Abstract
During Drosophila embryogenesis the CNS midline cells have organizing activities that are required for proper elaboration of the axon scaffold and differentiation of neighboring neuroectodermal and mesodermal cells. CNS midline development is dependent on Single-minded (Sim), a basic-helix-loop-helix (bHLH)-PAS transcription factor. We show here that Fish-hook (Fish), a Sox HMG domain protein, and Drifter (Dfr), a POU domain protein, act in concert with Single-minded to control midline gene expression. single-minded,fish-hook, and drifter are all expressed in developing midline cells, and both loss- and gain-of-function assays revealed genetic interactions between these genes. The corresponding proteins bind to DNA sites present in a 1 kb midline enhancer from theslit gene and regulate the activity of this enhancer in cultured Drosophila Schneider line 2 cells. Fish-hook directly associates with the PAS domain of Single-minded and the POU domain of Drifter; the three proteins can together form a ternary complex in yeast. In addition, Fish can form homodimers and also associates with other bHLH-PAS and POU proteins. These results indicate that midline gene regulation involves the coordinate functions of three distinct types of transcription factors. Functional interactions between members of these protein families may be important for numerous developmental and physiological processes.
Keywords: Drosophila, CNS midline, bHLH/PAS, Sox, POU, slit, gene expression
During Drosophila embryogenesis, a group of specialized CNS midline neurons and glia provide signals that are essential for the differentiation of neighboring ectodermal and mesodermal cells (for review, see Crews, 1998). The CNS midline also serves as an intermediate axon guidance target that expresses chemotactic factors, including Slit, Commissureless, and D-Netrin, which control axon crossing at the midline (Harris et al., 1996; Mitchell et al., 1996; Tear et al., 1996;Kidd et al., 1999). In particular, Slit is a conserved epidermal growth factor (EGF)-repeat protein that is strongly expressed by midline glia and interacts with the Roundabout receptor to prevent commissural axons from recrossing the midline (for review, see Guthrie, 1999; Van Vactor and Flanagan, 1999; Zinn and Sun, 1999). Development of the entire CNS midline lineage requires the gene regulatory functions of Single-minded (Sim), a basic-helix-loop-helix (bHLH)-PAS transcription factor (Nambu et al., 1991). Sim protein forms functional heterodimers with Tango (Tgo), a bHLH-PAS protein that exhibits widespread embryonic expression (Sonnenfeld et al., 1997; Ward et al., 1998). Sim:: Tgo heterodimers act through the CNS midline element (CME), an ACGTG sequence motif present in the regulatory regions of slit and other midline-expressed genes (Wharton et al., 1994; Sonnenfeld et al., 1997;Kasai et al., 1998). A CME present in a 380 bp slitregulatory DNA fragment is essential for midline expression of a linked reporter gene (Wharton et al., 1994), although sequences in addition to the CME are required for high levels of gene expression (Wharton and Crews, 1993). Interestingly, results from Sim ectopic expression experiments suggest that in addition to Tgo, Sim functionally interacts with other factors more specifically expressed in the midline and lateral and cephalic neuroectoderm (Nambu et al., 1991; Muralidhar et al., 1993).
One candidate for such a Sim cofactor is the Sox HMG domain protein Fish-hook (Fish) (Nambu and Nambu, 1996; Russell et al., 1996). Fish is strongly expressed in the early neuroectoderm and is required for proper differentiation of midline and lateral CNS cells (Nambu and Nambu, 1996; Ma et al., 1998; Sánchez Soriano and Russell, 1998). The Fish HMG domain binds DNA sequences related to AACAAT and AACAAAG and induces strong DNA bending (Ma et al., 1998). This suggests that Fish may provide chromatin architectural functions to facilitate the assembly of higher-order protein/DNA complexes. In this regard, the closely related vertebrate Sox2 protein directly associates with the Oct3 POU domain protein to synergistically activate expression of the fgf4 gene (Yuan et al., 1995; Ambrosetti et al., 1997). fish mutants exhibit genetic interactions with mutations in Drosophila POU genes (Ma et al., 1998;Sánchez Soriano and Russell 1998), and one of these POU genes,drifter (dfr), is expressed in the CNS midline and is essential for proper midline glial migrations (Anderson et al., 1995).
In this study we use genetic, biochemical, and cell culture assays to show that Sim, Fish, and Drifter (Dfr) act together to regulate CNS midline development and gene expression. In particular, these proteins synergistically influence slit transcription by acting through a 1 kb midline regulatory region that contains a single CME, as well as binding sites for Fish and Dfr. Regulation of slitexpression appears to involve direct interactions between Fish, Sim, and Dfr. Fish directly associates with the Sim PAS domain and the Dfr POU domain, and the three proteins can form a ternary complex in yeast cells. These results identify novel regulatory interactions between bHLH-PAS, Sox, and POU proteins.
MATERIALS AND METHODS
Fly strains. The following strains were used in this study: the fish87 null mutant (Nambu and Nambu, 1996), thedfrE82 lethal mutant (Anderson et al., 1995), P[1.0slit-lacZ] (Nambu et al., 1991;Wharton et al., 1994), P[UAS-fish] (Mukherjee et al., 2000), P[UAS-sim] (Xiao et al., 1996), and P[GMR-Gal4] (obtained from the Bloomington Drosophila Stock Center (Bloomington, IN); generated by M. Freeman). AdfrE82-fish87/TM3, P[ftz-lacZ] strain was generated via meiotic recombination.
Immunostaining of Drosophila embryos. For single-label immunostaining experiments, embryos were collected from wild-type and mutant strains, as well as from genetic crosses, and fixed in PEM buffer (0.1 m PIPES, pH 6.9, 2 mmEGTA, 1 mm MgSO4) with 4% formaldehyde (Patel, 1994). The embryos were incubated overnight with primary antibodies in PTN (1× PBS, 0.2% Triton X-100, 5% normal horse serum). Vectastain biotinylated secondary antibodies and streptavidin/horseradish peroxidase reagents (Vector Laboratories, Burlingame, CA) were used with H202/diaminobenzedine histochemistry to detect primary antibody binding. The monoclonal antibody (mAb) BP102 (Developmental Studies Hybridoma Bank, Iowa City, IA) was used at a 1:4 dilution. A monoclonal antibody against β-galactosidase (β-Gal) (Promega, Madison, WI) was used at a 1:500 dilution. Stained embryos were dehydrated through an ethanol series, mounted in methyl salicylate and Permount (Fisher), and examined via Nomarski optics using a Nikon Optiphot compound microscope.
For double-label fluorescence immunostaining experiments, embryos were fixed in PEM buffer with 4% formaldehyde. They were labeled by following previously described protocols (Mitchison and Sedat, 1983;Johnson, 1992). β-Gal expression was detected using either a rabbit polyclonal antiserum (Cappel, West Chester, PA) at a 1:500 dilution or a mouse mAb 40–1a (Developmental Studies Hybridoma Bank) at a 1:3 dilution. Pre-absorbed rat anti-Dfr serum was used at a final dilution of 1:3000 and rabbit anti-Fish serum at a dilution of 1:1000. After repeated washes in PBT (1× PBS, 0.5% bovine serum albumin, 0.2% Triton X-100), embryos were incubated with FITC-conjugated anti-rat (Jackson ImmunoResearch Laboratories, West Grove, PA) and rhodamine-conjugated anti-mouse (Jackson ImmunoResearch Laboratories) or anti-rabbit (Biosource, Camarillo, CA) secondary antibodies at 1:200 dilution. Stained embryos were mounted in Vectashield (Vector Laboratories) and observed using a Bio-Rad MRC-1024 confocal microscope.
DNA sequence analysis. DNA sequencing of the slit1 kb midline regulatory region was performed by Retrogen, Inc. (San Diego, CA). The DNA fragment was sequenced by primer walking approaches, and the sequences from both strands were obtained in multiple runs. The analysis revealed this fragment to be 970 bp in length and flanked by two HinDIII sites.
Gel mobility shift assays. For Fish gel mobility shift assays, two sets of complementary 26 mer oligonucleotides corresponding to Sox consensus DNA binding sites from theslit regulatory fragment were synthesized and annealed in 1× T4 polynucleotide kinase buffer (Promega). The sequence of the upper strand of each pair of oligonucleotides is shown below, and the underlined sequence corresponds to Sox consensus DNA binding sites (see Fig. 4A for location of probe sequences): TACAAT probe—5′-ACTATACTATATTGTATTATGCACAG-3′; TTCAAT probe—5′-ACTGTATTCAATTTCATTGAAACAAA-3′.
Fig. 4.
