Abstract
Background
In this research, an meta-analysis was performed for assessment of the associations between O6-methyguanine-DNA methyltransferase (MGMT) promoter hypermethylation possessing low-grade intraepithelial lesion (LSIL), high-grade intraepithelial lesion (HSIL), cervical cancer (CC), and clinicopathological characters of CC.
Methods
Literature selection were conducted through searching PubMed, Web of science, EMBASE, China National Knowledge Infrastructure and Wanfang databases (up to November 2018). An assessment of associations between MGMT methylation and LSIL, HSIL, CC risk and clinicopathological characteristics was performed through pooled odds ratios (ORs) with relevant 95% confidence intervals (CIs). Subgroup analyses, meta-regressions and Galbraith plots were conducted to conduct an exploration on the possible sources of heterogeneity. The genome-wide DNA methylation array studies were extracted from Gene Expression Omnibus (GEO) databases for validation of these outcomes.
Results
In this meta-analysis of 25 published articles, MGMT hypermethylation gradually elevated the rates among control group (12.16%), LSIL (20.92%), HSIL (36.33%) and CC (41.50%) specimens. MGMT promoter methylation was significant associated with the increased risk of LSIL by 1.74-fold (P<0.001), HSIL by 3.71-fold (P<0.001) and CC by 7.08-fold (P<0.001) compared with control. A significant association between MGMT promoter methylation with FIGO stage was also found (OR = 2.81, 95% CI: 1.79–4.41, p<0.001). The results of GEO datasets showed that 5 CpG sites in MGMT with a great diagnostic value for the screening of cervical cancer.
Conclusion
The meta-analysis indicated the association between MGMT promoter hypermethylation and squamous intraepithelial lesion and cervical cancer. MGMT methylation detection might have a potential value to be an epigenetic marker for the clinical diagnosis of cervical cancer.
Introduction
Cervical cancer continues to be the 2st commonest gynecologic carcinoma worldwide[1], which causes approximately 528,000 new cases and 266,000 deaths per year[2]. Squamous intraepithelial lesion (SIL), a precursor of cervical cancer[3], which is featured as a progressive process from low-grade squamous intraepithelial lesion (LSIL) to high-grade squamous intraepithelial lesion (HSIL) and eventually to invasive carcinoma[4]. Human papillomavirus (HPV) infection has been widely famous to have a key function in the process of cervical carcinoma, however, merely a limited number of HPV-induced lesions eventually progress to SILs or invasive cancer[5], indicating that there were other biomolecular mechanisms in the progress of cervical carcinoma.
DNA methylation, an epigenetic modification in genes that primarily causes transcriptional silencing of genes, which played a key role in regulating transcription, embryonic development, genomic imprinting, genome stability and chromatin structure. Aberrant DNA methylation of CpG islands is comparatively seldom seen in normal cells, based on which the detection of promoter hypermethylation of tumor suppressor genes (TSGs) in bodily fluids could serve as good biomarkers for screening and prognosis of tumorigenesis development[6]. O6-methyguanine-DNA methyltransferase (MGMT) is a DNA repair enzyme removing mutagenic and cytotoxic adducts out of O6-guanine in DNA. Methylation of discrete regions of the CpG island of MGMT becomes the main cause of gene silencing and decreased expression of MGMT in tumor tissues and cell lines[7]. It has been proved that the gene silencing of MGMT is related to elevated carcinogenic risk and sensitivity to therapeutic methylating agents. Thus, the promoter methylation of MGMT was widely regarded as a promising biomarker for the detection of early carcinoma. Previous researches concentrated upon this in other cancers like esophageal cancer[8, 9], lung cancer[10–12], glioma[13, 14], colon cancer[15–18], gastric cancer[19], neck squamous cell carcinoma[20, 21], ovarian cancer[22] and breast cancer[23]. In 2001, Virmani et al.[24] first revealed an important association between the aberrant methylation of MGMT with the risk and histological type of cervical carcinoma. Thereafter, an increasing number of researches were conducted to investigate the association between MGMT hypermethylation with the process of cervical carcinogenesis. Nevertheless, the sample sizes of the studies are still small, resulting in inconsistent outcomes and a wide range of MGMT methylation rates in cervical carcinoma tissues. Even the opposite conclusions have also been reported in a few studies, suggesting that MGMT gene is rarely methylated in cervical cancer. Moreover, a meta-analysis pooled the data of 28 studies to investigate the association between MGMT methylation and the risk of breast and gynecologic cancers, among which 11 studies focused on cervical cancer, indicating that MGMT methylation and cervical cancer were positively correlated[25]. However, there were still a lack of relevant comprehensive review that systematically appraised the effect of MGMT promoter hypermethylation upon diverse phases of cervical carcinogenesis that from LSIL to HSIL, and finally to invasive carcinoma, as well as their clinicopathological features.
Therefore, a meta-analysis was performed for comprehensive assessment of the association of MGMT promoter hypermethylation with squamous intraepithelial lesion, cervical cancer and their clinicopathological characteristics.
Methods
Literature search
The review was reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) 2009 guidelines (Table A in S1 File). The PubMed, EMBASE, Web of science, China National Knowledge Infrastructure (CNKI) and Wanfang databases were adopted to search candidate literature. We retrieved eligible literature updated before November 2018 by using following items “(MGMT) and (methylation or hypermethylation or epigene*) and (cervical cancer or cervical carcinoma or cervical tumor)”. Reference list in retrieved articles and relevant reviews were retrieved in a manual manner.
Eligibility criteria
Eligible studies were included when satisfying the following criteria: (1) the study should be an observation designing including cohort, case-control, case-only or cross-sectional study; (2) the study should evaluated the association between MGMT promoter methylation and LSIL, HSIL, CC or clinicopathological characteristics of CC; (3) the study should offer enough data for calculation of odds ratios (ORs) and 95% confidence intervals (CIs); (4) the study should be written in English or Chinese.
Exclusion criteria were as follows: (1) they were meeting abstracts, reviews, letters or case reports; (2) they regarding in vitro or ex vivo experiments of cell lines or animals.
Data extraction
The data of eligible studies were extracted by two independent authors. The following information of each eligible study were collected: the first author’s name, publication year, ethnicity, country, study design, sample size, methylation detection methods, materials, source of controls, involved diseases (LSIL, HSIL, CC), their clinicopathological characteristics (age at diagnose, HPV infection, histological type, FIGO stage, therapeutic response, histological grade, lymph node metastasis) and quality of studies.
Validation by GEO datasets
The genome-wide DNA methylation array studies were extracted from Gene Expression Omnibus (GEO) databases by using following items “Cervical cancer”, “Methylation” and “Homo sapiens”. Eligible criteria were as follows: (1) the quantitative methylation levels of datasets were detected by the Illumina HumanMethylation 27 or 450 k Beadchip; (2) datasets using cohort or case-control designs. Exclusion criteria were as follows: (1) datasets regarding in vitro or ex vivo experiments of cell lines or animals; (2) datasets without CpG number.
The following information of each eligible datasets were collected: submission and last update date, ethnicity, country, sample size, methylation detection methods, source of controls, involved diseases (LSIL, HSIL, CC).
Quality assessment of eligible studies and datasets
The quality of eligible studies and datasets was assessed by two independent authors (JH and JYL) in line with a preset system derived from the REMARK [26] and BRISQ [27] guidelines. 18 items were considered as quality components, including study design, study population, biospecimen information, methylation detection, clinicopathological characteristics and outcomes analysis (Table B in S1 File). Studies reporting exceeding 11 items were considered to be high-quality studies.
Statistical methods
The MGMT promoter hypermathylation rates in LSIL, HSIL and CC specimens were calculated by the inverse variance approach [28]. Cochran-Armitage (CA) trend test were used to compare the methylation frequency in control group, LSIL, HSIL and CC specimens. Pooled ORs and their 95% CIs were calculated to estimate the association between MGMT promoter methylation possessing LSIL, HSIL, CC and their clinicopathological characteristics. Heterogeneity across the included studies were assessed by the Cochran’s Q test and I2 statistic. I2 value of greater than 25%, 50% and 75% meant mild, moderate and high heterogeneity, respectively[29]. When significant heterogeneity (I2 value larger than 50% or PQ-test smaller than 0.1) was observed, the random-effect model was utilized to pool the results, otherwise, a fixed-effect was applied[30]. To further explore the potential source of heterogeneity, subgroup analyses and meta-regression were performed based on ethnicity, source of controls, materials, published year (≥2010 and <2010) and quality of studies. And, Galbraith plots were further depicted to seek the impact of individual studies of the overall heterogeneity. Moreover, sensitivity analysis was conducted through sequentially removing every study or heterogeneity spotted by Galbraith plots to further assess the stability of the pooled outcomes. In GEO datasets, the relationship between CpG sites of MGMT and cervical cancer were calculated by Mann-Whitney U test. Trial sequential analysis (TSA) was performed by TSA 0.9 software (Copenhagen Trial Unit, Center for Clinical Intervention Research, Denmark, http://www.ctu.dk/tsa/) with type I errors of 5%, type II errors of 20% and a statistical test power of 80%. Publication bias was evaluated by funnel plots and Egger’s test[31]. Funnel plot and PEgger≤0.05 indicated the presence of publication bias. All the statistical analysis above were undertaken by RevMan 5.3 (The Nordic Cochrane Centre, The Cochrane Collaboration) and Stata 15.0 (Stata, College, TX, USA).
