Abstract
The impact of oral commensal and pathogenic bacteria on peri‐implant mucosa is not well understood, despite the high prevalence of peri‐implant infections. Hence, we investigated responses of the peri‐implant mucosa to Streptococcus oralis or Aggregatibacter actinomycetemcomitans biofilms using a novel in vitro peri‐implant mucosa‐biofilm model. Our 3D model combined three components, organotypic oral mucosa, implant material, and oral biofilm, with structural assembly close to native situation. S. oralis induced a protective stress response in the peri‐implant mucosa through upregulation of heat shock protein (HSP70) genes. Attenuated inflammatory response was indicated by reduced cytokine levels of interleukin‐6 (IL‐6), interleukin‐8 (CXCL8), and monocyte chemoattractant protein‐1 (CCL2). The inflammatory balance was preserved through increased levels of tumor necrosis factor‐alpha (TNF‐α). A. actinomycetemcomitans induced downregulation of genes important for cell survival and host inflammatory response. The reduced cytokine levels of chemokine ligand 1 (CXCL1), CXCL8, and CCL2 also indicated a diminished inflammatory response. The induced immune balance by S. oralis may support oral health, whereas the reduced inflammatory response to A. actinomycetemcomitans may provide colonisation advantage and facilitate later tissue invasion. The comprehensive characterisation of peri‐implant mucosa‐biofilm interactions using our 3D model can provide new knowledge to improve strategies for prevention and therapy of peri‐implant disease.
Keywords: Aggregatibacter actinomycetemcomitans, dental implants, host modulation, organotypic oral mucosa, Streptococcus oralis
1. INTRODUCTION
Microorganisms are able to colonize oral surfaces, regardless of whether it is natural, for example, tooth enamel or mucosa, or artificial, for example, titanium implant, and form complex biofilms (G. N. Belibasakis, Charalampakis, Bostanci, & Stadlinger, 2015; Furst, Salvi, Lang, & Persson, 2007; Kolenbrander, Palmer, Periasamy, & Jakubovics, 2010). Different factors (immunodeficiency, systemic disease, environmental factors, and keystone pathogens) can induce a shift in species composition of oral biofilms incorporating more pathogenic bacteria (Graves, Correa, & Silva, 2019; G. Hajishengallis, 2014; G. Hajishengallis & Lamont, 2016). As a result, in people carrying dental implants, peri‐implant diseases might develop (G. N. Belibasakis, 2014; Berglundh et al., 2018). The reversible inflammation of the soft tissue around the implant is termed “peri‐implant mucositis.” The more severe form, which is termed peri‐implantitis, is irreversible and additionally characterised by loss of bone supporting the implant (Berglundh et al., 2018). Moreover, peri‐implant diseases are characterised by high prevalence. A recent meta‐analysis showed that 26% of patients with an implant function ≥5 years develop peri‐implantitis (Dreyer et al., 2018). One reason could be that dental implants are missing Sharpey's fibres and the periodontal ligament leading to a reduced physical barrier of the oral mucosa against bacterial invasion (G. N. Belibasakis, 2014). In order to expand the knowledge about the interaction of the peri‐implant mucosa and oral microbiome, physiologically relevant in vitro models are required. The in vivo situation is much better reflected in three dimensional (3D) organotypic models (Antoni, Burckel, Josset, & Noel, 2015). The co‐culture of organotypic oral mucosa models with planktonic bacteria, monospecies biofilms, or even multispecies biofilms facilitated in vitro studies, which explored the impact of host–microbe interactions (T. Ahlstrand et al., 2017; Andrian, Grenier, & Rouabhia, 2004; Bao, Papadimitropoulos, Akgul, Belibasakis, & Bostanci, 2015; Buskermolen et al., 2016; Diaz et al., 2012; Gursoy, Pollanen, Kononen, & Uitto, 2010; Pinnock, Murdoch, Moharamzadeh, Whawell, & Douglas, 2014). In order to study the soft‐tissue‐implant interface, Chai et al. developed an organotypic oral mucosa with an integrated implant. However, their model did not include an oral biofilm, which is a key element of the peri‐implant area (Chai et al., 2010; Chai et al., 2013; Chai, Brook, Palmquist, van Noort, & Moharamzadeh, 2012). To the best of our knowledge, an in vitro model to study the interactions between all three components, implant material, organotypic oral mucosa, and biofilm, is absent.
Balanced immune response maintains the host‐microbe homeostasis and confers oral health. The oral health‐associated symbiotic microbial community consists mainly of gram‐positive Streptococcus spp. and Actinomyces spp., and dozens of less studied species are present (G. Hajishengallis, 2015; Mombelli, Müller, & Cionca, 2012; Szafranski et al., 2015). The commensal Streptococcus oralis belongs to the initial colonizer and is one of the predominant Streptococcus spp. in the early biofilm (Diaz et al., 2006) and consequently should have a considerable impact on oral homeostasis. However, little is known about the mechanisms by which S. oralis interacts with the host. This knowledge would help to elucidate the role of this microbe in host‐microbiome homeostasis beyond biofilm initiation. The opportunistic pathogen Aggregatibacter actinomycetemcomitans is genetically diverse (Kittichotirat, Bumgarner, & Chen, 2016) and can be detected at peri‐implant disease sites (Rams, Degener, & van Winkelhoff, 2014; van Winkelhoff & Wolf, 2000). It expresses various virulence factors and has different strategies to evade host innate defence mechanisms, for example, migration through the epithelium, and binding of different human proinflammatory cytokines (T. Ahlstrand et al., 2017; Dickinson et al., 2011; Herbert, Novince, & Kirkwood, 2016). However, the overall impact including transcriptional response of A. actinomycetemcomitans on the oral mucosa remains unclear. Deciphering of how commensal and pathogenic bacteria, that is, S. oralis and A. actinomycetemcomitans, impact mucosal homeostasis would help to understand peri‐implant pathogenesis and to develop new therapeutic options.
The first aim of the present study was to develop an in vitro peri‐implant mucosa‐biofilm model combining the main three components: the organotypic oral mucosa, an implant material, and an oral biofilm (Figure 1). The second aim was to expand the knowledge about the species‐specific effect of commensals and opportunistic pathogens on the mucosal tissue, by studying the impact of either S. oralis or A. actinomycetemcomitans biofilms on the peri‐implant mucosa in our unique organotypic model.
Figure 1.
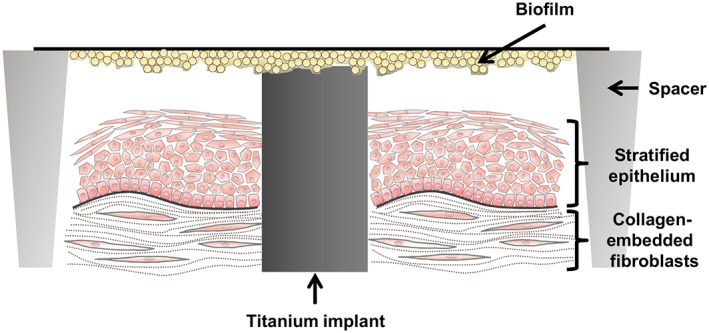
Schematic illustration of the peri‐implant mucosa‐biofilm model. The organotypic oral mucosa with an integrated implant was developed in culture inserts. Spacers with a ring form were placed around the tissue model, which allowed the disposition of the Streptococcus oralis or Aggregatibacter actinomycetemcomitans biofilm on top of the implant. Spacers and implant material have the same height keeping the biofilm planar
2. RESULTS
2.1. Characterisation of the peri‐implant mucosa model
The assembly of the three‐dimensional peri‐implant mucosa models had duration of 25 days. Briefly, a titanium disk (implant material) was integrated into collagen‐embedded human gingival fibroblasts (HGFs). On the top of the fibroblast‐collagen gel, oral keratinocytes (OKF6/TERT‐2) were added around the titanium and differentiated. The morphology of the peri‐implant mucosa model was evaluated by van Gieson staining and immunohistochemistry in order to confirm that the mucosal structure reflected the previously published engineered human oral mucosa (Dongari‐Bagtzoglou & Kashleva, 2006). The organotypic oral mucosa consisted of a differentiated stratified epithelium and the underlying connective tissue, including the HGF. Four different layers of the epithelium, the stratum basale, stratum spinosum, stratum granulosum, and the superficial keratinised layer were seen to be similar to native human gingival tissue (Figure S1A). The suprabasal epithelial layer was stained by cytokeratin 13, the basement membrane by collagen IV, and the keratinized superficial cells by cytokeratin 10 (Figure S1B–D). Sporadic proliferating cells were also detected—mainly at the basal layer—through Ki67 staining (Figure S1E). E‐cadherin and claudin staining confirmed the tight epithelial barrier (Figure S1F–G).
