Abstract
Quakingviable (qkv) is a well known dysmyelination mutation. Recently, the genetic lesion of qkv has been defined as a deletion 5′ to the qkI gene, which results in the severe reduction of the qkI-encoded QKI RNA-binding proteins in myelin-producing cells. However, no comprehensive model has been proposed regarding how the lack of QKI leads to dysmyelination. We hypothesized that QKI binds to myelin protein mRNAs, and the lack of QKI causes posttranscriptional misregulation, which in turn leads to the loss of the corresponding myelin proteins. To test this hypothesis, we developed an RNase protection assay to directly measure the mRNA isoforms encoding the myelin basic proteins (MBPs) in the brain. Our result suggested that isoform-preferential destabilization of MBP mRNAs in the cytoplasm was responsible for the reduced MBPs in the qkv/qkv brain during early myelination. In addition, we detected markedly reduced MBP mRNAs in the qkv/qkv myelin fraction with concomitant accumulation of MBP mRNAs associated with membrane-free polyribosomes. Presumably, the impaired localization of MBP mRNAs to the myelin membrane may cause insufficient incorporation of the newly synthesized MBPs into the myelin sheath. Finally, we observed interactions between QKI and MBP mRNAs, and removing MBP 3′UTR significantly reduced QKI-binding. Taken together, these observations suggest that misregulation at multiple posttranscriptional steps is responsible for the severe reduction of MBPs in qkvdysmyelination, presumably because of the lack of interactions between MBP mRNAs and the QKI RNA-binding proteins.
Keywords: myelination, myelin basic protein, mRNA degradation and localization, quakingviable, signal transduction activators of RNA, posttranscriptional regulation
Quakingviable (qkv) is an autosomal recessive mutation resulting in dysmyelination in the CNS and PNS (Hardy, 1998;Hogan and Greenfield, 1984). Recently, qkvhas been defined as a deletion 5′ to the qkI gene (Ebersole et al., 1996), leading to diminished expression of the selective RNA-binding protein QKI in myelin-producing cells (Hardy et al., 1996). QKI harbors amino acid domains characteristic of RNA-binding and interaction with Src homology 3 (SH3)-containing signaling molecules, therefore belongs to a fast-growing family denoted as signal transduction activators of RNA (STAR) (Vernet and Artzt, 1997). Several QKI isoforms, QKI-5, QKIK-6, and QKI-7, are derived from alternative splicing of the 3′ coding exon (Ebersole et al., 1996; Kondo et al., 1999). It is believed that the cytoplasmic QKI-6 and QKI-7 are the major isoforms involved in myelinogenesis (Ebersole et al., 1996; Cox et al., 1999). Indeed, QKI-6 and QKI-7 are lost in all the myelin-producing cells in the qkv/qkv mice (Hardy et al., 1996).
Presumably because of the lack of QKIs, qkv results in low expression of many myelin structural genes (for review, see Campagnoni, 1988; Campagnoni and Macklin, 1988; Hardy, 1998). The reduced myelin gene expression is unlikely because of death of oligodendrocytes (Friedrich, 1975). However, mechanisms regarding how the lack of QKIs leads to diminished myelin protein expression remain unclear. One of the most severely reduced myelin proteins in the qkv/qkv brain is the myelin basic protein (MBP), a major myelin component essential for forming compact myelin (for review, see Matthieu, 1993). In mice, four major MBP isoforms of 21.5, 18.5, 17.2, and 14 kDa are produced via alternate splicing of the primary MBP transcript (de Ferra et al., 1985; Newman et al., 1987). The amount of MBP in the qkv/qkv brain is only ∼5–20% of the normal level (Delassalle et al., 1981; Jacque et al., 1983), with the 14 kDa isoform most severely reduced (Carnow et al., 1984).
The selective RNA-binding feature of QKIs leads us to question whether QKIs interact with myelin protein mRNAs to regulate their homeostasis and whether qkv dysmyelination is a result of posttranscriptional misregulation because of the loss of QKIs. MBP is a good model system to test this hypothesis considering its important roles and sophisticated regulation in normal myelination, as well as its severe reduction in qkvdysmyelination (Carnow et al., 1984; Sorg et al., 1986, 1987). We observed significant isoform-preferential reduction of MBP mRNAs in the cytoplasm of qkv/qkvoligodendrocytes during early myelination without abnormalities in MBP transcription. In addition, the level of MBP mRNA was dramatically reduced in the qkv/qkvmyelin, presumably because of impaired localization of MBP mRNAs to the myelin membrane. Furthermore, we found that QKI interacts with MBP mRNAs. The MBP 3′UTR, which plays important roles in stabilization and localization of MBP mRNAs to the myelin sheath, is critical for such interaction. Thus, we propose that the interactions between MBP mRNAs and QKI may control the normal cellular fate of MBP mRNAs.
MATERIALS AND METHODS
Animals and RNA preparation. The qkv colony was purchased from Jackson Laboratory (Bar Harbor, ME). qkv/wt and qkv/qkvlittermates were produced by breeding qkv/qkvfemales with qkv/wt males. qkv/qkvanimals were distinguished from the qkv/wt littermates by the vigorous tremors developed at approximately postnatal day 10 (P10). Only male animals were used in our studies. At various ages, animals were killed by cervical dislocation. Cerebral cortices, brainstems, and cerebella were dissected immediately followed by total RNA extraction using Trizol according to manufacturer's protocol (Life Technologies, Gaithersburg, MD). The quantity of RNA from each sample was determined by OD260 reading and further confirmed by ethidium bromide-stained agarose gel electrophoresis.
Immunoblot analysis and antibodies. Brainstem was lysed by sonication of the tissue in 1× laemmli buffer containing 8m urea (Feng et al., 1995). The quantity of total protein in each sample was estimated by Bradford assay following the manufacturer's instructions (Bio-Rad, Hercules, CA). An equal amount of protein from each sample was subjected for SDS-PAGE analysis. The anti-MBP antibody was purchased from Chemicon (Temecula, CA), and the anti-lactate dehydrogenase (LDH) antibody was purchased from Sigma (St. Louis, MO). The anti-SP1 antibody was a gift from Dr. Xiaojiang Li.
RNase protection assay probes and analysis. A 377 bp MBP cDNA fragment containing exon 2–6 was generated by RT-PCR with primers of 5′-CAAGGTACCCCTGGCTAAGG and 5′-CCCAGCTTAAAGATTTTGG using total mouse brain RNA as templates. This MBP cDNA fragment was first cloned into TA-Cloning Vector (Invitrogen, Carlsbad, CA), and subcloned into Bluescript II KS (Stratagene, La Jolla, CA) to generate BSKSMBP. The sequence of the MBP cDNA was confirmed to be 100% identical, as previously published (de Ferra et al., 1985). Antisense strand riboprobe was generated by in vitrotranscription using T7 polymerase (Stratagene) in the presence of32P-UTP (Amersham, Arlington Heights, IL) using XhoI linearized BSKSMBP as DNA template. The glyceraldehyde phosphate dehydrogenase (GAPDH) cDNA construct was a gift generously provided by Dr. Sharie Ball (University of Alabama, Birmingham, AL). The GAPDH riboprobe was derived from in vitro transcription by T7 polymerase in the presence of32P-UTP using Sau3A linearized plasmid as DNA template. We hybridized 5 × 105cpm for each probe to RNA samples, as indicated in the corresponding figure legends, followed by RNase protection assay (RPA) analysis using previously published procedures (Feng et al., 1995). The antisense riboprobe of U3 snRNA was generated byin vitro transcription using T3 polymerase and used for Northern hybridization to demonstrate the nuclear integrity.
Nuclear run-on assay. Nuclei were isolated from the brainstem following the procedures described by Macklin et al. (1991). Freshly isolated nuclei were subjected to nuclear run-on assay immediately, and the 32P-labeled RNA was isolated by Trizol extraction (Life Technologies). The labeled RNA was hybridized to Zetaprobe membrane containing various cDNA constructs immobilized by slot blot following manufacturer's protocol (Bio-Rad). Hybridization and washes of the slot blot membrane were performed as described (Feng et al., 1995), followed by PhosphorImager analysis. The MBP construct contained the full-length cDNA encoding the 14 kDa MBP (Campagnoni et al., 1987). The proteolipid protein (PLP) cDNA was a 358 bp RT-PCR product derived by primers of 5′-GCAAGGGCCTGAGCGCAACG and 5′-GCAGATGGACAGAAGGTTGGAG. The GAPDH cDNA and the γ-actin cDNA was a full-length clone described previously (Feng et al., 1995). For nuclear RNA analysis, nuclei were isolated from brainstems followed by Trizole extraction to isolate total nuclear RNA for RPA as well as total nuclear protein to ensure the purity and integrity of the isolated nuclei (Life Technologies).
