherapy is regrettably reduced.3 Therefore, there is an urgent need to fully elucidate the therapy‐induced radioresistance mechanisms to improve the radiotherapy effect and prolong the survival of patients with SCLC.4
Ionizing radiation (IR) is one of the major modalities of SCLC treatment.5 It causes DNA damage by producing intermediate ions and oxygen free radicals, leading to tumor cells apoptosis.6 Although the DNA damage pathway plays an important role in radiation sensitivity, cell cycle checkpoint, and apoptosis pathways also play important roles in the susceptibility of SCLC to radiotherapy. Recently, scientists have devoted themselves to discover how IR attacks SCLC cells.7 Disappointingly, the relevant mechanisms remain obscure. SCLC radiotherapy is limited and ineffective due to SCLC radioresistance.8 Therefore, to explore effective and specific methods to enhance the radiosensitivity of SCLC is helpful to reduce radioresistance and side effects.9, 10
An increasing number of studies show that microRNAs (miRNAs) act as gene expression regulators in tumor initiation and procession, which cause translation inhibition by messenger RNA (mRNA) inactivation and degradation.11, 12 As negative regulators, miRNAs can act on essential signaling pathways, including cell response to IR.13 Therefore, regulating the expression of miRNAs has a significant impact on the clinical radiation response, as it enhances cell susceptibility.14 Although no miRNAs have been approved by the Food and Drug Administration as drugs, greater progress is being made in registering them as therapeutic agents.6 Moreover, the latest research has demonstrated that the efficacy of miRNA‐based therapeutic agents in preclinical models.15
Recently, miR‐22‐3p (miR‐22, for short), which is a 22 nucleotide noncoding RNA located on chromosome 17, has been found to regulate tumor‐related gene expression in different cancer models.16 However, whether miR‐22 controls tumor radioresistance in vivo and can be used as a tumor radiosensitizer remains unclear. In this study, we find the miR‐22 has significant effects on SCLC cell proliferation, migration, and apoptosis in an IR dose‐dependent manner. We further show that WRNIP1 is a direct target of miR‐22, and the related molecular mechanism of miR‐22 regulation of SCLC radiosensitivity is preliminarily explained. More importantly, our findings demonstrate the therapeutic utility of miR‐22 as a potential tumor radiosensitizer in a SCLC model. These results suggest that the miR‐22 cargo combined with radiotherapy may represent a new strategy for SCLC treatment.
2. MATERIALS AND METHODS
2.1. Cell culture
Cell lines were used in this paper. Human normal lung epithelial cell line BEAS‐2B, human SCLC cell line NCI‐H446 and human embryonic kidney cell line HEK293T were purchased from the Cell Bank of Shanghai Institute of Cell Biology, CAS. The cells were cultured in high glucose Dulbecco's modified Eagle's medium and Roswell Park Memorial Institute 1640 (Gibco, UK) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin (100 U/mL)‐streptomycin (100 μg/mL; Gibco) and were maintained in a 37°C incubator with a humidified, 5% CO2 atmosphere.
2.2. Cell transfection
Cells were transfected with vector controls and miRNA compounds by Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol. After 24 to 48 hours of transfection, cell samples were collected and subjected to transfection‐efficiency testing.
2.3. miR‐22 mimics/nc and inhibitors/nc oligonucleotides
Four individual products (GenePharma, China) were synthesized (Table 1). Cells were transfected with 100 nM of the indicated oligonucleotide separately using Lipofectamine 2000. Then, 24 to 48 hours after transfection, the resultant cells were used for functional assays, and the remaining cells were harvested for quantitative polymerase chain reaction (qPCR) analysis.
Table 1.
The oligo sequences of miR‐22 mimics and inhibitors
| Name | Sequence (5′‐3′) |
|---|---|
| miR‐22‐mimics | AAGCUGCCAGUUGAAGAACUGU |
| AGUUCUUCAACUGGCAGCUUUU | |
| miR‐22‐mimics nc | UUCUCCGAACGUGUCACGUTT |
| ACGUGACACGUUCGGAGAATT | |
| miR‐22‐inhibitors | ACAGUUCUUCAACUGGCAGCUU |
| miR‐22‐inhibitors nc | CAGUACUUUUGUGUAGUACAA |
2.4. Overexpression cell construct
The miR‐22 oligos were cloned into the pLKO.1 vector according to the manufacturer's instructions (Figure 1). The cells were transfected with the pLKO.1 control and miR‐22‐pLKO.1 plasmids, followed by drug screening and qPCR analysis. The primer sequences are shown in Table 2.
Figure 1.
The map of pLKO.1 vector
Table 2.
miRNAs stem‐loop primer sequences
| Name | Sequence (5′‐3′) |
|---|---|
| miR‐22 stem‐loop primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAGTT |
| U6 stem‐loop primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATA |
Abbreviation: miRNAs, microRNAs
2.5. RNA extraction and real‐time PCR analysis
Total RNA was extracted using the RNAiso Plus Kit (TaKaRa, Japan). For complementary DNA (cDNA) synthesis, total RNA was involved in the cDNA amplification by the HiScriptII Reverse Kit (Vazyme, China), and for qPCR analysis, the AceQ real‐time (RT)‐qPCR Kit (Vazyme) was used, all according to the manufacturer's instructions. The mRNA levels were normalized by U6 and β‐actin. The primer sequences are shown in Table 3.