The organotypic oral mucosa was attached to the implant and created an intact implant‐mucosa interface (Figure 2). If titanium disks free of fibroblasts were inserted, the epithelial cells grew apically along the titanium disk, deep into the collagen. This apical epithelial migration created an elongated junctional epithelium covering a considerable area of the implant surface (Figure 2a–c). In contrast, the use of fibroblast‐colonized titanium disks hindered such deep epithelial cell migration into the collagen (Figure 2d–f). The staining at the upper part of the implant is related to the fibroblasts, which grew around it prior to insertion into the tissue. The peri‐implant mucosa model with a fibroblast‐colonized titanium disk built an intact mucosa‐implant interface, with only minimal apical epithelial migration along the titanium disk and was used for the following co‐cultures.
Figure 2.
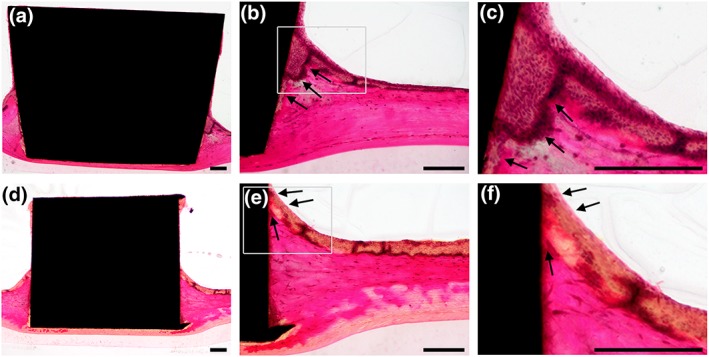
Histological sections of the peri‐implant mucosa model. The inserted implant was integrated into the organotypic mucosa and was non‐colonized or colonized with fibroblasts. Non‐colonized integrated implant: (a) overview of the mucosa‐implant interface, (b), (c) right site at higher magnifications. Fibroblast‐colonized integrated implant: (d) overview of the mucosa‐implant interface, (e), (f) right site at higher magnifications. Arrows indicate epithelial layer growth at the implant‐mucosa interface. The ground sections were stained according to van Gieson. Representative pictures of three independent experiments. Scale bars: 200 μm
2.2. S. oralis and A. actinomycetemcomitans biofilms formation
The developed peri‐implant mucosa should be challenged with either S. oralis or A. actinomycetemcomitans cells grown as biofilms. Therefore, reproducible and viable sessile communities of these two species were required. Three incubation times were tested for S. oralis biofilms: 48, 72, and 96 hr. The biofilm volume, determined by live/dead staining and confocal scanning laser microscopy (CLSM), increased over time, with the maximum detected after 96 hr (Figure 3a). However, at this time point, the proportion of dead bacteria was the highest (Figure 3b). To balance the biofilm volume and viability, we chose the 72‐hr S. oralis biofilm (Figure 3e) for the following co‐culture experiments. A viable and thick biofilm of A. actinomycetemcomitans was formed on the supporting material after 24 hr of culture (Figure 3c–d). Both cell morphotypes, fimbriated, and non‐fimbriated (corresponding to the rough and smooth colony morphotypes, respectively) were visible in the A. actinomycetemcomitans biofilm (Figure 3f).
Figure 3.
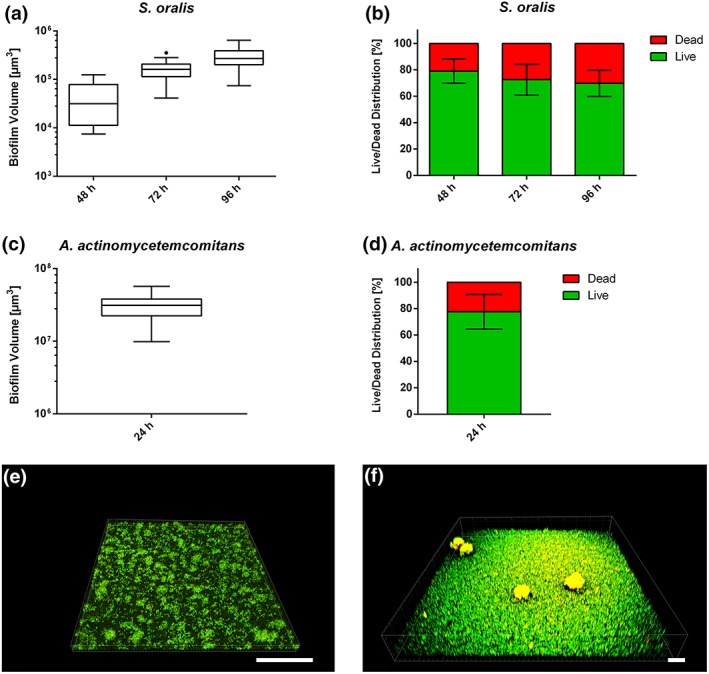
Biofilm formation on supporting material. (a) The Box and Whiskers graphs with Tukey error bars show the total biofilm volumes for Streptococcus oralis after 48, 72, or 96 hr of culture. (b) The bar graphs show the percentages of live and dead bacteria within the S. oralis biofilm. Data of three independent experiments were used for the S. oralis biofilm volume and live/dead distribution. (c) The Box and Whiskers graph with Tukey error bars shows the total biofilm volume for Aggregatibacter actinomycetemcomitans. (d) The bar graph shows the percentages of live and dead bacteria within the A. actinomycetemcomitans biofilm. Data of two independent experiments were used for the A. actinomycetemcomitans biofilm volume and live/dead distribution. (e) Representative 3D image of three independent experiments demonstrating the S. oralis biofilm after 72 hr culture. (f) Representative 3D image of two independent experiments demonstrating the A. actinomycetemcomitans biofilm after 24 hr culture. Live bacteria are depicted in green and dead in red. Scale bars: 100 μm
2.3. Histology of the peri‐implant mucosa after biofilm challenge
Peri‐implant mucosa was exposed to either S. oralis or A. actinomycetemcomitans biofilm for 24 hr, and the effect was evaluated with histological analysis. Co‐cultures with the biofilms resulted in an intact implant‐mucosa interface (Figure 4d,g). The epithelium located directly at the implant was slightly loosened after challenge with the S. oralis biofilm (Figure 4d,e). In contrast, the epithelium at a distance from the implant was histologically similar (Figure 4f) to the control tissue (Figure 4a–c). Thus, the effect of the S. oralis biofilm was restricted to the implant‐mucosa interface. Exposure to the A. actinomycetemcomitans biofilm (Figure 4g–i) had no visible histological effect on the mucosa. Immunohistological staining for adherent junctions (E‐cadherin) and proinflammatory factors (IL‐6, CXCL8, and TNF‐α) was similar for the control and for a tissue exposed to the S. oralis biofilm. However, the intensity of claudin staining for tight junctions appeared slightly diminished after co‐culture with the S. oralis biofilm (Figure S2).
Figure 4.