Sucrose gradient fractionation. Myelin membranes were isolated by a discontinuous sucrose gradient using the procedure described by Colman et al. (1982). A schematic representation of this procedure is illustrated in Figure 7A. Membrane-free polyribosomes were obtained as a pellet at the bottom of the centrifugation tube. Total RNA was isolated by Trizol extraction from the myelin fraction and the polyribosome pellet following the manufacturer's protocol (Life Technologies). Parallel 15–45% (w/v) linear sucrose gradients were used to fractionate cytoplasmic extracts derived from qkv/wt and qkv/qkvbrainstems to separate polyribosomes from nontranslating components, as described by Feng et al. (1997). To confirm the association of MBP mRNAs with polyribosomes, tissue lysates were prepared in the presence of 20 mm EDTA to dissociate ribosomes into subunits and release the mRNAs fractionated through sucrose gradient containing 1 mm EDTA.
Fig. 7.
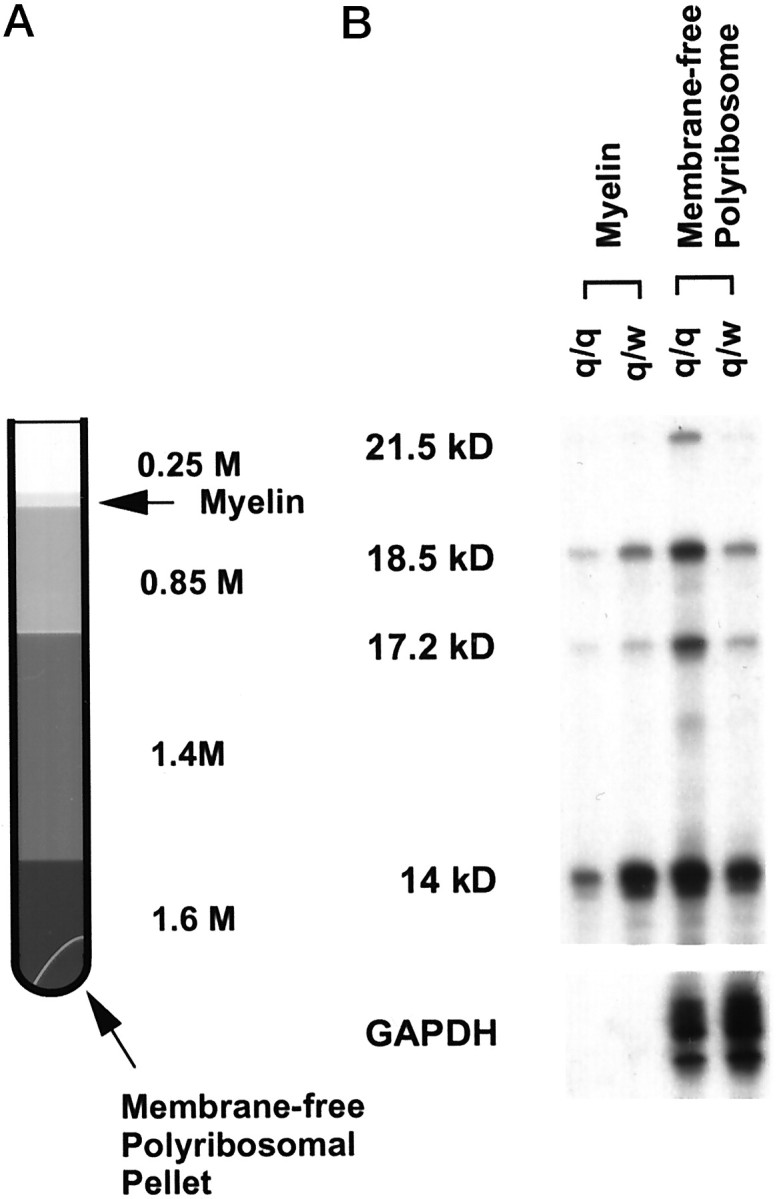
Reduced MBP mRNAs in adult qkv/qkv myelin. A,Schematic representation for isolating myelin membrane and membrane-free polyribosomes (indicated by arrows) by discontinuous sucrose gradient. B, Reduced MBP mRNAs in the myelin fraction with concomitant accumulation of MBP mRNAs in the membrane-free polyribosomes derived from qkv/qkv adult (P65). qkv/qkv and qkv/wt littermates were used for this experiment. Two brainstems were used in each fractionation. Two micrograms of RNA from each fraction was used for the RPA analysis. Note that the total MBP mRNA level at this age in qkv/qkv brain is ∼80% of that in the qkv/wt littermate (Fig. 2). Similar results were observed from three repeated experiments.
RNA-binding assay. QKI-7 cDNA was obtained by RT-PCR using total mouse brain RNA as template with primers described by Chen et al. (1997). The PCR product was cloned into TA-Cloning Vector (Invitrogen), and sequence analysis was conducted to confirm 100% identity to what was published. 35S-methionine-labeled QKI-7 was generated by TNT reaction containing T7 RNA polymerase (Promega, Madison, WI) before being exposed to biotinylated RNA synthesized by in vitro transcription (Stratagene). The Superscript brain cDNA library was purchased from Life Technologies. The M14, M18.5 and M21.5 cDNA constructs with and without full-length 3′UTR were generously provided by Dr. Campagnoni (University of California at Los Angeles, Los Angeles, CA). Plasmids were linearized by BamHI as transcription templates. Biotinylated RNAs were derived by in vitro transcription using T7 polymerase in the presence of biotin-UTP as described (Ashley et al., 1993). RNA binding was performed, and the bound QKI-7 was captured by streptavidin-conjugated Dynabeads (Ashley et al., 1993). The captured QKI-7 was fractionated on SDS-PAGE followed by PhosphorImager analysis or directly subjected to scintillation counting. For the competition experiments, various amounts of unlabeled transcripts of either M14 or sense strand β-globin were mixed with the biotinylated M14, as indicated in the corresponding figure legends before subjected to capture 35S-QKI as described above.
RESULTS
Isoform-preferential reduction of MBP mRNAs in the qkv/qkv brain during early myelination
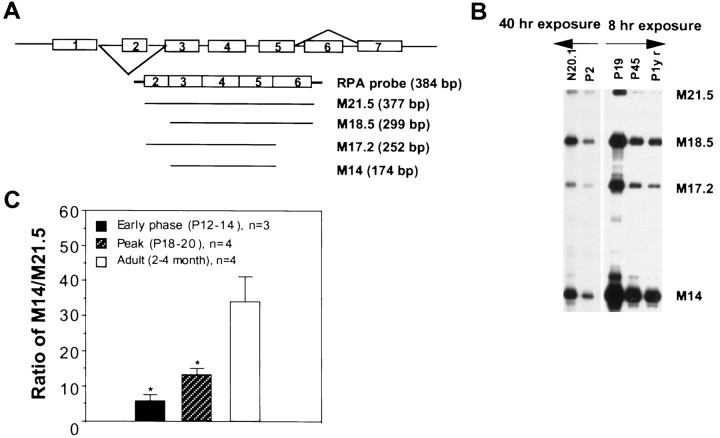
To directly evaluate the level of individual MBP mRNA isoforms, we developed an RPA, with which four major MBP mRNA isoforms (M21.5, M18.5, M17.2, and M14, encoding for the 21.5, 18.5, 17.2, and 14 kDa MBP) can be quantitatively measured simultaneously on a denaturing acrylamide gel (schematically represented in Fig.1A). Figure1B shows the developmental profile of MBP mRNA isoforms in normal mouse brainstem, with all four major MBP mRNA isoforms detected as early as P2. In addition, these MBP mRNA isoforms were also detected in the immortalized oligodendrocyte cell line N20.1. The highest level for each isoform was detected at the peak of myelination (P18–P20). The amount of each MBP mRNA gradually decreased toward adulthood. For all developmental stages examined, the relative quantity of MBP mRNA isoforms follows the order of M14 > M18.5 > M17.5 > M21.5. Interestingly, the percentage of M14 in the total MBP mRNA pool continuously increased during development, whereas the percentage of M21.5 and M17.2 decreased reciprocally (Table1). Such changes of MBP mRNA isoforms at least partly contribute to the accumulation of the 14 kDa MBP and the reduction of the 21.5 kDa MBP in the mature myelin, as described by previous reports (Barbarese et al., 1978; Campagnoni et al., 1978). A significant increase in the ratio of M14/M21.5 was observed when comparing the immature myelin (P12 and P20) to the adult myelin (2–4 month) (Fig. 1C), which can be used as an index for myelin maturation.
