Abstract
The shape of dendritic trees and the density of dendritic spines can undergo significant changes during the life of a neuron. We report here the function of the small GTPases Rac and Rho in the maintenance of dendritic structures. Maturing pyramidal neurons in rat hippocampal slice culture were biolistically transfected with dominant GTPase mutants. We found that expression of dominant-negative Rac1 results in a progressive elimination of dendritic spines, whereas hyperactivation of RhoA causes a drastic simplification of dendritic branch patterns that is dependent on the activity of a downstream kinase ROCK. Our results suggest that Rac and Rho play distinct functions in regulating dendritic spines and branches and are vital for the maintenance and reorganization of dendritic structures in maturing neurons.
Keywords: Rac, Rho, dendritic spines, biolistic transfection, pyramidal neurons, effector domain mutants, ROCK, Y-27632, PSD-95
Perhaps the most striking features of many mammalian neurons are their complex dendritic trees. Dendrites are the principal site where neurons receive, process, and integrate inputs from their multiple presynaptic partners. These functions are primarily influenced by the branching pattern of the dendritic tree (Rall, 1964). In addition to complex dendritic arbors, some mammalian neurons, including cerebellar Purkinje cells and cortical and hippocampal pyramidal neurons, have dendritic specializations called spines. These spines, which protrude from the dendritic branches, are the primary site of excitatory synapses and may function as the basic unit of synaptic integration (for review, see Harris and Kater, 1994; Yuste and Tank, 1996). Both the shape of dendritic trees and the density and shape of their spines can undergo significant changes during the development and life of a neuron. For instance, dendritic branches have been shown to undergo significant remodeling in adult superior cervical ganglion neurons in vivo (Purves and Hadley, 1985; Purves et al., 1986). Recent studies have also shown that dendritic filopodia and spines undergo dynamic changes in response to synaptic activity (Engert and Bonhoeffer, 1999; Maletic-Savatic et al., 1999; Toni et al., 1999) and neurotrophin overexpression (Horch et al., 1999).
Several extracellular molecules have been identified as potential regulators of dendrite and spine development (Purves et al., 1988;Snider, 1988; Lein et al., 1995; McAllister et al., 1995, 1997; Nedivi et al., 1998; Guo et al., 1999; Horch et al., 1999). To induce dendritic growth, branching, or retraction, as well as the formation, elaboration, or elimination of dendritic spines, these extracellular factors must eventually exert their effects by signaling to the cytoskeleton. Actin is one of the main components of the dendritic cytoskeleton and is highly enriched in dendritic spines (seeFischer et al., 1998). The Rho family of small GTPases, including Rho, Rac, and Cdc42, regulates various aspects of the actin cytoskeleton (for review, see Hall, 1994; Van Aelst and D'Souza-Schorey, 1997), and these GTPases are therefore good candidates for mediating these signals. In agreement with their differential effects in fibroblasts (for review, see Van Aelst and D'Souza-Schorey, 1997), both in vivo and in vitro studies have shown that these GTPases appear to have distinct effects on different aspects of neuronal morphogenesis (for review, see Luo et al., 1997; Mueller, 1999).
Although many studies illustrate the roles of Rho family GTPases in the establishment of neuronal processes, it remains unclear whether they also function in later stages of neuronal development or in mature neurons to regulate dendritic reorganization and dynamic changes of dendritic spines. To address these questions, we used biolistic transfection (Arnold et al., 1994; Lo et al., 1994) to introduce dominant mutants of Rac1 and RhoA acutely into organotypic hippocampal slices at a stage when pyramidal neurons have established dendritic arbors and possess dendritic spines yet still express the GTPases. We found that Rac1 is required for the maintenance of dendritic spines, whereas the elevation of RhoA activity leads to pronounced simplification of dendritic trees. We also identified candidate downstream effector pathways that mediate these morphological changes. These studies demonstrate that signaling pathways used in the early development of neuronal processes are also used in more mature neurons for the maintenance and reorganization of dendritic structures.
MATERIALS AND METHODS
In situ hybridizations
Sense and antisense S35-riboprobes were generated to the C-terminal ends of rat Rac1 and RhoA by the following steps: 5′-Phosphorylated primers were designed from published sequences to amplify base pairs 259–577 of the mouse Rac1 ORF and base pairs 226–551 of the rat RhoA ORF. These regions were PCR amplified from an embryonic day 13 rat cDNA library, and the bands were purified and ligated into pBLUESCRIPT II (SK+) digested with EcoRV. Plasmids containing insertions of both orientations were linearized with EcoRI and transcribed with T7 RNA polymerase to generate the sense and antisense riboprobes.
Freshly dissected brains of postnatal day 8 (P8) Long–Evans rats were submerged in ornithine carbamyltransferase and frozen in an ethanol and dry ice bath and then stored at −80°C. Twenty micrometer cryostat sections were prepared, fixed, washed in 1× PBS, and serially dehydrated before being returned to −80°C in a desiccated, sealed box. Sections were hybridized and processed for autoradiography according to the protocol used by Frantz et al. (1994).
Digoxigenin (DIG)-labeled in situ hybridizations were performed according to the protocol used by Wright and Snider (1995).
Molecular biology
p-chicken β actin–mouse CD8. pUC–mouse CD8 (mCD8) (Liaw et al., 1986) was digested with XhoI andBamHI, and the end nucleotides were filled in using Klenow to give a 900 bp fragment encoding mCD8. The p-chicken β actin (pCA)–GAP–enhanced green fluorescent protein (EGFP) vector (Okada et al., 1999) was digested with BamHI and NotI, and the end nucleotides were filled to generate a 5.1 kb pCA backbone. The mCD8 fragment and the pCA backbone were ligated together, generating the 6 kb pCA–mCD8.
pCA–hRac1V12. pBS–hRac1V12 (Luo et al., 1996) was digested with Asp718, blunted with Klenow, and cut again with NotI to yield a 600 bp fragment encoding hRac1V12. This fragment was ligated into the 5.1 kb pCA backbone generated by digesting pCA–GAP–EGFP withSmaI and NotI, yielding the 5.7 kb pCA–hRac1V12.
pCA–myc–hRac1WT. pUHD–myc–hRac1WT (Qiu et al., 1995a) was digested with EcoRI, and the resulting fragment encoding myc–hRac1WT (600 bp) was ligated into the compatible site of pNN03. pNN03–myc–hRac1WT was then digested with EcoRV and NotI, and the resulting fragments were ligated into the pCA backbone as described above to yield pCA–myc–hRac1WT.
pCA–myc–hRac1N17, pCA–myc–hRhoAV14, and pCA–myc–hRhoAN19. pEXV–myc–hRac1N17, pEXV–myc–hRhoAN19, and pEXV–myc–hRhoAV14 (Qiu et al., 1995a,b) were digested withEcoRI, and the resulting fragments encoding myc–hRac1N17 (600 bp), myc–hRhoAN19 (1 kb), and myc–hRhoAV14 (1 kb) were ligated into the compatible site of pBLUESCRIPT II (SK+). pBS–myc–hRac1N17, pBS–myc–hRhoAN19, and pBS–myc–hRhoAV14 were digested withEcoRV and NotI, and the resulting fragments were then ligated into the 5.1 kb pCA backbone generated by digesting pCA–GAP–EGFP with SmaI and NotI. The resulting plasmid pCA–myc–hRac1N17 is 5.7 kb in length, whereas pCA–myc–hRhoAV14 and pCA–myc–hRhoAN19 are 6.1 kb.
pCA–myc–hRac1L61, pCA–myc–hRac1L61K40, and pCA–myc–hRac1L61A37. pRK5–myc–hRac1L61, pRK5–myc–hRac1L61K40, and pRK5–myc–hRac1L61A37 (Lamarche et al., 1996; Nikolic et al., 1998) were digested with ClaI andEcoRI. The resulting 600 bp fragments encoding myc–hRac1L61, myc–hRac1L61K40, and myc–hRac1L61A37 were ligated into the compatible sites of pBLUESCRIPT II (SK+). These plasmids were then digested with XhoI, blunted with Klenow, and then digested withNotI. The resulting 600 bp fragments representing the myc-tagged Rac1 mutants were isolated and ligated into the 5.1 kb pCA backbone generated by digesting pCA–GAP–EGFP with SmaI andNotI. The resulting pCA–myc–hRac1L61, pCA–myc–hRac1L61K40, and pCA–myc–hRac1L61A37 are 5.7 kb in length.
Preparation of DNA-coated gold particles
Qiagen-Midi prepped plasmids were precipitated onto 1.6 μm gold beads (Bio-Rad, Hercules, CA) at a concentration of 2 μg of each plasmid per milligram of gold beads, according to the manufacturer's instructions. Briefly, DNA was precipitated onto gold beads that were subsequently dried to the sides of plastic tubing. This tubing was chopped into 0.5 inch fragments known as “bullets.” When a gold bead was to carry two plasmids simultaneously (cotransfection experiments), both plasmids (2 μg/mg of gold each) were first mixed and coprecipitated using a standard ethanol precipitation protocol before precipitation onto gold beads. For “dual gold” experiments, in which a single bullet contains beads carrying different DNAs, the two populations of gold beads were prepared separately and mixed just before precipitation onto the plastic tubing.