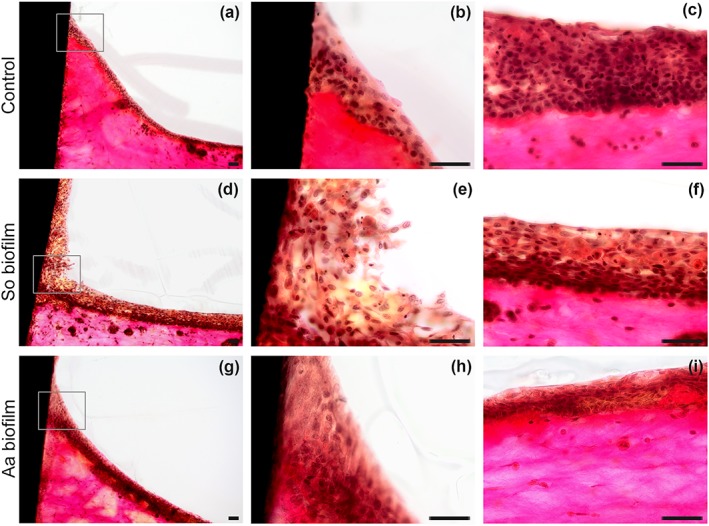
Histological sections of the peri‐implant mucosa‐biofilm model after 24 hr. An overview of the implant‐mucosa interface is shown for the control (a), Streptococcus oralis (d) and Aggregatibacter actinomycetemcomitans (g) biofilm challenged groups. An intact implant‐mucosa interface was observed in the control (b) at higher magnification. The epithelium at the implant site was slightly loosed, after S. oralis biofilm challenge (e), whereas an intact implant‐mucosa interface was observed after co‐culture with the A. actinomycetemcomitans biofilm (h). The adjacent tissues of the control (c), S. oralis (f) and A. actinomycetemcomitans (i) biofilm challenged group were intact. The ground sections were stained according to van Gieson. Representative pictures of three independent experiments. So = S. oralis and Aa = A. actinomycetemcomitans. Scale bars: 50 μm
2.4. Transcriptional response of the peri‐implant mucosa to biofilms
Transcriptional activity of the mucosa was measured by microarrays after 24 hr exposure to biofilms. After co‐culture with the S. oralis biofilm, 83 genes were differentially expressed in the peri‐implant mucosa compared with the unexposed tissue. Thirty six genes were upregulated whereas 47 genes were downregulated (Figure 5a). Most of the upregulated genes belonged to the heat‐shock proteins 70 (HSP70). These genes are involved in mitogen‐activated protein kinase signalling and antigen processing and presentation pathways (Table 1). In addition, some genes from the chemokine signalling pathway (i.e., CCL20, CCL8, and PIK3R5) were upregulated. Genes coding for the invariant alpha chain HLA‐DRA of the major histocompatibility complex Class II were downregulated (Table 2). Major histocompatibility complex Class II can be induced by IFN‐γ and is involved in antigen processing and presentation. Challenge of the peri‐implant mucosa with the A. actinomycetemcomitans biofilm led to regulation of 101 genes: 32 upregulated and 69 downregulated (Figure 5b). Upregulated genes were related either to cell division (FIGN, HMGA2, CDC25A, and ERCC6L) or to DNA repair/damage (CLSPN, POLQ, and FANCA; Table S3). No particular pathway was upregulated. The pathway analysis of the downregulated genes revealed the PI3K‐Akt signalling pathway (Table 3), including genes related to this signal transduction (MDM2, IL2RG, TLR4, and F2R).
Figure 5.
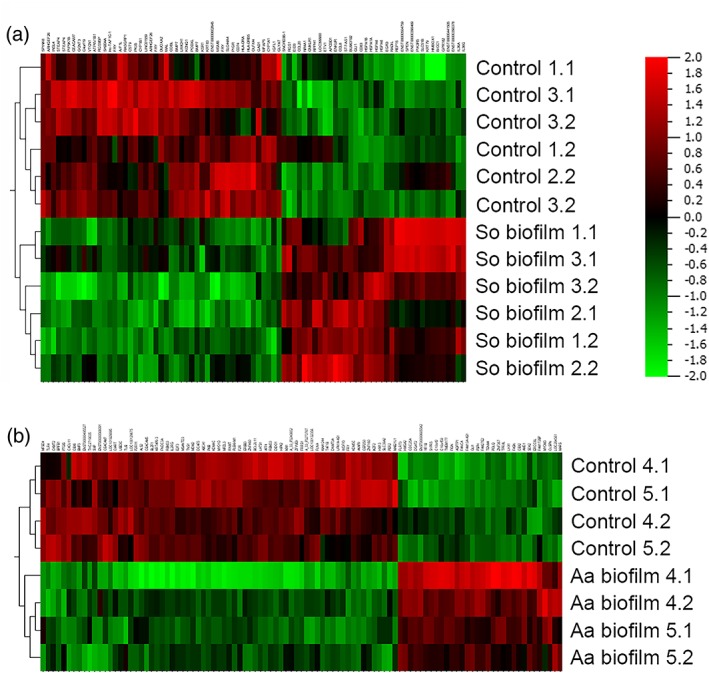
Heat maps of the global gene expression profiles from the peri‐implant mucosa comparing the control and biofilm challenged groups after 24 hr. Results for the Streptococcus oralis (a) and the Aggregatibacter actinomycetemcomitans (b) biofilm are shown in separate heat maps. The heat maps show the hierarchical clustering of the experimental groups and the differentially regulated genes. Data from two‐three independent experiments and duplicates were used. Red indicates upregulation and green downregulation after biofilm co‐culture. So = S. oralis and Aa = A. actinomycetemcomitans
Table 1.
Enriched and relevant pathways of upregulated genes in the peri‐implant mucosa‐biofilm model after 24‐hr Streptococcus oralis biofilm challenge
| Pathway | % | P | Genes |
|---|---|---|---|
| Antigen processing and presentation | 10 | .006212 | HSPA6, HSPA1A, HSPA1B |
| MAPK signalling pathway | 13.33333 | .006514 | HSPA6, NR4A1, HSPA1A, HSPA1B |
| Spliceosome | 10 | .018203 | HSPA6, HSPA1A, HSPA1B |
| Protein processing in endoplasmic reticulum | 10 | .028529 | HSPA6, HSPA1A, HSPA1B |
| Chemokine signalling pathway | 10 | .034067 | CCL20, CCL8, PIK3R5 |
Table 2.
Enriched and relevant pathways of downregulated genes in the peri‐implant mucosa‐biofilm model after 24‐hr Streptococcus oralis biofilm challenge
| Pathway | % | P | Genes |
|---|---|---|---|
| Intestinal immune network for IgA production | 7.5 | .006521 | HLA‐DRB5, PIGR, HLA‐DRA |
| Antigen processing and presentation | 7.5 | .016438 | HLA‐DRB5, CD74, HLA‐DRA |
| Cell adhesion molecules (CAMs) | 7.5 | .052171 | VTCN1, HLA‐DRB5, HLA‐DRA |
| Phagosome | 7.5 | .057529 | HLA‐DRB5, ATP6V1B1, HLA‐DRA |
| Tryptophan metabolism | 5 | .099767 | CYP1B1, CYP1A1 |
Table 3.
Enrichment and relevant pathways of downregulated genes in the peri‐implant mucosa‐biofilm model after 24‐hr Aggregatibacter actinomycetemcomitans biofilm challenge
| Pathway | % | P | Genes |
|---|---|---|---|
| PI3K‐Akt signalling pathway | 5.434783 | .060915 | MDM2, IL2RG, TLR4, BCL2L11, F2R |
2.5. Cytokine secretion
The cytokine levels in the collected supernatants were measured by using a Luminex‐based multiplex assay. The results showed that S. oralis biofilm challenge caused a significant increase in TNF‐α level in the peri‐implant mucosa compared with the unchallenged tissue (Figure 6). In contrast, the levels of IL‐6, CXCL8, and CCL2 were significantly reduced after stimulation with the S. oralis biofilm. Challenge of the peri‐implant mucosa with the A. actinomycetemcomitans biofilm led to significant lower levels of CXCL1, CXCL8, and CCL2 (Figure 6). The secretion level of CXCL2 was not affected by any of the studied biofilms.
Figure 6.