Fig. 1.
MBP mRNA isoforms detected by the RPA analysis.A, Schematic representation of MBP mRNA isoforms in relation to the RPA probe. MBP mRNA isoforms encoding MBPs of 21.5, 18.5, 17.2, and 14 kDa are denoted as M21.5, M18.5, M17.2, and M14, respectively, with the predicted size for protected fragments indicated at the right. B, The RPA pattern of major MBP mRNA isoforms during development of normal C57/B6 mice and in an immortalized oligodendrocyte cell line N20.1. Five micrograms of total RNA was used in each RPA reaction. C, Increased ratio of M14/M21.5 during normal development. M14/M21.5 is significantly increased in adult animals (p < 0.001, one-way ANOVA), with SE indicated for each developmental point. *p < 0.05 in comparison to the adult.
Table 1.
Percentage of MBP mRNA isoforms during development
| n | % M21.5 | % M18.5 | % M17.2 | % M14 | |
|---|---|---|---|---|---|
| Early phase | 3 | 10.00 ± 4.65* | 22.80 ± 2.70 | 17.53 ± 2.51* | 49.67 ± 9.05* |
| Peak phase | 4 | 5.90 ± 1.48* | 22.33 ± 3.59 | 13.93 ± 3.34* | 57.73 ± 8.25* |
| Adult | 5 | 2.07 ± 0.75 | 17.94 ± 4.98 | 8.06 ± 1.69 | 71.80 ± 4.19 |
The percentage of each MBP mRNA isoform in the total MBP mRNA pool was calculated based on PhosphorImager reading of the corresponding isoform in the RPA analysis. Early phase, P12–P14; Peak phase, P18–P20; Adult, 2–4 month. n indicates number of experiments. Statistic analysis was conducted using one-way ANOVA.
indicates p < 0.05 compared with adult group.
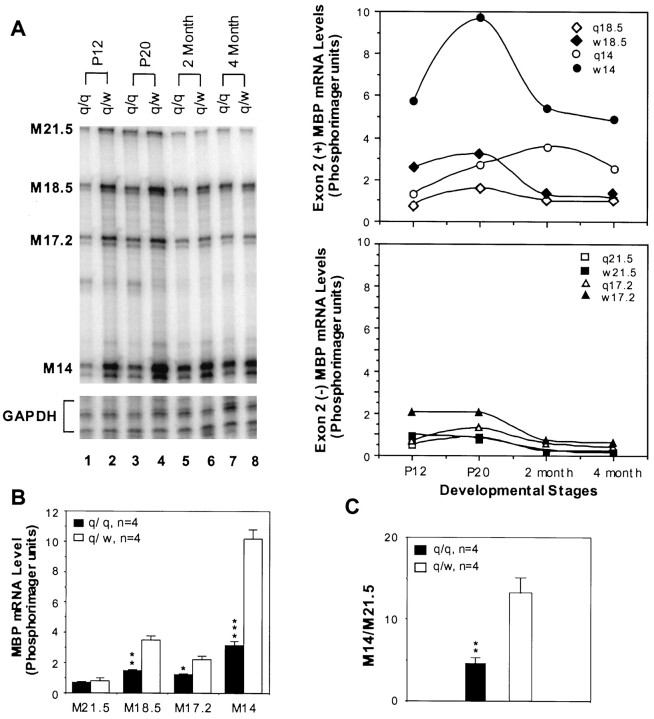
The RPA also allowed us to directly estimate and compare the quantity of each MBP mRNA isoform in the qkv/qkv brain with that in the qkv/wt littermate during development. Because qkv/wt mice do not display dysmyelination phenotype, such a comparison should yield a close estimate for the minimal reduction of MBP mRNAs that results in the diminished MBPs in qkv dysmyelination. Figure 2A is a representative RPA gel for the developmental profile of MBP mRNAs in the brainstem derived from qkv/qkv and qkv/wt littermates. The profile of qkv/wt MBP mRNAs closely mimics that in wild-type animals. In the qkv/qkvbrain, all four major MBP mRNA isoforms were expressed throughout development. However, the amount of MBP mRNAs was markedly reduced at P12–P20 in comparison to that in the qkv/wt littermate (Fig.2A). In general, the reduction of MBP mRNA isoforms containing exon 2 (M18.5 and M14) was more severely reduced in comparison to the ones lacking exon 2 (M21.5 and M17.2). PhosphorImager analysis indicated that the normal peak of myelination ∼P20 almost disappeared in the qkv/qkvanimals. At this age, the abundant MBP mRNA isoforms, including M18.5, M17.2, and M14 were significantly reduced in the qkv/qkvbrainstem (Fig. 2B). Among the MBP mRNA isoforms, M14 was the most severely affected, whereas M21.5 was not changed. As an end result, the ratio of M14 to M21.5 at P20 was significantly reduced in the qkv/qkvbrainstem (Fig. 2C). Toward later development, MBP mRNAs continue to accumulate in the qkv/qkv brain, with most of the isoforms expressed at a comparable level in qkv/qkv and qkv/wt brain. M14 still remains reduced in the qkv/qkvbrain, although the scale of reduction was much less as compared to that in early myelination. A similar result of isoform-preferential reduction of MBP mRNAs during development was observed in cerebral cortices and cerebella of the qkv/qkv mice (data not shown).
Fig. 2.
Isoform-preferential reduction of MBP mRNAs in the qkv/qkv brain during development.A, Developmental profile of MBP mRNA expression in the brainstem of qkv/qkv and qkv/wt littermates. Five micrograms of total RNA was used for each reaction. The left panel shows the phosphorimage of the RPA gel. GAPDH was used as a loading control. Note the severe reduction of MBP mRNAs at P12 and P20, with M14 most severely reduced (lanes 1, 3). The rightpanel illustrates the plot of PhosphorImager reading of exon 2 (+) and exon-2 (−) MBP transcripts normalized to GAPDH. Note the exon 2 (+) transcripts (M14 and M18.5) are more severely affected in the qkv/qkv brainstem, and the most severe reduction of M14 is at P20. B, Comparing the quantity of MBP mRNA isoforms in qkv/qkv and qkv/wt brainstem at the peak of myelination. Brainstem total RNA was isolated from paired qkv/qkv and qkv/wt littermates at P20 (n = 4). Five micrograms of total RNA was used in each reaction. The RPA signal of each MBP mRNA isoform was quantitatively measured by a PhosphorImager and normalized to GAPDH. SE for each group is indicated. *p < 0.05; **p < 0.01; ***p < 0.001 for comparison between littermates (paired t test). C, Preferential reduction of M14 in the brainstem at the peak of myelination as represented by the ratio of M14 over M21.5 (n = 4; **p < 0.01 based on paired t test). SE for each group is indicated.