Preparation of rat hippocampal organotypic cultures
Hippocampal slices were prepared from P8 Long–Evans rats as described previously (Stoppini et al., 1991). Briefly, the hippocampus was dissected in ice-cold, sterile dissection medium: 1× MEM (Hank's salts, 25 mm HEPES, without l-glutamine; from Life Technologies, Grand Island, NY) and 100 units/ml each penicillin–streptomycin (Life Technologies). Hippocampi were sliced transversely at a thickness of 400 μm on a tissue chopper (Stoelting, Wood Dale, IL) and separated from one another in filtered and preincubated (37°C; 5% CO2) culture medium: 0.5× MEM, 0.25× HBSS (Life Technologies), 0.25× horse serum (defined, heat-inactivated; from HyClone, Logan, UT), 100 units/ml each penicillin–streptomycin, and 1 mml-glutamine (Life Technologies). Slices were immediately plated onto Millicell CM membrane inserts (Millipore, Bedford, MA) in Petri dishes containing 1 ml of preincubated culture media. Slices were kept under 5% CO2 at 37°C, with media changes at 1 din vitro (DIV), 3 DIV, and every 3 d thereafter. In experiments using Y-27632 (gift from Yoshitomi Pharmaceuticals, Tokyo, Japan) slices were transferred to Petri dishes containing 100 μm Y-27632 in 1 ml of culture media just before biolistic transfection. Control slices were similarly transferred, but to Petri dishes containing media supplemented with the same volume of sterile water instead of Y-27632. Animals were treated in accordance with the animal safety protocols of the host institution.
Biolistic transfection
To identify optimal transfection and culture conditions, slices were prepared from P8 rats, transfected at various DIV, and fixed for immunocytochemistry 24 hr later. The number of healthy pyramidal neurons relative to those that were dead or dying (characterized by fragmented processes or blebbing processes, respectively) increased significantly when transfection occurred at 2 DIV, compared with transfection at 0 or 1 DIV. When transfection occurred at >2 DIV, the number of transfected glia increased significantly, often obscuring the dendritic arbors we wished to quantify. Thus, to maximize the number of healthy pyramidal cells transfected and to minimize interfering glia, experiments throughout this study were performed on a standard preparation: hippocampal slices prepared from P8 rats and cultured for 2 d before transfection.
After 2 DIV, slice inserts were removed from the incubator briefly for biolistic transfection using the Gene Gun (Bio-Rad). Gold beads containing expression plasmids were propelled from plastic tubing bullets into slices with a rapid helium burst of 160 psi. The gold beads exited the gun ∼3.5 cm above the slices, which were plated toward the periphery of the insert to avoid “ground zero” of the gold blast.
Immunocytochemistry
Slices were fixed according to the protocol used by McAllister et al. (1995) on the insert membrane for 1.5 hr in 2.5% formaldehyde and 4% sucrose in 1× PBS and then soaked in 30% sucrose (w/v of water) for at least 2 hr. After a quick freeze on dry ice, slices were thawed, rinsed in 1× PBS for 5 min, and incubated overnight (O/N) at 4°C in blocking solution: 10% normal goat serum and 0.25% Triton X-100 in 0.1 m phosphate buffer. Blocking solution was replaced with primary antibodies diluted in blocking solution: 1:50 mouse anti-myc (Santa Cruz Biotechnology, Santa Cruz, CA), 1:100 rat anti-mCD8 (Caltag, Burlingame, CA), or both before another O/N incubation at 4°C. The slices were then washed three times for 20 min each with fresh blocking solution and incubated O/N at 4°C in secondary antibody, diluted into blocking solution: 1:200 FITC anti-mouse (Jackson ImmunoResearch, West Grove, PA), 1:1000 indocarbocyanine (Cy3) anti-rat (Jackson ImmunoResearch), or both (in which the low cross-reactivity secondaries were used). Slices were subsequently washed three times for 15 min each in 1× PBS, with the first wash containing 1.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI), and mounted in SlowFade according to the manufacturer's specifications (Molecular Probes, Eugene, OR).
Image analysis and quantification
Transfected pyramidal neurons were identified by their typical morphology as well as their cell body locations in the CA1 and CA3 pyramidal layers as indicated by DAPI staining (see Fig.2A). Individual neurons were imaged using a Zeissmicroscope attached to a Bio-Rad MRC-1024 scanning laser confocal microscope. For quantification of dendritic branch segments, a stack of confocal images (Z steps of 1 μm) taken with a 16× objective and comprising the entire cell were merged using NIH Image 1.62 (National Institutes of Health, Bethesda, MD) and printed. From these printouts, the number of dendritic segments was derived by counting the number of dendrite branch points and dendrite terminal ends. To obtain the Sholl profiles of dendritic arbors (Sholl, 1953), printouts were placed under a clear sheet printed with concentric circles with increasing radii of 25 μm. To minimize the effect of cell body shape variation, we positioned the center of the circles at the base of the apical dendrite for analysis of apical dendrites. In addition, because CA1 and CA3 pyramidal neurons exhibited different Sholl profiles (data not shown), all Sholl analysis was with only CA1 neurons. The center of the circles was placed at the cell body edge, opposite the apical dendrite when analyzing basal dendrites. The number of dendrites crossing each concentric circle was then counted. If a branch point fell on a line, it was counted as two crossings.
Fig. 2.
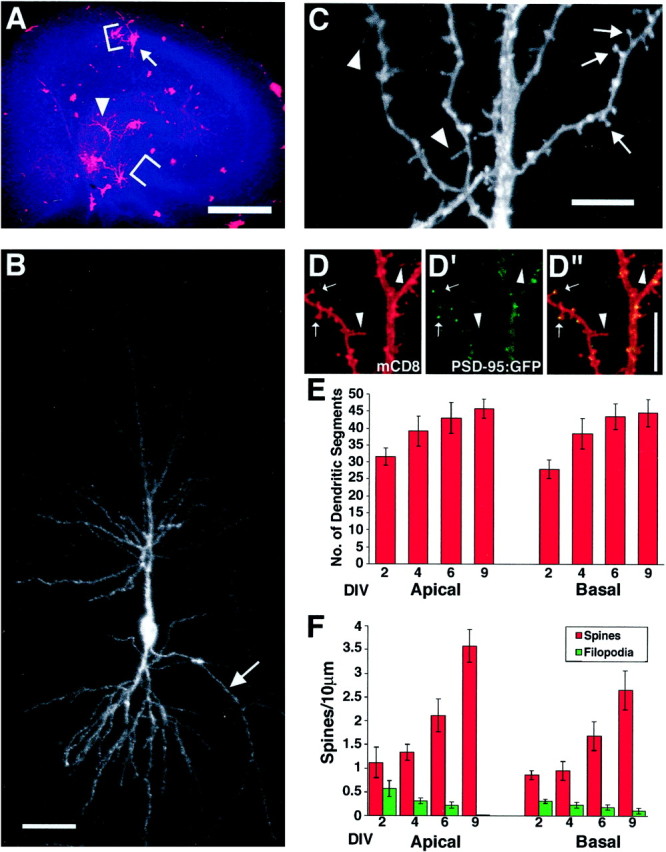
Development of hippocampal pyramidal neurons in cultured slices. A–C, Representative images of transfected hippocampal neurons show the developmental stage at the onset of experiments in our standard preparation. For these images only, transfection was performed at 1 DIV, and slices were fixed 24 hr later. A, Low magnification is shown of a hippocampal slice biolistically transfected with mCD8 (red), with brackets defining the DAPI-labeled (blue) pyramidal cell layers of CA1 and CA3.Pink labeling (from overlappingred and blue signals) represents transfected cells, including a granule cell in the dentate gyrus (arrowhead) and a pyramidal neuron overlapped by glia in the CA1 cell layer (arrow). B, A CA3 pyramidal neuron (composite confocal image using a 16× objective) is shown. The immunocytochemical detection of mCD8 reveals the structure of both apical and basal dendrites, as well as the axon (arrow). C, The spines on the apical dendrites of the neuron pictured in B (composite confocal image using a 100× objective) are shown.Arrows point to spines with characteristic head and neck morphology, whereas arrowheads show filopodial protrusions. D, Double labeling of mCD8 and PSD-95: GFP proteins in apical dendrites (composite confocal images using a 40× objective with a digital zoom factor of 3) is shown. mCD8 labeling is in red (D, D′′), and PSD-95:GFP labeling (PSD-95:GFP) is ingreen (D′, D′′).Arrows indicate spines with a head (PSD-95:GFP positive), whereas arrowheads indicate filopodial protrusions (PSD-95:GFP negative). E, Dendritic branch segments gradually increase on both apical and basal dendrites with successive days in culture (apical, n = 10, 13, 8, 10; basal, n = 10, 14, 7, 9 for 2, 4, 6, 9 DIV, respectively). F, Spine density increases over successive days in culture, whereas filopodial protrusions decrease. Spines and filopodia were counted from stacked confocal images collected at 40× with a zoom of three, from a stereotyped region of the dendrite as defined in Materials and Methods (apical,n = 10, 19, 17, 12; basal, n = 11, 14, 17, 11 for 2, 4, 6, 9 DIV, respectively). Scale bars:A, 1 mm; B, 50 μm; C, D, 10 μm.