Results
Characteristics of included studies
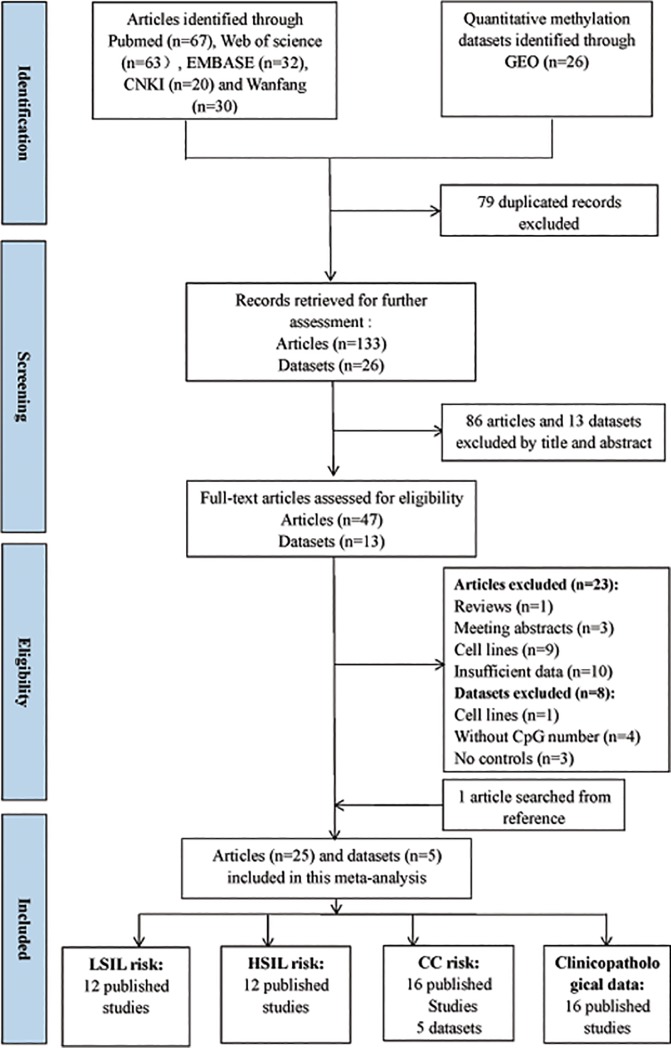
Based on the definitions of the 2001 Bethesda System[32], the category of LSIL encompassed cytopathic effects of HPV, cervical intraepithelial neoplasia (CIN) 1 and mild dysplasia. The category of HSIL contained moderate or extensive dysplasia and CIN 2 or 3. CC contained squamous cell carcinoma (SCC) and adenocarcinoma (AdC). Upon the basis of such definitions and the literature search, 47 full-text articles were initially selected and assessed for eligibility. Then, 23 articles were excluded because of reviews (n = 1), meeting abstracts (n = 3), cell lines (n = 9) and insufficient data (n = 10). Manual search of reference cited in the published articles spotted one additional study[33]. Finally, a total of 25 articles [33–55] were included in this meta-analysis. Of such studies, all studies were eligible to calculate the hypermethylation rates of MGMT. A total of 2933 patients with SIL or CC from 25 studies (6 case-only studies[34, 43, 46, 48, 49, 51] and 19 case-control studies) were eligible to estimate the association of MGMT methylation status with the clinicopathological features. For most of these 25 studies (17 of 25), the detection of MGMT promoter methylation was performed by methylation-specific PCR (MSP). Besides, two studies performed by HRM, only one study performed by pyrosequencing[46], one study performed by MS-MLPA[48], two studies performed by QMSP[45, 53], as well as two studies performed by MSP and sequencing[51, 55]. Among these 25 studies, 11 studies used exfoliated cells of cervical samples to detect MGMT methylation status, while other 10 studies involved cervical tissues, 3 studies involved tissue and plasma and one study only involved plasma. Regarding the type of ethnicity, eighteen studies were carries out on Asian, seven studies on Caucasians. The flow diagram for the process of included articles in this meta-analysis was revealed in Fig 1. The detailed characteristics of included articles were listed in Table 1.
Fig 1. Flow diagram for the procedures of eligible studies selection in this meta-analysis.
Table 1. Characteristics of included studies in this meta-analysis.
| No. | Author | Year | Country | Ethnicity | Study design | Sample size | Methylation detection method | Materials | Source of controls | Involved clincopathological features | Quality scores | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | CC | HSIL | LSIL | |||||||||||
| 1 | Lodi | 2018 | Germany | Caucasian | Case-only | - | 143 | - | - | MSP | Tissue | - | Therapeutic response | 10 |
| 2 | L.-L Sun | 2018 | China | Asian | Case-control | 45 | 5 | - | - | MS-HRM | Exfoliated cells | B | - | 11 |
| 3 | Wanga | 2018 | China | Asian | Case-control | 138 | - | 98 | 107 | MSP | Exfoliated cells | B | - | 10 |
| 4 | Nana | 2016 | China | Asian | Case-control | 156 | - | 134 | 147 | MSP | Exfoliated cells | H | - | 11 |
| 5 | Yin Sun | 2015 | China | Asian | Case-control | 48 | 45 | 103 | 54 | MS-HRM | Exfoliated cells | B | - | 10 |
| 6 | Banzai | 2014 | Japan | Asian | Case-control | 24 | 53 | - | - | MSP | Tissue | B | Histological type | 10 |
| 7 | Lua | 2014 | China | Asian | Case-control | 20 | 50 | 100 | 50 | MSP | Exfoliated cells | B | FIGO stage,Histological grade | 11 |
| 8 | Sun | 2012 | China | Asian | Case-control | 336 | - | 37 | 68 | MSP | Exfoliated cells | H | Age,HPV | 14 |
| 9 | Jiana | 2012 | China | Asian | Case-control | 30 | 52 | - | - | MSP | Tissue | B | FIGO stage, Histological grade | 11 |
| 10 | Spathis | 2011 | Greece | Caucasian | Case-control | 57 | 18 | 101 | 164 | MSP | Exfoliated cells | H | Histological type | 12 |
| 11 | Liua | 2011 | China | Asian | Case-only | - | 183 | - | - | MSP | Plasma | - | HPV,histological type | 12 |
| 12 | Kim | 2010 | Korea | Asian | Case-control | 41 | 69 | 67 | 32 | MSP | Exfoliated cells | B | - | 12 |
| 13 | Iliopoulos | 2009 | Greece | Caucasian | Case-control | 27 | 61 | 12 | 15 | QMSP | Exfoliated cells | H | FIGO stage | 12 |
| 14 | Flatley | 2009 | UK | Caucasian | Case-control | 45 | 42 | 102 | 49 | MSP | Exfoliated cells | H | - | 10 |
| 15 | Lee | 2008 | Korea | Asian | Case-only | - | 34 | - | - | Pyro- sequencing | Tissue | - | Histological type, FIGO stage,lymph node metastasis | 10 |
| 16 | Chena | 2008 | China | Asian | Case-control | 38 | 86 | - | - | MSP | Tissue and plasma | H | FIGO stage, histological grade,lymph node metastasis | 12 |
| 17 | Henken | 2007 | Nether- lands | Caucasian | Case-only | - | 29 | - | - | MS-MLPA | Tissue | - | Histological type | 11 |
| 18 | Hoenil | 2007 | Korea | Asian | Case-only | - | 82 | - | - | MSP | Tissue | - | - | 10 |
| 19 | Gaoa | 2007 | China | Asian | Case-control | 15 | 38 | 15 | 5 | MSP | Tissue | B | Histological grade,lymph node metastasis, FIGO stage | 10 |
| 20 | Yang | 2006 | China | Asian | Case-only | - | 127 | - | - | MSP and sequencing | Tissue | - | FIGO stage, histological grade, histological type, therapeutic response | 13 |
| 21 | Lin | 2005 | Korea | Asian | Case-control | 20 | 67 | 20 | 10 | MSP | Tissue | H | Histological type | 11 |
| 22 | Reesink-Peters | 2004 | Netherlands | Caucasian | Case-control | 41 | 48 | - | - | QMSP | Exfoliated cells | B | - | 10 |
| 23 | Yang | 2004 | China | Asian | Case-control | 100 | 85 | - | - | MSP | Tissue and plasma | A | Histological type,FIGO stage, histological grade | 13 |
| 24 | Dong | 2001 | Korea | Asian | Case-control | 24 | 53 | - | - | MSP and sequencing | Tissue | B | Histological type,FIGO stage, histological grade,age | 13 |
| 25 | Virmani | 2001 | USA | Caucasian | Case-control | 22 | 19 | 17 | 37 | MSP | Tisses,blood lymphocytes and buccal epithelial | H | - | 13 |
Abbreviations: CC, cervical cancer; LSIL, low-grade squamous intra-epithelial lesion; HSIL, high-grade squamous intra-epithelial lesion; MSP, methylation-specific PCR; H, healthy controls; B, controls with benign gynecological diseases; A, autologous controls.
Notes
a Studies written in Chinese
Pooled rates of MGMT hypermethylation in patients with LSIL, HSIL and CC
Altogether 1227 controls, 738 LSIL, 827 HSIL and 791 CC specimens were included in this meta-analysis. As shown in Table 2, the pooled rates of MGMT hypermethylation demonstrated a progressively increased trend (p<0.001) from control group (12.16%, 95% CI: 4.43–20.81%) to LSIL (20.92%, 95% CI: 9.22–32.62%), to HSIL (36.33%, 95% CI: 24.95–47.72%) and eventually to CC (41.50%, 95% CI: 28.19–54.81%) specimens.