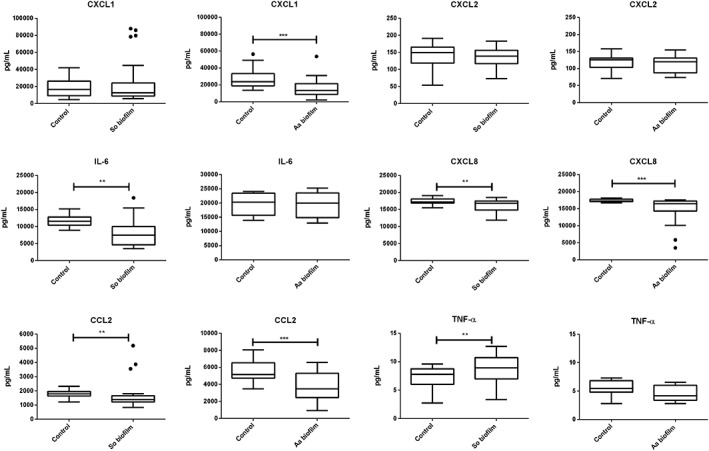
Cytokine levels in the co‐culture supernatants of the peri‐implant mucosa‐biofilm model after 24 hr. The groups challenged with the Streptococcus oralis or Aggregatibacter actinomycetemcomitans biofilm were compared with the control groups for their CXCL1, CXCL2, IL‐6, CXCL8, CCL2, and TNF‐α levels. The cytokines were measured using the luminex technology and a Bio‐Plex Kit. The Box and Whiskers graphs with Tukey error bars represent the measured data points. The S. oralis biofilm group includes 42 measured data points from 14 samples and four independent experiments. The A. actinomycetemcomitans biofilm group includes 29 measured data points from seven samples and two independent experiments. So = S. oralis and Aa = A. actinomycetemcomitans. The statistical significance was determined using the Mann–Whitney method, with P = .05. Single asterisk indicates P < .05 and double asterisk P ≤ .01
3. DISCUSSION
Peri‐implantitis is a disease with high prevalence (Dreyer et al., 2018). Development of a successful prevention or therapy strategy requires comprehensive understanding of the host–microbe interactions at the peri‐implant site. Here, by applying an organotypic model, we investigated the impact of either commensal S. oralis or the pathogenic A. actinomycetemcomitans biofilms on the human mucosa. An in vitro model, which reflects well the clinical situation including the three main compartments, human soft‐tissue, biofilm, and implant material, has been missing until now, and we addressed this problem with our peri‐implant mucosa‐biofilm model (Figure 1). The colonization of the implants with HGF hindered the intensive apical epithelial migration along the implant and thereby the elongated junctional epithelium covering most of the implant surface (Figure 2a–c). As a result, we obtained an organotypic oral mucosa attached to the implant with minimal apical epithelial growth (Figure 2d–f). Intensive apical epithelial migration is a characteristic during severe oral inflammation (Pollanen, Laine, Ihalin, & Uitto, 2012). The resulting implant‐mucosa interface reflected the in vivo histology and physiology of the healthy mucosa attached to the implant (Atsuta et al., 2005; Schupbach & Glauser, 2007).
The S. oralis biofilm led to a slight tissue loosening, which was restricted to the mucosa‐implant interface (Figure 4d,e). Claudin expression seemed to be less after co‐culture with S. oralis biofilm (Figure S2). Bacterial stimulation modified tight junctions in lungs, without major disruption of the epithelial barrier, and this correlated with transmigration of polymorphonuclear neutrophils (PMNs; Chun & Prince, 2009). Tissue loosing might facilitate a fast transmigration of PMNs to tissue‐biofilm interface and build a barrier against microbial invasion controlling the bacterial load (Darveau, 2009; G. Hajishengallis & Lamont, 2016). Our model could benefit from future implementation of PMNs to study their transmigration and bacterial clearance. In contrast to S. oralis, the exposure to A. actinomycetemcomitans biofilm did not disturb the mucosa (Figure 4g–i), which was also observed by others (T. Ahlstrand et al., 2017; A. Paino et al., 2012). Probably the S. oralis biofilm supports the immune cell migration through tissue loosening, in contrast to the A. actinomycetemcomitans biofilm that has no impact on the tissue structure.
Transcriptional analysis revealed a broader gene response to the A. actinomycetemcomitans biofilm compared with the S. oralis biofilm (Figure 5). Hasegawa et al. compared the transcriptional response of keratinocytes with commensals or opportunistic pathogens and detected similar differences as reported here (Handfield et al., 2005; Hasegawa et al., 2007). The weak transcriptional response to commensal bacteria supports the adaptive coevolution theory of commensal bacteria with the oral mucosa (Handfield, Baker, & Lamont, 2008; Hooper & Gordon, 2001). The overall response to S. oralis at the transcriptional level was related to protective response. Pathways related to tissue protection were upregulated (Table 1) including CCL20 and genes grouped in HSP70, with functions in mucosal homeostasis (Comerford et al., 2010; Pleguezuelos, Dainty, Kapas, & Taylor, 2005; Schutyser, Struyf, & Van Damme, 2003). In addition, the adaptive immune response was suppressed as indicated by the downregulation of antigen presentation and processing (Table 2). These might lead to a state of unresponsiveness—with decreased both humoral and cell‐mediated immune response (Han et al., 2003; Hasegawa et al., 2007). Hyporesponsiveness induced by commensals probably plays a role in protection from tissue destruction induced by inflammatory response (Pollanen et al., 2012). Compared with the S. oralis biofilm, transcriptional response to A. actinomycetemcomitans was broader without targeting pathways. Upregulated genes were related to DNA damage, DNA repair, and cell division suggesting general stress response. Analysis of the downregulated genes revealed a single enriched pathway: the PI3K‐Akt signalling (Table 3). Attenuation of this pathway by P. gingivalis can promote its invasion and colonisation of the mucosal tissue (Nakayama, Inoue, Naito, Nakayama, & Ohara, 2015). Similarly, our observed changes may promote colonization and survival of A. actinomycetemcomitans. In summary, the transcriptional profiles of the peri‐implant mucosa revealed a tissue protective response to the S. oralis biofilm and a stress response to the A. actinomycetemcomitans biofilm.
The classical proinflammatory cytokine IL‐6 and the neutrophil recruiting chemokines CXCL8 and CCL2 were found at lower levels in the supernatants after challenge with the S. oralis biofilm (Figure 6). Corresponding to our results, different studies could show that commensal bacteria reduce the proinflammatory cytokines, IL‐6 and CXCL8 (Cosseau et al., 2008; Hasegawa et al., 2007; Twetman et al., 2009; Zhang, Chen, & Rudney, 2008). Therefore, S. oralis biofilm might attenuate the proinflammatory response, which is consistent with our observations on gene expression. TNF‐α was increased in response to the S. oralis biofilm (Figure 6). It is one of the main inflammation mediators (Groeger & Meyle, 2015) and is present at low levels in the gingival crevicular fluid in healthy patients (Darveau, 2010; Petkovic‐Curcin, Matic, Vojvodic, Stamatovic, & Todorovic, 2011). Probably, cytokines controlled by commensal bacteria are involved in limiting biofilm development and consequently in maintaining gingival health (Darveau, 2010; Dickinson et al., 2011; Rouabhia, 2002). After challenge with the A. actinomycetemcomitans biofilm, the levels of CXCL1, CXCL8, and CCL2 were lower than in the control (Figure 6). Previously, it was found that A. actinomycetemcomitans can sense and bind cytokines; among them was CXCL8 (T. Ahlstrand et al., 2017; T. Ahlstrand et al., 2018; T. Ahlstrand, Kovesjoki, Maula, Oscarsson, & Ihalin, 2019). Lower metabolic activity of biofilms induced by cytokine binding could lead to higher resistance (A. Paino et al., 2011) and could reduce the production of virulence factors providing an explanation why the peri‐implant mucosa was not impaired after challenge with the A. actinomycetemcomitans biofilm. The lower levels of chemotactic cytokines may reduce immune cell recruitment leading to a colonization advantage by A. actinomycetemcomitans. Interleukin depletion may act in concert with virulence factors, like adhesion and toxins, and compromise tissue integrity at later infection (Henderson, Ward, & Ready, 2010; Szafranski et al., 2017). In summary, the S. oralis biofilm attenuated the proinflammatory response of the peri‐implant mucosa; nevertheless, basic awareness was maintained through increased TNF‐α level. On the contrary, A. actinomycetemcomitans diminished proinflammatory response creating a colonization advantage and potentially facilitates biofilm expansion.
The in vitro peri‐implant mucosa‐biofilm model reflected the local response of the host to the biofilms. The in vivo host–microbe interactions include also immune cells (Pollanen et al., 2012), which were not present in our model. However, our results are in line with in vivo studies, which showed that commensals and pathogens alone do not induce inflammation, in contrast to their co‐infections (Diaz et al., 2012; Ramsey & Whiteley, 2009; Whitmore & Lamont, 2011; Xu et al., 2014) suggesting that microbial synergy plays an important role in the pathogenesis. Within the limitations of our study, responses on transcription and cytokine levels were uncovered that may explain why monospecies commensal and pathogenic biofilms do not cause inflammatory response. Noteworthy, the reactions from the peri‐implant mucosa to these monospecies biofilms were in accordance with previous in vivo observations.
In conclusion, our novel peri‐implant mucosa‐biofilm model promises enormous experimental potential to investigate the interaction of three key components: mucosa, biofilm, and implant. Our 3D model reflected that commensal streptococci induce a balanced immune response of the soft tissue including specific transcriptional response and attenuated pro‐inflammatory cytokines. This subtle effect could preserve the oral health. Furthermore, the colonization advantage of opportunistic pathogens by suppression of inflammatory reaction could favour dysbiosis. We showed that species‐specific molecular reactions of the peri‐implant mucosa to biofilm can be successfully studied in our peri‐implant mucosa‐biofilm model.
