Abstract
Five novel peptides were identified in the brains of mice lacking active carboxypeptidase E, a neuropeptide-processing enzyme. These peptides are produced from a single precursor, termed proSAAS, which is present in human, mouse, and rat. ProSAAS mRNA is expressed primarily in brain and other neuroendocrine tissues (pituitary, adrenal, pancreas); within brain, the mRNA is broadly distributed among neurons. When expressed in AtT-20 cells, proSAAS is secreted via the regulated pathway and is also processed at paired-basic cleavage sites into smaller peptides. Overexpression of proSAAS in the AtT-20 cells substantially reduces the rate of processing of the endogenous prohormone proopiomelanocortin. Purified proSAAS inhibits prohormone convertase 1 activity with an IC50 of 590 nmbut does not inhibit prohormone convertase 2. Taken together, proSAAS may represent an endogenous inhibitor of prohormone convertase 1.
Keywords: carboxypeptidase E, prohormone convertase, 7B2, secretogranin, chromogranin, neuropeptide
Most neuroendocrine peptides are initially synthesized as larger precursors that require proteolytic processing to generate the bioactive moiety. Typically, the precursors contain multibasic amino acid cleavage sites with the consensus Lys/Arg-Xn-Arg, where X is any amino acid except Cys, and n = 0, 2, 4, or 6 (Devi, 1991; Lindberg and Hutton, 1991). These multibasic sites are often cleaved by prohormone convertases 1 and 2 (PC1 and PC2) (Zhou et al., 1999). The production of active PC2 requires the chaperone protein 7B2, which also serves as an endogenous inhibitor of this enzyme (Braks and Martens, 1994; Martens et al., 1994; Benjannet et al., 1995; Zhu and Lindberg, 1995). Except for the pro region of PC1 (Boudreault et al., 1998a), no endogenous inhibitor of PC1 has been previously reported. After the action of the endopeptidases, most peptide processing intermediates contain C-terminal basic residues that generally need to be removed before the peptides are bioactive (Fricker, 1988, 1991). In some cases, additional post-translational modifications occur, such as acetylation, amidation, sulfation, phosphorylation, and glycosylation (Bennett, 1991; Bradbury and Smyth, 1991).
Carboxypeptidase E (CPE) is responsible for removing C-terminal basic residues from the majority of peptide intermediates (Fricker, 1988,1991). This enzyme is restricted to neuroendocrine tissues, but within these tissues is broadly distributed (Fricker et al., 1989; Schafer et al., 1993; Zheng et al., 1994). A naturally occurring mutation inCpefat/Cpefatmice causes CPE to be inactive and to be rapidly degraded soon after synthesis (Naggert et al., 1995). Mice lacking CPE activity are still capable of a reduced level of neuropeptide processing, which is presumably attributable to the activity of carboxypeptidase D (CPD) (Song and Fricker, 1995; Fricker et al., 1996). However, CPD can only partially compensate for the defective CPE in theCpefat/Cpefatmice because CPD levels in brain are lower than those of CPE (Song and Fricker, 1996). Furthermore, CPD is primarily present in thetrans-Golgi network and not the late secretory pathway where the majority of peptide processing occurs (Varlamov and Fricker, 1998;Varlamov et al., 1999).Cpefat/Cpefatmice show greatly elevated levels of peptide intermediates containing C-terminal basic residues, whereas these intermediates are virtually undetectable in tissues from control mice (Naggert et al., 1995;Fricker et al., 1996; Rovere et al., 1996). A comparison of peptides fromCpefat/Cpefatmouse brains with those isolated from control brains revealed numerous differences (L. D. Fricker and L. Yan, unpublished observations). Mass spectrometric sequence analysis of the most abundant of these peptides identified dozens of known neuropeptide intermediates as well as fragments of other proteins present in the secretory pathway such as chromogranins A and B, secretogranin II, and others (L. Fricker, H. Li, L. Yan, P. Courchesne, and S. Patterson, unpublished).
In this report, we describe the identification of five peptides that are substrates for CPE, based on the accumulation of C-terminally extended forms of the peptides inCpefat/Cpefatmouse brain. cDNA cloning established that all five peptides are encoded by the same precursor protein. Overexpression of the novel protein in a pituitary cell line blocks processing of an endogenous prohormone, proopiomelanocortin (POMC). Furthermore, the novel protein directly inhibits PC1, but not PC2, in an in vitro assay. Taken together, the novel protein appears to function as an endogenous inhibitor of prohormone convertase 1, although other functions are also possible.
MATERIALS AND METHODS
Mice (Cpefat/Cpefatand wild-type littermates) were obtained from The Jackson Laboratory (Bar Harbor, ME) and killed at 9–10 weeks of age. Brain, pituitary, and other tissues were removed and frozen on dry ice until analysis. Before freezing, a portion of the cerebral cortex was removed and used for purification of CPE to confirm the genotyping; only wild-type animals contain CPE that binds to the substrate affinity resin (Fricker et al., 1996; Varlamov et al., 1996). Brains from 10–20Cpefat/Cpefatmice or wild-type littermates were pooled and homogenized in 10 ml of boiling 0.1 m acetic acid. The homogenate was boiled for 10 min and then centrifuged at 50,000 × gfor 30 min at 4°C. The supernatant was filtered through a Centriplus-10 membrane (Amicon, Beverly, MA), and the flow through was combined with sodium acetate to give a final pH of 5.0. The material was applied to a 0.5 ml column containing immobilized anhydrotrypsin (Panvera). The column was washed with 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid in pH 5 sodium acetate, and then with sodium acetate alone. Bound peptides were eluted with 2 ml of 5 mm HCl, as described (Kumazaki et al., 1987).
The column eluate was lyophilized and resuspended in 50 μl of 15 mm Tris(2-carboxyethyl)phosphine. After 30 min at room temperature, 55 μl of 1% formic acid was added and aliquots injected onto an HPLC (HP1100; Hewlett-Packard, Palo Alto, CA) connected on-line to a Finnigan LCQ ion trap mass spectrometer (Finnigan Thermoquest, San Jose, CA) operated using previously described instrument parameters (Courchesne et al., 1998). Ions that were detected in the extracts fromCpefat/Cpefatmice and were not present in the extracts from control mice were further analyzed. MS/MS spectra from five peptides enriched inCpefat/Cpefatmouse brain did not match any known protein, as determined using a variety of computer programs (MS-Tag; Sherpa). One of these, a peptide with a mass of 2125, gave the partial MS/MS sequence [200]-SAASAP-I/L-VE-[1098] (note: Ile and Leu have the same mass and cannot be distinguished by MS/MS sequencing). Other peptides enriched inCpefat/Cpefatmouse brain gave the following partial sequences: parent mass = 3058, partial sequence = [1363]-SAP-I/L-VETS-[911]; parent mass = 2600, partial sequence = [714]-PEVPP-[1367]; parent mass = 1335, partial sequence = L/I-EN-[805]-R; and parent mass = 952, partial sequence = [227]-PV-K/Q-EPR (note: Lys and Gln have very similar masses and are difficult to distinguish by MS/MS sequencing).
Cloning and sequence analysis of proSAAS cDNA.Nucleotide databases were searched using the partial amino acid sequence derived from MS/MS fragmentation of the peptide with a mass of 2125. This search resulted in the identification of dozens of mouse, rat, and human expressed sequence tags (ESTs) encoding the predicted amino acid sequences. Assembly of contiguous ESTs, followed by 5′ and 3′ rapid amplification of cDNA ends allowed for the initial determination of full-length cDNA sequences encoding mouse, rat, and human SAAS.
PCR (using brain cDNA as a template source) with oligonucleotides to the 5′ and 3′ ends of predicted mouse, rat, and human SAAS cDNA was then performed to isolate and confirm the nucleotide sequence of full-length SAAS cDNA. The resulting cDNA fragments (∼1 kb in length) were subcloned into pBluescript SK-, and 8–12 independent clones from each species were subjected to nucleotide sequence analysis. Consensus nucleotide sequences were then derived representing full-length mouse, rat, and human SAAS mRNA. The nucleotide sequences of mouse, rat, and human proSAAS have been submitted to GenBank (accession numbersAF181560, AF181561, and AF181562, respectively).
Northern blot analysis. Northern blots containing various human poly(A) mRNAs were purchased from Clontech (Cambridge, UK). The membranes were prehybridized for 1 hr at 65°C in ExpressHyb (Clontech). The prehybridization buffer was discarded and replaced with fresh hybridization buffer containing full-length32P-radiolabeled human SAAS cDNA probe generated using the Prime-It RmT kit (Stratagene, La Jolla, CA). After hybridization for 16 hr at 65°C, membranes were washed with 0.1× SSC containing 0.1% SDS and 5 mm EDTA at 68°C for 1–4 hr, dried, and exposed to Kodak (Eastman Kodak, Rochester, NY) XAR-5 film.
In situ hybridization. Tissue was obtained from adult male Sprague Dawley rats (Charles River Laboratories, Wilmington, MA; 250–350 gm) that were quickly decapitated. The brains were removed and frozen in isopentane at −30°C for 30 sec, then stored at −80°C until they were cut on a Micron cryostat maintained at −20°C. Ten micrometer coronal sections were cut through the pituitary, adrenal cortex, spinal cord, and the rostrocaudal extent of the brain; sections were thaw-mounted onto polylysine-subbed slides and stored at −80°C until used for in situ hybridization.