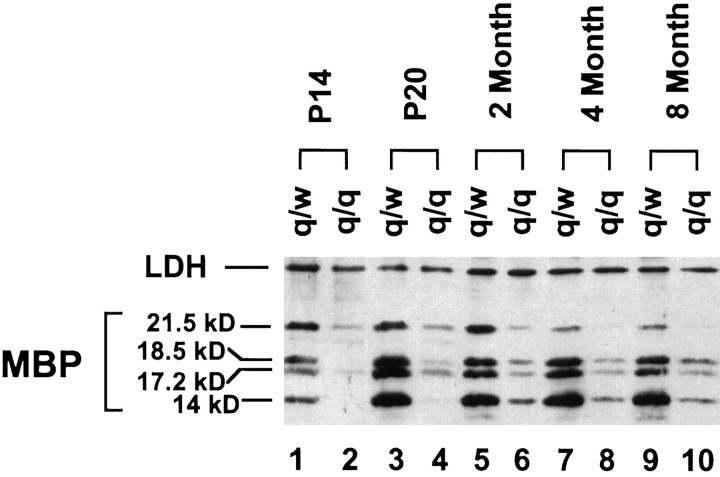
Using a monoclonal antibody that recognizes a common epitope in all MBP protein isoforms, we analyzed the developmental profile of MBP proteins in the brainstems derived from qkv/wt and qkv/qkvlittermates by high-resolution SDS-PAGE immunoblot (Fig.3). In qkv/wt animals, the 14 kDa MBP continuously accumulated over the rest of the MBP isoforms with the 21.5 kDa MBP decreased concomitantly by age. In the qkv/qkvbrain, a severe reduction of all MBP isoforms was observed, with the 14 kDa MBP most severely reduced and the 21.5 kDa MBP least severely reduced, reminiscent of that for the qkv/qkv MBP mRNA isoforms in early myelination (Fig. 2A). Although the MBP protein level was gradually increased in the qkv/qkv brain toward adulthood, the reduction of MBP proteins in the qkv/qkv brain was far more severe at all developmental stages examined as compared to that at the mRNA level. Thus, deficits in addition to the reduction of MBP mRNAs must exist that contribute to the loss of MBP in the qkv/qkvmyelin especially at the adult stage (discussed in later sections).
Fig. 3.
Isoform-preferential reduction of MBPs in the qkv/qkv brain during development. Whole-cell lysates of brainstems prepared from qkv/qkv and qkv/wt littermates at various ages were subjected to SDS-PAGE immunoblot analysis. The blot was sequentially probed by anti-MBP and anti-LDH antibody, with the housekeeping protein LDH as a loading control. The age and the genotype of each animal are depicted on top of the corresponding lanes, and protein signals detected are indicated on the left. Note the more severe reduction of the 14 kDa MBP at P14 and P20 (lanes 2, 4). Similar results were observed in three repeated experiments.
The reduction of MBP mRNAs appears to result from cytoplasmic destabilization
It is important to clarify whether the reduction of MBP mRNAs during early myelinogenesis is attributable to transcriptional or posttranscriptional deficits. We used a nuclear run-on assay to directly estimate the transcription level in the brainstem derived from qkv/qkv and qkv/wt mice. As shown in Figure4, transcription of MBP was nearly identical in the qkv/qkv and qkv/wt brainstem at the peak of myelination, despite the significant difference in the amount of MBP mRNAs between qkv/qkv and qkv/wt littermates (Fig.2B). This experiment clearly demonstrated that posttranscriptional destabilization is the primary deficit for the reduction of MBP mRNAs during early myelinogenesis in the qkv/qkvbrain. In addition, normal transcription of the PLP gene was also detected in the qkv/qkv brain despite the marked reduction of PLP mRNAs (Sorg et al., 1986, 1987) (Feng et al., unpublished results). These observations support our hypothesis that the low expression of myelin protein mRNAs in qkv dysmyelination is attributable to posttranscriptional destabilization.
Fig. 4.
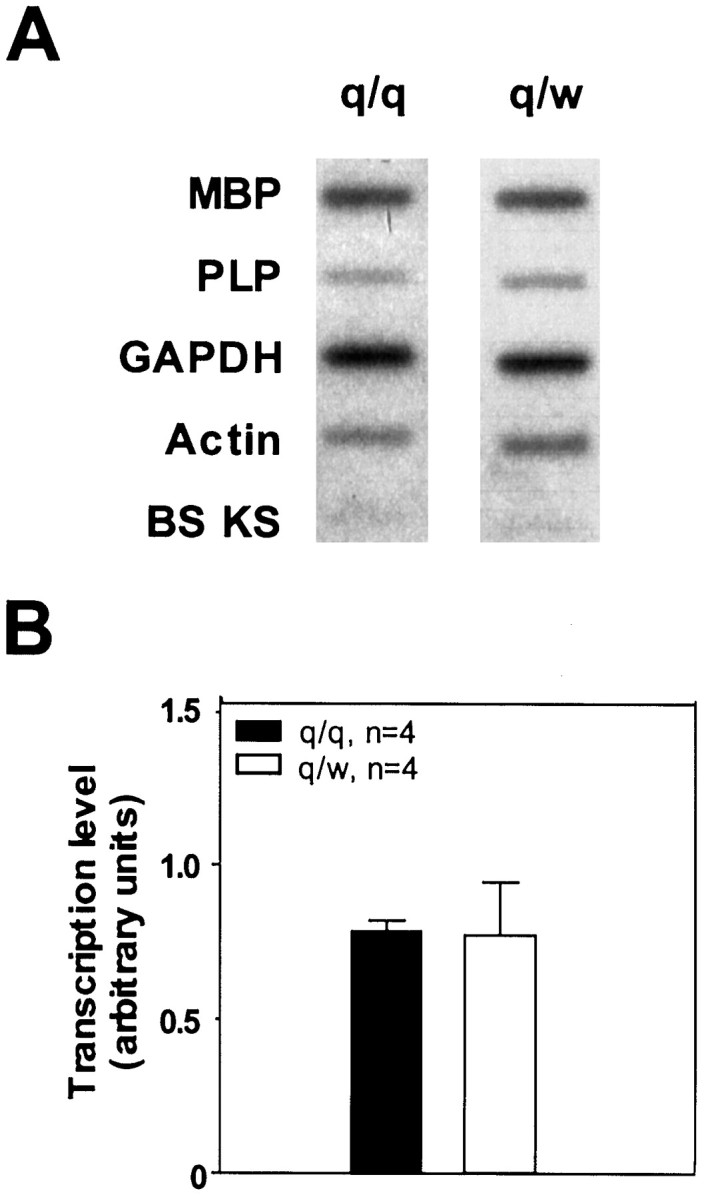
Comparable MBP transcription level at P17 in qkv/qkv and qkv/wt brain estimated by the nuclear run-on assay.A, Representative phosphorimage of a nuclear run-on experiment. Plasmids containing various cDNA or empty Bluescript vector (10 μg each), as indicated on the left, were immobilized on the membrane. B, Transcription level of MBP in qkv/qkv and qkv/wt littermates at P17–P18 represented by PhosphorImager reading of MBP signal normalized to that of GAPDH (n = 4). SE is indicated for each group.
To further explore whether destabilization of MBP mRNAs may occur in the nucleus or in the cytoplasm of qkv/qkvoligodendrocytes, we measured the MBP mRNA isoforms in the nuclei and in the total RNA pool in qkv/qkv and qkv/wt brain during early myelination. To minimize possible variations between individual animals, the nuclear RNA was prepared from one hemisphere, and the total RNA was isolated from the other hemisphere derived from the same animal. As shown in Figure 5A, comparable levels of MBP mRNA isoforms were detected in qkv/qkv and qkv/wt nuclei. In contrast, all the MBP mRNA isoforms were reduced in the total RNA from the qkv/qkvbrain, with M14 more severely affected than the others. A similar result was observed in the brainstem (data not shown). Further analysis indicated the absence of the cytoplasmic protein LDH in the nuclear lysates by SDS-PAGE immunoblot (Fig. 5B), suggesting negligible contamination of cytoplasmic component in the isolated nuclei. In addition, nuclear snRNA U3 was predominantly retained in the isolated nuclei (Fig. 5B), demonstrating the integrity of the isolated nuclei (Fig. 5C). Taken together, the isoform-preferential reduction of MBP mRNAs in the qkv/qkv brain is most likely attributable to accelerated degradation in the cytoplasm of oligodendrocytes.
Fig. 5.