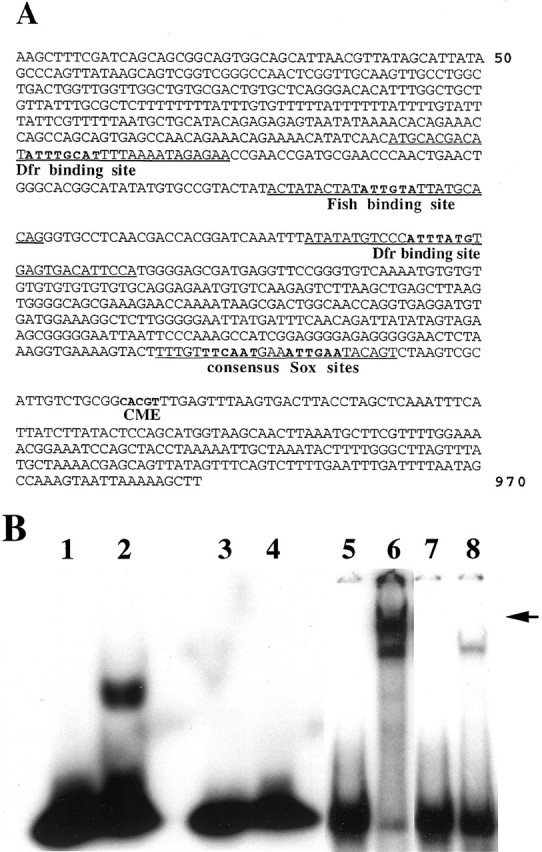
The slit 1 kb midline regulatory region contains binding sites for Sim:: Tgo, Fish, and Dfr.A, Nucleotide sequence of aHinDIII/HinDIII restriction enzyme fragment from a slit gene intron (Wharton and Crews, 1993) that contains CNS midline regulatory elements. Indicated inbold are the single CME site through which Sim:: Tgo heterodimers act, two closely linked TTCAAT consensus Sox sites, a TACAAT Fish binding site, and ATGCAAAT and CATAAAT Dfr binding sites. Sequences corresponding to the double-stranded oligonucleotide probes used in gel mobility shift assays with the Fish HMG domain or Dfr protein areunderlined. B, Gel mobility shift assays performed with purified Fish HMG domain or Dfr protein and sequences from the slit 1 kb regulatory region. Lane 1, Free TACAAT probe. Lane 2, TACAAT probe and Fish HMG domain protein. Note strong binding of Fish protein.Lane 3, Free TTCAAT probe. Lane 4, TTCAAT probe and Fish HMG domain protein. Note lack of detectable binding by Fish protein. Lane 5, Free ATGCAAAT probe. Lane 6, ATGCAAAT probe and Dfr protein. Note that Dfr binds strongly to this sequence as both an apparent monomer and a dimer (arrow). Lane 7, Free CATAAAT probe.Lane 8, CATAAAT probe and Dfr protein. Note that Dfr also binds to this sequence, although less strongly than to the ATGCAAAT sequence.
The annealed oligonucleotides were end-labeled using T4 polynucleotide kinase (Life Technologies, Gaithersburg, MD) and γ-32P-ATP (New England Nuclear) and gel-purified. Gel mobility shift assays were performed using a purified 6XHis-HMG Fish fusion protein as described previously (Ma et al., 1998).
For Dfr gel mobility shift assays, probes were generated by end-labeling double stranded oligonucleotides with γ-32P-ATP (ICN Biochemicals, Costa Mesa, CA) and T4 polynucleotide kinase (New England Biolabs). The sequence of the upper strands of each pair of oligonucleotides is shown below, and the underlined sequence corresponds to consensus Dfr binding sites (see Fig. 4A for location of probe sequences): ATGCAAAT probe—5′-ATGCACGACAT ATTTGCATTTTAAAATAGAGAA-3′; CATAAAT probe—5′-ATATATGTCCC ATTTATGTG- AGTGACATTCCA-3′.
Full-length Dfr protein was expressed in bacteria as a glutathione-S-transferase (GST) fusion protein and purified over glutathione-Sepharose beads (Amersham Pharmacia, Arlington Heights, IL) as previously described (Certel et al., 1996). While on beads, the GST affinity tail was cleaved by using PreScission protease (Amersham Pharmacia) as per manufacturer's suggestions. End-labeled probes were incubated with full-length Dfr protein at room temperature for 15 min in binding buffer (25 mmHEPES, pH 7.6, 12.5 mm MgCl2, 0.1 mm EDTA, 100 mm KCl, 10% glycerol, 1 mm DTT). Immediately after incubation, the complexes were resolved on 5% native polyacrylamide gels at room temperature in 1× TBE buffer. Gels were dried and exposed to Kodak X-OMAT film.
Yeast two-hybrid assays. The yeast two-hybrid vectors pEG202 and pJG4–5 (Origene) were used to generate bait and prey constructs. Baits contained the LexA DNA binding domain fused to the protein of interest, and the prey contained fusions to the B42 transcriptional activation domain. A chromosomally integrated LexAop-LEU2reporter gene was used that contains six LexA operator sites fused to the LEU2 gene. The bait and prey constructs were cotransformed into EGY48 host yeast cells (Matα trp1 his3 ura3 leu2:: :6lexAop-LEU2) according to the supplier's instructions (Origene). The transformed cells were plated on YNB/Leu/Ura (-His, -Trp) glucose medium and incubated at 30°C for 3–4 d. Resulting transformants were then plated on the YNB/Ura (-His, -Leu, -Trp) galactose/raffinose medium and incubated at 30°C for 3–7 d to assay for the presence of colonies that indicated interactions.
Bait constructs that express full-length Fish, or truncated versions consisting of the N-terminal 141 amino acids, the 79 amino acid HMG domain, the COOH-terminal 164 amino acids, the NH2+HMG, the HMG+COOH, and the NH2+COOH, are described in Ma et al. (1998). To generate a full-length Fish prey construct, the entire Fish 382 amino acid open reading frame was amplified via PCR from full-lengthfish cDNA clone 2–5 (Nambu and Nambu, 1996) using the following primers: 5′-AGAGAATTCATGGCCACCTTATCGACACACCC-3′; 5′-TGTGAATTCCTACTAATAGAGCACCGGAACCGGTCGCCT-3′.
The resulting DNA fragment was digested with EcoRI, purified via agarose gel electrophoresis, and cloned into the pJG4–5 vector.
Prey constructs that express full-length Dfr or the Dfr POU domain were generated via PCR using a full-length dfr p128 cDNA clone (Anderson et al., 1995) and the following primers: full-length Dfr—5′-CATGGAATTCCCCACGTCCGATGATCTGGAGGCC-3′, 5′-CATGCTCGAGTTACTAGTGGGCCGCCAACTGATGCGCCGC-3′; Dfr POU domain (amino acids 210–362)—5′-CCCCGAATTCACGTCCGATGATCTGGAGGCC-3′, 5′-GGGGCTCGAGCGTCATGCGCTTCTCCTTCTG-3′.
The PCR products were digested with EcoRI andXhoI, purified via agarose gel electrophoresis, and subcloned into the pJG4–5 vector.
A prey construct expressing the POU domain of Pdm-1 (amino acids 420–601) was generated via PCR from the full-length C616A cDNA clone (Billin et al., 1991), kindly provided by Steve Poole (University of California Santa Barbara), and the following primers: 5′- CATGGAA-TTCCCGGAGGAAACCACCGATCTAGAA-3′; 5′-CATGCT CGA- GTTACTAGGGACTGTCCAGGG-AGGGATTGAT-3′.
A similar pdm-2 POU domain (amino acids 283–457) prey construct was generated via PCR from the full-length C9A cDNA clone (Billin et al., 1991) and the following primers: 5′- CATGGAATTCGAACAATCGCCGGAAGAGACCACC-3′; 5′-CATGCTCGAGTTACTAGTCCAGATCCAGCGAGGGATTGAT-3′.
Both pdm-1 and pdm-2 PCR products were digested with EcoRI and XhoI, purified via agarose gel electrophoresis, and subcloned into the pJG4–5 vector.
A prey construct that expresses the full-length mouse Oct3 protein was generated via PCR from a full-length pGEX-2-Oct3 cDNA clone, kindly provided by Lisa Dailey (New York University) (Yuan et al., 1995), and the following primers: 5′-GGGGAATTCATGTTCGAGAAGGTGGAACCAA-3′; 5′-GGGCTCGAGTCACCCTGTAGCCTCATACT- CTT-3′.
The PCR product was digested with EcoRI and XhoI, purified via agarose gel electrophoresis, and subcloned into the pJG4–5 vector.
The following Sim and Trachealess (Trh) bait or prey constructs (in the pEG202 and pJG4–5 vectors) were used: full-length Sim (amino acids 1–673), Sim bHLH-PAS (amino acids 1–461), Sim bHLH-PAS-A (amino acids 1–183), Sim bHLH (amino acids 1–92), Sim PAS (amino acids 58–370), full-length Trh (amino acids 1–929), and Trh bHLH-PAS (amino acids 1–689) (Sonnenfeld et al., 1997; M. Sonnenfeld and S. T. Crews, unpublished results). The Period (Per) bait (Per-C2) construct containing the PAS domain and a COOH-extended region (amino acids 233–685) was kindly provided by Michael Rosbash (Brandeis University) (Huang et al., 1995).
To express native full-length Fish protein in yeast, the entire Fish open reading frame was amplified via PCR from fish cDNA clone 2–5 using the following primers: 5′-GGGGGAAGCTTATGGCCACCTTATCGACA-3′; 5′-CCCCCAAGCTTCTAATAGAGGACCGGAAC-3′.
