Abstract
Nonretinal/nonpineal photosensitivity has been found in the brain of vertebrates, but the molecular basis for such a “deep brain” photoreception system remains unclear. We conducted an extensive search for brain opsin cDNAs of the zebrafish (Danio rerio), a useful animal model for genetic studies, and we have isolated a partial cDNA clone encoding an ortholog of vertebrate ancient (VA) opsin, the function of which is unknown. Subsequent characterization revealed the occurrence of two kinds of mRNAs encoding putative splicing variants, VA and VA-Long (VAL) opsin, the latter of which is a novel variant of the former. Both opsins shared a common core sequence in the membrane-spanning domains, but VAL-opsin had a C-terminal tail much longer than that of VA-opsin. Functional reconstitution experiments on the recombinant proteins showed that VAL-opsin with bound 11-cis-retinal is a green-sensitive pigment (λmax ∼500 nm), whereas VA-opsin exhibited no photosensitivity even in the presence of 11-cis-retinal. Immunoreactivity specific to this functionally active VAL-opsin was localized at a limited number of cells surrounding the diencephalic ventricle of central thalamus, and these cells were distributed over ∼200 μm along the rostrocaudal axis. Taken together with the previous study on the locus of the teleost brain photosensitivity (von Frisch K, 1911), it is strongly suggested that the VAL-positive cells in the zebrafish brain represent the deep brain photoreceptors. The VAL-specific immunoreactivity was also detected in a subset of non-GABAergic horizontal cells in the zebrafish retina. The existence of VAL-opsin, a new member of the rhodopsin superfamily, in these tissues may indicate its multiple roles in visual and nonvisual photosensory physiology.
Keywords: VAL-opsin, VA-opsin, deep brain photoreceptor, horizontal cell, alternative splicing, zebrafish
Various nonmammalian vertebrates have photoreceptor cells not only in the retina (rod and cone) but also in the extraretinal tissues such as pineal complex, deep brain, and skin (Yoshikawa and Oishi, 1998). These nonvisual photoreceptor cells seem to play important roles in diverse physiological responses including photoentrainment of circadian rhythms, detection of seasonal changes in the photoperiod, and regulation of body color. Several lines of evidence suggested that opsin-type molecules contribute to the nonvisual photoreception of vertebrates (Deguchi, 1981; Lythgoe et al., 1984; Foster et al., 1985), and in fact, opsin-type molecules have been identified in the pineal complex (Okano et al., 1994; Max et al., 1995;Blackshaw and Snyder, 1997; Mano et al., 1999), dermal melanophores (Provencio et al., 1998), and the deep brain region of the pigeon (Wada et al., 1998), frogs (Provencio et al., 1998; Yoshikawa et al., 1998), and mouse (Blackshaw and Snyder, 1999). Despite evident expression of opsin-type molecules in the deep brain region, their roles in light-regulated physiological responses have not been precisely determined because of (1) no clear evidence for physiological responses attributable to the deep brain photoreceptors (in amphibians and mammals) or (2) infeasibility of genetic approaches (in birds) or both. With respect to these points, the zebrafish (Danio rerio) is a useful animal model for future genetic studies on the deep brain photoreceptive functions, which have been well characterized especially in teleosts (von Frisch, 1911; Scharrer, 1928; Oksche and Hartwig, 1975; van Veen et al., 1976; Hartwig and van Veen, 1979;Kavaliers, 1980; Tabata et al., 1988, 1989).
The pioneering study on the vertebrate deep brain photoreception (von Frisch, 1911) demonstrated that a light-induced change in skin color of a teleost (European minnow) is not abolished by the removal of the eyes and pineal complex, and the light sensitivity was ascribed to the “deep brain photoreceptor” located at the ependyma of the diencephalic ventricle. Despite this clear evidence, the molecular identity and detailed location of the teleost deep brain photoreceptor remain unknown. In the present study, we find a gene expression of VA-Long (VAL) opsin, a novel variant of vertebrate ancient (VA) opsin in the zebrafish brain. VA-opsin was originally identified in the salmon as an ocular opsin (λmax: 451 nm), with its function unknown, and the gene expression has been detected in a subset of horizontal and amacrine cells in the retina (Soni and Foster, 1997;Soni et al., 1998). Here we report that VAL protein is localized to a limited number of diencephalic cells and retinal horizontal cells, the location of the former being consistent with that of the deep brain photoreceptor described by von Frisch (1911). Together with the light sensitivity reconstituted with 11-cis-retinal in vitro, it is strongly suggested that VAL-opsin contributes to the deep brain photosensitivity as well as retinal physiology.
MATERIALS AND METHODS
PCR-based cDNA cloning. The RNA fraction extracted from the brains of light-adapted zebrafish (Danio rerio) was used for oligo(dT)-primed synthesis of the cDNA, and it was applied to PCR with the degenerate primers Op-Fw and Op-Rv (Yoshikawa et al., 1998), which were designed to amplify DNA fragments encoding various vertebrate rhodopsin family members. This PCR amplified two kinds of partial cDNA fragments: one encoded a pineal opsin termed exo-rhodopsin (Mano et al., 1999), and the other (Z9–20) encoded the zebrafish ortholog of VA-opsin (Soni and Foster, 1997). In the subsequent 3′ rapid amplification of cDNA end (RACE) of Z9–20, two kinds of cDNA fragments (V14E9 and V14E3) were obtained. The sequences of V14E9 and V14E3 were identical to each other in 119 nucleotides of their 5′-ends, but they showed a divergence in the rest of the 3′-ends. In contrast, 5′-RACE, by using internal primers of Z9–20 with the zebrafish brain cDNA template, amplified a single DNA fragment (V16E23). On the basis of the sequences of Z9–20, V14E9, V14E3, and V16E23, we assumed the presence of two kinds of VA-opsin mRNAs (VA and VAL) in the zebrafish brain. Finally, the two variants of VA-opsin mRNAs were identified by amplification of the entire coding sequences of VA and VAL, using the zebrafish brain cDNA as a template. The primers used were (1) the internal primers of V16E23 [5′-TCAGGGAGA(C/A)ATC(T/C)GAACTG-3′] and V14E9 (5′-AACTGAAATCAAATGCTCACTC-3′) for amplification of VA sequence or (2) the internal primers of V16E23 [5′-TCAGGGAGA(C/A)ATC(T/C)GAACTG-3′] and V14E3 (5′-CACTGAGATGAAGACTCTGC-3′) for amplification of VAL sequence. In this final series of PCR, four independent clones from each of the three independent amplifications of either VA or VAL were sequenced on both DNA strands. The nucleotide sequence data have been deposited to the DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank nucleotide sequence databases with the accession numbers AB035276 (VAL-opsin) and AB035277 (VA-opsin).