Table 3.
qPCR primer sequences
| Name | Sequence (5′‐3′) |
|---|---|
| miR‐22‐F | AGGGTCCGAGGTATTCGCA |
| miR‐22‐R | AGCGAAGCTGCCAGTTGAAG |
| U6‐F | CTCGCTTCGGCAGCACA |
| U6‐R | AACGCTTCACGAATTTGCGT |
| WRNIP1‐F | ATTGATGAGATTCATCGGTTCAA |
| WRNIP1‐R | GGCTAGAGTCTAGGACGTGGATTC |
| β‐Actin‐F | CTGGACTTCGAGCAAGAGAT |
| β‐Actin‐R | GATGTCCACGTCACACTTCA |
| GPM6A‐F | GTTTATTGTGGCACTTGCTGGA |
| GPM6A‐R | TGGCAGACAGAACCATAAGGTAGTG |
| STC1‐F | AAATGCATCGCCAACGGG |
| STC1‐R | TTCATCACATTCCAGCAGGCTT |
| KLK8‐F | GAAGTGTGAGGATGCTTACCCG |
| KLK8‐R | ATGTGATGCCCTGGAGTGC |
| PC‐F | CTGCGGTCCATCTTGGTCAA |
| PC‐R | CCATGGGTGAGGTCACCAC |
| SCUBE1‐F | AACTCATAGAGGACATCGTGCG |
| SCUBE1‐R | CGCTCCCCCCGGTTATTT |
Abbreviations: F, forward; qPCR, quantitative polymerase chain reaction; R, reverse.
2.6. Lentivirus preparation
The cells were inoculated into six‐well plates at a density of 2 × 106 cells per well and were cultured overnight in an incubator at 37°C. The cells were infected with pLKO.1 vector control or miR‐22, carrying lentiviral particles for 48 hours. After selection with 10 μg/mL puromycin, overexpression efficiency was verified by qPCR analysis.
2.7. Western blotting
Cells were collected and resuspended in 0.5 mL ice‐cold radioimmunoprecipitation assay lysis buffer (Solarbio, China). Cell lysates were lysed by vortexing with acid‐washed glass beads for 2 minutes, then placed on ice for 2 minutes; this cycle was repeated 10 times. After centrifugation at 12 000 rpm for 30 minutes, the sample protein concentration was determined using Nanotrop (Thermo Fisher Scientific). Lysate samples (50 μg) were electrophoresed on 8% or 10% polyacrylamide gels and transferred onto polyvinylidene difluoride membranes. The samples were blocked with 5% nonfat milk in Tris buffered saline solution containing 0.1% to 0.2% Tween‐20 at room temperature for 2 hours and then probed with primary antibodies (anti‐WRNIP1, 1:1000 and anti‐β‐actin, 1:1000; Santa Cruz), as indicated, overnight at 4°C; the samples were then incubated with horseradish peroxidase‐conjugated secondary antibodies (anti‐mouse, 1:2000; Santa Cruz). The bands were visualized using an ECL chemiluminescence detection kit (Thermo Fisher Scientific). Multiplication of the intensity and area of protein bands indicated the relative levels of protein expression.
2.8. γ‐Ray treatment
The prepared cells were divided into three parts and sent to the Institute of Radiological Medicine of the Chinese Academy of Medical Sciences for γ‐irradiation with doses of 0, 2, and 4 Gy, respectively.
2.9. miR‐22 expression levels
qRT‐PCR was performed to detect the miR‐22 expression levels in cells after γ‐irradiation. The experimental method was the same as in section 2.5.
2.10. 3‐(4,5‐dimethylthiazol‐2‐yl)‐ 5‐(3‐carboxymethoxyphenyl)‐ 2‐(4‐sulfophenyl)‐2H‐tetrazolium assay
The transfected cells were detected by using the CellTiter 96 AQ MTS Reagent Powder kit (Promega), according to the manufacturer's protocol. The 3‐(4,5‐dimethylthiazol‐2‐yl)‐5‐(3‐carboxymethoxyphenyl)‐ 2‐(4‐sulfophenyl)‐2H‐tetrazolium, inner salt (MTS) activity was determined by measuring absorbance at 490 nm.
2.11. Colony formation assay
In general, the concentration of 1 × 103 cells was inoculated into six‐well plates and gently shaken in the dish in a cross direction to disperse the cells evenly. The cells were cultured in a 37°C incubator with a humidified, 5% CO2 condition for 7 to 10 days. The cells were stained with Giemsa for 10 to 15 minutes and images were obtained. Colonies consisting of 100 or more cells were counted. The survival fraction was calculated as the mean number of colonies/(cells seeded × plating efficiency).
2.12. Flow cytometry analysis
Cell apoptosis was detected using an Annexin V/propidium iodide (PI) staining kit (Biolegend), according to the manufacturer's protocol. Cell proliferation was detected using a PE Mouse Anti‐Human Ki‐67 Set kit (BD Biosciences), according to the manufacturer's protocol. The cell cycle was detected using PI (50 μg/mL; Sangon Biotech, China) staining. The cells were predisposed to ethanol fixation and RNase A treatment.
2.13. Site‐directed mutation
The cDNA templates were changed to the locus of miR‐22 and WRINP1‐ 3′‐untranslated region (3′‐UTR), according to TargetScan (http://www.targetscan.org) and PICTAR (http://www.pictar.mdc‐berlin.de) predictions (Figure 2). The WRINP1‐ 3′UTR sequence is CTGGCAGCT, which binds to miR‐22. The mutation sequence is CTGCCTGGT, according to the mutation sites; primers were designed, as shown in Table 4, followed by WRNIP1‐3′UTR‐Mut‐psiCHECK2 construction and DNA sequencing.
Figure 2.

Site‐directed mutation of WRNIP1‐3′‐UTR‐psiCHECK2 mutant recombinant plasmid. 3′‐UTR, 3′‐untranslated region
Table 4.