For spine quantification, images of apical dendrites were taken just distal to the first apical branch point (25–100 μm from the cell body). Basal spines were imaged at the point of the first basal branch (25–75 μm from the cell body). Serial confocal images (Z steps of 0.5 μm) were taken of Cy3 fluorescence (detecting mCD8) with a 40× objective with a digital zoom factor of three. Sections were merged using NIH Image, and the number of spines and filopodia was tallied on the screen. Spines were defined as a headless dendritic protrusion 1–3 μm long or a headed protrusion of any length up to 3 μm. Filopodia were defined as headless protrusions >3 μm but not long enough to be visible on the 16× printout. The length of all dendrites in the fields used to quantify spine density was measured using NIH Image. The average dendritic diameter for each cell was based on three measurements (at the start, middle, and end) of every dendritic segment within the images used to analyze dendritic spine density.
Identical procedures for acquiring images and measurements of dendritic length were also used to analyze GFP-tagged postsynaptic density-95 (PSD-95:GFP) clustering with Rac1N17 expression. To count PSD-95:GFP clusters optimally in relation to spine profiles as defined above, images were placed into channels in Photoshop 4.0 (Adobe Systems, San Jose, CA). GFP clusters were counted independent of location, and the spine profiles with GFP clusters either within the spine head or just external to the head outline were also tallied. The dendrites and their spines were outlined using the magic wand tool in the channel corresponding to the myc label, which fills the entire dendritic tree including the spines (see Fig. 3).
Fig. 3.
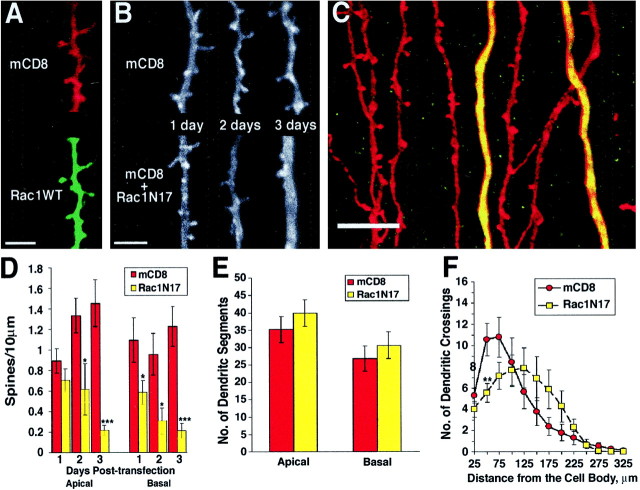
Dominant-negative Rac1 expression results in a progressive reduction of the dendritic spine density and mild changes in the dendritic-branching pattern. A,Transfected myc-tagged wild-type Rac1 protein is distributed along the dendrites and in dendritic spines (green, anti-myc; red, anti-mCD8; composite confocal image using 40× objective; digital zoom factor of 3). B,Representative images are shown of apical dendrites that were transfected with the marker mCD8 alone (top) or with mCD8 and myc-tagged Rac1N17 (bottom) for 1, 2, or 3 d (composite confocal images of mCD8 staining using 100× objective).C, Apical dendrites from neighboring pyramidal neurons are shown 3 d after expressing mCD8 alone (red) or expressing both mCD8 (red) and myc-tagged Rac1N17 (green) and therefore appearingyellow (composite confocal image using a 40× objective with a digital zoom factor of 3). Although it appears that Rac1N17 dendrites have thicker dendrites in this image, quantification of the average diameter of dendrites within every image used to measure dendritic spine density does not reveal any significant difference (paired t test, apical, p = 0.19; Rac1N17 = 1.50 ± 0.14 μm; control = 1.22 ± 0.51 μm; n = 18, 9, respectively; basal,p = 0.12; Rac1N17 = 0.83 ± 0.1 μm; control = 1.05 ± 0.08 μm; n = 9, 8, respectively). D, Rac1N17 expression progressively reduces the number of spines on apical and basal dendrites (for 1, 2, 3 d, apical mCD8, n = 16, 19, 10; apical Rac1N17, n = 19, 11, 24; basal mCD8,n = 15, 14, 10; basal Rac1N17,n = 18, 11, 17, respectively). E,Quantification of dendritic branch segments after 3 d of Rac1N17 expression is shown (n = 17, 23, 18, 21 for apical mCD8, apical Rac1N17, basal mCD8, basal Rac1N17, respectively).F, Sholl profiles of the basal dendrites of Rac1N17-expressing and control CA1 pyramidal neurons (3 d after transfection) illustrate the slight change in dendritic-branching pattern with Rac1N17 expression (n = 14, 15 for basal mCD8, Rac1N17, respectively). Posthoct tests reveal that the only significant difference occurs at 50 μm (paired t test, **p < 0.01). Scale bars: A, B, 5 μm; C, 10 μm.
Graphs throughout represent averages and SEM. Statistical comparisons of Sholl segments were done with StatView 5.0.1 (SAS Institute, Cary, NC), and all post hoc two-tailedt tests assuming equal variances were done using Microsoft Excel 98 (Microsoft, Seattle, WA).
RESULTS
Rac1 and RhoA are expressed in developing hippocampal pyramidal neurons
Although Rac1 and RhoA are ubiquitously expressed (for review, seeHall, 1994), the expression pattern of these GTPases in the hippocampus has not been described. To identify Rac1- and RhoA-expressing cells in the hippocampus, in situ hybridizations were performed on coronal P8 rat brain sections, the same stage at which our slice cultures were produced. Antisense S35-labeled riboprobes were generated to the C-terminal portions of each mRNA, where the nucleic acid sequence identities between Rac and Rho are lowest (50% identical). As shown in Figure 1, Rac1 (Fig.1A) and RhoA (Fig. 1B) antisense riboprobes labeled CA1 and CA3 pyramidal cell layers and the dentate granular cell layer, with the CA3 layer being the most intense. Rac and Rho transcripts are also widely distributed in other brain areas (data not shown). The sense riboprobes to both GTPases gave only diffuse background signal, similar to that shown for Rac1 (Fig. 1C).In situ hybridization using DIG-labeled probes was performed on sections made from P10 hippocampi (equivalents to the day slices were transfected), with similar results (data not shown). No obvious dendritic localization of the two mRNAs was observed.
Fig. 1.
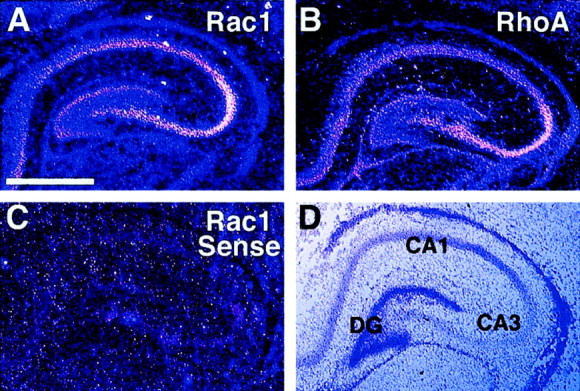
Rac1 and RhoA are expressed in developing hippocampus. A, Dark-field image of a coronally sectioned P8 rat hippocampus hybridized with an antisense S35-riboprobe against Rac1, showing distribution of Rac1 mRNA in the dentate gyrus and CA1 and CA3 pyramidal cell layers.B, Dark-field image of antisense riboprobe showing distribution of RhoA mRNA. C, Dark-field image of the Rac1 sense riboprobe showing background staining. D,Bright-field image of the same hippocampus shown in Cwith the CA1, CA3, and dentate gyrus (DG) labeled. Scale bar, 2 mm.
Pyramidal neurons maintain in vivo characteristics while in slice culture
We have used particle-mediated gene transfer (biolistic transfection) (Arnold et al., 1994; Lo et al., 1994) of mCD8 cDNA, and subsequent immunocytochemical detection of the protein, to visualize the dendritic arbors and spines of pyramidal neurons in hippocampal slice cultures. mCD8 is a cell-surface marker shown previously to label the entire morphology and fine structures of Drosophilaneurons without toxic side effects (Lee and Luo, 1999). As shown in Figure 2, delivery of a single gold particle carrying mCD8 under the control of the chicken β-actin promoter resulted in the intense labeling of not only the axon and the entire dendritic arbor (Fig. 2B) but individual spines as well (Fig. 2C). In addition to pyramidal neurons, many other hippocampal cell types were readily transfected, including glia, the granule cells of the dentate gyrus, and interneurons (Fig.2A).
In our standard preparation, hippocampal slices were obtained from postnatal day 8 rat pups and cultured for 2 d before transfection (see Materials and Methods). To determine the developmental state of pyramidal neurons at the time of transfection, 1 d in vitro cultures were transfected with mCD8 and fixed 24 hr later (2 DIV). By this time, pyramidal neurons had already acquired their characteristic dendritic-branching pattern, including well differentiated apical and basal dendrites (Fig. 2B). Counting the number of dendritic branch segments (see Materials and Methods) gave us a quantitative measure of dendritic complexity and revealed that over the first few days after transfection, there is a gradual increase in the number of dendritic segments (Fig.2E). At each time point examined, we found no statistically significant difference between the number of dendritic segments for apical and basal dendrites or between CA1 and CA3 neurons (data not shown).