Riboprobes complementary to rat SAAS mRNA were generated by labeling with 250 μCi of 35S-UTP (Amersham, Arlington Heights, IL) using 1 μl of the appropriate RNA polymerase in a 25 μl transcription reaction containing 5 μl of 5× transcription buffer, 2 μl of 0.1 m dithiothreitol, 12 μl of sterile water, 1 μl of linearized plasmid containing the ∼780 bp open reading frame of rat SAAS (1 μg/μl), 1 μl each of 10 mm stocks of ATP, CTP, and GTP, and 1 μl of RNasin. The transcription reaction was incubated for 2 hr at 37°C, and then the labeled probe was separated from unincorporated nucleotides on a Sephadex-G50 column.
Before hybridization, the sections were removed from storage at −80°C and placed in 4 gm% of paraformaldehyde for 1 hr at room temperature (RT). The slides were washed 3 times in 2× SSC (300 mm NaCl, 30 mm sodium citrate, pH 7.2), rinsed in water, and acetylated in triethanolamine and acetic anhydride (0.25%) for 10 min at RT. After acetylation, the slides were rinsed in 2× SSC and dehydrated through graded alcohols. Hybridization and posthybridization steps for the radioactive in situhybridization studies were as previously described (Curran and Watson, 1995). Briefly, the 35S-riboprobes were diluted in hybridization buffer to a final concentration of 1–2 × 106 dpm/35 μl. The hybridization buffer contained 75% formamide, 10% dextran sulfate, 3× SSC, 50 mm phosphate buffer, pH 7.4, 1× Denhardt's solution (0.02% polyvinyl pyrrolidone, 0.02% Ficoll, 0.02% bovine serum albumin), 0.1 mg/ml yeast tRNA, and 10 mmdithiothreitol. The tissue sections were apposed to the diluted probe with coverslips and then placed in a hybridization box containing Whatman filter paper moistened with 75% formamide. The hybridization boxes were then sealed and placed in a 55°C oven overnight. The next day the coverslips were removed with 2× SSC. The sections were washed 3 times in 2× SSC, treated with 200 μg/ml RNase A at 37°C for 1 hr, rinsed one time for 5 min each in 2× SSC, 1× SSC, and 0.5× SSC, washed for 1 hr at 70°C in 0.1× SSC, rinsed for 5 min in water at RT, and then dehydrated through graded alcohols. Dried slides were apposed to x-ray film for 1–10 d. These films were used to examine the macroscopic distribution of SAAS. After apposition to x-ray film, each slide was dipped in NTB-2 emulsion, dried, and stored at 4°C. Dipped slides were developed according to times determined by test slides, counterstained with cresyl violet (brain and spinal cord) or hematoxylin and eosin (adrenals and pituitary), and used to examine the cellular distribution of each mRNA.
Overexpression of rat proSAAS in AtT-20 cells. Full-length rat proSAAS was subcloned into the EcoRI/NotI sites of the pcDNA3 vector (Invitrogen, San Diego, CA), and the plasmid was transfected into AtT-20 cells using lipofectamine reagent (Life Technologies, Gaithersburg, MD). The stable cell lines were selected using 0.7 mg/ml geneticin (G418). The overexpressed proSAAS protein was identified by Western blot analysis using rabbit antisera raised to rat proSAAS 221–242 or 245–260. These antisera and an antiserum to rat proSAAS 42–59 were raised against synthetic peptides containing an additional N-terminal Cys residue that was conjugated to maleimide-activated keyhole limpet hemocyanin. For examination of immunocytochemistry, overexpressing proSAAS cell lines and wild-type AtT-20 cells were plated on glass growth-supporting coverslips (Fisher Scientific, Houston, TX), cultured for 2–3 d, and fixed in 4% paraformaldehyde for 10 min. The cells were rinsed in PBS, permeabilized in 0.1%Triton X-100 in PBS for 15 min, and then blocked in 5% bovine serum albumin for 1 hr. After blocking, the cells were immunostained for 1 hr with antiserum to proSAAS 245–260 (1:1000 dilution), cells were washed, and then fluorescein-labeled goat anti-rabbit IgG (1:200 dilutions) was added, and the cells were incubated in the dark for 1 hr. The coverslips were extensively washed, mounted on glass slides in 50% glycerol in PBS, and examined using a Bio-Rad (Hercules, CA) confocal microscope. The images represent a single plane of focus.
The ABI PRISM 7700 sequence detection system was used to quantitate levels of endogenous mouse SAAS, POMC, and PC1 mRNA, and exogenous rat SAAS mRNA in a variety of clonal AtT-20 cells line stably expressing transformed rat SAAS cDNA. PCR probe and primer oligonucleotides specific for the detection of the aforementioned species of mRNA were designed using Primer Express 1.0 software from ABI. Standard curves were generated using plasmid DNA containing the various cDNA inserts. Standard curve correlation coefficients of 0.995–1.000 were routinely observed over a 6 order of magnitude range of plasmid concentrations (representing from 50 to 50,000,000 molecules of target DNA sequence). Total RNA was isolated from control AtT-20 cells and four clonal cell lines stably expressing rat proSAAS from the pcDNA3 expression vector. cDNA was then generated and used as a sample source for quantitation of mRNAs noted above. Comparison of sample values with those from the plasmid standard curve allowed for the precise determination of mRNA copy number within the various AtT-20 cell lines.
Examination of regulated secretion. To examine whether proSAAS undergoes regulated secretion, four AtT-20 cell clones overexpressing proSAAS were cultured in 6-well plates to 90% confluency. The medium was removed, the cells were washed three times with PBS and then incubated in DMEM containing no addition or with either 10 μm forskolin or 5 mm8-Br-cAMP. After 30 min of incubation, the medium was removed and subjected to Western blot analysis on 10% polyacrylamide Tris-Tricine glycerol gels using antisera to proSAAS 245–260 (1:1000 dilution) or to the CPE C-terminal region (Varlamov et al., 1996) and the enhanced chemiluminescence procedure (Amersham).
Medium from the proSAAS-expressing cells and from wild-type cells was collected after secretagogue treatment with 5 mm 8-Br-cAMP, acidified with 0.1% trifluoroacetic acid (TFA), and applied to a C18 reverse phase Sep-Pak cartridge (Waters). The cartridge was washed with 0.1% TFA and then eluted with 80% acetonitrile in 0.1% TFA. The eluate was dried in a vacuum centrifuge, resuspended in 50 μl of water, and then an aliquot was combined with α-cyano-4-hydroxycinnamic acid and used for matrix-assisted laser desorption ionization time of flight mass spectrometry on a Perkin-Elmer (Norwalk, CT) DE-STR mass spectrometer.
Pulse chase analysis of AtT-20 cells. To examine the processing of proSAAS, AtT-20 cells expressing rat proSAAS were incubated in leucine-deficient medium for 1 hr at 37°C and then labeled (pulse) with [3H]leucine for 20 min. After the labeling, the cells were washed twice with PBS and incubated in DMEM for 0, 15, 30, or 90 min. Media and cells were then subjected to immunoprecipitation, using antisera to either the N-terminal region (residues 42–59) or the C-terminal region (residues 245–260) of rat proSAAS.
To examine whether the overexpression of proSAAS has an effect on POMC processing, wild-type AtT-20 cells and several clones expressing high levels of proSAAS were incubated in methionine-deficient medium for 1 hr and labeled (pulse) with [35S]methionine (150 μCi/ml) for 10 min. The cells were then washed twice with PBS and incubated with DMEM for 0, 30, or 90 min. Media and cells were then subjected to immunoprecipitation, using an antiserum to ACTH (JH 93) or an antiserum to β-endorphin (JH 2); the antisera were the generous gift of Drs. Richard Mains and Betty Eipper (Johns Hopkins University, Baltimore, MD). Immunoprecipitated peptides were analyzed on 10% polyacrylamide Tris-Tricine glycerol gels.
Inhibition of PC1 by proSAAS. Full-length rat proSAAS cDNA was subcloned into the bacterial expression plasmid pGEX2T (Pharmacia, Piscataway, NJ). Oligonucleotides were synthesized to create aBglII site on the 5′ end of the proSAAS cDNA, immediately downstream of the signal peptide cleavage site, and an EcoRI site on the 3′ end after the stop codon. After PCR with these oligonucleotides, the product was subcloned into the pGEX2T vector, and the nucleotide sequence was confirmed. The resulting vector contains glutathione S-transferase (GST) with proSAAS residues 34–260 attached to the C terminus of GST (GST-proSAAS). Both GST-proSAAS and GST alone were expressed in bacteria and purified using glutathione agarose columns, as described (Smith and Johnson, 1988). After elution of the protein from the glutathione agarose using glutathione, the protein was concentrated using centricon 10 (Amicon), and the buffer was changed to 10 mm TrisCl, pH 7.4. The material was then applied to a monoQ FPLC column, with a gradient from 0 to 500 mm NaCl in 10 mm Bis Tris, pH 6.0, buffer. The GST eluted at ∼50 mm NaCl, whereas the GST-proSAAS eluted at ∼250 mm NaCl. The purified GST ran as a single band of ∼27 kDa, and the purified GST-proSAAS as a single band of ∼52 kDa after denaturing PAGE. The concentration of each protein was determined from the absorption at 280 nm and the extinction coefficients of the two proteins (based on the number of Tyr, Phe, and Trp).
