Abstract
The dentate gyrus of the hippocampus is uniquely organized with a displaced proliferative zone that continues to generate dentate granule cells throughout life. We have analyzed the expression of Notch receptors, Notch ligands, and basic helix-loop-helix (bHLH) genes during dentate gyrus development to determine whether the need to maintain a pool of undifferentiated precursors is reflected in the patterns of expression of these genes. Many of these genes are expressed diffusely throughout the cortical neuroepithelium at embryonic days 16 and 17 in the rat, just preceding the migration of newly born granule cells and dentate precursor cells into the dentate anlage. However, at this time, Mash1, Math3, and Id3 expression are all concentrated in the area that specifically gives rise to granule cells and dentate precursor cells. Two days later, at the time of migration of the first granule cells and dentate precursor cells, cells expressing Mash1 are seen in the migratory route from the subventricular zone to the developing dentate gyrus. Newly born granule cells expressing NeuroD are also present in this migratory pathway. In the first postnatal week, precursor cells expressing Mash1 reside in the dentate hilus, and by the third postnatal week they have largely taken up their final position in the subgranular zone along the hilar side of the dentate granule cell layer. After terminal differentiation, granule cells born in the hilus or the subgranular zone begin to express NeuroD followed by NeuroD2. This study establishes that the expression patterns of bHLH mRNAs evolve during the formation of the dentate gyrus, and the precursor cells resident in the mature dentate gyrus share features with precursor cells found in development. Thus, many of the same mechanisms that are known to regulate cell fate and precursor pool size in other brain regions are likely to be operative in the dentate gyrus at all stages of development.
Keywords: NeuroD, basic helix-loop-helix protein, Mash1, dentate gyrus, hippocampus, precursor cell
The dentate gyrus is the primary afferent pathway into the hippocampus and thus has a crucial role in the circuitry of the hippocampal formation. The dentate gyrus is one of only two locations, along with olfactory interneurons generated in the subventricular zone, where new neurons are added to the existing neural circuitry throughout life (Altman, 1969; Altman and Das, 1965a,b, 1966;Bayer, 1980; Kempermann et al., 1997a,b; Cameron and McKay, 1998). This phenomenon has been observed in a variety of species, including humans living into the eighth decade (Kuhn et al., 1996; Gould et al., 1997,1999b; Eriksson et al., 1998; Kempermann et al., 1998a,b). The dentate gyrus is constructed using a plan unique within the mammalian nervous system. Like other regions of the cortex, the dentate gyrus is initially populated by Cajal–Retzius cells and radial glia that are likely to help direct its morphogenesis (Rickmann et al., 1987; Del Rio et al., 1997; Alcantara et al., 1998; Borrell et al., 1999). However, unlike other cortical regions, mitotic precursor cells that retain the ability to generate neurons migrate from the subventricular zone of the dentate neuroepithelium into the dentate anlage. There they establish an ectopic proliferative zone in the dentate hilus that persists for the first 2 weeks of life in rodents (Altman and Das, 1965a,b; Altman and Bayer, 1990a,b). In the second half of the first postnatal month the precursor cells settle in a region along the hilar side of the granule cell layer termed the subgranular zone (SGZ) (Altman and Das, 1965a,b; Altman and Bayer, 1990a,b). There is an ongoing turnover of older granule cells and production of new granule cells throughout life. This unusual mode of development of the dentate gyrus probably requires unique strategies to maintain the long-term proliferative capacity and undifferentiated state of dentate precursor cells.
An important set of molecules that are likely to govern differentiation are the Notch receptors and their downstream modulators and ligands, which are known to regulate the timing of neurogenesis in mammals (de la Pompa et al., 1997;Artavanis-Tsakonas et al., 1999). The proteins downstream of Notch include a large number of molecules that share common motifs, including a basic transcriptional regulatory domain and a helix-loop-helix protein interaction domain (Kageyama et al., 1997; Lee, 1997). These proteins are collectively called basic helix-loop-helix (bHLH) proteins (Kageyama et al., 1997; Lee, 1997). Anti-neurogenic members of this family include the mammalian homologs of the Drosophila hairy and enhancer-of-split (Hes1 and Hes5) andextramacrochaete (Id1, Id2, Id3, and Id4) genes. The Hes genes are upregulated by Notch activation and block commitment to a neuronal fate (Benezra et al., 1990; Ishibashi et al., 1994, 1995; Jen et al., 1996; Tomita et al., 1996; Jen et al., 1997; Kageyama et al., 1997; Lyden et al., 1999).
The pro-neurogenic bHLH genes include homologs of Drosophila atonal and have been divided into two groups (Kageyama et al., 1997; Lee, 1997). The first group, termed neuronal determination genes, is involved in the initial commitment to a neuronal fate and includes Neurogenin1 (Ngn1) and Ngn2 (Gradwohl et al., 1996; Ma et al., 1996, 1997, 1998; Sommer et al., 1996; Fode et al., 1998). The second group, termed the neuronal differentiation genes, comprises otheratonal homologs including NeuroD/BETA2, NeuroD2/NDRF, Math2/Nex1, and Math3. This group is thought to regulate later events in neuronal differentiation (Bartholomä and Nave, 1994; Lee et al., 1995; Naya et al., 1995; McCormick et al., 1996; Kageyama et al., 1997; Lee, 1997; Schwab et al., 1998; Tsuda et al., 1998).
Mash1, a pro-neurogenic bHLH protein and the only homolog ofDrosophila achaete-scute found to be expressed in the nervous system of mammals appears to have a different role than the atonal homologs in nervous system development (Johnson et al., 1990; Lo et al., 1991). Mash1 mutants fail to develop autonomic neurons, some enteric neurons, and neurons in the olfactory epithelium (Guillemot et al., 1993). Anderson, Guillemot, and colleagues have shown that cells with a neuronal precursor phenotype appear in the autonomic and enteric nervous systems of these mice, but the cells fail to progress to a more differentiated state and then die (Sommer et al., 1995; Blaugrund et al., 1996). Whether this is a defect in survival or a block in the further differentiation of these precursor cells is not clear. In the CNS Mash1 is also thought to be critical for the maintenance of precursor cells (Casarosa et al., 1999; Horton et al., 1999; Torii et al., 1999; Tuttle et al., 1999).
The unique features of the development of the dentate gyrus, including the persistence of neural precursor cells for extended periods, led us to analyze in detail the patterns of expression of the Notch receptors, their ligands, and downstream bHLH factors in this region during embryogenesis and postnatal life. Using in situhybridization we show that specific bHLH mRNAs are expressed in a manner consistent with their having a regulatory role in the various stages of dentate gyrus development. Furthermore, some of these factors are expressed in the persistent SGZ precursor population.
MATERIALS AND METHODS
Tissue preparation. All animals were treated according to protocols for animal care established by the University of California, San Francisco and the National Institutes of Health. For embryonic time points, either timed or untimed pregnant Sprague Dawley rats were anesthetized with pentobarbitol, and the embryos were removed by laparotomy. The brains were removed and immersion-fixed overnight in 4% paraformaldehyde (PFA) in PBS, pH 7.4. For postnatal ages, rats were anesthetized with pentobarbitol, perfused with PBS, and then perfused with 4% PFA, and the brains were post-fixed overnight. All brains, embryonic and postnatal, were equilibrated in 30% sucrose in PBS, frozen, and sectioned at 15–30 μm using a cryostat. Sections were melted onto Fisher Superfrost Plus slides and stored at −20°C until use. The estimated gestational ages of rat embryos were based on comparison to sections in an atlas (Altman and Bayer, 1995). Three to five animals from at least two different litters were used for each time point studied.
Nonradioactive in situ hybridization.Nonradioactive in situ hybridization was performed essentially as described using a protocol obtained from Dr. David J. Anderson (California Institute of Technology) and is modified from published protocols (Schaeren-Wiemers and Gerfin-Moser, 1993; Ma et al., 1997). Briefly, sections were pretreated with Proteinase K before prehybridization for 3 hr at 65°C in a solution containing yeast tRNA, 50% formamide, and Denhardt's solution. After prehybridization, sections were hybridized overnight at 65°C with digoxigenin-labeled probes at a final concentration of 1 μg/ml. The next day, the slides were washed at high stringency and incubated with sheep anti-digoxigenin Fab fragments conjugated to alkaline phosphatase (diluted 1:2000) for 2–3 hr at room temperature. After washes, the slides were incubated in buffer containing nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate until developed. Controls included sense probes for NeuroD and Mash1 and antisense probes for molecules with specific distributions in the hippocampal formation not including the dentate granule cells (e.g., Math2).