Our model permits future investigations of health‐related or dysbiotic multispecies biofilms as well as phages, bacterial viruses (Preus, Olsen, & Namork, 1987; Szafranski, Winkel, & Stiesch, 2017). The influence of various implant materials and surface functionalisation on biofilm formation and tissue reaction are additional factors that will be analyzed in the future. Accordingly, the findings will provide new opportunities for future strategies of disease prevention and treatment as well as for implant improvement.
4. EXPERIMENTAL PROCEDURES
4.1. Cell culture
HGFs (121 0412, Provitro GmbH) were cultured in Dulbecco's modified Eagle's medium (DMEM, FG0435, Biochrom AG), supplemented with 10% fetal bovine serum (FBS, P30‐3309, PAN‐Biotech GmbH), 100‐U/ml penicillin, and 100‐μg/ml streptomycin (A2212; Biochrom AG). The immortalized human oral keratinocyte cell line (OKF6/TERT‐2; Dickson et al., 2000) was cultured in KerSFM medium (10725‐018, Gibco Lifetechnologies), supplemented with 0.3‐mM CaCl2, 0.2‐mg/ml EGF, 25‐μg/ml BPE, 100‐U/ml penicillin, and 100‐μg/ml streptomycin. The cells were incubated at 37°C in a 5% CO2 humidified atmosphere.
4.2. Peri‐implant mucosa model
The peri‐implant oral mucosa model assembly was based on the protocol of Dongari‐Bagtzoglou and Kashleva (Dongari‐Bagtzoglou & Kashleva, 2006) and reproduced with slight modifications. Briefly, bovine type I collagen (2‐mg/ml PureCol®, 5005‐100ML, Advance Biomatrix) was mixed with FBS, L‐glutamine (G7513, Sigma‐Aldrich), 10 x DMEM (P03–01510, Pan‐Biotech), and a DMEM reconstitution buffer (2.2‐mg/ml sodium bicarbonate, 2‐mM HEPES, and 0.0062 N NaOH in DMEM P03‐01510). HGFs (passage 9 or 10) were then added to the collagen mixture and poured into culture inserts (PIHA 03050, Merck Millipore or 3414, Corning B.V. Life Sciences). Each model contained 4 × 105 HGFs in the collagen. Titanium disks (3 mm diameter, 2.3 mm height, Grade 4, machined surface) were colonized with HGF (1 × 106 cells/ml). Both were cultivated and submerged in fully supplemented DMEM at 37°C in a humidified atmosphere with 5% CO2. At day 5, a titanium disk with or without HGF colonization was integrated. For this purpose, the models were punched with a 2.5 mm diameter biopsy punch. The titanium disks were placed in the resulting holes. After 3 days, 1.2 × 106 oral keratinocytes (OKF6/TERT‐2, Passages 19–26) were seeded on the top of each fibroblast‐collagen gel. At day 12, the models were raised to an air‐liquid interface and cultivated for 13 days with a specific Airlift (AL) medium (3:1 DMEM [P04‐03591, Pan‐Biotech] and Ham's F‐12 [P04‐14559, Pan‐Biotech], 5‐μg/ml insulin, 0.4‐μg/ml hydrocortisone, 2 × 10−11 M 5‐triiodo‐L‐thyronine, 1.8 × 10−5 M adenine, 5‐μg/ml transferrin, 10−10 M cholera toxin, 2 mM L‐glutamine, 10% v/v FBS, 1% v/v penicillin/streptomycin).
4.3. Biofilm formation and quantification
S. oralis (DSM 20627, German collection of microorganisms and tissue culture cells) was cultivated in tryptone soya broth supplemented with 10% yeast extract (TSBY) for 18 hr under stirring at 37°C in an anaerobic atmosphere. For biofilm formation, S. oralis was diluted in Brain Heart Infusion (BHI) supplemented with 5% w/v sucrose to an optical density (600 nm) of 0.06, corresponding to 8.7 × 107 CFU/ml. The biofilm was cultured on a hydrophilic polyethersulfone membrane (GPWP04700, Merck Millipore) for 48, 72, or 96 hr at 37°C in a humidified atmosphere with 5% CO2. The medium was replaced each day with fully supplemented BHI. A. actinomycetemcomitans JP2 strain (HK1651, CCUG 56173, Culture Collection, University of Göteborg) isolated from aggressive juvenile periodontitis was cultivated in Todd‐Hewitt broth supplemented with 10% yeast extract (THBY) for 72 hr at 37°C in a humidified atmosphere with 5% CO2. Smooth and rough colony morphotypes were isolated and cultured separately. For biofilm formation, the A. actinomycetemcomitans strain was diluted in THBY to an optical density (600 nm) of 0.6 for smooth and 0.05 for rough type, corresponding to 3.25 × 107 CFU/ml and 2.3 × 106 CFU/ml. The two A. actinomycetemcomitans morphotypes were mixed at equal volumes and cultured on glass coverslips (18 mm diameter, CBAD00180RA120MNZ#0, Thermo Scientific Menzel) for 24 hr at 37°C in a humidified atmosphere with 5% CO2, to build a biofilm.
The S. oralis or A. actinomycetemcomitans biofilms, which were formed on the supporting material, were stained with the LIVE/DEAD®BacLight™ Bacterial Viability Kit (L7012, Life Technologies). After phosphate buffered saline (PBS) wash, the biofilms were fluorescently stained with SYTO9 and propidium iodide as a 1:1,000 dilution in PBS for 30 min. The biofilms were washed with PBS and subsequently fixed in 2.5% glutardialdehyde in PBS. The stained S. oralis biofilms were imaged at 40‐fold magnification through CLSM (Leica TCS SP2). Three random positions were scanned by creating z‐plane images. The stained A. actinomycetemcomitans biofilms were imaged at 10‐fold magnification through CLSM (Leica TCS SP8). Five random positions were scanned by creating z‐plane images. For both biofilms, 3D images were reconstructed by the Imaris® × 64 6.2.1 software package (Bitplane) and used to calculate the volume in the surpass mode. Finally, the percentages of live and dead cells were calculated.
4.4. Co‐culture of the peri‐implant mucosa with the biofilms
The co‐cultures were conducted in AL‐medium without any antibiotics. The peri‐implant oral mucosa model was used and washed with PBS prior to co‐culture. Either the 72‐hr‐old S. oralis or A. actinomycetemcomitans biofilm was washed five times with PBS and was placed on spacers with the biofilm side facing the peri‐implant oral mucosa model with direct contact to the integrated titanium disk (Figure 1). The co‐cultures were performed for 24 hr at 37°C in a 5% CO2 humidified atmosphere.
4.5. RNA extraction and microarray data analysis
The supernatants were collected after co‐culture for subsequent analysis of the secreted cytokines. The tissues were stored in RNAlater™ Solution (AM7020, Invitrogen) overnight at 4°C. Tissue RNA was isolated according to the manufacturer's protocol using the RNeasy® Mini Kit (74104, Qiagen). Briefly, the tissues were lysed with a microcentrifuge pestle in RLT buffer with 1% v/v β‐mercaptoethanol and vortexed. The lysates were homogenized using the QIAshredder (79654, Qiagen), and the RNA was isolated by using the RNeasy® Mini Kit (74104, Qiagen). The RNA was stored at −80°C for later analysis.
For the gene expression analysis, total RNA was reverse‐transcribed into double strand cDNA, and the Cy3‐labelled cRNA was synthesized by using the Quick Amp Labeling Kit, One Colour (Agilent) according to the manufacturer's instructions. The cRNA was purified with the RNeasy® Mini Kit (Qiagen). For cRNA fragmentation, hybridization, and washing, the One‐Colour Microarray‐Based Gene Expression Analysis Protocol V5.7 (Agilent) was used. The Cy3‐labelled cRNA (2,500 ng) was hybridized on the refined 026652QM_RCUG_HomoSapiens microarray (34,127 genes), which was developed at the Research Core Unit Genomics of the Hannover Medical School, for 17 hr at 65°C. The Agilent Micro Array Scanner G2565CA was used for scanning the slides. The raw data were extracted with the Feature Extraction Software V10.7.3.1 (pixel resolution 3 μm, bit depth 20) and imported into Qlucore Omics Explorer software under default import settings for Agilent One‐Colour Microarray for further transcriptomic analysis. Biofilm‐challenged and control tissues were compared with Student's t test under the conditions of log2 ratio > 2 and P < .01. The false discovery rate for tissues challenged with S. oralis and A. actinomycetemcomitans biofilm were q = 0.009321 and q = 0.408989, respectively.