These 2 DIV neurons also displayed numerous protrusions from their apical and basal dendrites. Although a minority of these protrusions were filopodial in shape (Fig. 2C, arrowheads), many more had the well defined neck and head structure characteristic of mature spines found in adults (Fig. 2C,arrows). To verify independently the maturity of these dendritic protrusions in our culture, we cotransfected GFP-tagged PSD-95 as a marker for postsynaptic density (Arnold and Clapham, 1999). We found that PSD-95:GFP labeling was concentrated in most of the heads of the protrusions but was absent from longer filopodia (Fig.2D). Using the morphological criterion described above (see Materials and Methods for details), we quantified the density of spine-like protrusions and filopodial protrusions. We found that spine density increased with the time spent in culture, whereas the number of filopodial protrusions gradually diminished (Fig.2F). The spine densities found in our cultures are similar to those reported previously for similarly aged cultures [P12 equivalent = 2 spines/10 μm by EM (Boyer et al., 1998)]. In accordance with Drakew et al. (1996), we did not find a significant difference in spine density between the apical and basal dendrites at any time point examined (data not shown). Additionally, we did not observe a significant difference in spine density between CA1 and CA3 pyramidal dendrites (data not shown). Because CA1 and CA3 neurons did not differ in either their branch segment number or their spine density, neurons from both areas have been combined in our quantification of branch segments and spine densities.
Expression of dominant-negative Rac1 results in a progressive reduction of spine density
To study the function of Rho family GTPases in the maintenance of dendritic structures, we cotransfected pyramidal neurons in our standard preparation with both mCD8 and dominant mutants of the GTPases. Cotransfection was accomplished by the introduction of gold beads coated with plasmids for both genes, each under the control of the chicken β-actin promoter. Although analogous experiments with two marker genes (mCD8 and GAP–EGFP) showed a high cotransfection rate (>85%), quantitative variations in the relative expression levels of the two constructs were observed. Therefore, we independently monitored the expression of the dominant mutants by using myc epitope-tagged GTPases, analyzing only those cells in which both plasmids were strongly expressed. As an internal control and to minimize variation among hippocampal slices, most experiments were performed using dual gold preparations. In these experiments, slices were transfected with a mixed population of gold beads, some carrying the mCD8 and GTPase plasmids and some the mCD8 plasmid alone. This enabled us to visualize both control (mCD8 alone) and experimental (mCD8 and GTPase) neurons from the same slice (see Fig.3C). We found that transfected wild-type Rac1, as well as all mutant proteins, was distributed throughout the entire dendritic tree, including dendritic spines and other fine processes (see Figs. 3A,C,4B–D,F–H).
Fig. 4.
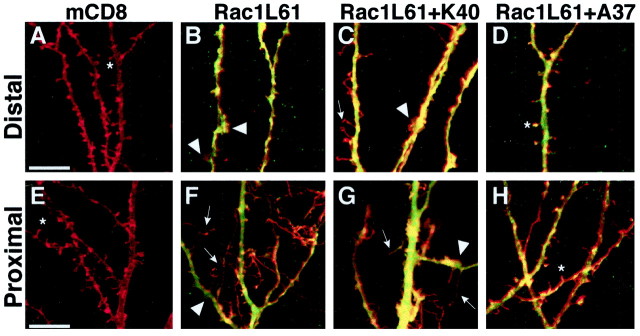
Effects of activated Rac1 expression on dendritic morphology. A–H, Representative distal (A–D) and proximal (E–H, justabove cell bodies) apical dendrites and spines of hippocampal pyramidal neurons, 24 hr after being transfected with mCD8 only (A, E), mCD8 and Rac1L61 (B, F), mCD8 and Rac1L61K40 (C, G), or mCD8 and Rac1L61A37 (D, H) (composite confocal images using a 40× objective with a digital zoom factor of 3).Red staining in all images represents anti-mCD8 immunoreactivity, and green staining inB–D and F–H represents anti-myc immunoreactivity. All mutant Rac proteins were expressed at comparable levels and were distributed throughout the entire neuron on the basis of their myc staining (see yellow labeling inB–D, F–H because of overlappinggreen signal for myc and red signal for mCD8). Asterisks represent mature spines with heads,arrowheads indicate ruffle-like structures, andarrows point to long and thin filopodial-like protrusions. Scale bars, 10 μm.
To test whether endogenous Rac1 is necessary for the maintenance of dendritic spine and branch morphology, we cotransfected pyramidal neurons with gold beads carrying mCD8 and myc-tagged dominant-negative Rac1 (Rac1N17). This allele is thought to exert its dominant-negative effect by sequestering rate-limiting GDP–GTP nucleotide exchange factors necessary for activation of endogenous Rac1 (Ridley et al., 1992). Expression of Rac1N17 resulted in a significant time-dependent loss of pyramidal dendritic spines from both apical and basal dendrites. Whereas neurons expressing mCD8 alone displayed an increasing trend of spine density with successive days after transfection, those also expressing Rac1N17 possessed fewer and fewer spines on both their apical and basal dendrites (Fig. 3B—D; see also Fig. 2F). Only 1 d after transfection, the basal dendritic spine density of Rac1N17 neurons was significantly reduced compared with that of neurons expressing mCD8 alone (68% fewer; p < 0.05), whereas the apical spine density was significantly lower than that of controls 2 d after transfection (54% fewer; p < 0.05). An example of a dual gold experiment is shown in Figure 3C. After 3 d of expression, mCD8-transfected pyramidal dendrites (red) displayed adult-like spines, whereas the dendrites from Rac1N17-expressing cells (yellow because ofred labeling of mCD8 and green labeling of myc-tagged Rac1N17) were nearly devoid of spines.
To address whether the corresponding synapse number is decreased with the loss of the dendritic spine profile in Rac1N17-transfected neurons, we cotransfected PSD-95:GFP with Rac1N17. In agreement with the loss of dendritic spine density, we observed a significant decrease in PSD-95:GFP clusters with 3 d of Rac1N17 expression (p < 0.05; Rac1N17 = 2.14 ± 0.35; mCD8 = 3.84 ± 0.52; units are PSD-95:GFP clusters per 10 μm; n = 9, 8, respectively).
To determine whether Rac1 may play a role in the maintenance of dendritic growth and branching, we quantified the number of dendritic segments after 3 d of Rac1N17 expression. As shown in Figure3E, there is no statistically significant difference between Rac1N17 and control cells (apical, p = 0.38; basal,p = 0.48). As another measure of dendritic complexity, a standard Sholl analysis (Sholl, 1953), which counts the number of dendritic crossings at 25 μm concentric circles, was performed. Although the Sholl profiles for the apical dendrites of Rac1N17-expressing neurons and control neurons are similar (repeated measures ANOVA, p = 0.40), their basal profiles are slightly different (Fig. 3F; repeated measures ANOVA,p < 0.05). There are more branches concentrated in the proximal regions in control neurons compared with Rac1N17 neurons, which appear to have more distally distributed branches (Fig.3F). This mild effect of Rac1N17 on the dendritic-branching pattern of pyramidal neurons is in contrast to its pronounced effect on spine density, suggesting a preferential role for endogenous Rac1 in the maintenance of dendritic spines.
Activated Rac1 disrupts normal spine morphology
To elucidate how Rac1 activity affects spine morphogenesis, we transfected pyramidal cells with a constitutively activated form of Rac1 that is deficient in GTP hydrolysis (hereafter referred to as activated Rac1). Previous transgenic expression of the activated allele Rac1V12 in Purkinje cells results in a slight simplification of dendritic complexity, a loss of normal dendritic spines, and the formation of supernumerary smaller protrusions that were only detectable with EM (Luo et al., 1996). We found that pyramidal neurons expressing Rac1L61, another activated Rac1 allele, for 1, 2, or 3 d also showed similar defects in spine development (Fig.4B). With the exception of some distal dendrites, apical and basal dendrites lacked normal spines. In contrast to our control neurons (Fig. 4A), which have even dendritic branches that are regularly punctuated by spines, Rac1L61 dendrites were irregular in thickness because of the presence of regions of numerous overlapping bumps and ruffle-like structures (Fig. 4B,F, arrowheads). Additionally, numerous long and fine processes were found on the cell soma and proximal dendritic shafts (Fig. 4, compare F,arrows, E). Although these fine processes prevented quantification of dendritic segments, 70% of Rac1L61-expressing neurons had an otherwise normal dendritic-branching pattern despite their perturbed dendritic spines. These findings suggest that dendritic spines are more sensitive to the hyperactivation of Rac1 than are the dendritic branches themselves. Expression of another constitutively active form of Rac1, Rac1V12, in hippocampal neurons gave indistinguishable results compared with Rac1L61 expression (data not shown). Although normal dendritic spines are lost with the expression of either dominant-negative or constitutively active Rac1, Rac1L61 expression resulted in a net increase in dendritic protrusion exemplified by an increase in filopodial-like structures and membrane ruffling, which were not seen with Rac1N17 expression (Fig.3B,C).