Preparation of recombinant proteins and spectroscopic measurements. The zebrafish VA- and VAL-opsin were produced in human embryonic kidney cells 293S as described previously (Kojima et al., 1996), with the aid of a eukaryotic expression vector pUSRα (Kayada et al., 1995) derived from pUC-SRα (Shimamoto et al., 1993). Briefly, five 10-cm-diameter dishes of 293S cells were co-transfected with the opsin expression vector and pRSV-TAg and cultured for 48 hr at 37°C. Then, the cells expressing VA or VAL were collected in 400 μl of buffer Pm (50 mm HEPES-NaOH, 140 mm NaCl, 3 mm MgCl2, 1 mmdithiothreitol, 1 μg/ml aprotinin, 1 μg/ml leupeptin, pH 6.6, at 4°C). Subsequent procedures were performed under a dim red light (wavelengths >660 nm) or in the dark. The collected cells were mixed with 4 nmol of 11-cis-retinal and incubated at 4°C for 5 hr, and proteins were extracted by the addition of 500 μl of 2% dodecyl-β-D-maltoside dissolved in buffer Pm. Then the extract was mixed with neutralized hydroxylamine (at a final concentration of 50 mm) and incubated at 0°C for 2 hr for conversion of free 11-cis-retinal into retinal-oxime. A difference absorption spectrum before and after bleaching of the sample with an orange light (>520 nm) at 0°C for 60 sec was recorded with a spectrophotometer (model MPS-2000; Shimadzu, Kyoto, Japan).
RT-PCR. For detection of VA and VAL mRNAs in the zebrafish tissues, RT-PCR was performed by using TaqGold polymerase (PE Biosystems, Foster City, CA) and oligo(dT)-primed cDNA template (synthesized as described in the previous section). The primers used were (1) 5′-TATGTGTTCATGAACAAACAG-3′ and 5′-ACAAGTCAGTTTTAATGATGC-3′ to amplify a 236 base pair fragment derived from VA mRNA or (2) 5′-TATGTGTTCATGAACAAACAG-3′ and 5′-GACACACTTTGTTCTCAGG-3′ to amplify a 238 base pair fragment from VAL mRNA. The cycling protocol used was 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec for 35 cycles.
Preparation of antibodies. For preparation of region-specific antibodies, three kinds of fusion proteins were designed: (1) MBP-VC, which is composed of maltose-binding protein (MBP) and the C-terminal region of VAL (termed VC region; Q303-M377) (see Fig. 1A,C), (2) GST-VC, composed of glutathione S-transferase (GST) and VC, and (3) GST-VN, composed of GST and the N-terminal region common to VA and VAL (termed VN region; M1-S31) (see Fig.1A,C). MBP-VC was expressed inEscherichia coli BL21 strain with the aid of a prokaryotic expression vector pMAL-c2 (New England Biolabs, Beverly, MA) and purified by affinity chromatography using amylose resin (New England Biolabs). GST-VC, GST-VN, and GST were expressed in BL21 with the aid of a prokaryotic expression vector pGEX-5X-1 (Amersham Pharmacia Biotech, Tokyo, Japan) and were purified by affinity chromatography using glutathione-Sepharose 4B (Amersham Pharmacia Biotech). Five female mice (BALB/c strain, 5 weeks old) were immunized with purified MBP-VC or GST-VN. For affinity purification of antisera, GST-VC, GST-VN, and GST were separately immobilized to glutathione-Sepharose 4B according to the method reported by Bar-Peled and Raikhel (1996). Then, the antibodies specific to the VC region (VC antibody) were purified by using the GST-VC-immobilized Sepharose to eliminate anti-MBP activity. As for the antisera to GST-VN, anti-GST fractions were removed by passing through a GST-immobilized Sepharose column, and the antibodies specific to the VN region (VN antibody) were purified by using GST-VN-immobilized Sepharose.
Fig. 1.
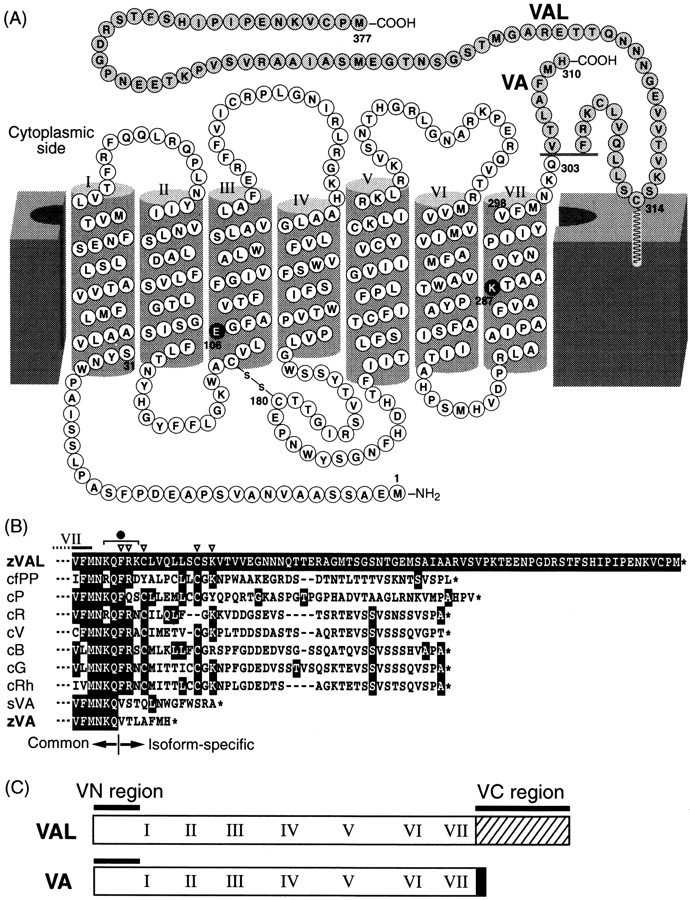
The deduced amino acid sequences of zebrafish VA- and VAL-opsin. A, A secondary structural model for VA- and VAL-opsin. VA- and VAL-opsin have a common core sequence (M1-Q303) and diverged C-terminal tails, which are shown on the gray background. Putative transmembrane segments (helices I–VII) are indicated with roman numerals. The lysine residue at 287 (K287) and the glutamic acid at 106 (E106) are the putative retinal binding site and the counter ion, respectively (white characters on the black background).B, Comparison of C-terminal amino acid sequences of zebrafish VAL-opsin (zVAL; V293-M377) with those of other opsins: catfish parapinopsin (cfPP; GenBank accession number AF028014), chicken pinopsin (cP;U15762), chicken red (cR; X57490), chicken violet (cV; M92039), chicken blue (cB; M92037), chicken green (cG; M92038), chicken rhodopsin (cRh; D00702), salmon VA-opsin (sVA;AF001499), and zebrafish VA-opsin (zVA). The multiple alignment was conducted with the Clustal W 1.8 program (Thompson et al., 1994). The residues identical to that of zVAL at each position are shown with white characters on black background. The C termini of the sequences are shown withasterisks. C, A schematic drawing of VN and VC regions in VA/VAL-opsin. VN region (M1-S31) and VC region (Q303-M377 in VAL-opsin) were used for raising antibodies (see Materials and Methods).