The reaction mixture for the PC1 assay contained pGlu-Arg-Thr-Lys-Arg-7-amino-4-methylcoumarinamide (RTKR-AMC, Peptides International) as the substrate in 100 mm sodium acetate buffer, pH 5.5, containing 5 mmCaCl2, 0.1% Triton X-100, and an inhibitor cocktail consisting of 1 μm pepstatin, 0.1 mmcaptopril, 0.3 mm tosylphenylalanylchloromethyl ketone, 1 μm trans-epoxysuccinic acid (E-64), and 0.15 mm tosyllysylchloromethyl ketone; this cocktail does not significantly affect PC1 activity. The reaction was initiated by the addition of PC1 to the mixture of substrate, buffer, inhibitor cocktail, and the indicated concentration of purified GST or GST-proSAAS. Conditioned medium from Chinese hamster ovary (CHO) cells expressing PC1 (a generous gift of Iris Lindberg, Louisiana State University Medical Center, New Orleans, LA), was used as the source of PC1 (Zhou and Lindberg, 1993). Similar results were obtained when the experiments were performed with medium from Sf9 cells infected with PC1-expressing baculovirus (a generous gift of Nabil Seidah; Clinical Research Institute of Montreal, Montreal, Québec Canada), prepared as described (Boudreault et al., 1998b). The fluorescence released was measured using a Perkin-Elmer spectrofluorimeter at various time intervals (30–300 min), and fluorescence units were quantitated using 7-amino-4-methylcoumarinamide (AMC) as the standard.
The PC2 assay was performed essentially as that of the PC1 assay, using medium from CHO cells expressing PC2 (Lamango et al., 1996), a generous gift of Iris Lindberg. Duplicate tubes were incubated with 1 μm CT peptide, a highly selective inhibitor of PC2 (Zhu et al., 1996). The difference in fluorescence between tubes without and with CT peptide was used as a measure of PC2 activity.
RESULTS
Several peptides were identified during a mass spectrometry-based screen for peptides that were enriched inCpefat/Cpefatmouse brain compared to the brains of control animals. Computer searches of mouse EST databases with the deduced partial amino acid sequences of the peptides revealed dozens of overlapping cDNA clones that encoded a single precursor protein containing five peptides that were enriched in fat/fat mouse brain (Fig.1). This protein has been termed “proSAAS” because one of the fat/fat mouse brain peptides contained the deduced partial amino acid sequence Ser-Ala-Ala-Ser. All five peptides that were identified by the mass spectrometric analysis are bracketed in the proSAAS precursor by basic amino acid proteolytic processing sites or by the predicted N-terminal signal peptide cleavage site. Proteolytic cleavage at a KxxR site within proSAAS gives rise to proSAAS 34–41 and 42–61. Detection of the peptide proSAAS 34–61 by mass spectrometry indicates that proteolytic cleavage at this KxxR site is not complete in theCpefat/Cpefatmouse brain.
Fig. 1.

Top, Nucleotide and deduced amino acid sequence of mouse proSAAS. The amino acid sequences of the peptides found to be elevated in tissues fromCpefat/Cpefatmice are underlined (double underlineindicates those peptides found in both a short and long form).Bottom, Amino acid identity among human, rat, and mouse proSAAS. Asterisks below the sequence denote residues conserved in all three species. Open arrowhead, Signal peptide cleavage site. Double arrows, Paired basic cleavage sites (RR, KR) that are used in theCpefat/Cpefatmouse. Single arrows, KxxR cleavage site used in theCpefat/Cpefatmouse. Half arrows, Additional predicted cleavage sites. The sequences of the peptides that correspond to those found in theCpefat/Cpefatmice (but without the C-terminal basic residues that would be removed by active CPE) are indicated by lines.
The full-length mouse proSAAS cDNA sequence was used to screen EST databases to obtain partial cDNA sequences for human and rat proSAAS. PCR was then used to generate full-length proSAAS cDNA for nucleotide sequence analysis. The amino acid sequence alignment of human, rat, and mouse proSAAS is shown in Figure 1 (bottom). The presence of a predicted 33 amino acid N-terminal signal sequence is conserved among the three species, as well as the endoprotease processing sites. Also conserved are several potential protein convertase cleavage sites (denoted by the half arrow), although there is currently no evidence to suggest that they are used in vivo. Overall amino acid sequence conservation is ∼97% between rat and mouse and 84% between human and rat/mouse. The central region of proSAAS (from ∼65–200) shows the highest degree of conservation across species, whereas the regions representing the five identified peptides (amino acids 34–61 and 221–256) show a lower degree of conservation. ProSAAS from all three species is enriched in Ala, Leu, Pro, and acidic residues (17, 16, 14, and 13%, respectively). The protein lacks Met and Cys residues (after removal of the N-terminal signal sequence) and has very few aromatic groups (<1% Tyr, Phe and Trp).
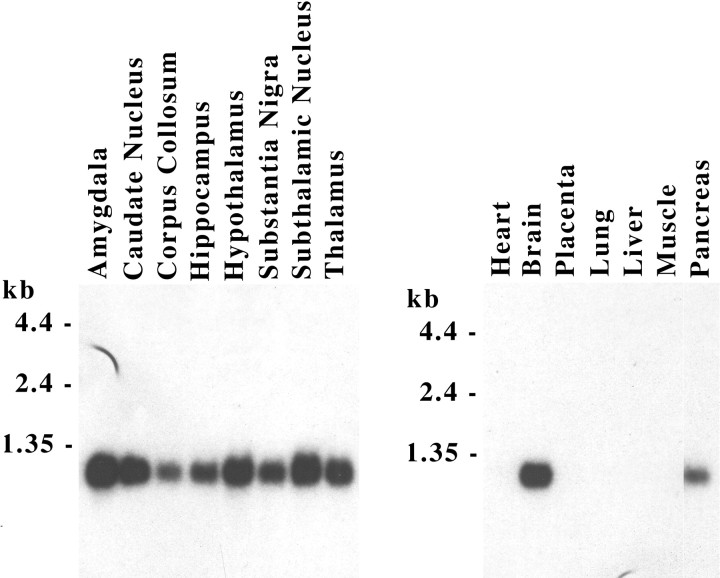
Northern blot analysis was used to determine the tissue expression pattern of the proSAAS transcript. The results shown in Figure2 reveal that proSAAS mRNA is widely distributed throughout the human brain and is also expressed in the pancreas. Northern analysis with a wide array of rat tissue RNAs (data not shown) also shows widespread expression throughout the CNS, as well as expression in endocrine tissues such as the pancreas, pituitary, and adrenal. Low but detectable levels of proSAAS mRNA were also observed in the rat eye, lung, and kidney, and in human kidney as well. Thus, proSAAS mRNA shows a distinct neuroendocrine pattern of expression. The ∼1.2 kb proSAAS mRNAs observed in both human (Fig. 2) and rat (data not shown) are consistent with the sizes of the corresponding full-length cDNA sequences isolated from human, rat, and mouse. A variety of mammalian neuroendocrine cell lines were also found to express proSAAS mRNA, including GH3 (rat pituitary somatotroph), PC12 (rat adrenal pheochromocytoma), and AtT-20 (mouse pituitary corticotroph) cells. In contrast, no proSAAS expression was observed in non-neuroendocrine fibroblast cell lines, including NIH-3T3, baby hamster kidney-21, CHO, or human embryonic kidney-293. Genomic Southern analysis (data not shown) using a radiation hybrid panel mapped the human proSAAS gene to marker SHGC-17255 (logarithm of the likelihood for linkage, 1000) on chromosome Xp11.3.
Fig. 2.
Northern blot analysis of proSAAS mRNA in human tissues. Northern blots containing poly(A) RNA (Clontech) were probed with 32P-labeled human proSAAS cDNA, as described in Materials and Methods, and were exposed to x-ray film for 2 hr at −80°C.
The distribution of proSAAS mRNA in the adult rat brain and endocrine tissues was further examined by in situ histochemistry (Fig.3A–H). In brain (Fig.3A–D), proSAAS mRNA shows a very widespread pattern of expression, with silver grains present in every major structural region. The medial hypothalamus, arcuate nucleus, supraoptic nucleus, and hippocampus are particularly robust with regard to proSAAS expression. In the adrenal gland (Fig. 3E,F), expression of proSAAS is localized to the medulla, with only background hybridization observed in the adrenal cortex. In the spinal cord (Fig.3G,H), proSAAS expression is widespread throughout both the dorsal and ventral horn. Furthermore, expression is limited to neuronal cells, with no silver grains observed over glial cells. Analysis of emulsion-dipped sections from brain also showed a cellular distribution that was restricted to neurons (data not shown). In the adult rat pituitary, proSAAS expression was seen in all cell types within the anterior and intermediate lobe, with no detectable expression seen in the posterior lobe (data not shown).
Fig. 3.