WRNIP1‐3′‐UTR‐Mut primer sequences
| Name | Sequence (5′‐3′) |
|---|---|
| WRNIP1‐3′‐UTR‐mut‐F | AAAATACCTGCCTGGTTTGTGCAATGAATTAATGT |
| WRNIP1‐3′‐UTR‐mut‐R | CAAACCAGGCAGGTATTTTCATAAGCATAACCG |
Abbreviation: 3′‐UTR, 3′‐untranslated region.
2.14. Transcriptome sequencing
The miR‐22 overexpressing SCLC cell line NCI‐H449 stably transfected miR‐22‐NCI‐H449 and the empty control pLKO.1‐NCI‐H446 were sequenced. The cells were cultured to a concentration of 1 × 107, which met the sequencing concentration requirement. The confirmatory samples were sent to Novegene Biotechnology Co, Ltd for transcriptome sequencing. The sequencing data generated was 5 Gb per sample. Bioinformatics analysis was carried out according to the raw reads and clean reads.
2.15. Luciferase assay
The cells were seeded in 96‐well plates at a density of 5 × 104 per well. Luciferase activity was detected using a Dual‐Luciferase Reporter Assay System kit (Promega), according to the manufacturer's protocol. A microplate reader was used to determine relative luminescence.
2.16. Bioinformatics analysis
The human genome and vector control were considered as the reference genome and control sequence, respectively. The expression abundance of the corresponding clean reads gene in the samples was obtained, and the differentially expressed genes (DEGs) were found. Then, the obtained DEGs were analyzed by Gene Ontology and Kyoto Encyclopedia of Genes and Genomes.
2.17. Statistical analysis
All results of this study were presented as the mean ± SEM, and statistical significance was examined by unpaired two‐tailed Student t test. The P < .05 was considered as statistically significant and indicated with *, P < .01 = **and P < .001 = ***.
2.18. Data availability
The authors declare that all the data supporting the findings of this study are available within the article and from the corresponding author on reasonable request.
3. RESULTS
3.1. The establishment of miR‐22 overexpression and knock down models
To investigate the possible function of miR‐22 in the regulation of SCLC, we first examined the mRNA expression level of miR‐22 in cell lines BEAS‐2B and NCI‐H446 by qPCR. Interestingly, the miR‐22 expression was significantly decreased in the SCLC cell line (Figure 3A). Presumably, it is possible to associate miR‐22 expression with SCLC treatment.
Figure 3.
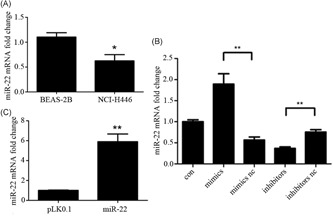
The establishment of miR‐22 overexpression and knockdown models. A‐C, Cells were cultured in the corresponding medium for the indicated times. Efficiency was determined by quantitative PCR and measured as the ratio of miR‐22 relative to the internal reference target U6 gene. *,**,***Significant differences between the tested and the control strains, P < .05, P < .01, and P < .001, respectively. PCR, polymerase chain reaction
To confirm our hypothesis, we established the miR‐22 overexpression and knockdown models. As a result, the transfected cells carrying miR‐22 mimics/nc and inhibitors/nc are considered ideal tools for miR‐22 research. As shown in Figure 3B, the miR‐22 expression is apparently higher than the vector control upon the transfection of mimics. Otherwise, once we transfected the miR‐22 inhibitors, the miR‐22 expression was expectedly lower than that in the other group, indicating that miR‐22 mimics and inhibitors have a remarkable effect on miR‐22 overexpression and knockdown. Meanwhile, the stable miR‐22 overexpressed cell line was successfully created by lentivirus infection and drug screening (Figure 3C).
3.2. miR‐22 enhances radiosensitivity by targeting tumor development
Recently, cancer cells, which are defined operationally as tumor‐initiating and tumor‐inducing cells, have been found to enhance radioresistance during DNA damage response activation.17 Therefore, we suspected that this miR‐22 may be negatively associated with radioresistance in SCLC. Then, we detected the effect of miR‐22 expression levels and miR‐22 on the cell proliferation of NCI‐H446 under different doses of γ‐irradiation. Indeed, with the increasing dose of γ‐irradiation, the miR‐22 expression was significantly increased in miR‐22 mimic‐transfected cells (Figure 4A), accompanied by the inhibition of cell proliferation in the miR‐22 overexpression group compared to the control. The proliferation level of miR‐22 mimic‐transfected cells was lower than in the NC group during 4 Gy irradiation. Conversely, cell proliferation in the miR‐22‐knockdown was notably promoted and the proliferation of miR‐22 inhibitors was higher than that under the same condition. It was concluded that miR‐22 expression may affect the sensitivity of SCLC cells to γ‐rays and play an important role in the radiosensitivity of SCLC (Figure 4B).
Figure 4.