Preparation of tissue sections. Tissue sections were prepared as described (Barthel and Raymond, 1990) with some modifications. Adult zebrafish were maintained in light (14 hr)/dark (10 hr) cycles, and decapitated 6 hr after the onset of the light phase. Then, the heads including the eyes and brain were immersion-fixed in 4% paraformaldehyde, 5% sucrose in phosphate buffer (PB; 0.1 m Na-phosphate buffer, pH 7.4) for 2 hr at room temperature (23°C). After they were rinsed with 5% sucrose in PB, the tissues were cryoprotected with increasing concentrations of sucrose (10, 15, and 20%) in PB, embedded in a solution of 2:1 mixture of 20% sucrose (in PB) and the OCT mounting medium (Sakura, Tokyo, Japan), frozen by using liquid nitrogen, and stored at −80°C until use. Finally, 10-μm-thick sections were cut out from the embedded tissues, mounted on gelatin-coated glass slides, and air-dried.
Immunofluorescence analyses of tissue sections. The sections on glass slides were pretreated with a blocking solution [1.5% horse or goat normal serum, 0.3% Triton X-100 in PBS (10 mmNa-phosphate buffer, 140 mm NaCl, 1 mmMgCl2, pH 7.4)], and then incubated with a primary antibody diluted in the blocking solution at 4°C for 3 d. After they were rinsed with PBS, the sections were treated with a secondary antibody for 20 hr at 4°C and again washed with PBS. Then, the sections were coverslipped with a solution of 1:1 mixture of 50% glycerol (in PBS) and Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA). The primary antibodies used were VC antibody (diluted to 0.6 μg/ml), VN antibody (diluted to 0.4 μg/ml), AS-Rh [(Kawata et al., 1992) a mouse antiserum raised against purified bovine rhodopsin, diluted 1:10,000], and GAD antibody (AB108, Chemicon, Temecula, CA; a rabbit antiserum raised against feline glutamic acid decarboxylase, diluted 1:1000). For a control staining, the primary antibody was eliminated or replaced by a mouse IgG fraction (diluted to 0.6 μg/ml) purified from preimmune sera by Protein G-Sepharose column chromatography, or replaced by rabbit preimmune serum (diluted 1:1000). The secondary antibodies used were the horse anti-mouse IgG antibody conjugated either with FITC (diluted to 7.5 μg/ml) or with Texas Red (diluted to 15 μg/ml), and the goat anti-rabbit IgG antibody conjugated with FITC (diluted to 7.5 μg/ml), all of which were purchased from Vector Laboratories. TO-PRO-3 (Molecular Probes, Eugene, OR), diluted to 0.2 μm, was used for staining of cell nuclei.
RESULTS
Two isoforms of zebrafish VA-opsin ortholog
To identify an opsin-type photosensitive molecule expressed in the zebrafish brain, we conducted an extensive PCR-based screening of cDNAs prepared from the whole-brain RNA fraction (see Materials and Methods). Then we isolated a cDNA clone (Z9–20) encoding a part of a putative opsin, of which the deduced amino acid sequence was highly similar (75% identical) to that of salmon VA-opsin (Soni and Foster, 1997). To obtain a full-length cDNA, 5′- and 3′-RACE were used, resulting in isolation of two distinct cDNA variants. The deduced amino acid sequences (and the corresponding nucleotide sequences) of these variants were identical to each other within the first 303 residues, and they showed a divergence in the C-terminal sequence (Fig.1A). These two forms were probably derived from alternative RNA splicing from a single gene (see Discussion), and the common core sequence (M1-Q303) showed higher similarity (73.4% identity) to salmon VA-opsin (Soni and Foster, 1997) than to any other known opsin sequences (<50% identity). Interestingly, one of the isoforms had an extremely short cytoplasmic tail, just like salmon VA-opsin, and hence this isoform is most likely to represent a counterpart of salmon VA-opsin. The cytoplasmic tail of the other isoform was 67 amino acids longer than that of VA-opsin, and thus we named it VAL-opsin (VA-Long-opsin). Such a longer cytoplasmic tail is a common feature among all the members of rhodopsin family except for VA-opsin. Furthermore, the proximal region in the cytoplasmic tail of VAL-opsin had several characteristic residues (Fig.1B, open triangles), which are well conserved among the members of vertebrate rhodopsin family except for salmon and zebrafish VA-opsin. These suggest a functional difference between VA- and VAL-opsin.