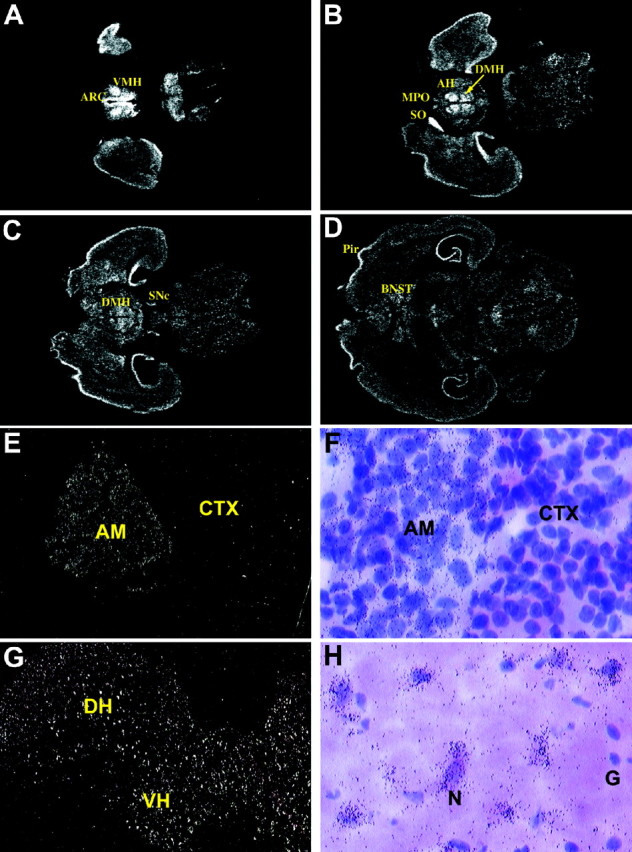
In situ hybridization analysis of proSAAS mRNA in rat brain (A–D), adrenal gland (E, F), and spinal cord (G, H). For the adrenal and spinal cord, the left panels (E, G) show autoradiograms from x-ray film, and the right panels (F, H) show emulsion-dipped sections. AH, Anterior hypothalamus; AM, adrenal medulla; ARC, arcuate nucleus;BNST, bed nucleus of the stria terminalus;CTX, adrenal cortex; DH, dorsal horn;DMH, dorsal medial hypothalamus; G, glia;MPO, medial preoptic area; N, neuron;Pir, piriform cortex; SNc, substantia nigra pars compacta; SO, supraoptic nucleus;VH, ventral horn; VMH, ventral medial hypothalamus.
When AtT-20 cells were transfected with rat proSAAS in the pcDNA3 vector, and stable clones were selected, a protein of 26 kDa was detected after Western blot analysis of the transfected cells (Fig.4A). The level of the immunoreactive 26 kDa protein is much higher in the transfected cells than in wild-type AtT-20 cells. No signal was detectable when duplicate blots were probed with preimmune antisera (data not shown). The intact 26 kDa protein is secreted into the media from transfected cells, and this secretion can be stimulated by the addition of secretagogues (Fig.4A). Similar results were obtained with four separate proSAAS-expressing clones (data not shown). Media from untransfected cells show no detectable signal for proSAAS-immunoreactive protein (data not shown). The subcellular distribution of proSAAS in the stably transfected AtT-20 cells was examined by immunofluorescence (Fig.4B). ProSAAS immunoreactivity shows a perinuclear distribution similar to the previously reported trans-Golgi network localization of CPE and CPD in this cell line (Varlamov and Fricker, 1998). In addition, immunoreactivity is detected in the tips of the AtT-20 cells, consistent with localization to the mature secretory vesicles.
Fig. 4.
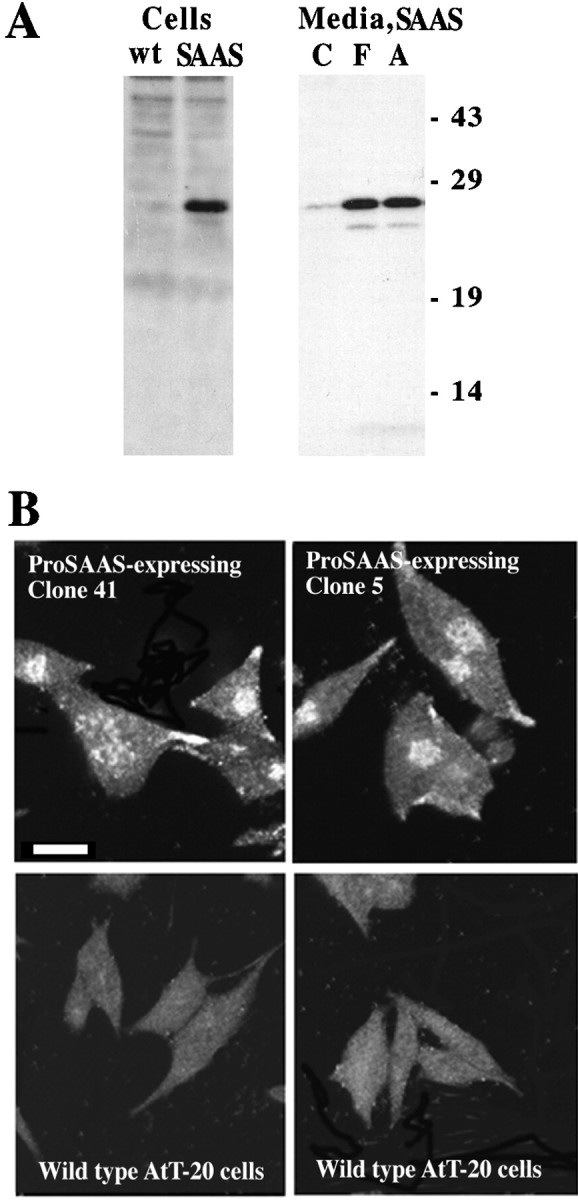
Expression of rat proSAAS in AtT-20 cells.A, Western blot analysis of expression of proSAAS in AtT-20 cells. Left, Wild-type (wt) cells or cells transfected with proSAAS cDNA (SAAS) in the pcDNA3 vector were analyzed on a Western blot and probed with an antiserum directed against proSAAS 245–260. Right,Media were collected from proSAAS-expressing cells treated for 30 min with control medium (C), or with media containing 10 μm forskolin (F) or 5 mm 8-Br-cAMP (A). The media were then analyzed on a Western blot with antiserum to proSAAS 245–260. The position and molecular mass (in kilodaltons) of prestained molecular weight markers (Life Technologies) are indicated. B,Confocal microscopy of immunoreactive SAAS in AtT-20 cells overexpressing proSAAS. Cells were cultured, fixed, permeabilized with 0.1% Triton X-100, and stained with rabbit antiserum to proSAAS 245–260, as described in Materials and Methods. Bound antiserum was detected with fluorescein-labeled anti-rabbit antiserum. Three proSAAS-expressing cell lines were examined with similar results. Scale bar, 10 μm.
The biosynthesis of proSAAS was examined with radiolabeled leucine because the protein does not contain methionine residues. Immediately after the 20 min labeling period, several forms of proSAAS are detected in cells using N- and C-terminally directed antisera (Fig.5A, left). Similar analysis with preimmune antiserum shows no detectable bands (data not shown). The 26 kDa form detected with both N- and C-terminally directed antisera presumably represents proSAAS (Fig. 5A). The C-terminally directed antiserum also detects forms of 25 and 23 kDa, which presumably represent forms lacking the N-terminal peptides that had been previously detected in mouse brain. A 4 kDa fragment is also detected with the C-terminal antiserum; this form may represent the C-terminal peptide from 221–260. The N-terminal antiserum detects a major band of 22 kDa, which presumably represents the N-terminal fragment resulting from release of the C-terminal 4 kDa 221–260 fragment. In addition to these major bands, minor fragments of 12 and 14 kDa are detected with the C-terminal and N-terminal antisera, respectively, and are likely to represent the two fragments formed from cleavage at an internal site that does not match the consensus for prohormone convertases. Analysis of the media from cells incubated for different chase times shows that the predominant secreted products that are detected with the antisera are the 4 kDa C-terminal fragment and the 22 kDa N-terminal fragment (Fig. 5A, right). Longer exposure of the gel to film reveals a faint signal of the 26 kDa proSAAS, consistent with the detection of this protein by Western blot analysis (Fig. 4).
Fig. 5.