Table 2. Pooled hypermethylation rates of MGMT in LSIL, HSIL and CC specimens.
| Comparisons | Studies | Specimens | Methylation rates (%) | 95% CI (%) |
|---|---|---|---|---|
| Control | 19 | 1227 | 12.16% | 4.43–20.81 |
| LSIL | 12 | 738 | 20.92% | 9.22–32.62 |
| HSIL | 12 | 828 | 36.33% | 24.95–47.72 |
| CC | 16 | 791 | 41.50% | 28.19–54.81 |
The relationship between MGMT promoter hypermethylation and LSIL risk
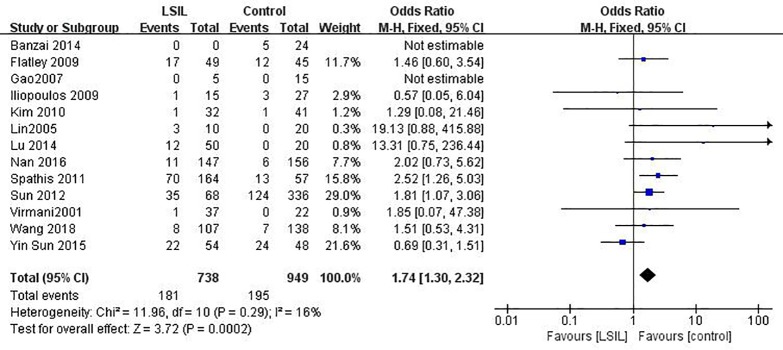
Twelve published studies including 738 patients with LSIL risk and 925 controls were included to estimate the effect of MGMT promoter hypermethylation on LSIL risk (Fig 2). MGMT promoter hypermethylation conferred a 1.75-fold (95% CI: 1.30–2.32) elevated risk of LSIL and a p value of <0.001 (Table 3). In ethnicity based subgroup analysis, there were no significant differences in methylation rates between Asians (OR: 1.65, 95% CI: 1.16–2.35) and Caucasians (OR: 1.93, 95% CI: 1.15–3.22). Such association was still significant in most subgroups except for the “non-healthy”, “tissue”, “publication year before 2010” and “Quality of studies lower than 11”. There was no significant heterogeneity in all comparisons (I2: 0–39%).
Fig 2. Funnel plots for associations of MGMT promoter hypermethylation with the risk of LSIL.
The squares represent the ORs for individual studies. The size of the square reflects the weight of included studies. Bars represent the 95% confidence intervals (CIs). The center of the diamond represents the summary effect size. LSIL, low-grade intra-epithelial lesion.
Table 3. Pooled results for the association of MGMT promoter hypermethylation with LSIL risk.
| Comparisons | Studies (N) | Sample size (LSIL/controls) | Heterogeneity | Modela | Effect size | ||
|---|---|---|---|---|---|---|---|
| I2(%) | P Q-text | OR (95% CI) | P | ||||
| Total | 12 | 738/925 | 16 | 0.29 | F | 1.74(1.30–2.32) | <0.001 |
| Ethnicity | |||||||
| Asian | 8 | 473/774 | 37 | 0.14 | F | 1.65(1.16–2.35) | 0.005 |
| Caucasian | 4 | 265/151 | 0 | 0.58 | F | 1.93(1.15–3.22) | 0.010 |
| Source of controls | |||||||
| Healthy | 7 | 490/663 | 0 | 0.65 | F | 1.97(1.40–2.78) | <0.001 |
| Non-healthyb | 5 | 248/262 | 39 | 0.18 | F | 1.23(0.70–2.15) | 0.480 |
| Materials | |||||||
| Tissue | 3 | 52/57 | 5 | 0.31 | F | 6.71(0.78–57.61) | 0.080 |
| Exfoliated cells | 9 | 686/868 | 16 | 0.30 | F | 1.68(1.25–2.25) | <0.001 |
| Publication year | |||||||
| ≥ 2010 | 7 | 622/796 | 31 | 0.19 | F | 1.75(1.27–2.40) | <0.001 |
| < 2010 | 5 | 116/129 | 9 | 0.35 | F | 1.70(0.82–3.53) | 0.160 |
| Quality of studies | |||||||
| High (>11) | 7 | 376/523 | 0 | 0.48 | F | 2.24(1.52–3.29) | <0.001 |
| Low (≤11) | 5 | 362/402 | 10 | 0.35 | F | 1.22(0.78–1.91) | 0.390 |
Abbreviations: N, number; LSIL, low squamous intra-epithelial lesion; F, fixed-effects model; R, random-effects model.
Notes
a When significant heterogeneity was found (I2≥50% or PQ-test≤0.1), a random-effects model with the inverse variance method was used to pool the results; otherwise, a fixed-effects model was applied.
b Non-healthy controls included autologous controls and controls with benign gynecological diseases.
The relationship between MGMT promoter hypermethylation and HSIL risk
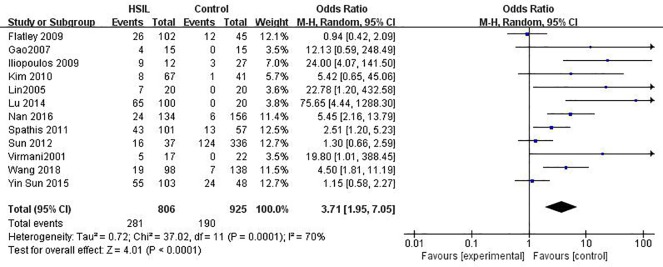
A total of 828 patients with HSIL and 949 controls from 12 studies were eligible to assess the association of MGMT promoter methylation status with HSIL risk (Fig 3). Overall, MGMT promoter hypermethylation was associated with a 3.37-fold (95% CI: 1.86–6.14) increased risk of HSIL and a p value of <0.001 (Table 4). The association was still significant in all subgroups as shown in Table 3.
Fig 3. Funnel plots for associations of MGMT promoter hypermethylation with the risk of HSIL.
The squares represent the ORs for individual studies. The size of the square reflects the weight of included studies. Bars represent the 95% confidence intervals (CIs). The center of the diamond represents the summary effect size. HSIL, high-grade intra-epithelial lesion.
Table 4. Pooled results for the association of MGMT promoter hypermethylation with HSIL risk.
| Comparisons | Studies (N) | Sample size (HSIL/controls) | Heterogeneity | Modela | Effect size | ||
|---|---|---|---|---|---|---|---|
| I2(%) | P Q-text | OR (95% CI) | P | ||||
| Total | 12 | 806/925 | 70 | <0.001 | R | 3.71(1.95–7.05) | <0.001 |
| Ethnicity | |||||||
| Asian | 8 | 574/774 | 70 | 0.001 | R | 3.93(1.73–8.90) | 0.001 |
| Caucasian | 4 | 232/151 | 78 | 0.003 | R | 3.89(1.05–14.35) | 0.040 |
| Source of controls | |||||||
| Healthy | 7 | 423/663 | 73 | 0.001 | R | 3.46(1.52–7.86) | 0.003 |
| Non-healthyb | 5 | 383/262 | 74 | 0.004 | R | 4.95(1.36–17.98) | 0.010 |
| Materials | |||||||
| Tissue | 3 | 74/81 | 0 | 0.950 | F | 17.97(3.23–9.90) | 0.001 |
| Exfoliated cells | 9 | 754/868 | 74 | <0.001 | R | 3.04(1.57–5.87) | <0.001 |
| Publication year | |||||||
| ≥ 2010 | 7 | 640/796 | 69 | 0.003 | R | 2.94(1.50–5.74) | 0.002 |
| < 2010 | 5 | 166/129 | 77 | 0.002 | R | 8.57(1.28–57.53) | 0.030 |
| Quality of studies | |||||||
| High (>11) | 7 | 234/483 | 67 | 0.020 | R | 4.00(1.46–10.92) | 0.007 |
| Low (≤11) | 5 | 572/442 | 76 | <0.001 | R | 3.84(1.49–9.93) | 0.005 |
Abbreviations: N, number; HSIL, high squamous intra-epithelial lesion; F, fixed-effects model; R, random-effects model.
Notes
aWhen significant heterogeneity was found (I2≥50% or PQ-test≤0.1), a random-effects model with the inverse variance method was used to pool the results; otherwise, a fixed-effects model was applied.
b Non-healthy controls included autologous controls and controls with benign gynecological diseases.
Due to observation of moderate heterogeneity in the overall comparison (I2 = 70%), subgroup, meta-regression and Galbraith plot analyses were performed to explore the potential sources of heterogeneity. In ethnicity based subgroup analysis, there were no significant differences in methylation rates between Asians (OR: 3.93, 95% CI: 1.73–8.90) and Caucasians (OR: 3.89, 95%CI: 1.05–14.35). Moderate heterogeneity remained in most of the subgroups, except for the “tissues” subgroup (I2 = 0%). The outcomes of meta-regression analyses illustrated that ethnicity (p = 0.978), source of controls (p = 0.999), materials (p = 0.513), publication year (p = 0.340) and quality of studies (p = 0.752) were all not main sources of heterogeneity (Table C in S1 File). Furthermore, a Galbraith plot was further depicted, spotting four outliers [33, 42, 56, 57] as major sources of heterogeneity (Figure A in S1 File). Such four studies were all classified into “exfoliated cells” studies, and exclusion of such four studies caused a decline in I2 value from 68 to 4%, followed by an apparent association between the methylation of MGMT with increased HSIL risk (OR = 8.51, 95% CI: 5.02–14.42, p<0.001).
The relationship between MGMT promoter hypermethylation and CC risk
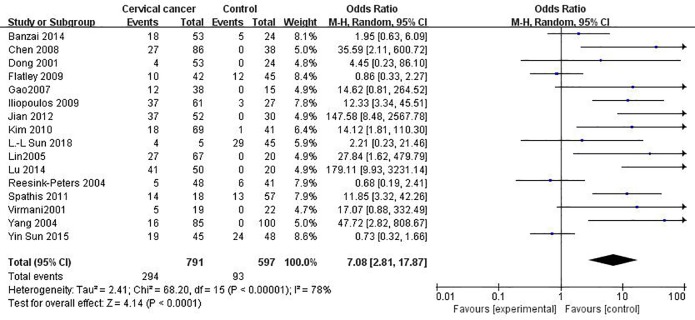
16 published studies including 791 CC patients and 597 controls were included to assess the effect of MGMT promoter hypermethylation upon CC risk (Fig 4). MGMT promoter hypermethylation conferred a 7.08-fold (95% CI: 2.81–17.87) increased risk of CC and a p value of <0.001 (Table 5).