Combined bromodeoxyuridine labeling and in situhybridization. For bromodeoxyuridine (BrdU) labeling, animals were injected intraperitoneally with doses of 100 mg/kg body weight at the times indicated in Results. After the in situhybridization was complete, slides were treated with 2N HCl for 30 min at 37°C, washed with PBS, and incubated with anti-BrdU monoclonal antibody for either 2 hr at room temperature or overnight at 4°C (Boehringer Mannheim, Indianapolis, IN; diluted 1:500–1:1000). The primary antibody was detected using the Vectastain Elite ABC-peroxidase kit (Vector Laboratories, Burlingame, CA) and diaminobenzidine (DAB).
Mash1-βgal mice. Mash1-βgal transgenic mice were obtained from Dr. Jane Johnson and bred as a homozygous line. Tissue was prepared as above and stained with X-gal according to published protocols (Verma-Kurvari et al., 1996)
Immunohistochemistry. Tissue sections on slides were incubated overnight at 4°C with anti-Mash1 rabbit antisera (kindly provided by Dr. Jane Johnson, University of Texas, Southwestern) diluted 1:1000. The next day the slides were washed and fixed with 4% PFA, treated with 2N HCl for 30 min at 37°C, and incubated with anti-BrdU antibody and biotinylated anti-rabbit antibody (Vector Laboratories; diluted 1:200). These were detected with Alexa 594-conjugated streptavidin (Molecular Probes, Eugene, OR), Cy5-conjugated donkey anti-mouse IgG (The Jackson Laboratory, Bar Harbor, ME), and sytox-Green (Molecular Probes; 1:25,000) to counterstain nuclei. The slides were mounted with ProLong Antifade (Molecular Probes), and images were obtained on a Bio-Rad (Hercules, CA) 1024 confocal microscope.
Probe preparation. The probes used for this study, along with their sources, are listed in Table1. Probes generated specifically for these studies were made by the PCR performed on reverse transcribed RNA generated either from embryonic day 13 (E13) rat embryos, postnatal day 7 (P7) rat hippocampus, or adult rat hippocampus. For probes obtained from other investigators, PCR primers were designed to confirm the presence of mRNAs encoding those gene products in the P7 hippocampus or adult hippocampus (Table2). PCR consisted of one cycle for 1 min at 94°C then 35 cycles of 1 min at 94°C, 2 min at the appropriate annealing temperature (see Table 2), and 2 min at 68°C. PolyA RNA was prepared using the micro Fast Track kit (Invitrogen, San Diego, CA) and reverse-transcribed with Superscript II (Life Technologies, Gaithersburg, MD). A typical first strand reaction used 0.5 μg of polyA RNA to generate a final volume of 30 μl of first strand. For PCR, 0.25 μl of this first strand was used. PCR fragments of the appropriate size were subcloned into the vector pCRII using the Topo-TA cloning kit (Invitrogen). The clones were then sequenced to check orientation and identity. Probes were generated using the Genius RNA labeling kit (Boehringer Mannheim) and purified with Clontech (Cambridge, UK) Chromaspin-100 columns packed in DEPC-treated water. Probes <300 bp were resuspended in hybridization buffer at 10 μg/ml. Probes >300 bp were resuspended in 40 mm NaHCO3/60 mm Na2CO3 at 60°C and hydrolyzed to produce 250 bp fragments. This was followed by ethanol precipitation and resuspension at 10 μg/ml in hybridization buffer.
Table 1.
Source of riboprobes used for in situhybridization
| Gene Product | Source |
|---|---|
| Notch 1 | Dr. Gerry Weinmaster |
| Notch 2 | RT-PCR |
| Notch 3 | RT-PCR |
| Delta 1 | Dr. Gerry Weinmaster |
| Delta 3 | RT-PCR |
| Jagged 1 | Dr. Gerry Weinmaster |
| Jagged 2 | RT-PCR |
| HES 1 | RT-PCR |
| HES 5 | Dr. Yoshiki Sasai |
| Id 1 | RT-PCR |
| Id 2 | RT-PCR |
| Id 3 | RT-PCR |
| Id 4 | Image Clone #400085 |
| Neurogenin 1 | Dr. David Anderson |
| Neurogenin 2 | Dr. David Anderson |
| Mash 1 | Dr. David Anderson |
| NeuroD | RT-PCR |
| NeuroD2(NDRF) | Dr. Stephen Tapscott |
| Ath2(Nex1) | RT-PCR |
| Ath3 | RT-PCR |
| Prox1 | RT-PCR |
These include other investigators, RT-PCR, or EST clones obtained from ATCC. Clones generated by RT-PCR and EST clones were all confirmed by DNA sequencing before use.
Table 2.
PCR primers and conditions for RT-PCR
| Gene product | Primers | Sequence | Conditions Tann = |
|---|---|---|---|
| Notch 1 | Notch 1 A/B | A-gcagccacagaacttacaaatccag | 62 |
| B-taaatgcctctggaatgtgggtgat | |||
| Notch 2 | Notch2 A/B | A-gctgtcctcttcatgctgca | 60 |
| B-agcagaagtcaagacagtc | |||
| Notch 3 | Notch3 C/D | C-aatggcttcagctgcacct | 64 |
| D-cagaagtacttgtccacca | |||
| Delta 1 | Delta1 A/B | A-atttgagctgaagctgcagg | 62 |
| B-aggtcgtcaggagaatctgt | |||
| Delta 3 | Delta3 E/F | E-atggtctctctgcaggtgtct | 61 |
| F-tcaggcagaccctgaagaaga | |||
| Jagged 1 | Jag1 C/D | C-tgcccactttgagtatcagat | 57 |
| D-ggatgcacttatcacagtaca | |||
| Jagged 2 | Jag2 A/B | A-tctgtgacaaagacctgaact | 57 |
| B-ttcacactcattggcgtcca | |||
| HES 1 | Hes1 C/D | C-aaattcctcgtcgccggtggct | 63 |
| D-acctcgttcatgcactcgctga | |||
| HES 5 | Hes5 C/D | C-aagtaccgtggcggtggagat | 63 |
| D-cgctggaagtggtaaagcagctt | |||
| Id 1 | Id1 A/B | A-catgaaggtcgccagtagcagt | 57 |
| B-taactatgctcctgagaagtaa | |||
| Id 2 | Id2 A/B | A-agcatgaaagccttcagtccggt | 59 |
| B-gccatttatttagccacagagta | |||
| Id 3 | Id3 A/B | A-tccaacatgaaggcgctgagcc | 64 |
| B-accgggtcagtggcaaaaactcc | |||
| Id 4 | Id4 A/J | A-atgaaggcggtg.agc.ccg.gt | 64 |
| J-ttcatatcgcactgcaggcacagc | (10%DMSO) | ||
| Neurogenin 1 | Ngn1 G/H | G-atgcctgcccctttggagacct | 66 |
| H-agtgcagcagcgcctcggaccgca | |||
| Mash 1 | Mash1 C/D | C-agcagctgctggacgagca | 60 |
| D-cctgcttccaaagtccattc | |||
| NeuroD | NeuroD A/B | A-ctcagttctcaggacgagga | 60 |
| B-tagttcttggccaagcgcag | |||
| NeuroD2 | NeuroD2 I/J | I-tactctatgcactactcggcgct | 59 |
| J-gacaaaggcaaagaaaaagagaggt | (5%DMSO) | ||
| Ath2 | Rath2 A/B | A-tgttaacactaccgtttgacg | 62 |
| B-agcatcattgaggccgtgcat | |||
| Ath3 | Rath3 A/B | A-tggcaaaaatgtatatgaaatccaa | 56 |
| B-gtcgaagagtctctatcttg | |||
| Prox1 | Prox1 A/B | A-tcttaagccggcaaaccaagagga | 64 |
| B-ttgctcctggaaaaggcatcatgg |
RT-PCR was used to generate probes and confirm the expression of the molecules studied. The sequences of the forward and reverse primers are shown as well as the annealing temperatures. In some cases DMSO was needed to help amplify GC-rich regions.