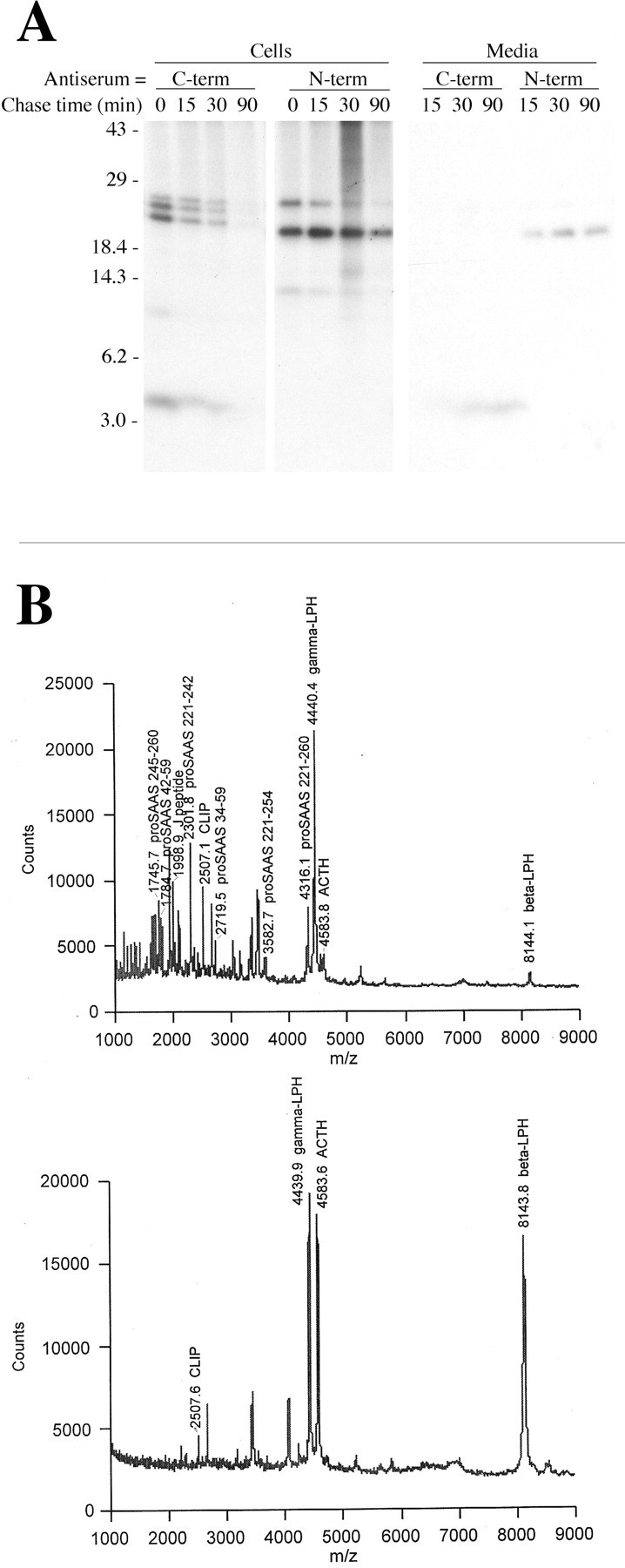
A, Pulse/chase analysis of proSAAS processing in AtT-20 cells overexpressing rat proSAAS. Cells were labeled with [3H]Leu for 20 min and then chased for 0, 15, 30, or 90 min in unlabeled Leu, and then the media and cell extracts were immunoprecipitated with antisera to the C-terminal region (245–260) or the N-terminal region (42–59) of pro- SAAS. The immunoprecipitates were fractionated on a denaturing polyacrylamide gel that was treated with Fluro-hance (Research Products International) and exposed to film for 3 d. The position and molecular mass (in kilodaltons) of prestained molecular weight markers (Life Technologies) are indicated. B,Analysis of media from wild-type AtT-20 cells (bottom) and rat proSAAS overexpressing cells (top) using matrix-assisted laser desorption ionization time of flight mass spectrometry. Predicted POMC- and proSAAS-derived peptides that are within 1 mass unit of the observed peaks are indicated.LPH, lipotropin; ACTH, adrenocorticotropic hormone; CLIP, corticotropin-like immunoreactive peptide.
When the medium from the rat proSAAS-overexpressing cells was examined by mass spectrometry, a number of peptides corresponding to the mass of the predicted fragments were detected (Fig. 5B, top panel). These peptides were not abundant in the medium from wild-type AtT-20 cells (Fig. 5B, bottom panel). Several of the observed peptides correspond to those initially found inCpefat/Cpefatmouse brain (but lacking the C-terminal basic residues, as expected because AtT-20 cells contain active CPE). Similar results were observed with media from three independent proSAAS-expressing cell lines.
The levels of ACTH in the media from the wild-type and proSAAS-overexpressing AtT-20 cells appeared different, relative to the signal for γ-lipotropin (LPH) (Fig. 5B). To quantitatively examine the extent of POMC processing, pulse chase analysis was performed on the wild-type cells and on proSAAS-overexpressing cells. Immediately after a 10 min pulse with 35S-Met, only the 29 kDa form of POMC is detected in the cells (data not shown). After 90 min of chase, the wild-type cells show extensive processing of POMC into smaller products, and these products are detected in the medium (Fig.6). In contrast, cells overexpressing proSAAS show greatly reduced processing of POMC into either ACTH, β-lipotropin, or β-endorphin (Fig. 6). Furthermore, the secretion of intact POMC showed an increase between 30 and 90 min in the proSAAS expressing cells, but not in the wild-type AtT-20 cells (Fig. 6). Similar results were found for two different proSAAS-expressing AtT-20 cell lines (data not shown).
Fig. 6.
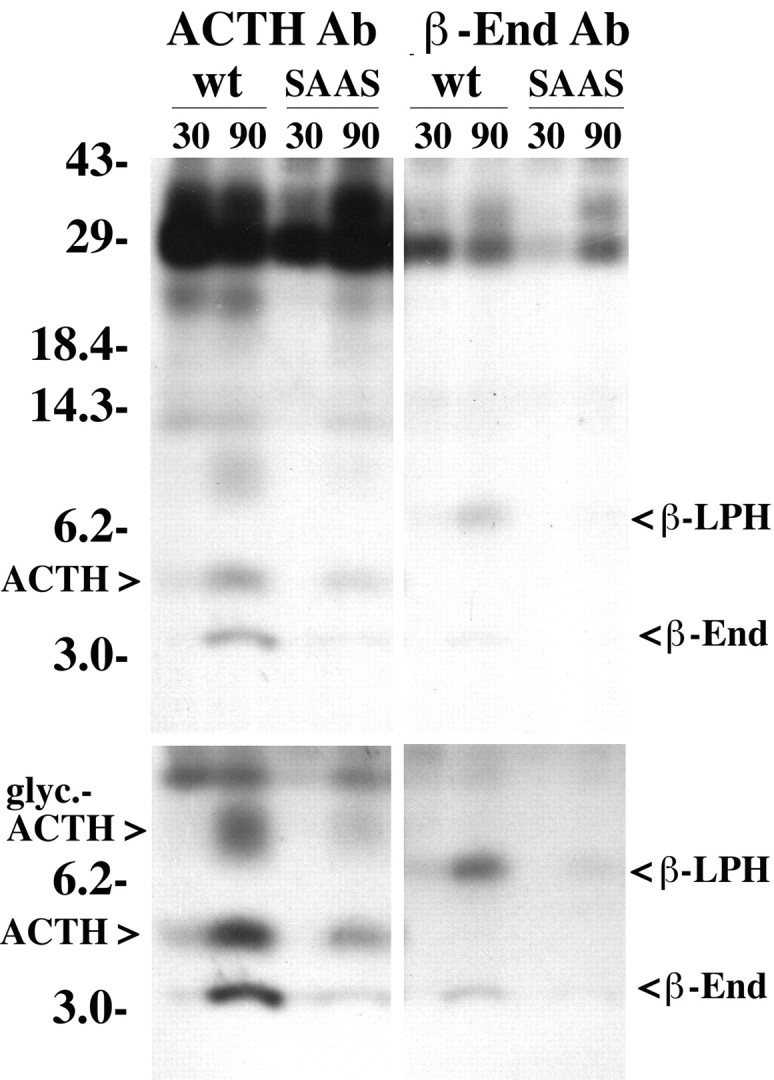
Pulse/chase analysis of proopiomelanocortin processing in wild-type AtT-20 cells (wt) and in AtT-20 cells overexpressing proSAAS (SAAS). Cells were labeled with 35S-Met for 10 min and then chased for 30 or 90 min in unlabeled Met, and then the media were immunoprecipitated with antisera to ACTH (left) or β-endorphin (right).Top, Exposure of the gel to x-ray film for 14 hr.Bottom, Exposure for 4 d.
The mRNA levels of rat proSAAS, endogenous mouse proSAAS, mouse POMC, and mouse PC1 were quantitated using the ABI PRISM 7700 sequence detection system (data not shown). This analysis revealed that POMC, PC1, and endogenous mouse proSAAS mRNA levels were relatively similar in the rat proSAAS-expressing cells compared to wild-type cells. The levels of rat proSAAS in the two cell lines expressing the highest levels of this mRNA were 40- to 50-fold higher than the level of endogenous mouse proSAAS mRNA in the same cell lines but generally similar to the level of endogenous mouse POMC mRNA. Thus, substantial inhibition of proteolytic processing of endogenous POMC is achieved at a level of proSAAS mRNA that is comparable to that of POMC mRNA.
To test whether proSAAS is directly able to inhibit prohormone convertase 1 and 2, proSAAS was expressed in bacteria as a GST fusion protein. Purified GST-proSAAS or GST alone were combined with PC1 or PC2 activity under assay conditions selective for each enzyme. PC1 activity was substantially inhibited by GST-proSAAS, with an IC50 of 590 nm (Fig.7A). This concentration is well below the level of substrate in the assay (100 μm). PC1 activity was slightly inhibited by GST alone, but the degree of inhibition was significantly lower than the inhibition with GST-proSAAS, even when the concentration of GST was twice that of the maximum concentration of GST-proSAAS tested (Fig.7A). In contrast to the effects of GST-proSAAS on PC1 activity, PC2 was not significantly inhibited by concentrations of GST-proSAAS up to 2.7 μm (Fig.7A).