Fig 4. Funnel plots for associations of MGMT promoter hypermethylation with the risk of cervical cancer.
The squares represent the ORs for individual studies. The size of the square reflects the weight of included studies. Bars represent the 95% confidence intervals (CIs). The center of the diamond represents the summary effect size.
Table 5. Pooled results for the association of MGMT promoter hypermethylation with CC.
| Comparisons | Studies (N) | Sample size (CC/controls) | Heterogeneity | Modela | Effect size | ||
|---|---|---|---|---|---|---|---|
| I2(%) | P Q-text | OR (95% CI) | P | ||||
| Total | 16 | 791/597 | 78 | <0.001 | R | 7.08(2.81–17.87) | <0.001 |
| Ethnicity | |||||||
| Asian | 11 | 603/405 | 79 | <0.001 | R | 11.12(2.95–41.95) | <0.001 |
| Caucasian | 5 | 188/192 | 82 | <0.001 | R | 3.67(0.87–15.46) | 0.08 |
| Source of controls | |||||||
| Healthy | 6 | 293/209 | 76 | <0.001 | R | 8.92(2.14–37.28) | 0.003 |
| Non-healthyb | 10 | 498/388 | 80 | <0.001 | R | 6.41(1.77–23.27) | 0.005 |
| Materials | |||||||
| Tissue | 8 | 453/273 | 55 | 0.03 | R | 15.38(4.06–58.30) | <0.001 |
| Exfoliated cells | 8 | 338/324 | 83 | <0.001 | R | 3.87(1.18–12.67) | 0.03 |
| Publication year | |||||||
| ≥ 2010 | 7 | 292/265 | 84 | <0.001 | R | 7.84(1.76–34.93) | 0.007 |
| < 2010 | 9 | 499/ | 74 | <0.001 | R | 6.88(1.89–25.00) | 0.003 |
| Quality of studies | |||||||
| High (>11) | 10 | 560/379 | 0 | 0.59 | F | 23.96(12.41–46.20) | <0.001 |
| Low (≤11) | 6 | 231/218 | 21 | 0.27 | F | 1.16(0.74–1.84) | 0.52 |
Abbreviations: N, number; CC, cervical cancer; F, fixed-effects model; R, random-effects model.
Notes
aWhen significant heterogeneity was found (I2≥50% or PQ-test≤0.1), a random-effects model with the inverse variance method was used to pool the results; otherwise, a fixed-effects model was applied.
b Non-healthy controls included autologous controls and controls with benign gynecological diseases.
Due to observation of extensive heterogeneity was observed in the overall comparison (I2 = 78%), subgroup, meta-regression and Galbraith plot analyses were conducted for seeking the possible sources of heterogeneity. In subgroup analyses, such association was till significant in nearly every subgroups in addition to the low-quality studies. However, moderate or extensive heterogeneity was still in most of the subgroups, except for the subgroups involving high-quality studies (I2 = 0%). The outcomes of meta-regression analyses illustrated that ethnicity (p = 0.248), source of controls (p = 0.880), materials (p = 0.654) and publication year (p = 0.570) were not main sources of heterogeneity (Table D in S1 File). Only the quality of studies was the major source of heterogeneity (p<0.001). Moreover, a Galbraith plot was further depicted, spotting four outliers [33, 37, 53, 56] as major sources of heterogeneity (Figure B in S1 File). These four studies were all classified into low-quality studies, and exclusion of such four studies caused a decline in I2 value from 78 to 0%, followed by a significant association between the methylation of MGMT with increased CC risk (OR = 20.31, 95% CI: 11.02–37.41, p<0.001), which further providing support to the outcomes of meta- regression.
The relationship between MGMT promoter hypermethylation and clinicopathological feature of cervical cancer
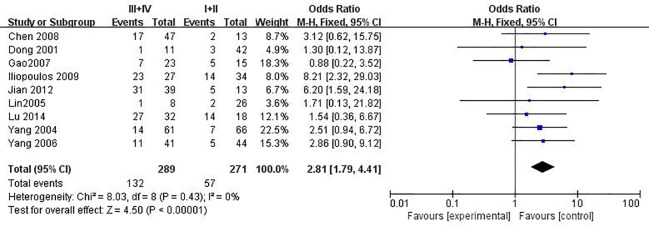
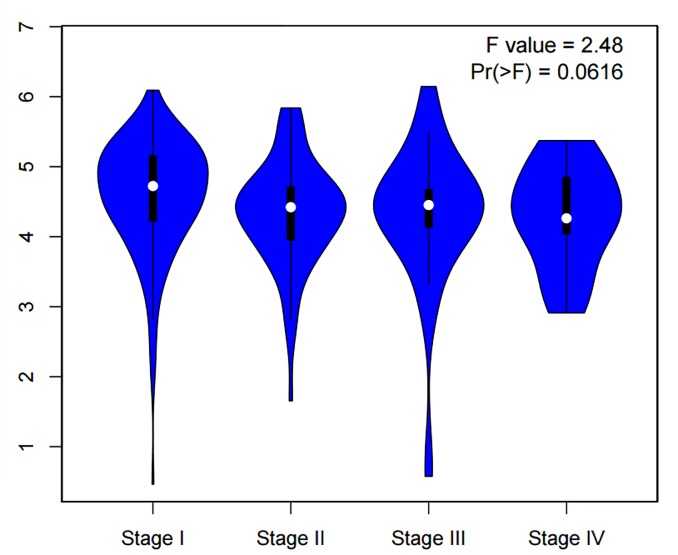
Through combination of the methylation data from 16 published studies including 1676 SIL or CC patients, we evaluated the relationship between MGMT promoter methylation and clinicopathological features including histological types, advanced International Federation of Gynecology and Obstetrics (FIGO) stage, histological grade, HPV infection, therapeutic response, age at diagnoses and lymph node metastasis. As shown in Table 6 and Fig 5, MGMT promoter methylation was significantly associated with FIGO stage (OR = 2.81, 95% CI:1.79–4.41, p<0.001), but not with histological types (see Figure C in S1 File), histological grade (see Figure D in S1 File), HPV infection, therapeutic response, age and lymph node metastasis. Furthermore, the results from the Gene Expression Profiling Interactive Analysis (GEPIA) databases demonstrated that there were no significant association between MGMT expression and FIGO stage (p>0.05) (Fig 6).
Table 6. Poole results for the associations between MGMT promoter hypermethylation and clincopathological features of CC.
| Clincopathological features | Studies (N) | Patients (N) | Heterogeneity | Modela | Effect size | ||
|---|---|---|---|---|---|---|---|
| I2(%) | PQ-test | OR (95% CI) | P | ||||
| Histological types (SCC vs. AdC) | 8 | 475 | 0 | 0.73 | F | 0.73(0.43–1.26) | 0.26 |
| FIGO stage (I+II vs. III+IV) | 9 | 560 | 0 | 0.43 | F | 2.81(1.79–4.41) | <0.001 |
| Histological grade(G3 vs. G1+G2) | 7 | 433 | 54 | 0.04 | R | 1.15(0.49–2.68) | 0.74 |
| HPV infection(Positive vs. Negative) | 2 | 850 | 96 | <0.001 | R | 17.24(0.02–190.55) | 0.43 |
| Therapeutic response (Yes vs. No) | 2 | 206 | 0 | 0.85 | F | 1.65(0.80–3.38) | 0.17 |
| Age at diagnoses (<50 vs. ≥50) | 2 | 720 | 0 | 0.98 | F | 1.40(0.93–2.10) | 0.11 |
| Lymph node metastasis (Yes vs. No) | 2 | 66 | 1 | 0.32 | R | 4.91(1.56–15.42) | 0.007 |
Abbreviations: N, number; SCC, squamous cell carcinoma; AdC, adenocinoma; F, fixed-effects model; R, random-effects model.
Notes
aWhen significant heterogeneity was found (I2≥50% or PQ-test≤0.1), a random-effects model with the inverse variance method was used to pool the results; otherwise, a fixed-effects model was applied.
Fig 5. Funnel plots for associations of MGMT promoter hypermethylation with the FIGO stage of cervical cancer.
The squares represent the ORs for individual studies. The size of the square reflects the weight of included studies. Bars represent the 95% confidence intervals (CIs). The center of the diamond represents the summary effect size.
Fig 6. The levels of MGMT expression in different FIGO stage of cervical cancer (p>0.05) from GEPIA databases.