RESULTS
The dentate neuroepithelium is a distinct subdomain of the cortical neuroepithelium
Using nonradioactive in situ hybridization, we determined if any of the three known Notch receptors and four known Notch ligands are differentially distributed in the dentate neuroepithelium. (For the sake of brevity, all descriptions of expression patterns refer to the expression of mRNA unless otherwise indicated.) At E16–E17, before the migration of precursor cells or newly born granule cells to the dentate gyrus, we found that all three Notch receptors and their ligands Delta1, Delta3, and Jagged 2 were ubiquitously expressed within the entire hippocampal neuroepithelium (Fig. 1). Expression of Jagged1 was not seen at this time point in the cortical neuroepithelium. At the cellular level, the expression pattern for the Notch receptors and the Notch ligands was similar to that described previously (Lindsell et al., 1996; Ma et al., 1997). Notch1 was expressed homogeneously in all cells in the ventricular zone while the Notch ligands were expressed in a more variegated pattern (Fig. 1). These patterns of expression are consistent with the model of lateral inhibition that has been proposed to regulate the emergence of differentiating cells from a pool of undifferentiated cells (Artavanis-Tsakonas et al., 1999). Interestingly, Notch2 and Notch3 were also expressed in a variegated pattern implying that their role in lateral inhibition may be distinct from that of Notch1.
Fig. 1.
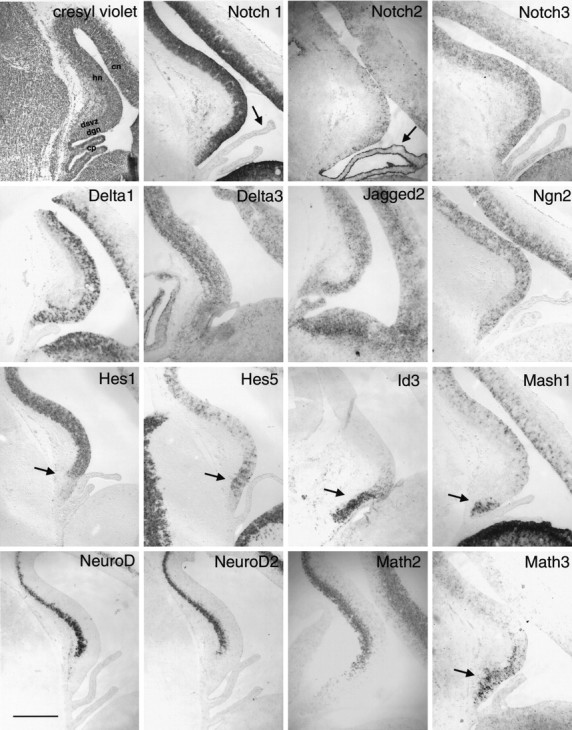
Expression of Notch receptors, ligands, and bHLH factors at E16–E17. Nonradioactive in situhybridization for probes as labeled in the figure. Note that the staining for Notch1 is fairly homogenous throughout the ventricular zone, whereas the staining for Notch2 and Notch3, the notch ligands Delta1, Delta3, and Jagged2 and the bHLH Ngn2 is patchy. This is consistent with the hypothesis that lateral inhibition regulates the specification of neurons within the ventricular zone. As described previously (Higuchi et al., 1995), Notch2 is expressed very strongly in the choroid plexus of the lateral ventricle (arrow), whereas the other Notch receptors are not (see arrow inNotch1 panel). Arrows also indicate the regions in the Hes5, Id3, Mash1, and Math3 panels that correspond to the region of neuroepithelium giving rise to the dentate gyrus and fimbria. Hes1 is less abundant in this region of neuroepithelium (arrow). Note that transcripts for the neural differentiation genes NeuroD, NeuroD2, and Math2 are not present in the neuroepithelium or nascent dentate gyrus at this time. The cresyl violet panel is labeled to provide orientation. cp,Choroid plexus; dgn, dentate gyrus neuroepithelium;dsvz, dentate subventricular zone; hn,hippocampal neuroepithelium; cn, cortical neuroepithelium. Scale bar, 300 μm.
Also consistent with this model is the widespread but patchy expression of the anti-neurogenic genes Hes1 and Hes5, both of which may be upregulated after Notch activation (Fig. 1). Interestingly, Hes1 was expressed at a relatively low level below the dentate notch, whereas Hes5 was expressed at higher levels in the same region (Fig. 1). In the E16–E17 dentate neuroepithelium, Id3 was specifically expressed in the dentate neuroepithelium and dentate subventricular zone (Fig. 1). In contrast to this localized expression of Id3, other Id genes were diffusely distributed in the hippocampal neuroepithelium, as described by others (Jen et al., 1997). These findings imply a distinct function for Id3 in the dentate granule cell lineage, perhaps acting analogously to Id2 in the neural crest to regulate the induction of particular differentiation pathways through the specific inactivation of other bHLH proteins (Martinsen and Bronner-Fraser, 1998).
Ngn2 was expressed throughout the E16–E17 hippocampal neuroepithelium in a pattern very similar to the Notch ligand Delta 1 (Fig. 1). This is consistent with the expression pattern of these bHLH proteins described previously in other regions of the rodent neuroepithelium and supports the idea that the neuronal determination genes begin to be expressed once their suppression by the Notch receptors is relieved (Sommer et al., 1996; Ma et al., 1997). In this model the expression of the Notch ligands accompanies the expression of neuronal determination genes, and the expression of both of these is blocked by Notch receptor activation.
Overall, the neuronal differentiation genes were not strongly expressed in the ventricular or subventricular zone at E16–E17. NeuroD, NeuroD2, and Math2 expression was observed in neurons in the hippocampal plate (Fig. 1). However, there was a region of increased Math3 expression that appeared specific for the dentate subventricular zone, suggesting a potential role in regulation of the dentate granule cell lineage (Fig. 1).
Because Mash1 has been implicated in the maintenance of precursor cells in the basal forebrain, autonomic nervous system, and olfactory epithelium (Guillemot et al., 1993; Sommer et al., 1995; Casarosa et al., 1999; Horton et al., 1999; Torii et al., 1999), we wanted to determine the relationship of Mash1 expression to the production of granule cells or their mitotic precursors. At E16–E17, in addition to its expression in the ventral telencephalon as previously reported (Ma et al., 1997), Mash1 was abundant in the neuroepithelium from the dentate notch to the fimbria but was at relatively low levels throughout the remainder of the cortical neuroepithelium (Fig. 1). The Mash1-expressing cells were predominantly in the subventricular zone, although some were at the ventricular surface. Altman and Bayer (1990a,b) and others (Rickmann et al., 1987; Sievers et al., 1992) have described this area as the fimbrial neuroepithelium and suggested that it gives rise to the glial cells that ultimately reside in the fimbria and dentate gyrus. The expression of Mash1 in this region suggests that this region might be the source of the precursor cells that populate the dentate gyrus and generate granule cells postnatally.
Distinct expression patterns of bHLH factors in precursor cells and immature granule cells migrating to the dentate anlage
At E18, cells begin to migrate from the subventricular zone just above and below the dentate notch to populate the dentate gyrus. Before this time, the cellular elements of the dentate gyrus consist of Cajal–Retzius cells, interneurons, and radial glial fibers (Soriano et al., 1986; Rickmann et al., 1987; Alcantara et al., 1998). At E19–E20 the migration is well under way, and it is possible to examine the expression of markers in the migrating cells. Previous analyses of this phase of dentate development have shown numerous newly born neurons, presumed to be granule cells, that migrate to the dentate anlage and form the scaffolding of the granule cell layer (Altman and Bayer, 1990a,b). There is also a distinct population of mitotic precursor cells that migrate to the dentate anlage and set up the tertiary matrix (Altman and Bayer, 1990a,b).