Fig. 7.
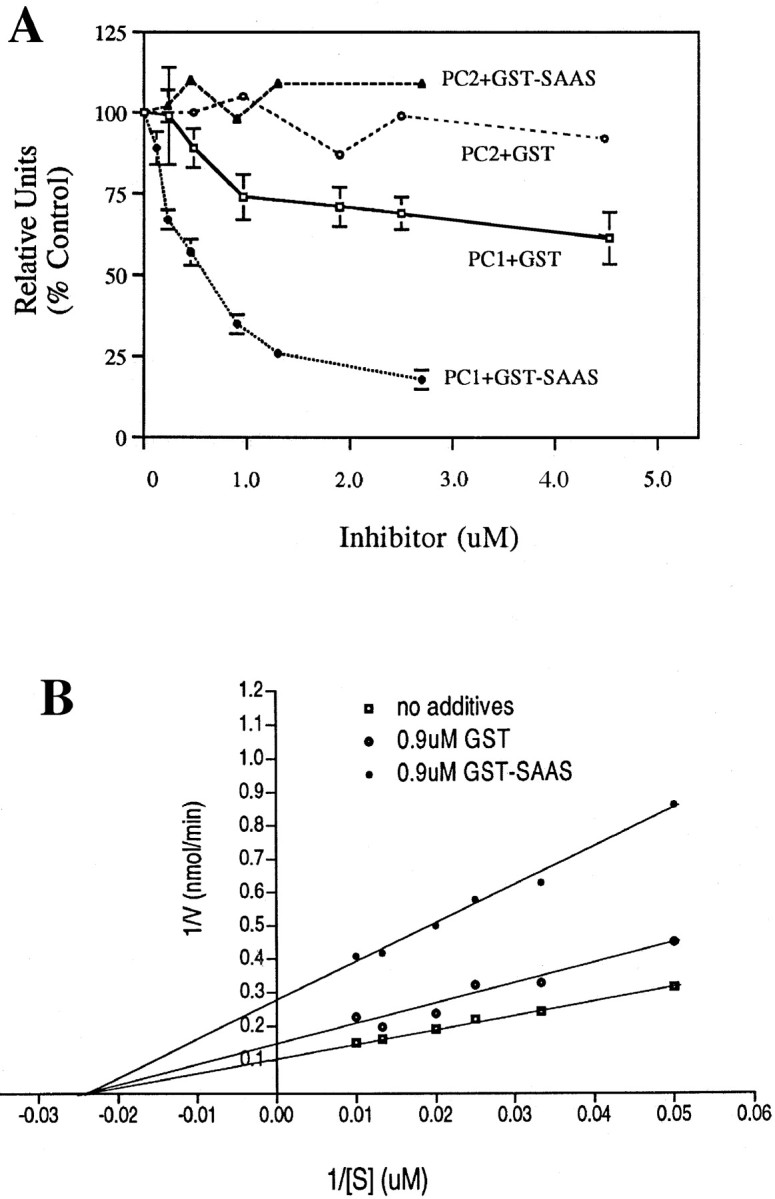
Inhibition of PC1 by proSAAS. A,Purified GST-proSAAS or GST at the indicated concentrations was combined with PC1 or PC2 and 100 μm RTKR-AMC substrate, as described in Materials and Methods. The results represent mean ± SEM from three determinations. The experiment was performed with two independent preparations of GST and GST-proSAAS, with similar results.B, Purified GST-proSAAS or GST at the indicated concentrations were combined with PC1 and various concentrations of RTKR-AMC substrate, as described in Materials and Methods. Kinetic constants were determined from linear regression analysis of plots of the inverse of the reaction velocity (with GST-proSAAS or GST) versus the inverse of the substrate concentration (Lineweaver–Burk plot). The experiment was performed with two independent preparations of GST-proSAAS and GST with similar results.
The inhibition of PC1 by GST-proSAAS was further evaluated with a range of substrate concentrations. PC1 alone or with 0.9 μm GST showed a straight line on a double reciprocal plot with a Km of ∼40 μm (Fig.7B), which is comparable to reported values (Jean et al., 1995). The addition of 0.9 μm GST-proSAAS shifted the apparent Vmax, without affecting the Km for substrate hydrolysis (Fig. 7B). This suggests that proSAAS is either a noncompetitive inhibitor or a tight binding competitive inhibitor with a slow off-rate.
DISCUSSION
In our studies on peptides that are not correctly processed in theCpefat/Cpefatmouse, we identified several peptides that did not match any sequences present in the various protein and nucleotide databases (excluding EST databases). Despite the absence of these sequences from computerized databases, a reviewer of this manuscript noticed a similarity between proSAAS and two peptides previously identified in bovine adrenal medulla chromaffin vesicles (Sigafoos et al., 1993). These two peptides (ARPVKEP) and (SLGAASPPLAEASXHRRF) presumably represent the bovine homolog of the two N-terminal peptides found in mouse brain (Fig. 1). Altogether, we identified five proSAAS-derived peptides that appear to be CPE substrates because of the presence of C-terminal basic residues in mice lacking CPE activity (and the absence of these C-terminally extended peptides in control mice). Many CPE substrates are neuropeptides, and it is possible that the proSAAS-derived peptides function in this capacity. Numerous orphan receptors have been identified (Marchese et al., 1999), and it will be of interest to test whether the novel peptides described in the present study are able to interact with specific receptors. Injection of the five proSAAS-derived peptides into mouse and rat brain (1–20 μg/d, i.c.v., either daily injections or continuous infusion) failed to show obvious changes in body weight, food intake, meal patterns, or serum levels of glucose, cholesterol, triglycerides, total protein, albumin, thyroxine, β-hydroxybutyrate, or nonesterified fatty acids (N. Levin, unpublished observations). However, these negative studies do not rule out an extracellular function for the proSAAS-derived peptides because many neuropeptides do not show dramatic behavioral and/or physiological effects after administration. Alternatively, it is possible that proSAAS or its peptide fragments perform another function in the cell. Although proSAAS does not have homology to any previously reported protein, there are some general similarities with the granin family of proteins. This family includes chromogranin A and B, secretogranin II, 7B2, and others (Huttner et al., 1991; Ozawa and Takata, 1995). Except for homology between chromogranin A and B, there is no sequence similarity among the family members. However, all granins have an N-terminal signal peptide, are hydrophilic with many acidic residues, and contain several cleavage sites that are recognized by the prohormone convertases (Huttner et al., 1991; Ozawa and Takata, 1995). Also, the granins have a broad neuroendocrine distribution and are present in the regulated secretory pathway. Based on these similarities, it is likely that proSAAS is a member of the granin family.
The functions of the various granins is not entirely clear. There is evidence that chromogranin A, B, and secretogranin II are precursors of bioactive peptides (Natori and Huttner, 1994). The chromogranins and secretogranin II may also be involved with the sorting of proteins into the regulated secretory pathway (Bauerfeind and Huttner, 1993; Ozawa and Takata, 1995). Several lines of evidence suggest that 7B2 is both an inhibitor and chaperone of PC2 (Braks and Martens, 1994; Martens et al., 1994; Benjannet et al., 1995; Zhu and Lindberg, 1995). Inhibition is caused by a C-terminal region of 7B2 that is slowly cleaved by PC2 (van Horssen et al., 1995; Zhu et al., 1996). Interestingly, CPE removes the C-terminal basic residues from the PC2-cleaved 7B2 and eliminates the inhibitory potency of the 7B2 fragment (Zhu et al., 1996); this could partially explain the defective endoprotease processing of prohormones seen inCpefat/Cpefatmice (Naggert et al., 1995; Fricker et al., 1996). 7B2 presumably performs functions in addition to those involving PC2 because knock-out mice that lack 7B2 protein develop Cushing's disease and die during adolescence (Westphal et al., 1999), whereas mice lacking PC2 are fully viable (Furuta et al., 1997).
It is likely that proSAAS functions as an endogenous PC1 inhibitor based on our studies of cells overexpressing proSAAS and on our studies with purified proSAAS and PC1. Although overexpression of an exogenous prohormone has been shown to interfere with endogenous prohormone processing (Dickerson et al., 1987), proSAAS is unique in several ways. First, the levels of exogenous rat proSAAS mRNA in the two AtT-20 cell lines expressing the highest levels of this transgene are comparable to the levels of endogenous mouse POMC mRNA in these cells, based on quantitation of the mRNA levels by PRISM analysis. If the two mRNA species are translated with the same efficiencies, then a large degree of inhibition of endogenous POMC processing is achieved with levels of proSAAS that are no higher than POMC. Second, the studies with purified proSAAS show inhibition of PC1 but not PC2, even though both enzymes are predicted to cleave proSAAS. Relatively small amounts of proSAAS (mid- to high nanomolar range) effectively compete with 100 μm substrate (Fig. 7). Furthermore, Lineweaver–Burke plots show that proSAAS is either a noncompetitive PC1 inhibitor or a competitive inhibitor with a slow dissociation rate. Third, the distribution of proSAAS mRNA in rat brain (Figs. 2, 3) is very broad and is generally similar to that of PC1 (Seidah et al., 1991; Schafer et al., 1993). Taken together, these observations are consistent with proSAAS being an endogenous inhibitor of PC1 and not merely a competitive substrate.