Pathway analysis was performed by DAVID (Database for Annotation, Visualisation and Integrated Discovery; Huang da, Huang, Sherman, & Lempicki, 2009a; Huang da, Huang, Sherman, & Lempicki, 2009b) using default settings for the upregulated and downregulated gene lists (Table S1 and S2) after biofilm challenge, which were analyzed separately.
4.6. Detection of cytokines in the supernatant
Cytokine (CXCL1, CXCL2, IL‐6, CXCL8, CCL2, and TNF‐α) quantification was performed using a customized all in one Bio‐Plex Pro Human Chemokine 6plx EXP kit (17002259, Bio‐Rad). The cytokines in the collected supernatants were measured by the Luminex‐based multiplex technique according to the manufacturer's instructions (Bio‐Rad). The concentrations were calculated with Bio‐Plex Manager 6.0 by comparison with the standard curves. The detection sensitivity ranged between 1 pg and 40 μg of protein per 1 ml.
4.7. Histological analysis
The peri‐implant oral mucosa models were fixed in a 4% buffered formalin solution for 24 hr. The samples were watered, dehydrated by using an ethanol gradient, and embedded in Technovit 9100. The embedded samples were either cut into 5‐μm slides for implant‐free sections or were grinded to 22–36‐μm slides for the peri‐implant ground sections according to the cutting‐grinding technique by Donath K. Prior staining, the Technovit 9100 was removed by rinsing the slides in acetone. Afterwards, the slides were rehydrated by using an ethanol gradient. Finally, the slides were stained according to van Gieson or specific antibodies. For van Gieson staining, the slides were rinsed for 10 min in ferric haematoxylin, and then washed once with tap water and twice with hydrochloric acid alcohol. After rinsing in tap water for 10 min, they were added into the van Gieson solution for 3 min, subsequently washed with 96% ethanol, 100% ethanol, xylol, and finally mounted. For immunohistochemical staining, the slides were washed with distilled water, washing buffer, antigen retrieval buffer, and washing buffer. The slides were incubated overnight with the primary antibody. All primary antibodies were against human epitopes. The rabbit polyclonal anti‐claudin 1 (359‐14), mouse monoclonal anti‐collagen Type IV CIV22 (239 M‐15), rabbit monoclonal anti‐cytokeratin 10 EP97 (410R‐14), rabbit monoclonal anti‐cytokeratin 13 EP69 (AC‐0066A), and mouse monoclonal anti‐Ki67 MIB‐1 (ILM 9252 C01) were purchased from medac GmBH. Mouse monoclonal anti‐interleukin 6 (ABIN2469708) and mouse monoclonal anti‐interleukin 8 (ABIN1724413) were purchased from antibodies‐online GmbH. The slides were rinsed in wash buffer, peroxide block, and washing buffer prior to incubation with the corresponding secondary detection antibody. The secondary antibodies, Histofine Simple Stain MAX PO goat anti‐mouse Ig F (ab`)‐fragments (414131F) and Histofine Simple Stain MAX PO goat anti‐rabbit Ig F (ab`)‐fragments (414141F) were purchased from medac GmBH. The slides were then rinsed in washing buffer, incubated for 10 min with DAB (957D‐50) and washed with distilled water. They were counterstained in haematoxylin (Leica), rinsed in distilled water, dehydrated using an ethanol gradient, washed with xylol, and mounted. All slides were examined under the Olympus CX41 microscope.
4.8. Statistical analysis
All presented data were derived from two to three independent experiments.
Statistical evaluation of the cytokine levels was performed using GraphPad Prism 7. A Mann–Whitney test was used to analyze the statistical differences between the controls and biofilm groups. Differences were considered statistically significant at P < .05.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Data S1: Supplementary Information
ACKNOWLEDGEMENTS
Microarray data used or referred to in this publication were generated by the Research Core Unit Genomics at Hannover Medical School. Especially, we would like to thank Dr. Oliver Dittrich‐Breiholz and Heike Schneider for support and advice. The spacers for the co‐cultures were designed by Richard Werth. We would also like to thank Prof. Anna Dongari‐Bagtzoglou and Angela Thompson for their technical advice on engineered human oral mucosa development. Ground sections were performed at the HIK Hannover by Maren Hasper. The authors would like to thank Ronja Hagemeier and Charlotte Kreuzkamp for their technical assistance.
Ingendoh‐Tsakmakidis A, Mikolai C, Winkel A, et al. Commensal and pathogenic biofilms differently modulate peri‐implant oral mucosa in an organotypic model. Cellular Microbiology. 2019;21:e13078 10.1111/cmi.13078
Contributor Information
Alexandra Ingendoh‐Tsakmakidis, Email: ingendoh-tsakmakidis.alexandra@mh-hannover.de.
Carina Mikolai, Email: mikolai.carina@mh-hannover.de.
REFERENCES
- Ahlstrand, T. , Kovesjoki, L. , Maula, T. , Oscarsson, J. , & Ihalin, R. (2019). Aggregatibacter actinomycetemcomitans LPS binds human interleukin‐8. Journal of Oral Microbiology, 11(1), 1549931 10.1080/20002297.2018.1549931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlstrand, T. , Torittu, A. , Elovaara, H. , Välimaa, H. , Pöllänen, M. T. , Kasvandik, S. , … Ihalin, R. (2018). Interactions between the Aggregatibacter actinomycetemcomitans secretin HofQ and host cytokines indicate a link between natural competence and interleukin‐8 uptake. Virulence, 9(1), 1205–1223. 10.1080/21505594.2018.1499378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlstrand, T. , Tuominen, H. , Beklen, A. , Torittu, A. , Oscarsson, J. , Sormunen, R. , Pöllänen, M.T. , Permi, P. and Ihalin, R. (2017). A novel intrinsically disordered outer membrane lipoprotein of Aggregatibacter actinomycetemcomitans binds various cytokines and plays a role in biofilm response to interleukin‐1beta and interleukin‐8. Virulence , 8(2), 115‐134. https://10.1080/21505594.2016.1216294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrian, E. , Grenier, D. , & Rouabhia, M. (2004). In vitro models of tissue penetration and destruction by Porphyromonas gingivalis. Infection and Immunity, 72(8), 4689–4698. 10.1128/IAI.72.8.4689-4698.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni, D. , Burckel, H. , Josset, E. , & Noel, G. (2015). Three‐dimensional cell culture: a breakthrough in vivo. International Journal of Molecular Sciences, 16(3), 5517–5527. 10.3390/ijms16035517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsuta, I. , Yamaza, T. , Yoshinari, M. , Goto, T. , Kido, M. A. , Kagiya, T. , … Tanaka, T. (2005). Ultrastructural localization of laminin‐5 (gamma2 chain) in the rat peri‐implant oral mucosa around a titanium‐dental implant by immuno‐electron microscopy. Biomaterials, 26(32), 6280–6287. S0142‐9612(05)00298‐X [pii] [DOI] [PubMed] [Google Scholar]
- Bao, K. , Papadimitropoulos, A. , Akgul, B. , Belibasakis, G. N. , & Bostanci, N. (2015). Establishment of an oral infection model resembling the periodontal pocket in a perfusion bioreactor system. Virulence, 6(3), 265–273. 10.4161/21505594.2014.978721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belibasakis, G. N. (2014). Microbiological and immuno‐pathological aspects of peri‐implant diseases. Archives of Oral Biology, 59(1), 66–72. 10.1016/j.archoralbio.2013.09.013 [DOI] [PubMed] [Google Scholar]
- Belibasakis, G. N. , Charalampakis, G. , Bostanci, N. , & Stadlinger, B. (2015). Peri‐implant infections of oral biofilm etiology. Advances in Experimental Medicine and Biology, 830, 69–84. 10.1007/978-3-319-11038-7_4 [DOI] [PubMed] [Google Scholar]