Functional reconstitution of VAL-opsin with 11-cis-retinal
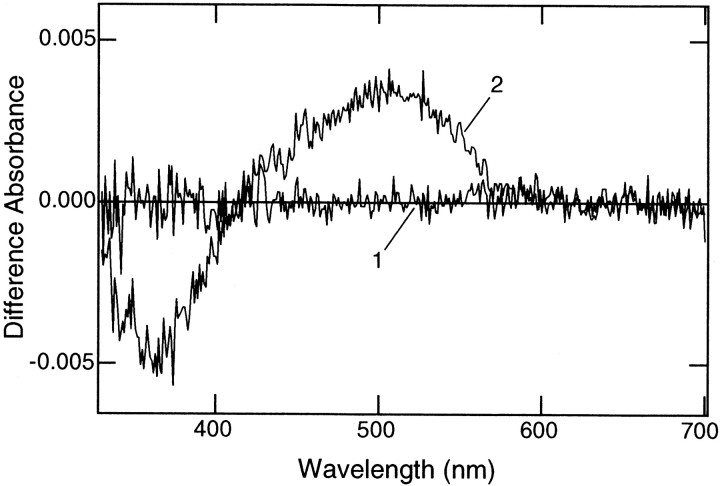
A possible functional difference between the two isoforms was examined by preparing recombinant proteins produced in the human embryonic kidney cells 293S. After being reconstituted with 11-cis-retinal, which is a common chromophore of the rhodopsin family, VA- and VAL-opsin were subjected to photobleaching experiments in the presence of hydroxylamine (Fig.2). On light irradiation, VAL-opsin with 11-cis-retinal showed a significant change in the absorption spectrum, and the difference absorption spectrum before and after complete bleaching had its maximum at ∼500 nm (Fig. 2, curve2). This represents photobleaching of a green-sensitive pigment (VAL-opsin with bound 11-cis-retinal) into opsin plus all-trans-retinal-oxime. On the other hand, no photobleaching signal was detected in the VA-opsin sample (curve1). These data indicated that only VAL-opsin can bind 11-cis-retinal and form a functional (light-sensitive) pigment under the conditions.
Fig. 2.
Light-induced changes in absorption spectra of reconstituted VA- and VAL-opsin. The recombinant proteins of zebrafish VA- and VAL-opsin produced in the 293S cells were incubated with an excess amount of 11-cis-retinal for reconstitution. Then the sample was irradiated with an orange light (>520 nm) in the presence of 50 mm hydroxylamine (see Materials and Methods). Shown are the difference absorbance spectra before and after complete bleaching of VA-opsin (curve 1) and VAL-opsin (curve 2). Equivalent sample from mock-transfected cells showed no spectral change on irradiation (data not shown).
VAL-opsin expression in the diencephalon
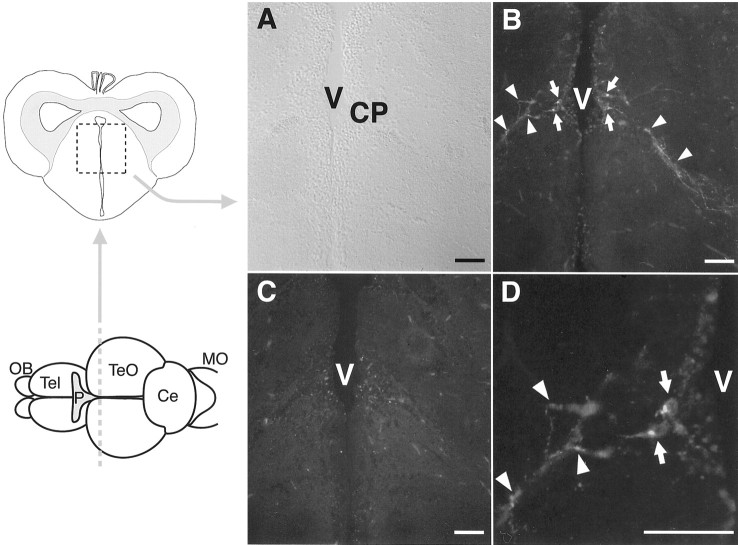
To investigate localization of functionally active VAL-opsin in the zebrafish brain, we prepared an antibody recognizing the C-terminal tail of VAL-opsin (Fig. 1C, VC region). Judging from the structure, this VC antibody is specific to VAL-opsin because the VC region shows no significant homology in sequence to any part of VA-opsin and any known opsins in the zebrafish. The specificity was confirmed by an immunoblot experiment, in which the VC antibody immunoreacted to the recombinant VAL-opsin (∼50 kDa band) expressed in the 293S cells but not to the recombinant VA-opsin (data not shown). An extensive search for VC immunoreactivity in the cross sections of whole brain of the zebrafish revealed the existence of a limited number of VC-positive cells in the diencephalic region, or more exactly in the central posterior thalamic nucleus (Fig.3B, CP), which was proximal to the diencephalic ventricle of central thalamus. Noticeably, their cell bodies and axon-like fibers were both immunoreactive (Fig.3D), a pattern reminiscent of the opsin immunoreactivity in the cerebrospinal fluid-contacting neurons in the pigeon deep brain (Wada et al., 1998). Our analysis of the serial sections demonstrated that the VC-positive neurons in CP are distributed over ∼200 μm along the rostrocaudal axis. These neurons could represent the deep brain photoreceptor cells in the zebrafish (see Discussion).
Fig. 3.
Immunohistochemical localization of VAL-opsin in the zebrafish brain. A, A Nomarski image of the zebrafish brain cross-section including the central posterior thalamic nucleus (CP), which is located near the diencephalic ventricle (V).B, Immunofluorescence labeling of the CP region with VC antibody. The VC immunoreactivity was monitored as fluorescence signals. The positive signals in the cell bodies and axon-like fibers are indicated by arrows and arrowheads, respectively. C, A control section. The section of the CP region was immunolabeled as in B without the primary antibody. Background fluorescence signals with week intensities should be ascribed to the endogenous fluorescence. D, A magnified image of B. Ce, Cerebellum;MO, medulla oblongata; OB, olfactory bulb; P, pineal gland; Tel, telencephalon; TeO, tectum opticum. Scale bar, 50 μm.