From E18 until birth, in situ hybridization shows the presence of numerous cells expressing Mash1 and Notch1 in the migratory route from the dentate neuroepithelium to the dentate anlage (Fig.2). In addition, there were fewer cells expressing Id3 in the same distribution (Fig. 2). Cells expressing these molecules were scattered over an area stretching from the pial boundary to just under the nascent CA3 pyramidal layer. These migrating cells did not express Ngn2, Notch2, Notch3, or Hes1 (Fig. 2; data not shown). It is unlikely that these cells represent a homogenous population of cells expressing all of these markers, because there appear to be larger numbers of cells expressing Mash1 and Notch1 than the other markers. Whether this represents the migration of several distinct lineages of cells or the migration of one group of precursor cells at distinct developmental stages remains to be established.
Fig. 2.
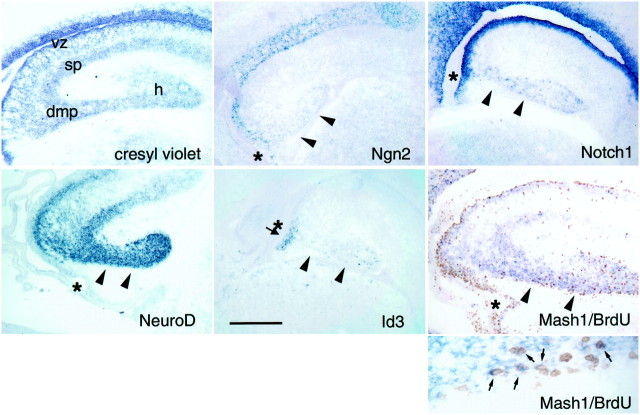
Migration of precursor cells and immature dentate granule cells to the dentate gyrus. Cresyl violet and nonradioactivein situ hybridization for probes in E20–E21 rat embryos as labeled in the figure. Note that Ngn2 is not expressed in the migrating cells but continues to be expressed in the neuroepithelium, whereas Notch1, Id3, and Mash1 are expressed in cells in the migratory route to the dentate gyrus. Id3 continues to have a localized expression domain in the neuroepithelium below the dentate notch at this time (an arrow indicates Id3 expression domain, andasterisks indicate the dentate notch in all the panels). The Mash1 panel shows double-labeling with anti-BrdU and anti-Mash1 antibodies, and the higher magnification panel shows several examples of Mash1/BrdU double-labeled cells (arrows). In these experiments the BrdU was administered 4 hr before killing. The NeuroD panel shows the large number of cells (presumably including immature granule cells) that are in the migratory pathway to the dentate gyrus. Arrowheads are used to mark the presumed migratory route of immature granule cells and precursors. Scale bar, 300 μm. vz, Ventricular zone; sp,stratum pyramidale; h, hilus; dmp,dentate migratory pathway.
In addition to these cells, many NeuroD-expressing cells were seen (Fig. 2). NeuroD is thought to be expressed at the time of terminal neuronal differentiation, whereas Hes5, Notch1, and Mash1 are expressed at high levels in cells before terminal differentiation. Thus, the NeuroD-expressing cells in the migratory stream may be granule cells born at the ventricular zone migrating to the dentate to form the granule cell layer. In contrast, most of the Mash1-expressing cells are likely to be mitotic precursor cells migrating to the dentate to form the tertiary matrix. To test this idea we determined the pattern of BrdU labeling after administration of BrdU 4 hr before killing. This time frame will label the actively dividing or very recent, postmitotic populations. The results showed that the pathway taken by migrating BrdU-labeled cells overlaps largely with the expression of Mash1 in the migratory route to the dentate gyrus, and there are numerous examples of double-labeled cells (Fig. 2). Thus, Mash1 is likely to be expressed in actively dividing cells as they migrate to the dentate. In principle, these mitotic cells could be limited precursors for granule cells or glial cells or they might be multipotential precursors for granule cells, glia, and other cell types. Because there is such a large number of Mash1-expressing cells in the migratory pathway, we also cannot exclude the possibility that newly born granule cells express Mash1 at this stage of development.
Continued presence of Mash1-expressing precursor cells in the tertiary matrix and SGZ
During dentate development the tertiary matrix stage is particularly crucial because the largest proportion of granule cells is born during this period. This stage involves several unique steps, including the ongoing migration of neuronal precursor cells to the dentate anlage, the formation of a transient ectopic proliferative zone, and the movement of the neuronal progeny of the precursors to their appropriate laminar position in the granule cell layer. From 2 weeks through adulthood the tertiary matrix gives way to the SGZ, where precursor cells continue to reside and retain the ability to generate new neurons for most of the remainder of the organism's life.
Throughout the first postnatal week, there were abundant Mash1-expressing cells in the tertiary matrix (Fig.3). When expression patterns of Mash1 and BrdU were compared using double labeling for BrdU and either Mash1 mRNA or Mash1 immunoreactivity, it was clear that virtually all the acutely BrdU-labeled cells (i.e., 4 hr after BrdU injection) expressed Mash1 (Fig. 4). Thus, at the tertiary matrix stage, dentate precursor cells expressed Mash1. The fates of the progeny of these precursors are not known because there have been no clonal analyses of dividing cells in the postnatal dentate gyrus. It remains to be determined whether these various progeny arise from different pools of precursor cells and whether Mash1-expressing precursors are the source of all of the cell types born during this period.
Fig. 3.
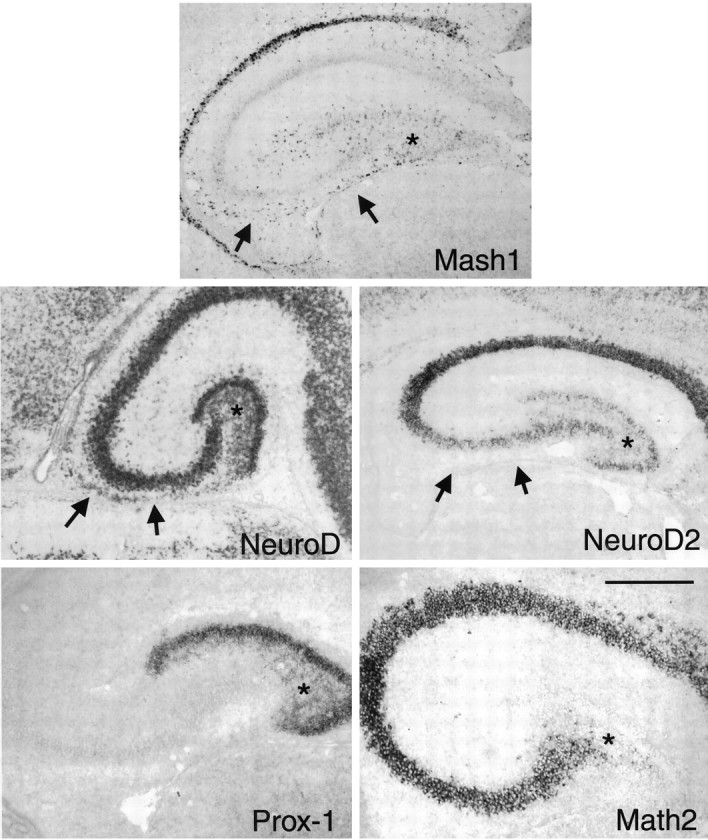
The tertiary matrix stage. Nonradioactive in situ hybridization of P4 rat hippocampal coronal sections with probes as shown in the panels. Note the continued presence of cells in the migratory pathway (indicated byarrows) expressing NeuroD and Mash1 but not Prox1 or NeuroD2. NeuroD and Prox1 are expressed throughout the tertiary matrix and developing dentate granule cell layer, whereas NeuroD2 is expressed only in the granule cell layer itself. Math2 is specifically not expressed in the granule cell layer or the newly born granule cells in the tertiary matrix. The asterisks mark the location of the tertiary matrix. Scale bar, 300 μm.
Fig. 4.