The resulting DNA fragment was digested with HindIII, purified via agarose gel electrophoresis, and cloned into the pDB-20 yeast expression vector (Becker et al., 1991). pDB20 permits high level of transgene expression driven by the ADC1 (ADH1) promoter and contains the selectable URA3 marker.
To analyze interactions between Fish, Sim, and Dfr, full-length Sim bait, full-length Dfr prey, and full-length native Fish constructs were transformed into EGY48 host cells. The cells were grown on YNB/Leu (-His, -Ura, -Trp) glucose plates and incubated at 30°C for 3–4 d. Transformed colonies were then streaked on YNB (-His, -Leu, -Trp, -Ura) galactose/raffinose plates and incubated at 30°C for 4 d. For controls, full-length Sim bait and native Fish or full-length Dfr prey and native Fish constructs were transformed into EGY48 cells that were grown on YNB/Trp/Leu (-His, -Ura) glucose plates at 30°C for 3–4 d. Transformed colonies were then streaked on the YNB/His (-Leu, -Trp, -Ura) or YNB/Trp (-His, -Leu, -Ura) galactose/raffinose plates and incubated at 30°C for 4 d.
GST pulldown assays. Full-length35S-labeled Sim protein was generated fromsim cDNA clone F1 (Nambu et al., 1991) using an in vitro transcription/translation reaction (Promega) and35S-methionine (Amersham Pharmacia). For the GST–Fish fusion protein, a DNA fragment encoding the full-length Fish protein, was generated by PCR using fish cDNA clone 2–5 as a template and the following two oligonucleotide primers: 5′-GGCCGAATTCATGCCACCTTATCGACACACCCCAAT-3′; 5′-CCGGGAATTCTTACTAATAGAGCACCGGAACCGG-3′.
The resulting PCR product was digested with EcoRI, purified via agarose gel electrophoresis, and subcloned into the pGEX-2T vector (Amersham Pharmacia) to generate an in-frame fusion with GST. The resulting construct was transformed into Escherichia coliBL-21 (Amersham Pharmacia), and expression of GST–Fish or GST alone was induced in 500 ml cultures via isopropyl-1-thio-β-d-galactopyranoside. Induced cells were pelleted and lysed in 4 ml of B-Per solution (Pierce, Rockford, IL). The debris was pelleted by centrifugation, and 200 μl of supernatant (containing ∼2 μg of GST–Fish or GST protein) was incubated with 10 μl of glutathione-Sepharose-4B beads (Amersham Pharmacia) and 5 μl of 35S-labeled Simin vitro translation mix for 2 hr at 4°C. The beads were washed five times with BC100N buffer (Ambrosetti et al., 1997) and then boiled with 2 μl of SDS gel loading buffer and 8 μl of H2O. The supernatants were electrophoresed on a 10% SDS-polyacrylamide gel along with 1 μl of the in vitro translation mixture (20% of the input used in the binding assays). The gel was dried and analyzed via autoradiography.
To test Fish dimerization, 1–4 μg of purified GST–Fish was incubated with glutathione-Sepharose-4B beads in NETN (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, and 0.5% NP40) at 4°C for 2 hr. This was followed by several washes with NETN.35S-labeled Fish protein was generated via an in vitro transcription/translation reaction (Promega) using fish cDNA clone 2–5 as template. Five microliters of35S-labeled Fish were separately incubated with glutathione-Sepharose-4B beads in NETN at 4°C for 2 hr. The supernatant containing 35S-labeled Fish was then incubated for 2 hr at 4°C with the GST–Sepharose-4B beads, which had been preincubated with GST–Fish. The beads were washed several times with NETN, and bound protein was eluted with buffer containing reduced glutathione (Amersham Pharmacia). These samples, along with 2 μl of the in vitro translation mixture (40% of the input used in the binding assays), were mixed with SDS sample buffer, boiled, and electrophoresed on an 8% SDS-polyacrylamide gel. The gel was fixed in methanol/acetic acid (1:1), dried, and analyzed via autoradiography.
Schneider line 2 cell transient expression assays. Drosophila Schneider line 2 (S2) cells were cotransfected with the P[1.0slit-lacZ] reporter plasmid or the lacZ control vector C4PLZ along with combinations of plasmids that provide a source of Sim, Tgo, Fish, and Dfr proteins. The reporter plasmid contained the slit 1 kb midline glial enhancer fragment cloned into the C4PLZ enhancer/tester vector (Wharton and Crews, 1993). Expression plasmids pAct-sim, pAct-tgo, pAct-fish, and pAct-dfr were made by subcloning cDNA fragments containing the complete coding regions into pAct5C (sim, tgo, dfr) or pAct5CSRS (fish), each containing the Drosophilaactin5C promoter and poly(A) site (Han et al., 1989; Sonnenfeld et al., 1997; K. Burtis, personal communication). S2 cells were transiently transfected using Ca2PO4 (Fehon et al., 1990). Transfections were performed in triplicate or more using 5 μg/plasmid and were normalized using 2.5 μg of copia-luc, which has a luciferase (luc) reporter (pGL3-Basic; Promega) under the control of the copia long terminal repeat (LTR) promoter. The cells were lysed 48 hr after transfection, and β-Gal and luciferase assays were performed using a LacZ/β-Gal Quantitation Kit (Molecular Probes, Eugene, OR) and Luciferase Assay System (Promega), respectively.
RESULTS
sim, fish, and dfr are expressed together in developing CNS midline cells
To address whether the sim, fish, anddfr genes might functionally interact to regulate development of the embryonic CNS midline, we first analyzed whether they exhibit overlapping expression in developing midline cells. This was accomplished using anti-Fish and anti-Dfr sera, as well as a P[3.7sim-lacZ] marker that mimics sim midline expression (Nambu et al., 1991; Kasai et al., 1998). P[3.7sim-lacZ] embryos were immunostained using anti-β-gal and either anti-Fish or anti-Dfr sera. Prominent overlapping expression was detected between Sim and Fish in developing CNS midline cells from stage 8 throughout the remainder of germ band extension. Overlap was also detected in a subset of prospective foregut cells. Similar overlapping expression was also detected between Sim and Dfr. Midline coexpression of Fish and Dfr was detected by immunostaining wild-type embryos with anti-Fish and anti-Dfr sera. Both genes are expressed together in the CNS midline throughout germ band extension. Examples of this overlapping expression in germ band-extended embryos are presented in Figure1. In germ band-retracted embryos, Fish exhibited overlapping expression with Sim and Dfr in the midline glia (data not shown). Fish and Dfr were also detected together in lateral cells of the thoracic ganglia and a subset of ventral epidermal cells (data not shown). Consistent with previous studies (Ma et al., 1998), Fish protein was not detected in the trachea, a prominent site of Dfr expression (Anderson et al., 1995; Certel et al., 1996). These analyses indicated that sim, fish, and dfr are coexpressed in developing CNS midline cells. The midline expression of these three genes also overlaps that of the slit gene, which is a downstream target of Sim (Wharton and Crews, 1993; Wharton et al., 1994).
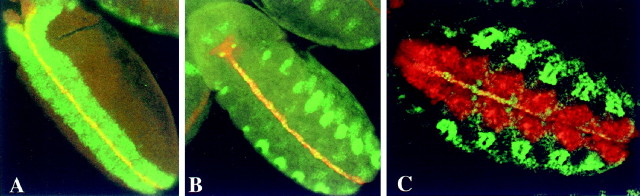
Fig. 1.
Coexpression of Sim, Fish, and Dfr in embryonic CNS midline cells detected via double-label immunostaining of P[3.7sim-lacZ] (A, B) and wild-type (C) embryos and confocal microscopy. A, A stage 9 P[3.7sim-lacZ] embryo immunostained with anti-β-gal (red) and anti-Fish (green) sera. Note strong overlapping expression (yellow) in the CNS midline cells. Fish is also strongly expressed in the lateral and cephalic neuroectoderm. B, A stage 10 P[3.7sim-lacZ] embryo immunostained with anti-β-gal (red) and anti-Dfr (green) sera. Note strong overlapping expression (yellow) in the CNS midline cells. Dfr is also strongly expressed in developing tracheal cells. C, A wild-type stage 11 embryo immunostained with anti-Fish (red) and anti-Dfr (green) sera. Note overlapping expression (yellow) in the CNS midline cells. Fish expression is not detected in tracheal cells. All views are ventral with anterior to left.
Genetic interactions reveal cooperative functions betweensim, fish, and dfr
Both loss-of-function and gain-of-function assays were used to detect genetic interactions between sim, fish, and dfr. Significantly, a previous study had revealed genetic interactions between fish (also calledDichaete) and dfr (also called ventral veins lacking) mutants in CNS midline differentiation and Slit protein expression (Sánchez Soriano and Russell, 1998). We have extended these studies to analyze potential cooperative interactions between sim, fish, and dfr in regulating slit gene transcription through use of a P[1.0slit-lacZ] marker. This reporter contains a portion of a slit intron that drives lacZ expression mimicking that of the native slit gene in developing midline glia; P[1.0slit-lacZ] expression is first detected in germ band-extended stage 11 embryos and is maintained throughout the remainder of embryogenesis (Fig.2A) (Nambu et al., 1991; Wharton and Crews, 1993; Wharton et al., 1994). fishnull mutant embryos exhibited a misplacement and loss of midline glia, as detected via anti-β-gal immunostaining (Fig.2B). P[1.0slit-lacZ] was expressed normally in stage 11 fish mutant embryos, but during germ band retraction the number of midline glia became reduced from wild type, and many cells were located at aberrant ventral positions within the nerve cord. Similar, although less severe, defects were observed indfr mutant embryos, where some midline glia were displaced from their normal positions (Fig. 2C). Notably, β-gal-expressing midline glia were still detected in bothfish and dfr mutants, indicating that unlike Sim, Fish and Dfr are not absolutely required for P[1.0slit-lacZ] expression or midline glial development.