The F37A effector domain mutant abolishes the activated Rac1 phenotype
Rac1 binds to different effectors to activate distinct downstream signal transduction pathways. Previous in vitro experiments have characterized several effector domain mutants that separate Rac1's role in lamellipodia formation from its role in activation of the Jun kinase cascade and nuclear signaling (Joneson et al., 1996;Lamarche et al., 1996). The Y40K mutation abolishes the binding of several CRIB-containing proteins [e.g., p21-activated kinase (Pak)] and when combined with an activating Rac1 mutation blocks the stimulation of the Jun kinase (JNK) pathway without affecting membrane ruffling and lamellipodia formation. Conversely F37A mutation does not bind to Rho-associated kinase (ROCK) and prevents the induction of membrane ruffling and lamellipodia by an activating mutation of Rac (Lamarche et al., 1996), yet it still binds to Pak and activates the JNK pathway. These data suggest that effectors dependent on tyrosine residue 40 for binding to activated Rac (e.g., Pak and other CRIB-containing proteins) are required for the activation of Jun kinase, whereas effectors dependent on phenylalanine residue 37 (e.g., ROCK) for binding are candidate mediators for Rac1's regulation of lamellipodia formation.
We attempted to identify the pathway responsible for the activated Rac1 phenotype in hippocampal pyramidal neurons using these same effector domain mutants. Rac1L61K40 expression resulted in irregular, bumpy dendrites containing numerous thin processes (Fig. 4C,G) that were indistinguishable from Rac1L61 dendrites (Fig.4B,F), suggesting that effectors dependent on tyrosine 40 for binding (e.g., Pak) are not necessary to mediate the effects of activated Rac1. In contrast, the dendritic morphology and spine density of pyramidal neurons expressing Rac1L61A37 (Fig.4D,H) resembled those of control neurons (Fig.4A,E), implying a requirement for effectors dependent on phenylalanine 37 to mediate the activated Rac1 phenotype. ROCK is one such protein (Joneson et al., 1996; Lamarche et al., 1996). We attempted to mimic the Rac1L61A37 rescue by treating Rac1L61-transfected slices with 100 μm Y-27632, a compound that specifically inhibits ROCK activity without affecting other similar kinases (see Uehata et al., 1997; Madaule et al., 1998). Although this treatment effectively blocked the activated RhoA phenotype (see below), the Rac1L61 effect persisted, suggesting that ROCK is not responsible for mediating the effect of activated Rac1.
Expression of activated RhoA results in marked simplification of the dendritic tree
Although expression of activated Rac1 had a mild effect on the overall dendritic branch complexity (discounting the filopodia-like thin processes), expression of activated RhoA (RhoAV14, analogous mutation to Rac1V12) caused a drastic simplification of the dendritic tree compared with those of neurons expressing mCD8 alone (Fig.5A,B). This effect was evident 1 d after transfection and persisted for 2 and 3 d after transfection. Branch segment quantification (Fig. 5E) indicated a highly penetrant and significant simplification of both the apical and basal dendrites as early as 1 d after transfection of RhoAV14 (t test, p < 0.001 for both apical and basal). The Sholl profile indicates that both the length and branching pattern of RhoAV14-expressing neurons are affected (Fig.5G; repeated measures ANOVA, p < 0.001). Interestingly, there appeared to be a recovery of dendritic simplification after 3 d of RhoAV14 transfection, as seen by quantification of both apical and basal dendritic segment numbers (Fig.5E), despite the constant level of RhoAV14 expression as judged by myc staining (data not shown).
Fig. 5.
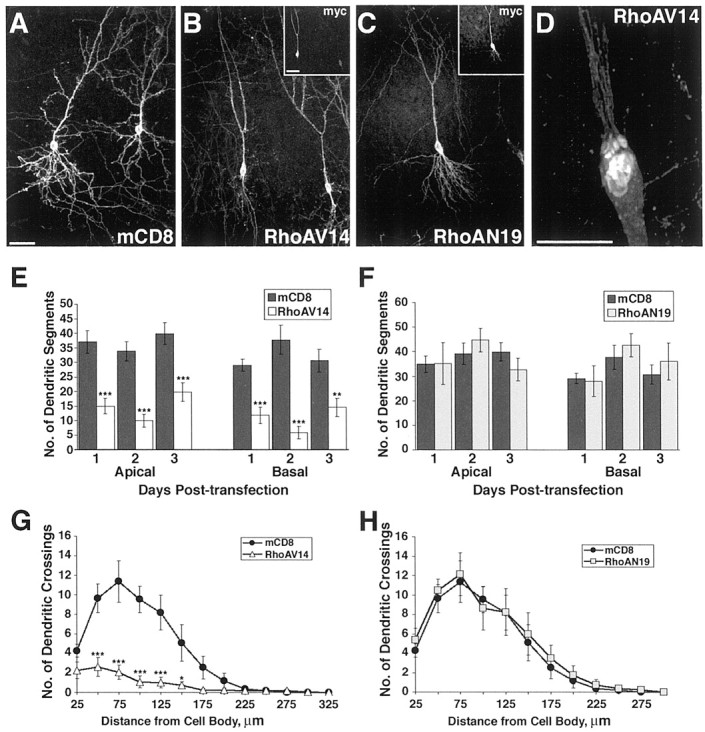
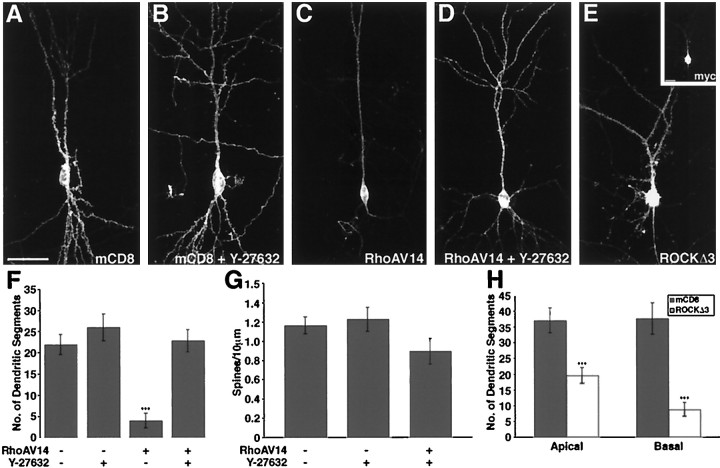
Expression of activated RhoA results in dendritic simplification. A–C, Representative images of pyramidal neurons that have expressed the marker mCD8 alone (A), mCD8 and myc-tagged RhoAV14 (B), or mCD8 and myc-tagged RhoAN19 (C) for 1 d (composite confocal images using 16× objective). Insets, Myc immunostaining for RhoAV14 (B) and RhoAN19 (C).D, Tip of an apical dendrite from a pyramidal neuron expressing RhoAV14 for 2 d (composite confocal image using 40× objective, with a digital zoom factor of 3). The soma is toward thebottom of the image. E, Quantification of dendritic branch segments after 1, 2, and 3 d of RhoAV14 expression. Both apical and basal dendrites exhibit a reduced number of dendritic segments (see Materials and Methods) with RhoAV14 expression compared with that of neurons expressing mCD8 alone (***p < 0.001; **p < 0.01; for 1, 2, 3 d, apical mCD8, n =14, 13, 17, and RhoAV14,n = 25, 19, 23; basal mCD8, n = 11, 14, 18, and RhoAV14, n = 25, 18, 22). F,Quantification of dendritic branch segments after 1, 2, and 3 d of RhoAN19 expression. Neurons expressing RhoAN19 do not show a change in dendritic branch segment number (for 1, 2, 3 d, apical,n = 9, 10, 7; basal, n = 7, 11, 7, respectively). G, H, Sholl profiles for basal dendrites of CA1 neurons 2 d after expressing mCD8 alone (n = 11) or expressing RhoAV14 (G;n = 16) or RhoAN19 (H;n = 8). Scale bars: A–C andinsets, 50 μm; D, 10 μm.
Strikingly, many neurons expressing activated RhoA (80%) had unusual structures at the end of their reduced dendrites: threads of extremely thin processes trailing behind the shortened dendrites (Fig.5D) resembling certain aspects of retracting axon terminals (for review, see Bernstein and Lichtman, 1999). These shortened dendrites often exhibited a fattened portion that appeared to have an increased aggregation of cytoplasm and membranous structures on the basis of the intensity of the cytoplasmic-myc and plasma membrane-localized CD8 labeling.
Perturbation of endogenous RhoA activity with transfection of the dominant-negative RhoA construct (RhoAN19, analogous mutation as Rac1N17) did not affect the dendritic morphology of our pyramidal neurons (Fig. 5C,F,H) despite their high level of expression (see Fig. 5C, inset). In addition, expression of RhoAN19 did not significantly affect the spine density when compared with control neurons 2 d after transfection (p = 0.56; n = 15 and 13 for RhoAN19 and mCD8 alone, respectively), supporting the specificity of the spine reduction effect seen with dominant-negative Rac1N17 expression.
The ROCK inhibitor blocks RhoAV14-induced dendritic simplification
RhoA acts via many different effector molecules to regulate diverse cellular functions, including organization of the actin cytoskeleton. To investigate the pathway by which RhoA regulates dendritic complexity, we tested the involvement of ROCK, because ROCK inhibition has been shown to block Rho-induced cell rounding and process retraction in cultured neuroblastoma cells (Hirose et al., 1998). We applied the ROCK inhibitor Y-27632 (100 μm) (Uehata et al., 1997) in culture media at the time of transfection. We limited our quantification to the basal dendrites of neurons 2 d after transfection because the dendritic simplification was most robust at this time, and both apical and basal dendrites were equally affected by RhoAV14 expression (Fig. 5E).