Normal nuclear MBP mRNAs with severe reduction of MBP mRNAs in the total RNA pool in the qkv/qkv brain. A pair of qkv/qkv and qkv/wt littermates at P14 was used for this experiment. The nuclear RNA and nuclear protein were isolated from the nuclei derived from one hemisphere, whereas the total RNA and protein were isolated from the other. A, Normal nuclear MBP transcripts in the qkv/qkv brain. Five micrograms of RNA from each sample was used for the RPA reaction. Similar results were observed in repeated experiments as well as in brainstem derived from qkv/qkvand qkv/wt littermates. B, The isolated nuclei do not contain detectable contamination of cytoplasmic protein. SDS-PAGE immunoblot was performed on nuclear protein and total protein using antibodies against the cytoplasmic protein LDH and the transcription factor SP1. Note LDH is not detected in the nuclear preparation. C, snRNA U3 is retained in the isolated nuclei. Northern hybridization analysis was performed on nuclear and total RNA using antisense riboprobe derived from U3 cDNA. Note the enriched signal in the nuclear RNA pool in comparison to the total RNA pool.
Normal association of qkv/qkv MBP mRNAs with translating polyribosomes
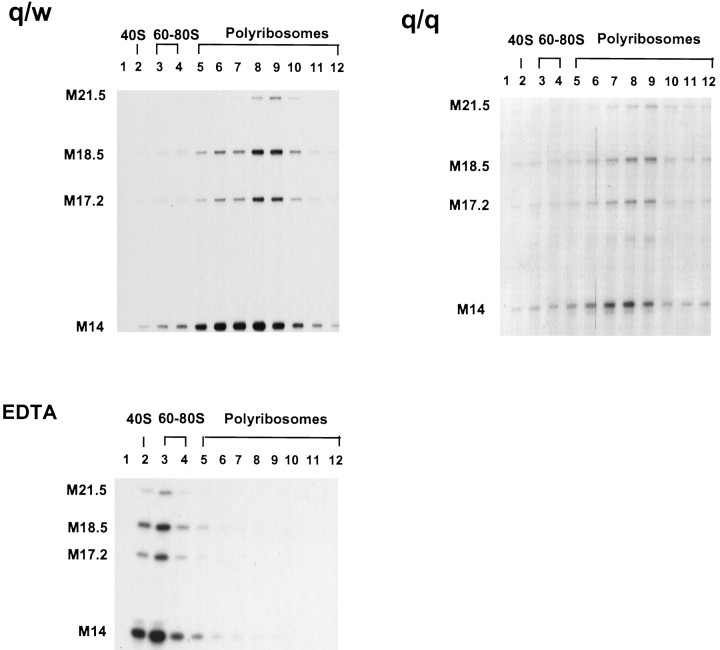
Because the reduction of MBP proteins is more severe than that of MBP mRNAs, one needs to test whether MBP mRNAs are translated abnormally in the qkv/qkvbrain, especially considering that changes in translation activity are often coupled with mRNA stability (Oliveira and McCarthy, 1995; Linz et al., 1997). Thus, we compared the polyribosome profile of MBP mRNA isoforms in the qkv/qkv and qkv/wt brainstem at the peak of myelination where MBP mRNAs were severely reduced. In qkv/wt brain, all MBP mRNA isoforms were found in the fractions containing translating polyribosomes, with negligible amount of MBP mRNA detected in the nontranslating pool (Fig.6). An almost identical translation profile for MBP mRNA isoforms in qkv/qkv brain was observed in comparison to what was derived from the qkv/wt littermate, although the amount of MBP mRNAs detected in the qkv/qkvgradient was significantly lower. The predominant distribution of MBP mRNAs in the polyribosome pool represents the association of MBP mRNAs with translating polyribosomes, because dissociation of polyribosomes by EDTA treatment completely shifted MBP mRNAs to the top of the gradient. This result suggested that MBP mRNAs can be actively translated in the qkv/qkvoligodendrocytes, and degradation of MBP mRNAs most likely occurred before their engagement of translation.
Fig. 6.
Translation profile of MBP mRNAs in the qkv/wt and qkv/qkv littermate brainstem. Cytoplasmic extract was prepared from one P20 brainstem derived from each littermate and fractionated through parallel linear sucrose gradients (15–45% w/w) to separate polyribosomes and nontranslating ribosomal components, as depicted on top of the corresponding lanes. In addition, EDTA was used to dissociate polyribosomes into ribosomal subunits and to release the mRNAs. Twelve fractions were obtained from each gradient followed by RPA analysis.
Reduced localization of MBP mRNAs to the adult qkv/qkv myelin membrane
In the adult, MBP mRNAs in the qkv/qkvbrainstem were ∼80% of the level detected in the qkv/wt littermate control (Fig.2A). However, the quantity of MBP proteins was still <10% of normal level (Fig. 3). The normal association of MBP mRNAs with translating polyribosomes (Fig. 6) and the rapid incorporation of radioactive-labeled amino acids into MBPs in the qkv/qkv brain (Brostoff et al., 1977) suggested efficient synthesis of MBP proteins. Thus, the newly synthesized MBP proteins must be degraded in the qkv/qkvbrain. One hypothesis is that the newly synthesized MBPs are degraded because of the inefficient incorporation into the myelin sheath. Because deposition of MBP mRNAs has been proposed as an important means for incorporation of MBP into the myelin membrane (Greenfield et al., 1977; Colman et al., 1982), we examined whether inefficient MBP mRNA localization to the myelin sheath may occur in qkv/qkvbrain.
RNA was isolated from purified myelin membrane derived from adult qkv/qkv and qkv/wt brainstems (∼2 months of age) as illustrated in Figure 7A. As shown in Figure 7B, MBP mRNA isoforms were clearly detected in the myelin fraction. M14 is the major isoform in the myelin fraction, whereas M21.5 was detected only after prolonged exposure. The purity of isolated myelin in this experiment was demonstrated by the absence of the GAPDH mRNA or the PLP mRNA (data not shown) that are known to be restricted in the cell body (Verdi et al., 1989). Interestingly, the quantity of MBP mRNAs, especially M14, in the qkv/qkvmyelin fraction was markedly reduced (Fig. 7B, comparelane 1, lane 2), with concomitant accumulation of MBP mRNAs in the membrane-free polyribosomes (Fig. 7B, comparelane 3, lane 4). The amount of GAPDH mRNA detected in the polyribosomal fractions further confirmed the increased MBP mRNAs in the qkv/qkvpolyribosomes presumably derived from the membrane-free cytoplasm from the cell body. This result provides in vivo evidence for mislocalization of MBP mRNAs in the qkv/qkvoligodendrocytes, which may lead to inefficient incorporation of MBP into the myelin sheath and the subsequent degradation of MBP protein in the qkv/qkvoligodendrocytes.
Interaction of QKI with MBP mRNAs
The above observations suggest that abnormalities in mRNA metabolism and subcellular localization contribute to the severe reduction of MBP proteins in qkv/qkvoligodendrocytes in which QKI proteins are lost. This raises the intriguing possibility that QKI may interact with MBP mRNAs, and such an interaction may be required for the normal cellular fate of MBP mRNAs. A well established RNA-binding assay that has been used to define RNA-target specificity for a number of RNA-binding proteins was used to test whether QKI may interact with MBP mRNAs.35S-labeled QKI-7 generated by a transcription–translation-coupled reaction was mixed with biotin-labeled mRNAs derived from various MBP cDNA constructs in the presence of excess amount of yeast tRNA to prevent nonspecific binding. The integrity and quantity of the various MBP transcripts were shown in Figure 8A. The protein–RNA complexes were captured by streptavidin-magnetic beads followed by SDS-PAGE, and the amount of captured QKI in each sample was quantitatively measured by a PhosphorImager.
Fig. 8.