Fig. 2.
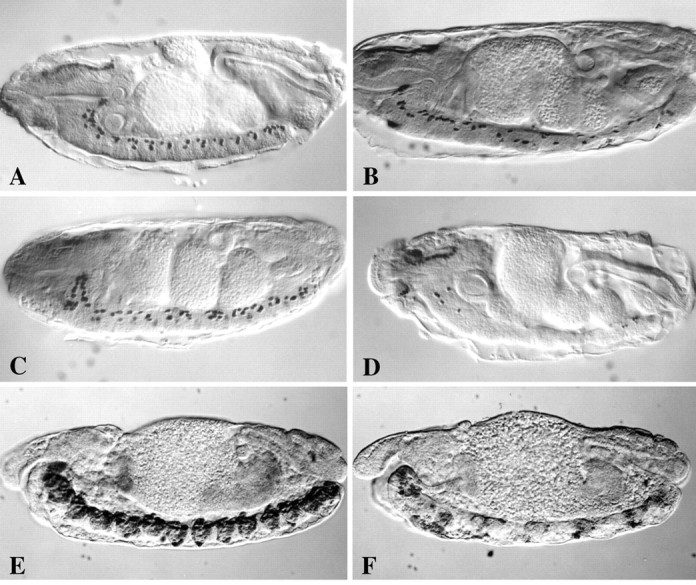
Genetic interactions between fishand dfr in CNS midline gene expression and development. Anti-β-gal immunostaining of stage 15 wild-type (A), fish87(B), dfrE82(C), anddfrE82-fish87double mutant (D) embryos carrying the P[1.0slit-lacZ] marker. In addition, anti-β-gal immunostaining was also performed on stage 15 wild-type (E) anddfrE82-fish87double mutant (F) embryos carrying the P[3.7sim-lacZ] marker. A, Note the predominantly dorsal positions of P[1.0slit-lacZ]-expressing midline glia in each segment of the ventral nerve cord in a wild-type embryo.B, In fish87 mutant embryos there is a loss and disorganization of P[1.0slit-lacZ]-expressing midline cells.C, In dfrE82 mutant embryos there is only modest misplacement of P[1.0slit-lacZ]-expressing midline cells.D, IndfrE82-fish87double mutant embryos there is a dramatic loss of P[1.0slit-lacZ]-expressing cells. This phenotype is much more severe than that seen in eitherfish87 (B) ordfrE82 (C) single mutant embryos. E, In wild-type embryos the CNS midline cells express sim and are organized into segmentally reiterated clusters of cells along the ventral nerve cord.F, IndfrE82-fish87double mutant embryos there is a severe decrease in simexpression and disorganization of midline cells. All views are sagittal with anterior to left.
We then used a dfr–fish double mutant strain to examine whether fish and dfr might act together to regulate midline gene expression. Embryos mutant for bothfish and dfr were shown to exhibit much more severe defects in P[1.0slit-lacZ] expression than eitherfish or dfr single mutants. Although P[1.0slit-lacZ] was activated normally in stage 11dfr-fish double mutant embryos, there was a striking loss of midline P[1.0slit-lacZ] expression during germ band retraction (Fig. 2D). This synergistic effect strongly suggests that Fish and Dfr function together to regulateslit transcription. These functions may be mediated directly through Fish and Dfr binding sites present in the slit 1 kb regulatory region (see below). Another, nonexclusive possibility is that Fish and Dfr might indirectly control slittranscription by regulating the expression of sim. To address this possibility we examined P[3.7sim-lacZ] expression in wild-type and dfr–fish embryos. Compared with wild-type embryos, dfr–fish double mutants exhibited a severe decrease in P[3.7sim-lacZ] expression, a phenotype that first became apparent during germ band retraction (Fig.2E,F). Thus, fishand dfr also influence sim expression and hence may indirectly influence the expression of a wide array of midline genes.
Because homozygous sim mutants exhibit severe CNS midline defects, it was not informative to analyze the phenotypes offish–sim or dfr–sim double mutants. Instead, we examined potential interactions between fish andsim via a gain-of-function approach using the Gal4/UAS targeted gene expression system (Brand and Perrimon, 1993). A P[GMR-Gal4] strain that drives Gal4 expression in and behind the morphogenetic furrow in the developing eye imaginal disk was crossed to P[UAS-fish] and P[UAS-sim] strains. P[GMR-Gal4]/+;P[UAS-fish]/+ animals exhibited a moderate eye roughening with disruption of ommatidia organization and loss of mechanosensory bristles (Mukherjee et al., 2000) (Fig.3A,D). In contrast, ectopic sim expression resulted in essentially normal eye morphology (Fig. 3B,E). The effects of fish and sim coexpression revealed a nonadditive phenotype; there was a stronger disorganization of ommatidia and mechanosensory bristles than seen in flies expressingfish or sim alone, and there was also a dramatic loss of eye pigmentation (Fig.3C,F). These results indicated that ectopic expression of fish and simsynergistically alters normal eye development, and along with other data described below supports the hypothesis that these genes can interact functionally.
Fig. 3.
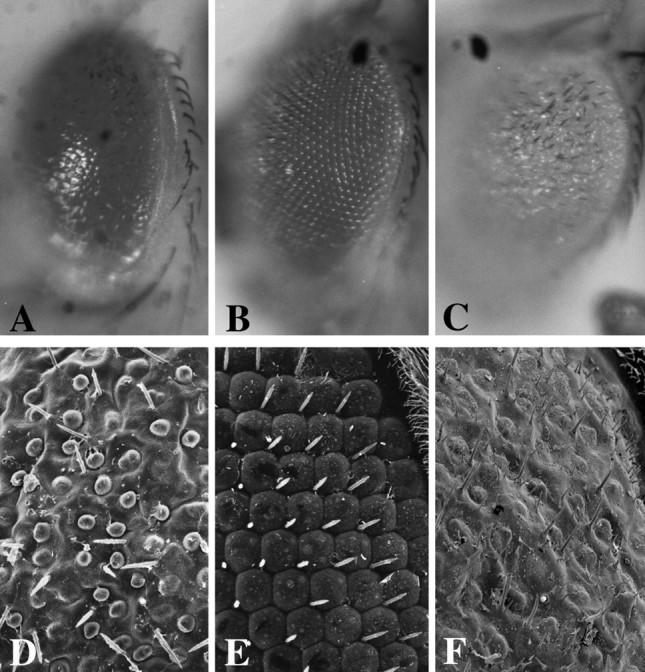
Interactions between Fish and Sim detected via gain-of-function assay. Light microscope (A–C) or scanning electron microscope (D–F) analysis of eyes from adults in which ectopic Fish and/or Sim expression was driven in developing eye imaginal disks via P[GMR-Gal4]. A,D, A P[GMR-Gal4]/+; P[UAS-fish]/+ animal. Note that the eye is roughened, and there is a disorganization of ommatidia and loss of most mechanosensory bristles. There is a slight loss of eye pigmentation. B, E, A P[GMR-Gal4]/+; P[UAS-sim]/+ animal. Note essentially normal organization of ommatidia and mechanosensory bristles and uniform eye pigmentation. C, F, A P[GMR-Gal4]/+; P[UAS-sim]/P[UAS-fish] animal. Note more severe disorganization of ommatidia and mechanosensory bristles as well as a strong loss of eye pigmentation. A–C, Light microscope images at 20× magnification. D–F, Scanning electron microscope images at 1000× magnification.