The treatment of slices with Y-27632 alone did not result in a significant change in dendritic complexity (Fig.6B,F; p= 0.30). Remarkably, Y-27632 treatment completely blocked the dendritic simplification associated with RhoAV14 expression (Fig. 6, compareD, C). As is quantified in Figure6F, although RhoAV14 expression caused a fivefold decrease in dendritic segments, application of Y-27632 restored the number of dendritic segments to the level of our control neurons. This result suggests that ROCK is a mediator of RhoAV14-induced dendritic simplification.
Fig. 6.
Function of ROCK in the maintenance of dendritic branches. A–E, Representative images of neurons 2 d after transfection and 100 μm Y-27632 treatment are shown. Neurons were transfected with mCD8 alone (A, B) or cotransfected with RhoAV14 (C, D) or ROCKΔ3 (E). E, Inset, The myc immunostaining for ROCKΔ3 is shown. Additionally, neurons inB and D were treated with 100 μm Y-27632 at the time of transfection (composite confocal images using 16× objective). F, Quantification of basal dendritic branch segment numbers of mCD8- and of mCD8 plus RhoAV14-expressing neurons with or without Y-27632 treatment is shown. Y-27632 treatment alone does not affect dendritic segment number (p = 0.31; control treatment,n = 23; Y-27632 treatment, n = 23). Y-27632 treatment blocks RhoAV14-associated dendritic segment reduction (***p < 0.001; RhoAV14 + control,n = 17; RhoAV14 expression + Y-27632 treatment,n = 24). G, Y-27632 application does not alter the basal dendritic spine density of neurons expressing mCD8 alone (p = 0.65; n = 29, 21 for control, Y-27632 treatment, respectively). Y-27632 treatment is capable of restoring the spine density of neurons expressing RhoAV14 close to control level (p = 0.08;n = 16 for Y-27632 treatment and RhoA V14 expression). H, Activated ROCKΔ3 expression results in significant reduction of dendritic segments compared with that in mCD8 alone (p < 0.001 for both apical and basal;n = 19, 20, respectively, for ROCKΔ3). Scale bars, 50 μm.
The significant dendritic simplification induced with RhoAV14 expression impeded our ability to quantify the density of dendritic spines. On a qualitative level it appeared that expression of activated RhoA resulted in a loss of adult-like dendritic spines. Interestingly, although Y-27632 alone does not affect spine density, it also restores spine density of RhoAV14-expressing neurons close to the control level (Fig. 6G; p = 0.08).
ROCK activation is sufficient to induce dendritic simplification
We next tested whether activation of ROCK is sufficient to induce dendritic simplification. Pyramidal neurons were transfected with a truncation mutant, ROCKΔ3, which eliminates the negative regulatory domain in the C terminal of the ROCK protein and has been shown to behave as a constitutively active version of ROCK in cultured cells (Ishizaki et al., 1997). We found that ROCKΔ3 expression resulted in the reduction of dendritic complexity to an extent similar to that of RhoAV14 expression (Fig. 6E,H), suggesting that ROCK activation is not only necessary but also sufficient in inducing the pruning of dendritic branches.
DISCUSSION
Using biolistic transfection to introduce dominant mutants of Rac1 and RhoA acutely in hippocampal pyramidal neurons with established dendritic arbors and some adult-like dendritic spines, we have shown that these GTPases play important roles in the maintenance and reorganization of dendritic structures. Rac1 seems to have preferential roles in regulating spine morphogenesis, whereas RhoA is implicated in limiting the growth of dendritic branches. The fact that expression of analogous dominant-negative or activated mutants of these similar GTPases gave qualitatively different phenotypes argues for the relative specificity of these perturbation experiments.
Generation and maintenance of dendritic spines
Although dendritic spines are important sites of synaptic input and plasticity (Harris and Kater, 1994; Yuste and Tank, 1996), little is known about the molecular mechanisms that regulate their morphogenesis (Harris, 1999). We have shown previously that transgenic expression of activated Rac1 in mouse Purkinje cells results in drastic changes in dendritic spine morphology (Luo et al., 1996). In those experiments, transgenes were expressed before the onset of dendritic growth and remained active thereafter. Although we concluded that hyperactivation of Rac affects the development of dendritic spines, we could not determine whether regulation of Rac activity is important for the dynamic changes of dendritic spines in more mature neurons, which may contribute to the morphological plasticity of neurons underlying learning and memory (Harris and Kater, 1994).
In this study, we have extended the previous findings in several new directions. First, because expression of activated Rac1 mutants in hippocampal pyramidal neurons results in a spine perturbation phenotype analogous to transgenic expression of activated Rac1 in Purkinje cells, it seems that the effect of increased Rac1 activity in dendritic spine morphogenesis is not specific to Purkinje cells. A notable difference between the two experimental conditions, however, is the presence of profuse long filopodial-like extensions in this study (Fig. 4) that were not observed in the previous transgenic studies (Luo et al., 1996). This could be caused by the differences in cell types, transgene expression level, and the relative timing of transfection versus neuronal differentiation. For instance, in the transgenic experiments, neurons may have adapted to the constant high level of Rac1 activity over a long period of time, such that the filopodial response is diminished. A second new insight gained from the current study is that activated Rac1 expression perturbs the maintenance of spines, because at the time of transfection these pyramidal neurons already possess dendritic spines.
Third and more importantly, our current study finds that expression of dominant-negative Rac1 resulted in a progressive reduction of spine number in both apical and basal dendrites. Taken together with the expression of Rac1 mRNA in the hippocampal pyramidal neurons at the time of our experiments, these results suggest that endogenous Rac1 is used for the maintenance of spine density in maturing neurons. Because of the slow but significant turnover of dendritic spines at analogous developmental stages reported from several recent live-imaging studies (Engert and Bonhoeffer, 1999; Horch et al., 1999), we cannot distinguish between the following two possibilities. (1) Rac1 is required for the generation of new spines. The net spine loss over time is a result of the failure of new spine formation while existing spines naturally turn over. (2) Rac1 is required for the maintenance of existing spine structure. Reduction in Rac activity speeds up the turnover rate of existing spines. Future time-lapse observation of dominant-negative Rac1-transfected neurons may help to distinguish between these two possibilities.
Interestingly, the number of PSD-95:GFP clusters, markers for postsynaptic density, decreases with Rac1N17 expression compared with controls. Although it is not possible to conclude whether loss of Rac1 activity causes a loss of PSD-95 clustering before spine structure loss or vice versa, this result strongly suggests that synapses are lost with concomitant loss of dendritic spines with Rac1N17 expression. It will be interesting to determine whether Rac1 activity directly regulates the development and maintenance of the postsynaptic density or whether it acts on the overall integrity of the dendritic spine itself.
Rac has been shown to regulate a number of biological processes including lamellipodia formation, transcription regulation, and cell cycle progression via activation of distinct effectors and downstream signaling pathways (Van Aelst and D'Souza-Schorey, 1997). We attempted to define the downstream signal transduction pathways by using an activated Rac1 with specific effector domain mutations shown previously to bind to a subset of effectors and have a subset of biological effects (Joneson et al., 1996; Lamarche et al., 1996). We found that the effector domain mutant shown previously to block the ability of Rac1L61 to induce lamellipodial formation in fibroblasts (Rac1L61A37) eliminates Rac1L61-induced morphological changes in pyramidal neurons. The similar properties of Rac1L61A37 on spine morphogenesis and lamellipodial formation, along with the fact that Rac proteins are found throughout the dendritic tree (Fig. 3A), suggest that Rac's role in spine morphogenesis is more likely via local regulation of the actin cytoskeleton.
Generation and maintenance of dendritic branches
Previous studies have implied that the Rho GTPases have different effects on the growth of axons and neuronal processes from neuronal cell lines and primary neuronal cultures. It has been generally thought that although Rac and Cdc42 have positive effects on process extension, Rho has a negative effect on process outgrowth or a positive effect on process retraction. Thus Rho activation opposes the effect of Rac (seeKozma et al., 1997; Van Leeuwen et al., 1997). An exception to this generalization was reported by Threadgill et al. (1997), who showed similar effects of Rho and that of Cdc42 and Rac on dendritic as well as axonal growth in dissociated mammalian cortical neurons in culture.
We investigated the effects of Rac1 and RhoA in the regulation of dendritic branch dynamics in neurons that have a relatively mature and established dendritic tree (Fig. 2B). Expression of dominant-negative and constitutively active Rac1, although profoundly affecting dendritic spines, had a relatively mild effect on the overall dendritic tree complexity (Figs. 3, 4). In contrast, expression of activated RhoA resulted in a drastic reduction of dendritic branches (Fig. 5B,E,G).