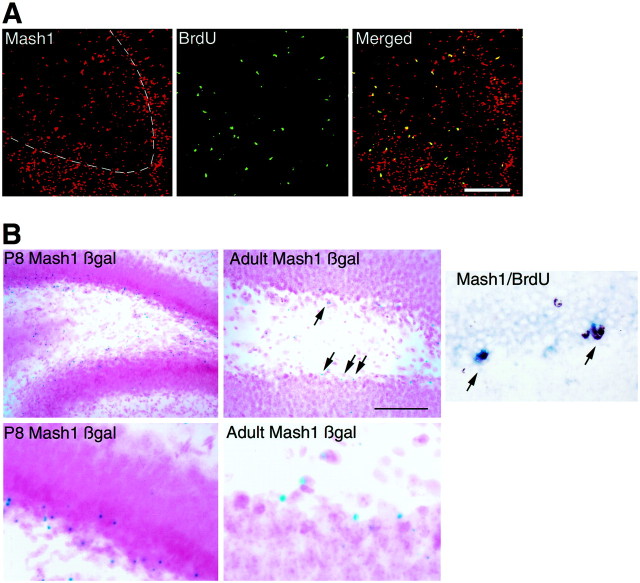
Expression of Mash1 protein and activity of the Mash1 promoter in the postnatal dentate gyrus. A, Mash1 protein and BrdU double-labeling in the P4 rat dentate gyrus. The merged image shows the overlap in the two patterns. Note that the great majority of dentate precursor cells in the tertiary matrix are double-labeled and thus appear yellow. The BrdU for this experiment was administered 4 hr before killing. The dashed line denotes the inner border of the granule cell layer.B, β-gal expression using X-gal histochemistry in coronal sections of the dentate gyrus taken from P8 and adult Mash1-βgal transgenic mice. Note that βgal expression in the P8 tissue is distributed throughout the tertiary matrix and granule cell layer, whereas in the adult it is largely confined to the SGZ. Cells expressing βgal are frequently seen in clusters in the SGZ. Scale bars: A, 55 μm; B, 150 μm for the low-power P8 Mash1βgal panel, 40 μm for the high-power P8 Mash1 βgal panel, 75 μm for the low-power adult Mash1 βgal panel, 25 μm for the high-power adult Mash1 βgal panel, and 60 μm for theMash1/BrdU panel.
During the second postnatal week, Mash1-expressing precursor cells began to appear predominantly in the SGZ at the hilar border of the granule cell layer (Fig. 4). The Mash1-expressing cells tended to be found in small clusters similar to the distribution of dentate precursor cells described in previous reports (Parent et al., 1997). Because the expression of Mash1 protein in the SGZ was lower than in the tertiary matrix (almost undetectable with the available antibodies), we wanted to confirm that the pattern of expression of Mash1 mRNA was authentic. We therefore used a transgenic mouse line engineered to express β-gal from the full-length Mash1 promoter. This transgenic line has been shown previously to have faithful expression of β-gal in cells expressing Mash1 mRNA and protein (Verma-Kurvari et al., 1996, 1998). Analysis of these mice using β-gal histochemistry showed that the distribution of β-gal expression was entirely consistent with our in situhybridization results, i.e., Mash1 is expressed in the tertiary matrix and SGZ at P8 and is limited primarily to the SGZ (and not in the dentate granule cell layer itself) in adulthood (Fig. 4). We also performed double labeling for actively dividing cells expressing Mash1 using nonradioactive in situ hybridization combined with BrdU labeling in P35 rats (4 hr after BrdU injection) (Fig. 4). This analysis showed at least one BrdU-labeled nucleus in most of the clumps of Mash1-stained cells in the subgranular zone of the mature dentate gyrus.
A cascade of bHLH proteins marks the birth of granule cells in the tertiary matrix and SGZ
NeuroD and Prox1, a divergent homeobox gene specifically expressed in dentate granule cells in the postnatal rodent brain (Oliver et al., 1993; Liu et al., 2000), were expressed widely throughout the tertiary matrix in newborn granule cells, whereas NeuroD2 began to be expressed at high levels only once the granule cells reached their final location in the granule cell layer (Fig. 3). This distinction between NeuroD/Prox1 and NeuroD2 expression patterns suggests that NeuroD is expressed in more immature granule cell populations, whereas NeuroD2 is expressed somewhat later. Expression of Math2, another atonal homolog, was excluded from the dentate granule cells (Fig. 3).
Once the SGZ forms, the mitotic precursor cells reside primarily in the SGZ, at the border of the hilus and the granule cell layer. As granule cells are born they migrate the short distance to the inner aspect of the granule cell layer. Because of this mode of production, there is thought to be a gradient of newly born to more mature granule cells from the inner granule cell layer to the outer granule cell layer. As mentioned above, Mash1 was expressed in the SGZ precursors. In contrast, NeuroD was expressed in the full thickness of the granule cell layer with the highest levels of expression in the more immature granule cells at the inner portion of the granule cell layer and somewhat lower expression in more mature granule cells (Fig.5). This was more apparent at P16, when the entire inner half of the granule cell layer has only recently been generated. At P35, only a few granule cells at the inner border of the granule cell layer have this higher level of NeuroD expression (Fig.5). NeuroD2 was expressed evenly throughout the granule cell layer at all ages without the gradient of expression seen for NeuroD (Fig.5). These data suggest that as newly born granule cells begin to express high levels of NeuroD and enter the granule cell layer they turn off expression of Mash1 and begin to express NeuroD2 as well. The specificity for high levels of Math2 for the pyramidal cells and the lack of expression in granule cells in the adult (either newly born or older cells) implies that Math2 does not play as significant a role in controlling granule cell differentiation as do the otheratonal homologs, NeuroD and NeuroD2. Math2 and NeuroD2 are also expressed at higher levels than NeuroD in pyramidal cells, suggesting a specific role for these two in pyramidal cell differentiation and function (Fig. 5).
Fig. 5.
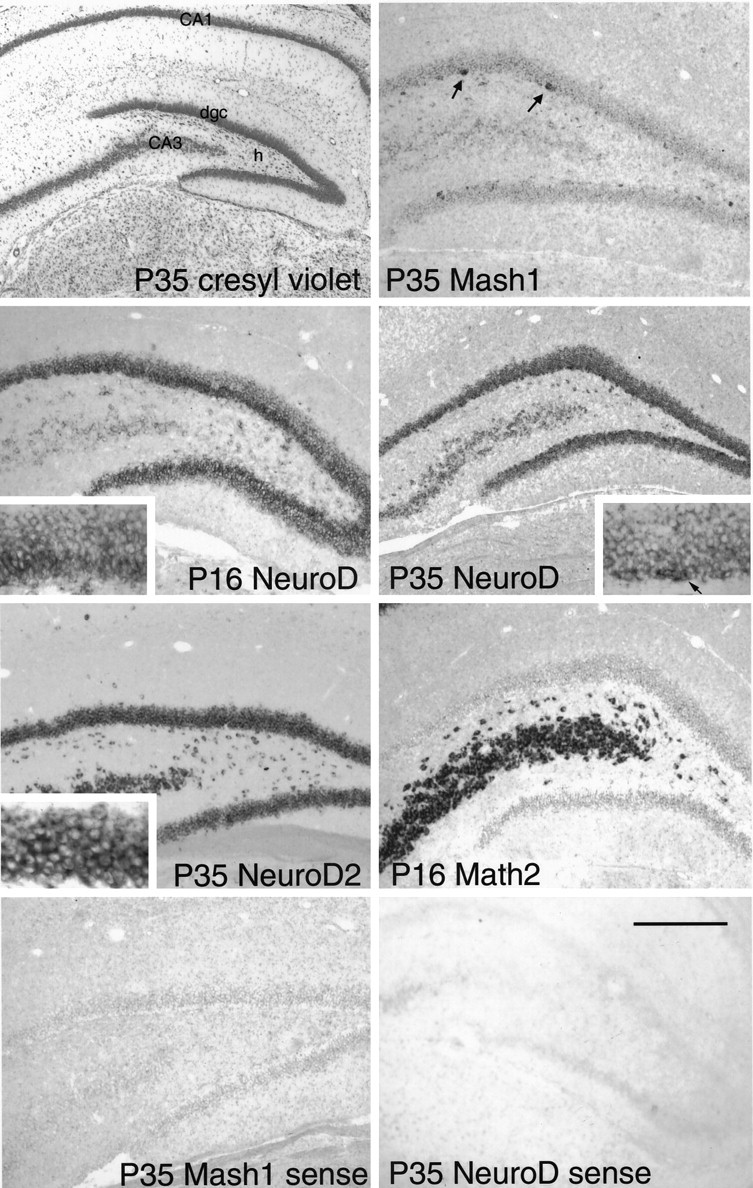
The mature dentate gyrus. Cresyl violet and nonradioactive in situ hybridization of probes as shown in the panels. Note the Mash1 is primarily expressed in small clusters of cells in the SGZ (arrows). There is some hybridization in the dentate granule cell layer that is considered to be nonspecific signal. NeuroD is expressed at highest levels in granule cells in the inner half of the granule cell layer at P16 (also see higher power inset), but at P35, this gradient of expression is largely gone except for an increased level of expression in some granule cells at the border of the SGZ and granule cell layer (arrow). These are presumably the most newly born granule cells. NeuroD2 is expressed throughout the granule cell layer, and Math2 is excluded from the granule cell layer. The twobottom panels show results of sense hybridization of tissue with two sense probes. Note the nonspecific signal that is most evident in the densely packed hippocampal cell layers. Scale bar:cresyl violet panel, 600 μm; other panels, 300 μm; and high-power insets, 100 μm.