The techniques of Western blot analysis, immunoprecipitation, and mass spectrometry showed differences in the relative intensities of the various forms of proSAAS-derived peptides in the AtT-20 cells and medium. The Western blot analysis revealed primarily proSAAS using the C-terminally-directed antiserum and no signal using the N-terminally directed antiserum. It is possible that the smaller fragments do not bind well to nitrocellulose and thus go undetected in this technique. The matrix-assisted laser desorption ionization mass spectrometry with medium from proSAAS-expressing AtT-20 cells revealed only the 4.3 kDa and smaller proSAAS-derived peptides, even though the full mass range was scanned, and several matrices that are optimal for larger peptides were used. The failure to detect larger proSAAS-derived peptides may be attributable to the abundance of acidic residues in the middle region of the protein. The immunoprecipitation of radiolabeled cell extracts and medium detected both high- and low-molecular weight peptides. Based on the immunoprecipitation analysis, it appears that a major cleavage site is located ∼4 kDa from the C terminus at the furin consensus site RLRR-A. It is possible that the antisera do not recognize the various fragments with equal affinity, and so the results from this analysis must also be interpreted with caution. For example, processing of proSAAS clearly occurs at N-terminal sites based on the immunoprecipitation with C-terminal antisera and the mass spectrometry, and the failure to detect the N-terminal fragments by immunoprecipitation with the N-terminally directed antiserum may reflect the specificity of the antiserum. Taken together, these three techniques show that proSAAS is secreted from the AtT-20 cells, but that it is also extensively processed at the consensus cleavage sites.
The genomic position of human proSAAS on chromosome X maps within regions associated with several different X-linked mental retardation syndromes (Gedeon et al., 1994; Stevenson et al., 1998). In addition, a susceptibility haplotype for insulin-dependent diabetes mellitus has been mapped to chromosome Xp13–11, which contains the proSAAS locus (Cucca et al., 1998). However, these various syndromes encompass large regions of the X chromosome, and so proSAAS is one of many candidate genes. Interestingly, two obesity quantitative trait loci genes (Bw1 and Bw3) map to the syntenic region of mouse chromosome X (Dragani et al., 1995). Further studies will be required to test if either Bw1 or Bw3 maps to the proSAAS locus.
Although the proSAAS-derived peptides were initially found as processing intermediates in theCpefat/Cpefatmice, it is not clear if these peptides contribute to obesity in this model.Cpefat/Cpefatmice lack functional CPE and thus produce only low levels of fully processed peptides, and so it is hypothesized that the absence of a peptide that reduces body weight is the cause of the obesity in these mice. Obesity has been found to be associated with mutations in the human PC1 gene (Jackson et al., 1997). Thus, increased levels of a PC1 inhibitor would be expected to produce the same phenotype as the loss of PC1 activity. Further studies are needed to examine whether the incomplete processing of proSAAS-derived peptides in theCpefat/Cpefatmice plays a role in the development of obesity.
Footnotes
This work was supported primarily by Amgen and also by National Institutes of Health Grants R01 DA-04494, K02 DA-00194 (L.D.F.), R01 NS-26880, and K04 NS-01788 (L.A.D.). Some of the mass spectrometry was performed in the Laboratory for Macromolecular Analysis of the Albert Einstein College of Medicine, which is supported in part by the Cancer Center Core grant CA13330 and by the Diabetes Research Training Center Core grant DK20541. Confocal microscopy was performed in the Analytical Imaging Facility of the Albert Einstein College of Medicine. Wei Fan is gratefully acknowledged for performing the SAAS gene-mapping studies. Iris Lindberg and Nabil Seidah generously provided PC1 expression systems and Michael McCaleb, Ruth Angeletti, Ed Nieves, and Hong Li provided helpful advice.
Correspondence should be addressed to Dr. Lloyd Fricker, Department of Molecular Pharmacology, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461. E-mail: fricker@aecom.yu.edu.
REFERENCES
- 1.Bauerfeind R, Huttner WB. Biogenesis of constitutive secretory vesicles, secretory granules and synaptic vesicles. Curr Opin Cell Biol. 1993;5:628–635. doi: 10.1016/0955-0674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 2.Benjannet S, Savaria D, Chretien M, Seidah NG. 7B2 is a specific intracellular binding protein of the prohormone convertase PC2. J Neurochem. 1995;64:2303–2311. doi: 10.1046/j.1471-4159.1995.64052303.x. [DOI] [PubMed] [Google Scholar]
- 3.Bennett HPJ. Glycosylation, phosphorylation, and sulfation of peptide hormones and their precursors. In: Fricker LD, editor. Peptide biosynthesis and processing. CRC; Boca Raton: 1991. pp. 111–140. [Google Scholar]
- 4.Boudreault A, Gauthier D, Lazure C. Proprotein convertase PC1/3-related peptides are potent slow tight-binding inhibitors of murine PC1/3 and Hfurin. J Biol Chem. 1998a;273:31574–31580. doi: 10.1074/jbc.273.47.31574. [DOI] [PubMed] [Google Scholar]
- 5.Boudreault A, Gauthier D, Rondeau N, Savaria D, Seidah NG, Chretien M, Lazure C. Molecular characterization, enzymatic analysis, and purification of murine proprotein convertase-1/3 (PC1/3) secreted from recombinant baculovirus-infected insect cells. Protein Expr Purif. 1998b;14:353–366. doi: 10.1006/prep.1998.0964. [DOI] [PubMed] [Google Scholar]
- 6.Bradbury AF, Smyth DG. Modification of the N- and C-termini of bioactive peptides: amidation and acetylation. In: Fricker LD, editor. Peptide biosynthesis and processing. CRC; Boca Raton: 1991. pp. 231–250. [Google Scholar]
- 7.Braks JAM, Martens GJM. 7B2 is a neuroendocrine chaperone that transiently interacts with prohormone convertase PC2 in the secretory pathway. Cell. 1994;78:263–273. doi: 10.1016/0092-8674(94)90296-8. [DOI] [PubMed] [Google Scholar]
- 8.Courchesne PL, Jones MD, Robinson JH, Spahr CS, McCracken S, Bentley DL, Luethy R, Patterson SD. Optimization of capillary chromatography ion-trap mass spectrometry for identification of gel-separated proteins. Electrophoresis. 1998;19:956–967. doi: 10.1002/elps.1150190611. [DOI] [PubMed] [Google Scholar]
- 9.Cucca F, Goy JV, Kawaguchi Y, Esposito L, Merriman ME, Wilson AJ, Cordell HJ, Bain SC, Todd JA. A male-female bias in type 1 diabetes and linkage to chromosome Xp in MHC HLA-DR3-positive patients. Nat Genet. 1998;19:301–302. doi: 10.1038/995. [DOI] [PubMed] [Google Scholar]
- 10.Curran EJ, Watson SJ., Jr Dopamine receptor mRNA expression patterns by opioid peptide cells in the nucleus accumbens of the rat: a double in situ hybridization study. J Comp Neurol. 1995;361:57–76. doi: 10.1002/cne.903610106. [DOI] [PubMed] [Google Scholar]
- 11.Devi L. Peptide processing at monobasic sites. In: Fricker LD, editor. Peptide biosynthesis and processing. CRC; Boca Raton: 1991. pp. 175–198. [Google Scholar]
- 12.Dickerson IM, Dixon JE, Mains RE. Transfected human neuropeptide Y cDNA expression in mouse pituitary cells. J Biol Chem. 1987;262:13646–13653. [PubMed] [Google Scholar]
- 13.Dragani TA, Zeng ZB, Canzian F, Gariboldi M, Ghilarducci MT, Manenti G, Pierotti MA. Mapping of body weight loci on mouse chromosome X. Mamm Genome. 1995;6:778–781. doi: 10.1007/BF00539002. [DOI] [PubMed] [Google Scholar]
- 14.Fricker LD. Carboxypeptidase E. Annu Rev Physiol. 1988;50:309–321. doi: 10.1146/annurev.ph.50.030188.001521. [DOI] [PubMed] [Google Scholar]
- 15.Fricker LD. Peptide processing exopeptidases: amino- and carboxypeptidases involved with peptide biosynthesis. In: Fricker LD, editor. Peptide biosynthesis and processing. CRC; Boca Raton: 1991. pp. 199–230. [Google Scholar]
- 16.Fricker LD, Adelman JP, Douglass J, Thompson RC, von Strandmann RP, Hutton J. Isolation and sequence analysis of cDNA for rat carboxypeptidase E [EC 3.4.17.10], a neuropeptide processing enzyme. Mol Endocrinol. 1989;3:666–673. doi: 10.1210/mend-3-4-666. [DOI] [PubMed] [Google Scholar]