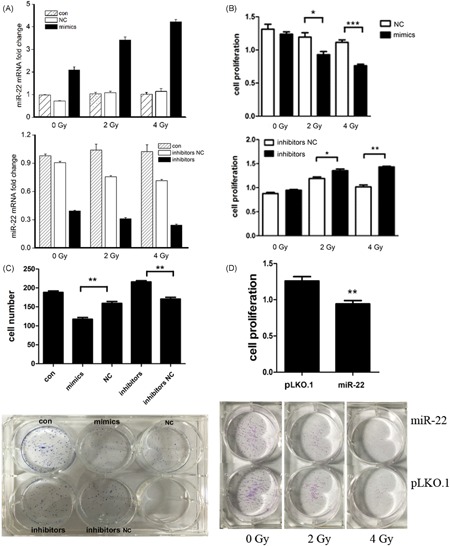
miR‐22 inhibits the SCLC cells proliferation under different doses of γ‐irradiation. A, miR‐22 expression in cells were detected by qPCR. B, Cells were stimulated with the indicated concentrations of MTS for the indicated conditions, cell viability was determined by microplate reader detection. CB‐DC, Cells were cultured in 96‐well plates for the indicated conditions, cell number was determined by Giemsa staining. *,**,***Significant differences between the tested and the control strains, P < .05, P < . 01, and P < . 001, respectively. MTS, 3‐(4,5‐dimethylthiazol‐2‐yl)‐ 5‐(3‐carboxymethoxyphenyl)‐ 2‐(4‐sulfophenyl)‐2H‐tetrazolium; NC, negative contriol; qPCR, quantitative polymerase chain reaction; SCLC, small‐cell lung cancer
In previous results, we found that miR‐22 caused significant inhibition of SCLC cell proliferation. Afterward, we studied the effects of miR‐22 on cell growth and colony formation by colony‐formation assay. The results revealed that the number of miR‐22 mimic‐transfected cells was significantly lower than in the NC group. In addition, after transfecting miR‐22 inhibitors, the number of cells was significantly higher than in the inhibitors NC group, and this result was basically consistent with the MTS results. In addition, we found the cell colony in mimics transfection was significantly less than in the NC group; transfected inhibitors were the opposite (Figure 4C). Thus, miR‐22 inhibited colony formation in NCI‐H446.
We further investigated the cell colony‐forming ability under γ‐irradiation conditions using the miR‐22 overexpressed cell line. Consistent with the previous analysis, the number of miR‐22 cells was lower than control cells in the condition of γ‐ray irradiation, and the trend was enhanced with the increasing γ‐ray irradiation dose (Figure 4D). These results showed that miR‐22 inhibited SCLC cell growth and increased the irradiation sensitivity to γ‐rays.
Since miR‐22 expression may affect the mitosis‐related Ki‐67 expression in SCLC cells,18, 19 we further investigated Ki‐67 expression after transfection with the miR‐22 mimics. At first, we detected Ki‐67 expression in transfected cells under nonirradiation conditions. The results are shown in Figure 5A. After mimics transfection, the positive rate of Ki‐67 was 61.0%. The NC transfection group showed a significant decrease. However, there was no significant difference compared to the inhibitors NC group. It was primarily shown that miR‐22 overexpression could inhibit Ki‐67 expression in SCLC samples.
Figure 5.
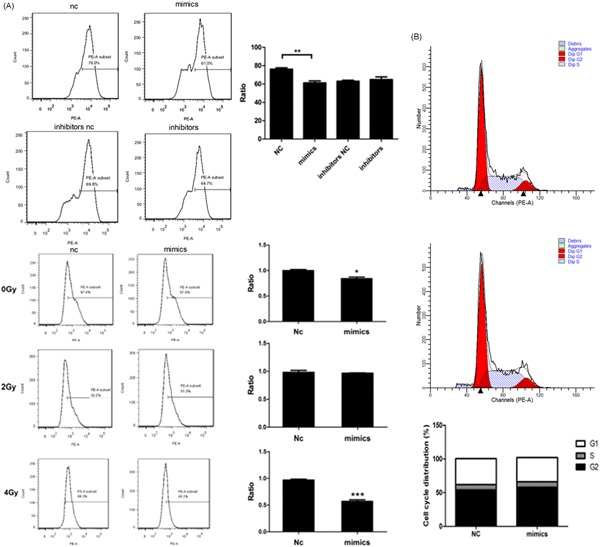
miR‐22 affects the Ki‐67 expression upon γ‐irradiation (A, B) Cells were cultured in 96‐well plates for the indicated conditions, Ki‐67 expression radio, and cell cycle was determined by flow cytometry analysis. *,**,***Significant differences between the tested and the control strains, P < .05, P < .01, and P < .001, respectively. NC, negative control
We explored Ki‐67 expression in miR‐22 overexpressed cells under different doses of γ‐irradiation. The data showed that, with an increasing dose of γ‐ray irradiation, the expected decreasing Ki‐67 expression gradually increased in the miR‐22 overexpression group. During 4 Gy irradiation, the positive rate of Ki‐67 in miR‐22 overexpressed cells was 48.2%, which was lower than that in control. We inferred that miR‐22 could inhibit Ki‐67 expression in NCI‐H446, leading to a boycott of cell proliferation, and this inhibition was more pronounced under high dose radiation conditions. Compared with the NC control, there was no significant difference in the proportion of the cell cycle after transfection with the miR‐22 mimics, indicating that the miR‐22 expression level may not affect the cell cycle of SCLC (Figure 5B).
To explore the effect of miR‐22 on SCLC cell migration, we carried out the scratching trial. The results showed that the migration ability of miR‐22 overexpression obviously decreased compared to control, revealing that miR‐22 overexpression could significantly inhibit SCLC cell migration (Figure 6A).
Figure 6.