Prognostic role of MGMT expression in cervical cancer
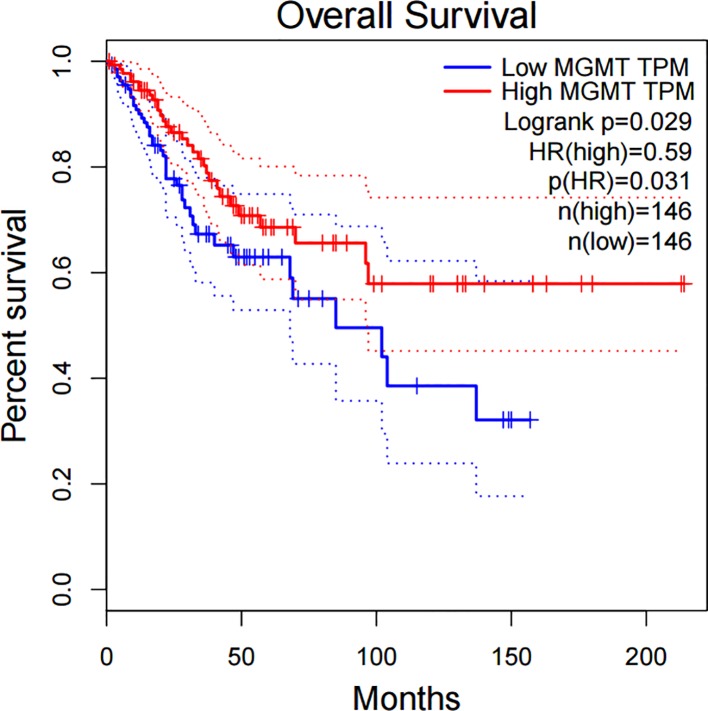
The further analysis from GEPIA databases illustrated that MGMT expression was related to overall survival (OS) in 292 patients with cervical cancer (p<0.05), as shown in Fig 7.
Fig 7. The correlation between MGMT expression and overall survival in cervical cancer (p<0.05) from GEPIA databases.
Validation by quantitative methylation data from GEO databases
The genome-wide DNA methylation array studies were extracted from GEO databases to validate the results. Totally, 5 datasets (GSE99511, GSE46306, GSE41384, GSE36637 and GSE30760) involved genome-wide DNA methylation array of 67 controls, 63 patients with cervical cancer, as shown in Fig 7. A total of 7 CpG sites (cg00904483, cg02381948, cg02803836, cg02941816, cg03271907, cg04473030, cg07453748) in promoter region of MGMT were included. 5 of 7 CpG sites (cg00904483, cg02381948, cg03271907, cg04473030, cg07453748) showed significance results with p-values<0.001 when methylation level of cervical cancer compared with that of controls (as shown in Fig 8, Table 7). Besides, these 5 CpG sites showed a great diagnostic value for cervical cancer with AUC from 0.779 to 0.818, specificities from 0.682 to 0.848, sensitivities from 0.700 to 0.829, which were shown in bold in Table 8.
Fig 8. Significant differences of methylation level in 5 CpG sites of MGMT between cervical cancer and controls in GEO dataset.
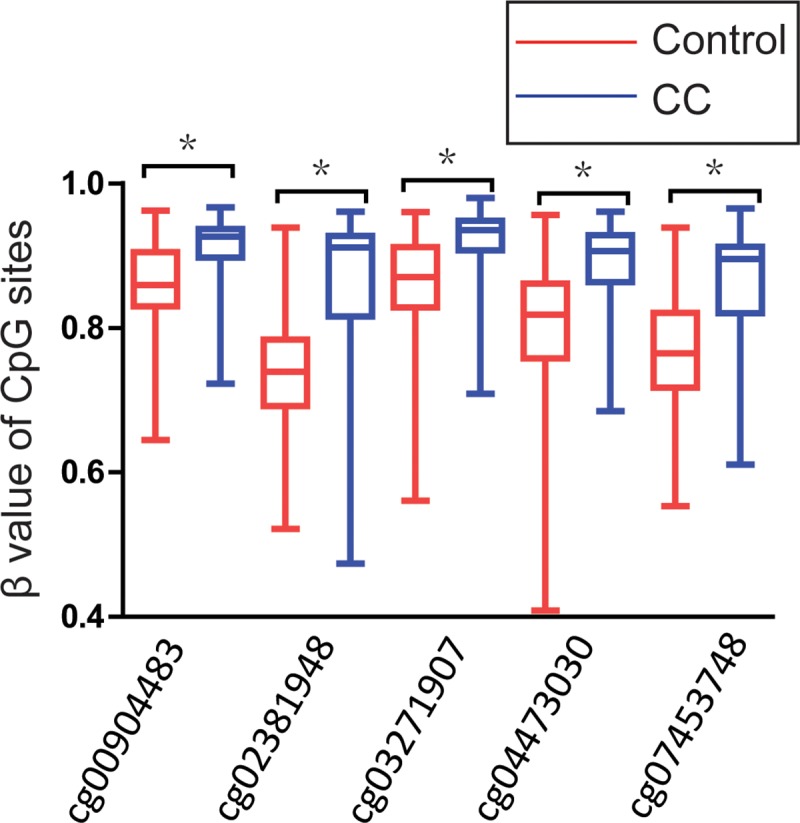
P-value were calculated by the Mann-Whitney U test. *P<0.001.
Table 7. Diagnostic value of 7 CpG sites of MGMT promoter for cervical cancer.
| CpG sites | CpG island | CpG island location | Diagnostic value of CpG sites in CC | p-value | |||
|---|---|---|---|---|---|---|---|
| Cut-off value | Specificity | Sensitivity | AUC | ||||
| cg00904483 | TRUE | chr10:131244704–131245359 | 0.878 | 0.682 | 0.829 | 0.776 | 3.39E-07 |
| cg02381948 | TRUE | chr10:131244704–131245359 | 0.795 | 0.788 | 0.814 | 0.817 | 1.33E-10 |
| cg02803836 | TRUE | chr10:131302456–131302963 | 0.914 | 0.606 | 0.657 | 0.548 | 0.390 |
| cg02941816 | TRUE | chr10:131154808–131155770 | 0.053 | 0.591 | 0.714 | 0.606 | 0.295 |
| cg03271907 | TRUE | chr10:131302456–131302963 | 0.900 | 0.742 | 0.800 | 0.802 | 7.96E-08 |
| cg04473030 | TRUE | chr10:131244704–131245359 | 0.879 | 0.833 | 0.714 | 0.818 | 5.99E-09 |
| cg07453748 | TRUE | chr10:131244704–131245359 | 0.862 | 0.848 | 0.700 | 0.797 | 4.26E-09 |
Abbreviations: CC, cervical cancer; AUC, area under the curve.
Table 8. Characteristics of included GEO datasets in this meta-analysis.
| Author | Year | Country | Ethnicity | Sample size | Methylation detection method | Materials | Source of controls | Quality scores | |
|---|---|---|---|---|---|---|---|---|---|
| Control | CC | ||||||||
| GSE99511 | 2017–2019 | Netherlands | Caucasian | 28 | 4 | Illumina HumanMethylation450 BeadChip | B | 11 | |
| GSE46306 | 2013–2019 | Sweden | Caucasian | 20 | 6 | Illumina HumanMethylation450 BeadChip | H | 13 | |
| GSE41384 | 2012–2015 | Colombia | Mix | 3 | 3 | Illumina HumanMethylation27 BeadChip | H | 13 | |
| GSE36637 | 2012–2015 | Belgium | Caucasian | 4 | 5 | Illumina HumanMethylation27 BeadChip | H | 11 | |
| GSE30760 | 2011–2015 | United Kingdom | Caucasian | 15 | 48 | Illumina HumanMethylation27 BeadChip | M | 12 | |
Abbreviations: CC, cervical cancer; HSIL, high-grade squamous intra-epithelial lesion; LSIL, low-grade squamous intra-epithelial lesion; B, controls with benign cervical diseases; H, healthy controls; A, autologous controls; M, mixed controls.
Sensitivity analysis for evaluating the stable feature of pooled results
In sensitivity analyses as shown in Figure E in S1 File, sequential removal of each study produced no apparent influence upon the pooled results except one study[56].
Publication bias of meta-analyses
In all comparisons, the shapes of funnel plots (see Figure F in S1 File) were symmetric and the values of the Egger’s test were greater than 0.05, indicating the non-existence of significant publication bias in this meta-analysis.
Trial sequence analysis (TSA)
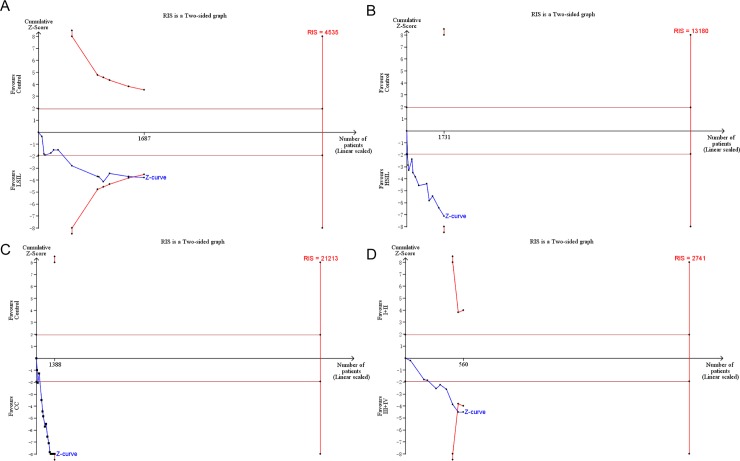
For statistical significance, trial sequence analysis (TSA) was conducted to estimate the required information size. Based on the a priori anticipated information size method, when LSIL (the estimated required sample size of 4535 cases: Fig 9A) and cervical cancer (the estimated required sample size of 21213 cases: Fig 9C) were compared with controls, and FIGO stage III /IV were compared with FIGO stage I/II (the estimated required sample size of 2741 cases: Fig 9D), the cumulative Z-curve crossed the conventional boundary and the trial sequential monitoring boundary but not crossed required information size, which indicated the size were sufficient and significant associations were observed. However, when HSIL were compared with controls (the estimated required sample size of 13180 cases: Fig 9B), the cumulative Z-curve crossed the conventional boundary but not crossed the trial sequential monitoring boundary or required information size, which indicated that there still need more studies with large sample sizes in the future.
Fig 9.
Trial sequential analysis estimating the required sample information in relation to LSIL (A), HSIL (B), CC (C) compared to controls and FIGO stage III or IV compared to FIGO stage I or II (D). Abbreviations: RIS, required information size.