Sim, Fish, and Dfr directly regulateslit transcription
Previous DNA sequence analysis of a 380 bp slit midline regulatory fragment indicated the presence of a single CME, through which Sim:: Tgo heterodimers act (Wharton et al., 1994;Sonnenfeld et al., 1997). The CME is located within 300 bp from the distal end (farther from the promoter in the native slitgene) of this fragment (Wharton et al., 1994). We additionally noted the presence of an inverted TTCAAT repeat (TTCAATTTCATTGAA) located 20 bp proximal to the CME. This sequence resembles a (A/T)(A/T)CAAT consensus binding site for Sox proteins, although to our knowledge, binding of Sox proteins to a TTCAAT sequence has not been reported. Because sequences present in an extended 1 kb slit DNA fragment are required for normal levels of slit expressionin vivo (Wharton and Crews, 1993), we obtained additional DNA sequences. This analysis indicated that no other CMEs are present in the 1 kb slit DNA fragment. However, we did identify two perfect Dfr consensus binding sites (Certel et al., 1996), ATGCAAAT and CATAAAT, located within 500 bp of DNA proximal to the CME (Fig.4A). These two Dfr binding sites are separated by ∼150 bp and flank a consensus Fish binding site, TACAAT (Fig. 4A). These data suggest that Fish, Sim, and Dfr may all bind to sites present in the 1 kbslit regulatory DNA fragment. To test this possibility, DNA gel mobility shift assays were performed using the Fish HMG domain and full-length Dfr protein on double-stranded oligonucleotide probes corresponding to sequences from the slit 1 kb fragment. The Fish HMG domain bound strongly to a 26 mer probe containing the TACAAT site (Fig. 4B). In contrast, Fish did not bind consistently to a 26 mer probe containing both TTCAAT sites, suggesting that Fish can distinguish between closely related DNA sequences. Dfr protein bound very strongly to a 33 mer probe that contained the ATGCAAAT site, and less strongly to a 32 mer probe containing the CATAAAT site (Fig. 4B). Dfr bound the ATGCAAAT site both as an apparent monomer and a dimer, because two distinct bands with reduced mobilities were detected. The 1 kb slitfragment thus may integrate the actions of at least three different types of regulatory proteins, represented by Sim, Fish, and Dfr.
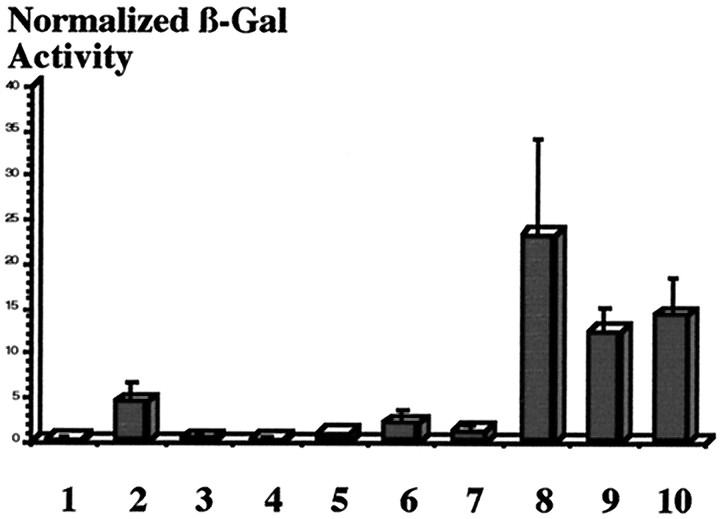
We next examined the ability of Fish, Dfr, Sim, and Tgo to directly control slit transcription using transient transcription assays in cultured Drosophila S2 cells (Fig.5). The P[1.0slit-lacZ] construct was used as a reporter with various combinations of plasmids that express Fish, Dfr, Sim, or Tgo. Fish modestly activated P[1.0slit-lacZ] transcription (4 units of β-gal activity), indicating that in both yeast (Ma et al., 1998) and fly cells, Fish can function as a direct transcriptional activator. Dfr resulted in little if any activation of P[1.0slit-lacZ] (<1 unit), and Dfr and Fish together did not exhibit any increased activation over the levels observed for Fish alone. Neither Sim nor Tgo alone was able to activate the P[1.0slit-lacZ] reporter, because only background levels of expression were detected (<0.4 units of activity). Furthermore, Sim and Tgo together yielded only minimal P[1.0slit-lacZ] activation (1 unit). These results imply that although Sim:: Tgo heterodimers strongly activate expression of a P[6XCME-lacZ] reporter (>150 units) that contains six multimerized CMEs (Sonnenfeld et al., 1997), additional factors are required to achieve high levels of P[1.0slit-lacZ] expression. Significantly, the combination of either Fish and Sim:: Tgo or Dfr and Sim:: Tgo both resulted in relatively high levels of P[1.0slit-lacZ] activation (23 units for Fish and Sim:: Tgo and 12 units for Dfr and Sim:: Tgo). Thus, both Fish and Dfr strongly enhanced the ability of Sim:: Tgo heterodimers to activate slittranscription. Comparable levels of P[1.0slit-lacZ] activation (14 units) were observed when all four proteins were expressed together. Taken together, the DNA binding and transcriptional activation assays provide additional evidence that regulation ofslit expression in the midline glia requires functional interactions between Fish, Dfr, Sim, and Tgo.
Fig. 5.
Fish and Dfr enhance Sim:: Tgo transcription of the P[1.0slit-lacZ] transgene in cultured Drosophila S2 cells. S2 cells were cotransfected with P[1.0slit-lacZ] reporter alone (1) or P[1.0slit-lacZ] with various combinations of the expression plasmids pAct-sim, pAct-tgo, pAct-fish, and pAct-dfr(2–10). Transfection efficiencies were normalized using a copia-luc plasmid. Cells were lysed 48 hr after transfection, and β-gal and luciferase activity were assayed. Normalized β-gal activity is expressed in arbitrary fluorescence units as the mean (SEM of 3–5 independent transfections). (1) 1.0slit-lacZ; (2) 1.0slit-lacZ + Fish; (3) 1.0slit-lacZ + Tgo; (4) 1.0slit-lacZ + Sim; (5) 1.0slit-lacZ + Dfr; (6) 1.0slit-lacZ + Fish + Dfr; (7) 1.0slit-lacZ + Sim + Tgo; (8) 1.0slit-lacZ + Sim + Tgo + Fish; (9) 1.0slit-lacZ + Sim + Tgo + Dfr; (10) 1.0slit-lacZ + Sim + Tgo + Fish + Dfr.
Interactions between Sim, Fish, and Dfr proteins
The ability of Sim, Fish, and Dfr to regulate P[1.0slit-lacZ] expression and the location of their respective DNA binding sites in the 1 kb slit DNA fragment suggests that their regulatory functions may involve direct protein–protein interactions. We initially addressed this possibility using yeast 2-hybrid assays, in which combinations of bait (fusions to the LexA-DNA binding domain) and prey (fusions to the B42 activation domain) constructs that express full-length or truncated Sim, Fish, and Dfr proteins were tested for their ability to activate expression of aLEU2 reporter gene. Full-length Fish yielded strong interaction with both full-length Sim and full-length Dfr (Fig.6A). In addition, Fish exhibited the ability to self-associate, suggesting that it may form homodimers. We then analyzed which regions of Sim and Dfr associate with Fish. Interactions were detected between full-length Fish and Sim bHLH-PAS and PAS constructs, but not a Sim bHLH-only construct (Fig.6A). Thus, Fish specifically interacts with the PAS domain of Sim. Only a single PAS region was required for this interaction, because Fish also exhibited interactions with a Sim bHLH-PAS-A construct. Full-length Fish was also found to interact with the POU domain of Dfr (Fig. 6A). This is consistent with the ability of the vertebrate Sox2 protein to associate with the POU domain of Oct3 (Ambrosetti et al., 1997). In an attempt to map which region of Fish is responsible for interactions with Sim and Dfr, we used several bait constructs that express truncated versions of Fish, including the HMG domain, NH2-terminal region, COOH-terminal region, NH2+HMG, HMG+COOH, or NH2+COOH regions. None of these constructs exhibited consistent interaction with any Sim or Dfr prey construct. Although it is possible that structural determinants present only on full-length Fish are required for interactions with PAS or POU domains in the yeast 2-hybrid assay, on the basis of previous studies on Sox2 (Ambrosetti et al., 1997) it is likely that the Fish HMG domain is a key contributor to these interactions.
Fig. 6.
Fish directly associates with POU and PAS domain proteins. A, Interactions between Fish, Sim, and Drifter revealed via yeast 2-hybrid assays. The following combinations of bait and prey constructs were transformed into yeast and assayed for growth on Leu- medium: (1) Fish/TrhbHLH-PAS; (2) Fish/PerL; (3) Fish/Per; (4) Fish/Sim; (5) Fish/SimPAS; (6) Fish/SimbHLH; (7) Fish/SimbHLH-PAS; (8) Fish/SimbHLH-PASA; (9) Fish/Fish; (10) Fish/Drifter; (11) Fish/DrifterPOU; (12) Fish/Pdm-1; (13) Fish/Pdm-2; (14) Fish/Oct-3; (15) Drifter/Sim; (16) Drifter/SimbHLH-PAS; (17) DrifterPOU/Sim; (18) DrifterPOU/SimbHLH-PAS. Note that interactions were detected between Fish and the POU domain of Drifter, as well as the PAS domain but not the bHLH region of Sim. No interactions were detected between any Sim and Drifter constructs. B, Binding of GST–Fish fusion protein to 35S-labeled Sim generated via in vitro translation. Lane 1, In vitro-translated Sim analyzed via SDS-PAGE and autoradiography. Note a radiolabeled band at 73 kDa corresponding to full-length Sim protein, as well as a second band of lower molecular weight.Lane 2, In vitro-translated Sim does not bind GST. Sim protein was incubated with GST and glutathione-Sepharose resin. The mixture was washed, and the bound material was eluted and analyzed via SDS-PAGE and autoradiography. Note the absence of labeled Sim protein. Lane 3, In vitro-translated Sim binds to GST–Fish. Sim protein was incubated with GST–Fish and glutathione-Sepharose resin. The mixture was washed, and the bound material was eluted and analyzed via SDS-PAGE and autoradiography. Note the presence of labeled Sim proteins. C, Binding of GST–Fish fusion protein to 35S-labeled Fish generated viain vitro translation. Lane 1, In vitro-translated Fish analyzed via SDS-PAGE and autoradiography. Note a major radiolabeled band at ∼45 kDa corresponding to full-length Fish. Lane 2, In vitro-translated Fish does not bind efficiently to GST. Fish protein was incubated with GST and glutathione-Sepharose resin. The mixture was washed, and the bound material was eluted and analyzed via SDS-PAGE and autoradiography. Note the absence of labeled Fish protein.Lane 3, In vitro-translated Fish does bind GST–Fish. Fish protein was incubated with GST–Fish and glutathione-Sepharose resin. The mixture was washed, and the bound material was eluted and analyzed via SDS-PAGE and autoradiography. Note the presence of labeled Fish protein.
