Figure 1.
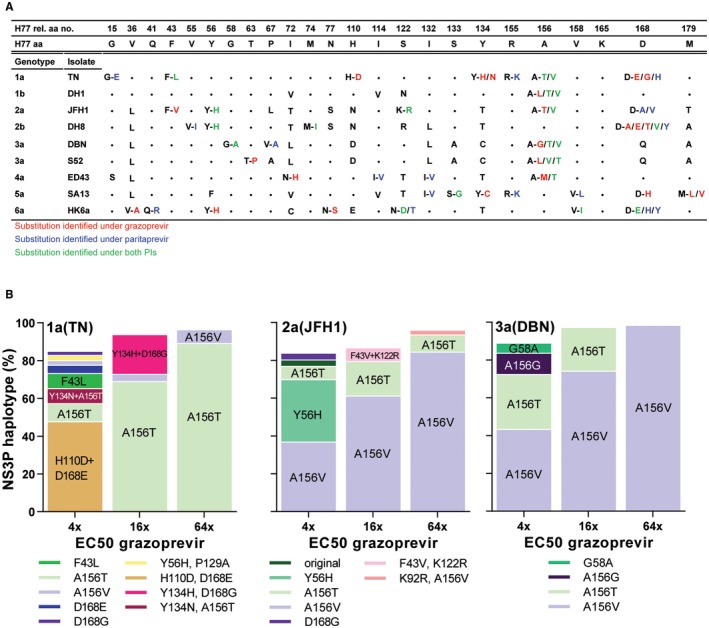
HCV genotype 1‐6 PI escape viruses harbored NS3P RASs with PI concentration‐dependent substitution patterns. (A) Putative RASs were identified by NGS. NS3P aa positions with putative RASs found in more than 5% of the viral population for at least one virus and one PI were included and numbered relative to NS3P of the H77 reference strain; H77 aa residues are specified. •, aa residues identical to that of H77; single letter, nonidentical aa residue; letters separated by dash, putative RASs indicated by the original and the mutated residues, color coded depending on the PI under which they were selected. For detailed data including NGS, see Supporting Figs. S3‐S11. (B) NS3P NGS substitution linkage analysis revealed haplotype distributions for 1a(TN), 2a(JFH1), and 3a(DBN) grazoprevir escape viruses (Supporting Figs. S3, S5, S8, S14A‐C). Haplotypes constituting greater than 2% of the viral population are included in bars; haplotypes greater than 5% are highlighted on bars.