Discussion
The carcinogenesis of cervical cancer involves promoter methylation or other epigenetic alterations, leading to the functional loss of TSGs[58, 59]. MGMT has been reported to be an important TSG that the promoter hypermethylated and silencing of MGMT were related to increased carcinogenic risk in several types of malignancy. Because of the inconsistent and controversial conclusions of MGMT promoter methylation in previous studies of cervical cancer based on different ethnicities, materials of sample and detection methods of methylation, we carried out a meta-analysis for comprehensively evaluating the association of MGMT promoter hypermethylation with cervical carcinogenesis.
In this meta-analysis, based upon the information of exceeding 3000 subjects from 25 relevant studies, we discovered that the methylated rates of MGMT progressively elevated with lesion severity, from 12.16% in control group, 20.92% in LSIL specimens, 36.33% in HSIL specimens to 41.50% in CC specimens, and that the promoter hypermethylation of MGMT was significant associated with the increased risk of LSIL by 1.74-fold, HSIL by 3.71-fold and CC by 7.08-fold. Besides, the results of validation by genome-wide DNA methylation array datasets extracted from GEO databases discovered that methylation of 5 loci in MGMT promoter CpG islands showed a great diagnostic value for the screening of cervical cancer. Such outcomes, in combination of earlier epidemiological evidence that MGMT promoter methylation was associated with the progression of squamous intraepithelial lesions and cervical cancer[33, 45], indicating that MGMT promoter hypermethylation may serve as an important biomarker for the progression of cervical carcinogenesis. Thus, the detection of the promoter methylation of MGMT gene could help clinicians to find out the progression of cervical carcinogenesis and even whether the patients with cervical disease is recovering or getting worse, which would improve the accuracy of diagnostic for cervical cancer.
Furthermore, whether MGMT aberrant promoter methylation was correlated with clinicopathological features were also analyzed based on the data presented in more than one study. It was found that MGMT promoter hypermethylation had significant association with the FIGO stage of cervical cancer, which was more common in advanced stage (FIGO stage III) than in low stage (FIGO stage I+II). Thus, the results further implicated that MGMT promoter methylation is likely to have a critical function in the progression of cervical cancer. This conclusion were consistent with previous studies in other malignant carcinoma such as follows. In Hengstler et al.’s research, it has been reported that MGMT expression is significantly associated with FIGO stages in ovarian tumors [60]. Fu et al. reported that MGMT methylation status exert a possible prognostic value in patients with duodenal adenocarcinoma in stage III [61]. Besides, in a meta-analysis by Chen et al., focused on the association between MGMT hypermethylation and non-small-cell-lung carcinoma (NSCLC), reported that MGMT methylation was observed to be specifically associated with NSCLC clinical stage.
Moreover, this meta-analysis was conducted to assess whether MGMT could be a biomarker for the prognosis of cervical cancer. Analysis from GEPIA databases showed that MGMT expression were associated with OS in cervical cancer, the survival percentage of patients with high MGMT expression is much higher than that of patients with low MGMT expression. We speculated that MGMT promoter hypermethylation could down-regulates MGMT mRNA expression, and progressively influenced the overall survival. This conclusion were consistent with previous studies in colorectal carcinoma, the results showed that hypermethylation of MGMT was an unfavourable prognostic markers in colorectal cancer[62]. However, there is still lack of related analysis between MGMT methylation and OS in cervical cancer. There is only one research[63] reported that MGMT does not seem to be implicated in OS of cervical cancer, at least not by promoter methylation-dependent mechanisms. Therefore, prospective studies could be focus on the impact of MGMT promoter hypermethylation on the prognosis of cervical cancer.
Moderate and extensive heterogeneity were observed in our meta-analysis for the association of MGMT methylation with HSIL and CC risk, respectively. Thus, such outcomes were firstly pooled by using a random-effect model, which conservatively estimates the study weights after adjusting for the inter-study variances[64]. Then, the potential sources of heterogeneity were explored by three statistical approaches, including subgroup analysis and meta-regression to identify the confounding factors associated with observed heterogeneity, and then Galbraith plots were depicted to explore the contributions of individual studies to overall heterogeneity. In the comparison between MGMT promoter hypermethylation and HSIL risk, the results of subgroup analysis showed that the specimen material not used by tumor tissues was probably the major origin of heterogeneity. And Galbraith plots spotted four outliers [33, 42, 56, 57] as major sources of moderate heterogeneity for the relationship between MGMT promoter hypermethylation and HSIL risk. Notably, these four studies collected exfoliated cells as biospecimen. In addition, the hypermethylation rates of control group in these four studies (22.81%, 26.67%, 36.90%, 50% respectively) were much higher than that of control group in overall (12.16%), suggesting the existence of inter-study differences. Besides, in the comparison between MGMT promoter hypermethylation and CC risk, the results of subgroup analysis and meta-regression both showed that the low quality of studies was probably the major origin of heterogeneity. Galbraith plots spotted four outliers [33, 37, 53, 56] as major sources of extensive heterogeneity, and two studies [33, 56] of them involving moderate heterogeneity for the relationship between MGMT promoter hypermethylation and HSIL risk. Notably, these four studies were all classified into low-quality studies, which is consistent with the conclusions of subgroup analysis and meta-regression. By appraising these four studies according to our quality scorning system, we found that the commonalities of these studies were as follows: lack of information of biospecimen characteristics, lack of blinding of laboratory staff, lack of clinical and pathological data. Besides, MGMT promoter hypermethylation was detected by MS-HRM in one56 of the two studies that regarded as both major sources of heterogeneity for the association of MGMT promoter hypermethylation with HSIL risk and cervical, which was the main difference in their study design from other studies.
In subgroup meta-analysis, the association between the promoter hypermethylation of MGMT with squamous intraepithelial lesions and cervical cancer remained significant in both Caucasian and Asian subgroups. Since no African study was available, more experiments should be performed to verify our observation in Africans together with other ethnicities in the future. Besides, in studies that regarded non-healthy as negative controls including autologous controls and controls with benign gynecological diseases, MGMT promoter methylation is significantly correlated with HSIL and CC risk, but not with LSIL risk, while MGMT methylation is significantly correlated with all SILs and cervical cancer in studies regarded healthy as controls. It could be due to the reason that the cytology of cervical cells in patients with benign gynecological disease and LSIL is similar [65].
In sensitivity analysis, sequential removal of each study had no significant impact on the pooled results except one study[56]. The methylation detection method of this study were performed by MS-HRM, which probably might be the major difference between this study and others.
However, several limitations were still needed to be mentioned in this meta-analysis. First, in most of included studies of this meta-analysis, detection of MGMT methylation were performed by MSP, a qualitative method relied on primer designs to guarantee the accuracy. However, different primers were designed to detect MGMT promoter methylation in the included studies, which may lead to the potential bias. Second, only full-text articles written in English or Chinese were included in this meta-analysis, while articles in other languages were excluded because of unreadable contents or insufficient data, thus resulting to a selection bias. Third, it is unfortunate that the pooled analyses of several clinicopathological features in this meta-analysis based on fewer than three studies, leading to an error or inaccurate conclusion in pooled results for the association between MGMT hypermethylation and these clinicopathological features including HPV infection, therapeutic response, age at diagnose and lymph node metastasis. Thus, more prospective clinical studies with detailed information are needed to confirm the association of MGMT hypermethylation with clinicopathological features in the future.
In this meta-analysis, we found that MGMT promoter hypermethylation was associated with squamous intra-epithelial lesion and cervical cancer. Moreover, MGMT promoter hypermethylation was also correlated with FIGO stage in patients with cervical cancer. Therefore, MGMT methylation detection might have a potential value to be an epigenetic marker for the clinical diagnosis of cervical cancer. And, it will help clinicians to decide whether to give complementary cytostatic drugs or not after the primary surgery. Besides, it also could make a contribution to explore the pathogenic mechanism of cervical cancer, as well as the preventive measures. However, prospective studies should be focus on the impact of MGMT methylation on the prognosis of cervical cancer.
Supporting information
Table A. Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) 2009 Checklist. Table B. Definitions of 18 items in our quality scoring system. Table C. The univariate meta-regression results of the association of MGMT promoter methylation and HSIL risk. Table D. The univariate meta-regression results of the association of MGMT promoter methylation and CC risk. Figure A. Galbraith plot for the association of MGMT methylation and HSIL risk. Each number is the number of the respective study included in this meta-analysis (shown in Table 1). Figure B. Galbraith plot for the association of MGMT methylation and CC risk. Each number is the number of the respective study included in this meta-analysis (shown in Table 1). Figure C. Funnel plots for associations between MGMT promoter hypermethylation and histological type. Figure D. Funnel plots for associations between MGMT promoter hypermethylation and histological grade. Figure E. Sensitivity analyses in this meta-analysis. Figure F. Funnel plots in this meta-analysis.