DISCUSSION
In this study we have examined the patterns of expression of the Notch receptors, Notch ligands, and several bHLH genes during dentate gyrus development. We have focused on the dentate gyrus because of its unique developmental plan. Because the dentate gyrus requires the maintenance of a pool of undifferentiated precursors throughout life, we reasoned that these molecules, which have been shown to regulate the timing of differentiation and the maintenance of undifferentiated pools of cells in other systems, might regulate the phenotype of precursors in the dentate gyrus. We found that the region of neuroepithelium giving rise to granule cells and mitotic dentate precursor cells is molecularly distinct from other regions of cortical neuroepithelium starting at early stages in development. We also observed that the migration of cells to the dentate was comprised of a mixture of newly born neurons and precursor cells having distinct molecular profiles. In the tertiary matrix and SGZ stages we found that dentate precursor cells express Mash1, and there appears to be a cascade of bHLH expression in the dentate lineage from the precursor state to the mature granule cell stage.
The dentate neuroepithelium is a distinct structure
Altman and Bayer (1990a,b) recognized that the dentate neuroepithelium, which is centered around an anatomic structure known as the dentate notch, is distinct morphologically from the remainder of the hippocampal neuroepithelium. In the present study we have found that, in addition to these anatomical features, increased expression of Id2, Id3, Hes5, and Mash1 and reduced levels of Hes1 and Id4 characterize the entire dentate neuroepithelium. Because dentate granule cells are a relatively late-born population of cells, it is possible that factors evenly distributed in the neuroepithelium at earlier stages of development remain more highly expressed in the dentate neuroepithelium because of its relatively undifferentiated state. Given that development of the dentate requires production of a distinct population of migratory precursor cells, another possibility is that these negative factors are necessary to prevent the premature activation of neuronal determination genes expressed in this region. One such factor could be Ngn2, which is uniformly expressed throughout the hippocampal neuroepithelium. The granule cells born in the subventricular zone and migrating into the dentate gyrus with the precursor cells may escape the negative influences of Id3 and Hes5, and they begin to express NeuroD in the subventricular zone. One prediction of this model would be that persistent expression of these negative influences would inhibit dentate granule cell differentiation and perhaps bias cells to produce Mash1-expressing precursor cells. The Math3 expression domain in the dentate neuroepithelium is of unclear significance but it may be part of a combinatorial mechanism that, along with Ngn2 and NeuroD, results in the generation of immature granule cells near the ventricular surface.
Mash1 is expressed in dentate precursor cells at all stages of development
Our results show that mitotic precursor cells migrating into the dentate express Mash1. Mash1 continues to be expressed postnatally in mitotic precursor cells within the tertiary matrix and then the subgranular zone. During the migration of the precursor cells the Mash1-expressing cells may also express Notch1, Id2, Id3, and Hes5 (although multiple labeling studies will be required to prove this). However, the expression of these factors decreases once the cells are established in the tertiary matrix. Mash1 expression overlaps with acute BrdU labeling in the tertiary matrix, suggesting that Mash1 is a useful marker for mitotic precursor cells in this region. Many granule cells and glial cells are born in the tertiary matrix during the first 2 weeks of life. Because Mash1 expression overlaps extensively with BrdU expression in the dentate gyrus, it seems likely that Mash1-expressing precursor cells are multipotential, at least at this stage of development.
In other regions of the central and peripheral nervous systems, Mash1 is required for the successful maintenance of the precursor cell phenotype (Sommer et al., 1995; Casarosa et al., 1999; Torii et al., 1999). Thus, it is reasonable to hypothesize that Mash1 plays a role in maintaining the undifferentiated state of dentate gyrus precursor cells in the adult animal. This is significant because one of the key features of dentate gyrus organization is the presence of long-lived precursor cells that respond to environmental stimuli by changing the rate of production of granule cells (Cameron et al., 1995, 1998; Gould et al., 1997, 1999a; Kempermann et al., 1997b, 1998a,b; Parent et al., 1997, 1998; Liu et al., 1998; Tanapat et al., 1998; van Praag et al., 1999). Mice with induced mutations at the Mash1 locus do not have an obvious hippocampal phenotype (Guillemot et al., 1993; Casarosa et al., 1999; Torii et al., 1999). However, a dramatic phenotype might not be expected because these mice are not viable beyond P0, and the dentate is formed relatively late. Further analysis of these mice may show evidence of abnormalities in dentate precursor cells. Alternatively, there may be other genes that are able to compensate for Mash1 function in these cell lineages.
A sequential cascade of bHLH genes regulates dentate granule cell production
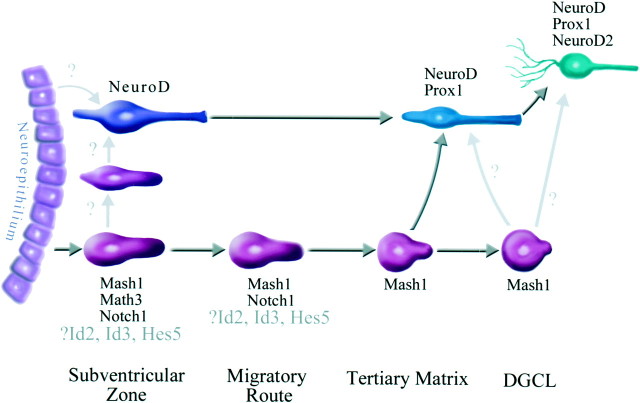
We have shown that the granule cell lineage expresses a variety of bHLH genes in a stage-specific manner. This, along with recent information gathered from analyses of NeuroD/BETA2 knock-out mice (Miyata et al., 1999; Liu et al., 2000), allows us to propose a model for the role of bHLH genes in granule cell production (Fig.6). The neuroepithelium destined to generate granule cells expresses Ngn2, Hes5, and Id3. In the subventricular zone, cells express Mash1 and Math3 depending on their location above or below the dentate notch, and the cells continue to express Id2, Id3, and Hes5. After leaving the SVZ, some granule cells are born and begin to express NeuroD. These cells are dependent on NeuroD expression for their proper specification, and without it they are unable to form the initial dentate granule cell layer (Liu et al., 2000). After reaching the dentate gyrus the cells express high levels of a granule cell-specific marker, Prox1. Other cells, retaining a precursor phenotype, continue to express Mash1 and form the tertiary matrix after arriving at the dentate anlage. In the tertiary matrix many more granule cells are generated from Mash1-expressing precursor cells, and these newly born granule cells migrate radially to the granule cell layer where they begin to express NeuroD followed by NeuroD2 and Prox1. This general pattern is maintained throughout adulthood.
Fig. 6.
Model of the dentate granule cell lineage. Initially, mitotic precursor cells expressing Mash1, Id3, Hes5, and Math3 are formed in the subventricular zone. It is also known that postmitotic granule cells expressing NeuroD are born in the subventricular zone (Miyata et al., 1999), but it is not clear whether these cells are born directly from the neuroepithelium or pass through the precursor cell stage. Immature NeuroD-expressing granule cells and mitotic precursor cells migrate to the tertiary matrix where the precursor cells continue to express Mash1 and the immature granule cells acquire the expression of high levels of Prox1. In the last stage, when the dentate granule cell layer is formed (DGCL), Mash1-expressing precursor cells are arrayed in the subgranular zone and generate granule cells that migrate to the inner portion of the granule cell layer.