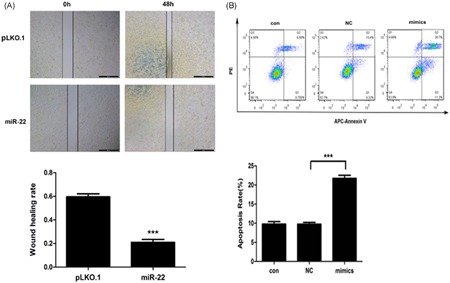
miR‐22 can inhibit the cell migration and promote the cell apoptosis in SCLC (A, B) Cells were cultured in 96‐well plates for the indicated conditions, wound healing rate was determined by measurement, apoptosis was determined by flow cytometry analysis. *,**,***Significant differences between the tested and the control strains, P < .05, P < .01, and P < .001, respectively. SCLC, small‐cell lung cancer
miR‐22, as a tumor suppressor, can cause cell apoptosis to some extent.20 As a result, we next explored the effect of miR‐22 on SCLC cell apoptosis by APC Annexin V/PI double staining. The results showed that the apoptosis rate of the miR‐22 mimic transfection group was significantly higher than in the NC group (Figure 6B), illustrating that miR‐22, as a potential tumor suppressor, could induce SCLC cell apoptosis.
3.3. miR‐22 is a negative regulator of WRNIP1 expression
To further explore the mechanism of miR‐22 affecting SCLC radiosensitivity, we predicted the target genes of miR‐22 and preliminary screened the delineated genes by bioinformatics analysis. The results showed that the TargetScan database predicted 611 target genes and the PICTAR database predicted 285 ones. From Figure 7A, we can see 112 common target genes were predicted by both of the above databases. Through NCBI and a literature search, WRNIP1, as a candidate target gene, was found to be associated with DNA damage repair, which is helpful in elucidating the mechanism of miR‐22 in SCLC cell radiosensitivity. Therefore, we selected WRNIP1 for further research.
Figure 7.
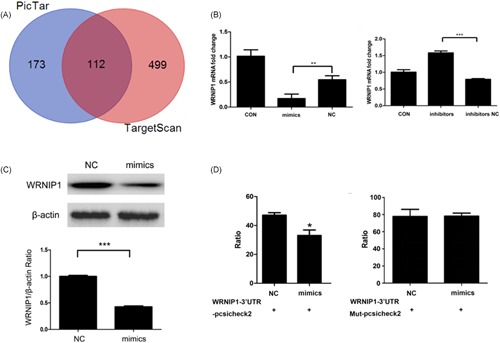
miR‐22 can negatively regulate the WRNIP1 expression in SCLC. A, Samples were cultured in the corresponding medium for the indicated conditions. Orange represents screening for the targeted genes by TargetScan, blue represents screening for the targeted genes by PICTAR, red represents screening for the overlapped genes by two databases. B, Cells were cultured in the corresponding medium for the indicated conditions. mRNA level was determined by quantitative PCR and measured as the ratio of WRNIP1 relative to the internal reference target β‐actin gene. C, Cells were cultured in the corresponding medium for the indicated conditions. Protein level was determined by western blotting, and measured as the ratio of WRNIP1 relative to the internal reference target β‐actin protein. D, Cells were cultured in the corresponding medium for the indicated conditions. Luciferase ratio was determined by microplate reader detection. *, **, ***Significant differences between the tested and the control strains, P < .05, P < .01, and P < .001, respectively. mRNA, messenger RNA; PCR, polymerase chain reaction; SCLC, small‐cell lung cancer
Therefore, we determined whether miR‐22 negatively regulated WRNIP1 during γ‐ray irradiation. Thus, we utilized the miR‐22 mimics and inhibitors and compared these with the NC control in regard to transcription and translation levels. Collectively, these results indicated that miR‐22 inhibits WRNIP1 expression by qPCR and Western blotting (Figure 7B,C). It can be seen there may be a negative regulation relationship between miR‐22 and WRNIP1, and WRNIP1 may be a downstream target gene of miR‐22.
To confirm our conclusion, we next detected the luciferase activity in each group. The luciferase activity was significantly lower than in the NC group in miR‐22 mimics and WRNIP1‐3′‐UTR‐psiCHECK2 double‐transfection. The luciferase activity in cells with the miR‐22 mimics and WRNIP1‐3′‐UTR‐mut‐psiCHECK2 showed almost no significant difference from the NC group (Figure 7D), elucidating that miR‐22 could be bound to the WRNIP1‐3′UTR. We eventually confirmed that WRNIP1 is the direct downstream target gene of miR‐22. Furthermore, it was speculated that miR‐22 was likely to enhance SCLC cell radiosensitivity by inhibiting WRNIP1 expression.
Moreover, we screened the high‐throughput DEGs KLK8, PC, STC1, GPM6A, and SCUBE1, which are related to cell proliferation, migration, and apoptosis, to detect mRNA levels (Figure 8A). qPCR was performed using the miR‐22 overexpression transfected cell line and its blank control cDNA as template. The results showed that KLK8 was significantly downregulated, and the other four genes were significantly upregulated upon overexpressing miR‐22 (Figure 8B).
Figure 8.