- Berglundh, T. , Armitage, G. , Araujo, M. G. , Avila‐Ortiz, G. , Blanco, J. , Camargo, P. M. , … Zitzmann, N. (2018). Peri‐implant diseases and conditions: Consensus report of workgroup 4 of the 2017 World Workshop on the Classification of Periodontal and Peri‐Implant Diseases and Conditions. Journal of Clinical Periodontology, 45, S286–S291. 10.1111/jcpe.12957 [DOI] [PubMed] [Google Scholar]
- Buskermolen, J. K. , Reijnders, C. M. , Spiekstra, S. W. , Steinberg, T. , Kleverlaan, C. J. , Feilzer, A. J. , … Gibbs, S. (2016). Development of a full‐thickness human gingiva equivalent constructed from immortalized keratinocytes and fibroblasts. Tissue Engineering. Part C, Methods, 22(8), 781–791. 10.1089/ten.TEC.2016.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, W. L. , Brook, I. M. , Palmquist, A. , van Noort, R. , & Moharamzadeh, K. (2012). The biological seal of the implant‐soft tissue interface evaluated in a tissue‐engineered oral mucosal model. Journal of the Royal Society, Interface, 9(77), 3528–3538. 10.1098/rsif.2012.0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, W. L. , Moharamzadeh, K. , Brook, I. M. , Emanuelsson, L. , Palmquist, A. , & van Noort, R. (2010). Development of a novel model for the investigation of implant‐soft tissue interface. Journal of Periodontology, 81(8), 1187–1195. 10.1902/jop.2010.090648 [DOI] [PubMed] [Google Scholar]
- Chai, W. L. , Moharamzadeh, K. , van Noort, R. , Emanuelsson, L. , Palmquist, A. , & Brook, I. M. (2013). Contour analysis of an implant–soft tissue interface. Journal of Periodontal Research, 48(5), 663–670. 10.1111/jre.12062 [DOI] [PubMed] [Google Scholar]
- Chun, J. , & Prince, A. (2009). TLR2‐induced calpain cleavage of epithelial junctional proteins facilitates leukocyte transmigration. Cell Host & Microbe, 5(1), 47–58. 10.1016/j.chom.2008.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford, I. , Bunting, M. , Fenix, K. , Haylock‐Jacobs, S. , Litchfield, W. , Harata‐Lee, Y. , … McColl, S. R. (2010). An immune paradox: How can the same chemokine axis regulate both immune tolerance and activation?: CCR6/CCL20: A chemokine axis balancing immunological tolerance and inflammation in autoimmune disease. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 32(12), 1067–1076. 10.1002/bies.201000063 [DOI] [PubMed] [Google Scholar]
- Cosseau, C. , Devine, D. A. , Dullaghan, E. , Gardy, J. L. , Chikatamarla, A. , Gellatly, S. , … Hancock, R. E. (2008). The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host‐microbe homeostasis. Infection and Immunity, 76(9), 4163–4175. 10.1128/IAI.00188-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau, R. P. (2009). The oral microbial consortium's interaction with the periodontal innate defense system. DNA and Cell Biology, 28(8), 389–395. 10.1089/dna.2009.0864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau, R. P. (2010). Periodontitis: A polymicrobial disruption of host homeostasis. Nature Reviews. Microbiology, 8(7), 481–490. 10.1038/nrmicro2337 [DOI] [PubMed] [Google Scholar]
- Diaz, P. I. , Chalmers, N. I. , Rickard, A. H. , Kong, C. , Milburn, C. L. , Palmer, R. J. Jr. , & Kolenbrander, P. E. (2006). Molecular characterization of subject‐specific oral microflora during initial colonization of enamel. Applied and Environmental Microbiology, 72(4), 2837–2848. 72/4/2837 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, P. I. , Xie, Z. , Sobue, T. , Thompson, A. , Biyikoglu, B. , Ricker, A. , … Dongari‐Bagtzoglou, A. (2012). Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infection and Immunity, 80(2), 620–632. 10.1128/IAI.05896-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, B. C. , Moffatt, C. E. , Hagerty, D. , Whitmore, S. E. , Brown, T. A. , Graves, D. T. , & Lamont, R. J. (2011). Interaction of oral bacteria with gingival epithelial cell multilayers. Molecular Oral Microbiology, 26(3), 210–220. 10.1111/j.2041-1014.2011.00609.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, M. A. , Hahn, W. C. , Ino, Y. , Ronfard, V. , Wu, J. Y. , Weinberg, R. A. , … Rheinwald, J. G. (2000). Human keratinocytes that express hTERT and also bypass a p16(INK4a)‐enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Molecular and Cellular Biology, 20(4), 1436–1447. 10.1128/MCB.20.4.1436-1447.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongari‐Bagtzoglou, A. , & Kashleva, H. (2006). Development of a highly reproducible three‐dimensional organotypic model of the oral mucosa. Nat. Protocols, 1(4), 2012–2018. 10.1038/nprot.2006.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer, H. , Grischke, J. , Tiede, C. , Eberhard, J. , Schweitzer, A. , Toikkanen, S. E. , … Stiesch, M. (2018). Epidemiology and risk factors of peri‐implantitis: A systematic review. Journal of Periodontal Research, 53(5), 657–681. 10.1111/jre.12562 [DOI] [PubMed] [Google Scholar]
- Furst, M. M. , Salvi, G. E. , Lang, N. P. , & Persson, G. R. (2007). Bacterial colonization immediately after installation on oral titanium implants. Clinical Oral Implants Research, 18(4), 501–508. CLR1381 [pii]. 10.1111/j.1600-0501.2007.01381.x [DOI] [PubMed] [Google Scholar]
- Graves, D. T. , Correa, J. D. , & Silva, T. A. (2019). The oral microbiota is modified by systemic diseases. Journal of Dental Research, 98(2), 148–156. 10.1177/0022034518805739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeger, S. E. , & Meyle, J. (2015). Epithelial barrier and oral bacterial infection. Periodontology 2000, 69(1), 46–67. 10.1111/prd.12094 [DOI] [PubMed] [Google Scholar]
- Gursoy, U. K. , Pollanen, M. , Kononen, E. , & Uitto, V. J. (2010). Biofilm formation enhances the oxygen tolerance and invasiveness of Fusobacterium nucleatum in an oral mucosa culture model. Journal of Periodontology, 81(7), 1084–1091. 10.1902/jop.2010.090664 [DOI] [PubMed] [Google Scholar]
- Hajishengallis, G. (2014). Immunomicrobial pathogenesis of periodontitis: Keystones, pathobionts, and host response. Trends in Immunology, 35(1), 3–11. 10.1016/j.it.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis, G. (2015). Periodontitis: From microbial immune subversion to systemic inflammation. Nature Reviews. Immunology, 15(1), 30–44. 10.1038/nri3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis, G. , & Lamont, R. J. (2016). Dancing with the stars: How choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends in microbiology, 24(6), 477–489. 10.1016/j.tim.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, D. C. , Huang, G. T. , Lin, L. M. , Warner, N. A. , Gim, J. S. , & Jewett, A. (2003). Expression of MHC Class II, CD70, CD80, CD86 and pro‐inflammatory cytokines is differentially regulated in oral epithelial cells following bacterial challenge. Oral Microbiology and Immunology, 18(6), 350–358. 094 [pii]. 10.1046/j.0902-0055.2003.00094.x [DOI] [PubMed] [Google Scholar]
- Handfield, M. , Baker, H. V. , & Lamont, R. J. (2008). Beyond good and evil in the oral cavity: Insights into host‐microbe relationships derived from transcriptional profiling of gingival cells. Journal of Dental Research, 87(3), 203–223. 87/3/203 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handfield, M. , Mans, J. J. , Zheng, G. , Lopez, M. C. , Mao, S. , Progulske‐Fox, A. , … Lamont, R. J. (2005). Distinct transcriptional profiles characterize oral epithelium‐microbiota interactions. Cellular Microbiology, 7(6), 811–823. CMI513 [pii]. 10.1111/j.1462-5822.2005.00513.x [DOI] [PubMed] [Google Scholar]
- Hasegawa, Y. , Mans, J. J. , Mao, S. , Lopez, M. C. , Baker, H. V. , Handfield, M. , & Lamont, R. J. (2007). Gingival epithelial cell transcriptional responses to commensal and opportunistic oral microbial species. Infection and Immunity, 75(5), 2540–2547. IAI.01957‐06 [pii]. 10.1128/IAI.01957-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, B. , Ward, J. , & Ready, D. (2010). Aggregatibacter (Actinobacillus) actinomycetemcomitans: A triple A* Periodontopathogen? Periodontology 2000, 54(1), 78–105. 10.1111/j.1600-0757.2009.00331.x [DOI] [PubMed] [Google Scholar]