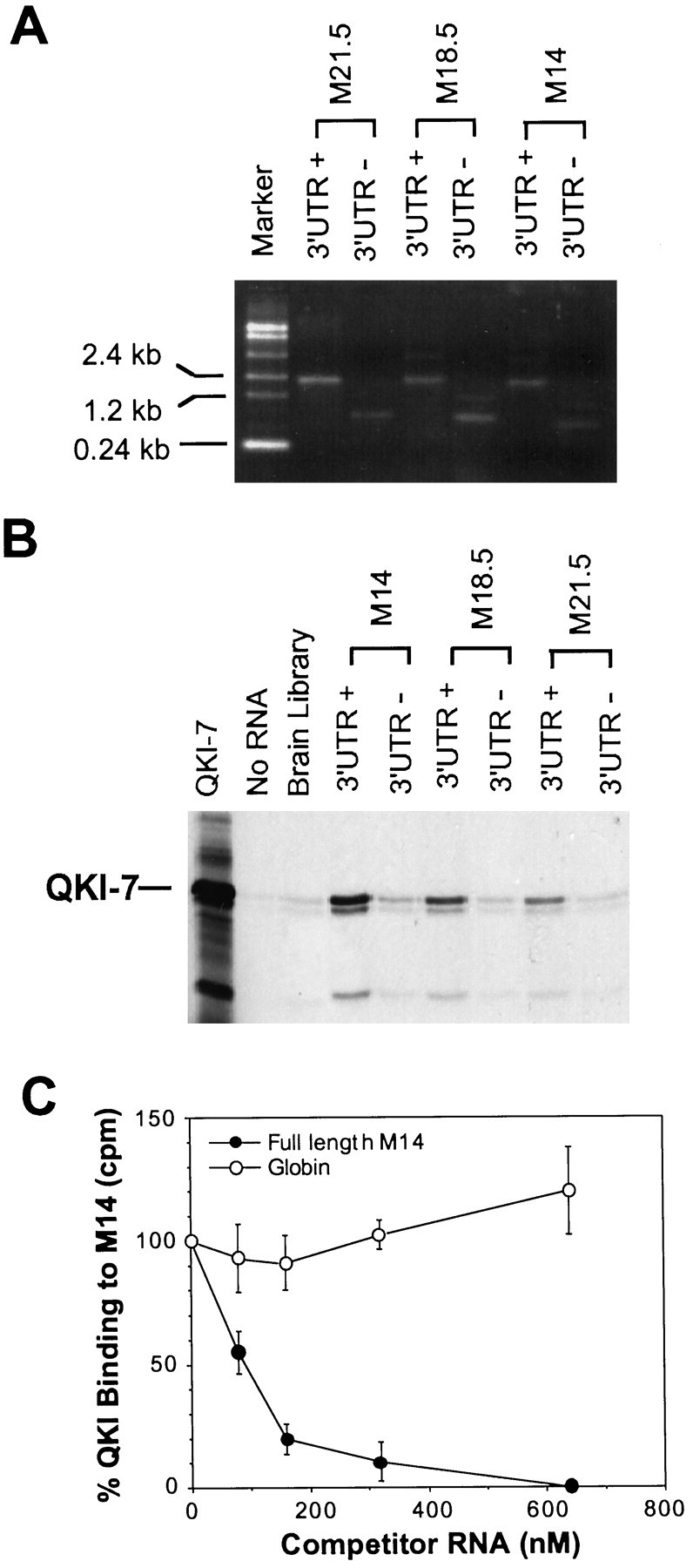
Association of MBP mRNAs with QKI-7.A, Biotinylated MBP mRNAs, with or without the full-length 3′UTR (+ and −) were derived by in vitrotranscription. The size and quantity of each MBP mRNA is visualized on ethidium bromide-stained agarose gel. B, Selective interaction of MBP mRNAs with 35S-Labeled QKI-7 derived byin vitro translation. We mixed 0.25 pmol of RNA with 3 μl of 35S-QKI-7 in each RNA-binding reaction. The RNA–protein complex was captured by streptavidin-conjugated magnetic beads followed by SDS-PAGE analysis. The RNA used in each reaction is depicted on top of the corresponding lanes, and the captured QKI-7 is marked on the left. The minor low molecular weight QKI bands are translation products from internal translation initiation. Similar results were observed in three repeated experiments. C, Specific competition of QKI binding by M14 but not by β-globin transcript. Increasing amounts of competitors were included in the binding reaction containing 35S-QKI-7 and biotin-labeled M14 as described in B, and the concentration of competitor RNAs were depicted at the bottom of the graph. The captured QKI-7 in each reaction was subjected to scintillation counting (counts per minute). The amount of QKI-7 captured by biotin M14 with no competitor is defined as 100%. The SE for each competitor dose is indicated in the curve.
As shown in Figure 8B, QKI was indeed cocaptured by various isoforms of full-length MBP mRNA, including M14, M18.5, and M21.5. When total brain transcripts were used as RNA target, very low level of QKI-binding was detected, suggesting that QKI may selectively bind a small subclass of brain mRNAs, including MBP mRNAs. Interestingly, the QKI-binding activity among different MBP mRNA isoforms followed the order of M14 > M18.5 > M21.5, indicating a good correlation with the severity of reduction of the corresponding MBP mRNA isoforms in the qkv/qkvbrain. In addition, removing the MBP 3′UTR significantly reduced the QKI-binding activity for all the MBP mRNA isoforms tested, suggesting that the MBP 3′UTR is important for the interaction of MBP mRNAs with QKI. The selectivity for the interactions of QKI with MBP mRNAs is indicated by the fact that the cold M14, but not β-globin transcript, can efficiently compete with biotin-labeled M14 in binding QKI (Fig.8C).
DISCUSSION
Previous studies have suggested that the expression of myelin protein genes are regulated by both transcriptional and posttranscriptional mechanisms (Kumar et al., 1989; Macklin et al., 1991; Umemori et al., 1999). Our nuclear run-on assay detected normal transcription of MBP and PLP, despite the severe reduction of the corresponding mRNAs in the qkv/qkvbrain. This study provides the first direct evidence that the severe reduction of myelin structural proteins, including MBPs, in qkv dysmyelination results from posttranscriptional abnormalities. Furthermore, destabilization and mislocalization of MBP mRNAs, presumably because of the lack of interactions with the QKI RNA-binding proteins, appear to be responsible for the diminished MBPs in qkvdysmyelination.
Isoform-preferential expression of MBP is largely determined by the quantity of the corresponding mRNA isoforms
It is known for years that the 14 kDa MBP is preferentially accumulated during myelin maturation (for review, see Campagnoni, 1988;Campagnoni and Macklin, 1988). In addition, in qkv dysmyelination, the 14 kDa MBP is more severely reduced (Carnow et al., 1984). Extensive studies have been conducted to estimate MBP mRNA level based on either hybridization to mixed MBP transcripts in polysomal RNA or indirect measurement ofin vitro translation of polysomal MBP transcripts (for review, see Campagnoni, 1988; Campagnoni and Macklin, 1988). However, because of the lack of tools to directly analyze MBP mRNA isoforms, mechanisms leading to the isoform-preferential expression of MBP remained unclear. This becomes a critical issue in understanding the molecular regulation of myelin development as well as in elucidating how the lack of QKI RNA-binding proteins leads to impaired myelin structural protein expression. Our RPA analysis directly measures the quantity of individual MBP mRNA isoforms, indicating that the isoform-preferential expression of MBP during development in normal and the qkv/qkvmice is largely reflected by the level of MBP mRNA isoforms.
It is clear that the percentage of M14 was raised with a concomitant decline of M21.5 in the total MBP mRNA pool toward late development in normal and qkv/wt brain (Table 1, Figs.1C, 2). The trends for such isoform-specific change were observed at both the mRNA and the protein level (Figs.2A, Fig. 3). In the qkv/qkvbrain, the relative severity of reduction for MBP mRNA isoforms correlated with that for the corresponding MBP protein isoforms. Thus, the quantity of MBP mRNA isoforms is a critical determinant for the isoform-preferential changes of MBP proteins during normal myelin development and in qkv dysmyelination.
Cytoplasmic destabilization of MBP mRNAs in the qkv/qkv brain significantly contributes to the reduced MBP levels during early myelinogenesis
The level of all the abundant MBP mRNAs, M14, M18.5, and M17.2, was significantly reduced in the qkv/qkv brain up to the peak of myelination (Fig. 2). However, no change was detected in MBP transcription in the qkv/qkv brain (Fig. 4). In addition, transcription of PLP was also at normal level, despite the dramatic reduction of PLP mRNA in the qkv/qkv brain (Sorg et al., 1986, 1987). Thus, the severe reduction of myelin protein mRNAs in qkv dysmyelination, including MBP mRNAs, appears to result from posttranscriptional destabilization. Therefore, maintaining the stability of myelin protein mRNAs is an essential mechanism for myelination. In contrast to the severe reduction of the MBP mRNAs in the total RNA pool derived from the qkv/qkvbrain, the quantity of each MBP mRNA isoform in the qkv/qkvnuclei was close to normal level (Fig. 5). Thus, MBP mRNA destabilization in the qkv/qkv brain most likely occurs in the cytoplasm of oligodendrocytes. The normal distribution of MBP mRNAs in polyribosome profile in the qkv/qkv brain (Fig. 6) suggests that degradation of MBP mRNAs most likely occurs before their engagement in translation elongation.