VAL-opsin expression in the retina
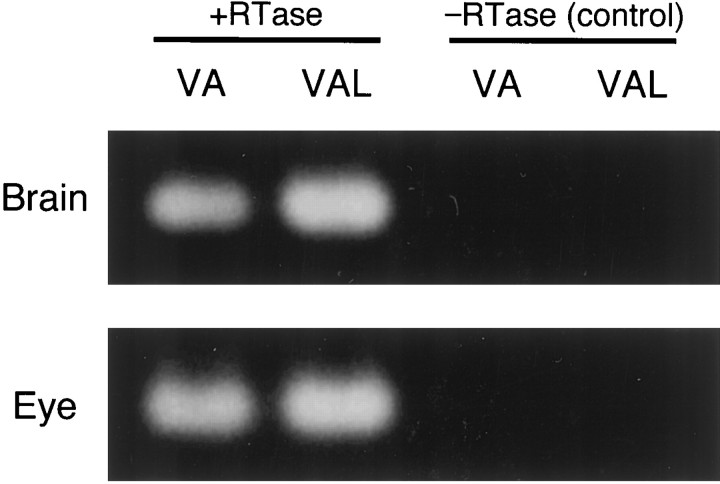
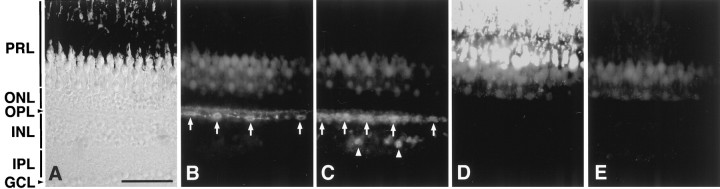
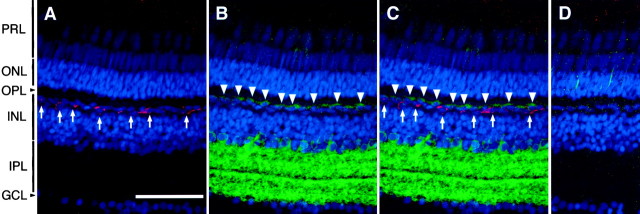
The RT-PCR-based examination of tissue-specific gene expression of VA- and VAL-opsin indicated that both were expressed not only in the brain but also in the eye (Fig. 4,+RTase). We detected no amplified product when the reverse transcriptase treatment was eliminated (Fig. 4, −RTase), confirming no detectable contamination of genomic DNA. Then, we investigated localization of VA and VAL proteins in the zebrafish eye (Fig. 5). The VC antibody labeled a subset of horizontal cells in the distal margin of the inner nuclear layer of the retina, but not the rod/cone photoreceptor layer (Fig.5B). The VAL-opsin localization in the retina was confirmed by using another antibody, VN, which was raised against the N-terminal region common to VA- and VAL-opsin (Fig. 1C). The VN antibody consistently labeled a subset of horizontal cells. This antibody also immunoreacted to a small fraction of amacrine cells in the inner nuclear layer (Fig. 5C). These amacrine cells were not detected by the VC antibody, suggesting the specific localization of VA-opsin.
Fig. 4.
RT-PCR analysis of VA- and VAL-opsin gene expression in the zebrafish brain and eye. The amplification reaction was performed after incubation with (+RTase) or without (−RTase) reverse transcriptase (see Materials and Methods).
Fig. 5.
Immunohistochemical localization of VA/VAL-opsin in the zebrafish retina. A, A Nomarski image of the zebrafish retinal section. B, Immunofluorescence labeling with the VC antibody. The VC immunoreactivity was detected as fluorescence signals in subpopulation of horizontal cells (arrows) in the inner nuclear layer (INL). C, Immunofluorescence labeling with the VN antibody. The VN antibody labeled not only the horizontal cells (arrows) but also the subpopulation of amacrine cells (arrowheads) in the inner nuclear layer (INL). D, Immunofluorescence labeling with AS-Rh, an antibody against bovine rhodopsin. E, A control section. The retinal section was immunolabeled as inD with replacement of the primary antibody by mouse preimmune serum. Background fluorescence signals in the photoreceptor layer (PRL) with weak intensities were attributed to the endogenous fluorescence, because similar signals were detected even without treatment of the secondary antibody. ONL, Outer nuclear layer; OPL, outer plexiform layer;IPL, inner plexiform layer; GCL, ganglion cell layer. Scale bar, 50 μm.
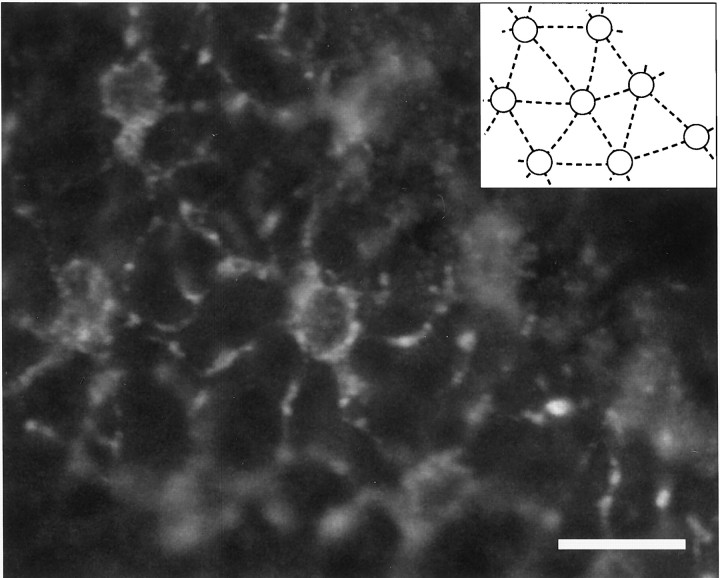
The horizontal cells in teleosts have been physiologically and histologically classified into several classes (Dowling, 1987). One of the good histochemical markers for the classification is glutamic acid decarboxylase (GAD), which is responsible for synthesis of GABA (Lam et al., 1979). By using an antibody to GAD, we examined whether the VAL-opsin-expressing cells are GABAergic. The GAD antibody labeled a subset of horizontal cells in addition to a subset of amacrine cells (Fig. 6B), just as reported previously (Connaughton et al., 1999). As shown in Figure6C, however, the GAD- and the VC-immunoreactive horizontal cells were mutually exclusive, indicating the presence of VAL-opsin in non-GABAergic horizontal cells. Judging from their location and non-GABAergic feature, the VAL-opsin-expressing cells seem to represent the rod-type horizontal cells, which are generally found in the teleost retina (Dowling, 1987). In these analyses, we noticed that the VAL-opsin-expressing horizontal cells were regularly distributed in the layer (Fig. 7) and that their relative signal intensity showed a dorsoventral gradient, i.e., the signal intensities in the horizontal cells are stronger in the dorsal part of zebrafish retina (data not shown). The cellular distribution of VAL-opsin in the zebrafish retina is not random but seems to be spatially regulated for an unknown function (see Discussion).
Fig. 6.
Double-immunofluorescence labeling of the zebrafish retinal section with the VC and GAD antibodies. The VC antibody, specific to VAL-opsin, labeled a subset of horizontal cells (A, red), and the GAD antibody labeled horizontal and amacrine cells (B, green).C is a merged image of A andB, demonstrating the mutually exclusive distribution of the VC-positive cells (arrows) and the GAD-positive horizontal cells (arrowheads). D is a control section that was treated with both mouse and rabbit preimmune sera instead of the primary antibodies. In each section, cell nuclei were stained by TO-PRO-3 (blue), and images were viewed with a confocal laser scanning microscopy (Leica, TCS-NT). Scale bar, 50 μm.