NeuroD is expressed in newly born granule cells at all stages of development
NeuroD is widely expressed in newly postmitotic neurons in the brain, and its expression decreases during neuronal maturation. In the dentate granule cell layer, where neurons continue to be added at later ages, NeuroD continues to be expressed and is at its highest levels in the most immature neurons. This implies that NeuroD regulates the expression of genes involved in the maturation of granule cells and their integration into the existing neuronal network. Some of the most interesting unanswered questions about ongoing granule cell neurogenesis are how newly born cells are able to extend axons that reach their appropriate targets and how these cells receive appropriate input from entorhinal cortex afferents that provide the dominant input into the hippocampus. Recent studies have shown that learning paradigms and environmental stimuli may have a dramatic effect on the rate of birth of granule cells and their subsequent survival (Kempermann et al., 1997b; Gould et al., 1999a; van Praag et al., 1999). Perhaps these stimuli act in part via genes that are regulated by the expression of Mash1 in precursor cells or by NeuroD in newly born granule cells.
Footnotes
This work was supported by a Howard Hughes Medical Institute (HHMI) postdoctoral fellowship for physicians (S.J.P.), an HHMI medical student fellowship (A.E.C.), and grants from the March of Dimes and National Institute of Neurological Diseases and Stroke (D.H.L). We thank Drs. David Anderson, Jane Johnson, Virginia Lee, Gerry Weinmaster, Stephen Tapscott, Yoshiki Sasai, Songli Wang, and Ben Barres for various probes, antibodies, and other reagents that were invaluable to this work. We also thank Dr. Jack Parent, Dr. Robert Elliott, Anil Bagri, Dr. Marc Tessier-Lavigne, and members of the Tessier-Lavigne laboratory for many helpful discussions. Erin Browne created the artwork for Figure 6.
S.J.P. and A.E.C. contributed equally to this work.
Correspondence should be addressed to Dr. Daniel Lowenstein, Harvard Medical School, Building A, Room 103, 25 Shattuck Street, Boston, MA 02115. E-mail: daniel_lowenstein@hms.harvard.edu.
REFERENCES
- 1.Alcantara S, Ruiz M, D'Arcangelo G, Ezan F, de Lecea L, Curran T, Sotelo C, Soriano E. Regional and cellular patterns of reelin mRNA expression in the forebrain of the developing and adult mouse. J Neurosci. 1998;18:7779–7799. doi: 10.1523/JNEUROSCI.18-19-07779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman J, Bayer SA. Migration and distribution of two populations of hippocampal granule cell precursors during the perinatal and postnatal periods. J Comp Neurol. 1990a;301:365–381. doi: 10.1002/cne.903010304. [DOI] [PubMed] [Google Scholar]
- 3.Altman J, Bayer SA. Mosaic organization of the hippocampal neuroepithelium and the multiple germinal sources of dentate granule cells. J Comp Neurol. 1990b;301:325–342. doi: 10.1002/cne.903010302. [DOI] [PubMed] [Google Scholar]
- 4.Altman J, Bayer SA. Atlas of prenatal rat brain development. CRC; Boca Raton, FL: 1995. [Google Scholar]
- 5.Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965a;124:319–335. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- 6.Altman J, Das GD. Post-natal origin of microneurones in the rat brain. Nature. 1965b;207:953–956. doi: 10.1038/207953a0. [DOI] [PubMed] [Google Scholar]
- 7.Altman J, Das GD. Autoradiographic and histological studies of postnatal neurogenesis. I. A longitudinal investigation of the kinetics, migration and transformation of cells incorporating tritiated thymidine in neonate rats, with special reference to postnatal neurogenesis in some brain regions. J Comp Neurol. 1966;126:337–389. doi: 10.1002/cne.901260302. [DOI] [PubMed] [Google Scholar]
- 8.Altman J. Autoradiographic and histological studies of postnatal neurogenesis. IV. Cell proliferation and migration in the anterior forebrain, with special reference to persisting neurogenesis in the olfactory bulb. J Comp Neurol. 1969;137:433–457. doi: 10.1002/cne.901370404. [DOI] [PubMed] [Google Scholar]
- 9.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 10.Bartholomä A, Nave KA. NEX-1: a novel brain-specific helix-loop-helix protein with autoregulation and sustained expression in mature cortical neurons. Mech Dev. 1994;48:217–228. doi: 10.1016/0925-4773(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 11.Bayer SA. Development of the hippocampal region in the rat. I. Neurogenesis examined with 3H-thymidine autoradiography. J Comp Neurol. 1980;190:87–114. doi: 10.1002/cne.901900107. [DOI] [PubMed] [Google Scholar]
- 12.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 13.Blaugrund E, Pham TD, Tennyson VM, Lo L, Sommer L, Anderson DJ, Gershon MD. Distinct subpopulations of enteric neuronal progenitors defined by time of development, sympathoadrenal lineage markers and Mash-1-dependence. Development. 1996;122:309–320. doi: 10.1242/dev.122.1.309. [DOI] [PubMed] [Google Scholar]
- 14.Borrell V, Del Rio JA, Alcantara S, Derer M, Martinez A, D'Arcangelo G, Nakajima K, Mikoshiba K, Derer P, Curran T, Soriano E. Reelin regulates the development and synaptogenesis of the layer-specific entorhino-hippocampal connections. J Neurosci. 1999;19:1345–1358. doi: 10.1523/JNEUROSCI.19-04-01345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron HA, McKay R. Stem cells and neurogenesis in the adult brain. Curr Opin Neurobiol. 1998;8:677–680. doi: 10.1016/s0959-4388(98)80099-8. [DOI] [PubMed] [Google Scholar]
- 16.Cameron HA, Tanapat P, Gould E. Adrenal steroids and N-methyl-d-aspartate receptor activation regulate neurogenesis in the dentate gyrus of adult rats through a common pathway. Neuroscience. 1998;82:349–354. doi: 10.1016/s0306-4522(97)00303-5. [DOI] [PubMed] [Google Scholar]
- 17.Cameron HA, McEwen BS, Gould E. Regulation of adult neurogenesis by excitatory input and NMDA receptor activation in the dentate gyrus. J Neurosci. 1995;15:4687–4692. doi: 10.1523/JNEUROSCI.15-06-04687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 19.de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J, Conlon RA. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 20.Del Rio JA, Heimrich B, Borrell V, Förster E, Drakew A, Alcantara S, Nakajima K, Miyata T, Ogawa M, Mikoshiba K, Derer P, Frotscher M, Soriano E. A role for Cajal-Retzius cells and reelin in the development of hippocampal connections. Nature. 1997;385:70–74. doi: 10.1038/385070a0. [DOI] [PubMed] [Google Scholar]
- 21.Eriksson PS, Perfilieva E, Björk-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 22.Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20:483–494. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- 23.Gould E, McEwen BS, Tanapat P, Galea LA, Fuchs E. Neurogenesis in the dentate gyrus of the adult tree shrew is regulated by psychosocial stress and NMDA receptor activation. J Neurosci. 1997;17:2492–2498. doi: 10.1523/JNEUROSCI.17-07-02492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gould E, Beylin A, Tanapat P, Reeves A, Shors TJ. Learning enhances adult neurogenesis in the hippocampal formation. Nat Neurosci. 1999a;2:260–265. doi: 10.1038/6365. [DOI] [PubMed] [Google Scholar]
- 25.Gould E, Reeves AJ, Fallah M, Tanapat P, Gross CG, Fuchs E. Hippocampal neurogenesis in adult Old World primates. Proc Natl Acad Sci USA. 1999b;96:5263–5267. doi: 10.1073/pnas.96.9.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gradwohl G, Fode C, Guillemot F. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev Biol. 1996;180:227–241. doi: 10.1006/dbio.1996.0297. [DOI] [PubMed] [Google Scholar]