Although QKIs have been speculated to function in regulating mRNA splicing (Vernet and Artzt, 1997), the preferential reduction of M14 is unlikely attributable to abnormal splicing against M14, because no accumulation of other MBP-splicing products can compensate the reduction of M14 quantitatively. Northern hybridization of total RNA to full-length MBP probe did not reveal detectable size difference of MBP transcripts when comparing qkv/qkv and qkv/wt brain (data not shown). The normal nuclear level and the severe reduction of M14 in the total RNA pool further suggests that cytoplasmic destabilization is responsible for the loss of M14. It is important to point out that qkv results in the complete loss of the cytoplasmic isoforms of QKI (QKI-6 and QKI-7) in all oligodendrocytes, whereas the nuclear QKI (QKI-5) is only reduced in a subclass of oligodendrocytes (Hardy et al., 1996). Thus, it is conceivable that the loss of cytoplasmic QKIs results in the cytoplasmic destabilization of MBP mRNAs. Interestingly, a minor MBP-specific RPA product was slightly increased in the qkv/qkv brain (Fig. 5). This RPA fragment may be a result of abnormal splicing caused by the reduced QKI-5 in the most severely affected oligodendrocytes.
Inefficient localization of MBP mRNAs to the myelin sheath may explain the failure of incorporation of MBP into qkv/qkv myelin
Although a similar isoform-preferential reduction was observed in the qkv/qkvbrain for the MBP mRNAs and the MBP proteins (Figs. 2, 3), the quantitative loss of MBP proteins is much more severe than that of the corresponding MBP mRNAs (Fig. 3). Thus, other deficits must exist in the qkv/qkvoligodendrocytes in addition to MBP mRNA destabilization. It has been proposed that inefficient incorporation of MBPs into the myelin sheath may cause degradation of MBPs in qkv/qkvoligodendrocytes (Brostoff et al., 1977; Greenfield et al., 1977;Hardy, 1998). Because localization of MBP mRNAs to the myelin sheath enables locally translated MBP to be directly inserted into myelin (Colman et al., 1982; Trapp et al., 1987; Brophy et al., 1993), a failure in targeting MBP mRNAs to the oligodendrocyte membrane may contribute to the loss of MBP in the qkv/qkvmyelin.
The quantitative RPA allowed us to detect marked reduction of MBP mRNAs in the purified myelin derived from adult qkv/qkv brain (Fig. 7B), whereas the total MBP mRNA is close to the level in the qkv/wt littermates (Fig.2A). This result extended the previous observation regarding the failure in translocation of MBP transcripts to the distal process of primary cultured qkv/qkvoligodendrocytes (Barbarese, 1991), demonstrating that impaired localization of MBP mRNAs to the qkv/qkvmyelin sheath occurred in vivo, perhaps more prominent in late development. The concomitant accumulation of MBP mRNAs in the membrane-free polyribosomes in the qkv/qkv brain further confirmed the mislocalization of MBP mRNAs. The impaired localization of MBP mRNAs to the qkv/qkvmyelin presumably will lead to accumulation of the newly synthesized MBPs in the soma of oligodendrocytes therefore more susceptible for degradation.
Interaction of QKI with MBP mRNA may determine the cytoplasmic fate of MBP mRNAs
The above observations suggest that misregulation of the cellular fate of MBP mRNAs occurs in the cytoplasm of qkv/qkvoligodendrocytes, presumably because of the lack of the cytoplasmic isoforms of the QKI RNA-binding proteins. A simple model is that QKI directly associates with MBP mRNAs in normal oligodendrocytes, which is required to protect MBP mRNAs from nuclease attack and to direct MBP mRNA to translocate to the myelin membrane. Thus, the absence of QKI results in deprotection of MBP mRNAs and retention of MBP mRNAs in the soma of qkv/qkvoligodendrocytes. Our results clearly demonstrated selective association of QKI-7 with full-length MBP transcripts, and removing MBP 3′UTR significantly reduced such interaction (Fig.8B). Interestingly, although all the MBP mRNA isoforms share the same 3′UTR, various MBP mRNA isoforms displayed different QKI-binding activities. It is particularly intriguing that the QKI-binding activity and the severity of reduction for various MBP mRNA isoforms in the qkv/qkv brain follows the same order: M14 > M18.5 > M21.5. The differential QKI-binding activity by various MBP mRNA isoforms could be explained by interactions of the MBP 3′UTR with the coding region in the corresponding mRNA isoform, which may mask the binding element for QKI to various degrees.
MBP 3′UTR has been shown to play critical roles in stabilizing MBP mRNAs (Ueno et al., 1994). Thus, interaction of MBP 3′UTR with QKI is likely an important factor in maintaining the quantity of MBP mRNAs via controlling the mRNA degradation rate. In addition, well defined cis-acting elements in MBP 3′UTR are responsible for translocation of MBP mRNAs to the oligodendrocyte process and for their further localization to the myelin sheath (Ainger et al., 1997). The fact that QKI-6 and QKI-7 localize to the processes of normal oligodendrocytes (Hardy et al., 1996) suggests the potential roles that QKIs may play in targeting MBP mRNAs to the myelin membrane, presumably via interactions with MBP 3′UTR. It is intriguing to mention that both the stability and the localization of MBP mRNA are potentially coupled with its translation status (Ueno et al., 1994a,b; Ainger et al., 1997), and QKI has been reported recently to act as a translation suppressor (Saccomanno et al., 1999). Taken together, interactions of MBP mRNAs with QKI may influence MBP expression at multiple posttranscriptional levels, including mRNA turnover, translation, and subcellular localization.
Footnotes
This work is supported by the PhRMA faculty development award, Emory University URC funds, and National Institutes of Health Grant 5 PO1 HD35576 (Project V) to Y.F. We thank Dr. A. Campagnoni for providing MBP cDNA constructs, Dr. M. Terns for providing U3 cDNA construct, and Dr. Ball for providing the GAPDH cDNA construct.
Z.L. and Y.Z. contributed equally to this work.
Correspondence should be addressed to Dr. Yue Feng, Department of Pharmacology, Emory University School of Medicine, 1510 Clifton Road, Atlanta, GA 30322. E-mail: yfeng@emory.edu.