To verify the interactions between Fish and Sim and the self-association of Fish, we additionally performed GST pulldown assays. Fish and Sim interaction was tested using bacterially expressed GST or GST–Fish fusion protein, and35S-labeled Sim generated via in vitro translation. The 35S-labeled Sim migrated on an SDS polyacrylamide gel as a doublet, with one band near the predicted molecular weight of Sim protein (73 kDa) and the second at a somewhat lower apparent molecular weight (Fig.6B). Equal amounts of labeled Sim protein were incubated with either GST or GST–Fish, and the mixtures were subjected to glutathione-Sepharose chromatography followed by SDS-PAGE and autoradiography. As expected, Sim did not bind to the GST because no labeled bands were detected (Fig. 6B). However, Sim did associate with GST–Fish, because both of the labeled Sim bands were observed (Fig. 6B). These data confirm the results of the yeast 2-hybrid assays and indicate that Fish and Sim are capable of direct physical association. Similar GST pulldown assays were performed to verify Fish self-association using GST–Fish and35S-labeled in vitro-translated Fish protein. The major band present in the35S-labeled Fish reaction migrated at ∼45 kDa (Fig. 6C), close to the predicted molecular weight of Fish protein (40 kDa). This 45 kDa band was specifically bound by GST–Fish but not GST alone (Fig. 6C). This result also confirmed the yeast 2-hybrid data indicating that Fish is able to self-associate.
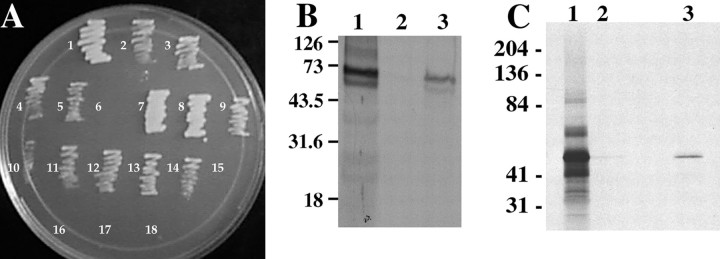
No interactions were detected between Sim bait and Dfr prey constructs in the yeast 2-hybrid assays (Fig. 6A), indicating that at least in yeast, they do not associate directly. Because both proteins did associate with Fish, we tested whether Fish might facilitate interactions between Sim and Dfr. This was pursued using Sim bait, Dfr prey, and a pDB20-fish construct to express full-length native Fish in yeast (see Materials and Methods). The combinations of pDB20-fish and either Sim bait or Dfr prey did not result in any detectable interaction, as indicated by the inability of transformed yeast cells to activate LEU2reporter gene expression and grow on leu−medium (Fig. 7). However, the simultaneous presence of pDB20-fish, Sim bait, and Dfr prey constructs did permit growth on leu−medium (Fig. 7), indicating that Sim, Fish, and Dfr are able to form a ternary complex in yeast cells.
Fig. 7.
Fish, Sim, and Dfr can form a functional ternary complex in yeast cells. Combinations of full-length Sim bait, full-length Dfr prey, and full-length Fish expression constructs were tested for their abilities to interact in yeast cells.A, A combination of Sim bait and native Fish did not activate LEU2 expression, as indicated by the absence of cell growth on medium lacking leucine. B, A combination of Dfr prey and native Fish did not activate LEU2expression, as indicated by the absence of cell growth on medium lacking leucine. C, A combination Sim bait, Dfr prey, and Fish did result in activation of LEU2 expression, as indicated by significant cell growth on medium lacking leucine. Fish can facilitate interactions between Sim bait and Dfr prey in yeast cells.
Additional yeast 2-hybrid assays were performed to examine potential interactions between Fish and other PAS and POU proteins. Full-length Fish interacted with the POU domains of both DrosophilaPdm-1 and Pdm-2, which are involved in embryonic segmentation and nervous system development (Bhat et al., 1995; Yeo et al., 1995; Ma et al., 1998), as well as full-length mouse Oct3 (Fig.6A). Fish also interacted with the bHLH-PAS region ofDrosophila Trachealess (Trh), a protein required for formation of several types of tubular tissues (Wilk et al., 1996). As well, Fish interacted with a PAS domain-containing fragment ofDrosophila Per (Per-C2) (Huang et al., 1995), a PAS-only protein that associates with the Timeless protein and plays a key role in regulating biological rhythms (for review, see Hardin, 1998; Young, 1998). Because Fish also interacted with PerL, a mutant version of Per that exhibits reduced ability to bind Timeless (Gekakis et al., 1995), it appears that distinct residues are important for Per/Fish versus Per/Timeless interactions. Together, these data indicate that Fish can associate with multiple POU and PAS domain proteins and suggest that functional interactions between Sox, PAS, and POU proteins may be important in diverse developmental and physiological processes.
DISCUSSION
Functions of Sim, Fish, and Dfr in the regulation of CNS midline gene expression
The Drosophila slit gene encodes a large extracellular protein that is required for normal midline glial migration and axon projection patterns (Rothberg et al., 1988, 1990). slitexpression in the CNS midline glia was mimicked by a P[1.0slit-lacZ] transgene that is first expressed in fully germ band-extended embryos (Wharton and Crews, 1993). This expression is completely dependent on the functions of the bHLH-PAS protein Sim, because sim mutant embryos exhibit a complete absence of P[1.0slit-lacZ] expression (Nambu et al., 1991). In this study, we show that the combined functions of the Sox protein Fish and the POU domain protein Dfr are also essential for P[1.0slit-lacZ] expression, because dfr–fishdouble mutant embryos exhibit a dramatic loss of P[1.0slit-lacZ] expression in germ band-retracted embryos. Additionally, Fish and Dfr significantly enhanced the ability of Sim and Tgo to activate P[1.0slit-lacZ] expression in culturedDrosophila S2 cells.
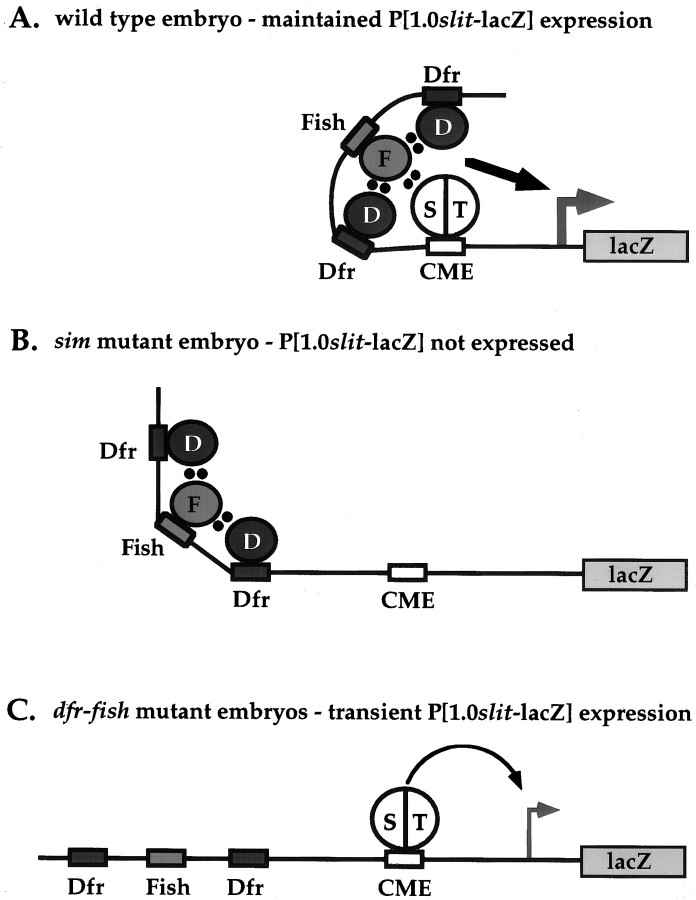
The 1 kb slit midline regulatory region was found to contain binding sites for Fish and Dfr, as well as a single CME through which Sim:: Tgo heterodimers function. Fish bound strongly to a TACAAT site and Dfr bound to two sites, ATGCAAAT and CATAAAT, that flank the Fish site within a 150 bp interval. Removal of the Fish and Dfr binding sites reduces slit expression, because a P[380slit-lacZ] strain that contains the CME but not the Fish or Dfr binding sites exhibits decreased levels of embryonic midline expression compared with P[1.0slit-lacZ] (Wharton and Crews, 1993). Fish did not consistently bind to two TTCAAT sequences located close to the CME; however, given the proximity of these sites, it will be of interest to determine whether Fish and Sim:: Tgo might bind these sequences cooperatively. We also determined that Fish protein directly associates with the PAS domain of Sim and the POU domain of Dfr. In addition, all three proteins were able to form a ternary transcriptional regulatory complex in yeast. Together, these results provide strong evidence that these proteins act together to directly regulate midline gene expression. One model for the functions of these proteins is that although Sim:: Tgo heterodimers are sufficient to activate embryonic expression of P[1.0slit-lacZ], Fish and Dfr are required to maintain high levels of LacZ expression (Fig.8).