- Herbert, B. A. , Novince, C. M. , & Kirkwood, K. L. (2016). Aggregatibacter actinomycetemcomitans, a potent immunoregulator of the periodontal host defense system and alveolar bone homeostasis. Molecular Oral Microbiology, 31(3), 207–227. 10.1111/omi.12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, L. V. , & Gordon, J. I. (2001). Commensal host‐bacterial relationships in the gut. Science (New York, N.Y.), 292(5519), 1115–1118. [DOI] [PubMed] [Google Scholar]
- Huang, D. W. , Sherman, B. T. , & Lempicki, R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research, 37(1), 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D. W. , Sherman, B. T. , & Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Kittichotirat, W. , Bumgarner, R. E. , & Chen, C. (2016). Evolutionary Divergence of Aggregatibacter actinomycetemcomitans . Journal of Dental Research, 95(1), 94–101. 10.1177/0022034515608163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander, P. E. , Palmer, R. J. Jr. , Periasamy, S. , & Jakubovics, N. S. (2010). Oral multispecies biofilm development and the key role of cell‐cell distance. Nature Reviews. Microbiology, 8(7), 471–480. 10.1038/nrmicro2381 [DOI] [PubMed] [Google Scholar]
- Mombelli, A. , Müller, N. , & Cionca, N. (2012). The epidemiology of peri‐implantitis. Clinical Oral Implants Research, 23, 67–76. 10.1111/j.1600-0501.2012.02541.x [DOI] [PubMed] [Google Scholar]
- Nakayama, M. , Inoue, T. , Naito, M. , Nakayama, K. , & Ohara, N. (2015). Attenuation of the phosphatidylinositol 3‐kinase/Akt signaling pathway by Porphyromonas gingivalis gingipains RgpA, RgpB, and Kgp. The Journal of Biological Chemistry, 290(8), 5190–5202. 10.1074/jbc.M114.591610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paino, A. , Lohermaa, E. , Sormunen, R. , Tuominen, H. , Korhonen, J. , Pöllänen, M. T. , & Ihalin, R. (2012). Interleukin‐1β is internalised by viable Aggregatibacter actinomycetemcomitans biofilm and locates to the outer edges of nucleoids. Cytokine, 60(2), 565–574. 10.1016/j.cyto.2012.07.024 [DOI] [PubMed] [Google Scholar]
- Paino, A. , Tuominen, H. , Jaaskelainen, M. , Alanko, J. , Nuutila, J. , Asikainen, S. E. , … Ihalin, R. (2011). Trimeric form of intracellular ATP synthase subunit beta of Aggregatibacter actinomycetemcomitans binds human interleukin‐1beta. PloS One, 6(4), e18929 10.1371/journal.pone.0018929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic‐Curcin, A. , Matic, S. , Vojvodic, D. , Stamatovic, N. , & Todorovic, T. (2011). Cytokines in pathogenesis of peri‐implantitis. Vojnosanitetski Pregled, 68(5), 435–440. 10.2298/VSP1105435P [DOI] [PubMed] [Google Scholar]
- Pinnock, A. , Murdoch, C. , Moharamzadeh, K. , Whawell, S. , & Douglas, C. W. I. (2014). Characterisation and optimisation of organotypic oral mucosal models to study Porphyromonas gingivalis invasion. Microbes and Infection, 16(4), 310–319. 10.1016/j.micinf.2014.01.004 [DOI] [PubMed] [Google Scholar]
- Pleguezuelos, O. , Dainty, S. J. , Kapas, S. , & Taylor, J. J. (2005). A human oral keratinocyte cell line responds to human heat shock protein 60 through activation of ERK1/2 MAP kinases and up‐ regulation of IL‐1beta. Clinical and Experimental Immunology, 141(2), 307–314. CEI2860 [pii]. 10.1111/j.1365-2249.2005.02860.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollanen, M. T. , Laine, M. A. , Ihalin, R. , & Uitto, V. J. (2012). Host‐bacteria crosstalk at the dentogingival junction. International Journal of Dentistry, 2012, 821383 10.1155/2012/821383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preus, H. R. , Olsen, I. , & Namork, E. (1987). The presence of phage‐infected Actinobacillus actinomycetemcomitans in localized juvenile periodontitis patients. Journal of Clinical Periodontology, 14(10), 605–609. 10.1111/j.1600-051X.1987.tb01523.x [DOI] [PubMed] [Google Scholar]
- Rams, T. E. , Degener, J. E. , & van Winkelhoff, A. J. (2014). Antibiotic resistance in human peri‐implantitis microbiota. Clinical Oral Implants Research, 25(1), 82–90. 10.1111/clr.12160 [DOI] [PubMed] [Google Scholar]
- Ramsey, M. M. , & Whiteley, M. (2009). Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proceedings of the National Academy of Sciences of the United States of America, 106(5), 1578–1583. 10.1073/pnas.0809533106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouabhia, M. (2002). Interactions between host and oral commensal microorganisms are key events in health and disease status. The Canadian Journal of Infectious Diseases = Journal Canadien des Maladies Infectieuses, 13(1), 47–51. 10.1155/2002/580476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach, P. , & Glauser, R. (2007). The defense architecture of the human periimplant mucosa: A histological study. The Journal of Prosthetic Dentistry, 97(6 Suppl), S15–S25. 10.1016/S0022-3913(07)60004-3 [DOI] [PubMed] [Google Scholar]
- Schutyser, E. , Struyf, S. , & Van Damme, J. (2003). The CC chemokine CCL20 and its receptor CCR6. Cytokine & Growth Factor Reviews, 14(5), 409–426. S1359610103000492 [pii]. 10.1016/S1359-6101(03)00049-2 [DOI] [PubMed] [Google Scholar]
- Szafranski, S. P. , Deng, Z. L. , Tomasch, J. , Jarek, M. , Bhuju, S. , Rohde, M. , … Wagner‐Döbler, I. (2017). Quorum sensing of Streptococcus mutans is activated by Aggregatibacter actinomycetemcomitans and by the periodontal microbiome. BMC Genomics, 18(1), 238 238‐017‐3618‐5. 10.1186/s12864-017-3618-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski, S. P. , Winkel, A. , & Stiesch, M. (2017). The use of bacteriophages to biocontrol oral biofilms. Journal of Biotechnology, 250, 29–44. S0168‐1656(17)30011‐1 [pii] [DOI] [PubMed] [Google Scholar]
- Szafranski, S. P. , Wos‐Oxley, M. L. , Vilchez‐Vargas, R. , Jauregui, R. , Plumeier, I. , Klawonn, F. , … Wagner‐Dobler, I. (2015). High‐resolution taxonomic profiling of the subgingival microbiome for biomarker discovery and periodontitis diagnosis. Applied and Environmental Microbiology, 81(3), 1047–1058. 10.1128/AEM.03534-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twetman, S. , Derawi, B. , Keller, M. , Ekstrand, K. , Yucel‐Lindberg, T. , & Stecksen‐Blicks, C. (2009). Short‐term effect of chewing gums containing probiotic Lactobacillus reuteri on the levels of inflammatory mediators in gingival crevicular fluid. Acta Odontologica Scandinavica, 67(1), 19–24. 10.1080/00016350802516170 [DOI] [PubMed] [Google Scholar]
- van Winkelhoff, A. J. , & Wolf, J. W. (2000). Actinobacillus actinomycetemcomitans‐associated peri‐implantitis in an edentulous patient. A case report. Journal of Clinical Periodontology, 27(7), 531–535. 10.1034/j.1600-051x.2000.027007531.x [DOI] [PubMed] [Google Scholar]
- Whitmore, S. E. , & Lamont, R. J. (2011). The pathogenic persona of community‐associated oral streptococci. Molecular Microbiology, 81(2), 305–314. 10.1111/j.1365-2958.2011.07707.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Sobue, T. , Thompson, A. , Xie, Z. , Poon, K. , Ricker, A. , … Dongari‐Bagtzoglou, A. (2014). Streptococcal co‐infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cellular Microbiology, 16(2), 214–231. 10.1111/cmi.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , Chen, R. , & Rudney, J. D. (2008). Streptococcus cristatus attenuates Fusobacterium nucleatum‐induced interleukin‐8 expression in oral epithelial cells. Journal of Periodontal Research, 43(4), 408–416. 10.1111/j.1600-0765.2007.01057.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Supplementary Information