Several lines of evidence suggest that the dendritic simplification associated with RhoAV14 expression is caused by the retraction of existing dendritic branches. First, at the time of transfection the dendrites are more complex than after 24 hr of RhoAV14 expression (compare Figs. 2B, 5B). The dynamics of dendrite addition and elimination (Dailey and Smith, 1996) is too slow for the simple blockage of new dendrite formation to explain the degree of simplification observed after 1 d of RhoAV14 expression. Second, if the elimination of existing dendrites were caused by local degeneration of dendrites, one would expect to catch remnants of such events. Although we occasionally observed this in all transfection conditions when the slice culture was not healthy, we observed no increase of such remnants above background associated with RhoAV14-transfected neurons. Finally, in most of the RhoAV14-expressing neurons, we observed a unique enlargement at the end of their dendrites. It appeared that the dendrites have an aggregation of cytoplasm and plasma membrane at the tip of the simplified dendrites. The detailed process involved in the reduction of the dendritic branches and the identity of the enlarged end structure remain to be characterized by live imaging and electron microscopy, respectively.
Several different effectors have been identified for RhoA signaling to the actin cytoskeleton (for review, see Narumiya, 1996; Van Aelst and D'Souza-Schorey, 1997). In particular, a multidomain serine/threonine kinase ROCK has been shown to mediate RhoA's effect on cell rounding and process retraction from cultured neuroblastoma cells (Hirose et al., 1998). This RhoA-mediated retraction is likely to result from RhoA regulation, via ROCK, of myosin light chain phosphorylation and actomyosin contractility (Jalink et al., 1994; Kimura et al., 1996;Hirose et al., 1998). We have extended these cell culture studies into hippocampal pyramidal neurons in slice cultures that retain a relatively native environment. We found that treatment with the ROCK inhibitor Y-27632 (Uehata et al., 1997) effectively blocked the dendritic simplification caused by RhoAV14 expression, indicating that ROCK is necessary for Rho-mediated dendritic branch reduction. The fact that expression of an activated ROCK mutant mimics the effect of RhoAV14 expression further supports the importance of the RhoA–ROCK pathway in mediating dendritic branch elimination.
Bito et al. (2000) have demonstrated recently that the RhoA–ROCK pathway is vital for axonogenesis in cerebellar granule cells. Despite the expression of RhoA mRNA and the highly penetrant dendritic simplification phenotype with either activated RhoA or ROCK, we did not detect any significant effect of inhibiting the activity of RhoA (by expressing dominant-negative RhoAN19) or ROCK (by application of Y-27632) on dendritic growth and branching complexity. One explanation is that RhoAN19 is not sufficient to block endogenous Rho activity, although similar treatment has been shown to be effective in blocking axonogenesis (Bito et al., 2000). We favor the hypothesis that the RhoA–ROCK pathway, although intact in these neurons, is primarily inactive during normal physiological conditions, thus allowing for the maintenance of the dendritic tree (and their spines). Inhibition of a primarily inactive pathway would not result in detectable consequences. The fact that inhibition of ROCK activity by itself does not result in any significant change of dendritic complexity, but can potently inhibit the effect of RhoA activation, strongly supports this possibility. Such an intact yet dormant signaling pathway may be useful in reorganizing local dendritic branches in response to local activation of RhoA.
In a separate study, we showed that removal of RhoA activity by null mutations of RhoA in Drosophila mushroom body neurons in mosaic animals resulted in an overextension of dendrites. Conversely, expression of activated RhoA in mushroom body neurons resulted in a significant reduction of dendritic field volume and complexity (Lee et al., 2000). Taken together, these studies indicate that RhoA plays an evolutionarily conserved role in limiting dendritic growth and complexity.
Functional significance of dendritic structural changes in normal physiology and in mental retardation
One implication from our study is that molecules and mechanisms used for initial dendritic morphogenesis may be reused in maturing neurons for the continuous reorganization of neuronal cytoarchitecture, possibly mediating plasticity in response to environmental changes. We found that similar to their effect in development, Rac1 had a positive effect on dendritic outgrowth (at the level of dendritic spines and filopodia-like extensions), although hyperactivation of Rac could lead to disruption of normal spine morphology. On the other hand, RhoA hyperactivation had a negative effect on the maintenance of dendritic branches. Recent findings demonstrate that dendritic spines can undergo dynamic changes in response to synaptic stimulation under conditions that generate long-term potentiation (Engert and Bonhoeffer, 1999; Toni et al., 1999) or to local increases in the neurotrophin BDNF (Horch et al., 1999). Estrogen treatment also increases dendritic spine densityin vivo (Woolley and McEwen, 1993). Regulation of Rac GTPase activity may be one way these extracellular factors and synaptic activity are able to exert their effects on dendritic spines. It is noteworthy that links between RhoA and neurotrophin receptor p75NTR and NMDA have been suggested by two recent studies (Yamashita et al., 1999; Li et al., 2000).
One consequence of how the misregulation of the Rho family of GTPases may adversely affect the physiology of neurons comes from the recent identification of genes responsible for nonspecific X-linked mental retardation (for review, see Chelly, 1999). Two of the first three identified genes encode components of the Rho GTPase-signaling pathways, oligophrenin (Billuart et al., 1998) and Pak3 (Allen et al., 1998). Oligophrenin possesses a GAP domain for Rho GTPases and has GAP activity in vitro for Rho, Rac, and Cdc42 (Billuart et al., 1998). Loss of GAP activity would increase the activity of Rho GTPases and would lead to disruption of spine morphology or pruning of dendritic branches according to our present study. Pak3 belongs to a family of p21-activated kinases that was identified as effectors of Rac and Cdc42 (Manser et al., 1994). Although our effector domain mutant analysis did not implicate Pak as directly responsible for mediating the effect of activated Rac1 on the disruption of spine morphology, Pak has also been shown recently to act upstream of Rac (Obermeier et al., 1998). Pak can also act downstream of Rac1 indirectly via association with the p35/Cdk5 complex, whose binding to Rac1L61 is abolished by both F37A and Y40K mutations (Nikolic et al., 1998). Dominant-negative Cdk5 has been shown to block the effects of Rac1 in axonogenesis (Ruchhoeft et al., 1999). Taken together, these findings suggest that the regulation of Rho GTPase-signaling pathways is important for the generation and maintenance of neuronal morphology that may eventually contribute to proper mental function.
Footnotes
McKnight, Klingenstein, and Sloan Fellowships to L.L. supported this study. A.Y.N. is a Howard Hughes Medical Institute predoctoral fellow. We are grateful to Larry Katz for introducing us to the biolistic transfection method and to Matt Scott for use of his equipment. We thank members of the laboratories of Sue McConnell, Stephen Smith, and Dan Madison and in particular Jim Weimann, Ami Okada, Aparna Desai, Jack Waters, and Eric Schaible for sharing experimental expertise essential for our study. We also thank Marc Symons, Rong-Guo Qiu, Li-Huei Tsai, Alan Hall, Jun-ichi Miyazaki, Morgan Sheng, Haruhiko Bito, and Shuh Narumiya for plasmids and Yoshitomi Pharmaceuticals for the Y-27632 compound. We thank Ben Barres, Stephen Smith, Li-Huei Tsai, and members of the Luo laboratory for stimulating discussions and their comments on this manuscript. We thank Haruhiko Bito and Shuh Narumiya for communicating results before publication.
A.Y.N. and M.B.H. contributed equally to this work.
Correspondence should be addressed to Dr. Liqun Luo, Department of Biological Sciences, Stanford University, 371 Serra Mall, Herrin Labs 144A, Stanford, CA 94305-5020. E-mail: lluo@stanford.edu.