Fig. 8.
A model for regulation of P[1.0slit-lacZ] expression in the CNS midline via interactions between Sim, Tgo, Fish, and Dfr. In wild-type embryos (A) the binding of a Sim:: Tgo heterodimer to the CME and Fish and Dfr to nearby sites results in the formation of a ternary regulatory complex that permits sustained midline P[1.0slit-lacZ] expression. In the absence of Sim (B), Fish and Dfr are not sufficient to activate expression of P[1.0slit-lacZ]. In the absence of both Fish and Dfr (C), the initial presence of Sim is sufficient to activate midline P[1.0slit-lacZ] expression; however, this expression is not maintained in germ band-retracted dfr-fish mutant embryos.D, Drifter; F, Fish; S, Sim; T, Tgo. Dots indicate direct protein–protein interactions.
Functional interactions between Sim, Fish, and Dfr may also regulate the midline expression of other genes, including sim andbreathless (btl). Thus, sim has autoregulatory functions (Nambu et al., 1991), and we have shown that the combined functions of dfr and fish are also required for sustained midline sim expression. In addition, a 2.8 kb interval in the P[3.7sim-lacZ] transgene used in this study contains six evolutionarily conserved CMEs (Kasai et al., 1998) as well as several consensus Fish and Dfr binding sites.btl encodes an FGF receptor homolog whose expression in the CNS midline and tracheal cells has been shown to depend, respectively, on Dfr as well as Sim and Tgo, or Trh and Tgo (Anderson et al., 1996;Ohshiro and Saigo, 1997). A 200 bp btl midline/tracheal regulatory region contains three evolutionarily conserved CMEs (Ohshiro and Saigo, 1997). Inspection of this region also revealed the presence of a conserved consensus ATCAAT Fish binding site located in a 40 bp interval between CME2 and CME3, as well as a conserved consensus GATAAAT Dfr binding site (Anderson et al., 1996) located 40 bp downstream of CME3. Thus, functional interactions between Sim, Fish, and Dfr could be a general mechanism to regulate gene transcription during CNS midline development.
Like other Sox proteins, Fish can influence transcription through DNA bending, direct transcriptional activation, and protein–protein interactions. It will be of interest to determine the relative contributions of these activities in regulating slitexpression. In this regard, Sox proteins may act in distinct capacities depending on the presence of other regulatory proteins and the organization of control elements in a specific target gene. For example, close range interactions between Sox2 and Oct3 are required to synergistically activate a distal enhancer element from the vertebratefgf4 gene (Yuan et al., 1995; Ambrosetti et al., 1997). Increasing the distance between the Sox2 and Oct3 binding sites from 3 to 6 bp abolishes the ability of these proteins to activate transcription (Ambrosetti et al., 1997). In contrast, Sox2 represses Oct4-induced activation of a preimplantation enhancer from the mouseosteopontin gene by acting at a site 39 bp away from the Oct4 binding site (Botquin et al., 1998). This is similar to the interactions between Fish and Dfr through nonadjacent binding sites in the slit 1 kb midline enhancer. These findings indicate that there are different mechanisms through which Sox and POU domain proteins can interact to regulate transcriptional processes. Significantly, in the native fgf4, osteopontin, and slit genes, the relevant regulatory regions are all located several kilobases downstream of the promoter. This suggests that in addition to local DNA bending activities, the in vivo functions of Sox proteins may also involve DNA looping events (Lamb and Rizzino, 1998). Because Sry and presumably other Sox proteins bind DNA as a monomer (Werner et al., 1995), DNA looping could be mediated via Sox protein dimerization, as detected for Fish and LSox5 and Sox6 proteins (Lefebvre and de Crombrugghe, 1998), as well as through association between Sox and PAS or POU domain proteins.
Conserved PAS, Sox, and POU regulatory interactions in nervous system development?
There are several examples of functional association between vertebrate HMG domain and POU domain proteins (Zwilling et al., 1995;Ambrosetti et al., 1997; Botquin et al., 1998; Kuhlbrodt et al., 1998), and our studies have revealed similar association betweenDrosophila Sox and POU proteins. In addition, we have identified novel interactions between Sox and PAS domain proteins. Given the considerable sequence divergence between the PAS domains of Sim or Trh and Per, the ability of Fish to directly associate with each of these proteins suggests that interactions between Sox and PAS domain proteins may also be widespread. In this regard we have found that the mouse Sox2 protein, which contains an HMG domain highly related to that of Fish, also associates with Sim and Per (Y. Gao and J. R. Nambu, unpublished results). In addition, midline-targeted expression of Sox2 has been shown to partially rescue the axon scaffold defects infish mutant embryos (Sánchez Soriano and Russell, 1998). Although the precise mechanism of PAS/Sox association is not clear, our results indicate that a single PAS region is sufficient for binding to a Sox protein, and they suggest that the HMG domain is crucial for this interaction. Further analyses of the regulatory interactions between Sim, Fish, and Dfr may ultimately provide a useful paradigm for a better understanding of the widespread functions of PAS, Sox, and POU genes in vertebrate neural development, and their involvement in specific human congenital disorders that result in deafness, obesity, and mental retardation (Pevny and Lovell-Badge, 1997; Crews, 1998; Crews and Fan, 1999; Latchman, 1999; McEvilly and Rosenfeld, 1999; Wegner, 1999).
Footnotes
This work was supported by grants from the March of Dimes and National Institutes of Health (NIH) to J.R.N., NIH Grant NS28743 to W.A.J., and grants from the National Institute of Child Health and Human Development and National Science Foundation to S.T.C. K.C. was supported by an NIH Genetics Training Grant. We are indebted to Lisa Dailey for providing us with Sox2 and Oct3 cDNA clones, Steve Poole for providing us with pdm-1 and pdm-2 cDNA clones, and Michael Rosbash for providing period yeast 2-hybrid constructs. In addition, we gratefully acknowledge Merrill Shaffer, Xiaoliang Shan, and Kahlin Clark for assistance in generating yeast 2-hybrid constructs.
Correspondence should be addressed to Dr. John R. Nambu, Biology Department, Morrill Science Center, University of Massachusetts, Amherst, MA 01003. E-mail: jnambu@bio.umass.edu.
E. Niemitz's present address: Predoctoral Training Program in Human Genetics, Johns Hopkins University School of Medicine, Baltimore, MD 21205.