Fig. 7.
A regular geometric pattern of VAL-opsin-expressing horizontal cells in the zebrafish retina. A section tangential to the retinal surface through the distal margin of the inner nuclear layer was immunolabeled with the VC antibody, specific to VAL-opsin, as in Figure 5B. The regular arrangement of the VC-positive horizontal cells is represented in theinset, where their cell bodies are depicted byopen circles. Scale bar, 20 μm.
DISCUSSION
The present study demonstrated that VAL-opsin, a newly identified variant of VA-opsin, can bind 11-cis-retinal in vitro and form a green-sensitive pigment (Fig. 2) and that VAL-opsin is localized to a limited number of cells in CP of the zebrafish deep brain (Fig. 3). Because 11-cis-retinal, a common chromophore of vertebrate opsins, has been detected in the deep brain regions such as telencephalon (Foster et al., 1993) and diencephalon (Masuda et al., 1994), our reconstitution experimentsin vitro strongly suggest the photosensitivity of VAL-opsin-expressing cells in the zebrafish CP. To our knowledge, little is known about the neuronal function of CP in teleosts, and a role for transmitting auditory information is only suggested (Echteler, 1984, 1985; Striedter, 1991). On the other hand, von Frisch (1911)demonstrated that a fish deep brain photoreceptor responsible for the regulation of skin color is present in the ependyma of the diencephalic ventricle. This location nicely fits the localization of VAL-positive cells in the ependyma or subependyma of the diencephalic ventricle of the zebrafish brain. Like the minnow (von Frisch, 1911), the zebrafish skin becomes pale in the dark and darkened on exposure to light. Therefore, one of the physiological roles of VAL-opsin in the zebrafish deep brain could be the skin color regulation that is dependent on environmental light conditions.
Our finding of the zebrafish VA/VAL-opsin provides the first example for alternative RNA splicing generating opsin isoforms with distinct amino acid sequences. This contrasts with the alternative splicing of mouse rhodopsin mRNAs in the 3′-untranslated sequence, leaving the protein sequence unaltered (Al-Ubaidi et al., 1990). The amino acid sequences of VA- and VAL-opsin are different from each other only in the C-terminal cytoplasmic tail (Fig. 1A), and this boundary between the common and the isoform-specific region (Fig.1B) corresponds exactly to the splice site conserved among vertebrate opsin genes (Nathans and Hogness, 1983; Nathans et al., 1986; Max et al., 1995). Our preliminary sequence data of the zebrafish VA/VAL-opsin gene (data not shown) indicate that the zebrafish VA-opsin mRNA is produced by an intron retention at this splice site, whereas the VAL-opsin mRNA is spliced at this conserved splice site. Because the cytoplasmic tail is different between zebrafish VA- and VAL-opsin, we anticipated a functional difference between the two isoforms. In the case of other G-protein-coupled receptors such as the prostaglandin EP3 receptor (Namba et al., 1993) and the endothelin ETBreceptor (Elshourbagy et al., 1996), similar alternative splicing produces multiple receptor isoforms with distinct cytoplasmic tails, each of which is coupled with a distinct set of G-protein subtypes. In contrast, the present results demonstrated all-or-none difference in ligand (11-cis-retinal) binding ability between the isoforms (Fig. 2), although it remains unclear why zebrafish VA-opsin formed no photopigment with 11-cis-retinal. A plausible explanation is that the recombinant VA-opsin was misfolded and thus failed to bind the chromophore, because the proximal region of the C-terminal tail (Fig.1B, closed circle), which includes the splice site of zebrafish VA- and VAL-opsin, has been shown to play a critical role in the proper folding and stability of bovine rhodopsin (Weiss et al., 1994).
Consistent with the previous study on salmon VA-opsin (Soni et al., 1998), we also detected the expression of VAL-opsin in a class of horizontal cells of the zebrafish retina. What is the role of VAL-opsin in the retinal horizontal cells? In spite of no evidence for the supply of 11-cis-retinal to retinal horizontal cells, the spatially well ordered distribution of VAL-positive cells in the inner nuclear layer seems to indicate its functional contribution to retinal physiology. In general, gap junctions between neighboring horizontal cells enable electrocoupling among the cells to form a large receptive field (Murakami et al., 1995). The pores of gap junctions in the horizontal cells are closed in the presence of background illumination, making the receptive field narrower (Piccolino, 1986). It has been accepted that such an effect of light on the teleost horizontal cell is mediated by dopamine released from a class of interplexiform cells, but the presence of an alternative pathway (dopamine independent) is also suggested (Baldridge and Ball, 1991; Umino et al., 1991). We can speculate that VAL-opsin in the horizontal cells may participate in the light-sensitive regulation of gap junctions. It is equally possible that VAL-opsin plays a role in resetting the phase of the retinal circadian clock, which regulates the rhythmic production of melatonin in the zebrafish retina (Cahill, 1996). Recent studies on transgenic mice lacking rods and cones (Freedman et al., 1999; Lucas et al., 1999) have demonstrated that non-rod/non-cone photoreceptors in the eyes can reset the circadian clock phase. Just like cryptochromes (Miyamoto and Sancar, 1998) and melanopsin (Provencio et al., 2000) found in the ganglion and inner nuclear layers, VAL-opsin present in the horizontal cells is a candidate for the circadian photoreceptor resetting the clock phase. In addition to VAL-opsin, VA-opsin also showed highly specific distribution in the zebrafish retina (Fig. 5), but its physiological role is totally unclear at present. The failure to reconstitute VA-opsin-based photosensitive pigment with 11-cis-retinal in vitro (Fig. 2) might indicate a nonphotoreceptive function in the zebrafish.
In summary, functionally active VAL-opsin, a novel variant of VA-opsin, is present in the diencephalic cells and in the retinal horizontal cells, suggesting its multiple roles in visual and nonvisual photoreceptive physiology. The identification of functional opsin in the zebrafish brain opens a new way to the molecular and genetic characterization of the deep brain photoreception system.
Footnotes
This work was supported in part by Grants-in-Aid from the Japanese Ministry of Education, Science, Sports and Culture. H. M. is supported by a research fellowship of the Japanese Society for the Promotion of Science for Young Scientists. We thank Prof. J. Nathans (Johns Hopkins University School of Medicine) for providing pRSV-TAg and 293S cells, Prof. M. Tachibana (The University of Tokyo) and Dr. M. Murata (National Institute for Physiological Sciences) for valuable discussion about VAL-positive cells, and Drs. T. Okano, K. Sanada, and T. Yoshikawa, and F. Shimizu (this laboratory) for helpful comments and providing PCR primers and other materials.