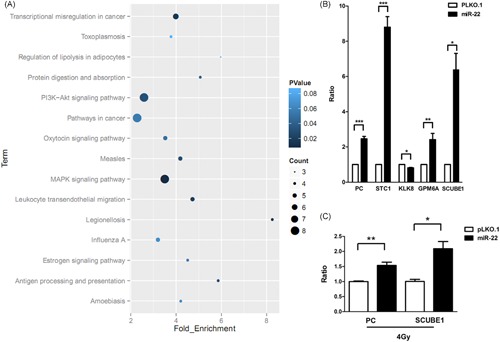
miR‐22 changes the tumor‐related genes expression in SCLC after γ‐irradiation. A, Samples were cultured in the corresponding medium for the indicated conditions. P value represents the clustering DEGs analysis. Cells were cultured in the corresponding medium for the indicated conditions. mRNA level was determined by quantitative PCR and measured as the ratio of PC and SCUBE1 relative to the internal reference target β‐actin gene. B, Cells were cultured in the corresponding medium for the indicated conditions. mRNA level was determined by quantitative PCR and measured as the ratio of PC, STC1, KLK8, GPM6A, and SCUBE1 relative to the internal reference target β‐actin gene. C, Cells were cultured in the corresponding medium for the indicated conditions. mRNA level was determined by quantitative PCR and measured as the ratio of PC and SCUBE1 relative to the internal reference target β‐actin gene. *, **, ***Significant differences between the tested and the control strains, P < .05, P < .01, and P < .001, respectively. DEG, differentially expressed genes; MAPK, mitogen‐activated protein kinases; mRNA, messenger RNA; PCR, polymerase chain reaction; P13K, phosphoinositide 3‐kinases; SCLC, small‐cell lung cancer
Under nonirradiation conditions, these five genes showed significant differences in transcription level, but their expression changed after γ‐irradiation. Only the changes of PC and SCUBE1 showed consistency in the nonirradiation and γ‐irradiation conditions, although the increasing trend was weakened (Figure 8C). In summary, the expression of these five genes are related to γ‐ray irradiation.
In conclusion, miRNAs are key participants and regulators in cancer treatment. miR‐22 can regulate tumor‐related gene expression, and it has a significant impact on LC diagnosis and treatment. Compared with the other common types of cancer, LC has a lower survival rate, which makes it the leading cause of cancer deaths worldwide. Tumor suppressors and carcinogenic factors miRNA are closely related to LC cell growth, development, and metastasis by changing the expression level. Otherwise, miRNAs are essential to the radioresistance and chemoresistance in LC. Furthermore, miRNAs also play an important role in cancer regulation. As a result, miRNAs may be used in LC clinical diagnosis, treatment, and prognosis in the future. With regard to miRNAs and cancer research, more samples and trials are needed to reveal the function of miRNAs in cancer research. Scientists have to further study the relationship between miRNAs and tumor radiosensitivity. As a result, miRNAs will become a promising tool for tumor prevention and treatment.
4. DISCUSSION
Radiotherapy is one of the main treatments for tumors.21 The efficacy of radiotherapy is affected by the sensitivity of tumor cells to radiotherapy.22 The radiosensitivity of different individual tumors is significantly different.23 Recently, with the in‐depth study of miRNAs, radiation conditions can induce changes in the expression of miRNAs, and some miRNAs participate in the regulation of tumor radiosensitivity by regulating the expression of target genes and vital signaling pathways.24 Shi et al7 reported that miR‐200c enhances the sensitization of LC cells A549 to radiation by targeting the VEGF‐VEGFR2 pathway. Hu et al4 performed qPCR detection for the expression of miRNAs in 102 patients with cervical cancer undergoing standardized treatments. They identified five miRNAs, miR‐9, miR‐21, miR‐200a, miR‐218, and miR‐203, that may be associated with cervical cancer radiosensitivity.
In the present study, we showed that the miR‐22 expression in NCI‐H446 was significantly lower than that in BEAS‐2B. It has previously been reported that miR‐22 plays an important role in multiple types of cancer development.25 Ling et al15 reported that miR‐22 expression in human LC tissues was significantly decreased compared to normal control tissues. Compared with the normal ovarian tissue, Wyman et al17 found that the miR‐22 expression was downregulated in ovarian cancer. Reports on gastric cancer also showed that miR‐22 expression was rare in gastric cancer samples.26 However, there are few studies on miR‐22 in SCLC cells.
In our study, we studied the biological function of miR‐22 in an SCLC model for tumor progression and metastasis. Combined with the results of MTS and colony‐forming assays, we infer that miR‐22 has a significant inhibitory effect on tumor growth and proliferation in SCLC cells. According to previous research, researchers found that miR‐22 acted as a tumor suppressor, which had an inhibitory effect on the development of tumors.27, 28 Yang et al19 reported that miR‐22 can significantly inhibit the cell proliferation, migration, and invasiveness of esophageal squamous cancer cells. Others found that miR‐22 overexpression in ovarian granulosa cells could aggravate apoptosis and inhibit cancer cell growth.29
Ki‐67 is an antigen expressed in proliferating cells and a common marker of cell proliferation; thus, it has been used as the most reliable indicator of tumor cell proliferation.30 We detected Ki‐67 expression in miR‐22 mimic‐ and inhibitor‐transfected cells and then explored the effect of miR‐22 on apoptosis in SCLC by APC Annexin V/PI staining. These results indicated that miR‐22, as a tumor suppressor, could change cell proliferation and promote apoptosis in SCLC cells, and the results of our study were consistent with the previous outcome. In subsequent studies, we also need to detect the effects of miR‐22 on the expression of apoptosis‐related proteins (Bcl‐2, Bax, and Caspase‐3)15 to further explain the specific molecular mechanism of miR‐22 promoting cell apoptosis in SCLC.
Leuzzi et al9 revealed that WRNIP1 can act on intracellular arrested replication forks and cooperate with RAD51 to protect the integrity of replication forks. In the experiment, WRNIP1‐deficient cells exhibited significant DNA damage and chromosomal aberrations. Therefore, WRNIP1 is considered as a protector of replication forks. Another publication suggests that WRNIP1 can recruit DNA polymerase to DNA damage, which plays an important role in DNA damage repair and genome stability maintenance.22 Radiotherapy can cause severe, irreparable DNA damage and cell cycle arrest, leading to tumor death and cell apoptosis.19 It is speculated that miR‐22 is likely to increase the radiosensitivity of SCLC cells by inhibiting WRNIP1 expression.