REFERENCES
- 1.Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson MG, Perkins GL, Chittick P, Shrigley RJ, Johnson WA. drifter, a Drosophila POU-domain transcription factor, is required for correct differentiation and migration of tracheal cells and midline glia. Genes Dev. 1995;9:123–137. doi: 10.1101/gad.9.1.123. [DOI] [PubMed] [Google Scholar]
- 3.Anderson MG, Certel, Certel K, Lee T, Montell DJ, Johnson WA. Function of the Drosophila POU domain transcription factor drifter as an upstream regulator of breathless receptor tyrosine kinase expression in developing trachea. Development. 1996;122:4169–4178. doi: 10.1242/dev.122.12.4169. [DOI] [PubMed] [Google Scholar]
- 4.Becker DM, Fikes JD, Guarente L. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc Natl Acad Sci USA. 1991;88:1968–1972. doi: 10.1073/pnas.88.5.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhat KM, Poole SJ, Schedl P. The miti-mere and pdm1 genes collaborate during specification of the RP2/sib lineage in Drosophila neurogenesis. Mol Cell Biol. 1995;15:4052–4063. doi: 10.1128/mcb.15.8.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billin AN, Cockerill KA, Poole SJ. Isolation of a family of Drosophila POU domain genes expressed in early development. Mech Dev. 1991;34:75–84. doi: 10.1016/0925-4773(91)90045-8. [DOI] [PubMed] [Google Scholar]
- 7.Botquin V, Hess H, Fuhrmann G, Anastassiadis C, Gross MK, Vriend G, Scholer HR. New POU dimer configuration mediates antagonistic control of anosteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998;12:2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 9.Certel K, Anderson MG, Shrigley RJ, Johnson WA. Distinct variant DNA-binding sites determine cell-specific autoregulated expression of the Drosophila POU domain transcription factor drifter in midline glia or trachea. Mol Cell Biol. 1996;16:1813–1823. doi: 10.1128/mcb.16.4.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crews ST. Control of cell lineage-specific development and transcription bybHLH-PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- 11.Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr Opin Genet Dev. 1999;9:580–587. doi: 10.1016/s0959-437x(99)00003-9. [DOI] [PubMed] [Google Scholar]
- 12.Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MAT, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- 13.Gekakis N, Saez L, Delahaye-Brown AM, Myers MP, Sehgal A, Young MW, Weitz CJ. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 14.Guthrie S. Axon guidance: starting and stopping with slit. Curr Biol. 1999;9:R432–435. doi: 10.1016/s0960-9822(99)80274-7. [DOI] [PubMed] [Google Scholar]
- 15.Han K, Levine MS, Manley JL. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 1989;56:573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- 16.Hardin PE. Activating inhibitors and inhibiting activators: a day in the life of a fly. Curr Opin Neurobiol. 1998;8:642–647. doi: 10.1016/s0959-4388(98)80093-7. [DOI] [PubMed] [Google Scholar]
- 17.Harris R, Sabatelli LM, Seeger MA. Guidance cues at the Drosophila CNS midline: identification and characterization of two Drosophila Netrin/UNC-6 homologs. Neuron. 1996;17:217–228. doi: 10.1016/s0896-6273(00)80154-3. [DOI] [PubMed] [Google Scholar]
- 18.Huang ZJ, Curtin KD, Rosbash M. PER protein interactions and temperature compensation of a circadian clock in Drosophila. Science. 1995;267:1169–1172. doi: 10.1126/science.7855598. [DOI] [PubMed] [Google Scholar]
- 19.Johnson WA. Characterization of neuron-specific transcription factors in Drosophila melanogaster. Methods Neurosci. 1992;9:362–380. [Google Scholar]
- 20.Kasai Y, Stahl S, Crews ST. Specification of the Drosophila CNS midline cell lineage: direct control of single-minded transcription by dorsal/ventral patterning genes. Gene Exp. 1998;7:171–189. [PMC free article] [PubMed] [Google Scholar]
- 21.Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 22.Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M. Cooperative function of POU proteins and SOX proteins in glial cells. J Biol Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- 23.Lamb KA, Rizzino A. Effects of differentiation on the transcriptional regulation of the FGF-4 gene: critical roles played by a distal enhancer. Mol Reprod Dev. 1998;51:218–224. doi: 10.1002/(SICI)1098-2795(199810)51:2<218::AID-MRD12>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Latchman DS. POU family transcription factors in the nervous system. J Cell Physiol. 1999;179:126–133. doi: 10.1002/(SICI)1097-4652(199905)179:2<126::AID-JCP2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre V, Li P, de Crombrugghe BA. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Niemitz EL, Nambu PA, Shan X, Sackerson C, Fujioka M, Goto T, Nambu JR. Gene regulatory functions of Drosophila Fish-hook, a high mobility group domain Sox protein. Mech Dev. 1998;73:169–182. doi: 10.1016/s0925-4773(98)00050-1. [DOI] [PubMed] [Google Scholar]
- 27.McEvilly RJ, Rosenfeld MG. The role of POU domain proteins in the regulation of mammalian pituitary and nervous system development. Prog Nucleic Acid Res Mol Biol. 1999;63:223–255. doi: 10.1016/s0079-6603(08)60724-2. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell KJ, Doyle JL, Serafini T, Kennedy TE, Tessier-Lavigne M, Goodman CS, Dickson BJ. Genetic analysis of Netrin genes in Drosophila: Netrins guide CNS commissural axons and peripheral motor axons. Neuron. 1996;17:203–215. doi: 10.1016/s0896-6273(00)80153-1. [DOI] [PubMed] [Google Scholar]
- 29.Mitchison TJ, Sedat J. Localization of antigenic determinants in whole Drosophila embryos. Dev Biol. 1983;99:261–264. doi: 10.1016/0012-1606(83)90275-0. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee A, Shan X, Mutsuddi M, Ma Y, Nambu JR. The Drosophila Sox gene, fish-hook, is required for postembryonic development. Dev Biol. 2000;217:91–106. doi: 10.1006/dbio.1999.9506. [DOI] [PubMed] [Google Scholar]
- 31.Muralidhar MG, Callahan CA, Thomas JB. Single-minded regulation of genes in the embryonic midline of the Drosophila central nervous system. Mech Dev. 1993;41:129–138. doi: 10.1016/0925-4773(93)90043-w. [DOI] [PubMed] [Google Scholar]
- 32.Nambu JR, Lewis JO, Wharton KA, Jr, Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein which acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 33.Nambu PA, Nambu JR. The Drosophila Fish-hook gene encodes a HMG domain protein essential for segmentation and CNS development. Development. 1996;122:3467–3475. doi: 10.1242/dev.122.11.3467. [DOI] [PubMed] [Google Scholar]
- 34.Ohshiro T, Saigo K. Transcriptional regulation of breathless FGF receptor gene by binding of TRACHEALESS/dARNT heterodimers to three central midline elements in Drosophila developing trachea. Development. 1997;124:3975–3986. doi: 10.1242/dev.124.20.3975. [DOI] [PubMed] [Google Scholar]
- 35.Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: practical uses in cell and molecular biology, methods in cell biology, Vol 44. Academic; New York: 1994. pp. 446–485. [DOI] [PubMed] [Google Scholar]
- 36.Pevny LH, Lovell-Badge R. Sox genes find their feet. Curr Opin Genet Dev. 1997;7:338–344. doi: 10.1016/s0959-437x(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 37.Rothberg JM, Hartley DA, Walther Z, Artavanis-Tsakonas S. slit: an EGF-homologous locus of D. melanogaster involved in the development of the embryonic central nervous system. Cell. 1988;55:1047–1059. doi: 10.1016/0092-8674(88)90249-8. [DOI] [PubMed] [Google Scholar]
- 38.Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S. slit: an extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes Dev. 1990;4:2169–2187. doi: 10.1101/gad.4.12a.2169. [DOI] [PubMed] [Google Scholar]
- 39.Russell SR, Sánchez-Soriano N, Wright CR, Ashburner M. The Dichaete gene of Drosophila melanogaster encodes a Sox-domain protein required for embryonic segmentation. Development. 1996;122:3669–3676. doi: 10.1242/dev.122.11.3669. [DOI] [PubMed] [Google Scholar]
- 40.Sánchez Soriano N, Russell R. The Drosophila SOX-domain protein Dichaete is required for the development of the central nervous system midline. Development. 1998;125:3989–3996. doi: 10.1242/dev.125.20.3989. [DOI] [PubMed] [Google Scholar]
- 41.Sonnenfeld M, Ward M, Nystrom G, Mosher J, Stahl S, Crews ST. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development. 1997;124:4571–4582. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- 42.Tear G, Harris R, Sutaria S, Kilomanski K, Goodman CS, Seeger MA. commissureless controls growth cone guidance across the CNS midline in Drosophila and encodes a novel membrane protein. Neuron. 1996;16:501–514. doi: 10.1016/s0896-6273(00)80070-7. [DOI] [PubMed] [Google Scholar]
- 43.Van Vactor D, Flanagan JG. The middle and the end: slit brings guidance and branching together in axon pathway selection. Neuron. 1999;22:649–652. doi: 10.1016/s0896-6273(00)80723-0. [DOI] [PubMed] [Google Scholar]
- 44.Ward MP, Mosher JT, Crews ST. Regulation of bHLH-PAS protein subcellular localization during Drosophila embryogenesis. Development. 1998;125:1599–1608. doi: 10.1242/dev.125.9.1599. [DOI] [PubMed] [Google Scholar]
- 45.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 47.Wharton KA, Jr, Crews ST. CNS midline enhancers of the Drosophila slit and Toll genes. Mech Dev. 1993;40:141–154. doi: 10.1016/0925-4773(93)90072-6. [DOI] [PubMed] [Google Scholar]
- 48.Wharton KA, Jr, Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation. Development. 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- 49.Wilk R, Weizman I, Shilo BZ. trachealess encodes a bHLH-PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes Dev. 1996;10:93–102. doi: 10.1101/gad.10.1.93. [DOI] [PubMed] [Google Scholar]
- 50.Xiao H, Hrdlicka L, Nambu JR. Alternate functions of the single-minded and rhomboid genes in development of the Drosophila ventral neuroectoderm. Mech Dev. 1996;58:65–74. doi: 10.1016/s0925-4773(96)00559-x. [DOI] [PubMed] [Google Scholar]
- 51.Yeo SL, Lloyd A, Kozak K, Dinh A, Dick T, Yang X, Sakonju S, Chia W. On the functional overlap between two Drosophila POU homeo domain genes and the cell fate specification of a CNS neural precursor. Genes Dev. 1995;9:1223–1236. doi: 10.1101/gad.9.10.1223. [DOI] [PubMed] [Google Scholar]
- 52.Young MW. The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu Rev Biochem. 1998;67:135–152. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- 53.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 54.Zinn K, Sun QV. Slit branches out: a secreted protein mediates both attractive and repulsive axon guidance. Cell. 1999;97:1–4. doi: 10.1016/s0092-8674(00)80707-2. [DOI] [PubMed] [Google Scholar]
- 55.Zwilling S, Konig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]