- 17.Fricker LD, Berman YL, Leiter EH, Devi LA. Carboxypeptidase E activity is deficient in mice with the fat mutation: Effect on peptide processing. J. Biol. Chem. 1996;271:30619–30624. doi: 10.1074/jbc.271.48.30619. [DOI] [PubMed] [Google Scholar]
- 18.Furuta M, Yano H, Zhou A, Rouille Y, Holst JJ, Carroll R, Ravazzola M, Orci L, Furata H, Steiner DF. Defective prohormone processing and altered pancreatic islet morphology in mice lacking active SPC2. Proc Natl Acad Sci USA. 1997;94:6646–6651. doi: 10.1073/pnas.94.13.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gedeon AK, Donnelly AJ, Mulley JC, Kerr B, Turner G. How many X-linked genes for non-specific mental retardation (MRX) are there? Am J Med Genet. 1994;64:158–162. doi: 10.1002/(SICI)1096-8628(19960712)64:1<158::AID-AJMG26>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 20.Huttner WB, Gerdes HH, Rosa P. The granin (chromogranin/secretogranin) family. Trends Biochem Sci. 1991;16:27–30. doi: 10.1016/0968-0004(91)90012-k. [DOI] [PubMed] [Google Scholar]
- 21.Jackson RS, Creemers JWM, Ohagi S, Raffin-Sanson M, Sanders L, Montague CT, Hutton JC, O'Rahilly S. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat Genet. 1997;16:303–306. doi: 10.1038/ng0797-303. [DOI] [PubMed] [Google Scholar]
- 22.Jean F, Boudreault A, Basak A, Seidah NG, Lazure C. Fluorescent peptidyl substrates as an aid in studying the substrate specificity of human prohormone convertase PC1 and human furin and designing a potent irreversible inhibitor. J Biol Chem. 1995;270:19225–19231. doi: 10.1074/jbc.270.33.19225. [DOI] [PubMed] [Google Scholar]
- 23.Kumazaki T, Terasawa K, Ishii S. Affinity chromatography on immobilized anhydrotrypsin: general utility for selective isolation of C-terminal peptides from protease digests of proteins. J Biochem. 1987;102:1539–1546. doi: 10.1093/oxfordjournals.jbchem.a122202. [DOI] [PubMed] [Google Scholar]
- 24.Lamango NS, Zhu X, Lindberg I. Purification and enzymatic characterization of recombinant prohormone convertase 2: stabilization of activity by 21 kDa 7B2. Arch Biochem Biophys. 1996;330:238–250. doi: 10.1006/abbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- 25.Lindberg I, Hutton JC. Peptide processing proteinases with selectivity for paired basic residues. In: Fricker LD, editor. Peptide biosynthesis and processing. CRC; Boca Raton: 1991. pp. 141–174. [Google Scholar]
- 26.Marchese A, George SR, Kolakowski LF, Lynch KR, O'Dowd BF. Novel GPCRs and their endogenous ligands: expanding the boundaries of physiology and pharmacology. Trends Pharmacol Sci. 1999;20:370–375. doi: 10.1016/s0165-6147(99)01366-8. [DOI] [PubMed] [Google Scholar]
- 27.Martens GJ, Braks JA, Eib DW, Zhou Y, Lindberg I. The neuroendocrine polypeptide 7B2 is an endogenous inhibitor of prohormone convertase PC2. Proc Natl Acad Sci USA. 1994;91:5784–5787. doi: 10.1073/pnas.91.13.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naggert JK, Fricker LD, Varlamov O, Nishina PM, Rouille Y, Steiner DF, Carroll RJ, Paigen BJ, Leiter EH. Hyperproinsulinemia in obese fat/fat mice associated with a point mutation in the carboxypeptidase E gene and reduced carboxypeptidase E activity in the pancreatic islets. Nat Genet. 1995;10:135–142. doi: 10.1038/ng0695-135. [DOI] [PubMed] [Google Scholar]
- 29.Natori S, Huttner WB. Peptides derived from the granins (chromogranins/secretogranins). Biochimie. 1994;76:277–282. doi: 10.1016/0300-9084(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 30.Ozawa H, Takata K. The granin family - its role in sorting and secretory granule formation. Cell Struct Funct. 1995;20:415–420. doi: 10.1247/csf.20.415. [DOI] [PubMed] [Google Scholar]
- 31.Rovere C, Viale A, Nahon J, Kitabgi P. Impaired processing of brain proneurotensin and promelanin-concentrating hormone in obese fat/fat mice. Endocrinology. 1996;137:2954–2958. doi: 10.1210/endo.137.7.8770919. [DOI] [PubMed] [Google Scholar]
- 32.Schafer MK-H, Day R, Cullinan WE, Chretien M, Seidah NG, Watson SJ. Gene expression of prohormone and proprotein convertases in the rat CNS: a comparative in situ hybridization analysis. J Neurosci. 1993;13:1258–1279. doi: 10.1523/JNEUROSCI.13-03-01258.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seidah NG, Marcinkiewicz M, Benjannet S, Gaspar L, Beaubien G, Mattei MG, Lazure C, Mbikay M, Chretien M. Cloning and primary sequence of a mouse candidate prohormone convertase PC1 homologous to PC2, furin, and Kex2: Distinct chromosomal localization and messenger RNA distribution in brain and pituitary compared to PC2. Mol Endocrinol. 1991;5:111–122. doi: 10.1210/mend-5-1-111. [DOI] [PubMed] [Google Scholar]
- 34.Sigafoos J, Chestnut WG, Merrill BM, Taylor LCE, Diliberto EJ, Viveros OH. Novel peptides from adrenomedullary chromaffin vesicles. J Anat. 1993;183:253–264. [PMC free article] [PubMed] [Google Scholar]
- 35.Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 36.Song L, Fricker LD. Purification and characterization of carboxypeptidase D, a novel carboxypeptidase E-like enzyme, from bovine pituitary. J Biol Chem. 1995;270:25007–25013. doi: 10.1074/jbc.270.42.25007. [DOI] [PubMed] [Google Scholar]
- 37.Song L, Fricker LD. Tissue distribution and characterization of soluble and membrane-bound forms of metallocarboxypeptidase D. J Biol Chem. 1996;271:28884–28889. doi: 10.1074/jbc.271.46.28884. [DOI] [PubMed] [Google Scholar]
- 38.Stevenson RE, Arena JF, Ouzts E, Gibson A, Shokeir MH, Vnencak-Jones C, Lubs HA, May M, Schwartz CE. Renpenning syndrome maps to Xp11. Am J Hum Genet. 1998;62:1092–1101. doi: 10.1086/301835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Horssen AM, van den Hurk WH, Bailyes EM, Hutton JC, Martens GJ, Lindberg I. Identification of the region within the neuroendocrine polypeptide 7B2 responsible for the inhibition of prohormone convertase PC2. J Biol Chem. 1995;270:14292–14296. doi: 10.1074/jbc.270.24.14292. [DOI] [PubMed] [Google Scholar]
- 40.Varlamov O, Fricker LD. Intracellular trafficking of metallocarboxypeptidase D in AtT-20 cells: localization to the trans-Golgi network and recycling from the cell surface. J Cell Sci. 1998;111:877–885. doi: 10.1242/jcs.111.7.877. [DOI] [PubMed] [Google Scholar]
- 41.Varlamov O, Leiter EH, Fricker LD. Induced and spontaneous mutations at Ser202 of carboxypeptidase E: effect on enzyme expression, activity, and intracellular routing. J Biol Chem. 1996;271:13981–13986. doi: 10.1074/jbc.271.24.13981. [DOI] [PubMed] [Google Scholar]
- 42.Varlamov O, Eng FJ, Novikova EG, Fricker LD. Localization of metallocarboxypeptidase D in AtT-20 cells: potential role in prohormone processing. J Biol Chem. 1999;274:14759–14767. doi: 10.1074/jbc.274.21.14759. [DOI] [PubMed] [Google Scholar]
- 43.Westphal CH, Muller L, Zhou A, Zhu X, Bonner-Weir S, Schambelan M, Steiner DF, Lindberg I, Leder P. The neuroendocrine protein 7B2 is required for peptide hormone processing in vivo and provides a novel mechanism for pituitary Cushing's disease. Cell. 1999;96:689–700. doi: 10.1016/s0092-8674(00)80579-6. [DOI] [PubMed] [Google Scholar]
- 44.Zheng M, Streck RD, Scott REM, Seidah NG, Pintar JE. The developmental expression in rat of proteases furin, PC1, PC2, and carboxypeptidase E: implications for early maturation of proteolytic processing capacity. J Neurosci. 1994;14:4656–4673. doi: 10.1523/JNEUROSCI.14-08-04656.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou A, Webb G, Zhu X, Steiner DF. Proteolytic processing in the secretory pathway. J Biol Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y, Lindberg I. Purification and characterization of the prohormone convertase PC1 (PC3). J Biol Chem. 1993;268:5615–5623. [PubMed] [Google Scholar]
- 47.Zhu X, Lindberg I. 7B2 facilitates the maturation of proPC2 in neuroendocrine cells and is required for the expression of enzymatic activity. J Cell Biol. 1995;129:1641–1650. doi: 10.1083/jcb.129.6.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu X, Rouille Y, Lamango NS, Steiner DF, Lindberg I. Internal cleavage of the inhibitory 7B2 CT peptide by PC2: a potential mechanism for its inactivation. Proc Natl Acad Sci USA. 1996;93:4919–4924. doi: 10.1073/pnas.93.10.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]