(DOCX)
(XLSX)
Acknowledgments
The authors thank all the anonymous reviewers and editors for their suggestions, which will be helpful for the authors to improve their paper.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87–108. Epub 2015/02/06. 10.3322/caac.21262 . [DOI] [PubMed] [Google Scholar]
- 2.Di J, Rutherford S, Chu C. Review of the Cervical Cancer Burden and Population-Based Cervical Cancer Screening in China. Asian Pacific journal of cancer prevention: APJCP. 2015;16(17):7401–7. Epub 2015/12/03. 10.7314/apjcp.2015.16.17.7401 . [DOI] [PubMed] [Google Scholar]
- 3.Chen JY, Wang ZL, Wang ZY, Yang XS. The risk factors of residual lesions and recurrence of the high-grade cervical intraepithelial lesions (HSIL) patients with positive-margin after conization. Medicine. 2018;97(41):e12792 Epub 2018/10/14. 10.1097/MD.0000000000012792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vale DB, Westin MC, Zeferino LC. High-grade squamous intraepithelial lesion in women aged <30 years has a prevalence pattern resembling low-grade squamous intraepithelial lesion. Cancer cytopathology. 2013;121(10):576–81. Epub 2013/06/15. 10.1002/cncy.21312 . [DOI] [PubMed] [Google Scholar]
- 5.Guan P, Howell-Jones R, Li N, Bruni L, de Sanjose S, Franceschi S, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. International journal of cancer. 2012;131(10):2349–59. Epub 2012/02/11. 10.1002/ijc.27485 . [DOI] [PubMed] [Google Scholar]
- 6.Robertson KD. DNA methylation and human disease. Nature reviews Genetics. 2005;6(8):597–610. Epub 2005/09/02. 10.1038/nrg1655 . [DOI] [PubMed] [Google Scholar]
- 7.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer research. 1999;59(4):793–7. Epub 1999/02/24. . [PubMed] [Google Scholar]
- 8.Rehman AU, Saikia S, Iqbal MA, Ahmad I, Sadaf, Anees A, et al. Decreased expression of MGMT in correlation with aberrant DNA methylation in esophageal cancer patients from North India. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2017;39(6):1010428317705770 Epub 2017/06/18. 10.1177/1010428317705770 . [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Tong T. Clinical Significance of O-6-Methylguanine-DNA-Methyltransferase Promoter Methylation in Patients with Esophageal Carcinoma: A Systematic Meta-Analysis. Digestive diseases (Basel, Switzerland). 2018;36(2):89–97. Epub 2017/12/20. 10.1159/000481342 . [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Hua H, Han C, Cheng Y, Cheng Y, Wang Z, et al. Prognosis value of MGMT promoter methylation for patients with lung cancer: a meta-analysis. International journal of clinical and experimental pathology. 2015;8(9):11560–4. Epub 2015/12/01. [PMC free article] [PubMed] [Google Scholar]
- 11.Wang BH, Li YY, Han JZ, Zhou LY, Lv YQ, Zhang HL, et al. Gene methylation as a powerful biomarker for detection and screening of non-small cell lung cancer in blood. Oncotarget. 2017;8(19):31692–704. Epub 2017/04/14. 10.18632/oncotarget.15919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Li F. O-6-methylguanine-DNA methyltransferase gene promoter methylation and lung cancer risk: A meta-analysis. Journal of cancer research and therapeutics. 2016;12(Supplement):C233–c6. Epub 2017/02/24. 10.4103/0973-1482.200745 . [DOI] [PubMed] [Google Scholar]
- 13.Switzeny OJ, Christmann M, Renovanz M, Giese A, Sommer C, Kaina B. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response. Clinical epigenetics. 2016;8:49 Epub 2016/05/10. 10.1186/s13148-016-0204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banan R, Christians A, Bartels S, Lehmann U, Hartmann C. Absence of MGMT promoter methylation in diffuse midline glioma, H3 K27M-mutant. Acta neuropathologica communications. 2017;5(1):98 Epub 2017/12/17. 10.1186/s40478-017-0500-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alizadeh Naini M, Kavousipour S, Hasanzarini M, Nasrollah A, Monabati A, Mokarram P. O6-Methyguanine-DNA Methyl Transferase (MGMT) Promoter Methylation in Serum DNA of Iranian Patients with Colorectal Cancer. Asian Pacific journal of cancer prevention: APJCP. 2018;19(5):1223–7. Epub 2018/05/29. 10.22034/APJCP.2018.19.5.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagci B, Sari M, Karadayi K, Turan M, Ozdemir O, Bagci G. KRAS, BRAF oncogene mutations and tissue specific promoter hypermethylation of tumor suppressor SFRP2, DAPK1, MGMT, HIC1 and p16 genes in colorectal cancer patients. Cancer biomarkers: section A of Disease markers. 2016;17(2):133–43. Epub 2016/08/20. 10.3233/cbm-160624 . [DOI] [PubMed] [Google Scholar]
- 17.Coppede F, Lopomo A, Spisni R, Migliore L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World journal of gastroenterology. 2014;20(4):943–56. Epub 2014/02/28. 10.3748/wjg.v20.i4.943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michailidi C, Theocharis S, Tsourouflis G, Pletsa V, Kouraklis G, Patsouris E, et al. Expression and promoter methylation status of hMLH1, MGMT, APC, and CDH1 genes in patients with colon adenocarcinoma. Experimental biology and medicine (Maywood, NJ). 2015;240(12):1599–605. Epub 2015/04/25. 10.1177/1535370215583800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Y, Yang Q, Wang B, Ye G, Tong X. The Correlation of MGMT Promoter Methylation and Clinicopathological Features in Gastric Cancer: A Systematic Review and Meta-Analysis. PloS one. 2016;11(11):e0165509 Epub 2016/11/09. 10.1371/journal.pone.0165509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai F, Xiao X, Niu X, Shi H, Zhong Y. Aberrant Methylation of MGMT Promoter in HNSCC: A Meta-Analysis. PloS one. 2016;11(9):e0163534 Epub 2016/09/23. 10.1371/journal.pone.0163534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutsimpelas D, Pongsapich W, Heinrich U, Mann S, Mann WJ, Brieger J. Promoter methylation of MGMT, MLH1 and RASSF1A tumor suppressor genes in head and neck squamous cell carcinoma: pharmacological genome demethylation reduces proliferation of head and neck squamous carcinoma cells. Oncol Rep. 2012;27(4):1135–41. Epub 2012/01/17. 10.3892/or.2012.1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao B, Zhang Z, Li Y. Association of MGMT promoter methylation with tumorigenesis features in patients with ovarian cancer: A systematic meta-analysis. Molecular genetics & genomic medicine. 2018;6(1):69–76. Epub 2017/12/02. 10.1002/mgg3.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An N, Shi Y, Ye P, Pan Z, Long X. Association Between MGMT Promoter Methylation and Breast Cancer: a Meta-Analysis. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;42(6):2430–40. Epub 2017/08/30. 10.1159/000480196 . [DOI] [PubMed] [Google Scholar]
- 24.Virmani AK, Muller C, Rathi A, Zoechbauer-Mueller S, Mathis M, Gazdar AF. Aberrant methylation during cervical carcinogenesis. Clinical Cancer Research. 2001;7(3):584–9. WOS:000167883100021. [PubMed] [Google Scholar]
- 25.Chen R, Zheng Y, Zhuo L, Wang S. Association between MGMT Promoter Methylation and Risk of Breast and Gynecologic Cancers: A Systematic Review and Meta-Analysis. Scientific Reports. 2017;7 10.1038/s41598-017-13208-3 WOS:000412492400009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS medicine. 2012;9(5):e1001216 Epub 2012/06/08. 10.1371/journal.pmed.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore HM, Kelly AB, Jewell SD, McShane LM, Clark DP, Greenspan R, et al. Biospecimen reporting for improved study quality (BRISQ). Cancer cytopathology. 2011;119(2):92–101. Epub 2011/03/25. 10.1002/cncy.20147 . [DOI] [PubMed] [Google Scholar]
- 28.Thakkinstian A, McEvoy M, Minelli C, Gibson P, Hancox B, Duffy D, et al. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. American journal of epidemiology. 2005;162(3):201–11. Epub 2005/07/01. 10.1093/aje/kwi184 . [DOI] [PubMed] [Google Scholar]
- 29.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical research ed). 2003;327(7414):557–60. Epub 2003/09/06. 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu XC, Yu W, Tao Y, Zhao PL, Li K, Tang LJ, et al. Contribution of transforming growth factor alpha polymorphisms to nonsyndromic orofacial clefts: a HuGE review and meta-analysis. American journal of epidemiology. 2014;179(3):267–81. Epub 2013/11/19. 10.1093/aje/kwt262 . [DOI] [PubMed] [Google Scholar]
- 31.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed). 1997;315(7109):629–34. Epub 1997/10/06. 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solomon D, Davey D, Kurman R, Moriarty A, O'Connor D, Prey M, et al. The 2001 Bethesda System: terminology for reporting results of cervical cytology. Jama. 2002;287(16):2114–9. Epub 2002/04/23. 10.1001/jama.287.16.2114 . [DOI] [PubMed] [Google Scholar]
- 33.Flatley JE, McNeir K, Balasubramani L, Tidy J, Stuart EL, Young TA, et al. Folate Status and Aberrant DNA Methylation Are Associated With HPV Infection and Cervical Pathogenesis. Cancer Epidemiology Biomarkers & Prevention. 2009;18(10):2782–9. 10.1158/1055-9965.EPI-09-0493 WOS:000270702100029. [DOI] [PubMed] [Google Scholar]
- 34.da Costa Lodi CT, Michelin MA, Miranda Lima MI, Candido Murta EF, Braga LdC, Montes L, et al. Predicting cervical intraepithelial neoplasia recurrence in HIV-infected and -noninfected women by detecting aberrant promoter methylation in the CDH1, TIMP3, and MGMT genes. Archives of Gynecology and Obstetrics. 2018;298(5):971–9. 10.1007/s00404-018-4899-x WOS:000446970300018. [DOI] [PubMed] [Google Scholar]
- 35.Wang, L. Interaction between polycyclic aromatic hydrocarbons exposure and CpG islands methylation of RAR-β and MGMT genes in cervical intraepithelial neoplasia. Thesis. Shanxi Medical University, (2018)
- 36.Nan, J. Effect of HPV, folate and related suppressor gene CpG island methylation in cervical intraepithelial neoplasias and related interaction. Thesis. Shanxi Medical University, (2016).