REFERENCES
- 1.Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, Carson JH. Transport and localization elements in myelin basic protein mRNA. J Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 3.Barbarese E. Spatial distribution of myelin basic protein mRNA and polypeptide in quaking oligodendrocytes in culture. J Neurosci Res. 1991;29:271–281. doi: 10.1002/jnr.490290302. [DOI] [PubMed] [Google Scholar]
- 4.Barbarese E, Carson JH, Braun PE. Accumulation of the four myelin basic proteins in mouse brain during development. J Neurochem. 1978;31:779–782. doi: 10.1111/j.1471-4159.1978.tb00110.x. [DOI] [PubMed] [Google Scholar]
- 5.Brophy PJ, Boccaccio GL, Colman DR. The distribution of myelin basic protein mRNAs within myelinating oligodendrocytes. Trends Neurosci. 1993;16:515–521. doi: 10.1016/0166-2236(93)90196-s. [DOI] [PubMed] [Google Scholar]
- 6.Brostoff SW, Greenfield S, Hogan EL. The differentiation of synthesis from incorporation of basic protein in Quaking mutant mouse myelin. Brain Res. 1977;120:517–520. doi: 10.1016/0006-8993(77)90404-8. [DOI] [PubMed] [Google Scholar]
- 7.Campagnoni AT. Molecular biology of myelin proteins from the central nervous system. J Neurochem. 1988;51:1–14. doi: 10.1111/j.1471-4159.1988.tb04827.x. [DOI] [PubMed] [Google Scholar]
- 8.Campagnoni AT, Macklin WB. Cellular and molecular aspects of myelin protein gene expression. Mol Neurobiol. 1988;2:41–89. doi: 10.1007/BF02935632. [DOI] [PubMed] [Google Scholar]
- 9.Campagnoni AT, Hunkeler MJ, Moskaitis JE. Translational regulation of myelin basic protein synthesis. J Neurosci Res. 1987;17:102–110. doi: 10.1002/jnr.490170203. [DOI] [PubMed] [Google Scholar]
- 10.Campagnoni CW, Carey GD, Campagnoni AT. Synthesis of myelin basic proteins in the developing mouse brain. Arch Biochem Biophys. 1978;190:118–125. doi: 10.1016/0003-9861(78)90258-8. [DOI] [PubMed] [Google Scholar]
- 11.Carnow TB, Carson JH, Brostoff SW, Hogan EL. Myelin basic protein gene expression in quaking, jimpy, and myelin synthesis-deficient mice. Dev Biol. 1984;106:38–44. doi: 10.1016/0012-1606(84)90058-7. [DOI] [PubMed] [Google Scholar]
- 12.Chen T, Damaj BB, Herrera C, Lasko P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colman DR, Kreibich G, Frey AB, Sabatini DD. Synthesis and incorporation of myelin polypeptides into CNS myelin. J Cell Biol. 1982;95:598–608. doi: 10.1083/jcb.95.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox RD, Hugill A, Shedlovsky A, Noveroske JK, Best S, Justice MJ, Lehrach H, Dove WF. Contrasting effects of ENU induced embryonic lethal mutations of the quaking gene. Genomics. 1999;57:333–41. doi: 10.1006/geno.1999.5804. [DOI] [PubMed] [Google Scholar]
- 15.de Ferra F, Engh H, Hudson L, Kamholz J, Puckett C, Molineaux S, Lazzarini RA. Alternative splicing accounts for the four forms of myelin basic protein. Cell. 1985;43:721–727. doi: 10.1016/0092-8674(85)90245-4. [DOI] [PubMed] [Google Scholar]
- 16.Delassalle A, Zalc B, Lachapelle F, Raoul M, Collier P, Jacque C. Regional distribution of myelin basic protein in the central nervous system of quaking, jimpy, and normal mice during development and aging. J Neurosci Res. 1981;6:303–313. doi: 10.1002/jnr.490060305. [DOI] [PubMed] [Google Scholar]
- 17.Ebersole TA, Chen Q, Justice MJ, Artzt K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins [see comments]. Nat Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- 18.Feng Y, Lakkis L, Devys D, Warren ST. Quantitative comparison of FMR1 gene expression in normal and premutation alleles. Am J Hum Genet. 1995;56:106–113. [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedrich VL., Jr Hyperplasia of oligodendrocytes in quaking mice. Anat Embryol. 1975;147:259–271. doi: 10.1007/BF00315075. [DOI] [PubMed] [Google Scholar]
- 21.Greenfield S, Brostoff S, Hogan E. Evidence for defective incorporation of proteins in myelin of the quaking mutant mouse. Brain Res. 1977;120:507–515. doi: 10.1016/0006-8993(77)90403-6. [DOI] [PubMed] [Google Scholar]
- 22.Hardy RJ. Molecular defects in the dysmyelinating mutant quaking. J Neurosci Res. 1998;51:417–422. doi: 10.1002/(SICI)1097-4547(19980215)51:4<417::AID-JNR1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 23.Hardy RJ, Loushin CL, Friedrich VL, Jr, Chen Q, Ebersole TA, Lazzarini RA, Artzt K. Neural cell type-specific expression of QKI proteins is altered in quakingviable mutant mice. J Neurosci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogan EL, Greenfield S. Animal models of genetic disorders of myelin (Morrell P, ed). Plenum; New York: 1984. [Google Scholar]
- 25.Jacque C, Delassalle A, Raoul M, Baumann N. Myelin basic protein deposition in the optic and sciatic nerves of dysmyelinating mutants quaking, jimpy, Trembler, mld, and shiverer during development. J Neurochem. 1983;41:1335–1340. doi: 10.1111/j.1471-4159.1983.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 26.Kondo T, Furuta T, Mitsunaga K, Ebersole TA, Shichiri M, Wu J, Artzt K, Yamamura K, Abe K. Genomic organization and expression analysis of the mouse qkI locus. Mamm Genome. 1999;10:662–669. doi: 10.1007/s003359901068. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Cole R, Chiappelli F, de Vellis J. Differential regulation of oligodendrocyte markers by glucocorticoids: post-transcriptional regulation of both proteolipid protein and myelin basic protein and transcriptional regulation of glycerol phosphate dehydrogenase. Proc Natl Acad Sci USA. 1989;86:6807–6811. doi: 10.1073/pnas.86.17.6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linz B, Koloteva N, Vasilescu S, McCarthy JE. Disruption of ribosomal scanning on the 5′-untranslated region, and not restriction of translational initiation per se, modulates the stability of nonaberrant mRNAs in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272:9131–9140. doi: 10.1074/jbc.272.14.9131. [DOI] [PubMed] [Google Scholar]
- 29.Macklin WB, Gardinier MV, Obeso ZO, King KD, Wight PA. Mutations in the myelin proteolipid protein gene alter oligodendrocyte gene expression in jimpy and jimpymsd mice. J Neurochem. 1991;56:163–171. doi: 10.1111/j.1471-4159.1991.tb02576.x. [DOI] [PubMed] [Google Scholar]
- 30.Matthieu J-M. An introduction to the molecular basis of inherited myelin diseases. J Inherit Metab Dis. 1993;16:724–732. doi: 10.1007/BF00711904. [DOI] [PubMed] [Google Scholar]
- 31.Newman S, Kitamura K, Campagnoni AT. Identification of a cDNA coding for a fifth form of myelin basic protein in mouse. Proc Natl Acad Sci USA. 1987;84:886–890. doi: 10.1073/pnas.84.3.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliveira CC, McCarthy JE. The relationship between eukaryotic translation and mRNA stability. A short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:8936–8943. doi: 10.1074/jbc.270.15.8936. [DOI] [PubMed] [Google Scholar]
- 33.Saccomanno L, Loushin C, Jan E, Punkay E, Artzt K, Goodwim EB. The STAR protein QKI-6 is a translational repressor. Proc Natl Acad Sci USA. 1999;96:12605–12610. doi: 10.1073/pnas.96.22.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorg BJ, Agrawal D, Agrawal HC, Campagnoni AT. Expression of myelin proteolipid protein and basic protein in normal and dysmyelinating mutant mice. J Neurochem. 1986;46:379–387. doi: 10.1111/j.1471-4159.1986.tb12979.x. [DOI] [PubMed] [Google Scholar]
- 35.Sorg BA, Smith MM, Campagnoni AT. Developmental expression of the myelin proteolipid protein and basic protein mRNAs in normal and dysmyelinating mutant mice. J Neurochem. 1987;49:1146–1154. doi: 10.1111/j.1471-4159.1987.tb10005.x. [DOI] [PubMed] [Google Scholar]
- 36.Trapp BD, Moench T, Pulley M, Barbosa E, Tennekoon G, Griffin J. Spatial segregation of mRNA encoding myelin-specific proteins. Proc Natl Acad Sci USA. 1987;84:7773–7777. doi: 10.1073/pnas.84.21.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueno S, Handley VW, Byravan S, Campagnoni AT. Structural features of myelin basic protein mRNAs influence their translational efficiencies. J Neurochem. 1994a;62:1254–1259. doi: 10.1046/j.1471-4159.1994.62041254.x. [DOI] [PubMed] [Google Scholar]
- 38.Ueno S, Kotani Y, Kondoh K, Sano A, Kakimoto Y, Campahnoni AT. The 3′-untranslated region of mouse myelin basic protein gene increases the amount of mRNA in immortalized mouse oligodendrocytes. Biochem Biophys Res Commun. 1994b;204:1352–1357. doi: 10.1006/bbrc.1994.2612. [DOI] [PubMed] [Google Scholar]
- 39.Umemori H, Kadowaki Y, Hirosawa K, Yoshida Y, Hironaka K, Okano H, Yamamoto T. Stimulation of myelin basic protein gene transcription by Fyn tyrosine kinase for myelination. J Neurosci. 1999;19:1393–1397. doi: 10.1523/JNEUROSCI.19-04-01393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdi JM, Kampf K, Campagnoni AT. Translational regulation of myelin protein synthesis by steroids. J Neurochem. 1989;52:321–324. doi: 10.1111/j.1471-4159.1989.tb10935.x. [DOI] [PubMed] [Google Scholar]
- 41.Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]