REFERENCES
- 1.Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- 2.Arnold D, Feng L, Kim J, Heintz N. A strategy for the analysis of gene expression during neural development. Proc Natl Acad Sci USA. 1994;91:9970–9974. doi: 10.1073/pnas.91.21.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold DB, Clapham DE. Molecular determinants for subcellular localization of PSD-95 with an interacting K+ channel. Neuron. 1999;23:149–157. doi: 10.1016/s0896-6273(00)80761-8. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein M, Lichtman JW. Axonal atrophy: the retraction reaction. Curr Opin Neurobiol. 1999;9:364–370. doi: 10.1016/s0959-4388(99)80053-1. [DOI] [PubMed] [Google Scholar]
- 5.Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet MC, Zemni R, Crollius HR, Carrie A, Fauchereau F, Cherry M, Briault S, Hamel B, Fryns JP, Beldjord C, Kahn A, Moraine C, Chelly J. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;39:923–926. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- 6.Bito H, Furuyashiki T, Ishihara H, Shibasaki Y, Ohashi K, Mizuno K, Maekawa M, Ishizaki T, Narumiya S. A critical role for a Rho-associated kinase p160ROCK in determining axon outgrowth in mammalian CNS neurons. Neuron. 2000;26:431–441. doi: 10.1016/s0896-6273(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 7.Boyer C, Schikorski T, Stevens CF. Comparison of hippocampal dendritic spines in culture and in brain. J Neurosci. 1998;18:5294–5300. doi: 10.1523/JNEUROSCI.18-14-05294.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chelly J. Breakthroughs in the molecular and cellular mechanisms underlying X-linked mental retardation. Hum Mol Genet. 1999;8:1833–1838. doi: 10.1093/hmg/8.10.1833. [DOI] [PubMed] [Google Scholar]
- 9.Dailey ME, Smith SJ. The dynamics of dendritic structure in developing hippocampal slices. J Neurosci. 1996;16:2983–2994. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drakew A, Muller M, Gahwiler BH, Thompson SM, Frotscher M. Spine loss in experimental epilepsy: quantitative light and electron microscopic analysis of intracellularly stained CA3 pyramidal cells in hippocampal slice cultures. Neuroscience. 1996;70:31–45. doi: 10.1016/0306-4522(95)00379-w. [DOI] [PubMed] [Google Scholar]
- 11.Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- 12.Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 13.Frantz GD, Bohner AP, Akers RM, McConnell SK. Regulation of the POU domain gene SCIP during cerebral cortical development. J Neurosci. 1994;14:472–485. doi: 10.1523/JNEUROSCI.14-02-00472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo X, Chandrasekaran V, Lein P, Kaplan PL, Higgins D. Leukemia inhibitory factor and ciliary neurotrophic factor cause dendritic retraction in cultured rat sympathetic neurons. J Neurosci. 1999;19:2113–2121. doi: 10.1523/JNEUROSCI.19-06-02113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 16.Harris KM. Structure, development, and plasticity of dendritic spines. Curr Opin Neurobiol. 1999;9:343–348. doi: 10.1016/s0959-4388(99)80050-6. [DOI] [PubMed] [Google Scholar]
- 17.Harris KM, Kater SB. Dendritic spines: cellular specializations imparting both stability and flexibility to synaptic function. Annu Rev Neurosci. 1994;17:341–371. doi: 10.1146/annurev.ne.17.030194.002013. [DOI] [PubMed] [Google Scholar]
- 18.Hirose M, Ishizaki T, Watanabe N, Uehata M, Kranenburg O, Moolenaar WH, Matsumura F, Maekawa M, Bito H, Narumiya S. Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells. J Cell Biol. 1998;141:1625–1636. doi: 10.1083/jcb.141.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horch HW, Kruttgen A, Protbury SD, Katz LC. Destabilization of cortical dendrites and spines by BDNF . Neuron. 1999;23:353–364. doi: 10.1016/s0896-6273(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 20.Ishizaki T, Naito M, Fujisawa K, Maekawa M, Watanabe N, Saito Y, Narumiya S. p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett. 1997;404:118–124. doi: 10.1016/s0014-5793(97)00107-5. [DOI] [PubMed] [Google Scholar]
- 21.Jalink K, van Corven EJ, Hengeveld T, Morii N, Narumiya S, Moolenaar WH. Inhibition of lysophosphatidate- and thrombin-induced neurite retraction and neuronal cell rounding by ADP ribosylation of the small GTP-binding protein Rho. J Cell Biol. 1994;126:801–810. doi: 10.1083/jcb.126.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. RAC regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 23.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 24.Kozma R, Sarner S, Ahmed S, Lim L. Rho family GTPases and neuronal growth cone remodeling: relationships between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol Cell Biol. 1997;17:1201–1211. doi: 10.1128/mcb.17.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamarche N, Tapon N, Stowers L, Burbelo PD, Aspenstrom P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 26.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 27.Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- 28.Lein P, Johnson M, Guo X, Rueger D, Higgins D. Osteogenic protein-1 induces dendritic growth in rat sympathetic neurons. Neuron. 1995;15:597–605. doi: 10.1016/0896-6273(95)90148-5. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Van Aelst LV, Cline HT. Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat Neurosci. 2000;3:217–225. doi: 10.1038/72920. [DOI] [PubMed] [Google Scholar]
- 30.Liaw CW, Zamoyska R, Parnes JR. Structure, sequence, and polymorphism of the Lyt-2 T cell differentiation antigen gene. J Immunol. 1986;137:1037–1043. [PubMed] [Google Scholar]
- 31.Lo DC, McAllister AK, Katz LC. Neuronal transfection in brain slices using particle-mediated gene transfer. Neuron. 1994;13:1263–1268. doi: 10.1016/0896-6273(94)90412-x. [DOI] [PubMed] [Google Scholar]
- 32.Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- 33.Luo L, Jan LY, Jan YN. Rho family small GTP-binding proteins in growth cone signalling. Curr Opin Neurobiol. 1997;7:81–86. doi: 10.1016/s0959-4388(97)80124-9. [DOI] [PubMed] [Google Scholar]
- 34.Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- 35.Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 1999;283:923–927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- 36.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 37.McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 38.McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 39.Mueller BK. Growth cone guidance: first steps towards a deeper understanding. Annu Rev Neurosci. 1999;22:351–388. doi: 10.1146/annurev.neuro.22.1.351. [DOI] [PubMed] [Google Scholar]
- 40.Narumiya S. The small GTPase Rho: cellular functions and signal transductions. J Biochem. 1996;120:215–228. doi: 10.1093/oxfordjournals.jbchem.a021401. [DOI] [PubMed] [Google Scholar]
- 41.Nedivi E, Wu GY, Cline HT. Promotion of dendritic growth by CPG15, an activity-induced signaling molecule. Science. 1998;281:1863–1866. doi: 10.1126/science.281.5384.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikolic M, Chou MM, Lu W, Mayer BJ, Tsai LH. The p35/Cdk5 kinase is a neuron-specific Rac effector that inhibits Pak1 activity. Nature. 1998;395:194–198. doi: 10.1038/26034. [DOI] [PubMed] [Google Scholar]
- 43.Obermeier A, Ahmed S, Manser E, Yen SC, Hall C, Lim L. PAK promotes morphological changes by acting upstream of Rac. EMBO J. 1998;17:4328–4339. doi: 10.1093/emboj/17.15.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada A, Weimann JM, Fraser SE, McConnell SK. Imaging cells in the developing nervous system with retrovirus expressing modified green fluorescent protein. Exp Neurol. 1999;156:394–406. doi: 10.1006/exnr.1999.7033. [DOI] [PubMed] [Google Scholar]
- 45.Purves D, Hadley RD. Changes in the dendritic branching of adult mammalian neurones revealed by repeated imaging in situ. Nature. 1985;315:404–406. doi: 10.1038/315404a0. [DOI] [PubMed] [Google Scholar]
- 46.Purves D, Hadley RD, Voyvodic JT. Dynamic changes in the dendritic geometry of individual neurons visualized over periods of up to three months in the superior cervical ganglion of living mice. J Neurosci. 1986;6:1051–1060. doi: 10.1523/JNEUROSCI.06-04-01051.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purves D, Snider WD, Voyvodic JT. Trophic regulation of nerve cell morphology and innervation in the autonomic nervous system. Nature. 1988;336:123–128. doi: 10.1038/336123a0. [DOI] [PubMed] [Google Scholar]
- 48.Qiu RG, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995a;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 49.Qiu RG, Chen J, Kirn D, McCormick F, Symons M. A role for Rho in Ras transformation. Proc Natl Acad Sci USA. 1995b;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rall W. Theoretical significance of dendritic trees for neuronal input-output relations. In: Reiss RF, editor. Neural theory and modeling. Stanford UP; Palo Alto, CA: 1964. pp. 63–97. [Google Scholar]
- 51.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 52.Ruchhoeft ML, Ohnuma S, McNeill L, Holt CE, Harris WA. The neuronal architecture of Xenopus retinal ganglion cells is sculpted by Rho-family GTPases in vivo. J Neurosci. 1999;19:8454–8463. doi: 10.1523/JNEUROSCI.19-19-08454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- 54.Snider WD. Nerve growth factor enhances dendritic arborizations of sympathetic ganglion cells in developing mammals. J Neurosci. 1988;8:2628–2634. doi: 10.1523/JNEUROSCI.08-07-02628.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoppini L, Buchs DA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 56.Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 57.Toni N, Buchs PA, Nikonenko I, Bron CR, Muller D. LTP promotes formation of multiple spine synapses between a single axon terminal and a dendrite. Nature. 1999;402:421–424. doi: 10.1038/46574. [DOI] [PubMed] [Google Scholar]
- 58.Uehata M, Ishizaki T, Satoh H, Ono T, Kawahara T, Morishita T, Tamakawa H, Yamagami K, Inui J, Maekawa M, Narumiya S. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 59.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 60.Van Leeuwen FN, Kain HET, van der Kammen RA, Nichiels F, Kranenburg OW, Collard JG. The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J Cell Biol. 1997;139:797–807. doi: 10.1083/jcb.139.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woolley CS, McEwen BS. Roles of estradiol and progesterone in regulation of hippocampal dendritic spine density during the estrous cycle in the rat. J Comp Neurol. 1993;336:293–306. doi: 10.1002/cne.903360210. [DOI] [PubMed] [Google Scholar]
- 62.Wright DE, Snider WD. Neurotrophin receptor mRNA expression defines distinct populations of neurons in rat dorsal root ganglia. J Comp Neurol. 1995;351:329–338. doi: 10.1002/cne.903510302. [DOI] [PubMed] [Google Scholar]
- 63.Yamashita T, Tucker KL, Barde YA. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron. 1999;24:585–593. doi: 10.1016/s0896-6273(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 64.Yuste R, Tank DW. Dendritic integration in mammalian neurons, a century after Cajal. Neuron. 1996;16:701–716. doi: 10.1016/s0896-6273(00)80091-4. [DOI] [PubMed] [Google Scholar]