The psiCHECK2 vector utilizes Renilla luciferase as a reporter gene, and the target gene is cloned into a multiple cloning site, which is downstream of the Renilla luciferase translation termination codon. Additionally, the psiCHECK2 vector contains a second reporter gene, firefly luciferase, which was designed for end‐point cleavage assays to normalize Renilla luciferase expression, resulting in robust and reproducible data.11, 15 Since the psiCHECK2 vector contains two kinds of luciferases, it can reduce the internal reference luciferase plasmid introduction during the experiment, thereby avoiding the multiplasmid cotransfection systems emergence and providing convenience for cell research.13 In addition, the Renilla luciferase assay is more sensitive, more convenient, and more rapid in quantification.21 The experimental results show that miR‐22 can inhibit the reporter gene‐vector luciferase activity, indicating that WRNIP1 is a direct target gene downstream of miR‐22.
In this study, to elucidate the mechanism of miR‐22 affecting the radiosensitivity of SCLC cells, we analyzed the transcriptome using the miR‐22 overexpression and control samples. Bioinformatics analysis highlighted five DEGs (KLK8, PC, SCUBE1, STC1, and GPM6A) in miR‐22 overexpression and vector control cells, which were selected for quantitative detection.
STC1 is a glycoprotein found in the endocrine glands of the fish kidney, and it is considered a tumor cell apoptosis‐inducing factor.17 GPM6A is a transmembrane protein widely distributed on the neuronal cells surface in the central nervous system; it is believed that GPM6A is one of the pathogenic genes of human lymphatic leukemia and is closely related to apoptosis.13 Therefore, we chose STC1 and GPM6A for RT‐qPCR analysis. The results showed that STC1 and GPM6A expression in the miR‐22 overexpression cell line was significantly higher than that in the control group, which indicated that STC1 and GPM6A could be the apoptosis‐inducing factors in SCLC cells. Therefore, miR‐22 is likely to promote SCLC cell apoptosis by elevating the expression of apoptosis‐inducing factors STC1 and GPM6A in tumors.
The kallikrein‐related peptidase enzyme (KLK) family, located on human chromosome 19q13.4, is a serine subfamily. KLK8 is a member of this family, and abnormalities in KLK8 transcription or translation products may lead to the development of uterine, ovarian, and other cancers.25 Therefore, it is speculated that KLK8 might be a cancer‐promoting factor. In this study, we found that KLK8 expression in miR‐22 overexpression cells was significantly lower than another group, indicating that KLK8 may be a cancer‐promoting factor and negatively regulated by miR‐22. Therefore, we further speculate that miR‐22 inhibition of SCLC cell proliferation and migration may be related to the inhibition of KLK8 expression.
SCUBE1 is a glycoprotein secreted by platelet endothelium. Tokuda et al27 found that SCUBE1 had an inhibitory effect on tumor development to some extent.28 Our results show that SCUBE1 expression in miR‐22 overexpression samples is higher than in the control, indicating that the inhibitory effect of miR‐22 on tumor metastasis may be related to the upregulation of SCUBE1 expression.
Pyruvate carboxylase (PC) is a kind of nonsteroidal enzyme, which is important to the tricarboxylic acid cycle.6 In addition, there is evidence that PC is associated with tumor invasiveness and metastasis.14 In our study, we found that PC was significantly increased in miR‐22 overexpression cells, indicating that miR‐22 could inhibit SCLC cell migration by regulating PC expression.
Because miRNAs can negatively regulate target genes, the results show that STC1, GPM6A, PC, and SCUBE1 are also overexpressed in miR‐22 overexpression cells, so it is presumed that these genes are not the direct target genes of miR‐22. In addition to the changes in the expression of the five DEGs, it is important to detect the expression of STC1, GPM6A, PC, KLK8, and SCUBE1 in miR‐22 overexpression cells under γ‐irradiation conditions. The results showed that only the changes in PC and SCUBE1 were consistent, but the trend was weakening. Therefore, this study preliminarily explains the mechanism of miR‐22 effects on the radiosensitivity of SCLC cells, and the follow‐up needs more in‐depth study.
5. CONCLUSION
At present, although independent studies have shown that miRNAs can be used to evaluate the radiosensitivity of tumors, which can guide the development of clinical radiotherapy, this approach is seldom applied clinically. Therefore, in the follow‐up study, we also need to study the expression of miR‐22 in patients with SCLC undergoing radiotherapy and further explain the mechanism of miR‐22 regulation of radiosensitivity in SCLC cells. Thus, improving the curative effect of radiotherapy, reducing radiation injury and minimizing the side effects in patients will be possible. Summarily, miR‐22 may be widely used in the future as a means to evaluate tumor radiosensitivity and as a prognostic biomarker for patients receiving clinical treatment.
CONFLICT OF INTERESTS
The authors declare that there is no conflict of interests.
ACKNOWLEDGMENTS
We would like to thank all the members of our research group for their enthusiastic participation in this study. This work was supported by the Programs of National Natural Science Foundation of China (No.81702098) and Tianjin Municipal Health Bureau Science and Technology Foundation (No.2014KZ102).
Jiang W, Han X, Wang J, et al. miR‐22 enhances the radiosensitivity of small‐cell lung cancer by targeting the WRNIP1 . J Cell Biochem. 2019;120:17650‐17661. 10.1002/jcb.29032
Wenhua Jiang and Xuemei Han contributed equally to this study and should be considered as co‐first authors.