- 27.Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 28.Higuchi M, Kiyama H, Hayakawa T, Hamada Y, Tsujimoto Y. Differential expression of Notch1 and Notch2 in developing and adult mouse brain. Mol Brain Res. 1995;29:263–272. doi: 10.1016/0169-328x(94)00257-f. [DOI] [PubMed] [Google Scholar]
- 29.Horton S, Meredith A, Richardson JA, Johnson JE. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol Cell Neurosci. 1999;14:355–369. doi: 10.1006/mcne.1999.0791. [DOI] [PubMed] [Google Scholar]
- 30.Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 32.Jen Y, Manova K, Benezra R. Expression patterns of Id1, Id2, and Id3 are highly related but distinct from that of Id4 during mouse embryogenesis. Dev Dyn. 1996;207:235–252. doi: 10.1002/(SICI)1097-0177(199611)207:3<235::AID-AJA1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 33.Jen Y, Manova K, Benezra R. Each member of the Id gene family exhibits a unique expression pattern in mouse gastrulation and neurogenesis. Dev Dyn. 1997;208:92–106. doi: 10.1002/(SICI)1097-0177(199701)208:1<92::AID-AJA9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 34.Johnson JE, Birren SJ, Anderson DJ. Two rat homologues of Drosophila achaete-scute specifically expressed in neuronal precursors. Nature. 1990;346:858–861. doi: 10.1038/346858a0. [DOI] [PubMed] [Google Scholar]
- 35.Kageyama R, Ishibashi M, Takebayashi K, Tomita K. bHLH transcription factors and mammalian neuronal differentiation. Int J Biochem Cell Biol. 1997;29:1389–1399. doi: 10.1016/s1357-2725(97)89968-2. [DOI] [PubMed] [Google Scholar]
- 36.Kempermann G, Kuhn HG, Gage FH. Genetic influence on neurogenesis in the dentate gyrus of adult mice. Proc Natl Acad Sci USA. 1997a;94:10409–10414. doi: 10.1073/pnas.94.19.10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kempermann G, Kuhn HG, Gage FH. More hippocampal neurons in adult mice living in an enriched environment. Nature. 1997b;386:493–495. doi: 10.1038/386493a0. [DOI] [PubMed] [Google Scholar]
- 38.Kempermann G, Brandon EP, Gage FH. Environmental stimulation of 129/SvJ mice causes increased cell proliferation and neurogenesis in the adult dentate gyrus. Curr Biol. 1998a;8:939–942. doi: 10.1016/s0960-9822(07)00377-6. [DOI] [PubMed] [Google Scholar]
- 39.Kempermann G, Kuhn HG, Gage FH. Experience-induced neurogenesis in the senescent dentate gyrus. J Neurosci. 1998b;18:3206–3212. doi: 10.1523/JNEUROSCI.18-09-03206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 42.Lee JE. Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 43.Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Solway K, Messing RO, Sharp FR. Increased neurogenesis in the dentate gyrus after transient global ischemia in gerbils. J Neurosci. 1998;18:7768–7778. doi: 10.1523/JNEUROSCI.18-19-07768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu M, Pleasure SJ, Collins AE, Noebels JL, Naya FJ, Tsai M-J, Lowenstein DH. Loss of BETA2/NeuroD leads to malformation of the dentate gyrus and epilepsy. Proc Natl Acad Sci USA. 2000;97:865–870. doi: 10.1073/pnas.97.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo L, Sommer L, Anderson DJ. MASH1 maintains competence for BMP2-induced neuronal differentiation in post-migratory neural crest cells. Curr Biol. 1997;7:440–450. doi: 10.1016/s0960-9822(06)00191-6. [DOI] [PubMed] [Google Scholar]
- 47.Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 48.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 49.Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 51.Martinsen BJ, Bronner-Fraser M. Neural crest specification regulated by the helix-loop-helix repressor Id2. Science. 1998;281:988–991. doi: 10.1126/science.281.5379.988. [DOI] [PubMed] [Google Scholar]
- 52.McCormick MB, Tamimi RM, Snider L, Asakura A, Bergstrom D, Tapscott SJ. NeuroD2 and neuroD3: distinct expression patterns and transcriptional activation potentials within the neuroD gene family. Mol Cell Biol. 1996;16:5792–5800. doi: 10.1128/mcb.16.10.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 55.Oliver G, Sosa-Pineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-m. [DOI] [PubMed] [Google Scholar]
- 56.Parent JM, Yu TW, Leibowitz RT, Geschwind DH, Sloviter RS, Lowenstein DH. Dentate granule cell neurogenesis is increased by seizures and contributes to aberrant network reorganization in the adult rat hippocampus. J Neurosci. 1997;17:3727–3738. doi: 10.1523/JNEUROSCI.17-10-03727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parent JM, Janumpalli S, McNamara JO, Lowenstein DH. Increased dentate granule cell neurogenesis following amygdala kindling in the adult rat. Neurosci Lett. 1998;247:9–12. doi: 10.1016/s0304-3940(98)00269-9. [DOI] [PubMed] [Google Scholar]
- 58.Rickmann M, Amaral DG, Cowan WM. Organization of radial glial cells during the development of the rat dentate gyrus. J Comp Neurol. 1987;264:449–479. doi: 10.1002/cne.902640403. [DOI] [PubMed] [Google Scholar]
- 59.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 60.Shou J, Rim PC, Calof AL. BMPs inhibit neurogenesis by a mechanism involving degradation of a transcription factor. Nat Neurosci. 1999;2:339–345. doi: 10.1038/7251. [DOI] [PubMed] [Google Scholar]
- 61.Sievers J, Hartmann D, Pehlemann FW, Berry M. Development of astroglial cells in the proliferative matrices, the granule cell layer, and the hippocampal fissure of the hamster dentate gyrus. J Comp Neurol. 1992;320:1–32. doi: 10.1002/cne.903200102. [DOI] [PubMed] [Google Scholar]
- 62.Sommer L, Shah N, Rao M, Anderson DJ. The cellular function of MASH1 in autonomic neurogenesis. Neuron. 1995;15:1245–1258. doi: 10.1016/0896-6273(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 63.Sommer L, Ma Q, Anderson DJ. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 64.Soriano E, Cobas A, Fairéen A. Asynchronism in the neurogenesis of GABAergic and non-GABAergic neurons in the mouse hippocampus. Brain Res. 1986;395:88–92. doi: 10.1016/s0006-8993(86)80012-9. [DOI] [PubMed] [Google Scholar]
- 65.Tanapat P, Galea LA, Gould E. Stress inhibits the proliferation of granule cell precursors in the developing dentate gyrus. Int J Dev Neurosci. 1998;16:235–239. doi: 10.1016/s0736-5748(98)00029-x. [DOI] [PubMed] [Google Scholar]
- 66.Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- 67.Torii M, Matsuzaki F, Osumi N, Kaibuchi K, Nakamura S, Casarosa S, Guillemot F, Nakafuku M. Transcription factors Mash-1 and Prox-1 delineate early steps in differentiation of neural stem cells in the developing central nervous system. Development. 1999;126:443–456. doi: 10.1242/dev.126.3.443. [DOI] [PubMed] [Google Scholar]
- 68.Tsuda H, Takebayashi K, Nakanishi S, Kageyama R. Structure and promoter analysis of Math3 gene, a mouse homolog of Drosophila proneural gene atonal. Neural-specific expression by dual promoter elements. J Biol Chem. 1998;273:6327–6333. doi: 10.1074/jbc.273.11.6327. [DOI] [PubMed] [Google Scholar]
- 69.Tuttle R, Nakagawa Y, Johnson JE, O'Leary DD. Defects in thalamocortical axon pathfinding correlate with altered cell domains in Mash-1-deficient mice. Development. 1999;126:1903–1916. doi: 10.1242/dev.126.9.1903. [DOI] [PubMed] [Google Scholar]
- 70.van Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci. 1999;2:266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- 71.Verma-Kurvari S, Savage T, Gowan K, Johnson JE. Lineage-specific regulation of the neural differentiation gene MASH1. Dev Biol. 1996;180:605–617. doi: 10.1006/dbio.1996.0332. [DOI] [PubMed] [Google Scholar]
- 72.Verma-Kurvari S, Savage T, Smith D, Johnson JE. Multiple elements regulate Mash1 expression in the developing CNS. Dev Biol. 1998;197:106–116. doi: 10.1006/dbio.1998.8873. [DOI] [PubMed] [Google Scholar]