Correspondence should be addressed to: Dr. Yoshitaka Fukada, Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113–0033, Japan. E-mail: sfukada@mail.ecc.u-tokyo.ac.jp.
REFERENCES
- 1.Al-Ubaidi MR, Pittler SJ, Champagne MS, Triantafyllos JT, McGinnis JF, Baehr W. Mouse opsin: gene structure and molecular basis of multiple transcripts. J Biol Chem. 1990;265:20563–20569. [PubMed] [Google Scholar]
- 2.Baldridge WH, Ball AK. Background illumination reduces horizontal cell receptive-field size in both normal and 6-hydroxydopamine-lesioned goldfish retinas. Vis Neurosci. 1991;7:441–450. doi: 10.1017/s0952523800009731. [DOI] [PubMed] [Google Scholar]
- 3.Barthel LK, Raymond PA. Improved method for obtaining 3-μm cryosections for immunocytochemistry. J Histochem Cytochem. 1990;38:1383–1388. doi: 10.1177/38.9.2201738. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Peled M, Raikhel NV. A method for isolation and purification of specific antibodies to protein fused to the GST. Anal Biochem. 1996;241:140–142. doi: 10.1006/abio.1996.0390. [DOI] [PubMed] [Google Scholar]
- 5.Blackshaw S, Snyder SH. Parapinopsin, a novel catfish opsin localized to the parapineal organ, defines a new gene family. J Neurosci. 1997;17:8083–8092. doi: 10.1523/JNEUROSCI.17-21-08083.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackshaw S, Snyder SH. Encephalopsin: a novel mammalian extraretinal opsin discretely localized in the brain. J Neurosci. 1999;19:3681–3690. doi: 10.1523/JNEUROSCI.19-10-03681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahill GM. Circadian regulation of melatonin production in cultured zebrafish pineal and retina. Brain Res. 1996;708:177–181. doi: 10.1016/0006-8993(95)01365-2. [DOI] [PubMed] [Google Scholar]
- 8.Connaughton VP, Behar TN, Liu WL, Massey SC. Immunocytochemical localization of excitatory and inhibitory neurotransmitters in the zebrafish retina. Vis Neurosci. 1999;16:483–490. doi: 10.1017/s0952523899163090. [DOI] [PubMed] [Google Scholar]
- 9.Deguchi T. Rhodopsin-like photosensitivity of isolated chicken pineal gland. Nature. 1981;290:706–707. doi: 10.1038/290706a0. [DOI] [PubMed] [Google Scholar]
- 10.Dowling JE. The retina: an approachable part of the brain. Harvard UP; Cambridge, MA: 1987. [Google Scholar]
- 11.Echteler SM. Connections of the auditory midbrain in a teleost fish, Cyprinus carpio. J Comp Neurol. 1984;230:536–551. doi: 10.1002/cne.902300405. [DOI] [PubMed] [Google Scholar]
- 12.Echteler SM. Organization of central auditory pathways in a teleost fish, Cyprinus carpio. J Comp Physiol. 1985;A156:267–280. [Google Scholar]
- 13.Elshourbagy NA, Adamou JE, Gagnon AW, Wu HL, Pullen M, Nambi P. Molecular characterization of a novel human endothelin receptor splice variant. J Biol Chem. 1996;271:25300–25307. doi: 10.1074/jbc.271.41.25300. [DOI] [PubMed] [Google Scholar]
- 14.Foster RG, Follett BK, Lythgoe JN. Rhodopsin-like sensitivity of extra-retinal photoreceptors mediating the photoperiodic response in quail. Nature. 1985;313:50–52. doi: 10.1038/313050a0. [DOI] [PubMed] [Google Scholar]
- 15.Foster RG, Garcia-Fernandez JM, Provencio I, DeGrip WJ. Opsin localization and chromophore retinoids identified within the basal brain of the lizard Anolis carolinensis. J Comp Physiol. 1993;A172:33–45. [Google Scholar]
- 16.Freedman MS, Lucas RJ, Soni B, von Schantz M, Munoz M, David-Gray Z, Foster RG. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science. 1999;284:502–504. doi: 10.1126/science.284.5413.502. [DOI] [PubMed] [Google Scholar]
- 17.Hartwig HG, van Veen T. Spectral characteristics of visible radiation penetrating into the brain and stimulating extraretinal photoreceptors. J Comp Physiol. 1979;130:277–282. [Google Scholar]
- 18.Kavaliers M. Retinal and extraretinal entrainment action spectra for the activity rhythms of the lake chub, Couesius plumbeus. Behav Neural Biol. 1980;30:56–67. doi: 10.1016/s0163-1047(80)90884-5. [DOI] [PubMed] [Google Scholar]
- 19.Kawata A, Oishi T, Fukada Y, Shichida Y, Yoshizawa T. Photoreceptor cell types in the retina of various vertebrate species: immunocytochemistry with antibodies against rhodopsin and iodopsin. Photochem Photobiol. 1992;56:1157–1166. doi: 10.1111/j.1751-1097.1992.tb09740.x. [DOI] [PubMed] [Google Scholar]
- 20.Kayada S, Hisatomi O, Tokunaga F. Cloning and expression of frog rhodopsin cDNA. Comp Biochem Physiol. 1995;110B:599–604. doi: 10.1016/0305-0491(94)00179-x. [DOI] [PubMed] [Google Scholar]
- 21.Kojima D, Oura T, Hisatomi O, Tokunaga F, Fukada Y, Yoshizawa T, Shichida Y. Molecular properties of chimerical mutant of gecko blue and bovine rhodopsin. Biochemistry. 1996;35:2625–2629. doi: 10.1021/bi9511548. [DOI] [PubMed] [Google Scholar]
- 22.Lam DMK, Su YYT, Swain L, Marc RE, Brandon C, Wu J-Y. Immunocytochemical localisation of l-glutamic acid decarboxylase in the goldfish retina. Nature. 1979;278:565–567. doi: 10.1038/278565a0. [DOI] [PubMed] [Google Scholar]
- 23.Lucas RJ, Freedman MS, Munoz M, Garcia-Fernandez JM, Foster RG. Regulation of the mammalian pineal by non-rod, non-cone, ocular photoreceptors. Science. 1999;284:505–507. doi: 10.1126/science.284.5413.505. [DOI] [PubMed] [Google Scholar]
- 24.Lythgoe JN, Shand J, Foster RG. Visual pigment in fish iridocytes. Nature. 1984;308:83–84. [Google Scholar]