References
REFERENCES
- 1. Du YZ, Gu XH, Cheng SF, et al. The oncogenetic role of stanniocalcin 1 in lung adenocarcinoma: a promising serum candidate biomarker for tracking lung adenocarcinoma progression. Tumor Biol. 2016;37(4):5633‐5644. [DOI] [PubMed] [Google Scholar]
- 2. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522‐531. [DOI] [PubMed] [Google Scholar]
- 3. Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849‐864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hu X, Schwarz JK, Lewis JS, Jr , et al. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010;70(4):1441‐1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613‐626. [DOI] [PubMed] [Google Scholar]
- 6. Gu X, Fu C, Lin L, et al. miR‐124 and miR‐9 mediated downregulation of HDAC5 promotes neurite development through activating MEF2C‐GPM6A pathway. J Cell Physiol. 2018;233(1):673‐687. [DOI] [PubMed] [Google Scholar]
- 7. Shi Y, Zhang X, Tang X, Wang P, Wang H, Wang Y. miR‐21 is continually elevated long‐term in the brain after exposure to ionizing radiation. Radiat Res. 2012;177(1):124‐128. [DOI] [PubMed] [Google Scholar]
- 8. Chun‐Zhi Z, Lei H, An‐Ling Z, et al. MicroRNA‐221 and microRNA‐222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10(1):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leuzzi G, Marabitti V, Pichierri P, Franchitto A. WRNIP1 protects stalled forks from degradation and promotes fork restart after replication stress. EMBO J. 2016;35(13):1437‐1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li B, Song Y, Liu TJ, et al. miRNA‐22 suppresses colon cancer cell migration and invasion by inhibiting the expression of T‐cell lymphoma invasion and metastasis 1 and matrix metalloproteinases 2 and 9. Oncol Rep. 2013;29(5):1932‐1938. [DOI] [PubMed] [Google Scholar]
- 11. Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68(15):6416‐6424. [DOI] [PubMed] [Google Scholar]
- 12. Wang X, Zhang Z, Wang Y, et al. Increased miRNA‐22 expression sensitizes esophageal squamous cell carcinoma to irradiation. J Radiat Res. 2012;54(3):401‐408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xin M, Qiao Z, Li J, et al. miR‐22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget. 2016;7(28):44252‐44265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou J, Liu J, Cheng CJ, et al. Biodegradable poly (amine‐co‐ester) terpolymers for targeted gene delivery. Nat Mater. 2015;11(1):82‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ling B, Wang GX, Long G, Qiu JH, Hu ZL. Tumor suppressor miR‐22 suppresses lung cancer cell progression through post‐transcriptional regulation of ErbB3. J Cancer Res Clin Oncol. 2012;138(8):1355‐1361. [DOI] [PubMed] [Google Scholar]
- 16. Planque C, Choi YH, Guyetant S, Heuze‐Vourc'h N, Briollais L, Courty Y. Alternative splicing variant of kallikrein‐related peptidase 8 as an independent predictor of unfavorable prognosis in lung cancer. Clin Chem. 2010;56(6):987‐997. [DOI] [PubMed] [Google Scholar]
- 17. Wyman SK, Parkin RK, Mitchell PS, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLOS One. 2009;4(4):e5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang X, Wan G, Berger FG, He X, Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell. 2011;41(4):371‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang C, Ning S, Li Z, Qin X, Xu W. miR‐22 is down‐regulated in esophageal squamous cell carcinoma and inhibits cell migration and invasion. Cancer Cell Int. 2014;14(1):138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Templin T, Paul S, Amundson SA, et al. Radiation‐induced micro‐RNA expression changes in peripheral blood cells of radiotherapy patients. Int J Radiat Oncol Biol Phys. 2011;80(2):549‐557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kozomara A, Griffiths‐Jones S. miRbase: integrating microRNA annotation and deep‐sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152‐D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let‐7 microRNA family. Cell. 2005;120(5):635‐647. [DOI] [PubMed] [Google Scholar]
- 23. Phannasil P, Thuwajit C, Warnnissorn W, Wallace JC, MacDonald MJ, Jitrapakdee S. Pyruvate carboxylase is up‐regulated in breast cancer and essential to support growth and invasion of mda‐mb‐231 cells. PLOS One. 2015;10(6):e0129848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshimura A, Seki M, Enomoto T. The role of WRNIP1 in genome maintenance. Cell Cycle. 2017;16(6):515‐521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee SY, Jeon HM, Ju MK, et al. Wnt/snail signaling regulates cytochrome c oxidase and glucose metabolism. Cancer Res. 2012;72(14):3607‐3617. [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y, Li H, Liu Y. The function and mechanism of miRNA in tumor radiosensitivity. J Modern Oncol. 2016;24(2):314‐318. [Google Scholar]
- 27. Tokuda E, Horimoto Y, Arakawa A, et al. Differences in ki67 expressions between pre‐ and post‐neoadjuvant chemotherapy specimens might predict early recurrence of breast cancer. Hum Pathol. 2017;63:40‐45. [DOI] [PubMed] [Google Scholar]
- 28. Babar IA, Cheng CJ, Booth CJ, et al. Nanoparticle‐based therapy in an in vivo microRNA‐155 (miR‐155)‐dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109(26):10140‐10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Girardi C, De Pitta C, Casara S, et al. Analysis of miRNA and mRNA expression profiles highlights alterations in ionizing radiation response of human lymphocytes under modeled microgravity. PLOS One. 2012;7(2):723‐727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Babar IA, Czochor J, Steinmetz A, Weidhaas JB, Glazer PM, Slack FJ. Inhibition of hypoxia‐induced miR‐155 radiosensitizes hypoxic lung cancer cells. Cancer Biol Ther. 2011;12(10):908‐914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that all the data supporting the findings of this study are available within the article and from the corresponding author on reasonable request.