- 37.Banzai C, Nishino K, Quan J, Yoshihara K, Sekine M, Yahata T, et al. Promoter methylation of DAPK1, FHIT, MGMT, and CDKN2A genes in cervical carcinoma. International Journal of Clinical Oncology. 2014;19(1):127–32. 10.1007/s10147-013-0530-0 WOS:000331700200020. [DOI] [PubMed] [Google Scholar]
- 38.Lu H. et al. Relationship between methylation of MGMT gene promoter and cervical carcinoma. Zhejiang Medical Journal. 2014; 36: 1841–3. [Google Scholar]
- 39.Sun L-l, Cao D-y, Yang J-x, Li H, Zhou X-r, Song Z-q, et al. Population-based case-control study on DAPK1, RAR-beta 2 and MGMT methylation in liquid-based cytology. Archives of Gynecology and Obstetrics. 2012;285(5):1433–9. 10.1007/s00404-011-2149-6 WOS:000302814200035. [DOI] [PubMed] [Google Scholar]
- 40.Sun LL, Liu Y, Sun X, Pan L, Wu D, Wang YD. Limited Role of Promoter Methylation of MGMT and C13ORF18 in Triage of Low-Grade Squamous Intraepithelial Lesion. Chin Med J (Engl). 2018;131(8):939–44. Epub 2018/04/18. 10.4103/0366-6999.229896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jian Y. et al. Relationship between hypermethylation of DNA repair gene MGMT and cervical carcinoma. Journal of Qinghai Medical College. 2012; 33: 90–4. [Google Scholar]
- 42.Spathis A, Aga E, Alepaki M, Chranioti A, Meristoudis C, Panayiotides I, et al. Promoter methylation of p16(INK4A), hMLH1, and MGMT in liquid-based cervical cytology samples compared with clinicopathological findings and HPV presence. Infectious diseases in obstetrics and gynecology. 2011;2011:927861–. 10.1155/2011/927861 MEDLINE:21747645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, Y. MGMT promoter methylation status in serum of patients with squamous carcinoma of the cervix and the relation with hr-HPV infection. Thesis. Wenzhou Medical University. (2011).
- 44.Kim J-H, Choi YD, Lee JS, Lee JH, Nam JH, Choi C. Assessment of DNA methylation for the detection of cervical neoplasia in liquid-based cytology specimens. Gynecologic Oncology. 2010;116(1):99–104. 10.1016/j.ygyno.2009.09.032 WOS:000273108700019. [DOI] [PubMed] [Google Scholar]
- 45.Iliopoulos D, Oikonomou P, Messinis I, Tsezou A. Correlation of promoter hypermethylation in hTERT, DAPK and MGMT genes with cervical oncogenesis progression. Oncology Reports. 2009;22(1):199–204. 10.3892/or_00000425 WOS:000267259100028. [DOI] [PubMed] [Google Scholar]
- 46.Park N-H, 이승호. Promoter hypermethylation of MGMT (O6-methylguanine-DNA methyltransferase)in cervical squamous cell carcinoma promoter과메칠화에 대한 연구. Obstetrics & Gynecology Science. 2008;51(1):24–30. KJD:ART001213600. [Google Scholar]
- 47.Chen, S. (2008). Detection of aberrant methylation of the promoter of MGMT gene in the tissue and plasma in Uigur cervical cancer and its clinical significance. Thesis. Shihezi University.
- 48.Henken FE, Wilting SM, Overmeer RM, van Rietschoten JGI, Nygren AOH, Errami A, et al. Sequential gene promoter methylation during HPV-induced cervical carcinogenesis. British Journal of Cancer. 2007;97(10):1457–64. 10.1038/sj.bjc.6604055 WOS:000250956100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jo H, Kang S, Kim JW, Kang GH, Park NH, Song YS, et al. Hypermethylation of the COX-2 gene is a potential prognostic marker for cervical cancer. Journal of Obstetrics and Gynaecology Research. 2007;33(3):236–41. 10.1111/j.1447-0756.2007.00517.x WOS:000247321600001. [DOI] [PubMed] [Google Scholar]
- 50.Gao Y. et al. Promoter hypermethylation of DNA repair gene O6-Methylguanine DNA methylatransferase in cervical carcinoma. Prog Obset Gynecol. 2007: 569–72. [Google Scholar]
- 51.Yang H-J, Liu VWS, Wang Y, Tsang PCK, Ngan HYS. Differential DNA methylation profiles in gynecological cancers and correlation with clinico-pathological data. Bmc Cancer. 2006;6 10.1186/1471-2407-6-212 WOS:000240420200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Z, Gao M, Zhang X, Kim YS, Lee ES, Kim HK, et al. The hypermethylation and protein expression of p16 INK4A and DNA repair gene O6-methylguanine-DNA methyltransferase in various uterine cervical lesions. J Cancer Res Clin Oncol. 2005;131(6):364–70. Epub 2005/03/24. 10.1007/s00432-004-0657-5 . [DOI] [PubMed] [Google Scholar]
- 53.Reesink-Peters N, Wisman GBA, Jeronimo C, Tokumaru CY, Cohen Y, Dong SM, et al. Detecting cervical cancer by quantitative promoter hypermethylation assay on cervical scrapings: A feasibility study. Molecular Cancer Research. 2004;2(5):289–95. WOS:000221944600003. [PubMed] [Google Scholar]
- 54.Yang HJ, Liu VWS, Wang Y, Chan KYK, Tsang PCK, Khoo US, et al. Detection of hypermethylated genes in tumor and plasma of cervical cancer patients. Gynecologic Oncology. 2004;93(2):435–40. 10.1016/j.ygyno.2004.01.039 WOS:000221120500026. [DOI] [PubMed] [Google Scholar]
- 55.Dong SM, Kim HS, Rha SH, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clinical Cancer Research. 2001;7(7):1982–6. WOS:000169782400024. [PubMed] [Google Scholar]
- 56.Sun Y, Li S, Shen K, Ye S, Cao D, Yang J. DAPK1, MGMT and RARB promoter methylation as biomarkers for high-grade cervical lesions. International Journal of Clinical and Experimental Pathology. 2015;8(11):14939–45. WOS:000368140100133. [PMC free article] [PubMed] [Google Scholar]
- 57.Sun W, Zaboli D, Liu Y, Arnaoutakis D, Khan T, Wang H, et al. Comparison of promoter hypermethylation pattern in salivary rinses collected with and without an exfoliating brush from patients with HNSCC. PloS one. 2012;7(3):e33642 Epub 2012/03/23. 10.1371/journal.pone.0033642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ouadid-Ahidouch H, Rodat-Despoix L, Matifat F, Morin G, Ahidouch A. DNA methylation of channel-related genes in cancers. Biochimica et biophysica acta. 2015;1848(10 Pt B):2621–8. Epub 2015/02/24. 10.1016/j.bbamem.2015.02.015 . [DOI] [PubMed] [Google Scholar]
- 59.Huang T, Chen X, Hong Q, Deng Z, Ma H, Xin Y, et al. Meta-analyses of gene methylation and smoking behavior in non-small cell lung cancer patients. Sci Rep. 2015;5:8897 Epub 2015/03/11. 10.1038/srep08897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hengstler JG, Tanner B, Moller L, Meinert R, Kaina B. Activity of O(6)-methylguanine-DNA methyltransferase in relation to p53 status and therapeutic response in ovarian cancer. Int J Cancer. 1999;84(4):388–95. Epub 1999/07/15. . [DOI] [PubMed] [Google Scholar]
- 61.Fu T, Sharmab A, Xie F, Liu Y, Li K, Wan W, et al. Methylation of MGMT Is Associated with Poor Prognosis in Patients with Stage III Duodenal Adenocarcinoma. PloS one. 2016;11(9):e0162929 Epub 2016/09/20. 10.1371/journal.pone.0162929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nilsson TK, Lof-Ohlin ZM, Sun XF. DNA methylation of the p14ARF, RASSF1A and APC1A genes as an independent prognostic factor in colorectal cancer patients. Int J Oncol. 2013;42(1):127–33. Epub 2012/11/07. 10.3892/ijo.2012.1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lof-Ohlin ZM, Sorbe B, Wingren S, Nilsson TK. Hypermethylation of promoter regions of the APC1A and p16(INK4a) genes in relation to prognosis and tumor characteristics in cervical cancer patients. International Journal of Oncology. 2011;39(3):683–8. 10.3892/ijo.2011.1078 WOS:000293492900018. [DOI] [PubMed] [Google Scholar]
- 64.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemporary clinical trials. 2007;28(2):105–14. Epub 2006/06/30. 10.1016/j.cct.2006.04.004 . [DOI] [PubMed] [Google Scholar]
- 65.Lee H, Lee EJ. HPV infection and p16 promoter methylation as predictors of ASC-US/LSIL progression. Cancer cytopathology. 2016;124(1):58–65. Epub 2015/09/04. 10.1002/cncy.21615 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) 2009 Checklist. Table B. Definitions of 18 items in our quality scoring system. Table C. The univariate meta-regression results of the association of MGMT promoter methylation and HSIL risk. Table D. The univariate meta-regression results of the association of MGMT promoter methylation and CC risk. Figure A. Galbraith plot for the association of MGMT methylation and HSIL risk. Each number is the number of the respective study included in this meta-analysis (shown in Table 1). Figure B. Galbraith plot for the association of MGMT methylation and CC risk. Each number is the number of the respective study included in this meta-analysis (shown in Table 1). Figure C. Funnel plots for associations between MGMT promoter hypermethylation and histological type. Figure D. Funnel plots for associations between MGMT promoter hypermethylation and histological grade. Figure E. Sensitivity analyses in this meta-analysis. Figure F. Funnel plots in this meta-analysis.
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.