- 25.Mano H, Kojima D, Fukada Y. Exo-rhodopsin: a novel rhodopsin expressed in the zebrafish pineal gland. Mol Brain Res. 1999;73:110–118. doi: 10.1016/s0169-328x(99)00242-9. [DOI] [PubMed] [Google Scholar]
- 26.Masuda H, Oishi T, Ohtani M, Michinomae M, Fukada Y, Shichida Y, Yoshizawa T. Visual pigments in the pineal complex of the Japanese quail, Japanese grass lizard and bullfrog: immunocytochemistry and HPLC analysis. Tissue Cell. 1994;26:101–113. doi: 10.1016/0040-8166(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 27.Max M, McKinnon PJ, Seidenman KJ, Barrett RK, Applebury ML, Takahashi JS, Margolskee RF. Pineal opsin: a nonvisual opsin expressed in chick pineal. Science. 1995;267:1502–1506. doi: 10.1126/science.7878470. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto Y, Sancar A. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc Natl Acad Sci USA. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakami M, Miyachi E-I, Takahashi K-I. Modulation of gap junctions between horizontal cells by second messengers. Prog Retinal Eye Res. 1995;14:197–221. [Google Scholar]
- 30.Namba T, Sugimoto Y, Negishi M, Irie A, Ushikubi F, Kakizuka A, Ito S, Ichikawa A, Narumiya S. Alternative splicing of C-terminal tail of prostaglandin E receptor subtype EP3 determines G-protein specificity. Nature. 1993;365:166–170. doi: 10.1038/365166a0. [DOI] [PubMed] [Google Scholar]
- 31.Nathans J, Hogness DS. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983;34:807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- 32.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 33.Okano T, Yoshizawa T, Fukada Y. Pinopsin is a chicken pineal photoreceptive molecule. Nature. 1994;372:94–97. doi: 10.1038/372094a0. [DOI] [PubMed] [Google Scholar]
- 34.Oksche A, Hartwig HG. Photoneuroendocrine systems and the third ventricles. In: Knigge KM, Scott DE, Kobayashi H, Ishii S, editors. Brain-endocrine interaction II. Karger; Basel: 1975. pp. 40–53. [Google Scholar]
- 35.Piccolino M. Horizontal cells: historical controversies and new interest. Prog Retinal Eye Res. 1986;5:147–163. [Google Scholar]
- 36.Provencio I, Jiang G, DeGrip WJ, Hayes WP, Rollag MD. Melanopsin: an opsin in melanophores, brain and eye. Proc Natl Acad Sci USA. 1998;95:340–345. doi: 10.1073/pnas.95.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Provencio I, Rodriguez IR, Jiang G, Hayes WP, Moreira EF, Rollag MD. A novel human opsin in the inner retina. J Neurosci. 2000;20:600–605. doi: 10.1523/JNEUROSCI.20-02-00600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scharrer E. Die Lichtempfindlichkeit blinder Elritzen. I. Untersuchungen über das Zwischenhirn der Fische. Z Vergl Physiol. 1928;7:1–38. [Google Scholar]
- 39.Shimamoto A, Kimura T, Matsumoto K, Nakamura T. Hepatocyte growth factor-like protein is identical to macrophage stimulating protein. FEBS Lett. 1993;333:61–66. doi: 10.1016/0014-5793(93)80375-5. [DOI] [PubMed] [Google Scholar]
- 40.Soni BG, Foster RG. A novel and ancient vertebrate opsin. FEBS Lett. 1997;406:279–283. doi: 10.1016/s0014-5793(97)00287-1. [DOI] [PubMed] [Google Scholar]
- 41.Soni BG, Philp AR, Foster RG, Knox BE. Novel retinal photoreceptors. Nature. 1998;394:27–28. doi: 10.1038/27794. [DOI] [PubMed] [Google Scholar]
- 42.Striedter GF. Auditory, electrosensory, and mechanosensory lateral line pathways through the forebrain in channel catfishes. J Comp Neurol. 1991;312:311–331. doi: 10.1002/cne.903120213. [DOI] [PubMed] [Google Scholar]
- 43.Tabata M, Minh-Nyo M, Oguri M. Involvement of retinal and extraretinal photoreceptors in the mediation of nocturnal locomotor activity rhythms in the catfish, Silurus asotus. Exp Biol. 1988;47:219–225. [PubMed] [Google Scholar]
- 44.Tabata M, Minh-Nyo M, Oguri M. Thresholds of retinal and extraretinal photoreceptors measured by photobehavioral response in catfish, Silurus asotus. J Comp Physiol. 1989;A164:797–803. [Google Scholar]
- 45.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umino O, Lee Y, Dowling JE. Effects of light stimuli on the release of dopamine from interplexiform cells in the white perch retina. Vis Neurosci. 1991;7:451–458. doi: 10.1017/s0952523800009743. [DOI] [PubMed] [Google Scholar]
- 47.van Veen T, Hartwig HG, Müller K. Light-dependent motor activity and photonegative behavior in the eel (Anguilla anguilla L. ). J Comp Physiol. 1976;111:209–219. [Google Scholar]
- 48.von Frisch K. Beiträge zur Physiologie der Pigmentzellen in der Fischhaut. Pflügers Arch. 1911;138:319–387. [Google Scholar]
- 49.Wada Y, Okano T, Adachi A, Ebihara S, Fukada Y. Identification of rhodopsin in the pigeon deep brain. FEBS Lett. 1998;424:53–56. doi: 10.1016/s0014-5793(98)00138-0. [DOI] [PubMed] [Google Scholar]
- 50.Weiss ER, Osawa S, Shi W, Dickerson CD. Effects of carboxyl-terminal truncation on the stability and G protein-coupling activity of bovine rhodopsin. Biochemistry. 1994;33:7587–7593. doi: 10.1021/bi00190a011. [DOI] [PubMed] [Google Scholar]
- 51.Yoshikawa T, Oishi T. Extraretinal photoreception and circadian systems in nonmammalian vertebrates. Comp Biochem Physiol. 1998;119B:65–72. [Google Scholar]
- 52.Yoshikawa T, Okano T, Oishi T, Fukada Y. A deep brain photoreceptive molecule in the toad hypothalamus. FEBS Lett. 1998;424:69–72. doi: 10.1016/s0014-5793(98)00139-2. [DOI] [PubMed] [Google Scholar]